Abstract
Pharmacological activation of voltage-gated ion channels by ligands serves as the basis for therapy and mainly involves a classic gating mechanism that augments the native voltage-dependent open probability. Through structure-based virtual screening, we identified a new scaffold compound, Ebio1, serving as a potent and subtype-selective activator for the voltage-gated potassium channel KCNQ2 and featuring a new activation mechanism. Single-channel patch-clamp, cryogenic-electron microscopy and molecular dynamic simulations, along with chemical derivatives, reveal that Ebio1 engages the KCNQ2 activation by generating an extended channel gate with a larger conductance at the saturating voltage (+50 mV). This mechanism is different from the previously observed activation mechanism of ligands on voltage-gated ion channels. Ebio1 caused S6 helices from residues S303 and F305 to perform a twist-to-open movement, which was sufficient to open the KCNQ2 gate. Overall, our findings provide mechanistic insights into the activation of KCNQ2 channel by Ebio1 and lend support for KCNQ-related drug development.

Similar content being viewed by others
Main
Voltage-gated ion channels (VGICs), including voltage-gated sodium (NaV), calcium (CaV) and potassium (Kv) channels, are a large and specific class of ion channels. Our understanding of VGICs’ physiological and pharmacological functions has been greatly advanced by the development of molecular modulators capable of manipulating VGICs’ activities. Small-molecule modulators that target and regulate VGICs have emerged as a remarkably effective therapeutic strategy1,2,3,4,5,6. So far, more than 10% of approved drugs exert their therapeutic effects by acting on VGICs, including the World Health Organization (WHO)’s list of essential medicines such as phenytoin (a NaV inhibitor used for seizures), nifedipine (a CaV inhibitor used for hypertension) and amiodarone (a broad-spectrum Kv channel inhibitor used for arrhythmias)2,6. However, small-molecule drugs that achieve specific selectivity are still lacking for many VGICs. Efforts to discover and expand the repertoire of channel-specific pharmaceutical agents and delineate the structural basis for ligand recognition will facilitate rational design and the development of drugs to treat a wide range of diseases with a high unmet medical need.
Among the 12 subfamilies of Kv channels, KCNQ (KCNQ1–KCNQ5, Kv7.1–Kv7.5) channels are expressed in several tissues. KCNQ2 and KCNQ3 subunits form heteromeric channels conducting the M current, a main potassium current throughout the nervous systems7,8,9. The KCNQ2 channel is prominently expressed at the axon initial segment and at nodes of Ranvier, and act as a gatekeeper to regulate neuronal firing and transmission10,11. Loss-of-function mutations in KCNQ2 result in various types of epileptic seizure12,13,14,15, including benign familial neonatal convulsions and developmental and epileptic encephalopathy. Additionally, because KCNQ2 is implicated in a broad range of phenotypes characterized by hyperexcitability, modulating KCNQ2 presents a potential therapeutic strategy for treating diseases associated with hyperexcitability-associated disorders, such as pain, aged-related sleep disruption, depressive disorder, schizophrenia and amyotrophic lateral sclerosis16,17,18,19,20.
Efforts have been dedicated to identifying small molecules that enhance KCNQ2 currents, particularly at subthreshold voltages, in which increased KCNQ2 activity can effectively prevent abnormal or excessive neuronal firing21,22. Several small-molecule modulators, both endogenous and exogenous, directly target KCNQ2 channel at various sites23,24,25,26,27. Activators such as retigabine (RTG (1), ezogabine) have been exploited for pharmacological applications by negatively shifting channel activation voltage28, thereby augmenting the open probability (PO) of the channel at more negative membrane potentials than usual. Consequently, these ligands substantially boost the current at subthreshold potentials. This mechanism has been supported by our previously reported cryogenic-electron microscopy (cryo-EM) structures of the human KCNQ2 complexed with the activators RTG and ztz240 (ref. 24). The same principle underlying ligand activation also applies to other VGICs, such as CaV and NaV, as well as TRP channels29,30.
Here, we identified the small molecule Ebio1 (2) as a potent and selective KCNQ2 activator. Whole-cell electrophysiology and single-channel patch-clamp recordings revealed that Ebio1 activates KCNQ2 by increasing both the PO and the conductance of the channel, which is distinct from that of the anti-epileptic drug RTG. We confirmed this mechanism by solving the cryo-EM structure of KCNQ2–Ebio1, revealing an extended pore size in the intracellular bundle-crossing gate. To further confirm the activation mechanism, we also synthesized two analogs of RTG (RTG-S1 (3)) and Ebio1 (Ebio1-S1 (4)) as chemical probes and successfully resolved the structure of KCNQ2 in complex with Ebio1-S1. Combining cryo-EM structures, computational simulations, mutagenesis and patch-clamp recordings, we revealed the structural mechanisms for gate opening, involving a twist-to-open motion by the S6 helix.
Results
Ebio1 is a new KCNQ2 channel activator
Using the structure of the KCNQ2–RTG complex (Protein Data Bank (PDB) accession code 7CR2) as an entry point24, we conducted structure-based virtual screening of the Specs database and the ChemDiv database to seek new KCNQ2 channel activators (Fig. 1a). Compounds with poor drug-like properties or pan-assay interference fragments were first filtered, and then the remaining compounds (roughly 1.8 million) were docked to the KCNQ2 structure with a high-throughput virtual screening method. The compounds were refined to form an intermediate library of 1,000 top-ranked compounds using docking simulations involved application of standard precision and extra precision modes. Following filtration based on drug-likeness prediction and clustering analysis, 15 compounds were selected for further KCNQ2 activation activity evaluation (Extended Data Fig. 1a).
a, 1.8 million molecules were docked into the complex structure of KCNQ2 (PDB 7CR2)24. The RTG (purple sphere) binding pocket was chosen as a druggable pocket for docking. b, Chemical structures of RTG and Ebio1. c, Representative macroscopic current traces of KCNQ2 channel before (left traces) and after (right traces) application of 10 μM RTG or Ebio1. The holding potential was −80 mV. The KCNQ2 current was elicited by a series of voltage steps from −90 to +60 mV in 10 mV increments. d, Concentration-dependent curves of RTG and Ebio1 effects on outward current amplitude of KCNQ2 channel at +50 mV. e, Concentration-dependent curves of RTG and Ebio1 effects on the half-activation voltage shift (ΔV1/2) for KCNQ2 channel. n = 3 biological replicates (d,e). Data are presented as the means ± s.e.m.
To test the impact of the selected 15 compounds on the channel, both current amplitude-dependent (I/Icontrol) and voltage-dependent (GV) activation were examined in Chinese hamster ovary cell line K1 (CHO-K1) overexpressing human KCNQ2, using the whole-cell patch-clamp recording. Three compounds enhanced the KCNQ2 current amplitudes and produced a hyperpolarization shift of the voltage-dependent activation curve. Among them, compound 7 (renamed Ebio1) exhibited the most potent KCNQ2 activation activity (Extended Data Fig. 1b,c). Compared with the RTG molecule, Ebio1 contains a completely different scaffold with a nonidentified function (Fig. 1b) and displays a different potentiation activity. Ebio1 dramatically enhanced the amplitude of KCNQ2 outward current in a dose-dependent manner, with a half-maximal effective concentration (EC50) of 247.30 ± 7.90 nM, n ≥ 3 (Fig. 1c,d). Figure 1c shows an example, in which 10 μM Ebio1 produced an enhanced current by an average factor of 5.95 ± 0.35 (n ≥ 3). However, RTG had little effect on the maximal current amplitude, with no clear dose-dependent relationship (Fig. 1c,d). The enhancement in the KCNQ2-mediated current was also produced by a shift in the GV trace of the channel in the hyperpolarizing direction with increasing Ebio1 concentration. The saturation concentration of Ebio1 (10 μM) dramatically caused up to 34.32 ± 2.00 mV left shift of the half-maximal activation voltage (V1/2), from −16.65 to −50.97 mV, n ≥ 3 (Supplementary Fig. 1a). The ΔV1/2 was fitted to a Hill equation with an EC50 of 0.86 ± 0.25 μM (n ≥ 3) with a slope value of 0.65, compared to that of the anti-epileptic drug RTG (EC50 = 1.66 ± 0.04 μM, n ≥ 3) with a slope value of 1.83 (Fig. 1e). Notably, the slope value for RTG is almost three times the slope value for Ebio1, which might indicate that Ebio1 uses a different mechanism of action. In addition to the hyperpolarizing shift of V1/2, Ebio1 also noticeably slows the kinetics of KCNQ2 activation and deactivation. The activation and deactivation rate were increased from 115.12 ± 5.99 to 204.10 ± 15.07 ms (n ≥ 3) and 12.92 ± 1.99 to 65.64 ± 5.13 ms (n ≥ 3) by Ebio1 at +50 mV, respectively (Supplementary Fig. 1b,c). Collectively, the electrophysiological test data support that Ebio1 is a potent activator ligand for the KCNQ2 channel.
Ebio1 engages KCNQ2 activation by raising P O and conductance
KCNQ1–KCNQ5 belong to the KCNQ subfamily and exhibit high sequence and structure similarity. To assess the potential selectivity of Ebio1, we evaluated its activation effect on KCNQ1 to KCNQ5 homomeric channels and KCNQ2/3 heteromeric channel expressed in CHO-K1 cells using the whole-cell voltage-clamp technique. Ebio1 (1 μM) exhibited no effect on the voltage sensitivity of KCNQ1, KCNQ3, KCNQ4 and KCNQ5 (Fig. 2a). External application of 1 μM Ebio1 was ineffective on KCNQ1 and KCNQ3 K+ currents, with values of 0.97 ± 0.01 (n ≥ 3) and 0.98 ± 0.03 (n ≥ 3) for I/I0 at +50 mV, respectively, compared to 4.61 ± 0.17 (n ≥ 3) for KCNQ2. The same experiments carried out with KCNQ2/3, KCNQ4 and KCNQ5 revealed moderately increased currents by 2.62-, 1.85- and 1.63-fold (Fig. 2b and Supplementary Fig. 2a–e), respectively. Although Ebio1 showed moderate activity for the KCNQ2/3, KCNQ4 and KCNQ5 channels, its selectivity for KCNQ2 was substantially higher than that for these two channels. Ebio1 exhibited an activation effect on KCNQ2 with an EC50 of 247.30 ± 7.90 nM, making it tenfold selective for KCNQ2/3, KCNQ4 and KCNQ5, with EC50 values of 2.16 ± 0.33 (n ≥ 3), 2.43 ± 0.19 (n ≥ 3) and 2.27 ± 0.95 μM (n ≥ 3), respectively (Fig. 2c). In the presence of Ebio1, the V1/2 of the KCNQ2/3 channel shifts toward hyperpolarization, similar to activation effect of the KCNQ2 channel. Whereas, Ebio1 has weak or no activation effect of V1/2 on KCNQ4 and KCNQ5 channels (Fig. 2d). These data suggested that Ebio1 was a potential subtype activator of the KCNQ2 channel. Additionally, Ebio1 exhibited minimal to negligible activation effects on other tested Kv, NaV, CaV and K2P channel members, including hERG, BK, NaV1.1, CaV2.1 and TREK1 (Extended Data Fig. 2a–f). These findings robustly indicate that Ebio1 acts as a potent and subtype-selective activator for the KCNQ2 channel.
