Abstract
In mouse peritoneal and other serous cavities, the transcription factor GATA6 drives the identity of the major cavity resident population of macrophages, with a smaller subset of cavity-resident macrophages dependent on the transcription factor IRF4. Here we showed that GATA6+ macrophages in the human peritoneum were rare, regardless of age. Instead, more human peritoneal macrophages aligned with mouse CD206+ LYVE1+ cavity macrophages that represent a differentiation stage just preceding expression of GATA6. A low abundance of CD206+ macrophages was retained in C57BL/6J mice fed a high-fat diet and in wild-captured mice, suggesting that differences between serous cavity-resident macrophages in humans and mice were not environmental. IRF4-dependent mouse serous cavity macrophages aligned closely with human CD1c+CD14+CD64+ peritoneal cells, which, in turn, resembled human peritoneal CD1c+CD14−CD64− cDC2. Thus, major populations of serous cavity-resident mononuclear phagocytes in humans and mice shared common features, but the proportions of different macrophage differentiation stages greatly differ between the two species, and dendritic cell (DC2)-like cells were especially prominent in humans.
Similar content being viewed by others
Main
Mouse peritoneal macrophages have distinct transcriptional features and functional roles in immunity and tissue repair1,2,3,4,5,6,7,8,9. In homeostasis, two distinct populations of peritoneal macrophages exist. The major population, called large peritoneal cavity macrophages (LCMs), are F4/80hiCD115+ICAM2+ cells that rely on the transcription factor GATA6 for development and persistence8,10,11,12,13,14. LCMs are not only defined by markers, but also by evidence that they initially arise from embryonic progenitors to seed body cavities before birth15,16. They are long-lived15 and locally sustained through proliferation2,15,17. In the absence of GATA6, they downregulate some molecules, such as CD73, and upregulate proteins such as CD206 and LYVE1 (refs. 8,10). Besides peritoneum, mouse LCMs occupy the pleural and pericardial cavities18,19, but are absent elsewhere8,10. Rather than anchoring in interstitial tissue, LCMs float within cavity fluid3,7. However, in response to inflammation or injury, LCMs facilitate containment of infectious organisms by aggregating on perturbed cavity surfaces or within fibrin clots that form de novo3,5,6,7. Many LCMs die by pyroptosis after aggregating6, such that local proliferation can be insufficient for LCM recovery, and repopulation from blood monocytes is needed15,20. During filarial nematode infection in C57BL/6 mice, pleural LCM numbers markedly increase, suppressing nematode persistence21. In BALB/c mice, pleural LCM repopulation and terminal differentiation after nematode infection is constrained, leading to chronic persistence of the infection9. Instead of complete maturation from monocytes to Gata6+ LCMs, LCMs accumulate in a transitional differentiation state characterized by a GATA6−CD206+LYVE1+ phenotype21 that associates with nematode persistence21. The GATA6−CD206+LYVE1+ transitional macrophage phenotype resembles that in Lyz2–Cre Gata6fl/fl mice, in which GATA6 is conditionally deleted in macrophages (hereafter Gata6 cKO mice)10.
A second population of F4/80loMHCII+ macrophages within serous body cavities is the small cavity macrophage (SCM)1,12,16,22. SCMs arise postnatally and turn over more rapidly than LCMs, with a developmental program that depends upon the transcription factor IRF4 (refs. 16,22). SCMs express a core gene signature that includes CD115, CD11c, which is variable in some mouse colonies13,16,22,23, high major histocompatibility complex (MHC) II, the costimulatory molecule CD226, and low expression of F4/80, a marker that is typically strongly expressed by macrophages1. The overall gene expression pattern of SCMs, including expression of CD115, supported their classification as macrophages in past broad profiling studies1. However, F4/80loMHCII+ SCMs also have features of dendritic cells (DCs), such that some members of the population are more DC-like and others are more like typical macrophages20,23.
It remains unclear which, if any, features of mouse LCMs and SCMs are shared by human cavity macrophages and to what extent. In this Resource, to compare the mouse and human peritoneal macrophage populations, we performed single-cell sequencing, imaging and flow cytometry on cells obtained from human adults and children undergoing peritoneal washes during laparoscopic surgery. We found that GATA6+ peritoneal macrophages were present but rare in humans. Instead, other macrophage populations dominated. All individuals, regardless of age or sex, had higher numbers of macrophages that closely resembled the transitional Mrc1+Lyve1+Gata6− LCMs observed in BALB/c mice after helminth infection21 or in Gata6 cKO C57BL/6 mice10 than they had GATA6+ macrophages. Analysis of mice fed a high-fat diet or mice captured in the wild rarely shifted away from the dominance of Gata6+ LCMs, suggesting that the species differences between humans and mice may not arise from their distinct environments or living conditions. We also identified many CD1c+ human peritoneal mononuclear phagocytes, including prominent numbers of bona fide CD1c+ cDC2s and CD1c+CD14+CD64+CD226+ cells bearing hybrid macrophage and DC features. Gene expression comparisons indicated that CD1c+CD14+CD64+CD226+ cells were close counterparts to mouse SCMs. Collectively, this work defines the human peritoneal landscape from childhood to adulthood and identifies how mouse and human macrophage and DC populations relate.
Results
Human GATA6+ peritoneal macrophages are rare
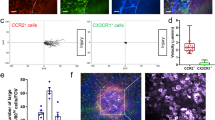
We collected peritoneal wash samples from seven adults undergoing laparoscopic surgery, including five obese patients receiving gastric bypass or related weight reduction procedures and two with normal body mass index having surgery for hernia repair or achalasia (Supplementary Table 1), conditions that would not be expected to involve direct inflammation in the peritoneal cavity. We sorted CD45+ peritoneal cells and carried out single-cell RNA sequencing (scRNA-seq) using the 10x Genomics platform. A combined analysis of myeloid cells in all seven samples separated distinct clusters of macrophages or DCs (Fig. 1a, Extended Data Fig. 1a and Supplementary Table 2). Each sample contained all clusters, except for the low abundance cluster 13, which originated from one sample only (Extended Data Fig. 1b). Few single macrophages expressed GATA6+ (Fig. 1b), and these localized to a TIMD4+ subcluster (Fig. 1b). In mice, the only tissue macrophage to express Selp messenger RNA, which encodes CD62P, is the Gata6+ LCM1. SELP+ cells were within the TIMD4+ subcluster of human macrophages and in cells that were also GATA6+ (Fig. 1b). Other clusters were identified as MRC1+LYVE1+CD163+ macrophages; GLUL+CD163++FABP5+ macrophages; MARCO+ macrophages; IL1B+ macrophages; interferon-stimulated genes (ISG)+ macrophages; CCR2+ macrophages; and CD1C+CD14+ macrophages (Fig. 1a, Extended Data Fig. 1a and Supplementary Table 2).
a, UMAP projection of mononuclear phagocytes (MNPs) from the peritoneal cavity of 7 adults (5 females, 2 males) forming 16 distinct clusters. Cluster names assigned based on inferred function or functional markers, including CCR2+ monocyte-like cavity macrophages (CCR2+ mono-CM), IL1B+ inflammatory cavity macrophages (IL1B inflam-CM). NK/NKT contaminants (NK/NKT contam) were present in a delineated cluster. b, Feature plots showing the expression of general marker genes for myeloid cells and peritoneal macrophages. Color scale represents the normalized gene expression. c, Confocal analysis of GATA6 and CD62P expression in human peritoneal CD14+ macrophages. Biological samples were analyzed over two independent experiments. Scale bars: 10, 5 and 10 μm, left to right. d, Bar plot delineating the proportions of GATA6+ peritoneal macrophages, identified by either RNA (n = 7 human samples) or protein expression assessed by immunofluorescence (IF) (n = 3 human samples and n = 4 mice). A one-way ANOVA and post-hoc comparisons using Tukey’s honestly significant difference were conducted to compare the means of the three groups. ****P < 0.0001. e, Pathway activity estimation of previously published converting macrophage and monocyte-like macrophage gene sets across MNPs in human peritoneal cavity. Color scale represents the scaled score calculated by GSVA. f, RNA velocity analysis of macrophage clusters in all seven adult patients, visualized on the predefined UMAP plot from a. Arrows denote velocity vectors illustrating potential differentiation paths. g, GSEA comparing LCM from Gata6 cKO mice to the gene signatures of the MRC1hi convMac or TIMD4hi LCM clusters from the human peritoneal macrophage dataset. Ticks below the line correspond to gene ranks. Statistical analysis was performed using a Kolmogorov–Smirnov test. Multiple testing correction was conducted automatically using the BH-FDR method. Normalized enrichment scores (NESs) and FDR q values are shown for each cluster.
We also analyzed the expression of GATA6 and CD62P proteins using an antibody that recognizes human and mouse GATA6 that was validated by lack of reactivity in Gata6−/− mouse macrophages10. CD62P and GATA6 were rare among CD14+ macrophages (Fig. 1c and Extended Data Fig. 1c,d), but when found, colocalized to the same cells (Fig. 1c and Extended Data Fig. 1c). GATA6 staining in human cells was confirmed in the GATA6+ HCT116 cell line (Fig. 1c). In mice, peritoneal CD115+ macrophages were GATA6+ (Fig. 1c). Thus, both immunofluorescence and scRNA-seq data indicated that GATA6+ CD62P+ macrophages were rare in the human peritoneal cavity (Fig. 1d).
