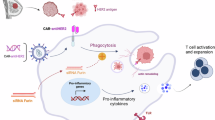
Abstract
Chimeric antigen receptor (CAR) T cell therapies have successfully treated hematological malignancies. Macrophages have also gained attention as an immunotherapy owing to their immunomodulatory capacity and ability to infiltrate solid tumors and phagocytize tumor cells. The first-generation CD3ζ-based CAR-macrophages could phagocytose tumor cells in an antigen-dependent manner. Here we engineered induced pluripotent stem cell-derived macrophages (iMACs) with toll-like receptor 4 intracellular toll/IL-1R (TIR) domain-containing CARs resulting in a markedly enhanced antitumor effect over first-generation CAR-macrophages. Moreover, the design of a tandem CD3ζ-TIR dual signaling CAR endows iMACs with both target engulfment capacity and antigen-dependent M1 polarization and M2 resistance in a nuclear factor kappa B (NF-κB)-dependent manner, as well as the capacity to modulate the tumor microenvironment. We also outline a mechanism of tumor cell elimination by CAR-induced efferocytosis against tumor cell apoptotic bodies. Taken together, we provide a second-generation CAR-iMAC with an ability for orthogonal phagocytosis and polarization and superior antitumor functions in treating solid tumors relative to first-generation CAR-macrophages.
Similar content being viewed by others
Main
Cancer immune cell therapies illustrated by chimeric antigen receptor (CAR) T cells have achieved great success in B cell leukemia/lymphoma and other hematologic malignancies1. Although therapeutic cells have been designed to precisely target solid tumor antigens, the immune cell therapy of solid tumors warrants further investigation2,3. To date, challenges including exhaustion in the immunosuppressive tumor microenvironment (TME)4, limited infiltration into dense extracellular matrix5, off-target effects6, antigen escape and heterogeneity within the tumor7,8 are widely thought to be the substantial obstacles to successful immune cell therapies against solid tumors.
Macrophages, a type of terminally differentiated monocytic phagocytes, come to the horizon of the tumor immunotherapy field due to their engulfing capacity, central roles in the crosstalk between adaptive and innate immune systems and critical roles in defending pathogens and combating cancer cells9,10. Accordingly, macrophages have become a potential target of immunotherapy10. An increasing body of evidence substantiates that tumor-associated macrophages (TAMs) reside in tumor tissues and account for most of the infiltrated leukocytes10,11. However, these TAMs are reprogramed toward the alternatively activated M2 phenotype and have a tumorigenic role in carcinomas through suppressing immune responses, such as downregulating immunological activation-related transcripts including IL-12A and adaptors involved in the toll-like receptor (TLR) signaling pathway, inducing angiogenesis and facilitating the escape of tumor cells12,13,14,15,16,17,18,19. Therefore, the introduction of pro-inflammatory or M1-like macrophages or conversion of M2- to M1-like macrophages represents a viable antitumor strategy (macrophage polarization is a continuum, with the M1 or M2 fully polarized cell at the extremes, and for simplicity, the macrophages on either end of the spectrum are named as M1-like or M2-like here)20,21,22. Over the years, substantial efforts have been made to target endogenous macrophages or TAMs23,24. Recent technology progress in genetically engineered macrophages enabled us to better harness macrophages as a type of therapeutic cell for adoptive transfer25,26,27. Currently, preclinical and clinical studies using genetically engineered macrophages are ongoing. With the large availability, stability and standardization of the engineered macrophages, induced pluripotent stem cell (iPSC)-derived macrophages (iMACs) have been used as a valuable alternative solution25,28,29,30. Previous work has verified the feasibility of producing CAR-iMACs and demonstrated the CAR-dependent activation of CAR-iMACs in treating blood cancers25. However, the first-generation CAR containing the CD3ζ activating domain that is responsible for the effector cell function of phagocytosis in macrophages was borrowed from CAR-T cells. In theory, it is not feasible to polarize macrophages toward the durable M1-like pro-inflammatory state25. Thus, designing a new macrophage-specific CAR (M-CAR) to confer CAR-iMACs with both phagocytosis abilities and polarization functions would contribute to the fight against solid cancers.
TLRs are pattern-recognition receptors (PRRs) involved in immune regulation by recognizing distinct pathogen-associated molecular patterns (PAMPs)31. TLR4 is a typical TLR member widely expressed by myeloid cells such as macrophages32,33. Once recognizing lipopolysaccharide (LPS), TLR4 interacts with adaptor molecules via its toll/IL-1R (TIR) signal transduction domain and leads to nuclear translocation of nuclear factor kappa B (NF-κB)/p65, promoting the expression of pro-inflammatory cytokines such as interleukin (IL)-1A, IL-1B, IL-6, IL-12, chemokine (C–C motif) ligand 8 (CCL8) and tumor necrosis factor (TNF)34,35,36,37,38,39,40. Based on the biological mechanisms of TLR4, we hypothesized that introducing the intracellular TIR domain of TLR4 into the CAR would stimulate and maintain the M1-like phenotype of iMACs upon engaging antigens and elevate antitumor efficacy.
In this work, we developed the second-generation M-CAR by integrating intracellular CD3ζ and TIR domains in tandem to construct an antigen-targeting M-CAR. Genetic modification of iMACs with the second-generation M-CAR substantially improved CAR- and antigen-dependent antitumor efficacy both in vitro and in vivo in two different solid tumor models. Further cellular and molecular analysis and comprehensive single-cell RNA-seq (scRNA-seq) assessment showed that the underlying mechanisms consisted of TIR promoting the M1-like pro-inflammatory state and suppressing the M2-like state in an NF-κB pathway-dependent manner. Notably, we also identify an antitumor mechanism of CAR-mediated efferocytosis of iMACs, consisting of induction of target tumor cell apoptosis and engulfment of apoptotic bodies from tumor cell debris by CAR-iMACs. Thus, we have established a proof-of-concept for second-generation CAR-iMACs and provided insights into mechanisms of action for future myeloid cell-based immune cell therapies for solid tumors.
Results
Construction of M-CARs and development of antigen-targeting CAR-iMACs
There is ample evidence that the CD3ζ signal transduction domain obtained from T cell-specific CAR can stimulate the first generation of human CAR-iMACs25,26,27. However, this domain may not enable stimulation of the M1-like state of macrophages in the TME25. Therefore, we designed M-CARs containing the intracellular TIR domain from TLR4, whose activation can drive M1-like macrophage polarization via interplay with TIR domain-containing adaptors. Meanwhile, we used EGFRvIII- and Glypican-3 (GPC3)-targeting single-chain fragment variable (scFv) as the extracellular domain and designated it TIR-CAR to distinguish it from the previous CD3ζ-CAR (Fig. 1a). Inspired by the evolutionary process of different generations of CARs in T cells (T-CARs), we simultaneously designed a second-generation M-CAR by integrating intracellular CD3ζ and TIR domains in tandem designated as CD3ζ-TIR-CAR (Fig. 1a). A truncated CAR without an intracellular domain was designed as the negative control (Fig. 1a). We then transduced the above CARs into human iPSCs with lentiviruses to construct CAR-expressing iPSCs. The CAR-expressing iPSCs were subsequently differentiated into CAR-iMACs via our established differentiation platform25 (Fig. 1b). Flow cytometric (FCM) analysis showed that all types of CAR-iMACs exhibited relatively high expression of enhanced green fluorescent protein (EGFP) (Fig. 1c) and achieved above 80% CAR transgene expression (Fig. 1d). We thus have established the iPSC lines for producing antigen-specific CAR-iMACs.
a, Constructs of EGFRvIII/GPC3-targeting CARs. The CARs mainly comprise an extracellular signal peptide, a scFv recognizing EGFRvIII/GPC3, a transmembrane (TM) domain and a hinge region from CD8α, and either without an intracellular domain (truncated CAR), or with an intracellular CD3ζ signaling domain (CD3ζ-CAR), or cytoplasmic TIR domain from TLR4 (TIR-CAR), or both CD3ζ and TIR domains in tandem (CD3ζ-TIR-CAR). The hinge was added between scFv and CD8α TM domains to endow the CARs with the flexibility to target antigens. All the CAR transgenes were linked to the EGFP-expressing sequence via a T2A element. b, Overview of the differentiation process of EGFRvIII-targeting CAR-iMACs from the CAR-expressing iPSCs. EGFP-labeled CAR-iPSCs were differentiated into CAR-iMACs by mimicking the in vivo process of mesoderm induction, hematopoietic stem cell specification and myeloid cell production. Confocal imaging was performed to observe the morphology of CAR-iMACs by detecting the EGFP protein. The experiment was performed more than five times. c, Flow cytometry analysis of EGFP-positive populations of EGFRvIII-CAR-iMACs. d, Flow cytometry analysis was performed to determine the transduction efficiency of the four types of EGFRvIII-CARs in iMACs by detecting the expression of CARs via an allophycocyanin (APC)-conjugated rabbit antibody specific to F(ab′)2 of human IgG. Results in c and d are representative plots from three biological replicates. The results were processed using FlowJo.
We subsequently analyzed the expression of TLR4 signaling pathway genes at different stages of wild-type (WT) myeloid/macrophage differentiation with RNA-seq data25. We found that key factors such as IRF7, RELA (encoding NF-κB P65), MYD88, TRAP6, TRAM and TAB2/3 were highly expressed on day 18 and declined on day 28 (Fig. 2a). This finding suggested that most downstream modulators of TLR4 were higher when the iMACs were still in a monocyte-like state, compared with the later stage where the iMACs were more matured. Furthermore, M1-polarized iMACs showed higher expression of TLR4 signaling pathway-associated genes compared to the naive or M2-polarized iMACs (Fig. 2b). Thus, the data suggested that the early differentiated iMACs resembled the M1-polarized iMACs in terms of higher expression of the TLR4 pathway genes and might reserve more potent signaling capacity upon TLR4 pathway activation. We thus chose to use the earlier-stage iMACs for the following studies.
a, A heatmap from RNA-seq showed dynamic changes in the expression levels of TLR4 pathway-related signal transduction genes along with the differentiation process from iPSCs to mature iMACs. The samples were taken from the undifferentiated iPSC population, EBs on day 2 and day 7 and iMACs on day 18 and day 28. b, Comparison of the expression level of TLR4 pathway-related signal transduction genes in the in vitro cultured naive iMACs, LPS/IFN-γ-polarized M1-like iMACs and IL-4/IL-10-polarized M2-like iMACs. All the above heatmaps were generated with Cluster 3.0 and Treeview software. c,d, After coculturing EGFRvIII-targeting truncated CAR-iMACs, CD3ζ-CAR-iMACs and TIR-CAR-iMACs with U87MGEGFRvIII cells at E/T of 3/1, 5/1 and 10/1 for 12 h (c) and 24 h (d), respectively, statistics of FCM analyses showed the proportion of residual U87MGEGFRvIII cells (the tdTomato+ tumor cells in the pregated EGFP− population of the cocultured cells; n = 3 biologically independent samples per group). Significance was calculated with two-way ANOVA analysis with multiple comparisons and is presented as mean ± s.e.m. e, Progressive anticancer cell activity shown by the proportion of residual U87MGEGFRvIII cells from 12 to 24 h incubation. f, Statistics of BLI from viable FFluc+ U87MGEGFRvIII cells after incubating with the three types of CAR-iMACs for 24 h. E/T ratios were set as 1/1, 3/1, 5/1 and 10/1. The experimental groups without CAR-iMACs treatment were designed as negative controls (NCs, n = 3 biologically independent samples per group). Significance was calculated with two-way ANOVA analysis with multiple comparisons and is presented as mean ± s.e.m. g–j, After 24 h of co-incubation of the three types of CAR-iMACs with U87MGEGFRvIII cells, respectively, ELISA assays were performed to detect the production of IL-6 (g), IL-12 (h), IL-23 (i) and TNF (j) in the medium of the individual experimental group (n = 3 biologically independent samples per group). Significance was calculated with multiple t tests analysis and is presented as mean ± s.e.m. All the statistical data above was shown by GraphPad Prism 8.2.1. *P <0.05; **P <0.01; ***P <0.001; ****P < 0.0001. ANOVA, analysis of variance; NS, not significant.
To establish a target cell line for the EGFRvIII-CAR-expressing iMACs, we cloned the human EGFRvIII sequence into a lentivirus vector (Extended Data Fig. 1a) and overexpressed the gene in U87MG cells to obtain a stable EGFRvIII-expressing tumor cell line (U87MGEGFRvIII). While basal expression of total EGFR was observed in both WT and transgenic U87MG cells, only the latter exhibited high expression of EGFRvIII protein (Extended Data Fig. 1b,c). We also observed that EGFRvIII was localized both on the cell membrane and in the cytoplasm due to overexpression (Extended Data Fig. 1d). As for GPC3, it is naturally expressed on the HepG2 cells. Thus, we have obtained tumor cell lines for assessing the effector functions of different antigen-targeting CAR-iMACs.