a, Histogram showing the effects of 1 μM Ebio1 on ΔV1/2 for different KCNQ isoforms. NA means the results are not available. Statistical analysis was a one-way ANOVA with Dunnett’s test. b, Histogram showing the effects of 1 μM Ebio1 on the outward current amplitude of different KCNQ isoforms at +50 mV. Statistical analysis was a one-way ANOVA with Dunnett’s test. c, The dose–response curve of Ebio1 effects on the outward currents of KCNQ2, KCNQ2/3, KCNQ4 and KCNQ5 channels. d, Concentration-dependent curves of Ebio1 effects on the half-activation voltage shift (ΔV1/2) for KCNQ2, KCNQ2/3, KCNQ4 and KCNQ5 channels. e, Representative single-channel recordings from inside-out patches of KCNQ2 at +50 mV in the absence and presence of 10 μM Ebio1, RTG or XE991 (left panel). The corresponding all-point amplitude histograms for the sweeps were fitted by Gaussian distributions (solid line in red) (right panel). f, Histogram showing the PO of KCNQ2 at +50 mV in the absence and presence of 10 μM Ebio1 and RTG. Statistical analysis was a two-tailed t-test. g, Single-channel conductance of KCNQ2 was fitted from two peak values in e. Statistical analysis was a two-tailed t-test. NS, not significant. n = 3, 4, 3, 3 and 3 (a), n = 6, 3, 5, 3, 3 and 3 (b), n = 3 (c,d), n = 4 (f,g) biological replicates. Data are presented as the means ± s.e.m.
To investigate the biophysical and target engagement of Ebio1 on KCNQ2 more directly, we recorded the single-channel activities of KCNQ2 at a +50 mV saturated voltage in CHO-K1 cells. In inside-out recordings, the single-channel PO for KCNQ2 increased in the presence of 10 μM Ebio1 (from 0.66 ± 0.04 to 0.93 ± 0.01, n = 4) or RTG (from 0.67 ± 0.05 to 0.88 ± 0.03, n = 4) (Fig. 2e, f). The single-channel activity was fully blocked by 10 μM XE991, a broad-spectrum KCNQ channel blocker31, indicating that target engagement of Ebio1 on KCNQ2 induces an increase in cell currents (Fig. 2e).
A significant increase in single-channel conductance and single-channel amplitude was observed in response to Ebio1 (from 18.15 ± 1.07 to 36.68 ± 4.78 pS for single-channel conductance and from 1.38 to 2.83 pA for single-channel amplitude; n = 4) (Fig. 2e,g). By contrast, little change in overall conductance was observed for the activator RTG (Fig. 2e,g). This finding aligns with previous research, demonstrating that RTG acts on KCNQ channels not by increasing single-channel conductance or single-channel amplitude but rather by a mechanism that causes V1/2 of KCNQ2 to shift to a more hyperpolarizing potential24,32. Collectively, these data suggest that Ebio1 induces the opening of KCNQ2 channel by a mechanism different from that of the KCNQ channel activator RTG. Meanwhile, Ebio1 did not alter the reversal potential or Rb+ or K+ permeability ratio of KCNQ2 channel, suggesting that K+ selectivity and permeability of the channel was not affected by Ebio1, thus supporting our central claim regarding Ebio1’s impact on single-channel conductance (Supplementary Fig. 3a,b).
Furthermore, in inside-out recordings, Ebio1 has no activity against the single-channel PO and single-channel conductance of KCNQ4 or KCNQ5 channels. While the open probability and single-channel conductance of KCNQ2/3 channel were increased in response to Ebio1 (from 0.74 ± 0.03 to 0.95 ± 0.02 for open probability and from 20.16 ± 0.91 to 32.12 ± 2.23 pS for single-channel conductance; n = 4) (Extended Data Fig. 3a–c). Therefore, Ebio1 may activate KCNQ2/3 channel with a similar mechanism to the KCNQ2 channel.
Cryo-EM structure reveals the detailed Ebio1-binding site
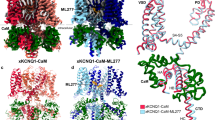
To explain the specific mechanism by which Ebio1 binds to and activates the KCNQ2 channel, we embarked on solving the cryo-EM structure of KCNQ2 in complex with Ebio1. The previously developed truncated human KCNQ2 sample (residues 64–674) and calmodulin (CaM) were coexpressed in human embryonic kidney 293S (HEK293S) cells. Then, the homogeneous sample was purified in the detergent glyco-diosgenin (GDN) and incubated with 150 μM Ebio1 and 1 mM PIP2 at pH 8.0. We conducted cryo-EM single-particle analysis on the purified sample of human KCNQ2–CaM and refined the C4 symmetry-imposed reconstruction map of Ebio1-bound human KCNQ2–CaM to roughly 3.4 Å. The density map was of high quality, allowing us to construct precise atomic models for KCNQ2, CaM and the ligand (Fig. 3a,b, Extended Data Fig. 4a–h, Supplementary Fig. 4 and Supplementary Table 1). The cryo-EM density map showed that the overall architecture of KCNQ2 was similar to that in previously reported maps24. The transmembrane domain of KCNQ2 adopts a canonical domain-swapped arrangement, in which the ion-conduction pore domain (PD) (formed by S4–S5 linker, transmembrane helices S5–S6 and pore helix (PH)) in the center is surrounded by four voltage-sensing domains (VSDs) (formed by transmembrane helices S1–S4) (Fig. 3b). Compared with the VSD in active-state KCNQ2 (PDB 7CR0)24 and intermediate-state KCNQ1 (PDB 6MIE)33, KCNQ2–Ebio1 also possessed an active-state VSD characterized by S4 in our structure in the ‘up’ position (Extended Data Fig. 5a,b). In addition, in the KCNQ2–Ebio1 complex, S6 and helix A (HA) form a continuous helix at the EKR motif, and the whole CaM rotates almost 180° along with HA and helix B (HB), resulting in CaM moving away from the VSD (Extended Data Fig. 5c), similar to that in the previously reported PIP2-bound open-state KCNQ1 and KCNQ4 structure26,34.
a, Cryo-EM reconstructions of human KCNQ2 in complex with Ebio1 (yellow). The four repeat subunits are colored by domain, and CaM is colored pink. The gray bars represent approximate boundaries of the cell membrane. b, Structure model of the KCNQ2–Ebio1 complex viewed parallel to the membrane (left panel) and top-down to the membrane (right panel) from the extracellular space. c, Cryo-EM density of Ebio1 (yellow stick). The gray mesh contoured at 3.4 σ. d, The Ebio1-bound site is shown with specific residues interacting with Ebio1 that are rendered as sticks and spheres. Ebio1 is shown as a yellow stick and sphere. The dashed line represents hydrogen bonding interaction between Ebio1 and hKCNQ2. e, Schematic diagram between Ebio1 and selected surrounding residues. A hydrogen bond with W236 and L299 is shown as a red dashed line. f, The dose–response curves of four mutations (W236A, L299A, I300A, S303A and F305A) on Ebio1 activity. n = 3 biological replicates. Data are presented as means ± s.e.m.
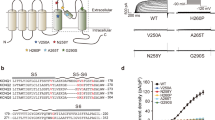
Four pieces of unambiguous density are located in a groove near the PD formed by the S4–S5 linkerI, S5I and S6I from one subunit and S6II from the neighboring subunit (Fig. 3a–d). Ebio1 neatly fits within this density, both sterically and electrostatically, and thus, we revealed the Ebio1-binding site in KCNQ2 (Fig. 3c). Ebio1 achieves stability through van der Waals contacts, hydrophobic interactions and hydrogen bonds, as indicated by the high-resolution map (Fig. 3d,e and Extended Data Fig. 4g). Two aromatic residues, F240 in S5 and F305 in S6, along with the side chain of L299 from the adjacent S6, play pivotal roles in creating space within the groove to accommodate the hydrophobic dihydroacenaphthylene group of Ebio1; the bulky residue W236 in S5 forms π–π stacking with the dihydroacenaphthylene group of Ebio1; the backbone carbonyl oxygen of L299 and indole of W236 form hydrogen bonds with the amide group; and L221 and V225 in the S4–S5 linker, L232 in S5 and L312 in S6 contribute to stabilizing the fluorobenzene group (Fig. 3d,e and Extended Data Fig. 4g). Next, to assess the precise interactions between Ebio1 and KCNQ2 observed in the cryo-EM structure, we performed electrophysiology assays to examine the influences of KCNQ2 mutations on the activation effects of Ebio1. Compared to the WT KCNQ2, the mutations W236A, L299A, I300A, S303A and F305A substantially decreased the potency of Ebio1, suggesting that these residues are crucial for Ebio1 binding (Fig. 3f and Supplementary Fig. 5a–e). For example, at +50 mV, 10 μM Ebio1 increased the current of WT KCNQ2 by sixfold, whereas it only increased the current of I300A by 2.7-fold (Fig. 3f).