In mice, before expression of Gata6 and Timd4, monocytes differentiate to an intermediate CD206+ (encoded by Mrc1) LYVE1+ state21. LYVE1 and MRC1 were not always co-expressed in the same human peritoneal cells, but their co-expression was evident in one cluster (Fig. 1b). While 202 genes defined this human cluster (Supplementary Table 2), we refer to it by the short designation ‘MRC1hi convMac’ (Fig. 1e,f). To investigate whether the cells within the MRC1hi convMac cluster, or any of the other human clusters, shared similarity with the Mrc1+Lyve1+ macrophages described in mice21, we analyzed published data21 to generate a gene signature for this subset of mouse peritoneal macrophages. The signature encompassed 330 genes, and, hereafter, we refer to it as the ‘converting cavity macrophage (CM)’ signature (Supplementary Table 3)21. Another 360 genes characterized a different signature specific for CCR2+ monocyte-like macrophages, which we refer to as ‘monocyte-like CM’ (Supplementary Table 3)21. We used gene set variation analysis (GSVA) to assign a ‘converting CM’ or a ‘monocyte-like CM’ score to each cell in the adult human dataset (Fig. 1e). The MRC1hi convMac human cluster had a substantially higher average ‘converting CM’ score and, thus, associated more with Mrc1+Lyve1+ macrophages in mice21 than any other human cluster (Fig. 1e and Extended Data Fig. 1e). RNA velocity analysis suggested that the GATA6+TIMD4+ cluster in humans arose, in part, through conversion from cells within the MRC1hi convMac cluster (Fig. 1f). As quality control, the proportion of spliced GATA6 transcripts was greatest in those with the most numerous GATA6 mRNA transcripts (Extended Data Fig. 1f). This analysis indicated that human MRC1hi convMacs, which co-expressed MRC1 and LYVE1, but not macrophages that expressed just one of these markers, aligned closely to murine Mrc1+Lyve1+ macrophages that represent a differentiation intermediate in the generation of monocyte-derived LCM21.
Next, we compared the human scRNA-seq analysis to three publicly available datasets8,10,11 of bulk-sorted wild-type or Gata6 cKO mouse LCMs (Supplementary Table 4). Principal component analysis (PCA) indicated that wild-type LCM and Gata6 cKO LCMs clustered distinctly by genotype (Extended Data Fig. 1g). Gene set enrichment analysis (GSEA) comparing the clusters of human macrophages to mouse wild-type or Gata6 cKO LCMs uncovered a strong relationship between mouse Gata6 cKO LCMs and the human MRC1hi convMac cluster, but not the more terminally differentiated TIMD4+ human macrophages (Fig. 1g). These data emphasize the scarcity of GATA6+ human peritoneal macrophages and identify the presence of a human MRC1hi convMac cluster that parallels mouse LCM lacking expression of GATA6.
scRNA-seq defines relationship between mouse and human peritoneal cells
To further investigate the relationship between species, we aligned the scRNA-seq for human-cavity macrophages with scRNA-seq analyses generated from three independent peritoneal cavity washes from C57BL/6 mice. The human orthologs of mouse genes were retrieved from the Ensembl database, and canonical correlation analysis (CCA)-based integration was then performed on either all immune cell types (Extended Data Fig. 2a) or myeloid cells only (Fig. 2a), culminating in separate uniform manifold approximation and projection (UMAP) plots of human and mouse cells (Fig. 2a and Extended Data Fig. 2b). Lymphocyte clusters, including CD19+ B cells in mice and CD3+ T cells and NKG7+ NK cells in humans, were evident (Extended Data Fig. 2a–d), as were myeloid clusters that globally expressed genes such as CD14 (Fig. 2b), ITGAM (encoding CD11b) and ITGAX (encoding CD11c) (Extended Data Fig. 2c). Focusing on myeloid cells, most mouse macrophages fell into three related clusters characterized by the expression of GATA6 and other genes such as CXCL13 and SELP (Fig. 2a–e and Extended Data Fig. 3a), with the separation between these three clusters being driven by the differential magnitude of expression of ICAM2, CD44 and APOE (Fig. 2a–c and Supplementary Table 5). Less than 5% of human macrophages were GATA6+ (Fig. 2a–c). In both species, GATA6 was co-expressed with SELP (Fig. 2b). LYVE1 overlapped with some GATA6+ clusters in humans, but co-expression of MRC1 and LYVE1 did not overlap with GATA6 or SELP (Extended Data Fig. 3b). Lyve1+ Mrc1+ macrophages in wild-type C57BL/6 mice were less abundant than in humans, but, when present, localized to the same cluster (Fig. 2b,d). A replot of the human clusters analyzed in Fig. 1 in a new analysis wherein human and mouse data were integrated indicated that the human MRC1hi convMac cluster was consistently identified, regardless of how the analysis was approached (Extended Data Fig. 3c). CCR2+ macrophages from humans and mice, respectively, overlaid within the same UMAP cluster, but were far more abundant in humans, as were GLUL+ macrophages (Fig. 2d), identifying some human macrophages with no clear mouse peritoneal cavity counterpart. The Glul gene in mice marks macrophages universally over other cell types1. When we charted genes associated with the S, G1 or G2M phases of the cell cycle, we identified macrophages in S phase, indicative of cell proliferation, in both species (Extended Data Fig. 3d). Macrophages in S phase were slightly fewer in humans than in mice (Extended Data Fig. 3d,e).
a, UMAP projection of MNPs as in Fig. 1 and three C57BL/6J mouse datasets (GSM7053956, GSM7053957 and GSM7053958) (left) and UMAP plots showing the distinct MNP composition of mouse (middle) and human (right) peritoneal cavity. Cluster names assigned on the basis of marker genes and clusters are color coded. b, Feature plots depicting the expression of marker genes of interest on macrophages in mouse and human. Color scale represents the normalized gene expression. c, Bar plot defining proportions of GATA6+ macrophages in the peritoneal cavity of human (n = 7) and mouse (n = 3) peritoneal cavity. Bars depict means with error bars representing standard deviation. Two-sided Student’s t-test, ****P < 0.0001 (P = 1.91326 × 10−9). d, Proportion of MNP clusters in total MNPs for each sample including those expressing GATA6, MRC1hi convMacs, GLUL+ CM, TIMD4+ LCM, CCR2+ mono-LCM, CD1C+CD14+ cells, CD1C+CD14− cells and cDC1 in human (n = 7) and mice (n = 3). Bars depict means with error bars representing standard deviation. Multiple two-tailed t-tests followed by false discovery rate (FDR) correction; *P < 0.05, **P < 0.01, ****P < 0.0001, n.s. not significant. The exact P values and FDR-corrected q values are reported as source data. e, Z-transformed mean mRNA expression intensity of the top 50 differentially expressed genes between the CD1C+CD14+ and CD1C+CD14− human cell clusters. f, GSEA analysis for the enrichment of published SCM gene signatures16 when compared to CD1C+CD14− or CD1C1+CD14+ human cells. Ticks below the line correspond to gene ranks. Statistical analysis was performed using a Kolmogorov–Smirnov test. Multiple testing correction was conducted automatically using the BH-FDR method. Normalized enrichment scores (NESs) and FDR q values are shown for each cluster.
Mouse CD115+F4/80loMHCII+ SCMs bear numerous genes involved in antigen processing and presentation, including costimulatory CD226 (ref. 16). Mouse CD226+ SCMs coclustered with human CD1C+ cells (Fig. 2b and Extended Data Fig. 2c), previously described in the human peritoneal cavity without an identified mouse counterpart24. CD1C+ human cells were relatively abundant and divisible into two clusters, 1 CD1C+CD14+ cell cluster that was enriched in FCGR1A encoding CD64, FCGR3A (encoding CD16) and MAFB, genes that traditionally associate with macrophages1,25,26, along with FABP5, CSF1R (encoding CD115), CD63 and CD163; and a second CD1C+CD14− population expressing genes more traditionally expressed by DCs, including LTB, FCER1A, CD37 and CD1E (Fig. 2e). GSEA comparison of the mouse SCM signature16 with human CD1C+ cells underscored strong enrichment between genes expressed in mouse SCM and human CD1C+CD14+ DCs, but not CD1C+CD14− DCs (Fig. 2f). Instead, the CD1c+CD14− DCs carried bona fide cDC2 features26 (Fig. 2e,f). Thus, our data identify human peritoneal CD1C+CD14+ cells that simultaneously express genes canonical for both macrophages and DCs, and these CD1C+CD14+ human cells emerge as counterparts to mouse SCMs established to have both macrophage and DC features.
