Abstract
The gut experiences daily fluctuations in microbes and nutrients aligned with circadian rhythms that regulate nutrient absorption and immune function. Group 3 innate lymphoid cells (ILC3s) support gut homeostasis through interleukin-22 (IL-22) but can convert into interferon-γ-producing ILC1s. How circadian proteins control this plasticity remains unclear. Here we showed that the circadian proteins REV-ERBα and REV-ERBβ maintain ILC3 identity. Their combined deletion promoted ILC3-to-ILC1 conversion, reduced energy metabolism and IL-22 production, increased interferon-γ production, and heightened susceptibility to Citrobacter rodentium infection. Single-cell multiomics and gene editing revealed that REV-ERBα/REV-ERBβ deficiency upregulated the transcription factor NFIL3, which repressed the expression of RORγt via a –2-kb cis-regulatory element in the Rorc gene, shifting cells toward a T-bet-driven state. Chromatin and metabolic analyses indicated that REV-ERBα/REV-ERBβ loss reprogrammed regulatory and metabolic circuits. Thus, REV-ERBα/REV-ERBβ safeguard gut integrity by regulating clock genes that control RORγt expression and preserve ILC3 identity and resistance to intestinal inflammation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
ChIP–seq data were previously published (GSE96724, GSE163341 and GSM1859213). ChIP–seq datasets for NFIL3 in T cells were provided by V. K. Kuchroo34. The original data, mapped to the mm9 genome, were converted to mm10 using the liftOver tool from UCSC utilities (https://genome.ucsc.edu/cgi-bin/hgLiftOver). RNA-seq, scRNA-seq and scATAC–seq data are deposited in the Gene Expression Omnibus under accession number GSE267792. Bacterial V4 16S rDNA amplicon sequencing data in raw format before processing and data analyses have been deposited at the NCBI SRA under accession number PRJNA1252108. Source data are provided with this paper.
Code availability
All software used in this study is published and cited either in the main text or Methods. No custom code was used for data processing or analysis.
References
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018).
Daussy, C. et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211, 563–577 (2014).
Yomogida, K. et al. Hobit confers tissue-dependent programs to type 1 innate lymphoid cells. Proc. Natl Acad. Sci. USA 118, e2117965118 (2021).
Friedrich, C. et al. Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues. Nat. Immunol. 22, 1256–1267 (2021).
Klose, C. S. et al. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature 494, 261–265 (2013).
Rankin, L. C. et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14, 389–395 (2013).
Sciume, G. et al. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209, 2331–2338 (2012).
Colonna, M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity 48, 1104–1117 (2018).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity 33, 736–751 (2010).
Stehle, C. et al. T-bet and RORα control lymph node formation by regulating embryonic innate lymphoid cell differentiation. Nat. Immunol. 22, 1231–1244 (2021).
Fiancette, R. et al. Reciprocal transcription factor networks govern tissue-resident ILC3 subset function and identity. Nat. Immunol. 22, 1245–1255 (2021).
Parker, M. E. et al. c-Maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J. Exp. Med. 217, e20191030 (2020).
Tizian, C. et al. c-Maf restrains T-bet-driven programming of CCR6-negative group 3 innate lymphoid cells. eLife 9, e52549 (2020).
Cella, M. et al. Subsets of ILC3–ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20, 980–991 (2019).
Li, Y. et al. Cell autonomous expression of BCL6 is required to maintain lineage identity of mouse CCR6+ ILC3s. J. Exp. Med. 220, e20220440 (2023).
Pokrovskii, M. et al. Characterization of transcriptional regulatory networks that promote and restrict identities and functions of intestinal innate lymphoid cells. Immunity 51, 185–197 (2019).
Wang, Q. & Colonna, M. Keeping time in group 3 innate lymphoid cells. Nat. Rev. Immunol. 20, 720–726 (2020).
Guan, D. & Lazar, M. A. Interconnections between circadian clocks and metabolism. J. Clin. Invest. 131, e148278 (2021).
Curtis, A. M., Bellet, M. M., Sassone-Corsi, P. & O’Neill, L. A. Circadian clock proteins and immunity. Immunity 40, 178–186 (2014).
Takahashi, J. S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017).