Ebio1 activates KCNQ2 via a twist-to-open mechanism
Comparisons of Ebio1- or RTG-bound KCNQ2 structures shed light on the entirely different mechanism of KCNQ2 activation by diverse ligands. Although the overall structure of our resolved KCNQ2–Ebio1 is similar to the previously reported KCNQ2–RTG structure, we noted substantial conformational alterations in both the KCNQ2–RTG and KCNQ2–Ebio1 structures. Overall, the structures exhibited root-mean square-deviation (RMSD) values of 1.86 and 12.02 Å, respectively, against the apo-state KCNQ2 structure, suggesting that the KCNQ2–Ebio1 structure is more different from the apo-state KCNQ2 structure24. In particular, the activation gate (inner gate) is opened to an extended state in the KCNQ2–Ebio1 structure (Fig. 4a–c), which was not observed in previous structural studies of the KCNQ2 channel24. Structurally, in the RTG-bound KCNQ2 channel, residues S314 and L318 in the S6 helices form two constrictions (4.4 Å at S314 and 6.0 Å at L318), whereas in the Ebio1-bound state, the S6 helices began to separate away from the central axis of the pore (Fig. 4b,c). The separation of helices results from an 8° clockwise twisting motion (viewed from the extracellular side) with respect to the S6 axis itself, which induces the C-terminal regions of S6 helices to expand translation by 0.4–5.6 Å with respect to the fourfold axis (from amino acids S303 to E330) (Fig. 4d–f). The twisted region starts from the hydrophilic residue S303, which is relatively conserved among VGICs and other ion channels (Supplementary Fig. 6). Based on the unique conformation of the S6 helix, the side chain of S303 on one side of the twist and the backbone oxygen of F305 on the other side move away from each other from 4.3 to 5.7 Å (Fig. 4g). In this case, both the side chains of S314 and L318 move apart from the ion pathway, leading to an extended central pore and creating the shortest adjacent atom-to-atom distance from approximately 4.4 to 12.4 Å (Fig. 4b,c,g).
a, Structural comparison between RTG-bound KCNQ2 and Ebio1-bound KCNQ2. b, Ion-conduction pores of KCNQ2 channel in the RTG-bound and Ebio1-bound states shown as gray surfaces. Only the PD are shown. Gating residues are shown in sticks and colored orange for S314 and L318. c, Pore radii calculated using the HOLE program as colored in b. Key residues restricting the pore and regions spanning the selectivity filter, pore cavity and inner gate are denoted. d, Overlay of the RTG-bound and Ebio1-bound cryo-EM structures of the KCNQ2 PD. The S6 helix is highlighted. The Cα of residues S303, S314 and E330 are shown as orange spheres. e, During the conformational changes, the vectors representing the amplitude and direction of residues were mapped on the S6 helix. f, Twist-to-open and translational motion regions are magnified from e for clarity. The thin black arrow indicates conformation shifts of key residues. The thick black arrow indicates conformation shifts of different regions. g, An extracellular view of the starting position of the twisted region (upper panels). The distance between S303 and F305 on the adjacent S6 helix is indicated. An intracellular view of the inner gate (bottom panels). The distance between S314 on the adjacent S6 helix is indicated. h, Comparison of two neighboring subunits in the RTG- and Ebio1-bound structures at their binding site. For clarity, the S4–S5 linker and S5 in the second subunit are removed. i, Close-up views of the activators binding site from the membrane plane (left panel) and from the top-down to the membrane (right two panels). Dashed lines represent hydrogen bonding interactions between activators and KCNQ2. RTG and Ebio1 are shown as sticks and spheres.
Although both RTG and Ebio1 fit snugly in the same pocket in KCNQ2 composed of a S5, S6 and S4–S5 linker (Fig. 4h,i), the differences in the chemical structures of RTG and Ebio1 may elicit the twist-to-open activation mechanism by Ebio1. The free –NH2 group of RTG faces toward the S6 helices, at which the group forms a hydrogen bond, like a lock, with the S303 side chain of one S6 helix and the backbone oxygen of F305 on the adjacent S6 helix (Fig. 4i). Compared with RTG, Ebio1 cannot form hydrogen bonds with either S303 or F305, thus making the S6 helix more dynamic and the channel activation gate more open (Fig. 4i). Therefore, with Ebio1 present, we captured the KCNQ2 structure with an expanded activation gate in a twist-to-open mechanism (Supplementary Fig. 7), consistent with the electrophysiology data that Ebio1 is a potent activator to enhance the channel open conductance.
MD of the KCNQ2 gate transition by ligands
High-resolution cryo-EM structures revealed that the gate of the RTG-bound KCNQ2 channel was in a relatively narrow state (renamed the RTG-dependent state), while the gate of the Ebio1-bound KCNQ2 channel was in a more extended state (renamed the Ebio1-dependent state). To gain a deeper understanding of the conformational dynamics of the S6 helix and pore gate and how they may be dependent on ligand binding, we conducted a series of molecular dynamics (MD) simulations under five systems (System I–V, below) derived from the cryo-EM structures of KCNQ2 bound to RTG and Ebio1. Considering the reduced computational costs and the truncated structure sufficient to capture the primary structural differences between the two conformations within the transmembrane region, CaM and the intracellular domain of KCNQ2 were omitted from the structure in these simulation systems. To assess reliability, three independent 500 ns simulations for each system were carried out.
In system I, apo-state KCNQ2 was simulated from the previously published RTG-bound KCNQ2 cryo-EM structure24, in which RTGs were removed from the complex. In the three independent apo-state KCNQ2 simulations, we found that a water molecule presented in the PD of KCNQ2 and formed a hydrogen bond network with residues S303 and F305, similar to the hydrogen bond network among S303–RTG–F305, which limits the expansion of the inner gate (Supplementary Fig. 8a–d).
In system II, the RTG–KCNQ2 complex was simulated from the previously published RTG-bound KCNQ2 cryo-EM structure24. We note that the RTGs are extremely stable in simulations, deviating only approximately 1.7 Å from the pose determined by cryo-EM (Extended Data Fig. 6a,b). Stable hydrogen bonds were observed between the –NH2 group of RTG and S303 or F305 throughout these simulations (Extended Data Fig. 6c). In addition, the pore gate of KCNQ2 was still stable in the RTG-dependent state during the simulation, as in the cryo-EM structure (Extended Data Fig. 6d).
In system III, the Ebio1–KCNQ2 complex from our cryo-EM structure in the paper was simulated. MD simulations reveal stable binding of Ebio1s in the pocket of KCNQ2 with RMSD values at 1.6 Å from the pose determined by cryo-EM (Extended Data Fig. 6e,f). In addition, hydrogen bonds between the ligand and S303 or F305 are abolished, as the hydrophobic dihydroacenaphthylene group of Ebio1 dissipates close to S303 and F305 (Extended Data Fig. 6g), and the pore gate of KCNQ2 remains stable in the Ebio1-dependent state during the simulation, as in the cryo-EM structure (Extended Data Fig. 6h).
In system IV, the RTG–KCNQ2 complex was simulated in which RTGs were docked into the Ebio1 position of the KCNQ2–Ebio1 cryo-EM structure (Fig. 5a). RTGs were stable in simulations initiated with a pose analogous to the cryo-EM pose of Ebio1 (Supplementary Fig. 9a,b). In simulations, the –NH2 group of RTG induced the wobble of S303 and then formed a hydrogen bond network among S303–RTG–F305, thereby closing the distance of S6 helices between two adjacent subunits (Fig. 5b and Supplementary Fig. 9c). Furthermore, the simulations also realized a transition process of the channel gate from an extended state to a narrow state, as the gate diameter of the channel decreased gradually from approximately 10 to 4.6 Å, representing the coordinates of the RTG-dependent KCNQ2 cryo-EM structure (PDB 7CR2)24 (Fig. 5c,d and Supplementary Fig. 9d).
a, Simulation system of RTG docked into the cryo-EM structure of KCNQ2–Ebio1. b,c, Representation of the distance between residues S303 and F305 from the adjacent subunit (b) or channel pore diameter (c) along the three independent repeats of RTG-dependent MD simulations. d, Typical KCNQ2 channel pore conformations of the two states (initial and final) from the RTG-dependent trajectories. e, Simulation system of Ebio1 docked into the cryo-EM structure of KCNQ2–RTG (ref. 24). f,g, Representation of the distance between residues S303 and F305 from the adjacent subunit (f) or channel pore diameter (g) along the three independent repeats of Ebio1-dependent MD simulations. h, Typical KCNQ2 channel pore conformations of the two states (initial and final) from the Ebio1-dependent trajectories.
In system V, the Ebio1–KCNQ2 complex in which Ebio1s were docked into the RTG position of the KCNQ2–RTG cryo-EM structure (PDB 7CR2)24 was simulated (Fig. 5e). Ebio1s were stable in simulations initiated with a pose analogous to the cryo-EM pose of RTG (Supplementary Fig. 9e,f). The movements of the S6 helices and the channel gate were analyzed. During the initial stages, the KCNQ2 channel remained in the RTG-dependent state with the S303–F305 distance at 4.3 Å and the gate diameter at 4.6 Å, and it then dynamically translated to the final state with the S303–F305 distance at 6.8 Å and the gate diameter at 10 Å, representing the coordinates of the Ebio1-dependent KCNQ2 cryo-EM structure (Fig. 5f–h and Supplementary Fig. 9g).
RTG forms a hydrogen bond with the side chain of S303. To test the effects of the hydrogen bond associated to S303 contributed to KCNQ2 activate, we introduced the mutation S303A and measured the effects of S303A on the activities of RTG and Ebio1. We found that removing the hydroxyl group in the side chain of S303 by S303A enhanced RTG activation effect and increased the open probability and the single-channel conductance of the KCNQ2S303A channel (Extended Data Fig. 7a–e), supporting our hypothesis that disruption of a hydrogen bond is essential for RTG-induced current increase. Conversely, when S303 was mutated to alanine, the Ebio1-induced activation effect was largely abolished (Extended Data Fig. 7a–e). To further understand how the hydrogen bond between RTG and S303 restricts the expansion of the KCNQ2 channel inner gate, we performed MD simulation of RTG–KCNQ2S303A. By analyzing all three independent MD trajectories, we observed that the ligand RTG in the S303A mutant was stable. Moreover, the channel inner gate was opened in the two of the three MD trajectories (Supplementary Fig. 10a–d). Therefore, our MD simulation also suggested that mutation S303A would enhance RTG activation, consistent with our patch-clamp recordings.
Overall, our MD simulations support the expectations from the electrophysiological and crystallographic observations, demonstrating that Ebio1 maintains the channel gate in an extended state (Figs. 2g and 4a–g). Furthermore, these computational data also reveal the ligand-dependent conformational transition process of the KCNQ2 activation gate between the RTG-dependent state and Ebio1-dependent state, confirming that the Ebio1-induced twist movement of S6 helices is sufficient to explore KCNQ2 gate activation.
