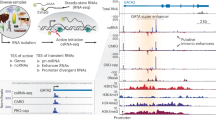
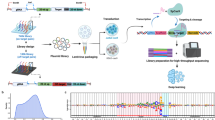
Abstract
Gene expression is modulated jointly by transcriptional regulation and messenger RNA stability, yet the latter is often overlooked in studies on genetic variants. Here, leveraging metabolic labeling data (Bru/BruChase-seq) and a new computational pipeline, RNAtracker, we categorize genes as allele-specific RNA stability (asRS) or allele-specific RNA transcription events. We identify more than 5,000 asRS variants among 665 genes across a panel of 11 human cell lines. These variants directly overlap conserved microRNA target regions and allele-specific RNA-binding protein sites, illuminating mechanisms through which stability is mediated. Furthermore, we identified causal asRS variants using a massively parallel screen (MapUTR) for variants that affect post-transcriptional mRNA abundance, as well as through CRISPR prime editing approaches. Notably, asRS genes were enriched significantly among a multitude of immune-related pathways and contribute to the risk of several immune system diseases. This work highlights RNA stability as a critical, yet understudied mechanism linking genetic variation and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Bru-seq/BruChase-seq from 16 human cell lines (GM12878, HCT116, HepG2, IMR-90, K562, MCF-7, PC-3, Panc1, PC-9, A673, MCF10A, Calu3, Caco-2, OCI-LY7, endothelial cell of umbilical vein (HUVEC) and mammary epithelial cell (HMEC)) were downloaded from the ENCODE data portal (https://www.encodeproject.org/). Accession IDs can be found in Supplementary Table 1b. The GRCh38 reference genome and gene annotation can be found at https://www.gencodegenes.org/human/release_36.html (filenames: GRCh38.primary_assembly.genome.fa.gz; gencode.v36.primary_assembly.annotation.gtf.gz). Significant GTEx cis-eQTLs were downloaded from the GTex portal (v.8 release) at https://www.gtexportal.org/home/datasets (GTEx_Analysis_v8_eQTL.tar). ABSOLUTE CNVs from CCLE can be obtained from https://depmap.org/portal/data_page/?release=CCLE+2019&file=CCLE_ABSOLUTE_combined_20181227.xlsx&tab=allData. Allele-specific binding sites were obtained from our previous work (Supplementary Data 2 from ref. 22). SNPs overlapping miRNA seed regions that create or disrupt miRNA binding sites were downloaded from miRNASNPv3 (ref. 25). eCLIP data for reproducible peaks (as determined from the irreproducible discovery rate approach50) were downloaded from the ENCODE portal. ActD RNA-seq data can be accessed on GEO (Series record GSE276016). MapUTR sequencing data can be accessed on GEO (Series record GSE298114). CRISPR editing results can be accessed on GEO (Series record GSE298112). All GWAS summary statistics used in this paper can be downloaded from the GWAS catalog (https://www.ebi.ac.uk/gwas/; accession codes in Supplementary Table 8a). GRCh38 genotype reference files from the 1000 Genomes project can be found at https://www.internationalgenome.org/data-portal/data-collection/grch38. Source data are provided with this paper.
Code availability
Code for reproducing the RNAtracker gene categorization results and other data analysis scripts is available via GitHub at https://github.com/gxiaolab/RNAtracker and via Zenodo at https://doi.org/10.5281/zenodo.15528784 (ref. 55). We used bbduk from the BBmap package (v.38.91) (https://sourceforge.net/projects/bbmap/) for read adapter trimming, STAR47 (v.2.7.8a) for read mapping, Picard Tools (https://broadinstitute.github.io/picard/) (v.1.94) to remove PCR duplicates and extract uniquely mapped reads, NeoloopFinder43 (v.0.3.0) for CNV predictions, rrvgo52 (v.1.6.0) for GO enrichment analysis, PLINK56 (v.1.9) to obtain tag SNPs and bedtools (v.2.30.0)57 to overlap genomic regions. Perbase (v.0.10.0) (https://github.com/sstadick/perbase) was used to obtain variant allelic counts in the CRISPR prime editing sequencing data. MPRAnalyze48 was used to identify functional variants in the MapUTR data. S-LDSC54 was used to estimate disease heritability. FUSION.compute_weights.R from http://gusevlab.org/projects/fusion/ was used to build gene prediction models. For analyzing whole-genome sequencing data, we used bwa mem58 (v.0.7.17) for read mapping and CNVpytor41 (v.1.3.1) for identifying CNV regions. The VGAM59 (v.1.1) R package was used to compute probability density values and simulate allelic counts. Genome coordinate conversions were performed using liftOver (https://www.bioconductor.org/packages/release/workflows/html/liftOver.html). Other R packages used for plotting include ComplexUpSet60, ComplexHeatmap61 and AllelicImbalance62.