The TIR domain contributes to CAR-iMACs anticancer activity
To evaluate whether the TIR signal domain can enhance CAR-iMACs’ phagocytosis of tumor cells in a CAR-dependent manner, we first incubated the single intracellular domain-containing CAR-iMACs cells with U87MGEGFRvIII cells in vitro. We stimulated WT-iMACs and EGFRvIII-targeting CAR-iMACs (truncated CAR-iMACs, CD3ζ-CAR-iMACs and TIR-CAR-iMACs) with interferon-γ (IFN-γ) and LPS, respectively, and then cocultured the cells with tdTomato-expressing (tdTomato+) U87MGEGFRvIII cells with different effect/target ratios (E/T) of 3/1, 5/1 and 10/1. After 4 h of coculturing, we observed the enhanced adhesion of all types of CAR-iMACs to the cancer cells compared to WT-iMACs (Extended Data Fig. 1e), suggesting that the CAR conferred iMACs with targeting capacity. Moreover, immunofluorescence (IF) assays showed that both CD3ζ-CAR-iMACs and TIR-CAR-iMACs more potently engulfed tdTomato-labeled components of tumor cells than WT-iMACs and truncated CAR-iMACs after 12 h of coculturing (Extended Data Fig. 1f), indicating that overexpression of CAR enhanced this function. Noteworthy, we observed that the CAR-iMACs tended to bind to tumor cells and induce their apoptosis followed by phagocytizing the debris. Next, FCM analysis showed that compared to both WT-iMACs and truncated CAR-iMACs, CD3ζ- and TIR-CAR-iMACs treatment led to less residual tumor cells when co-incubated with U87MGEGFRvIII cells at the same E/T ratio (Fig. 2c), suggesting their stronger antitumor capacity. We did not observe a significant difference between CD3ζ-CAR-iMACs and TIR-CAR-iMACs at 12 h of incubation (Fig. 2c).
To demonstrate the progressive antitumor effect of EGFRvIII-targeting CAR-iMACs, we performed the same experiments at 24 h of co-incubation. The antitumor capacity of CAR-iMACs was improved at all tested E/T ratios compared with 12 h of co-incubation (Fig. 2d), and the TIR domain conferred stronger activity than CD3ζ at this time point (Fig. 2d). When we plotted the residual tumor cells at both 12 and 24 h, we found that TIR-CAR-iMACs gradually exhibited a greater antitumor cell effect than CD3ζ-CAR-iMACs (Fig. 2e). To further confirm the cytolytic activity of CAR-iMACs, we overexpressed a firefly luciferase (FFluc) gene in U87MGEGFRvIII cells and then co-incubated the tumor cells with the three types of CAR-iMACs, respectively, for 24 h. Consistently, the BLI signal from tumor cells was significantly attenuated by both CD3ζ-CAR-iMACs and TIR-CAR-iMACs compared to truncated CAR-iMACs, and the optimal efficiency was achieved at an E/T ratio of 10/1 (Fig. 2f). Furthermore, to investigate whether the pro-inflammatory activity of CAR-iMACs was dependent on CAR activation, we measured the secreted TIR downstream cytokines released in the medium. The ELISA assay revealed that when incubated with U87MGEGFRvIII cells for 24 h at an E/T ratio of 10/1, TIR-CAR-iMACs secreted considerably higher levels of pro-inflammatory cytokines including IL-6 (Fig. 2g), IL-12 (Fig. 2h), IL-23 (Fig. 2i) and TNF (Fig. 2j) indicating higher pro-inflammatory activity of TIR-CAR-iMACs. Interestingly, the T cell-specific CD3ζ domain-containing CAR also promoted low expression of these cytokines in CAR-iMACs (Fig. 2g–i), suggesting that combining both domains might achieve the maximal pro-inflammatory activity. Together, these results demonstrated that both TIR and CD3ζ domains enhanced CAR-dependent cytotoxic activity of CAR-iMACs, but TIR-CAR-iMACs exhibited higher potential for persistent antitumor activity and potent pro-inflammatory activity.
Second-generation CAR-iMACs have robust antitumor function
In the above work, we established that TIR-CAR-iMACs exhibited the M1-like phenotype and both TIR and CD3ζ domains provided CAR-iMACs with CAR-dependent phagocytosis capacity. Based on the design of the second-generation T-CAR, we hypothesized that the integration of TIR and CD3ζ domains into one CAR structure could further promote the antitumor potency of CAR-iMACs. Thus, we developed a new EGFRvIII-targeting M-CAR containing intracellular CD3ζ and TIR domains in tandem (Fig. 1a) and then generated CD3ζ-TIR-CAR-iMACs (Fig. 1b). We first compared the expression of pro-inflammatory genes reportedly driven by TIR activation. As expected, the quantitative real-time PCR (qRT–PCR) analysis revealed that after incubation of the three types of CAR-iMACs with U87MGEGFRvIII cells for 24 h, expression of IL-1A/IL-1B, IL-6, IL-12A, IL-23, CCL8, CXCL8 and TNFA were significantly elevated in CD3ζ-TIR-CAR-iMACs compared to TIR- or CD3ζ-CAR-iMACs, all without IFN-γ/LPS pretreatment (Fig. 3a–h), revealing the synergistic effect of TIR and CD3ζ domains. Correspondingly, the ELISA assay confirmed that CD3ζ-TIR-CAR-iMACs mostly improved the production of IL-6, IL-12, IL-23 and TNF (Fig. 3i–l). These results further substantiated that the TIR domain contributed to the M1-like polarization of CAR-iMACs. Subsequently, we examined the cytotoxicity of the integrated CD3ζ-TIR-CAR-iMACs against FFluc+ U87MGEGFRvIII cells. Measurement of BLI from FFluc+ U87MGEGFRvIII cells indicated that compared to single CD3ζ or TIR domain-containing CAR-iMACs, 24h incubation with CD3ζ-TIR-CAR-iMACs led to the lowest number of viable tumor cells at an E/T of 10/1 (Fig. 3m), revealing that CD3ζ-TIR-CAR-iMACs conferred significantly higher cytotoxicity against tumor cells than single domain-containing CAR-iMACs. Subsequently, we co-incubated the CD3ζ-TIR-CAR-iMACs with FFlu+U87MGEGFRvIII cells as well as EGFRvIII-negative U87MG and HepG2 cells, respectively, at an E/T of 10/1 in vitro and found that CD3ζ-TIR-CAR-iMACs showed most evident cytotoxicity only against the EGFRvIII-positive cells (Fig. 3n), demonstrating the activity was antigen-dependent. Furthermore, to test whether the second-generation CAR-iMACs had antigen presentation capacity to activate T cells, we first cocultured the CD3ζ-TIR-CAR-iMACs with U87MGEGFRvIII cells for 48 h and then added the Jurkat T cells expressing a NY-ESO-1-specific T cell receptor (TCR) to the coculturing system for 24 h (Fig. 3o). CAR-iMACs along led to basal level of T cell activation, and the T cells were further activated in the presence of NY-ESO-1-overexpressing tumor cells (Fig. 3o).
a–h, qRT–PCR analysis showing the expression of immune-related factor genes IL-1A (a), IL-1B (b), IL-6 (c), IL-12A (d), IL-23 (e), CCL8 (f), CXCL8 (g) and TNFA (h) in EGFRvIII-CAR-iMACs, after coculturing with U87MGEGFRvIII cells for 24 h with an E/T ratio of 10/1 (n = 3 biologically independent samples per group). i–l, ELISA experiments showing the release of pro-inflammatory cytokines IL-6 (i), IL-12 (j), IL-23 (k) and TNF (l) from WT-iMACs and EGFRvIII-targeting CAR-iMACs in the medium, after 24 h incubating with U87MGEGFRvIII cells, respectively, with an E/T ratio of 10/1 (n = 3 biologically independent samples per group). The medium for the culturing of tumor cells without CAR-iMACs was used as an NC. m, Statistics of BLI from viable FFluc+ U87MGEGFRvIII cells after incubating with the CAR-iMAC for 24 h with an E/T ratio of 10/1 (n = 3 biologically independent samples per group). The experimental group containing the tumor cells without CAR-iMACs was used as an NC. The data above was shown by GraphPad Prism 8.2.1. Significance was calculated with two-tailed multiple t tests analysis and is presented as mean ± s.e.m. NS, not significant; *P <0.05; **P <0.01. n, The WT iMACs and EGFRvIII-targeting CAR-iMACs were incubated with the FFluc+ U87MGEGFRvIII cells as well as the EGFRvIII-negative FFluc+ U87MG cells and FFluc+ HepG2 cells with the E/T ratio of 10/1 to evaluate the antigen-dependent cytotoxicity of CAR-iMACs against tumor cells (n = 3 biologically independent samples per group). The BLI from viable tumor cells was captured after 48 h of incubation. o, Flow cytometry analysis was performed to count the activated T cells by detecting the CD69 expression on Jurkat cells expressing a NY-ESO-1-specific TCR, after 24 h of incubation with EGFRvIII-targeting CD3ζ-TIR-CAR-iMACs and U87MGEGFRvIII cells expressing NY-ESO-1 (U87MGEGFRvIII-NY) in vitro (n = 5 biologically independent samples per group). U87MGEGFRvIII expressing NY-ESO-1 and the cognate HLA was used as a positive control. p,q, After coculturing the truncated CAR-iMACs, CD3ζ-CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs with tdTomato-negative U87MGEGFRvIII cells with an E/T ratio of 10/1, flow cytometry analyses were performed to detect the proportion of CD80 or CD163-positive CAR-iMACs at the time points of day 1, day 2, day 3 and day 7, respectively. Statistical analyses showed the trends of the percentage of CD80-positive CAR-iMACs (p) or CD163-positive CAR-iMACs (q) over the course of 7 d. The statistical analysis from n to q was shown by GraphPad Prism 8.2.1. Significance was calculated with two-way ANOVA analysis with multiple comparisons and is presented as mean ± s.e.m. NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Substantial evidence suggests that the interaction with tumor cells leads to the exhaustion of T cells and promotes M2-like polarization of macrophages41,42,43,44. To explore whether the TIR-based CAR-iMACs could withstand immunosuppressive effects from tumor cells, we examined the polarization state of the four types of CAR-iMACs cocultured with tdTomato+ U87MGEGFRvIII cells with no IFN-γ/LPS pretreatment at an E/T ratio of 3/1. Subsequent FCM analysis showed that incubation with the tumor cells for 24 h stimulated higher expression of CD80 in the two TIR-based CAR-iMACs compared to truncated CAR-iMACs and CD3ζ-CAR-iMACs (Fig. 3p). Of note, prolonged periods of exposure to tumor cells resulted in a significant reduction of CD80+ populations to about 40–50% for truncated CAR-iMACs and CD3ζ-CAR-iMACs (Fig. 3p), whereas the percentage of CD80+ cells in both TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs was kept at a higher level of ~80% at day 7 (with a slight drop to 70% at day 3 for CD3ζ-TIR-CAR-iMACs), indicating that TIR enhanced the sustained stimulation of the M1 state. Besides, we also determined M2-like populations of the four types of CAR-iMACs by measuring CD163, a typical M2 marker. We found that prolonged exposure to the tumor cells led to various CD163 expression levels in the first three types of CAR-iMACs, whereas CD3ζ-TIR-CAR-iMACs exhibited the most resistance against M2 conversion on day 3 and remained low on day 7 (Fig. 3q). Based on the above results, we concluded that the combination of the TIR and CD3ζ domains induced M1 polarization of CAR-iMACs exposed to tumor cells and prevented the conversion from the M1 to the M2-like state.
We next investigated the antitumor potency of EGFRvIII-targeting CD3ζ-TIR-CAR-iMACs in vivo. This time, the four types of CAR-iMACs without IFN-γ/LPS pretreatment were used to treat FFluc+ U87MGEGFRvIII cell-bearing mice at an E/T ratio of 15/1 (Extended Data Fig. 2a). As expected, treatment by CD3ζ-TIR-CAR-iMACs alleviated tumor cell growth compared to truncated CAR-iMACs (Extended Data Fig. 2b,c) and prolonged survival of the tumor-bearing mice (Extended Data Fig. 2d). To assess whether TIR contributed to M1 polarization of CAR-iMACs in vivo, we isolated the immune cells from the mice after 2 d of treatment and measured CD80+ and CD163+ populations by FCM. More CD3ζ-TIR-CAR-iMACs exhibited CD80 positivity than CD3ζ-CAR-iMACs (Extended Data Fig. 2e,f), and lower CD163 expression was present in CD3ζ-TIR-CAR-iMACs (Extended Data Fig. 2e,f). Next, to mimic the clinical situation, we carried out another animal experiment with an orthotopic glioblastoma (GBM) model in which FFluc+ U87MGEGFRvIII cells were intracranially inoculated to NOD-SCID mice. IFN-γ/LPS-pretreated truncated CAR-iMACs and CD3ζ-TIR-CAR-iMACs were delivered into the intracranial GBM lesion after 6 d of transplantation of the FFluc+ U87MGEGFRvIII cells, followed by live imaging to monitor the tumor growth (Extended Data Fig. 3a). Similar to the above therapeutic outcome, CD3ζ-TIR-CAR-iMACs evidently restrained the progression of intracranial GBM compared to that of truncated CAR-iMACs (Extended Data Fig. 3b,c) and prolonged the survival of the GBM-bearing mice (Extended Data Fig. 3d). Notably, the expression of EGFRvIII persisted during the tumor growth (Extended Data Fig. 3e).