Ebio1 and RTG derivatives influence gate-related twisting
To further clarify the molecular mechanism underlying ligand-dependent gate opening, we designed and synthesized an analog of RTG (renamed RTG-S1) in which the –NH2 group from the middle phenyl ring of the RTG molecule was substituted for a hydrophobic –CH3 group to disrupt the hydrogen bond interaction with S303 and F305 of KCNQ2 (Fig. 6a,b and Supplementary Note). Whole-cell patch-clamp electrophysiology tests showed that RTG-S1 activated KCNQ2 channel in the same way as Ebio1 by increasing outward current amplitude in a dose-dependent manner (EC50 = 0.04 ± 0.02 μM, n ≥ 3) and enhancing the voltage sensitivity of KCNQ2 by shifting the GV curve to the left (EC50 = 0.88 ± 0.36 μM, n ≥ 3) (Fig. 6c,d), which is different from that of the RTG molecule. These data suggest that RTG-S1 stabilizes the channel gate in an extended gate. To test this idea directly, we built an RTG-S1–KCNQ2 complex simulation system by docking RTG-S1 into the RTG position of the KCNQ2–RTG cryo-EM structure. RTG-S1 molecules are stable during the 500 ns simulation and induce major conformational changes in the KCNQ2 gate from a narrow state (as in the RTG-dependent state) to an extended state (as in the Ebio1-dependent state) by promoting gate-related S6 helix twisting from the S303 residue (Extended Data Fig. 8a–f). The MD simulation results support the idea that the ligand-dependent S6 helix twist is central to opening the gate of the KCNQ2 channel.
a, Chemical structure of an Ebio1 analog, Ebio1-S1. b, The detailed binding site for Ebio1-S1 in KCNQ2 channel is shown. Key residues that restricted the motion of the S6 gate are shown as spheres and sticks. c, Concentration-dependent curves of Ebio1 and Ebio1-S1 effects on the outward current amplitude of KCNQ2 channel at +50 mV. d, Concentration-dependent curves of Ebio1 and Ebio1-S1 effects on the ΔV1/2 for KCNQ2 channel. e, Chemical structures of a RTG analog, RTG-S1. f, The detailed binding site for RTG-S1 in KCNQ2 channel is shown. Dashed lines represent hydrogen bonding interactions between RTG-S1 and KCNQ2. Key residues that restricted the motion of the S6 gate are shown as spheres and sticks. g, Concentration-dependent curves of RTG and RTG-S1 effects on the outward current amplitude of KCNQ2 channel at +50 mV. h, Concentration-dependent curves of RTG and RTG-S1 effects on the ΔV1/2 for KCNQ2 channel. i, Cryo-EM reconstructions of human KCNQ2 in complex with Ebio1-S1 indicated in blue. The four repeat subunits are colored by domain, and CaM is colored pink. Cryo-EM density for Ebio1-S1 (blue stick). The gray mesh is contoured at 4.5 σ. j, The Ebio1-S1-bound site is shown with specific residues interacting with Ebio1-S1 rendered stick-like. Ebio1-S1 is shown as a blue stick and sphere and superposed on the docked pose (pink stick and sphere). The dashed line represents the hydrogen bonding interaction between Ebio1-S1 and hKCNQ2. k, Ion-conduction pores of KCNQ2 channel in the Ebio1-S1-bound state are shown as gray surfaces. Only the PD are shown. Gating residues are shown in sticks and colored orange for S314 and L318. l, Pore radii calculated using the HOLE program. Key residues restricting the pore and regions spanning the selectivity filter, pore cavity and inner gate are denoted. n = 3 biological replicates (c,d,g,h). Data are presented as the means ± s.e.m.
Moreover, we also designed and synthesized an analog of Ebio1 (renamed Ebio1-S1) in which a –NH2 group was added to the dihydroacenaphthylene group of Ebio1 (Fig. 6e and Supplementary Note). We docked Ebio1-S1 into the KCNQ2–Ebio1 cryo-EM structure. Consistent with their similar chemical scaffolds, Ebio1-S1 shares a similar binding mode with that of Ebio1 (Fig. 6f). Compared with Ebio1, Ebio1-S1 forms additional hydrogen bond interactions with residues S303 and F305 by the –NH2 group, which stabilize its binding to KCNQ2 and may prevent the channel gate from the twisting and opening (Fig. 6f). Electrophysiological recording also showed that Ebio1 and Ebio1-S1 display different potentiation activities on KCNQ2. Ebio1-S1 minimally affects the maximal current amplitude of the KCNQ2 channel (Fig. 6g). Instead, it enhances KCNQ2 channel activation, achieved by shifting the V1/2 of the GV curve toward hyperpolarization, with an EC50 value of 0.68 ± 0.18 μM (n ≥ 3) (Fig. 6h).
We resolved the cryo-EM structure of human KCNQ2 bound to the high-affinity ligand Ebio1-S1 at a nominal resolution of 3.0 Å (Fig. 6i, Extended Data Fig. 9a–h, Supplementary Fig. 11 and Supplementary Table 1). Electron density maps confirmed the Ebio1-S1 docking predictions, with an RMSD value between the cryo-EM structure and the docked pose of 1.1 Å (Fig. 6i,j and Extended Data Fig. 9f). Hydrogen bond interactions between the –NH2 group of Ebio1-S1 and the side chain of S303 and the main chain carbonyls of F305 are also found in the cryo-EM complex structure (Fig. 6j). The Ebio1-S1 bound structure of KCNQ2 contains an activated VSD and a relatively narrow channel gate, which is similar to that in the RTG-bound KCNQ2 cryo-EM structure24, but it opened in the Ebio1-bound KCNQ2 cryo-EM structure (Fig. 6k,l and Extended Data Fig. 10a,b). These findings are consistent with our electrophysiological studies results that Ebio1-S1 may adopt a similar activation mechanism to RTG but not to Ebio1. To investigate the conformational variations of KCNQ2 on binding of Ebio1-S1, we conducted MD simulation of Ebio1-S1 bound to the Ebio1-dependent state KCNQ2 cryo-EM structure. MD simulation reveals stable binding of Ebio1-S1 in the KCNQ2 channel with the channel gate conformation translating from an extended state (as in the Ebio1-dependent state) to a relatively narrow state (as in the cryo-EM structure of the KCNQ2–Ebio1-S1 complex), which further validated the binding model and the ligand-dependent activation mechanism in our structures (Supplementary Fig. 12a–g).
Discussion
VGICs are integral in cellular signaling, involving a step-by-step process of voltage-dependent gating. This process includes detecting voltage changes across the membrane35,36,37, VSD activation leading to conformational rearrangements38,39 and the subsequent opening of the ion-conduction pore40. According to the structural and gating characteristics of VGICs, the most common activation mechanism of VGICs by small molecules is that small molecules increase the voltage sensitivity of the channel, either by stabilizing the S4 or S5 helix24,29 or enhancing VSD-pore coupling through direct interactions23,26,41,42. These small molecules cause a leftward shift in the voltage-dependent activation of VGICs, ultimately increasing VGICs’ PO at specific membrane potential. Theoretically, another putative activation strategy for small molecules to VGICs is directly influence the pore gate size of the channel to promote its openness to conduct ions. Here, we identified that the small molecule Ebio1 activates the KCNQ2 channel by directly opening the channel gate. Ebio1 not only increased the PO of the channel but also increased the channel conductance by approximately twofold. The channel conductance is a fundamental property influencing ion flow across cell membranes, affecting cellular excitability and information processing. Numerous factors contribute to the determination of the channel conductance, including the size and composition of the ion channel pore, the presence of ion selectivity filters and the presence of gating mechanisms43,44,45. Moreover, small molecules can exert their effects on unitary conductance through several mechanisms, including altering channel gating kinetics, affecting ion permeation properties or binding directly to specific channel domains46,47,48. Such modulation not only provides insights into the fundamental biophysics of ion channels but also offers opportunities for developing precision pharmacological interventions. The cryo-EM structure of KCNQ2 bound with Ebio1 and MD simulations revealed that Ebio1 increases the channel conductance by stabilizing the gate of the KCNQ2 channel in a more open state, offering a new class of ligands with a new mechanism of action.
Some ion channels may have intrinsic twist-to-open motions49,50,51,52,53. KirBac, an inwardly rectifying potassium channel from bacteria, is activated, which involves a ‘twisting engagement’ followed by a ‘locking rotation’ of the cytoplasmic and transmembrane domains essential for gate opening53,54. In this study, we demonstrate that small molecules could trigger twist-to-open motions for ion channels. Specifically, the Ebio1 or RTG-S1-induced opening initiates a rotation at the residue S303 of the S6 helix. This rotation motion is transmitted through the helix and eventually drives the opening of the inner gate in the S6 helix. Small molecules RTG and Ebio1-S1 form hydrogen bonds with the S303 side chain of one S6 helix and the residue F305 backbone of the adjacent S6 helix through the –NH2 group, locking S303’s movement and limiting the helix’s rotation. However, Ebio1 is unable to form hydrogen bonds, thus making the S6 helix more dynamic and further opening the channel gate. Notably, the residue S303 in S6 helix is conserved across multiple ion channels (Supplementary Fig. 6), suggesting the ligand-dependent twist-to-open activation mechanism of KCNQ2 gating reported here may serve as a door opener for developing modulators of other ion channels.
Small-molecule Ebio1 shows high selectivity for KCNQ2 over other subtypes, yet the underlying mechanism remains unclear, as it involves multiple factors. For instance, the microenvironment within the binding pockets55 and conformational dynamics56, along with distal residues57, may influence ligand binding and contribute to selectivity. Further, obtaining the structures of Ebio1 with other KCNQ subtypes may be helpful for investigating the precise mechanism of selectivity. In conclusion, we report the discovery of Ebio1, a new potent, selective chemical activator for the KCNQ2 channel. Through the development of target engagement assays, including mutagenesis and single-channel electrophysiology, we demonstrate that Ebio1 displays a pronounced on-target KCNQ2 channel. The cryo-EM structures and MD simulations of Ebio1 and two negative control compounds, Ebio1-S1 and RTG-S1, bound to the KCNQ2 channel highlight a unique activation mechanism by small molecules on VGICs. In addition, these results provide a template for discovering and clarifying the mechanism of channel-selective chemical tools, and their function can be probed to inform the development of new therapeutic agents.