Children have abundant peritoneal DC2 but not GATA6 + macrophages
To test whether peritoneal washes from children were more likely to be enriched in embryonic macrophages such that GATA6+ SELP+ macrophages might be more abundant, we extended the scRNA-seq done on the seven adults above by adding scRNA-seq analysis on sorted CD45+ cells from nine children (four females and five males), aged 14 months to 17 years (median age, 9 years) (Supplementary Table 1 and Extended Data Fig. 4a). These children underwent surgery for inguinal hernia repair, splenectomy, cholecystectomy or gastrostomy tube placement (Supplementary Table 1). Adult and pediatric samples aligned together into 25 UMAP clusters (Fig. 3a). Each pediatric or adult participant had cells that contributed to each of the 25 clusters in the combined dataset (Extended Data Fig. 4b). Genes of interest, including TIMD4, LYVE1, MRC1, MARCO and CCR2, were similarly distributed in pediatric and adult cohorts (Fig. 3a,b). Analysis of genes that defined different clusters revealed a subset of macrophages previously classified in humans as ‘TLF macrophages’27, a name arising from co-expression of TIMD4, LYVE1 and FOLR2 (Extended Data Fig. 5a and Supplementary Table 6). The proportions of key myeloid populations between children and adults (Fig. 3c,d) indicated that, while GATA6 and SELP mRNAs co-enriched in the same clusters (Fig. 3b), cells bearing these mRNAs were even less abundant in children than adults (Fig. 3c,d) (<1% of peritoneal mononuclear phagocytes in seven of the nine samples and 2–3% in the other two samples; Fig. 3c,d). LYVE1+MRC1+ macrophages were more abundant than GATA6+ macrophages, but, nonetheless, only 5% of pediatric mononuclear phagocytes (Fig. 3c,d). As with analysis of adults only, GLUL+ and CCR2+ macrophages were 10–20%, respectively. The frequency of various macrophage or DC clusters did not differ on the basis of sex (Extended Data Fig. 5b). While the frequency of human CD1C+CD14+ SCM-like human cells was similar between children and adults (Fig. 3c), human CD1C+CD14− DC2s were double the frequency in children over adults, representing >25% of total peritoneal mononuclear phagocytes in children, on average, rivaling CCR2+ macrophages as the most abundant peritoneal mononuclear phagocyte (Fig. 3c). The high abundance of CD1C+ cells in children increased the robustness of the GSEA match between human CD1C+CD14+ cells and mouse SCM (Fig. 3e), but the lack of a significant association between CD1C+CD14− DCs and mouse SCM in GSEA remained (Fig. 3e). In summary, GATA6+ macrophages were not more abundant in children. Instead, children bore a large proportion of CD1C+CD14+ cells and, especially, CD1C+CD14− DCs that outnumbered most peritoneal macrophage populations.
a, UMAP projection of live CD45+ cells from adult (n = 7) and pediatric (n = 9) peritoneal wash samples (left), or immune cell clustering in adult (middle) or pediatric (right) peritoneal wash samples. Cluster names assigned on the basis of marker genes. Abbreviations include mono-CM for monocyte-like cavity macrophages and ACT for activated T cells. b, Feature plots showing the expression of marker genes in adult and pediatric human peritoneal cavity macrophages as in a. Color scale represents the normalized gene expression. c, The frequency of each cluster, as defined in a, among total MNPs in each human adult (n = 7) and pediatric (n = 9) sample. GATA6+ cells are a subcluster and not depicted here. Bars depict means with error bars representing standard deviation. Multiple two-tailed t-tests followed by FDR correction; *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant. Both the exact P values and FDR-corrected q values are reported in Source Data Fig. 3. d, The percentage of GATA6+ cells as determined from the UMAP plots between adult (n = 7) and pediatric (n = 9) samples. Bars depict means with error bars representing standard deviation. Two-sided Student’s t-test was applied and P = 0.08. e, GSEA analysis evaluating enrichment of a published SCM gene signature16 in CD1C+CD14+ or CD1C+CD14− human cells. Ticks below the line correspond to gene ranks. Statistical analysis was performed using a Kolmogorov–Smirnov test. Multiple testing correction was conducted automatically using the BH-FDR method. Normalized enrichment scores (NESs) and FDR q values are shown for each cluster.
Flow cytometry identifies few CD62P+ but many CD1c+ mononuclear phagocytes
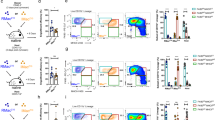
Next, we used flow cytometry gating on human and mouse peritoneal cells (Extended Data Fig. 6a,b) to validate and extend our findings using scRNA-seq. First, we analyzed peritoneal washes from seven adults (six females, one male), age range 24–55 years, and six children (one female, five males), age range 14 months to 14 years (Supplementary Table 1). Unsupervised analyses of mononuclear phagocytes in the combined samples after pre-gating on CD3−CD56−CD19− cells and gating out rare CD16+ high side-scatter neutrophils (Fig. 4a and Extended Data Fig. 6a) yielded results that resembled manual gating after staining for CD206, LYVE1, CD62P, TIMD4 and CCR2 (Extended Data Fig. 7a). Distinct distributions of the markers led to identification of seven different subpopulations (Fig. 4b–d), including the DC or DC-like subsets XCR1+ cDC1, CD123+ pDC and CD1c+CD14− cDC2, and CD1c+CD14+ cells (Fig. 4b). The major peritoneal cavity macrophage populations segregated as CCR2+ or CCR2− (Fig. 4a). CD206+ cells were also LYVE1+, and included CCR2− cells (Fig. 4a,b). A small subset of the CD206+LYVE1+ macrophages expressed CD62P and TIMD4 (Fig. 4a,b). A population of CD206+LYVE1+CD62P− cells seemed to correspond to the MRC1hi macrophage cluster identified using scRNA-seq (Fig. 4b,c), while CD62P+ macrophages were the closest equivalents to the scRNA-seq GATA6+ cavity macrophages (Extended Data Fig. 6a).
a, Unsupervised clustering based on multicolor flow cytometry of all MNPs from human adult (n = 7) and pediatric (n = 6) samples, projected on a UMAP space after cells from each patient were randomly downsampled to 10,000 events to generate UMAP dimensional reduction and concatenated, and UMAP was generated after negative selection of gating out dead cells, CD45− cells, CD3+ T cells, CD56+ NK cells, CD19+ B cells and CD16+ SSChi neutrophils. Clusters are identified on the basis of their unique surface markers. b, UMAP plots of the negatively selected MNPs showing the expression of CD14, CD11b, CD11c, CD64, CCR2, HLA-DR, LYVE1, CD206, TIMD4, CD62P, CLEC9A, CD163, CD1C and CD226. c, Frequency of CCR2+CD14+ CM, CD206+LYVE1+CCR2−CD62P− CM, CD62P+CCR2− CM, CD1c+CD14+CD64+ cells, CD1c+CD14−CD64− cells and cDC1 MNP subsets in adult (n = 7) and pediatric (n = 6) samples. Bars depict means with error bars representing standard deviation. Multiple two-tailed t-tests followed by FDR correction were applied; *P < 0.05, **P < 0.01, ****P < 0.0001, n.s. not significant. Both the exact P values and FDR-corrected q values are reported in Source Data Fig. 4. d, Frequency of ICAM2−CD11b+CD115+CD226+ SCMs, ICAM2−CD115−CD11c+MHC-II+XCR1+ cDC1, ICAM2−CD115−CD11c+MHC-II+Sirpα+ cDC2 and ICAM2−CD115−CD11cintMHC-IIintSirpα+Ly6C+ pDC among mouse MNPs (n = 12). Bars depict means with error bars representing standard error. A one-way ANOVA and post-hoc comparisons using Tukey’s honestly significant difference were conducted. ***P < 0.001 and ****P < 0.0001. e, Expression of TIMD4, LYVE1, CD163, CD14 and CD226 across human MNP subsets as in Extended Data Fig. 7a.
In agreement with the scRNA-seq data, the frequency of CD62P+ macrophages was very low in all pediatric (<0.4%) or adult (<2%) samples examined (Fig. 4c), whereas the CD206+ LYVE1+ macrophages were five to ten times more abundant (approximately 8% in adults, 3% in children), albeit less frequent than CCR2+ macrophages (nearly 40% in adults and 25% in children, Fig. 4c). CD1c+ cells were separated into CD1c+CD14+ SCM-like and CD1c+CD14− DCs, respectively (Fig. 4d). The frequency of CD1c+CD14− cDC2s was high, especially in pediatric samples, where they represented approximately 30% of mononuclear phagocytes in peritoneal fluid, while in adults they represented about 15% (Fig. 4c). In mice, the frequency of CD115+CD11b+ICAM2−MHCII+CD226+ SCMs (about 4%, Fig. 4d), CD115−CD11c+SIRPα+CD226+ cDC2 (about 1%, Fig. 4d), CD115−CD11c+XCR1+ cDC1 (<0.5%) and CD115−CD11b−CD11c+MHCIIint pDCs (<1%; gated as in Extended Data Fig. 6b) were lower compared to each population in humans (Fig. 4c,d).
Expression of CD206 was high in all CD1c+ cells, whether CD14+ or CD14− (Fig. 4b,e) but low in cDC1s (Fig. 4b,e). In each human sample, CD62P−CCR2− and CD62P+CCR2− macrophage subpopulations expressed CD206 and CD163 strongly, but were heterogeneous for LYVE1 and TIMD4 (Fig. 4e and Extended Data Fig. 7b). Within this heterogeneity, expression of LYVE1 and TIMD4 was always equal to or greater in CD62P+ macrophages than in LYVE1+CD206+CD62P− peritoneal macrophages in the same individual (Fig. 4e and Extended Data Fig. 7b), consistent with the RNA velocity data, suggesting that LYVE1+CD206+CD62P− macrophages were less mature, or have a shorter residence time, than CD62P+ macrophages. Overall, flow cytometry data indicated that human peritoneal lavage contained cDC1, cDC2, pDCs and DC2-like SCM, alongside substantial numbers of CCR2+ macrophages and fewer, but significant, numbers of CD206+LYVE1+CD62P− macrophages, which seemed to be less mature versions of rare GATA6+CD62P+ peritoneal human macrophages.