In summary, we confirmed that the second-generation CAR design by integrating intracellular CD3ζ and TIR domains into one CAR structure could enhance the cytotoxic activity against tumor cells, promote the antigen presentation capacity and maintain the polarization state of CAR-iMACs.
GPC3-targeting TIR-based CAR-iMACs have robust antitumor function
To consolidate the above proof-of-concept results with another type of tumor cell naturally expressing a targetable antigen, we next investigated whether GPC3-targeting CAR-iMACs could achieve similar efficacy in hepatocellular carcinoma (HCC) models using HepG2 cells. Similar as before, we obtained GPC3-targeting CAR-iMACs by differentiating the engineered iPSCs. Flow cytometry analysis showed that all types of GPC3-CAR-iMACs had high CAR expression of more than 80% (Extended Data Fig. 4a). To assess the antitumor activity of the CAR-iMACs against GPC3-expressing cells, we incubated the CAR-iMACs with tdTomato-expressing HepG2 cells at an E/T ratio of 10/1 (Fig. 4a) and found that both TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs led to remarkable inhibition of tumor cell growth compared to truncated CAR-iMACs over a time course of ~5 d (Fig. 4b and Extended Data Fig. 4b), strongly demonstrating that the TIR-containing CARs enhanced antitumor activity of CAR-iMACs against HepG2 in vitro. Moreover, luciferase assays revealed that GPC3-targeting CD3ζ-TIR-CAR-iMACs possessed significantly higher cytotoxicity on GPC3-expressing HepG2 cells but not on GPC3-negative U87MG cells (Fig. 4c), demonstrating the antigen-dependent cytotoxicity.
a, A sketch map of HepG2 cell-killing assay with GPC3-targeting CAR-iMACs in an Incucyte 2022A Live-Cell Analysis Instrument. b, The growth curve of tdTomato+ HepG2 cells in a time course of ~5 d after incubating GPC3-targeting truncated CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs with tdTomato+ HepG2 cells at an E/T ratio of 10/1 (n = 32 technical replicate wells per group). The GPC3-expressing HepG2 cells were identified via the tdTomato signal. Signal capturing was conducted every 2 h. c, The WT iMACs and GPC3-targeting CAR-iMACs were incubated with the FFluc+ HepG2 cells and GPC3-negative FFluc+ U87MG cells with the E/T ratio of 10/1, respectively, to assess the antigen-dependent cytotoxicity of the GPC3-targeting CAR-iMACs against tumor cells (n = 3 biologically independent samples per group). The bioluminescence (BLI) from residual tumor cells was captured after 48 h of incubation. The data above was shown by GraphPad Prism 8.2.1. Significance was calculated with two-way ANOVA analysis with multiple comparisons and is presented as mean ± s.e.m. NS, not significant; ****P < 0.0001. d, An overview of the antitumor activity experiment using GPC3-targeting CAR-iMACs in a FFluc+ HepG2 xenograft HCC model. e, Live animal imaging showing the time-dependent change of bioluminescence (BLI) from the HCC model after being treated by GPC3-targeting truncated CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs. f, The statistics of BLI from e (n = 5 mice from truncated CAR-iMAC group, n = 7 mice from TIR-CAR-iMAC group and n = 8 mice from CD3ζ-TIR-CAR-iMAC group). The data above was shown by GraphPad Prism 8.2.1. Significance was calculated with two-way ANOVA analysis with multiple comparisons and is presented as mean ± s.e.m. NS, not significant; **P < 0.01; ****P < 0.0001. g, The Kaplan–Meier survival curve of tumor-bearing mice from e. All the data above was shown by GraphPad Prism 8.2.1. Significance was calculated with two-tailed log-rank Mantel–Cox test (TIR-CAR-iMAC group versus truncated CAR-iMAC group, P = 0.0064; CD3ζ-TIR-CAR-iMAC group versus truncated CAR-iMAC group, P = 0.0024).
To prove the in vivo efficacy, we used a HepG2 xenograft models (Fig. 4d). Four days after seeding the tumor cells when tumors were established, GPC3-targeting truncated CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs were used to treat the tumor-bearing mice and animal imaging was conducted to monitor the tumor growth (Fig. 4e). TIR-based CAR-iMACs exhibited significant antitumor activity compared to truncated CAR-iMACs (Fig. 4e,f). Moreover, CD3ζ-TIR-CAR-iMACs achieved the strongest effect and led to complete remission in 6 of 8 mice. Correspondingly, CD3ζ-TIR-CAR-iMACs evidently prolonged the survival of HCC-bearing mice (Fig. 4g). Together, GPC3-targeting CAR-iMACs containing the intracellular TIR domain showed potent antitumor activity, and the CAR design by integrating CD3ζ and TIR domains further enhanced the potency.
Next, we tested whether a combination of CAR-iMACs and CD47 antibody could provide synergistic effects. Anti-HCC tumor assays with HepG2-bearing mice showed that TIR-based CAR-iMACs combined with a low dose of 0.5 μg CD47 antibody achieved superior antitumor efficacy compared to CD47 antibody alone, or CD47 antibody combined with the truncated CAR-iMAC control (Extended Data Fig. 4c,d). Notably, all the mice receiving the combination therapy of CD3ζ-TIR-CAR-iMACs and CD47 antibody showed significant remission after 10 d of treatment, with almost complete elimination of tumor signal on day 30. Moreover, the combined treatment showed no apparent effect on the weight of the mice (Extended Data Fig. 4e), indicating there was no toxicity associated with this treatment strategy.
To assess whether the second-generation CAR-iMACs could modify the TME, we designed a mouse-specific CAR (G2-CAR) containing the extracellular scFv sequence recognizing the human GPC3 protein and the intracellular mouse TIR and CD3ζ tandem domains, as well as the truncated control (G1-CAR; Extended Data Fig. 5a). Next, we transduced the G1- and G2-CARs into mouse bone marrow-derived macrophages (BMDMs) with lentivirus and obtained the CAR-BMDMs, even though the transduction efficiency was relatively low (Extended Data Fig. 5b–d). Afterward, we incubated the mouse CAR-BMDMs with human GPC3-expressing Hepa1-6 (Hepa1-6GPC3) cells in vitro and observed significantly stronger phagocytosis of the tumor cells by G2-CAR-BMDMs compared to G1-CAR-BMDMs (Extended Data Fig. 5e,f). For in vivo analysis, the progression of Hepa1-6GPC3-derived subcutaneous tumors was restrained after the treatment with the G2-CAR-BMDMs (Extended Data Fig. 5g,h), without affecting the weight of the mice (Extended Data Fig. 5i). Next, we assessed the changes of immune cells in the TME. We found an increased proportion of M1-like CD86+ macrophages in the G2-CAR-BMDMs treated group (Extended Data Fig. 5j), while the CD206+ macrophages (Extended Data Fig. 5k) and dendritic cells (DCs) (Extended Data Fig. 5l) showed no significant changes. Meanwhile, G2-CAR-BMDMs also increased the proportion of NK (Extended Data Fig. 5m) and T cells (Extended Data Fig. 5n), implying enhanced recruitment of these cells to tumors. Together, the CAR-BMDMs improved the TME for better elimination of tumor cells.
NF-κB/P65-mediated M1 polarization of TIR-based CAR-iMACs
The TIR domain is responsible for signal transduction from TLR4, and the downstream signaling liberates NF-κB/P65 from IκB (inhibitor of NF-κB) inhibition, followed by the translocation of NF-κB/P65 into the nucleus to transcriptionally activate pro-inflammatory target genes39,45. To investigate the molecular mechanism of whether NF-κB/P65 mediated the pro-inflammatory activity of TIR-based CAR-iMACs when encountering the antigen, we first measured the expression of NF-κB/P65 gene RELA with qRT–PCR analysis and found higher expression in both TIR domain-containing CAR-iMACs (Fig. 5a). Next, we monitored nuclear translocation of NF-κB/P65 by IF imaging when the control truncated CAR-iMACs were treated with LPS for 20 min (Extended Data Fig. 6a). Afterward, we cocultured the EGFRvIII-targeting truncated CAR-iMACs, TIR-CAR-iMACs, CD3ζ-CAR-iMACs and CD3ζ-TIR-CAR-iMACs with tdTomato+ U87MGEGFRvIII cells, respectively, at an E/T ratio of 3/1, followed by IF imaging to detect NF-κB/P65 subcellular localization. Confocal imaging showed that interaction between CAR-iMACs and U87MGEGFRvIII cells for 4 h triggered NF-κB/P65 expression in CD3ζ-TIR-CAR-iMACs (Extended Data Fig. 6b). When the incubation time was prolonged to 12 h, NF-κB/P65 exhibited a higher expression in TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs, with aggregation in the nuclei (Fig. 5b,c). As the positive control, all the IFN-γ/LPS-pretreated CAR-iMACs showed the accumulation of NF-κB/P65 in nuclei (Fig. 5d,e). Taken together, these results demonstrated that the NF-κB signaling pathway was activated in the TIR domain-containing CAR-iMACs.
a, qRT–PCR analysis showing the expression level of NF-κB/P65 encoding gene RELA in EGFRvIII-targeting truncated CAR-iMACs, CD3ζ-CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs after incubating the four different types of CAR-iMACs with U87MGEGFRvIII cells for 12 h without IFN-γ/LPS pretreatment (n = 5 biologically independent samples per group). b, IF staining indicating the expression of NF-κB/P65 protein in nuclei of the four CAR-iMACs that were incubated with U87MGEGFRvIII cells for 12 h in vitro without IFN-γ/LPS pretreatment. c, Statistical analysis of NF-κB/P65 protein expression level in nuclei of the four CAR-iMACs as in b (n = 10 samples in the random visual field from three independent experiments). The expression level of NF-κB/P65 protein in nuclei was detected with ImageJ software. d, IF staining showing the expression of NF-κB/P65 protein in nuclei of the four CAR-iMACs which were pretreated with IFN-γ/LPS and then incubated with U87MGEGFRvIII cells in vitro for 12 h. e, Statistical analysis of NF-κB/P65 protein expression level in nuclei of the four CAR-iMACs as in d (n = 10 samples in random visual field from three independent experiments). f, The residual luciferase-transduced tumor cells were determined by luciferase activity after 24 h of coculturing with CAR-iMACs (without IFN-γ/LPS pretreatment) in the presence of JSH23 or DMSO (n = 3 biologically independent samples per group). All the above data was shown by GraphPad Prism 8.2.1. Significance was calculated with two-tailed multiple t tests analysis and is presented as mean ± s.e.m. NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To further confirm the role of NF-κB/P65 in CAR-iMACs, we treated the same CAR-iMACs cocultured with FFluc/tdTomato+ U87MGEGFRvIII cells using an NF-κB/P65 inhibitor JSH23. As expected, blocking the nuclear translocation of NF-κB/P65 by JSH23 abrogated cytotoxicity of CAR-iMACs against FFluc/tdTomato+ U87MGEGFRvIII cells (Fig. 5f). Collectively, we revealed that immune activation and M1 polarization of TIR-based CAR-iMACs were through the NF-κB pathway.