Methods
Cell culture and transfection
CHO-K1 cells were cultured in DMEM/F12 (Gibco, catalog no. 11330-032) supplemented with 10% FBS (Gibco, catalog no. 16000-044) and 1% penicillin-streptomycin (Gibco, catalog no. 15140122). HEK293T and HEK293 cells were cultivated in DMEM (Gibco, catalog no. 11995500BT) contained 10% FBS and 1% penicillin-streptomycin. Cells were incubated at 37 °C in the presence of 5% CO2. When conducting plasmids of ion channels for electrophysiology experiments, the complementary DNA (cDNA) of human KCNQ1, KCNQ2 and KCNQ5 were inserted into a pIRES2-EGFP vector and the cDNAs of human KCNQ3, KCNQ4, BK and CaV2.1 (CaV2.1/CaVβ4/CaVα2δ1) were subcloned into pcDNA3.1 vector, respectively. Also, the cDNA of human TREK1 was ligated to a pEGFP-N1 vector. All plasmids and site-directed mutagenesis of KCNQ2 used in experiments were conducted and sequencing was confirmed by providers (Genewiz), except for CaV2.1 that was provided by ICE Bioscience Inc., and KCNQ3 and KCNQ4 that were provided by Youbio Biological Technology Co., Ltd. For further electrophysiological recordings, the TREK1, KCNQ1–5 and mutation of KCNQ2 expression plasmids were transiently transfected with Lipofectamine 3000 reagent (Invitrogen, catalog no. 11668019) into CHO-K1 cells seeded in six-well plates. In the same way, HEK293T and HEK293 cells were transiently transfected with the plasmids of BK and CaV2.1, respectively. Electrophysiological recordings were performed on the cells that had been transfected for 24–48 h.
Electrophysiological tests of hERG and NaV1.1 were performed in stable cell lines. Cells that stably expressed hERG potassium channels were provided by Creacell, whereas CHO-K1-NaV1.1 stable cell lines were generated by ICE Bioscience Inc. Cells that stably expressed hERG potassium channels were cultivated in DMEM with 10% FBS and 0.8 mg ml−1 G418 (GPC, catalog no. AK108) and CHO-K1-NaV1.1 cells grown in HAM’S/F12 (HyClone, catalog no. SH30026.01) containing 10% FBS, 100 μg ml−1 Zeocin (Solarbio, catalog no. Z8020) and 10 μg ml−1 Blasticidin (Solarbio, catalog no. B9300).
Electrophysiological recording
Whole-cell voltage-clamp recordings were performed in transiently transfected CHO-K1 cells at the room temperature (23–25 °C) by the amplifier EPC10 (HEKA). The patch pipettes were pulled from borosilicate glass capillaries (World Precision Instruments) and fire-polished to a resistance of 3–5 MΩ. For recordings of the KCNQ2 channel, pipettes were filled with the intracellular solution composed of the following: 150 mM KCl, 3 mM MgCl2, 5 mM EGTA and 10 mM HEPES (pH 7.3 with KOH). The bath solution contained 145 mM NaCl, 5 mM KCl, 1 mM CaCl2, 3 mM MgCl2 and 10 mM HEPES (pH 7.4 with NaOH). Patches were voltage clamped at a holding potential of −80 mV. Currents of KCNQ2 channel were elicited by a series of depolarization voltage stepping from −90 to +60 mV at 10 mV increments, followed by −120 mV to record the tail current. For the ion permeation experiments, 145 mM NaCl in the bath solution was replaced with 100 mM K+ or 100 mM Rb+, and the intracellular solution and the recording program were as same as those described above. When recording the reversal potential, pipettes were filled with the intracellular solution composed of the following: 150 mM KCl, 3 mM MgCl2, 5 mM EGTA and 10 mM HEPES (pH 7.3 with KOH). The bath solution contained 145 mM NaCl, 5 mM KCl, 1 mM CaCl2, 3 mM MgCl2 and 10 mM HEPES (pH 7.4 with NaOH). Patches were voltage clamped at a holding potential of −80 mV and elicited by a depolarization voltage at +60 mV, following by a series of stepping voltages from −120 to +60 mV.
The methods of recording the KCNQ1, KCNQ3, KCNQ4 and KCNQ5 channels were as same as the procedure for recording the KCNQ2 channel. The recording procedures of the TREK1 channel, BK channel and hERG channel were as same as we reported before in refs. 58,59. For NaV1.1 channel recording, the extracellular solution and the pipette solution were consistent with those for the NaV1.7 channel recording we have already reported in ref. 59. Cells expressing NaV1.1 channel were stimulated by two protocols at a holding potential of −120 mV. First, the depolarization to 0 mV for 500 ms was performed to test the resting state sodium current, and then the voltage was applied to a half inactivation voltage for 5 s, followed by 20 ms recovery at −120 mV. Second, to detect the half-inactivated state sodium current, the procedure for depolarizing to 0 mV for 50 ms was carried out. When performing the experiments on the CaV2.1 channel, the standard pipette solution contained 120 mM CsCl, 1 mM MgCl2•6H2O, 10 mM HEPES, 10 mM EGTA, 0.3 mM Na2-GTP, 4 mM Mg-ATP (pH 7.2, adjusted with CsOH) and the external solution contained 140 mM TEA-Cl, 2 mM MgCl2•6H2O, 10 mM CaCl2•2H2O, 10 mM HEPES and 5 mM d-glucose (pH 7.4, adjusted with TEA-OH). The depolarizing voltage lasting for 0.3 s was stepped from the holding voltage of −80 to +20 mV to elicit the inward current of the CaV2.1 channel (the test pulse was modified slightly based on the lead IV test). All the electrical signals were filtered at 2.9 kHz and digitized at 10 kHz.
Single-channel recordings were made 48 h after the transfection in the excised inside-out configuration of the patch clamp using EPC10 (HEKA) at 23–25 °C. The composition of intracellular solution was 144 mM NaCl, 2.5 mM KCl, 0.5 mM MgCl2, 2 mM MgCl2 and 5 mM HEPES (pH 7.4, adjusted with NaOH) and the patch pipette resistance was 7–15 MΩ. The bath solution contained 144 mM NaCl, 40 mM KCl, 0.5 mM MgCl2, 2 mM MgCl2 and 5 mM HEPES (pH 7.4, adjusted with NaOH). Records were digitized at 10 kHz and filtered at 2 kHz. Obtained records were filtered at 500 kHz to reduce baseline noise. All-point histograms were fitted with single- or muti-Gaussian curves to obtain the mean single-channel current (i) and the PO. The ratio of the area, whose peak corresponded to a bigger current, called the ‘open state’, to the total area under the entire Gaussian was considered to be PO, and the value of the peak of the open state was denoted i. Single-channel conductance (g) was obtained by the formula g = i/(V − VE), where V is the test potential and VE is the reversal potential of potassium.
Patch-clamp data analysis
Processed by Clampfit v.10.4 (Molecular Devices), patch-clamp data were analyzed in GraphPad Prism v.8.0.2 (GraphPad Software). Voltage-dependent activation curves were fitted with the Boltzmann equation, G = Gmin + (Gmax − Gmin)/(1 + exp(V − V1⁄2)/S), where Gmax is the maximum conductance, Gmin is the minimum conductance, V1⁄2 is the voltage at which 50% of the maximum conductance is reached and S is the slope factor. Hill’s equation was used to acquire the dose–response curves, E = Emax/(1 + (EC50/C)n), where Emax is the maximal effect, EC50 was defined previously and n is the Hill coefficient. Exponential equations containing one or two components were used to fit the activation and deactivation traces in Clampfit v.10.4.
Protein expression and purification
DNAs encoding human KCNQ2 (Homo sapiens NP_004509.2) and CaM (Homo sapiens NP_001734.1) were synthesized by GenScript. To improve the biochemical and thermal stability of KCNQ2, the N- and C-terminal regions were truncated, leaving a construct with residues 64–674. The gene containing a C-terminal strep-tag was cloned into a pEZT-BM expression vector. The human CaM gene was cloned into a pEZT-BM vector with a C-terminal histidine tag. The KCNQ2 and CaM complexes were heterologously expressed in HEK293S GnTl− suspension cells (Life Technologies) using the BacMam system (Thermo Fisher Scientific). Suspended HEK293S GnTl− cells at 3.0–3.5 × 106 cells per ml were infected with P2 baculovirus mixture of KCNQ2:CaM (6:1) generated in Sf9 cells (Life Technologies) at a molar ratio of 1:10 (virus to HEK293S, v/v). Then, 12 h after infection, 10 mM sodium butyrate was added to enhance protein expression. Cells were cultured at 30 °C for another 48–60 h before they were collected by centrifugation at 3,000g.
The cell pellet was resuspended in lysis buffer A (20 mM Tris, pH 8.0, 150 mM KCl) supplemented with a protease inhibitor cocktail (2 μg ml−1 DNase I (Sigma, catalog no. AMPD1), 0.5 μg ml−1 pepstatin (AMRESCO, catalog no. 26305-03-3), 2 μg ml−1 leupeptin (AMRESCO, catalog no. 103476-89-7), 1 μg ml−1 aprotinin (AMRESCO, catalog no. 9087-70-1), 1 mM phenylmethylsulfonyl fluoride (AMRESCO, catalog no. 329-98-6)) and homogenized by sonication on ice. KCNQ2 protein was extracted with 1.5% (w/v) n-dodecyl-β-d-maltopyranoside (DDM) (Anatrace, catalog no. D310) and 0.3% (w/v) cholesteryl hemisuccinate (CHS) (Anatrace, catalog no. CH210) by gentle agitation for 3 h at 4 °C. After ultracentrifugation at 48,000g, the supernatant was collected and then incubated gently with Strep-Tactin Sepharose resin (IBA) at 4 °C for 1.5 h. The collected resin was washed in buffer B (buffer A + 0.05% (w/v) DDM and 0.01% CHS) for 4 column volumes. Then, the detergent was changed to 0.02% GDN (Anatrace, catalog no. GDN101) for 16 column volumes. The KCNQ2 was eluted with 10 mM d-desthiobiotin in buffer C (buffer A + 0.02% GDN). The protein sample was further purified by size-exclusion chromatography with a Superose 6 10/300 GL column (GE Healthcare) equilibrated with buffer C. The peak fraction of KCNQ2 was collected and concentrated for cryo-EM analyses. For KCNQ2–CaM bound to Ebio1, the purified protein was incubated with 0.15 mM Ebio1 and 1 mM PIP2. For KCNQ2–CaM bound to Ebio1-S1, the purified protein was incubated with 0.3 mM Ebio1-S1 and 1 mM PIP2.
Cryo-EM sample preparation and data acquisition
Ebio1 was synthesized in the laboratory. The cryo-EM data of KCNQ2 structures was collected at the Center of Cryo-Electron Microscopy, Zhejiang University. For grid preparation, 3 μl concentrated protein was loaded onto glow-discharged R1.2/1.3 Quantifoil grids at 4 °C under 100% humidity. Grids were blotted for 4.5 s and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI). Micrographs were acquired on a Titan Krios microscope (FEI) operating at a voltage of 300 kV equipped with the energy filter and Falcon 4 detector (Thermo Fisher Scientific). A calibrated magnification of ×130,000 was used for imaging, yielding a pixel size of 0.93 Å on images. Each micrograph was dose-fractionated to 40 frames with a dose rate of 7.49 e−/pixel s−1, with a total exposure time of 6 s, corresponding to a total dose of about 52 e−/Å2.