Mouse peritoneal macrophage profile is unimpacted by environment
The human subjects enrolled in our study were selected using criteria that deemed peritoneal inflammation unlikely. However, participants had specific conditions, including obesity, which may or may not affect the peritoneal cavity. While robust inflammation depletes GATA6+ LCMs from the peritoneum in mice3, we tested whether CD206+LYVE1+ macrophages (MRC1hi convMacs) in humans might accumulate during inflammation. To do so, we capitalized on the fact that Gata6 expression in mouse macrophages correlates with high expression of F4/80 (F4/80hi), while Gata6− macrophages, which are CD206+LYVE1+ and in transition to GATA6+ macrophages21,28, are F4/80int. CD206+ and/or LYVE1+ cells were scarce at steady-state in C57BL/6 wild-type mice (Extended Data Fig. 8a). In CD115ERCreGata6fl/fl mice, macrophages that were F4/80intICAM2+LYVE1+CD206+ emerged between 8 and 16 days after tamoxifen-induced deletion of Gata6 (Fig. 5a–c and Extended Data Fig. 8b), confirming that deletion of Gata6 promotes accumulation of F4/80intCD206+LYVE1+ macrophages. Intraperitoneal injection of zymosan resulted in fewer F4/80intICAM2+LYVE1+CD206+ cells in Gata6 cKO mice (Fig. 5d), indicating that inflammation induced loss, rather than accumulation of LYVE1+CD206+ macrophages and suggesting that inflammation may not account for the accumulation of LYVE1+CD206+ macrophages.
a, Confocal microscopy examining Gata6 expression in peritoneal cells from Csf1rERCre+Gata6fl/fl and Gata6fl/fl mice 16 days post-tamoxifen administration. Scale bar, 20 μm. b, Kinetic of CD206+LYVE1+ LCM accumulation in Csf1rERCre+GATA6fl/fl (day 2, n = 2; day 8, n = 1 day 16, n = 3) and GATA6fl/fl (day 2, n = 2; day 8, n = 2; day 16, n = 3) mice at the indicated days post-tamoxifen administration orally as a percentage of large peritoneal macrophages (LPM). Data representative of two independent experiments. c, Quantification of LCMs in Csf1rERCre+Gata6fl/fl (n = 8) and Gata6fl/fl (n = 10) mice on day 16 post-tamoxifen administration. Biological samples were analyzed over two independent experiments. Two-sided Mann–Whitney (P = 0.0003), ***P < 0.001. d, Representative flow cytometry plots (left) and quantification (right) of F4/80+ICAM2+ LCMs in Lyz2Cre/+Gata6fl/fl (n = 6 mice per group) and Lyz2+/+Gata6fl/fl mice 3 h post-intraperitoneal injection of saline or 1 mg zymosan (saline, n = 3; zymosan, n = 4). Biological samples were analyzed in two independent experiments. Two-way ANOVA test, ***P < 0.001, ****P < 0.0001. e, Representative flow cytometry plots (top) and quantification (bottom) of total LCM (P = 0.9307) and LYVE1+CD206+ LCMs (P = 0.0043) in female C57BL/6 wild-type mice fed either chow diet (n = 6) or high-fat diet (n = 5) for 5 months to induce severe obesity. Mice were 8 weeks old at the onset of high-fat diet. Data representative of two similar experiments. Two-sided Mann–Whitney tests, **P < 0.01, n.s. not significant. f, Representative flow cytometry plots (left) and frequency (right) of F4/80hiCD206low LCMs and F4/80intCD206+ converting CM (conv-CM) in wild mice (n = 165). H-014 and H-019, identifier codes assigned to two wild mice with different macrophage profiles. Two-sided Mann–Whitney test; ****P < 0.0001. g, Correlation between proportions of converting CM, Trichuris burden and maturity index in wild mice using spearman’s rank-order correlation. In all bar graphs, each dot represents a single sample (n = 165 wild mice) and error band represents 95% confidence interval.
Because some human samples were collected from obese patients, we next investigated whether obesity promoted a shift toward the MRC1hi convMacs or the CD206+LYVE1+ cell phenotype in peritoneal macrophages. C57BL/6 mice fed a high-fat diet for 5 months weighed more than control-diet mice (Extended Data Fig. 8c). However, obese mice retained a high frequency of CD206−LYVE1− LCMs, with fewer CD206+LYVE1+ macrophages compared to lean mice (Fig. 5e and Extended Data Fig. 8d), suggesting that obesity did not shift the peritoneal macrophage phenotype.
Finally, we tested whether an unrestricted natural environment would induce a F4/80intCD206+ phenotype in peritoneal macrophages in mice. Accordingly, we profiled peritoneal macrophages from wild Mus musculus domesticus mice captured on the Isle of May near Scotland, UK29. Analysis of 165 wild-caught mice accounted for real-world pathogens and incorporated social behavior, dietary variations, ambient temperature and seasonal changes that might affect immunophysiology. Using F4/80hi and F4/80int as a surrogate for GATA6 status10,28, as described above, and a gating strategy that accounted for potentially confounding cells like F4/80+ eosinophils (Extended Data Fig. 8e), wild mice showed heterogeneity in peritoneal macrophages (compare mouse H-014 to H-019, Fig. 5f). However, most wild-caught mice had F4/80hiCD206lo LCMs, with few CD206+ macrophages, resembling SPF mice (Fig. 5f). Increased maturity in mice (age and growth) and increased nematode parasite (Trichuris muris) burden paralleled increased F4/80intCD206+ macrophages (Fig. 5g). Nonetheless, only 10/165 wild mice had more F4/80intCD206+ macrophages than F4/80hi macrophages in the same mouse (Fig. 5f). By comparison, all humans had more MRC1hiLYVE1+ peritoneal macrophages (usually about fivefold more) than GATA6+SELP+ (Fig. 3d,e) or surface protein CD62P+ macrophages (Fig. 4b). Thus, we did not identify mouse life-environmental conditions that rendered mice toward a higher frequency of peritoneal CD206+LYVE1+ macrophages that was typical in humans.
Discussion
Here we profiled human peritoneal washes under conditions that avoided frank peritonitis or malignancy. We found that few human peritoneal macrophages expressed GATA6, a marker that defines mouse LCMs1,8,10,11. More common were macrophages characterized by co-expression of CD206 and LYVE1, matching an analogous state in mouse LCMs that precedes expression of GATA6. In addition, other human macrophages, including CCR2+ or GLUL+ macrophages without a clear mouse counterpart, were uncovered. Furthermore, our profiling linked human CD1c+CD14+CD64+ DC2-like cells with mouse F4/80loMHCII+ SCM and underscored the marked abundance of CD1c+CD14−CD64− DC2s in the human peritoneum.
Human peritoneal macrophages were previously characterized in persons with cirrhotic liver disease30, peritoneal dialysis31, or laparoscopic surgery for benign gynecologic indications24. In common with our findings, one study observed that human peritoneal macrophages were more similar to Gata6 cKO macrophages than wild-type macrophages30, but the breadth and depth of our analysis was more extensive. Indeed, our findings now allow us to consider one of the other earlier studies with new insight. By tracking patients undergoing peritoneal dialysis serially for longer than 1 year and studying their peritoneal macrophages, CD206 and CD1c mononuclear phagocytes were recognized as prominent subpopulations respectively in humans, but not studied in relation to mice24. The authors concluded that the most mature macrophages were those expressing CD206 (ref. 24), in line with our study, and that CCR2+ cells were an earlier stage of differentiation linked to monocyte infiltration, especially in cases of peritonitis24. Although GATA6 or SELP expression were not analyzed, the study fits with our findings. Putative longevity markers like TIMD4 (refs. 32,33,34,35) were indeed more common in CD206+ macrophages in our datasets than in CCR2+CD206− macrophages, and a small fraction of CD62P+ macrophages, which coincides with GATA6 expression, in the same individuals had an even higher frequency of TIMD4+ cells. Thus, it appears that differentiation of some human peritoneal macrophages progresses along a similar program of differentiation as mouse LCMs. However, in humans, many macrophages seem to stall at the stage preceding GATA6 expression or die shortly thereafter.
A caveat is that we could not sample the peritoneal cavity of truly healthy individuals, as all were undergoing surgery for clinical indications. We avoided conditions involving diagnosed peritonitis, but the health status of individuals studied may impact results. We attempted to address the inclusion of obese patients by studying obese mice fed a high-fat diet. However, obesity and high-fat diet feeding in mice did not cause a shift toward the transitional CD206+LYVE1+ macrophage. We also profiled mice living in the wild to capture a fuller range of the variables that can affect peritoneal cell subsets. While some wild mice shifted to a CD206+ LCM phenotype (10/165 > 50% CD206+ 20/165 > 25% CD206+) in a manner that correlated with age and infection with the Trichuris parasite, most wild mice had a macrophage profile resembling specific pathogen-free mice. Our analysis of pediatric samples did not show an enrichment in GATA6+ LCM compared to adults, ruling out the possibility that aging accounts for the results.