References
Degner, J. F. et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394 (2012).
Farh, K. K.-H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Gaffney, D. J. et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 13, R7 (2012).
Liebhaber, S. A. mRNA stability and the control of gene expression. Nucleic Acids Symp. Ser. 36, 29–32 (1997).
Hollams, E. M., Giles, K. M., Thomson, A. M. & Leedman, P. J. MRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 27, 957–980 (2002).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Tani, H. et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 22, 947–956 (2012).
Courel, M. et al. GC content shapes mRNA storage and decay in human cells. eLife 8, e49708 (2019).
LaMarre, J., Gingerich, T. J., Feige, J.-J. & LaMarre, J. AU-rich elements and the control of gene expression through regulated mRNA stability. Anim. Health Res. Rev. 5, 49–63 (2004).
Agarwal, V., Bell, G. W., Nam, J.-W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015).
Wu, Q. et al. Translation affects mRNA stability in a codon-dependent manner in human cells. eLife 8, e45396 (2019).
Pai, A. A. et al. The contribution of RNA decay quantitative trait loci to inter-individual variation in steady-state gene expression levels. PLoS Genet. 8, e1003000 (2012).
Alkallas, R., Fish, L., Goodarzi, H. & Najafabadi, H. S. Inference of RNA decay rate from transcriptional profiling highlights the regulatory programs of Alzheimer’s disease. Nat. Commun. 8, 909 (2017).
Li, J.-R., Tang, M., Li, Y., Amos, C. I. & Cheng, C. Genetic variants associated mRNA stability in lung. BMC Genomics 23, 196 (2022).
Paulsen, M. T. et al. Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc. Natl Acad. Sci. USA 110, 2240–2245 (2013).
Bedi, K. et al. Co-transcriptional splicing efficiencies differ within genes and between cell types. RNA 27, 829–840 (2021).
The GTEx Consortium et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Salton, M. et al. Matrin 3 binds and stabilizes mRNA. PLoS ONE 6, e23882 (2011).
Zhang, G. et al. Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat. Commun. 13, 859 (2022).
Meyer, C. et al. The TIA1 RNA-binding protein family regulates EIF2AK2-mediated stress response and cell cycle progression. Mol. Cell 69, 622–635 (2018).
Kim, Y. K. & Maquat, L. E. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25, 407–422 (2019).
Yang, E.-W. et al. Allele-specific binding of RNA-binding proteins reveals functional genetic variants in the RNA. Nat. Commun. 10, 1338 (2019).
Zhang, J. et al. An integrative ENCODE resource for cancer genomics. Nat. Commun. 11, 3696 (2020).
Fabian, M. R., Sonenberg, N. & Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 (2010).
Liu, C.-J. et al. miRNASNP-v3: a comprehensive database for SNPs and disease-related variations in miRNAs and miRNA targets. Nucleic Acids Res. 49, D1276–D1281 (2021).
Fu, T. et al. Massively parallel screen uncovers many rare 3′ UTR variants regulating mRNA abundance of cancer driver genes. Nat. Commun. 15, 3335 (2024).
Griesemer, D. et al. Genome-wide functional screen of 3′ UTR variants uncovers causal variants for human disease and evolution. Cell 184, 5247–5260 (2021).
Chen, P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652 (2021).
Bresson, S. & Tollervey, D. Tailing off: PABP and CNOT generate cycles of mRNA deadenylation. Mol. Cell 70, 987–988 (2018).
Springer, T. A. Adhesion receptors of the immune system. Nature 346, 425–434 (1990).
González-Amaro, R., Diaz-González, F. & Sánchez-Madrid, F. Adhesion molecules in inflammatory diseases. Drugs 56, 977–988 (1998).
Ryter, S. W., Cloonan, S. M. & Choi, A. M. K. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cells 36, 7–16 (2013).
Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Prim. 1, 37–49 (2021).
Sollis, E. et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–D985 (2023).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Zhou, M. et al. Inhibition of Fam114A1 protects melanocytes from apoptosis through higher RACK1 expression. Aging 13, 24740–24752 (2021).
Subbaiah, K. C. V., Wu, J., Tang, W. H. W. & Yao, P. FAM114A1 influences cardiac pathological remodeling by regulating angiotensin II signaling. JCI Insight 7, e152783 (2022).
Imamachi, N. et al. BRIC-seq: a genome-wide approach for determining RNA stability in mammalian cells. Methods 67, 55–63 (2014).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Suvakov, M., Panda, A., Diesh, C., Holmes, I. & Abyzov, A. CNVpytor: a tool for copy number variation detection and analysis from read depth and allele imbalance in whole-genome sequencing. Gigascience 10, giab074 (2021).