Image processing
Motion correction and contrast transfer function (CTF) parameters estimation were performed with the MotionCorr2 (ref. 60) and the GCTF programs61, respectively. All image processing steps were carried out with RELION v.3.1 (ref. 62).
For the human KCNQ2 with Ebio1, 1,611 micrographs were collected and 715,625 particles were auto-picked and extracted with a binning factor of 3 for 2D classification. The following two rounds of 3D classification with 669,146 selected particles were performed using the map of human KCNQ2–CaM complex (PDB 7CR3) as a ref. 24. After 3D classification, selected particles were combined and re-extracted to the pixel size of 0.93 Å for 3D refinement with a C4 symmetry and Bayesian polishing via RELION v.3.1. The final resolution of the EM map by 3D reconstruction of 28,232 particles was 3.4 Å.
For the human KCNQ2 with Ebio1-S1, 5,683 micrographs were collected and 1,886,690 particles were auto-picked and extracted with a binning factor of 3 for 2D classification. The following two rounds of 3D classification with 1,187,036 selected particles were performed using the map of human KCNQ2–CaM complex (PDB 7CR3) as a ref. 24. After 3D classification, selected particles were combined and re-extracted to the pixel size of 0.93 Å for 3D refinement with a C4 symmetry and Bayesian polishing via RELION v.3.1. The final resolution of the EM map by 3D reconstruction of 151,312 particles was 3.0 Å.
The resolution was estimated by applying a soft mask around the protein density and the gold-standard Fourier shell correlation (FSC) = 0.143 criterion. Local resolution maps were calculated with RELION v.3.1 (ref. 62).
Structure determination, refinement, and validation
De novo atomic model building was performed in Coot based on the 3.4 Å resolution map of human KCNQ2–Ebio1 (ref. 63). An initial model of human KCNQ2–Ebio1 was performed using ligand-free KCNQ2 (PDB 7CR3) as a template24. PHENIX was used for model refinement against cryo-EM maps using real-space refinement64, with secondary structure restraints and noncrystallography symmetry applied. The MolProbity was used for model geometry statistics generation65. The data validation statistics are shown in Supplementary Table 1. Pore radii were calculated and analyzed by the HOLE and MOLE2 program66,67. Figures were produced using PyMOL (The PyMOL Molecular Graphics System, v.1.8 Schrödinger, LLC.), ChimeraX68.
Molecular docking and virtual screening
The cryo-EM structure of human KCNQ2 (PDB 7CR2)24 was used to perform structure-based virtual screening via a workflow application of Glide in Maestro (Schrödinger Release 2021-2). The structure was prepared in the protein preparation Wizard Workflow of Glide. The center of the three-dimensional receptor grid was generated according to the position of RTG in the structure via Receptor Grid Generation module.
The commercial Specs and the ChemDiv database were selected as screening databases. Compounds containing pan-assay interference compounds or violating ‘Lipinski’s Rule of Five’ were removed before virtual screening. The LigPrep module of Maestro was used to generate the three-dimensional (3D) conformations of the remaining about compounds. During the process, the remaining compounds were considered to be at pH 7.0 ± 2.0 to estimate their protonation state using the Epik module. At the very beginning, the compounds were screened using the high-throughput virtual screening module and the top-ranked 20,000 candidates were selected on the basis of the Glide G-score. Then, these candidates were redocked using the standard precision and extra precision mode in turn. Finally, 15 compounds with shape rationality and structural diversity were picked for electrophysiological assay.
The extra precision mode of Glide was used to predict the binding poses of Ebio1 and RTG analogs. First, the Ebio1 and RTG binding pockets were set as the grid center. Then, we set the inner box values to 10 × 10 × 10 Å3. In the final step, The Ebio1-S1 and RTG-S1 were docked into the Ebio1-binding site of Ebio1 and RTG (PDB 7CR2)24 cryo-EM structures, respectively. The maximum number output poses were set to 1,000. Subsequent screening details are shown in Supplementary Table 2.
Simulation systems preparation
A total of eight simulation systems of KCNQ2-bound ligands were built. For apo-state KCNQ2 simulation (system 1), the cryo-EM structure of the KCNQ2–RTG (PDB 7CR2)24 that removed the ligands was selected as the initial structure. For systems 2 and 3, the cryo-EM structures of the KCNQ2 transmembrane domain (residues 70–326) in complex with RTG or Ebio1 were selected as the initial structures, respectively. The missing residues from 185 to 194 in the KCNQ2–RTG and KCNQ2–Ebio1 cryo-EM structures (S3–S4 loop) were modeled and subjected to 500 rounds of very slow loop refinement assessed by DOPE scoring using Modeller v.9.24 software69. Simulations of KCNQ2-bound, RTG or Ebio1-S1 (system 4 and 5) were initiated with the ligands docked into the Ebio1 position of the KCNQ2–Ebio1 cryo-EM structure reported in this work. Based on system 1, the residue S303 was mutated to alanine and selected as the initial structure for system 6. Simulations of KCNQ2-bound, Ebio1 or RTG-S1 (systems 7 and 8) were initiated with the ligands docked into the RTG position of the KCNQ2–RTG cryo-EM structure (PDB 7CR2)24. Then, all the structure models were refined by the protein preparation Wizard Workflow integrated in Maestro (Schrödinger Release v.2021-2), including adding missing heavy atoms and all hydrogens, followed by full energy minimization. Titratable residues were left in their dominant state at pH 7.0 and all histidine side chains were represented with a hydrogen atom on the epsilon nitrogen. Protein terminal free-amino and free-carboxyl groups were capped with neutral acetyl and methylamine groups. Afterward, the refined structure models were embedded in a pre-equilibrated palmitoyloleoylphosphatidylcholine bilayer containing 521 lipid molecules by orienting the transmembrane properly in the membrane using the CHARMM-GUI website70. The generated systems were solvated by TIP3P waters and neutralized with 0.15 M KCl, leading to a total of around 182,000 atoms and a box size of 15 × 15 × 9.0 nm3.
MD simulations
All-atom MD simulations were performed using the GROMACS2020.2 package in the NPT ensemble with periodic boundary conditions and the CHARMM36m force field71. Parameters for ligand molecules were generated using the CGenFF program72, with additional optimization using quantum mechanics for ligands with high penalty scores. Before the final production run of 500 ns simulations, 50,000 steps of energy minimization were performed for each system followed by equilibration in the NPT ensembles for 5 ns, with positional restraints (1,000 kJ mol−1 nm−2) placed on heavy atoms of proteins. A second round of NTP equilibration for 20 ns was run with positional restraints (1,000 kJ mol−1 nm−2) on backbone atoms to allow for side-chain relaxation. Meanwhile, the positions of ligand molecules were also restrained during the NTP equilibration processes. System temperature was maintained at 300 K using the v-rescale method with a coupling time of 0.1 ps and pressure was maintained at 1 bar using the Berendsen barostat with a coupling time of 1.0 ps and compressibility of 4.5 × 10−5 bar−1 with semi-isotropic coupling. Simulations were performed with a 2 fs timestep and all bond lengths were constrained using LINCS. The restraints of ligands and protein were removed in the process of 500 ns MD simulation. Electrostatic interactions were computed using the particle mesh Ewald method with nonbonded interactions cut at 12.0 Å. Analysis of hydrogen bonds was performed by using criterion that distance (such as O–N distance in the case of an amino group of molecules to the side chain hydroxyl of S303 or the main chain of F305) less than 0.35 Å and an angle between the donor proton, the donor heavy atom and the acceptor heavy atom less than 30°. Distance analysis was performed by using standard GROMACS analysis tools.
Synthesis of Ebi1, Ebio-S1 and RTG-S1
Please see the Supplementary Note in the Supplementary Information.
Statistics
No statistical methods were used to determine the sample size in advance. Statistical analyses were conducted with GraphPad Prism v.8.0.1 (GraphPad Software). Parameters of statistic are shown in figures and figure legends. Concentration–response and GV curves were acquired by nonlinear fitting, as concentration–response was fitted by the standard four-parameter Hill equation and a GV curve was fitted by the Boltzmann equation. Under the assumption that all data from the samples satisfied the normal distribution, comparisons between two-group means were made by a two-tailed t-test, while comparisons for means among more than three groups was made by a one-way analysis of variance (ANOVA) with Dunnett’s test applied to account for multiple comparisons. Data were reported as mean ± s.e.m. (standard error of the mean).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 3D cryo-EM density maps of Ebio1- and Ebio-S1-bound KCNQ2 structures have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-35487 and EMD-38041, respectively. Atomic coordinates for the atomic models of Ebio1- and Ebio-S1-bound KCNQ2 structures have been deposited in the PDB under the accession numbers 8IJK and 8X43, respectively. All stable reagents generated in this study are available from the lead contact without restriction. Plasmids and strains are available from the authors upon request. Source data are provided with this paper.
References
Alsaloum, M., Higerd, G. P., Effraim, P. R. & Waxman, S. G. Status of peripheral sodium channel blockers for non-addictive pain treatment. Nat. Rev. Neurol. 16, 689–705 (2020).
Santos, R. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017).
Schewe, M. et al. A pharmacological master key mechanism that unlocks the selectivity filter gate in K+ channels. Science 363, 875–880 (2019).
Wulff, H. & Zhorov, B. S. K+ Channel modulators for the treatment of neurological disorders and autoimmune diseases. Chem. Rev. 108, 1744–1773 (2008).
Wulff, H., Castle, N. A. & Pardo, L. A. Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discov. 8, 982–1001 (2009).
Wulff, H., Christophersen, P., Colussi, P., Chandy, K. G. & Yarov-Yarovoy, V. Antibodies and venom peptides: new modalities for ion channels. Nat. Rev. Drug Discov. 18, 339–357 (2019).
Peters, H. C., Hu, H., Pongs, O., Storm, J. F. & Isbrandt, D. Conditional transgenic suppression of M channels in mouse brain reveals functions in neuronal excitability, resonance and behavior. Nat. Neurosci. 8, 51–60 (2005).
Wang, H. S. et al. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science 282, 1890–1893 (1998).
Soh, H., Springer, K., Doci, K., Balsbaugh, J. L. & Tzingounis, A. V. KCNQ2 and KCNQ5 form heteromeric channels independent of KCNQ3. Proc. Natl Acad. Sci. USA 119, e2117640119 (2022).