Mechanism of M1 polarization in TIR-based CAR-iMACs
We next sought to further reveal the mechanism by examining the TIR-based CAR-iMACs exposed to tumor cells with scRNA-seq analysis (Fig. 6a). IFN-γ/LPS-pretreated EGFRvIII-targeting CAR-iMACs were incubated with U87MGEGFRvIII cells for 24 h. Subsequently, we performed 10x Genomics scRNA-seq and found that all cells could be clustered into six main subpopulations (Extended Data Fig. 7a,c,e). According to the published in vivo scRNA-seq data of human hematopoietic development, C1, C2, C3, C5 and C6 clusters were mainly matched to the macrophage (Extended Data Fig. 7b,d,f). Meanwhile, the C4 cluster was primarily matched with the U87MG cancer cell phenotype in all three types of CAR-iMACs groups. Also, compared with truncated CAR-iMACs and TIR-CAR-iMACs groups, CD3ζ-TIR-CAR-iMACs treatment resulted in fewer remaining tumor cells (C4 cluster; Extended Data Fig. 7a,c,e), consistent with the cytotoxicity results above. In addition, the C5 cluster of all three groups also possesses (DC) characteristics (Extended Data Fig. 7a–f). Interestingly, the percentage of DC-feature cells in the C5 cluster showed a growing trend from truncated CAR-iMACs and TIR-CAR-iMACs to CD3ζ-TIR-CAR-iMACs (Extended Data Fig. 7a–f), implying that TIR-based CAR-iMACs, especially the second-generation ones, might have acquired enhanced DC cell capacity.
a, A schematic picture showing the procedures of the scRNA-seq experiment. b–d, Expression levels of CD80 (b), CD86 (c) and CD206 (d) in truncated CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs, respectively. e, The percentage of three types of CAR-iMACs expressing M1 state-associated genes. The CD80 gene here was used as a positive control. f–i, Expression levels of M1 state-associated genes in the three types of CAR-iMACs. j, The percentage of the three types CAR-iMACs expressing M2 state-associated genes. Similarly, the CD206 gene here was used as a positive control. k,l, Expression levels of the three types of CAR-iMACs expressing M2 state-associated genes. m, The PCA showing the relationship between CAR-iMACs and stimulated M1- and M2-iPSC-derived macrophages (IPSDMs). To more accurately distinguish polarized and unpolarized CAR-iMACs, we further divided CAR-iMACs into CD80-positive and CD80-absent (non-CD80) categories. n, A heatmap comparing activation levels of M1- and M2-related pathways in CD80-positive and CD80-absent (non-CD80) populations of the three types of CAR-iMACs. Oxidative phosphorylation and respiratory electron transport activity were linked to the M2-like state of macrophages. IFN-γ response was related to the M1-like state. The IPSDM MAC, IPSDM M1 and IPSDM M2 were obtained from the GEO database (GSE55536)46. For b–d, f–i, k and l, the center line is the median, the bottom of the box is the 25th percentile boundary, the top of the box is the 75th and the top and bottom of the vertical line define the boundary of the data that are not considered outliers, with outliers defined as greater/less than ±1.5× IQR. n (truncated CAR-iMAC group) = 10,615 cells, n (TIR-CAR-iMAC group) = 12,581 cells, n (CD3ζ-TIR-CAR-iMAC group) = 12,629 cells. P values are calculated by the two-tailed Wilcoxon signed-rank test (for paired samples) and the two-tailed Mann–Whitney U test (for independent samples). NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001. IQR, interquartile range.
Next, we performed a quantitative analysis of M1- or M2-like state-associated marker genes (M1 or M2 genes) using scRNA-seq data. Normalized gene expression showed that more TIR-CAR-iMACs exhibited high expression of CD80 and CD86, and CD3ζ-TIR-CAR further significantly boosted the expression levels of the two M1 marker genes (Fig. 6b,c), and consistently validated by the previous FCM analysis. Accordingly, the M2 marker gene CD206 exhibited the significantly lowest expression in CD3ζ-TIR-CAR-iMACs, followed by TIR-CAR-iMACs and truncated CAR-iMACs (Fig. 6d). Moreover, the percentage of CAR-iMACs that expressed other M1 genes, including CD83, CCL8, CXCL9 and CXCL11, increased incrementally from truncated CAR-iMACs to TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs (Fig. 6e). Accordingly, the normalized expression level of these genes exhibited the same trend (Fig. 6f–i). On the contrary, a smaller percentage of TIR-CAR-iMAC and CD3ζ-TIR-CAR-iMACs expressed M2 genes such as CCL13 and CD163 (Fig. 6j), or TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs presented significantly lower expression of these genes compared to truncated CAR-iMACs (Fig. 6k,l). Collectively, these results illustrated that integration of the TIR and CD3ζ domains facilitated M1 gene expression and simultaneously suppressed M2 gene expression at the single macrophage level.
Next, we investigated whether the global gene expression signature resembled M1-like macrophages. We divided CAR-iMACs into CD80-expressing and CD80-absent (non-CD80) populations, as the latter represented the iMACs that might not encounter tumor cells, and evaluated the discrepancy of their expression profile. Principal component analysis (PCA) using M1 genes identified in our previous study and a published work indicated that CD80-expressing TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs were positioned closer to the IFN-γ/LPS-stimulated M1-like iMACs, compared to CD80-positive truncated CAR-iMACs and non-CD80 CAR-iMACs from all types (Fig. 6m). Furthermore, pathway analysis revealed that CD80-expressing CAR-iMACs showed lower activation of M2-related pathways such as oxidative phosphorylation and respiratory electron transport than in non-CD80 CAR-iMACs, but a higher activation of M1-related pathway IFN-γ response (Fig. 6n). More importantly, within the CD80-expressing groups, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs showed significantly higher IFN-γ responses and lower oxidative phosphorylation (Fig. 6n), indicating that TIR-based CAR conferred the CAR-iMACs with more robust pro-inflammatory activity. Finally, hierarchical clustering analysis based on a defined set of M1 and M2 genes46 showed that both TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs clustered with IFN-γ/LPS-polarized iMACs, whereas truncated CAR-iMACs clustered with M2-polarized iMACs (Extended Data Fig. 8a,b). The same clustering trend was observed when using the CD80-expressing populations (Extended Data Fig. 8c,d). To sum up, the above scRNA-seq analysis corroborated the mechanism that TIR-based CARs contributed to the pro-inflammatory state of CAR-iMACs in a CAR-dependent manner when encountering target tumor cells.
CAR-iMACs-mediated efferocytosis of apoptotic tumor cells
To further dissect cytolytic mechanisms of the CAR-iMAC, we next examined the expression of apoptosis-related genes in U87MGEGFRvIII from Fig. 6a, due to the abovementioned apoptosis-inducing phenomenon of CAR-iMACs against tumor cells (Extended Data Fig. 1f). We found increased expression of genes related to apoptosis such as CASP3/CASP6/CASP7/CASP8 and APAF1, as well as genes involved in transducing cell death signals such as FAS, TNFRSF10A and TNFRSF25 in the CD3ζ-TIR-CAR-iMAC-treated U87MGEGFRvIII cells (Fig. 7a). These results implied an apoptosis-promoting role of CD3ζ-TIR-CAR-iMACs upon encountering targeting tumor cells. To verify it, we directly measured the apoptosis of target tumor cells after incubation with CAR-iMACs. FCM analysis with Annexin V showed that ~40% of U87MGEGFRvIII cells underwent apoptosis after 6 h of treatment by CD3ζ-CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs, significantly higher than a ~5% apoptosis in the treatment with the truncated CAR-iMAC control (Fig. 7b,c). Moreover, 12 h of incubation markedly increased the apoptotic rate of CD3ζ-TIR-CAR-iMAC-treated U87MG cells to more than 80% compared to a 24% rate for the truncated control (Fig. 7b,c). To strengthen these results, another experiment using GPC3-CAR-iMACs against HepG2 cells also showed stronger target cell apoptosis-inducing capacity of TIR-based CAR-iMACs, particularly with an apoptotic rate above 90% when treated with CD3ζ-TIR-CAR-iMACs (Fig. 7d,e). Macrophage-secreted cytokines such as TNF can lead to apoptosis of tumor cells. As TNF was markedly increased in the medium of the second-generation CAR-iMACs and tumor cell coculture (Fig. 3), we added the TNF-neutralizing antibody adalimumab to the coculture and found that the cytolytic activity of CAR-iMACs was largely abolished (Fig. 7f,g), demonstrating that the tumor cell apoptosis was contributed by CAR-mediated M1 polarization and secretion of cytokine TNF.
a, A heatmap showed the expression of apoptosis genes in U87MGEGFRvIII cells after co-incubation with EGFRvIII-targeting truncated CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs, respectively, in vitro for 24 h. b, FCM analysis with APC-conjugated annexin V showed the proportion of apoptotic U87MGEGFRvIII cells after co-incubation with EGFRvIII-targeting truncated CAR-iMACs, CD3ζ-CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs, respectively, in vitro for 6 and 12 h. c, Statistics of the proportion of apoptotic U87MGEGFRvIII cells from b (n = 3 biologically independent samples per group). d, FCM analysis with APC-conjugated annexin V showed the proportion of apoptotic HepG2 cells after co-incubation with GPC3-targeting truncated CAR-iMACs, TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs, respectively, in vitro for 6 and 12 h. e, Statistics of the proportion of apoptotic HepG2 cells from d (n = 3 biologically independent samples per group). All the FCM experiments were carried out in triplicates, and the results were processed using FlowJo. f, The proportion of apoptotic tumor cells was determined by FCM analysis of U87MGEGFRvIII tumor cells cocultured with CAR-iMACs in the presence and absence of an anti-TNF antibody adalimumab (n = 3 biologically independent samples per group). g, The proportion of apoptotic tumor cells was determined by FCM analysis of HepG2 tumor cells cocultured with CAR-iMACs in the presence and absence of an anti-TNF antibody adalimumab (n = 3 biologically independent samples per group). h, Confocal imaging showed efferocytosis of GPC3-targeting CAR-iMACs against HepG2 cells. i, Long-term super-resolution real-time imaging with an HIS-SIM system showed the process of HepG2 apoptosis induced by GPC3-targeting CD3ζ-TIR-CAR-iMACs followed by efferocytosis in the bright field over time. j, Long-term super-resolution real-time imaging with an HIS-SIM system showed the process of engulfment of CD3ζ-TIR-CAR-iMAC against the debris of HepG2 cells. k, Real-time imaging with a HIS-SIM system displayed that EGFP-marked CD3ζ-TIR-CAR-iMACs induced apoptosis of tdTomato-labeled U87MGEGFRvIII cells and subsequently engulfed the debris. l, A cartoon diagram illustrating the mechanism of CAR-iMAC efferocytosis against apoptotic tumor cell. All the above statistic data were shown by GraphPad Prism 8.2.1. The significance was calculated with a two-way ANOVA analysis with multiple comparisons and is presented as mean ± s.e.m. NS, not significant; *P < 0.05; **P < 0.01; ****P < 0.0001.
Next, we performed confocal imaging to visualize the antitumor process of GPC3-CAR-iMACs against HepG2 cells after 12 h of incubation and observed fragmentation of HepG2 cells as well as tumor cell debris-engulfing processes of both TIR-CAR-iMACs and CD3ζ-TIR-CAR-iMACs (Fig. 7h). To monitor the killing process of CAR-iMACs against tumor cells, we conducted long-term super-resolution imaging via a high sensitivity structured illumination microscope (HIS-SIM). Real-time imaging in a bright field revealed that GPC3-targeting CD3ζ-TIR-CAR-iMACs induced dismantling of HepG2 cells, followed by engulfing the debris over time (Fig. 7i,j and Supplementary Video 1), a process characteristic of efferocytosis47. Consistently, the EGFRvIII-targeting CD3ζ-TIR-CAR-iMACs induced the cell death process of tdTomato+ U87MGEGFRvIII cells followed by engulfing and digesting the tumor cell debris (Fig. 7k and Supplementary Video 2). Together, we illustrated that our CAR-iMACs display antitumor cell cytolytic activity via a sequential process, which is as follows: (1) engagement of tumor cell antigen and CAR to activate CAR-iMACs, (2) CAR-mediated M1 polarization and secretion of TNF to induce tumor cell apoptosis and (3) tumor cell apoptotic bodies clearance by CAR-iMACs through efferocytosis (Fig. 7l).
Discussion
Macrophages and genetically engineered CAR macrophages have drawn substantial attention, given their potential advantages in homing and infiltrating solid tumors and manifold roles in regulating the immunosuppressive TME12,48. More excitingly, the first clinical study in patients with a solid tumor has started49, and the proof-of-concept results will provide the foothold for using CAR macrophages as a new weapon to combat solid tumors.
The abovementioned progresses were based on the first-generation CAR macrophages, in which the intracellular signaling is mediated by a CD3ζ domain or a substitute effector domain from macrophages25,26,27,50. To integrate a polarization signal, we added a TIR domain into the CAR and thus provided an orthogonal signal of macrophage polarization (Extended Data Fig. 9). We demonstrated that the iPSC-derived second-generation CAR-iMACs exhibited enhanced antitumor activity both in vitro and in vivo, with a strong M1 polarization phenotype. Particularly, the second-generation GPC3-CAR-iMACs showed strong efficacy in that it led to complete remission either by itself or combined with a low dose of CD47 antibody, and the possible reasons for the stronger efficacy compared with the EGFRvIII-CAR include the different quality of the antibodies from which the CAR’s scFv was derived. We also confirmed the safety of the CAR-iMACs in the mouse model (Extended Data Fig. 4e). Even though the M1-like state cytokines such as IL-1 and IL-6 were induced, as CAR-iMACs were not significantly expanded in vivo25, and they tended to go into tissues, the accumulated amount of IL-1 or IL-6 in the circulation system might not necessarily lead to cytokine release syndrome that warrants further studies using humanized models. We also elucidated a surprising cytolytic mechanism of CAR-mediated efferocytosis of iMACs against apoptotic tumor cells. Our analysis indicated engulfment of apoptotic tumor cell bodies is one of the main mechanisms of tumor cell clearance. This mechanism of action will provide more molecular basis for moving the CAR-iMACs therapy toward the clinic.