Abdennur, N. & Mirny, L. A. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316 (2020).
Wang, X. et al. Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nat. Methods 18, 661–668 (2021).
Yan, J. et al. Improving prime editing with an endogenous small RNA-binding protein. Nature 628, 639–647 (2024).
Chow, R. D., Chen, J. S., Shen, J. & Chen, S. A web tool for the design of prime-editing guide RNAs. Nat. Biomed. Eng. 5, 190–194 (2021).
Nelson, J. W. et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410 (2022).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Ashuach, T. et al. MPRAnalyze: statistical framework for massively parallel reporter assays. Genome Biol. 20, 183 (2019).
Ormond, C., Ryan, N. M., Corvin, A. & Heron, E. A. Converting single nucleotide variants between genome builds: from cautionary tale to solution. Brief. Bioinform. 22, bbab069 (2021).
Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020).
Smedley, D. et al. BioMart–biological queries made easy. BMC Genomics 10, 22 (2009).
Sayols, S. rrvgo: a Bioconductor package for interpreting lists of Gene Ontology terms. microPubl. Biol. https://doi.org/10.17912/micropub.biology.000811 (2023).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
gxiaolab. Gxiaolab/RNAtracker: for publication. Zenodo https://doi.org/10.5281/zenodo.15528784 (2025).
Purcell, S. et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Human Genet. 81, 559–575 (2007).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Yee, T. W. The VGAM package for categorical data analysis. J. Stat. Softw. 32, 1–34 (2010).
Lex, A., Gehlenborg, N., Strobelt, H., Vuillemot, R. & Pfister, H. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992 (2014).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Gådin, J. R., van’t Hooft, F. M., Eriksson, P. & Folkersen, L. AllelicImbalance: an R/bioconductor package for detecting, managing, and visualizing allele expression imbalance data from RNA sequencing. BMC Bioinformatics 16, 194 (2015).
Acknowledgements
We thank members of the Xiao laboratory for helpful discussions and comments on this work. This work was supported in part by grants from the National Institutes of Health (U01HG009417 and R01AG075206 to X.X.). E.H. was supported by the Graduate Research Fellowship of the NSF under Grant No. DGE-2034835. T.F. was supported by the UCLA Hyde Fellowship and Dissertation Year Fellowship. K.A. was supported by the University of California-Historically Black Colleges and Universities (UC-HBCU) Fellowship. S.T. was supported by the NIH T32GM145388. M.L. was supported by NHGRI grant UM1 HG009382. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
E.H., L.Z. and X.X. designed the study with inputs from all other authors. E.H., L.Z., K.A., R.Y. and J.H. conducted the bioinformatics works. E.H., G.Y. and J.J.L. worked on the statistical modeling. T.F., S.T., T.L.N., C.G.-F., A.K., J.H.B. and R.V. conducted the molecular, cellular and biochemical experiments. M.T.P. and B.M. generated the Bru-seq/BruChase-seq data. X.X., J.J.L. and M.L. provided supervisory inputs. All authors contributed to the writing of the paper. All authors approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Michael Hagemann-Jensen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 RNAtracker facilities the classification of ASE genes.
a, Transcriptomic comparison of Bru-labeled vs. unlabeled K562 cells based on two-sided Pearson’s correlation test (p < 2.2 e-16). b, Allelic ratio distribution after copy-number variant (CNV) removal for the 11 cell lines with >100 genes eligible for classification. Allelic ratio (AR) is calculated by dividing the number of reference allelic counts by total counts per variant. c, Number of genes eligible for classification by RNAtracker in each cell line. d, Number of genes identified as asRS, asRT, or mixed.
Extended Data Fig. 2 Read coverage, gene half-life, and alternative-splicing contribute minimally to RNAtracker performance.
a, Coverage of asRS, asRT, and mixed genes. For each gene, we take the average coverage across all genic regions. b, Estimated half-lives of asRS, asRT, and mixed genes. In boxplots, minima/maxima represent least/greatest proportion values, bounds show 25th and 75th percentiles, and whiskers indicate values within 1.5 * the interquartile range. c, Number of genes with and without gene classification changes after removing SNVs in alternatively spliced regions.
Extended Data Fig. 3 RNAtracker exhibits high Precision and Recall in simulations.
a,b,c 1,000 allelic count datasets were simulated by sampling reads from beta-binomial models using hyperparameters representing 3 different allelic imbalance conditions to assess Precision (a), Recall (b), and Recall among genes passing classification thresholds only (c). In boxplots, minima/maxima represent least/greatest proportion values, bounds show 25th and 75th percentiles, and whiskers indicate values within 1.5 * the interquartile range.