Kuba, H., Yamada, R., Ishiguro, G. & Adachi, R. Redistribution of Kv1 and Kv7 enhances neuronal excitability during structural axon initial segment plasticity. Nat. Commun. 6, 8815 (2015).
Schwarz, J. R. et al. KCNQ channels mediate IKs, a slow K+ current regulating excitability in the rat node of Ranvier. J. Physiol. 573, 17–34 (2006).
Orhan, G. et al. Dominant-negative effects of KCNQ2 mutations are associated with epileptic encephalopathy. Ann. Neurol. 75, 382–394 (2014).
Vanoye, C. G. et al. High-throughput evaluation of epilepsy-associated KCNQ2 variants reveals functional and pharmacological heterogeneity. Jci. Insight 7, e156314 (2022).
Miceli, F. et al. Genotype-phenotype correlations in neonatal epilepsies caused by mutations in the voltage sensor of KV7.2 potassium channel subunits. Proc. Natl Acad. Sci. USA 110, 4386–4391 (2013).
Perucca, P. & Perucca, E. Identifying mutations in epilepsy genes: Impact on treatment selection. Epilepsy Res. 152, 18–30 (2019).
Lopez, J. P. et al. Ketamine exerts its sustained antidepressant effects via cell-type-specific regulation of Kcnq2. Neuron 110, 2283–2298 (2022).
Li, S. B. et al. Hyperexcitable arousal circuits drive sleep instability during aging. Science 375, eabh3021 (2022).
Tsuboi, D. et al. Dopamine drives neuronal excitability via KCNQ channel phosphorylation for reward behavior. Cell Rep. 40, 111309 (2022).
Huang, X. et al. Human amyotrophic lateral sclerosis excitability phenotype screen: target discovery and validation. Cell Rep. 35, 109224 (2021).
Maljevic, S., Wuttke, T. V. & Lerche, H. Nervous system KV7 disorders: breakdown of a subthreshold brake. J. Physiol. 586, 1791–1801 (2008).
Peretz, A. et al. Targeting the voltage sensor of Kv7.2 voltage-gated K+ channels with a new gating-modifier. Proc. Natl Acad. Sci. USA 107, 15637–15642 (2010).
Borgini, M., Mondal, P., Liu, R. & Wipf, P. Chemical modulation of Kv7 potassium channels. RSC Med. Chem. 12, 483–537 (2021).
Zhang, Q. et al. Dynamic PIP2 interactions with voltage sensor elements contribute to KCNQ2 channel gating. Proc. Natl Acad. Sci. USA 110, 20093–20098 (2013).
Li, X. et al. Molecular basis for ligand activation of the human KCNQ2 channel. Cell Res. 31, 52–61 (2021).
Abbott, G. W. KCNQs: ligand- and voltage-gated potassium channels. Front. Physiol. 11, 583 (2020).
Zheng, Y. et al. Structural insights into the lipid and ligand regulation of a human neuronal KCNQ channel. Neuron 110, 237–247 (2022).
Li, T. et al. Structural basis for the modulation of human KCNQ4 by small-molecule drugs. Mol. Cell 81, 25–37 (2021).
Stafstrom, C. E., Grippon, S. & Kirkpatrick, P. Ezogabine (retigabine). Nat. Rev. Drug Discov. 10, 729–730 (2011).
Catterall, W. A., Lenaeus, M. J. & El-Din, T. M. G. Structure and pharmacology of voltage-gated sodium and calcium channels. Annu. Rev. Pharmacol. Toxicol. 60, 133–154 (2020).
Diaz-Franulic, I., Poblete, H., Miño-Galaz, G., González, C. & Latorre, R. Allosterism and structure in thermally activated transient receptor potential channels. Annu. Rev. Biophys. 45, 371–398 (2016).
Zaczek, R. et al. Two new potent neurotransmitter release enhancers, 10, 10-bis(4-pyridinylmethyl)-9(10H)-anthracenone and 10, 10-bis(2-fluoro-4-pyridinylmethyl)-9 (10H)-anthracenone: comparison to linopirdine. J. Pharmacol. Exp. Ther. 285, 724–730 (1998).
Tatulian, L. & Brown, D. A. Effect of the KCNQ potassium channel opener retigabine on single KCNQ2/3 channels expressed in CHO cells. J. Physiol. 549, 57–63 (2003).
Taylor, K. C. et al. Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state. eLife 9, e53901 (2020).
Sun, J. & MacKinnon, R. Structural basis of human KCNQ1 modulation and gating. Cell 180, 340–347 (2020).
Jiang, Y. et al. X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 (2003).
Swartz, K. J. Sensing voltage across lipid membranes. Nature 456, 891–897 (2008).
Catterall, W. A. Structure and function of voltage-gated ion channels. Annu. Rev. Biochem. 64, 493–531 (1995).
Hou, P. et al. Two-stage electro-mechanical coupling of a KV channel in voltage-dependent activation. Nat. Commun. 11, 676 (2020).
Kalstrup, T. & Blunck, R. S4-S5 linker movement during activation and inactivation in voltage-gated K+ channels. Proc. Natl Acad. Sci. USA 115, E6751–E6759 (2018).
Jensen, M. Ø. et al. Mechanism of voltage gating in potassium channels. Science 336, 229–233 (2012).
Xiong, Q., Sun, H. & Li, M. Zinc pyrithione-mediated activation of voltage-gated KCNQ potassium channels rescues epileptogenic mutants. Nat. Chem. Biol. 3, 287–296 (2007).
Rodriguez-Menchaca, A. A. et al. PIP2 controls voltage-sensor movement and pore opening of Kv channels through the S4-S5 linker. Proc. Natl Acad. Sci. USA 109, E2399–E2408 (2012).
Naranjo, D., Moldenhauer, H., Pincuntureo, M. & Díaz-Franulic, I. Pore size matters for potassium channel conductance. J. Gen. Physiol. 148, 277–291 (2016).
Lansky, S. et al. A pentameric TRPV3 channel with a dilated pore. Nature 621, 206–214 (2023).
Henderson, S. W., Nourmohammadi, S., Ramesh, S. A. & Yool, A. J. Aquaporin ion conductance properties defined by membrane environment, protein structure, and cell physiology. Biophys. Rev. 14, 181–198 (2022).
Perszyk, R. E. et al. The negative allosteric modulator EU1794-4 reduces single-channel conductance and Ca2+ permeability of GluN1/GluN2A N-methyl-d-aspartate receptors. Mol. Pharmacol. 99, 399–411 (2021).
Carnevale, V. & Klein, M. L. Small molecule modulation of voltage gated sodium channels. Curr. Opin. Struct. Biol. 43, 156–162 (2017).
Canul-Sánchez, J. A. et al. Different agonists induce distinct single-channel conductance states in TRPV1 channels. J. Gen. Physiol. 150, 1735–1746 (2018).
Sansom, M. S. Ion-channel gating. Twist to open. Curr. Biol. 5, 373–375 (1995).
Taly, A., Hénin, J., Changeux, J. P. & Cecchini, M. Allosteric regulation of pentameric ligand-gated ion channels: an emerging mechanistic perspective. Channels 8, 350–360 (2014).
Valadié, H., Lacapcre, J. J., Sanejouand, Y. H. & Etchebest, C. Dynamical properties of the MscL of Escherichia coli: a normal mode analysis. J. Mol. Biol. 332, 657–674 (2003).
Yang, H. Y. et al. Inherent dynamics of the acid-sensing ion channel 1 correlates with the gating mechanism. PLoS Biol. 7, e1000151 (2009).
Clarke, O. B. et al. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell 141, 1018–1029 (2010).
Bavro, V. N. et al. Structure of a KirBac potassium channel with an open bundle crossing indicates a mechanism of channel gating. Nat. Struct. Mol. Biol. 19, 158–163 (2012).
Powers, A. S. et al. Structural basis of efficacy-driven ligand selectivity at GPCRs. Nat. Chem. Biol. 19, 805–814 (2023).
Hollingsworth, S. A. et al. Cryptic pocket formation underlies allosteric modulator selectivity at muscarinic GPCRs. Nat. Commun. 10, 3289 (2019).
Huang, X. S. et al. Structural basis for high-voltage activation and subtype-specific inhibition of human NaV1.8. Proc. Natl Acad. Sci. USA 119, e2208211119 (2022).
Liao, P. et al. Selective activation of TWIK-related acid-sensitive K+ 3 subunit–containing channels is analgesic in rodent models. Sci. Transl. Med. 11, eaaw8434 (2019).
Zhang, Q. et al. Inhibiting Hv1 channel in peripheral sensory neurons attenuates chronic inflammatory pain and opioid side effects. Cell Res. 32, 461–476 (2022).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. GCTF: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Sehnal, D. et al. MOLE 2.0: advanced approach for analysis of biomacromolecular channels. J. Cheminform. 5, 39 (2013).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Berendsen, H. J. C., van der Spoel, D. & van Drunen, R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995).
Vanommeslaeghe, K. & MacKerell, A. D. Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012).
Acknowledgements
This work is funded by grants from the National Key Research and Development Program of China (grant nos. 2022YFE0205600 to H.Y. and P.H.; 2018YFA0508100 to J.G. and Q.Z. and 2020YFA0908501 to J.G.), the National Natural Science Foundation of China (grant nos. 82273857 to Q.Z. and 32171221 to P.H.), the Joint Funding of the Macau Science and Technology Development Fund and the Ministry of Science and Technology of the People’s Republic of China (grant no. 0006/2021/AMJ to P.H.), the Innovative Research Team of High-level Local Universities in Shanghai (grant no. SHSMU-ZDCX20211201 to H.Y.) and the East China Normal University Medicine and Health Joint Fund (grant no. 2022JKXYD07001 to H.Y. and Z.C.). J.G. is supported by the MOE Frontier Science Center for Brain Science & Brain-Machine Integration, Zhejiang University. Single-particle cryo-EM data were collected at Center of Cryo-Electron Microscopy at Zhejiang University. We are also grateful for the support of the East China Normal University Multifunctional Platform for Innovation (001 and 011).