It is well established that macrophages are equipped with receptors called pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) and then release pro-inflammatory or anti-inflammatory cytokines to exert various functions in defense31,34. The TIR-containing chimera antigen receptor is a new engineered PRR that recognizes ‘antigen-associated molecular patterns,’ enabling macrophages with antigen-dependent capacity of M1 polarization, which in our study is an advantage in cancer immune cell therapy.
Methods
Cell culture
Human iPSCs were reprogrammed from peripheral blood mononuclear cells of a volunteer donor (approved by the Human Subjects Committee of the First Affiliated Hospital of Zhejiang University25 and were cultured in mTeSR™-1 medium (a feeder-free maintenance medium for human ES and iPS cells. STEMCELL Technologies, 85851) or ncTarget hPSC (human pluripotent stem cell) medium (Nuwacell Biotechnology, RP01020) with Matrigel Matrix (Corning, 354277) or Vitronectin (Nuwacell Biotechnology, RP01002) coated onto six-well cell culture plates. iPSCs were passaged using Versene (Gibco, 15040-066). Matured CAR-iMACs and WT-iMACs were maintained in StemSpan-XF medium (STEMCELL Technologies, 100-0073) containing M-CSF (PEPROTECH, 300-25; 100 ng ml−1) and GM-CSF (PEPROTECH, 300-3; 100 ng ml−1). HEK293T cells (ATCC, CRL-3216) and HepG2 cells were cultured in high-glucose DMEM (Gibco, 11965-065) containing 10% FBS (Gibco, 10099-141) and 1× Penicillin–Streptomycin (Pen–Strep; 10,000 U ml−1 penicillin and 10,000 µg ml−1 streptomycin; Gibco, 15140-122). U87MG cells (ATCC, HTB-14) and U87MGEGFRvIII cells were cultured in MEM (Hyclone, SH30024.01) medium supplemented with 10% FBS (Gibco, 10099-141), 1× nonessential amino acids (Gibco, 11140-050), 1 mM sodium pyruvate (Gibco, 11360-070) and 1× Pen–Strep. Mouse BMDM cells were cultured in RPMI 1640 medium (Meilunbio, MA0215) containing 10% inactive FBS (Gibco, 10099-141) and mouse M-CSF (PEPROTECH, 315-02).
Construction of CARs
We designed T-CAR or M-CARs according to the intracellular CD3ζ domain (amino acids 52–164) or TIR domain (amino acids 672–818), integrating hinge and transmembrane region of human CD8α (amino acids 183–206). The extracellular fragment of the CAR consisted of the humanized scFv nucleotide sequence, respectively, specific to EGFRvIII (139 scFv nucleotide sequence, patent: US 9,394,368 B2) and GPC3 (patent: US2016/0215261 A1). The integration of cytoplasmic parts of the CD3ζ and TIR was designed for the second-generation M-CAR. All CARs were synthesized by GenScript Biotech and cloned into the Lenti-EF1A-T2A-EGFP-PGK-Puro vector. We also designed GPC-targeting CAR containing intracellular mouse CD3ζ and TIR domains. All the sequences are listed in Supplementary Data.
Stable overexpression of CARs in iPSCs
CAR sequence-containing lentiviral vector (Lenti-EF1α-T2A-EGFP-PGK-Puro), psPAX2 and pMD2.G were transfected into HEK293T cells cultured in 10 cm dishes using Lipofectamine 2000 (Invitrogen, 11668). The lentivirus-containing medium was collected at 24 h, 36 h, 48 h and 60 h post-transfection and filtered with a 0.45 μm filter. The filtered medium was mixed with a half volume of 30% PEG8000 and centrifuged at 4,000 r.p.m. for 30 min at 4 °C. The pellet was resuspended with mTeSR medium (STEMCELL Technologies, 85851) containing 10 μg ml−1 polybrene (Yeasen Biotechnology, 40804ES76). The resuspended lentivirus was then added to prepare iPSCs in six-well plates. The medium was refreshed after 6 h postinfection with a new mTeSR medium. The infected cells were cultured for 24 h and then treated with 0.5 μg ml−1 puromycin for at least 48 h to select successfully transduced clones. Subsequently, the CAR-expressing iPSCs were expanded to obtain enough cells for differentiation.
Macrophage differentiation from iPSCs
WT-iPSCs and CAR-expressing iPSCs were treated with TrypLE (Gibco, 12604-021) for 1 min to disaggregate into individual cells. The cells were collected and transferred to low-attachment plates (Corning, 3471) to allow the formation of embryoid bodies (EBs) in mTeSR medium (STEMCELL Technologies, 85851) supplemented with Rock inhibitor Y27632 (STEMCELL Technologies, 72304). The process of formation of EBs and macrophage differentiation from EBs has been described in our previous work25.
Generation of stable tumor cell lines
The coding sequence of EGFRvIII was synthesized in GenScript Biotech and then cloned into the Lenti-EF1α-PGK-Puro vector. Lentivirus containing EGFRvIII coding sequence was collected from HEK293T cells and used to infect U87MG cells to develop the U87MGEGFRvIII cell line. The GPC3 coding sequence from human HepG2 cells was cloned into the Lenti-EF1α-PGK-Puro vector and overexpressed in Hepa1-6 cells using the abovementioned strategy to establish the Hepa1-6GPC3 cell line. The sequence cloning primers are listed in Supplementary Data.
FFluc and tdTomato genes were overexpressed in U87MGEGFRvIII cells, HepG2 cells and Hepa1-6GPC3 cells via PCDH-EF1α-tdTomato-FFluc2 plasmids to establish the FFluc/tdTomato-expressing cell lines.
Examination of CAR-iMAC cytotoxicity
To assess the cytotoxicity of CAR-iMACs against targeting cells, we cocultured CAR-iMACs with FFluc+ tumor cells at indicated E/T ratios in a black 96-well plate (Wohong Bio, WHB-96-02) in vitro and then added 50 μl 15 mg ml−1 luciferin buffer (GoldBio, LUCK-1G) into the culturing wells and captured the BLI signal from tumor cells through the microplate reader (Molecular Devices, MD M5).
To examine the antigen-dependent activation of CAR-iMACs, we incubated the EGFRvIII-targeting CAR-iMACs with 1 × 104 U87MGEGFRvIII cells, EGFRvIII-negative U87MG cells and HepG2 cells at E/T ratios of 10/1, respectively, for 48 h in a black 96-well plate (Wohong Bio, WHB-96-02) in vitro and then captured the BLI signal from tumor cells as described above.
Anti-HCC assay
To evaluate the inhibition effect of GPC3-targeting CAR-iMACs on HCC cells, we mixed CAR-iMACs with tdTomato+ HepG2 cells at an E/T ratio of 10/1 in a 24-well plate (NEST, 082721BH01) in vitro and then incubated the coculture in Incucyte 2022A Live-Cell Analysis Instrument that can monitor the dynamic activity of cells and capture the change of tdTomato fluorescence in a lively manner.
Detection of cytokines
We detected the cytokines released by CAR-iMACs with an ELISA kit (Elabscience). After incubating CAR-iMACs with U87MGEGFRvIII cells for 24 h, the medium was collected and used to quantify IL-6 (E-EL-H0102c), IL-12 (E-EL-H0150c), IL-23 (E-EL-H0107c) and TNF (E-EL-H0109c) levels. The fluorescence signals were detected by a microplate reader (Molecular Devices, MD M5).
In vivo antitumor assays
NOD-Prkdcem26Cd52/Gpt (NOD-SCID, stock: T001492) mice and C57BL/6 (stock: N000295) mice were obtained from Jiangsu Biocytogen and maintained under pathogen-free conditions at the animal center of the Zhejiang Academy of Medical Sciences. All mice were kept at 25 °C in a clean, pathogen-free and humid environment (typically 50%), with a 12-h dark/12-h light cycle, and fed with sufficient and sterile water and food (Xietong, 1010085). The protocols used were approved by the Ethics Committee of the Zhejiang Academy of Medical Sciences. The progression of tumor health status of mice was monitored carefully. The mouse with tumor size exceeding 2,000 m3 or tumor diameter exceeding 20 mm was killed in peace.
To assess the capability of CAR-iMACs targeting GBM in vivo, 4 × 105 FFluc+ U87MGEGFRvIII cells mixed with 200 μm 0.1% BSA in PBS were injected intraperitoneally into 6-week-old male NOD-SCID mouse. EGFRvIII-targeting CAR-iMACs were cultured with StemSpan-XF medium for 48 h before injection. Four hours later after intraperitoneal injection of the tumor cells, 4 × 106 above pretreated CAR-iMACs mixed with 200 μm 0.1% BSA in PBS were injected at the same position in the abdominal cavity of the tumor-bearing mouse. Two hours after CAR-iMACs injection, d-luciferin (GoldBio, LUCK-1G) was injected intraperitoneally into the mice at 150 mg kg−1 dosage, followed by live animal imaging through the animal imaging system (IVIS Lumina series III) to capture the BLI signal. Subsequently, live animal imaging was conducted at the same time point on days 1, 3, 7 and 15 to monitor tumor growth.
To evaluate the efficacy of the CAR-iMACs against intracranial GBM, 1 × 104 FFluc+ U87MGEGFRvIII cells were seeded into the corpus striatum of 6-week-old male NOD-SCID mice. After 6 d of good care, 2 × 105 EGFRvIII-targeting CAR-iMACs with no pretreatment were intracranially injected into the GBM lesion. Live animal imaging was conducted at the same time point on days 1, 2, 2, 5 and 15 after treatment.
To test the antitumor potency of GPC3-targeting CAR-iMACs against HCC in vivo, 5 × 105 FFluc+ HepG2 cells were intraperitoneally injected into a 6-week-old male B-NDG mouse. Four days after that, 2.5 × 107 cytokine-free medium pretreated GPC3-targeting CAR-iMACs were injected into HCC-bearing mice, followed by live animal imaging as performed above on days 0, 1, 2, 10, 14, 34 and 54. The treatment of HCC-bearing mice by GPC3-targeting CAR-iMACs combined with CD47 antibody (50 μg per mouse) was performed as the strategy above.
To assess the TME with syngeneic mouse models, 3 × 106 Hepa1-6GPC3 cells were inoculated subcutaneously into the left flank of 8-week-old male C57BL/6 mice on day 0. On day 3, the tumors were palpable, mice were treated intratumorally with C57BL/6 mice-derived G1-CAR-BMDM (1 × 107 cells), or G2-CAR-BMDM (1 × 107 cells), or with PBS. Before treatment, the CAR-BMDM cells were stimulated with 50 ng ml−1 IFNγ for 24 h and 100 ng ml−1 LPS for 30 min. The mice were monitored for tumor growth and body weight every 2 d starting on the day the CAR-BMDM cells were injected. Tumor volumes were measured by length (L) and depth (D) and calculated as tumor volume = (4/3) × π × (L/2) × (D/2) × (D/2). On day 10, the mice were sacrificed, and the tumors were collected and digested to obtain single-cell suspensions. The cells were incubated with IF-conjugated Abs in PBS containing 2% FBS for 30 min at room temperature after blockade by TruStain FcX PLUS (anti-mouse CD16/32) Antibody (BioLegend, CloneS17011E, 156603, Lot: B362118; 0.25 µg per 10−6 cells). The following Abs were used: Phycoerythrin (PE)/Cyanine7 anti-mouse CD3 (BioLegend, Clone: 17A2, 126408, Lot: B356287; 1:100), FITC anti-mouse CD45 (BioLegend, Clone: 30-F11, 103108, Lot: B363912; 1:100), PE anti-mouse CD4 (BioLegend, Clone: RM4-5, 100511, Lot: B359453; 1:100), PE anti-mouse CD69 (BioLegend, Clone: H1.2F3, 104514, Lot: B372693; 1:100), PE/Cyanine7 anti-mouse CD107a (BioLegend, Clone: 1D4B, 121619, Lot: B376215; 1:100), PE/Cyanine7 anti-mouse CD11b (BioLegen, Clone: M1/70, 101207, Lot: B279981; 1:100), APC anti-mouse CD11c (BioLegend, Clone: N418, 117310, Lot: B372999; 1:100), BV421 anti-mouse CD86 (BioLegend, Clone: GL-1, 105031, Lot: B261490; 1:100) and PE/Cyanine7 anti-mouse CD206 (BioLegend, Clone: C086C2, 141705, Lot: B347491; 1:100). The stained cell samples were analyzed on a flow cytometer (BD Biosciences) with FlowJo (Version 9).