Our data linked human CD1c+ mononuclear phagocytes that bear monocytic markers CD14 and CD64 with mouse F4/80lo MHCII+ SCM. Mouse SCM simultaneously express features of macrophages and DCs1,20,22,23,34. Future experiments will test whether these CD1c+CD14+CD64+ cells possess DCs functions such as homing to lymph nodes or presenting antigen26. The higher frequency in humans of CD1c+CD14+CD64+ cells and bona fide CD1c+CD14−CD64− cDC2, which are especially elevated in children, highlight generalized peritoneal orientation toward TH2 cell immunity. With few GATA6+ macrophages, which in mice support robust TH2 immunity and host defense against parasites9,21, expansion of human TH2-oriented cDC2 and CD1c+CD14+CD64+ cells may fill in to effect robust TH2-oriented host defense.
In summary, we provide a robust resource for exploring the human peritoneal immune compartment. Our analyses focused on the macrophage and DC populations. In-depth exploration of the lymphocyte and natural killer (NK) cell compartments in this dataset is still needed. These studies are foundational for translational research in the peritoneal cavity.
Methods
Mice
All C57BL/6J wild-type mice were purchased from The Jackson Laboratory. Gata6fl/fl mice (Gata6tm2.1Sad/J, Jax 008196) were backcrossed to C57BL6/J background and crossed to either Lyz2Cre (B6.129P2–Lyz2tm1(cre)Ifo/J, Jax 004781) and R26LSL–YFP (B6.129 × 1-Gt(ROSA)26Sortm1(EYFP)Cos/J, Jax 006148), or to Csf1rERCre (FVB–Tg(Csf1r–cre/Esr1*)1Jwp/J, Jax 019098) and R26LSL–tdTomato (B6.Cg–Gt(ROSA)26Sortm9(CAG–tdTomato)Hze/J, Jax 007909). In all experiments, cre+ mice were compared to cre− littermate controls. Both male and female mice were used in each condition of each experiment except for the high-fat diet experiment in which female mice were used. Mice were used between the ages of 12 and 24 weeks old, and all experiments were performed on age-matched cohorts. Mice were kept in specific pathogen-free conditions maintained by the Washington University School of Medicine Division of Comparative Medicine. Facilities were maintained at an ambient temperature of 23–24 °C with a 12/12 h light/dark cycle. Mice had access to food and water ad libitum. To induce recombinase activity, Csf1rERCre × Gata6fl/fl × R26TdTomato mice and Gata6fl/fl × R26TdTomato littermate controls received 2 mg tamoxifen (100 μl of a 20 mg ml−1 stock solution, diluted in corn oil) by oral gavage every other day for a total of three doses. Mice were analyzed at different time points after the last dose, as indicated on the corresponding figure. Diet-induced obesity was induced by feeding the mice with a 60 kcal% high-fat diet (Research Diets, D12492) for 5 months to create the model, while controls were placed on a regular equivalent chow diet. C57BL/6J wild-type peritoneal lavage scRNA-seq data were generated at Dartmouth College. All experiments and procedures were conducted in accordance with procedures and protocols approved by the Washington University in St. Louis or the Dartmouth College Institutional Animal Care and Use Committee.
Wild-caught house mice
The capture and study of wild house mice from the Isle of May off the coast of Scotland have been described, and data were reanalyzed from this previous study to focus on peritoneal macrophages30. Maturity index was defined as a relative age estimate base on the first principal component of a PCA including body length, tail length, weight and dry eye lens weight. This work on wild mice was approved by the University of Nottingham Animal Welfare and Ethical Review Body and complies with the UK’s Animals (Scientific Procedures) Act of 1986.
Human subjects
Patients were recruited from the Barnes-Jewish Hospital and the St. Louis Children’s Hospital in St. Louis, USA. All human studies were performed in accordance with ethical regulation and preapproved by the Washington University Human Research Protection Office and the Washington University Institutional Review Board (#201903051). Institutional review board-approved written, informed consent was obtained from patients above the age of 18 years, or from both the parents/guardians of participants under the age of 18 years. This was not a prospective study. Patients recruited involved those receiving laparoscopic surgeries including Roux-en-Y gastric bypass surgery, sleeve gastrectomy, splenectomy, gastrostomy tube placement, cholecystectomy or inguinal hernia repair without any other clinical indication of peritoneal pathological conditions. Consideration was made by the surgeon as to whether to approach a patient for consent was entirely based on the clinical indicators that would suggest potential suitability for the study and the expert judgment that the study would not compromise patient safety. Participants did not receive compensation for agreeing to contribute sample to the study. This study collected data from 28 participants (demographics in Supplementary Table 1) with ages ranging from 3 months to 60 years. Peritoneal wash was collected at the beginning of the procedure before it was affected by the surgical process. Briefly, about 1 liter (for adults) or 200 ml (for children) of sterile saline was flushed into the human peritoneal cavity, and the abdomen was massaged for 2 min for a thorough wash before the saline-containing peritoneal cells were vacuumed out. This protocol delivered approximately 20 ml kg−1 of normal saline up to 1 liter, per estimate. The cells were placed on ice immediately for further processing and preservation.
Sample processing and preservation
Right after the collection, cells were counted and then centrifuged at 350g for 10 min and resuspended in cell freezing and preservation media that contained 90% fetal bovine serum + 10% dimethyl sulfoxide. Cells were then preserved in liquid nitrogen at −160 °C for future experiments, including flow cytometry, multiplex imaging and both single-cell RNA and single-cell T cell receptor (TCR) sequencing.
Single-cell RNA profiling
Immediately following sorting of human cells, cells were placed on ice and centrifuged at 350g for 5 min. Supernatant was carefully removed, and the cells were resuspended in phosphate-buffered saline (PBS) + 0.04% bovine serum albumin (BSA) at a concentration of 1,000 cells µl−1. The cell concentration was then confirmed by the cell counter. The cell concentration was used to calculate the volume of single-cell suspension needed in the reverse-transcription master mix, aiming to achieve approximately 5,000–10,000 cells for each sample. The RNA sequencing (RNA-seq) libraries were generated using a Chromium Next GEM Single Cell 5′ Kit v2 (10x Genomics, product number 1000263), following the manufacturer’s protocol. Briefly, following capture, emulsions containing droplet-encapsulated single cells were reverse-transcribed and barcoded, emulsions broken and barcoded complementary DNA amplified by polymerase chain reaction according to the 10x Genomics V(D)J enrichment protocol (10x Genomics, product number 1000252). Amplified cDNA libraries were prepared following the standard 10x procedure to generate libraries for Illumina sequencing. Samples were uniquely barcoded, pooled and sequenced across multiple Illumina NovaSeq600 High Output runs to generate enough reads per cell for the gene expression library. Raw sequencing data were processed through the Cell Ranger v. cellranger-3.1.0, 6.1.1 or 7.0.1 pipeline (10x Genomics) using human reference genome GRCh38 to generate gene expression matrices for the single-cell 5′ RNA-seq data from each single cell. For mouse scRNA-seq, three different preparations of mouse peritoneal lavage from C57BL/6J individuals were checked for viability and then submitted for 10x Genomics scRNAseq profiling. Single cells were processed using the Chromium Next GEM Single Cell 3′ Platform (10x Genomics). Approximately 25,000 cells were loaded on each channel with an average recovery rate of 20,000 cells. Libraries were sequenced on NextSeq 500/550 (Illumina) with an average sequencing depth of 50,000 reads per cell.
scRNA-seq data processing
Human peritoneal wash scRNA-seq datasets were generated by coauthors at Washington University, and the mouse peritoneal and bronchoalveolar lavage scRNA-seq datasets were generated by coauthors at Dartmouth University. The unique molecular identifier (UMI) counts-based gene expression matrix was processed using the R package Seurat (v.4.0.0). To exclude low-quality cells, those with fewer than 200 genes detected, or more than 10% mitochondrial genes, were filtered from the dataset. To remove potential multiplets, cells with a total number of UMI counts higher than 50,000 or a number of genes detected higher than 6,000 were also removed. In analysis integrating the mouse and human gene expression matrixes, the human equivalent of the mouse genes were first retrieved from the Ensembl database using the getLDS function from the biomaRt interface. Optional: in analysis where the mouse sample had way more cells than the human samples, the mouse samples were first randomly downsampled 40–50% to match a similar number of cells as the human samples. The number of UMIs of all cells in each sample was then log-normalized, and the highly variable features were identified. A method to match (or ‘align’) shared cell populations across datasets was then applied to integrate the datasets from multiple experiments. Briefly, cross-dataset pairs of cells that are in a matched biological state (‘anchors’) were identified utilizing CCA based on the highly variable genes across different datasets. This method has been shown to both robustly correct for technical differences between datasets (that is batch-effect correction), and to perform comparative scRNA-seq analysis of across experimental conditions. PCA was then performed over the integrated dataset and clusters were identified using k-nearest-neighbor graph-based clustering, implemented in the R functions FindNeighbors and FindClusters, on the basis of the first 15 PCs. The same PCs were used to generate the UMAP projections for visualization.