Author information
Authors and Affiliations
Contributions
H.Y., Q.Z. and J.G. conceived the project, designed the research and supervised the study. S.Z. performed virtual screening. K.W., Y.L. and J.H. performed whole-cell electrophysiology tests. K.W. and J.L. performed single-channel data acquisition. S.Z. and K.W. performed electrophysiological experiment analysis. D.M., Z.Y. and X.L. performed sample preparation, collected the cryo-EM structural data and solved the structures. S.Z., Q.Z. and J.S. performed and analyzed MD simulations. Q.Z., L.M. and Y.Y. assisted in compounds preparation and synthesis. Z.C. and P.H. provided intellectual expertise and shared key methodologies. S.Z., D.M. and Q.Z. prepared the draft of the manuscript. H.Y., Q.Z. and J.G. wrote the manuscript. All authors read and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.Z., K.W., Y.L., L.M., Y.Y., J.S., J.G., Q.Z. and H.Y. are inventors of patent applications that cover the potential usage of Ebio1 and its derivatives. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Huaizong Shen and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Virtual screening and biophysical characterization of small-molecules on KCNQ2 channel.
a, Chemical structures of initial 15 docking compounds, each with a different scaffold. b, Histogram showing the outward current potentiation effects of 15 candidates on the KCNQ2 channel. The effects of 10 μM candidates are checked at +50 mV. The dash line indicates a potentiation level of 1 (that is no effect). c, The effects of 10 μM candidates on the ΔV1/2 of KCNQ2 channel. NA means the results are not available. n = 3, 3, 2, 2, 2, 3, 3, 3, 3, 2, 3, 6, 3, 5 and 5 (b), n = 2, 2, 2, 4, 2, 2, 2, 3, 6, 2, 3 and 2 (c) biological replicates. Data are presented as the means ± s.e.m.
Extended Data Fig. 2 The selectivity of Ebio1 on several ion channels.
a, Summary of the effects of Ebio1 on TREK1, BK, hERG, CaV2.1, and NaV1.1 channels. Statistical analysis: one-way ANOVA with Dunnett’s test. b-f, The representative current traces of TREK1 (b), BK (c), hERG (d), CaV2.1 (e), and NaV1.1 (f) channels in the absence and the presence 10 μM Ebio1 are shown. n = 3, 3, 3, 2, 2 and 2 biological replicates (a). Data are presented as the means ± s.e.m.
Extended Data Fig. 3 Effects of Ebio1 on the activity of single KCNQ2/3, KCNQ4 and KCNQ5 channels.
a, Representative single-channel recordings from inside-out patches of KCNQ2/3, KCNQ4 and KCNQ5 at +50 mV in the absence and presence of 10 μM Ebio1 (left panel). The corresponding all-point amplitude histograms for the sweeps were fitted by Gaussian distributions (solid line in red) (right panel). b, Histogram showing the PO of KCNQ2/3, KCNQ4 and KCNQ5 at +50 mV in the absence and presence of 10 μM Ebio1. Statistical analysis: two-tailed t-test. n.s., not significant. c, Single-channel conductance of KCNQ2/3, KCNQ4 and KCNQ5 was fitted from two peak values in (a). Statistical analysis: two-tailed t-test. n.s., not significant. n = 4, 4, 3, 3, 3 and 3 (b), n = 4, 4, 3, 3, 3 and 3 (c) biological replicates. Data are presented as the means ± s.e.m.
Extended Data Fig. 4 Structure determination of human KCNQ2-Ebio1 complex.
a, Size-exclusion chromatography of KCNQ2-CaM on Superose 6 and SDS-PAGE analysis of the final sample. n = 3 independent experiments. b, Representative cryo-EM micrograph of KCNQ2-Ebio1. n = 3 independent experiments. c, Representative 2D classes. d, Flowchart of image processing for KCNQ2-Ebio1 particles. e, The density map of KCNQ2-Ebio1 is colored by local resolution. The local resolution is estimated with RELION 3.1 and generated in Chimera. f, The Gold-standard FSC curves of the final 3D reconstruction of KCNQ2-Ebio1, and the FSC curve for cross-validation between the map and the model of KCNQ2-Ebio1. g, EM densities for Ebio1 in the KCNQ2 channel. Gray meshes represent EM densities for Ebio1 and its surrounding residues. h, The weak and isolated densities are observed between VSD and PD in the map of KCNQ2-Ebio1 complex. The EM densities are contoured at the level of 0.01 in UCSF ChimeraX.
Extended Data Fig. 5 Structural changes induced by Ebio1.
a, The VSD structure of the Ebio1-bound KCNQ2. Only the S2-S4 helices are shown for clarity. The side chain of gating charges in S4 and the gating charge transfer center residue are shown in sticks and spheres. b, Comparison of Ebio1-bound VSD of KCNQ2 with open-state VSD of KCNQ2 (PDB code: 7CR0)24 and intermediate-state VSD of KCNQ1 (PDB code: 6MIE)33, which colored cyan, gray and purple, respectively. The side chain of gating charges in S4 and the gating charge transfer center residue are shown in sticks. c, Conformational change of the KCNQ2 channel complex in one KCNQ2-CaM subunit after Ebio1 bound. The conserved ‘EKR’ motif is colored red, which undergo structural rearrangement from a loop to a helix. S6 and HA helices of KCNQ2 are colored cyan. CaM is shown as helix and surface with its N-lobe in purple and C-lobe in pink. The rotational motion of CaM after Ebio1 bound is represented by a cartoon with a dash arrow.
Extended Data Fig. 6 MD simulations of the Ebio1- and RTG-bound KCNQ2 channel.
a, Simulation system II, RTG/KCNQ2 complex from the RTG-bound KCNQ2 cryo-EM structure. b, RMSD of RTG molecules against simulation time in the three independent repeats MD simulations of simulation system II. c, The number of hydrogen bonds formed between the -NH2 group of RTG and residue S303 (upper) or F305 (bottom) in the three independent repeats MD simulations of simulation system II. d, Representation of the channel pore diameter along the three independent repeats of RTG/KCNQ2 MD simulations of simulation system II. e, Simulation system III, Ebio1/KCNQ2 complex from the Ebio1-bound KCNQ2 cryo-EM structure. f, RMSD of Ebio1 molecules against simulation time in the three independent repeats MD simulations of simulation system III. g,h, Representation of the distance between residue S303 and F305 from adjacent subunit (g) or channel pore diameter (h) along the three independent repeats of Ebio1/KCNQ2 MD simulations of simulation system III.
Extended Data Fig. 7 The hydrogen bond restrained pore gate opening by affecting S6 helix motion.
a, Representative macroscopic current traces of KCNQ2S303A channel before (left traces) and after (right traces) application of 10 μM RTG or Ebio1. The holding potential was −80 mV. The KCNQ2S303A current was elicited by a series of voltage steps from −90 mV to +60 mV in 10 mV increments. b, Histogram showing the effects of 10 μM RTG or Ebio1 on the outward current amplitude of KCNQ2S303A at +50 mV. Statistical analysis: two-tailed t-test. c, Representative single-channel recordings from inside-out patches of KCNQ2S303A channel at +50 mV in the absence and presence of 10 μM RTG or Ebio1 (left panel). The corresponding all-point amplitude histograms for the sweeps were fitted by Gaussian distributions (solid line in red) (right panel). d, Histogram showing the PO of KCNQ2S303A at +50 mV in the absence and presence of 10 μM RTG or Ebio1. Statistical analysis: two-tailed t-test. n.s., not significant. e, Single-channel conductance of KCNQ2S303A was fitted from two peak values in (c). Statistical analysis: two-tailed t-test. n.s., not significant. n = 6, 7, 3 and 3 (b), n = 7, 7, 3 and 3 (d), n = 7, 7, 3 and 3 (e) biological replicates. Data are presented as the means ± s.e.m.
Extended Data Fig. 8 Small molecules RTG-S1 induce the dynamic rearrangement of the pore region of KCNQ2 channel.
a, RMSD of RTG-S1 molecules against simulation time in the three independent repeats MD simulations. b, Ensemble plot of the RTG-S1 molecule in the binding pocket of KCNQ2 channel during the MD simulations. c,d, Representation of the distance between residues S303 and F305 from the adjacent subunit (c) or channel pore diameter (d) along the three independent repeats of RTG-S1-dependent MD simulations. e, Typical KCNQ2 channel pore conformations of the two states (initial and final) from the RTG-S1-dependent trajectories. f, Superimposition of the gate region of the RTG-S1-dependent final state and KCNQ2-RTG (left) or KCNQ2-Ebio1 (right) cryo-EM structures.
Extended Data Fig. 9 Structure determination of human KCNQ2-Ebio1-S1 complex.
a, Size-exclusion chromatography of KCNQ2-CaM on Superose 6 and SDS-PAGE analysis of the final sample. n = 3 independent experiments. b, Representative cryo-EM micrograph of KCNQ2-Ebio1-S1. n = 3 independent experiments. c, Representative 2D classes. d, Flowchart of image processing for KCNQ2-Ebio1-S1 particles. e, The density map of KCNQ2-Ebio1-S1 is colored by local resolution. The local resolution is estimated with RELION 3.0 and generated in Chimera. f, EM densities for Ebio1-S1 in the KCNQ2 channel. Gray meshes represent EM densities for Ebio1-S1 and its surrounding residues. g, The Gold-standard FSC curves of the final 3D reconstruction of KCNQ2-Ebio1-S1, and the FSC curve for cross-validation between the map and the model of KCNQ2-Ebio1-S1. h, The weak and isolated densities are observed between VSD and PD in the map of KCNQ2-Ebio1-S1 complex. The EM densities are contoured at the level of 0.01 in UCSF ChimeraX.
Extended Data Fig. 10 Cryo-EM structures of human KCNQ2-Ebio1-S1 complex.
a, Structure model of the KCNQ2-Ebio1-S1 complex viewed parallel to the membrane (left) and top-down to the membrane (right) from the extracellular space. b, Comparison of Ebio1-S1-bound VSD of KCNQ2 with Ebio1-bound VSD of KCNQ2, which colored yellow and blue, respectively. The side chain of gating charges in S4 and the gating charge transfer center residue are shown in sticks.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Figs. 1–12.
Supplementary Data 1
Statistical source data for Supplementary Figs. 1–3, 5, 8–10 and 12.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data
Source Data Extended .Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed gel.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed gel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, S., Ma, D., Wang, K. et al. A small-molecule activation mechanism that directly opens the KCNQ2 channel. Nat Chem Biol 20, 847–856 (2024). https://doi.org/10.1038/s41589-023-01515-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41589-023-01515-y
This article is cited by
-
Structural basis for the subtype-selectivity of KCa2.2 channel activators
Nature Communications (2026)
-
Accessing sulfonamides via formal SO2 insertion into C–N bonds
Nature Chemistry (2025)
-
The small molecule Ebio3 inactivates the KCNQ2 channel without blocking the pore
Nature Chemical Biology (2025)
-
Small molecule inhibits KCNQ channels with a non-blocking mechanism
Nature Chemical Biology (2025)
-
A new twist to increase ion flow
Nature Chemical Biology (2024)