Antigen cross-presentation assays
The U87MGEGFRvIII cells were transduced with lentiviral vectors to express NY-ESO-1 only or with HLA-A0201. The NY-ESO-1-specific Jurkat cells were generated by transducing the Jurkat TCRαβ-KO cell line with a lentivirus encoding the anti-NY-ESO-1-specific TCR(1G4). The human CAR-iMACs with HLA-A0201+ phenotype were pretreated by IFN-γ (20 ng ml−1) for 24 h, followed by LPS (100 ng ml−1) for 30 min. Next, 5 × 104 CAR-iMACs were cocultured with 1 × 104 U87MGEGFRvIII, U87MGEGFRvIII-NY and U87MGEGFRvIII-HLA0201-NY cells for 48 h in 96-well plates. Subsequently, 5 × 104 NY-ESO-1-specific Jurkat cells were added to each well. Twenty-four hours later, T cells were analyzed for activation marker CD69 using Flow Cytometry. The data were presented by GraphPad Prism 8.2.1.
IF assay
To detect the expression of EGFRvIII in the U87MGEGFRvIII cell line, the U87MGEGFRvIII cells attaching to 0.1% gelatin-coated glass slides were incubated with primary antibody against EGFRvIII (CST, 64952; 1:200) at 4 °C overlight, followed by incubation with a secondary antibody (Alexa Fluor 647 donkey anti-rabbit immunoglobulin G (IgG), Abcam, ab150075; 1:1,000), for 1 h in 37 °C. Afterward, the glass slides were mounted with an antifading mounting medium (with DAPI; Solarbio, S2110). Cells were observed with a Zeiss LSM800 fluorescence microscope at a ×63 oil objective. LSM800 with the Airyscan module was used to capture high-resolution pictures.
For detecting NF-κB/P65, the EGFP-marked CAR-iMACs were co-incubated with tdTomato+ U87MGEGFRvIII cells for 4 and 12 h, respectively, in 0.1% gelatin-coated glass slides. Afterward, the cells were incubated with Alexa Fluor 647-conjugated rabbit antibodies against NF-κB/p65 (CST, 8801; 1:100) at 4 °C overnight, followed by being mounted with an antifading mounting medium.
Detection of phagocytosis by CAR-iMACs/CAR-BMDMs
To capture phagocytosis of CAR-iMACs/BMDMs against target tumor cells, we incubated EGFP-labeled CAR-iMACs/BMDMs with tdTomato+ tumor cells at an E/T ratio of 10/1 in Glass Bottom Cell Culture Dish (NEST, 801001) for 12 h. Afterward, cells were observed under an Olympus FV3000 confocal microscope with the ×60 oil objective. Processing of images was conducted through ImageJ software.
HIS-SIM imaging
The procedure for Hessian-SIM imaging was performed by following the previous report51. A commercial HIS-SIM was used to acquire and reconstruct the cell images. To further improve the resolution and contrast in reconstructed images, sparse deconvolution was used by following the previous report52.
RNA isolation and qRT–PCR
Total RNA extraction was performed using Eastep Super Total RNA Extraction Kit (Shanghai Promega Bio, LS1040) according to the manufacturer’s instructions. cDNA was prepared using HiScript II Q Select RT SuperMix for qRT–PCR with gDNA wiper (Vazyme, R223-01). Gene expression was analyzed in triplicates using HiScript II One Step qRT–PCR SYBR Green Kit (Vazyme, Q221-04) and Bio-Rad PCR machine (CFX-96 Touch). The difference in cycle threshold values (ΔCT) of all genes tested was normalized to the ΔCT of glyceraldehyde-3-phosphate dehydrogenase, and the fold change in expression was expressed relative to WT iMACs. The primer sequences used in the qRT–PCR assay are listed in the Supplementary Data.
Immunoblotting (IB)
The IB assay was performed with conventional SDS–PAGE. The lysates were obtained from WT and EGFRvIII-expressing U87MG cells with RIPA buffer (Beyotime, P0013K) containing protease inhibitors (Roche, 04693124001). The antibodies used in these experiments are as follows: rabbit anti-human EGFR (CST, 4267; 1:2,000), rabbit anti-human EGFRvIII polyclonal antibody (Absin, abs124275; 1:1,000) and rabbit anti-human β-actin (CST, 4970; 1:10,000). Blots were incubated with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, 34577). Signal from blots was detected with ChemiDoc Touch Imaging System (Bio-Rad).
Fluorescence-activated cell sorting (FACS) analysis
Flow cytometry was performed on Beckman CytoFLEX LX (Version 9). FACS data were collected using CytExpert (Version 2.3) and were processed using FlowJo (Version 9). To detect the expression efficiency of CARs in CAR-iMACs, we performed flow cytometry by using rabbit anti-human IgG F(ab′)2/APC (Bioss, bs-0364R-APC; 1:100) that can recognize the F(ab′)2 fragment of EGFRvIII- and GPC3-CARs. The WT-iMACs stained or unstained with the APC-conjugated F(ab′)2 antibody were applied as the negative control.
To count the residual tumor cells after treatment by CAR-iMACs, tdTomato+ U87MGEGFRvIII cells were incubated with CAR-iMACs at E/T ratios of 3/1, 5/1 and 10/1 for 12 and 24 h, respectively, in vitro. The cells were digested using 0.25% trypsin-EDTA (Meilunbio, MA0233). The dissociated cells were resuspended with 0.1% BSA and then filtered into individual cells using a 300-mesh. The PE fluorescence channel was chosen to detect tdTomato+ U87MGEGFRvIII cells. The FITC channel was used to recognize EGFP-marked CAR-iMACs.
To assess dynamic changes in the polarization state of the CAR-iMACs in the presence of tumor cells in vitro, we first incubated EGFP-marked CAR-iMACs with U87MGEGFRvIII cells without tdTomato at an E/T ratio of 10/1 for 1, 2, 3 and 7 d, respectively. After that, the cells were collected as performed above and stained with PE fluorophore-conjugated antibodies recognizing human CD80 protein (PE-CD80; Biolegend, 305208, Lot: B330518; 1:200) or the human CD163-specific antibody conjugated with PE (PE-CD163; Biolegend, 333606, Lot: B347256; 1:200). Then, flow cytometry was performed to collect the EGFP-positive population of CAR-iMACs, followed by detection of CD80+ and CD163+ cells from EGFP-marked CAR-iMACs
To analyze the polarization state of CAR-iMACs in vivo, we first cultured EGFRvIII-targeting CAR-iMACs with a cytokine-free medium (for 48 h). This time, we created tumor models using U87MGEGFRvIII cells without tdTomato to eliminate interference from its signal. After 2 d of CAR-iMACs treatment, we isolated CAR-iMACs from the tumor-bearing mice using a dissociative buffer containing 0.02% collagenase IV (Solarbio, C8160), 0.01% hyaluronidase (Meilunbio, 37326-33-3) and 0.002% DNAase (Meilunbio, 9003-98-9), and the cells were stained using the PE-CD80 or PE-CD163 antibodies.
Apoptosis analysis
To analyze the apoptosis of target tumor cells, we incubated EGFP-labeled CAR-iMACs with tdTomato+ tumor cells at an E/T ratio of 10/1 in vitro for 6 and 12 h, respectively. Afterward, cells were collected for FCM analysis with an Annexin V-Alexa Fluor 647/PI Apoptosis Detection Kit (Yeasen, 40304ES50) on the Beckman CytoFLEX LX (Version 9). The FACS data were processed by FlowJo V10. To investigate the effect of TNF on tumor cell apoptosis, we treated the incubation system with 5 μg ml−1 Adalimumab (Targetmol, 331731-18-1) for 12 h and performed FCM analysis.
Generation of single-cell GEMs and sequencing libraries
We established single-cell GEMs (gel beads in emulsion) and sequencing libraries using methods described in our previous work25. In total, 10,000 cells (90–95% viability) were captured per sample using a 10x Chromium device using 10x V2 Single-Cell 3′ Solution reagents (10x Genomics) at a concentration of 1,000 cells per μl. The experiment was performed according to the manual instructions. After GEM-reverse–transcription (RT) incubation, barcoded cDNA was purified with DynaBeads cleanup mix, followed by ten cycles of PCR amplification (98 °C for 3 min; (98 °C for 15 s, 67 °C for 20 s, 72 °C for 1 min) × 10; 72 °C for 1 min). The total cDNA of single-cell transcriptomes was then fragmented, with double-size selection using solid-phase reversible immobilization (SPRI) beads (Beckman), followed by 12 cycles of sample index PCR amplification (98 °C for 45 s; (98 °C for 20 s, 54 °C for 30 s, 72 °C for 1 min) × 10; 72 °C for 1 min). Subsequently, another double-size selection with SPRI beads was performed before sequencing (Illumina NextSeq platform).
Public bulk RNA-seq data mapping and analysis
A publicly available bulk RNA-seq dataset of iPSC and macrophage cells was obtained from two previous studies46,53. All publicly available bulk RNA-seq reads were first trimmed using Trimmomatic (version 0.36) software with the parameters ‘ILLUMINACLIP: TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36’ and were further quality-filtered using the FASTX Toolkit (version 0.0.13, http://hannonlab.cshl.edu/fastx_toolkit/) with the minimum quality score 20 and a minimum of 80% of bases that have a quality score larger than this cutoff value. The high-quality reads were mapped to the GRCh38 genome by HISAT2, a fast and sensitive spliced alignment program for mapping RNA-seq reads, with the -dta parameter. PCR duplicate reads were removed using Picard tools (v2.18.2), and only uniquely mapped reads were kept for further analysis. The expression levels of genes were calculated by StringTie (v1.3.4d, with -e -B -G parameters) using Release 28 (GRCh38.p12) gene annotations downloaded from the GENCODE data portal. To obtain reliable and cross-sample comparable expression abundance estimation for each gene, reads mapped to the reference genome were counted as transcripts per million (TPM) reads based on their genome locations. We used ‘prcomp’ and pheatmap function with default parameters for PCA and hierarchical clustering, respectively. We also used high confident genes characterizing the M1 and M2 states (with the maximum TPM values among different samples larger than 1) for PCA and hierarchical clustering analysis.
scRNA-seq data analysis
The ‘cellrange count’ program, a subcommand included in the 10× single-cell gene expression analysis pipeline (https://support.10xgenomics.com/single-cell-gene-expression/), was used to produce a gene–cell barcode expression matrix. The single-cell gene expression matrix was analyzed with Seurat v3.2.1 (https://satijalab.org/seurat/). To guarantee the quality of genes and cells, we excluded genes expressed in fewer than three cells and cells expressing less than 200 genes. We also filtered the cells with the expressional percentages of mitochondrial genes larger than 20%. We then adopted the top 20 principal components for tSNE and clustering analysis with a cluster resolution of 0.1.
The known cell identities (types) of each cell cluster of gene expression data were further predicted by an entropy-based predictor with default parameters (http://scibet.cancer-pku.cn/; SciBet v1.0). To perform a comprehensive annotation of well-known cell types, we used an Atlas database of cell types during human fetal liver hematopoiesis and integrated them with 30 major human cell type databases from 42 scRNA-seq datasets54,55. To perform PCA, hierarchical clustering and differential gene expression analysis on bulk RNA-seq data, we grouped and summed up the normalized expression levels of our single-cell gene expression data to produce the synthetic bulk RNA-seq dataset. We used ‘prcomp’ function, pheatmap (https://cran.r-project.org/web/packages/pheatmap/index.html) and DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) packages with default parameters for PCA, hierarchical clustering and differential gene expression analysis, respectively. The IPSDM MAC, IPSDM M1 and IPSDM M2 used in PCA, and hierarchical clustering were obtained from the Gene Expression Omnibus (GEO) database (GSE55536)46. The bioinformatics data analysis and visualization of results were performed in R software (4.0.2)/Bioconductor (v3.13) using custom R scripts.
Statistical analysis
All statistical analyses for next-generation sequencing (NGS) data were performed with R software (4.0.2). The other statistical analyses were performed with GraphPad Prism 8.2.1 software. Details of individual tests are outlined within each figure legend, including the number of replications performed (n) and the reported error as standard error of the mean (mean ± s.e.m.). P values of statistical analyses for NGS data were as the following: NS, not significant, *P < 0.05, **P < 0.01 and ***P < 0.001, calculated by the Wilcoxon signed-rank test (for paired samples), Mann–Whitney U test (for independent samples). P values of statistical analyses performed with GraphPad Prism 8.2.1 software were as the following: NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, and were calculated by the two-way ANOVA, multiple t tests, two-tailed/non-paired t test and as described in the figure legends. Data distribution was assumed to be normal, but this was not formally tested. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications25,26. All the data collection was randomized. For animal studies, tumor-bearing mice were randomly assigned to the various experimental groups and received CAR-iMACs treatment. Data collection and analysis were not performed blind to the conditions of the experiments. No animals or data points were excluded from the analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
scRNA-seq generated in this study has been deposited in the National Center for Biotechnology Information GEO database under the accession code GSE214140. Previously published single-cell and bulk RNA-Seq data that were re-analyzed here are available under accession codes E-MTAB-7407 and GSE55536. Source data are provided with this paper.