Extended Data Fig. 4 Bias against asRT identification varies across cell lines.
a, Number of expressed genes with and without heterozygous genic single-nucleotide variants (SNVs). Genes without heterozygous SNVs are further categorized into whether they have intronic heterozygous SNVs or 0 heterozygous SNVs (even when introns are considered). To be considered expressed, a gene must have average base coverage ≥ 10 across all genic regions, across all 6 timepoint samples. b, Prevalence (top panel) and number (bottom panel) of intron-based asRT genes. Prevalence is calculated using the total number of genes that are testable based on having SNVs in intronic regions. c, Comparison of the prevalence of stability-regulated genes versus transcriptionally regulated genes (including and excluding intron-based asRT genes). Stability regulated genes include asRS and mixed genes. Transcriptionally regulated genes include asRT, intronic asRT, and mixed genes. To calculate prevalence, the number of genes falling under each of these categories is summed and divided by the total number of genes that were tested using RNAtracker in the cell line.
Extended Data Fig. 5 Low proportion of asRS sharing across cell lines can be attributed to their unique genetic backgrounds.
a, UpSet plot of asRS genes that are unique to or shared across cell lines. b,c UpSet plot of heterozygous single-nucleotide variants (SNVs) within genes classified by RNAtracker (b), as well as all (including intronic) heterozygous SNVs (c) that are unique to or shared across cell lines. Note that Calu3 is not shown because only the top 30 largest intersections are plotted. d, UpSet plot of all genes categorized by RNAtracker across cell lines. e, UpSet plot of asRS variants that are unique to or shared across cell lines. f, Pairs of cell lines that exhibited a significant difference between the expected and actual proportion of overlapping asRS variants. All 24 comparisons had significantly greater actual proportion than expected. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (two-sided binomial test).
Extended Data Fig. 6 Low proportion of asRT sharing across cell lines can be attributed to their unique genetic backgrounds.
a, UpSet plot of asRT genes shared across cell lines. b, UpSet plot of asRT variants shared across cell lines. c, Pairs of cell lines that exhibited a significant difference between the expected and actual proportion of overlapping asRT variants. 33 out of 33 of these comparisons had significantly greater actual proportion than expected. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (two-sided binomial test).
Extended Data Fig. 7 asRS and asRT events are both important contributors to gene expression.
a, Proportion of asRS and asRT variants in each cell line (n = 11) that overlapped expression quantitative trait loci (eQTLs) (p = 3.71e-20). P value was calculated via a two-sided Wilcoxon’s signed rank-test. In boxplots, minima/maxima represent least/greatest proportion values, bounds show 25th and 75th percentiles, and whiskers indicate values within 1.5 * the interquartile range. b, Enrichment (that is Fisher’s exact test odds ratio) of asRS genes that overlap eGenes (compared to asRT genes).*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. None of the comparisons in which the asRT overlap proportion was higher than the asRS overlap proportion were significant.
Extended Data Fig. 8 asRS variants may function by disrupting interactions with trans-regulatory factors.
a, RNA binding proteins (RBPs) with allele-specific binding (ASB) sites that overlap asRS variants. X axis shows percentage of each RBP’s ASB sites that overlap asRS variants. Number of ASB sites that overlap asRS variants is shown to the right of each bar. Purple: RBPs involved with RNA stability and decay according to previous manual literature curation. b, Proportion of miRNA target gain/loss SNPs that overlap asRS or control variants in each cell line (n = 11). In boxplots, minima/maxima represent least/greatest proportion values, bounds show 25th and 75th percentiles, and whiskers indicate values within 1.5 * the interquartile range.
Extended Data Fig. 9 Prime editing supports the causality of asRS variants.
a-c, Genomic DNA sequencing supports the successful genome editing of chr2:173364016:T > C (a), chr11:838672:C > T (b), and chr11:834745:G > T (c). d, Comparison of SNV normalized counts in Bru/BruChase-seq data (2 biological replicates per timepoint) for chr2:173364016:T > C in CDCA7. In boxplots, minima/maxima represent least/greatest proportion values, bounds show 25th and 75th percentiles, and whiskers indicate values within 1.5 * the interquartile range.
Extended Data Fig. 10 asRS and asRT genes are involved in various pathways.
a, Top 20 enriched Gene Ontology (GO) terms for asRT genes. P values were derived from an empirical Gaussian distribution of number of control genes containing each GO term (Methods).
Supplementary information
Supplementary Information
Supplementary Notes 1–7.
Supplementary Tables 1–9
Excel file housing Tables 1–9. Tabs are colored by Table number.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, E., Fu, T., Zhang, L. et al. Genetic variants affecting RNA stability influence complex traits and disease risk. Nat Genet 57, 2578–2588 (2025). https://doi.org/10.1038/s41588-025-02326-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02326-8