GSEA
We first defined comprehensive gene signatures for Gata6 KO and Gata6 wild-type peritoneal macrophages (Supplementary Table 4) using three microarray datasets (GSE56711, GSE47049 and GSE37448) of three independent studies, including peritoneal macrophages8,11,36. This approach enabled identification of a consensus gene list that differentiated GATA6 KO macrophages from wild type. Human orthologs of mouse genes were used. Quantile normalization37 was applied to re-scale the expression profiles at the probe level across all four datasets, and ComBat (from the sva package version 3.38.0)38 was utilized to integrate expression data into a single meta-dataset. The gene signatures for the GATA6 KO macrophages and the GATA6 wild-type macrophages were then identified by a two-way unpaired t-test, corrected for multiple comparisons by the Benjamini–Hochberg procedure. Adjusted P values less than 0.05 and average log2-fold gene expression change higher than 1 were used as the cutoffs to generate the gene signature. Both signatures were added to the ‘c2.all.v6.2.symbols’ gene sets collection from the MSigDB database (modified C2 dataset) for GSEA analysis39,40.
We then performed preranked GSEA using the GSEA_4.0.3 software on the clusters of interest from the scRNA-seq analysis. Differential expressed genes of each cluster of interest (scRNA-seq) were identified using the FindAllMarkers function in Seurat with parameters of log2 fold change (log2FC) of 0.001. The log2FC value was used as the ranking metric for preranked GSEA.
GSVA analysis for identifying the converting macrophages in human peritoneal cavity
scRNA-seq data from Finlay et al. (2023), which defined converting macrophages were used to identify a converting macrophage gene signature. Specifically, we applied FindAllMarkers function to the publicly available dataset-GSE189031_seurat.combined.Rdata, with log2FC higher than 0.2 to extract the marker genes from the ‘converting SCM’ cluster identified in the original manuscript21. Following the identification of this specific gene signature, we applied GSVA (version 1.38.2) to calculate a score for the converting macrophage gene signature across all single cells in our sequenced dataset. The scores were then scaled, and relative expression was visualized using feature plots. To pinpoint the cluster with the highest converting macrophage score, one-way analysis of variance (ANOVA) tests and multiple t-test comparisons were applied. This approach allowed us to rigorously assess the presence and behavior of converting macrophages in our experimental context, providing valuable insights into their role and function.
RNA velocity analysis
RNA velocity is widely used to infer the directionality and future state of individual cells by distinguishing unspliced and spliced mRNAs from scRNA-seq data. The spliced and unspliced UMIs for each gene in each cell were counted using the Python (version 3.7.7) package velocyto (version 0.17.17); subsequent analyses were performed by scanpy (version 1.9.1) and scvelo (version 0.2.4)41. Specifically, the count matrices of the top 2000 variable genes underwent normalization based on library size. Next, the top 30 principal components obtained from PCA conducted on the logarithmized spliced matrix were used to build a k-nearest-neighbor graph (where k was set to 30). For each individual cell, normalized spliced/unspliced counts were analyzed using the moments (mean and uncentered variances) of the 30 nearest neighbors through the scv.pp.moments function. By implementing the scv.tl.velocity function with mode set to ‘dynamical,’ RNA velocity estimation was made possible using the computed moments. The resulting velocities were then used to construct a velocity graph that represented transition probabilities, with the function scvelo.tl.velocitygraph. Finally, the velocity graph was used to embed RNA velocities in the UMAP map. This was accomplished using the scv.pl.velocity_embedding_grid function.
Flow cytometry analysis of mouse peritoneal cells
Peritoneal cells were retrieved by flushing the peritoneal cavity with 5 ml PBS containing 2 mM ethylenediaminetetraacetic acid (EDTA). For acute peritonitis, mice were injected intraperitoneally with 1 mg zymosan (Sigma cat. no. Z4250) 3 h before analysis. Cells were then centrifuged and resuspended in flow buffer (PBS containing 0.1% BSA and 2 mM EDTA). For quantification, cells were stained with acridine orange and counted on a Nexcellom Cellometer Auto X4 cell counter. For flow cytometry, cells were washed in PBS, stained with ZombieNIR, washed again, and resuspended with the antibody mix diluted in flow buffer with BD Horizon Brilliant Stain Buffer (cat. no. 563794) for 30 min at 4 °C. The panel consisted of ICAM2-AF647 (3C4), CD45-BUV805 (30-F11), F4/80-SparkNIR685 (BM8), CD11b-BUV737 (M1/70), MHC-II-BUV496 (M5/114.15.2), CD226-PE/Cy.7 (TX42.1), Lyve1-AF488 (ALY7), XCR1-BV421 (ZET), Sirpα-PE/Dazzle594 (P84), CD115-PE (AFS98), Ly6G-BV650 (1A8), Ly6C-BV570 (HK1.4), CD11c-BUV395 (N418), CD206-AF700 (C068C2). All antibodies were used with a 1:300 dilution. Analytical flow cytometry and analysis was run using Cytek Aurora, as described below.
Flow cytometry analysis of human peritoneal cells
Frozen human peritoneal wash samples were thawed in 37 °C water. Cells were incubated with FcR Blocking Reagent (Miltenyi, cat. no. 130-059-901) diluted 1/20 in flow buffer (PBS containing 1% BSA and 2 mM EDTA) for 15 min, washed twice with PBS and incubated 15 min with Zombie NIR Fixable Viability dye diluted 1/500 (BioLegend, cat. no. 423106). Cells were then washed twice with flow buffer and incubated 30 min on ice with the antibody mix diluted in flow buffer with BD Horizon Brilliant Stain Buffer (cat. no. 563794). The panel consisted of CD1c–PE/Dazzle594 (L161), CD14–BUV615 (M5E2), CD62L–BV785 (DREG-56), CD56–BV650 (5.1H11), CD45–APC/Fire810 (HI30), HLA–DR–BV750 (L243), XCR1–PE/Fire-810 (S15046E), CCR2–BB700 (LS132.1D9), CD11c–BUV737 (B-Ly6), CD163–BUV805 (Mac2.158), TIM4–PE (9F4), CD206–BUV563 (19.2), CD123–BUV661 (9F5), CD115–BV711 (G043H7), CD3–BV605 (OKT3), CD62P–PECy7 (AK4), CD226–BUV395 (DX11), CD11b–PE/Cy5, LYVE1–Dylight488 (PA5-22783), CD16–BV570 (3G8), CD64–AF700 (10.1) and CD19–PE/Fire-700 (HIB19). All antibodies were used with a 1:100 dilution except the LYVE1–Dylight488 antibody, which was diluted 1:50. Cells were then washed twice in flow buffer, fixed with 1% paraformaldehyde for 15 min, washed twice and acquired on a Cytek Aurora (5 laser configuration). All analysis was performed using FlowJo. Unsupervised analysis was conducted after gating out dead cells, doublets, CD45−, CD3+ and CD56+ cells. The DownSample plugin was used to randomly select 5,000 events per sample and all samples were then concatenated. Dimensionality reduction was performed using UMAP, and FlowSom was then used for cell clustering to generate 12 meta-clusters. ClusterExplorer was used to generate protein expression plots.
For cell sorting, frozen human peritoneal wash samples were thawed in a 37 °C water bath. Single-cell suspensions were incubated in FcR Blocking Reagent (Miltenyi, cat. no. 130-059-901, 1:20) in flow buffer (PBS + 0.1% BSA and 2 mM EDTA) for 15 min. For scRNA-seq, samples were then stained with 1:100 dilutions of antibodies to CD45 (Alexa Fluor 700-conjugated anti-human CD45 mAb clone HI30). Samples were stained in the dark for 30 min on ice, washed twice with flow buffer and stained with 4′,6-diamidino-2-phenylindole (DAPI) for dead cells. CD45+DAPI− cells were sorted through a BD FACSAria II sorter. Sorting was terminated when the total number of sorted cells reached 50,000.
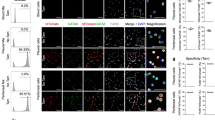
Immunofluorescence analysis of peritoneal cells
Peritoneal cells were seeded on Alcian blue-coated coverslips for 30 min in PBS. Cells were then fixed in 4% paraformaldehyde for 10 min, washed in PBS and stored in PBS until analysis. Cells were permeabilized using 0.1% Triton X-100 in PBS for 15 min and then washed in PBS containing 0.1% Tween-20. Cells were then saturated with rabbit serum for 2 h at room temperature and washed in PBS–Tween. Mouse samples were incubated with 1 μg ml−1 anti-ICAM2-AF647 (3C4, Invitrogen) and 1 μg ml−1 anti-GATA6-AF488 (D61E4, cellsignal) and human peritoneal samples were incubated with 4 μg ml−1 anti-CD14-AF488 (63D3, BioLegend), 5 μg ml−1 anti-CD62P-AF647 (AK4, BioLegend) and 4.93 μg ml−1 anti-GATA6 (D61E4, cellsignal) overnight at 4 °C. Cells were then washed twice in PBS–Tween, incubated 10 min in PBS–Tween containing 1 μg ml−1 DAPI, washed again twice in PBS–Tween and once in dH2O before being mounted using ProLong Glass Antifade Mountant (Invitrogen cat. no. P36984). Images were acquired using a Leica SP8 confocal microscope and processed using the Imaris 9 software.