Code availability
The codes used for the analysis reported in this study are freely available at https://github.com/huayu1111/carmProj.
Change history
18 December 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41590-023-01734-4
References
Gomes-Silva, D. et al. CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood 130, 285–296 (2017).
Chuntova, P., Downey, K. M., Hegde, B., Almeida, N. D. & Okada, H. Genetically engineered T-cells for malignant glioma: overcoming the barriers to effective immunotherapy. Front. Immunol. 9, 3062 (2018).
Ma, L. et al. Enhanced CAR-T cell activity against solid tumors by vaccine boosting through the chimeric receptor. Science 365, 162–168 (2019).
Lynn, R. C. et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019).
Adachi, K. et al. IL-7 and CCL19 expression in CAR-T cells improves immune cell infiltration and CAR-T cell survival in the tumor. Nat. Biotechnol. 36, 346–351 (2018).
Upadhyay, R. et al. A critical role for Fas-mediated off-target tumor killing in T-cell immunotherapy. Cancer Discov. 11, 599–613 (2021).
Hamieh, M. et al. CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature 568, 112–116 (2019).
Martinez, M. & Moon, E. K. CAR T cells for solid tumors: new strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front. Immunol. 10, 128 (2019).
Bonnardel, J. & Guilliams, M. Developmental control of macrophage function. Curr. Opin. Immunol. 50, 64–74 (2018).
Cassetta, L. & Pollard, J. W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17, 887–904 (2018).
Cortez-Retamozo, V. et al. Origins of tumor-associated macrophages and neutrophils. Proc. Natl Acad. Sci. USA 109, 2491–2496 (2012).
Qian, B. Z. & Pollard, J. W. Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010).
Gambardella, V. et al. The role of tumor-associated macrophages in gastric cancer development and their potential as a therapeutic target. Cancer Treat. Rev. 86, 102015 (2020).
Chen, Y., Zhang, S., Wang, Q. & Zhang, X. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J. Hematol. Oncol. 10, 36 (2017).
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382 (2019).
Vitale, I., Manic, G., Coussens, L. M., Kroemer, G. & Galluzzi, L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 30, 36–50 (2019).
Song, M. et al. Tumor derived UBR5 promotes ovarian cancer growth and metastasis through inducing immunosuppressive macrophages. Nat. Commun. 11, 6298 (2020).
Cioni, B. et al. Androgen receptor signalling in macrophages promotes TREM-1-mediated prostate cancer cell line migration and invasion. Nat. Commun. 11, 4498 (2020).
Huang, R. et al. CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating β-catenin/STAT3 signaling. Cell Death Dis. 11, 234 (2020).
Salvagno, C. et al. Therapeutic targeting of macrophages enhances chemotherapy efficacy by unleashing type I interferon response. Nat. Cell Biol. 21, 511–521 (2019).
Wiehagen, K. R. et al. Combination of CD40 agonism and CSF-1R blockade reconditions tumor-associated macrophages and drives potent antitumor immunity. Cancer Immunol. Res. 5, 1109–1121 (2017).
Kang, M. et al. Nanocomplex-mediated in vivo programming to chimeric antigen receptor-M1 macrophages for cancer therapy. Adv. Mater. 33, e2103258 (2021).
Ries, C. H. et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25, 846–859 (2014).
Akkari, L. et al. Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci.Transl. Med. 12, eaaw7843 (2020).
Zhang, L. et al. Pluripotent stem cell-derived CAR-macrophage cells with antigen-dependent anti-cancer cell functions. J. Hematol. Oncol. 13, 153 (2020).
Klichinsky, M. et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 38, 947–953 (2020).
Morrissey, M. A. et al. Chimeric antigen receptors that trigger phagocytosis. eLife 7, e36688 (2018).
Takata, K. et al. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity 47, 183–198 (2017).
Zhang, H. & Reilly, M. P. Human induced pluripotent stem cell-derived macrophages for unraveling human macrophage biology. Arter. Thromb. Vasc. Biol. 37, 2000–2006 (2017).
Lachmann, N. et al. Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Rep. 4, 282–296 (2015).
Kawai, T. & Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011).
Lancaster, G. I. et al. Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid-induced inflammation by reprogramming macrophage metabolism. Cell Metab. 27, 1096–1110 (2018).
Fan, W. et al. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 29, 4223–4236 (2010).
Fitzgerald, K. A. & Kagan, J. C. Toll-like receptors and the control of immunity. Cell 180, 1044–1066 (2020).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Kondo, T., Kawai, T. & Akira, S. Dissecting negative regulation of toll-like receptor signaling. Trends Immunol. 33, 449–458 (2012).
Schappe, M. S. et al. Chanzyme TRPM7 mediates the Ca2+ influx essential for lipopolysaccharide-induced toll-like receptor 4 endocytosis and macrophage activation. Immunity 48, 59–74 (2018).
Kagan, J. C. et al. TRAM couples endocytosis of toll-like receptor 4 to the induction of interferon-β. Nat. Immunol. 9, 361–368 (2008).
Yamamoto, M. et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 (2003).
Wermuth, P. J. & Jimenez, S. A. Gadolinium compounds signaling through TLR4 and TLR7 in normal human macrophages: establishment of a proinflammatory phenotype and implications for the pathogenesis of nephrogenic systemic fibrosis. J. Immunol. 189, 318–327 (2012).
Scharping, N. E. et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 22, 205–215 (2021).
Park, J. E. et al. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 38, 5158–5173 (2019).
Liu, Y. N. et al. Hypoxia induces mitochondrial defect that promotes T cell exhaustion in tumor microenvironment through MYC-regulated pathways. Front. Immunol. 11, 1906 (2020).
Xu, J. et al. Hypoxic glioma-derived exosomes promote M2-like macrophage polarization by enhancing autophagy induction. Cell Death Dis. 12, 373 (2021).
Moynagh, P. N. TLR signalling and activation of IRFs: revisiting old friends from the NF-κB pathway. Trends Immunol. 26, 469–476 (2005).
Zhang, H. et al. Functional analysis and transcriptomic profiling of iPSC-derived macrophages and their application in modeling Mendelian disease. Circ. Res. 117, 17–28 (2015).
Mehrotra, P. & Ravichandran, K. S. Drugging the efferocytosis process: concepts and opportunities. Nat. Rev. Drug Discov. 21, 601–620 (2022).
Franklin, R. A. et al. The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 (2014).
Chen, Y. et al. CAR-macrophage: a new immunotherapy candidate against solid tumors. Biomed. Pharmacother. 139, 111605 (2021).
Sadelain, M., Brentjens, R. & Rivière, I. The promise and potential pitfalls of chimeric antigen receptors. Curr. Opin. Immunol. 21, 215–223 (2009).
Huang, X. et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol. 36, 451–459 (2018).
Zhao, W. et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nat. Biotechnol. 40, 606–617 (2022).
Douvaras, P. et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Rep. 8, 1516–1524 (2017).
Popescu, D. M. et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019).
Liu, B. et al. An entropy-based metric for assessing the purity of single cell populations. Nat. Commun. 11, 3155 (2020).
Acknowledgements
We acknowledge Dan Kaufman (UCSD), Huang Zhu (UCSD) and Jin Zhang lab members for discussion of the manuscript. We also thank Chong Liu (Zhejiang University) for providing the U87MG cell line and valuable suggestions and Xuanming Yang (Shanghai Jiao Tong University) for providing the reagents such as the JurkatTCRαβ-KO cell line. This work was funded by the National Natural Science Foundation of China (82373238, 31871453 and 91857116 to J.Z.), the National Key Research and Development Program of China (2018YFA0107103 and 2018YFC1005002 to J.Z.), Zhejiang Innovation Team (grant 2019R01004 to J.Z.), the Zhejiang Natural Science Foundation (LR19C120001 to J.Z.), the Key Research and Development Program of Zhejiang Province (2023C03036) and research fund from CellOrigin and Qilu Pharmaceutical, the Zhejiang Natural Science Foundation of China (LQ22H160001 to A.L.) and the China Postdoctoral Science Foundation (2019M662035 to A.L.). We thank Guangzhou CSR Biotech for live-cell imaging by using their commercial super-resolution microscope (HIS-SIM), data acquisition, SR image reconstruction, analysis and discussion.
Author information
Authors and Affiliations
Contributions
J.Z. and A.L. designed the study. A.L. S.L., H.L., X.D., T.T., X.W., H.Z., S.S., M.Z., L.T., D. X., W.Z., S.Z., Y.C., W.X., L.Z., Y.Z. and J.Z. performed the experiments. H.Y. contributed to the NGS data analysis. W.J. contributed to the culture of U87MG cells. G.C., F.C. and Z.G. provided important discussions. J.Z. and A.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.Z. is a scientific cofounder of CellOrigin. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: N. Bernard, in collaboration with the Nature Immunology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Examining phagocytosis of CAR-iMACs against the antigen-expressing cancer cells.
a, Diagrammatic drawing for an EGFRvIII-expressing lentivirus vector. b, Western blotting showing the expression of EGFRvIII in wild-type U87MG (WT-U87MG) cells and lentivirus-infected U87MG cells (U87MGEGFRvIII). c, Western blotting showing the expression of the total EGFR in wild-type U87MG cells and lentivirus-infected U87MG cells. d, Immunofluorescence showing expression and localization of EGFRvIII in U87MGEGFRvIII cells. EGFRvIII exhibited prominently high expression and evident localization on the membrane of cells (indicated by the white arrows) of U87MGEGFRvIII. e, Co-incubating experiment showing that EGFP-marked EGFRvIII-targeting CAR-iMACs (truncated, CD3ζ- and TIR-CAR-iMACs) adhered to the tdTomato-expressed U87MGEGFRvIII cells more closely than EGFP-marked WT-iMACs after co-culturing for 4 hours. f, Both CD3ζ-CAR-iMACs and TIR-CAR-iMACs showed enhanced cancer cell phagocytosis activity against U87MGEGFRvIII cells compared to truncated CAR-iMACs and WT-iMACs after co-culturing for 12 hours.
Extended Data Fig. 2 CD3ζ-TIR-CAR-iMACs exhibited enhanced and persistent antitumor activity.
a, An overview of the antitumor assay of EGFRvIII-targeting CAR-iMACs in vivo. Four hours after injection of FFluc+U87MGEGFRvIII cells into NOD-SCID mice, EGFRvIII-targeting truncated CAR-iMACs, CD3ζ-CAR-iMACs, TIR-CAR-iMACs, and CD3ζ-TIR-CAR-iMACs without IFN-γ/LPS-polarization pretreatment were injected into the tumor-bearing mice. Two hours after the CAR-iMACs injection, animal imagings were conducted to detect the signal of BLI from tumor cells. On the following day 1, day 3, and day 7, bioluminescent imaging was performed at the same time during the days. b, Visual presentation of the tumor signal from the tumor-bearing mice after treatment by the four types of CAR-iMACs. c, Statistics analysis of tumor BLI signal from b (n = 5 mice from truncated CAR-iMAC group, n = 5 mice from CD3ζ-CAR-iMAC group, n = 10 mice from TIR-CAR-iMAC group, n = 10 mice from CD3ζ-TIR-CAR-iMAC group). Significance was calculated with a two-way ANOVA analysis and are presented as mean±s.e.m. p-value: ns, not significant, *P < 0.05, **P < 0.01. d, The Kaplan-Meier survival curve of CAR-iMACs treated mice over 29 days from b. Significance was calculated with two-tailed log-rank Mantel-Cox test (CD3ζ-CAR-iMAC group versus truncated CAR-iMAC group, P = 0.0999. TIR-CAR-iMAC group versus truncated CAR-iMAC group, P = 0.0003. CD3ζ-TIR-CAR-iMAC group versus truncated CAR-iMAC group, P = 0.0003) e, FCM analysis of CD80-positive population and CD163-positive population of EGFRvIII-targeting CD3ζ-CAR-iMACs, TIR-CAR-iMACs, and CD3ζ-TIR-CAR-iMACs isolated from the mice treated in the same way as in b for 48 hours. The FCM experiments were carried out in triplicates and the results were processed using FlowJo. f, Histograph analysis of e (n = 3 biologically independent samples per group). The significance was calculated with a two-way ANOVA analysis and are presented as mean±s.e.m. p-value: ns, not significant, *P < 0.05, **P < 0.01. All the statistical data above was shown by Graphpad Prism 8.2.1.
Extended Data Fig. 3 The CD3ζ-TIR-CAR-iMACs showed potent antitumor activity against intracranial GBM.
a, An overview of the anti-GBM assay of EGFRvIII-targeting CAR-iMACs in vivo. 1 × 104 FFluc+U87MGEGFRvIII cells were injected into the intracranial corpus striatum of NOD-SCID mice six days before immune cell therapy. On day -1, live animal imaging was conducted to monitor the formation of GBM. On day 0, EGFRvIII-targeting truncated CAR-iMACs and CD3ζ-TIR-CAR-iMACs with no IFN-γ/LPS-pretreatment were injected into the GBM lesion. On the following day 1, day 2, day 3, day 5, and day 10, the bioluminescent imaging was performed at the same time point to record the change of signal from the GBM. b, Visual presentation of the tumor signal from the intracranial GBM after treatment by the truncated CAR-iMACs and CD3ζ-TIR-CAR-iMACs. The PBS treatment group was designed as a negative control. c, Statistics analysis of tumor BLI signal from b (n = 5 mice from PBS group, n = 5 from truncated CAR-iMAC group, n = 5 mice from CD3ζ-TIR-CAR-iMAC group). The significance was calculated with the two-way ANOVA analysis and are presented as mean±s.e.m. p-value: ns, not significant, ***P < 0.001. d, The Kaplan-Meier survival curve of CAR-iMACs treated mice over 17 days from b. Significance was calculated with two-tailed log-rank Mantel-Cox test (CD3ζ-TIR-CAR-iMAC group versus PBS group, P = 0.0003. CD3ζ-TIR-CAR-iMAC group versus truncated CAR-iMAC group, P = 0.0003) All the statistic data above was shown by Graphpad Prism 8.2.1. e, The IHC staining showing the EGFRvIII protein expression of FFluc+U87MGEGFRvIII-derived GBM on day 8 and day 16 after the tumor cell inoculation, indicating the persistent expression of EGFRvIII antigen.
Extended Data Fig. 4 GPC3-targeting CAR-iMACs showed enhanced antitumor capacity against HCC.
a, The flow cytometry analysis was performed to determine the expression of GPC3-CARs on truncated CAR-iMACs, TIR-CAR-iMAC, and CD3ζ-TIR-CAR-iMACs by an APC-conjugated rabbit antibody specific to F(ab’)2 of human IgG. Shown is a representative result from three biological replicates. The results were processed using FlowJo. The numbers are mean±s.e.m. b, Real-time imaging by an Incucyte2022 A Live-Cell Analysis Instrument showed the signals of the tdTomato+ HepG2 cells (white arrow) during a time course of 138 hours after incubating GPC3-targeting truncated CAR-iMACs, TIR-CAR-iMAC, and CD3ζ-TIR-CAR-iMACs (bright field) with the tumor cells at the E/T ratio of 10/1. c, The TIR-based CAR-iMACs cooperating with CD47-antibody cured HepG2-derived hepatocellular neoplasm in tumor-bearing mice. 5 × 105 FFluc+HepG2 cells were intraperitoneally injected to B-NDG mouse. 4 days after that, GPC3-targeting truncated CAR-iMACs, TIR-CAR-iMAC, and CD3ζ-TIR-CAR-iMACs mixed with CD47-antibody were injected intraperitoneally into the tumor-bearing mice. 2 hours after the treatment, live animal imaging was performed to monitor the growth of HepG2 cells in vivo. The treatment group with only CD47-antibody was designed as a control. d, The statistics of change of tumor burden from the treated mice from c (n = 5 mice from CD47 antibody group, n = 5 mice from truncated CAR-iMAC + CD47 antibody group, n = 5 mice from TIR-CAR-iMAC + CD47 antibody group, n = 10 mice from CD3ζ-TIR-CAR-iMAC + CD47 antibody group). e, The statistics of change of weight of the mice from c. All the above data was shown by GraphPad Prism 8.2.1. The significance was calculated with a two-way ANOVA analysis with multiple comparisons and are presented as mean±s.e.m. p-value: ns, not significant, *P < 0.05,***P < 0.001,****P < 0.0001.
Extended Data Fig. 5 The second-generation CAR improved the TME.
a, The schematic diagram of constructs of GPC3-targeting CARs specific to mouse cells. Compared to the human-specific second-generation CAR, we replaced the intracellular signal transduction domain with mouse-specific TIR and CD3ζ domains in tandem, and then named the new CAR G2-CAR. The truncated CAR (G1-CAR) without an intracellular signal domain was designed as a negative control. b, The imaging by fluorescence microscope showed the successful transduction of G2-CAR into mouse BMDMs. c and d, The flow cytometry analysis was performed to detect the expression level of the EGFP protein (c) and the expression level of the CAR (d). The results were from one experiment and were processed using FlowJo e, Confocal imaging showed the phagocytosis of EGFP-labeled CAR-BMDMs against tdTomato+ Hepa1-6 cells expressing human GPC3 (Hepa1-6GPC3) in vitro. The experiment was performed for three times. f, The statistic of CAR-BMDMs that have phagocytized the tumor cells from e (n = 11 BMDM samples per group from three independent experiments). g, The solid tumors were harvested nine days after subcutaneous injection of Hepa1-6GPC3 cells in C57BL/6 mice and six days after intratumoral CAR-BMDM treatment. The PBS treatment group was designed as a negative control. h and i, The change of tumor size (h) and body weight (i) were recorded from day 3 (CAR-BMDM treatment) to day 9 (n = 6 samples from PBS group, n = 6 samples from G1-CAR-BMDM group, n = 6 samples from G2-CAR-BMDM group). The data was shown by GraphPad Prism 8.2.1. The significance was calculated with a two-way ANOVA analysis with multiple comparisons and are presented as mean±s.e.m. p-value: **P < 0.01. j–n, The population of CD86-positive macrophages (j), CD206-positive macrophages (k), DC cells (l), NK cells (m), and CD69-positive T cells (n) were detected respectively by flow cytometry analysis from tumor samples in g (n = 6 independent samples per group). The data was shown by GraphPad Prism 8.2.1. The significance was calculated with a two-tailed/non-paired t-test and are presented as mean±s.e.m. p-value: ns, not significant, *P < 0.05, **P < 0.01.
Extended Data Fig. 6 IF assays showed subcellular localization of NF-κB/P65 in CAR-iMACs.
a, NF-κB/P65 exhibited nuclear localization in response to LPS stimulation to EGFRvIII-targeting CAR-iMACs (indicated by the white arrow). The red circle indicating monocyte-like iMACs showed high expression of NF-κB/P65 in the absence of LPS stimulation. b, 4 hours of incubation of U87MGEGFRvIII cells with four types of CAR-iMACs without IFN-γ/LPS pretreatment showed limited distribution of NF-κB/P65 in the nuclei of CAR-iMACs (indicated by the white arrow). The experiment was performed three times.
Extended Data Fig. 7 scRNA-seq analysis revealed the features of sub-populations of CAR-iMACs.
a, A tSNE plot of 10× Genomic single-cell RNA-seq data showing six clusters from the EGFRvIII-targeting truncated CAR-iMACs group after being incubated with U87MGEGFRvIII cells for 24 hours in vitro. b, Cell type annotations of the different sub-populations or clusters from a obtained by an entropy-based computational approach (Liu et al., 2020). c, A tSNE plot of the six main clusters derived from the TIR-CAR-iMACs group after being incubated with U87MGEGFRvIII cells for 24 hours in vitro. d, Cell type annotation of different clusters from c using an entropy-based computational approach. e, A tSNE plot of six main clusters in the CD3ζ-TIR-CAR-iMACs group after being incubated with U87MGEGFRvIII cells for 24 hours in vitro. f, Cell type annotation of different clusters from e. All the scales in b, d, and f were designed to indicate the percentage of a specific cell type.
Extended Data Fig. 8 scRNA-seq analysis revealed similar expression profiles between TIR-based CAR-iMACs and M1-like macrophages.
a, The heatmap comparing the gene expression profile of a set of M1 and M2-associated genes for EGFRvIII-targeting truncated CAR-iMACs, TIR-CAR-iMACs, and CD3ζ-TIR-CAR-iMACs to that of stimulated M1-like, M2-like and unstimulated iMACs. b, Hierarchical cluster tree showing the relationship of the truncated CAR-iMACs, TIR-CAR-iMACs, and CD3ζ-TIR-CAR-iMACs with stimulated M1-like, M2-like, and unstimulated iMACs. c, The heatmap comparing the gene expression profile of a set of M1 and M2-associated genes in CD80+ and nonCD80 cells in truncated CAR-iMACs, TIR-CAR-iMACs, and CD3ζ-TIR-CAR-iMACs to that of stimulated M1-like, M2-like and unstimulated iMACs. d, Hierarchical cluster tree showing the relationship of CD80+ and nonCD80 cells in truncated CAR-iMACs, TIR-CAR-iMACs, and CD3ζ-TIR-CAR-iMACs with stimulated M1-like, M2-like, and unstimulated iMACs. The data of IPSDM MAC, IPSDM M1, and IPSDM M2 were obtained from the GEO database (GSE55536).
Extended Data Fig. 9 A schematic illustration of the second-generation CAR.
All the CARs comprise an extracellular scFv domain recognizing a specific epitope, a transmembrane (TM) domain and a hinge region from CD8α. The truncated CAR was designed as the negative control without the intracellular signaling domain. The CD3ζ-CAR containing a single intracellular CD3ζ signaling domain was defined as the first-generation CAR corresponding to the first-generation T-CAR. The TIR-CAR contains an intracellular TIR signal transduction domain. The CD3ζ-TIR-CAR containing CD3ζ and TIR in the intracellular domain was defined as the second-generation CAR and possesses higher proinflammatory activity than the first-generation CAR upon binding to a specific antigen.
Supplementary information
Supplementary Video 1
A real-time video reveals the events of apoptosis and efferocytosis of HepG2 cells induced by CD3ζ-TIR-CAR-iMACs. The highlighted white arrows and numbers indicate the following events: the 1 arrow indicates the anchoring event of a GPC3-targeting CD3ζ-TIR-CAR-iMAC cell to a HepG2 cell; the 2 arrow points to the aggregation of tumor cell apoptotic bodies, which are gradually engulfed by the adjacent CD3ζ-TIR-CAR-iMAC cell; the 3 arrow indicates the apoptosis process of a living HepG2 cell induced by the CD3ζ-TIR-CAR-iMAC cell, and the apoptotic bodies are gradually taken up by the CAR-iMAC cell. The imaging was performed every 2 min.
Supplementary Video 2
A real-time video shows the apoptosis and efferocytosis of U87MGEGFRvIII cells mediated by CD3ζ-TIR-CAR-iMACs. The highlighted white arrows and numbers indicate the following events: the 1 arrow indicates the anchoring event of a tdTomato-labeled U87MGEGFRvIII cell to an adjacent EGFP-marked EGFRvIII-targeting CD3ζ-TIR-CAR-iMAC cell; the 1 and 2 arrows point to the apoptosis event of the tumor cell indicated by the formed granules, and the tumor cell is eventually eliminated; the 3 arrow and the white box show the debris from tdTomato-labeled U87MGEGFRvIII cell taken up by the CD3ζ-TIR-CAR-iMAC cells; the 4 arrow indicates that the apoptotic bodies of tumor cells are eventually eradicated after in contact with a CAR-iMAC cell; the 5 arrow indicates that a WT iMAC without the CAR and EGFP expression moves around without being anchored. The imaging was performed every 2 min.
Supplementary Data
Sequence of oligonucleotides.
Source data
Source Data Fig. 1
Primary pictures and gating strategy of FCM.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data and gating strategy of FCM.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data and primary pictures of FCM.
Source Data Fig. 7
Statistical source data, gating strategy of FCM and primary pictures of FCM.
Source Data Extended Data Fig. 1
Unprocessed western blots of Extended Data Fig.1b,c.
Source Data Extended Data Fig. 1
Primary pictures of Extended Data Fig. 1d,f.
Source Data Extended Data Fig. 2
Statistical source data and gating strategy of FCM.
Source Data Extended Data Fig. 3
Statistical source data and gating strategy of FCM.
Source Data Extended Data Fig. 4
Statistical source data, gating strategy of FCM and primary pictures.
Source Data Extended Data Fig. 5
Statistical source data, gating strategy of FCM and primary pictures.
Source Data Extended Data Fig. 6
Primary pictures.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lei, A., Yu, H., Lu, S. et al. A second-generation M1-polarized CAR macrophage with antitumor efficacy. Nat Immunol 25, 102–116 (2024). https://doi.org/10.1038/s41590-023-01687-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-023-01687-8
This article is cited by
-
CAR-engineered cell therapies: current understandings and future perspectives
Molecular Biomedicine (2026)
-
tRNA m1A modification orchestrates STING translation in macrophages to enhance antitumor immunity and CAR-macrophage immunotherapy
Cellular & Molecular Immunology (2026)
-
Fasting-mimicking diet induces IFNβ secretion in tumor-associated macrophages via NRF1-mediated ubiquitin-dependent proteolysis of Trex1
British Journal of Cancer (2026)
-
m6A-modified CTC-297N7.9 inhibits hepatocellular carcinoma metastasis via epigenetically downregulating CCL2 and CD47
Cancer Cell International (2025)
-
Synergistic innate-adaptive immunity by NKG2D-specific CAR-macrophages drives durable remission in hepatocellular carcinoma
Molecular Cancer (2025)