Statistical analysis and reproducibility
All measurements were taken from distinct samples, and the number of subjects in each experiment or analysis was clearly indicated either in the text or in figure legends. Data distribution was assumed to be normal, but this was not formally tested. Significance was evaluated using the following statistical analyses: two-tailed Student’s t-test (with or without multiple comparison corrections), two-sided Mann–Whitney test, two-sided permutation test with multiple testing correction by the Benjamini–Hochberg procedure controlling false discovery rate method (BH-FDR), two-sided log-rank test, one-way ANOVA and two-way ANOVA. The calculation of differentially expressed genes between different clusters in the scRNA-seq data was performed using two-sided Wilcoxon rank-sum test, and P values were corrected for multiple comparisons with the Bonferroni method. Descriptive statistics and Spearman rank correlations were used to describe the correlations between the percentage of CMs with Trichuris total number or mouse maturity. These analyses were performed using R (v.4.0.3), Prism-GraphPad or the GSEA_4.0.3 software. P < 0.05 was considered statistically significant. No statistical method was used to predetermine sample sizes, but our experimental cohort size was based on technical manageability and our final sample sizes are similar to or greater than those reportedly used for RNA-seq in previous publications for human peritoneal lavage24,31 or for studies in mice1,30,41. Most samples were given a code name, and this was processed without reference to its cohort features until the end of the experiment. However, this single-blind method is a not a classic double-blind approach. In some experiments, blinding was not practical, as the phenotype (for example, obese mice) was evident whether blinded or not. Each human patient sample or data from a single mouse were considered an independent biological sample in the study. Data distribution was assumed to be normal, but this was not formally tested. Randomization of human samples was not relevant. Sequencing was using cryopreserved samples, with a few consecutive samples batched in order of surgery. For flow cytometry on human cells, cells were recovered from cryopreservation and run simultaneously after batch staining for final analysis after initial samples were used to develop the staining panels. In the selection of experimental cohorts of mice, randomization was not the dominant driver of the process. Most importantly, littermate controls were assigned appropriately to match mice that were genetically altered, so that controls were tested side by side with those bearing a different genotype or treatment. Experimental analysis was carried out so that for any given length of a protocol, all experimental cohorts were dealt with simultaneously. That is, we never processed one whole group first before tending to the next, but distributed the cohorts evenly throughout a procedure. No data points were excluded from the study. However, results of inconclusive or flawed preliminary experiments that ultimately helped us optimize staining panels, experimental conditions or experimental design for the best quality data were not included in the final set of data presented here.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
De-identified human scRNA-seq data that support the findings of this study are publicly available in the Gene Expression Omnibus (GEO) database under accession number: GSE228030. Publicly available browser tools to explore the data are found at: https://artyomovlab.wustl.edu/scn/?token=gjrandolph.fig1.adult_peritoneal_mnp; https://artyomovlab.wustl.edu/scn/?token=gjrandolph.fig2.adult_b6_peritoneal_mnp; https://artyomovlab.wustl.edu/scn/?token=gjrandolph.fig3.adult_pediatric_peritoneal_all_immune_cell. Mouse scRNA-seq data can be accessed under the code GSE225668. Human reference genome data are available at https://cf.10xgenomics.com/supp/cell-exp/refdata-gex-GRCh38-2020-A.tar.gz and mouse reference genome is available at https://cf.10xgenomics.com/supp/cell-exp/refdata-gex-mm10-2020-A.tar.gz. The previously published microarray datasets used to generate the Gata6 KO versus wild-type peritoneal macrophage gene signature for the GSEA analysis were accessible at Gene Expression Omnibus (GEO) under accession codes GSE56711, GSE47049 and GSE37448. The remaining gene sets used in the GSEA analysis are accessible at the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb). Source data are provided with this paper and can be found at https://doi.org/10.6084/m9.figshare.24319603. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
Customized code is available at https://github.com/TrmMelanoma/macrophages-and-dendritic-cells-between-mouse-and-human.
References
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Jenkins, S. J. et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288 (2011).
Zhang, N. et al. Expression of factor V by resident macrophages boosts host defense in the peritoneal cavity. J. Exp. Med. 216, 1291–1300 (2019).
Chow, A. et al. Tim-4+ cavity-resident macrophages impair anti-tumor CD8+ T cell immunity. Cancer Cell 39, 973–988 e979 (2021).
Jin, H. et al. Genetic fate-mapping reveals surface accumulation but not deep organ invasion of pleural and peritoneal cavity macrophages following injury. Nat. Commun. 12, 2863 (2021).
Vega-Perez, A. et al. Resident macrophage-dependent immune cell scaffolds drive anti-bacterial defense in the peritoneal cavity. Immunity 54, 2578–2594 e2575 (2021).
Zindel, J. et al. Primordial GATA6 macrophages function as extravascular platelets in sterile injury. Science 371, eabe0595 (2021).
Okabe, Y. & Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844 (2014).
Campbell, S. M. et al. Myeloid cell recruitment versus local proliferation differentiates susceptibility from resistance to filarial infection. eLife 7, e30947 (2018).
Gautier, E. L. et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J. Exp. Med. 211, 1525–1531 (2014).
Rosas, M. et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 344, 645–648 (2014).
Ghosn, E. E. et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl Acad. Sci. USA 107, 2568–2573 (2010).
Salm, L., Shim, R., Noskovicova, N. & Kubes, P. Gata6+ large peritoneal macrophages: an evolutionarily conserved sentinel and effector system for infection and injury. Trends Immunol. 44, 129–145 (2023).
Liu, M., Silva-Sanchez, A., Randall, T. D. & Meza-Perez, S. Specialized immune responses in the peritoneal cavity and omentum. J. Leukoc. Biol. 109, 717–729 (2021).
Epelman, S. et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104 (2014).
Kim, K. W. et al. MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes. J. Exp. Med. 213, 1951–1959 (2016).
Hashimoto, D. et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013).
Buechler, M. B. et al. A stromal niche defined by expression of the transcription factor WT1 mediates programming and homeostasis of cavity-resident macrophages. Immunity 51, 119–130 e115 (2019).
Deniset, J. F. et al. Gata6+ pericardial cavity macrophages relocate to the injured heart and prevent cardiac fibrosis. Immunity 51, 131–140 e135 (2019).
Bain, C. C. et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat. Commun. 7, ncomms11852 (2016).
Finlay, C. M. et al. T helper 2 cells control monocyte to tissue-resident macrophage differentiation during nematode infection of the pleural cavity. Immunity 56, 1064–1081 e1010 (2023).
Bain, C. C. et al. CD11c identifies microbiota and EGR2-dependent MHCII+ serous cavity macrophages with sexually dimorphic fate in mice. Eur. J. Immunol. 52, 1243–1257 (2022).
Goudot, C. et al. Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 47, 582–596 e586 (2017).
Liao, C. T. et al. Peritoneal macrophage heterogeneity is associated with different peritoneal dialysis outcomes. Kidney Int. 91, 1088–1103 (2017).
Miller, J. C. et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Dick, S. A. et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci. Immunol. 7, eabf7777 (2022).
Gundra, U. M. et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123, e110–e122 (2014).
Mair, I. et al. A lesson from the wild: the natural state of eosinophils is Ly6Ghi. Immunology 164, 766–776 (2021).
Irvine, K. M. et al. CRIg-expressing peritoneal macrophages are associated with disease severity in patients with cirrhosis and ascites. JCI Insight 1, e86914 (2016).
Ruiz-Alcaraz, A. J. et al. Characterization of human peritoneal monocyte/macrophage subsets in homeostasis: phenotype, GATA6, phagocytic/oxidative activities and cytokines expression. Sci. Rep. 8, 12794 (2018).
Scott, C. L. et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016).
Shaw, T. N. et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J. Exp. Med. 215, 1507–1518 (2018).
Louwe, P. A. et al. Recruited macrophages that colonize the post-inflammatory peritoneal niche convert into functionally divergent resident cells. Nat. Commun. 12, 1770 (2021).
Jenkins, S. J. & Allen, J. E. The expanding world of tissue-resident macrophages. Eur. J. Immunol. 51, 1882–1896 (2021).
Elpek, K. G. et al. The tumor microenvironment shapes lineage, transcriptional, and functional diversity of infiltrating myeloid cells. Cancer Immunol. Res. 2, 655–667 (2014).
Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Acknowledgements
We are very grateful to the patients and families who consented to this study. We thank the members of the Randolph laboratory for their suggestions and critical evaluation of the manuscript. We are indebted to M. Artyomov and M. Terekhova (Washington University, Department of Pathology) for facilitating user-friendly public access to the data. This work was principally funded by NIH grant R37AI049653 to G.J.R. and associated Primary Caregiver Award to E.J.O. J.H. was funded by NIH grant KOOCA264434. N.Z. was funded by NIH grant K99AI151198. R.L.M. is funded by NIH grant F30CA281124 and D.D.L. by NIH grant T32 HL007081. C.V.J. is funded by NIH grants R01 HL115334, R01 HL135001 and R35 HL155458. C.M.F. and J.E.A. were supported by MRC-UK (MR/V011235/1, MR/K01207X/2) and the Wellcome Trust (106898/A/15/Z). We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. We thank J. Huecker of the Division of Biostatistics for consultation on statistical analysis, a service funded through Washington University’s Institute for Clinical and Translational Sciences program (NIH grant UL1TR002345). Additional support included use of core services funded by the Digestive Diseases Research Core Center at Washington University # P30 DK052574. B.A.H. is currently affiliated with the Department of Surgical Oncology at the University of Texas MD Anderson Cancer Center in Houston, TX; E.J.O. is now affiliated with Children’s Hospital of Philadelphia; B.A.S. is now affiliated with University of Chicago. The work on the wild mouse population on the Isle of May, UK, was supported by a Joint Biotechnology and Biological Sciences Research Council (grant number BB/P018157/1) awarded to J. Bradley (University of Nottingham) and K.J.E. (University of Manchester). We thank Nature Scotland for permission to carry out work on the Isle of May; D. Steel (Nature Scotland), B. Outram (Nature Scotland) and M. Newell (Centre for Ecology and Hydrology) for support with fieldwork; other members of the full research team (J. Bradley, A. Lowe, A. Bennett, A. Muir and A. Wolfenden) as well as our fieldwork volunteers.
Author information
Authors and Affiliations
Contributions
E.C.E., S.R.E., B.A.H., G.J.R., E.J.O. and N.Z. developed and organized study infrastructure; J.H., E.J.O., B.A.H., S.R.E., F.M.D., B.A.S. and H.M.P. collected samples; B.A.H., J.H., A.G., E.C.E., I.M., X.L., N.Z., R.L.M. and D.D.L. conducted experiments and analyzed data; C.M.F. and J.E.A. advised analyses; M.M.C. and J.D.S. provided key resources; B.A.H., K.J.E., C.V.J., J.D.S. and G.J.R. provided supervision and obtained regulatory compliance and funding; J.H., A.G. and G.J.R. wrote the manuscript with editing input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Charlotte Scott and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Ioana Visan was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Human peritoneal macrophages are heterogenous and have limited population expression GATA6.
(a) Heatmap of differentially expressed genes (rows) of different clusters (columns). Heatmap colors indicate Z-transformed expression of genes in each row, with scale depicted in legend. Annotations (left) highlight representative genes with high differential gene expression within each cluster, relative to other clusters. Colors of gene names indicate corresponding clusters in Fig. 1a. (b) The proportion of cells from each patient contributed to each cluster. Color represents different human samples. (c) Confocal images showing the channels of DAPI and CD14, CD62P and GATA6, separately for the merged image in Fig. 1c. Biological samples were analyzed over two independent experiments. (d) Additional confocal images showing the expression GATA6 and CD62P in human adult peritoneal cells. Biological samples were analyzed over two independent experiments. (e) Violin plot showing the converting macrophage signature score (y-axis) in Fig. 1f for each cluster (x-axis) (n = 3977, 3537, 3504, 2169, 1895, 1571, 1551, 1518, 1001, 351, 347, 283,67,66,28,27 cells respectively). Red bars depict means with error bars representing standard deviation. A one-way ANOVA and post-hoc comparisons using Tukey′s HSD were conducted to compare the means of different groups, **** p < 0.0001. (f) The spliced ratio of GATA6 transcripts in GATA6+ cells versus GATA6- cells in 7 human adult samples. Bars depict means with error bars representing standard deviation. ****p < 0.0001, two-sided t-test. Exact p-value = 2.77028E-05 (g) Principal component analysis showing the distinct transcriptome profiles (microarray) between Gata6 KO and wild-type peritoneal macrophages integrated from three independent studies (GSE56711, GSE47049 and GSE37448).
Extended Data Fig. 2 Human and mouse peritoneal immune cell composition is different.
(a) UMAP projection of all CD45+ cells sequenced from the peritoneal cavity of 7 adults and 3 C57BL6 mice, forming 30 distinct clusters (colored as shown in the legend), with cluster names assigned based on inferred function. (b) UMAP plots showing the distinct immune cell composition of the mouse (right) and human (left) peritoneal cavity. Colors are coded as in a. (c) Feature plots demonstrating the expression of key immune cell markers, including markers for T and B lymphocytes and myeloid cells. Color scale represents the normalized gene expression. (d) Proportion of each cluster out of total immune cells for each sample, grouped by species (n = 7 human adult samples and n = 3 mouse samples). Bars depict means with error bars representing standard deviation. Multiple two-tailed t-tests followed by false discovery rate (FDR) correction; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. not significant. The exact p-values and FDR corrected q-values are reported as Source Data.
Extended Data Fig. 3 Further analysis of peritoneal MNPs between mouse and human.
(a) Heatmap of differentially expressed genes (rows) of different clusters (columns). Heatmap colors indicate Z-transformed expression of genes in each row, with scale depicted in legend. Annotations (left) highlight representative genes with high differential gene expression within each cluster, relative to other clusters. Colors of gene names indicate corresponding clusters in Fig. 2a. (b) Dot plot showing the average Z-transformed normalized expression of markers for large cavity-, converting- or monocyte like macrophages. The size of each dot indicates the fraction of cells expressing each gene; the color scale represents Z-transformed normalized expression. (c) The projection of cells from each cluster in Fig. 1 in the new UMAP space of Fig. 2. Colors correspond to Fig. 1a. (d) UMAP projection showing the different cell cycle phases of the peritoneal immune cell subsets in mouse or human separately. Colors representing the predicted classification of cell cycle phase based on the S and G2/M scores calculated by the CellCycleScoring function in the Seurat package. (e) The proportions of cells in each cell cycle phase of mouse and human (n = 7 human adult samples and n = 3 mouse samples). Bars depict means with error bars representing standard deviation. * represents p < 0.05, two-tailed t-test.
Extended Data Fig. 4 Summary of patient cohort and patient contributions to each cluster.
(a) Summary for the age, gender and operational procedure of patients we have collected and analyzed in this study. The table was organized by the different experimental approaches performed on the samples collected. (b) Pie charts depicting the proportion of cells from each patient to the total number of cells in each cluster (Fig. 3a). Each color represents one individual patient, with the total number of each cluster labeled at the bottom of each pie chart.
Extended Data Fig. 5 The frequency of various macrophage or DC clusters did not differ based on sex.
(a) Heatmap of differentially expressed genes (rows) of different clusters (columns). Heatmap colors indicate Z-transformed expression of genes in each row, with scale depicted in legend. Annotations (left) highlight representative genes with high differential gene expression within each cluster, relative to other clusters. Colors of gene names indicate corresponding clusters in Fig. 3a. (b) The percentage (x-axis) of each macrophage/DC cluster (y-axis) out of total MNPs in each patient (n = 7 human adult samples, n = 9 human pediatric samples). Black and white squares represent male and female patient, respectively. Bars depict means with error bars representing standard deviation. Multiple two-sided student′s t-test was applied and n.s. represents not significant.
Extended Data Fig. 6 Gating strategies for human and mouse macrophages and dendritic cells subpopulations.
(a) Gating strategy for the different human peritoneal macrophage and DC subsets in Fig. 4. (b) Gating strategy for the mouse counterparts of human peritoneal SCM, cDC1, cDC2 and pDC populations.
Extended Data Fig. 7 . Unsupervised clustering of human peritoneal macrophages based on flow cytometry validates the key macrophage populations.
Extended Data Figure 7 (a) Projection of manually gated CD14+ CCR2+ CM (blue), CD206+ LYVE1+ CCR2- CD62P- CM (orange), CD62P+ CM (red), CD1c+ CD64+ MNPs (green), CD1c+ CD64- MNPs (pink) and cDC1 (purple) over UMAP displayed in Fig. 4a. (b-c) Histograms representing TIMD4 and LYVE1 expression in CD14+ CCR2+ (blue), CD206+ CCR2- CD62P- (Orange) and CCR2- CD62P+ (red) CM across all adult (b, n = 7) or pediatric (c, n = 6) samples.
Extended Data Fig. 8 Gating strategies and flow cytometry plots for the studies in mice to assess impact of environmental triggers on macrophage phenotype.
(a) Gating strategy used for identification of mouse LCM populations based on LYVE1 and CD206 expression. (b) Representative plots of Fig. 5b showing expression of CD206 and LYVE1 by LCMs from control (Gata6fl/fl) and Csf1rERCre x Gata6fl/fl mice 16 days post-tamoxifen administration. (c) Body weight from female mice fed either normal chow diet (n = 6) or high-fat diet (n = 5) for 5 months to induce obesity. Data representative of two similar experiments. A two-sided Mann-Whitney test was used for statistical analysis (p = 0.0087). ** represents p < 0.01 and data are represented as mean value +/− SEM. (d) Isotype controls for CD206 and LYVE1, comparing to the expression of control or high-fat diet mice. (e) Gating strategy used for analysis of LCMs from wild-caught mice.
Supplementary information
Supplementary Table 1
Supplementary Table 1.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, J., Gallerand, A., Erlich, E.C. et al. Human serous cavity macrophages and dendritic cells possess counterparts in the mouse with a distinct distribution between species. Nat Immunol 25, 155–165 (2024). https://doi.org/10.1038/s41590-023-01688-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-023-01688-7
This article is cited by
-
Extracellular matrix dynamics in tumor immunoregulation: from tumor microenvironment to immunotherapy
Journal of Hematology & Oncology (2025)
-
Liver macrophages: development, dynamics, and functions
Cellular & Molecular Immunology (2025)
-
Peritoneal resident macrophages constitute an immunosuppressive environment in peritoneal metastasized colorectal cancer
Nature Communications (2025)
-
Biophysically interpretable inference of cell types from multimodal sequencing data
Nature Computational Science (2024)
-
Mouse and human macrophages and their roles in cardiovascular health and disease
Nature Cardiovascular Research (2024)