Wang, Q. et al. Circadian rhythm-dependent and circadian rhythm-independent impacts of the molecular clock on type 3 innate lymphoid cells. Sci. Immunol. 4, eaay7501 (2019).
Cho, H. et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012).
Bugge, A. et al. RER-ERBα and REV-ERBβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 (2012).
Dierickx, P. et al. SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc. Natl Acad. Sci. USA 116, 12147–12152 (2019).
Dierickx, P. et al. Circadian REV-ERBs repress E4bp4 to activate NAMPT-dependent NAD+ biosynthesis and sustain cardiac function. Nat. Cardiovasc. Res. 1, 45–58 (2022).
Jarade, A., Di Santo, J. P. & Serafini, N. Group 3 innate lymphoid cells mediate host defense against attaching and effacing pathogens. Curr. Opin. Microbiol. 63, 83–91 (2021).
Hepworth, M. R. et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature 498, 113–117 (2013).
Sato, F., Kohsaka, A., Bhawal, U. K. & Muragaki, Y. Potential roles of Dec and Bmal1 genes in interconnecting circadian clock and energy metabolism. Int. J. Mol. Sci. 19, 781 (2018).
Luby, A. & Alves-Guerra, M. C. UCP2 as a cancer target through energy metabolism and oxidative stress control. Int. J. Mol. Sci. 23, 15077 (2022).
Lee, M., Lee, S. Y. & Bae, Y. S. Functional roles of sphingolipids in immunity and their implication in disease. Exp. Mol. Med 55, 1110–1130 (2023).
Narni-Mancinelli, E. et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl Acad. Sci. USA 108, 18324–18329 (2011).
Wang, Y. et al. The transcription factors T-bet and RUNX are required for the ontogeny of pathogenic interferon-γ-producing T helper 17 cells. Immunity 40, 355–366 (2014).
Yu, X. et al. TH17 cell differentiation is regulated by the circadian clock. Science 342, 727–730 (2013).
Zhu, C. et al. An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat. Commun. 6, 6072 (2015).
Iwata, S. et al. The transcription factor T-bet limits amplification of type I IFN transcriptome and circuitry in T helper 1 cells. Immunity 46, 983–991 (2017).
Chang, D. et al. A cis-element at the Rorc locus regulates the development of type 3 innate lymphoid cells. Front. Immunol. 14, 1105145 (2023).
Chang, C. et al. The nuclear receptor REV-ERBα modulates TH17 cell-mediated autoimmune disease. Proc. Natl Acad. Sci. USA 116, 18528–18536 (2019).
Hall, J. A. et al. Transcription factor RORα enforces stability of the TH17 cell effector program by binding to a Rorc cis-regulatory element. Immunity 55, 2027–2043 (2022).
Allan, D. S. et al. An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol. 8, 340–351 (2015).
Kojetin, D., Wang, Y., Kamenecka, T. M. & Burris, T. P. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem. Biol. 6, 131–134 (2011).
Amir, M. et al. REV-ERBα regulates TH17 cell development and autoimmunity. Cell Rep. 25, 3733–3749 (2018).
Teng, F. et al. A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut. Sci. Immunol. 4, eaax1215 (2019).
Godinho-Silva, C. et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature 574, 254–258 (2019).
Ukai, H. & Ueda, H. R. Systems biology of mammalian circadian clocks. Annu. Rev. Physiol. 72, 579–603 (2010).
Huntington, N. D. The unconventional expression of IL-15 and its role in NK cell homeostasis. Immunol. Cell Biol. 92, 210–213 (2014).
Gerhart-Hines, Z. & Lazar, M. A. REV-ERBα and the circadian transcriptional regulation of metabolism. Diabetes Obes. Metab. 17, 12–16 (2015).
Solt, L. A. et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 (2012).
Zhang, Y. et al. Discrete functions of nuclear receptor REV-ERBα couple metabolism to the clock. Science 348, 1488–1492 (2015).
Talbot, J. et al. Feeding-dependent VIP neuron–ILC3 circuit regulates the intestinal barrier. Nature 579, 575–580 (2020).
Seillet, C. et al. The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat. Immunol. 21, 168–177 (2020).
Caballero-Flores, G., Pickard, J. M., Fukuda, S., Inohara, N. & Nunez, G. An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe 28, 526–533 (2020).
Schauer, D. B. & Falkow, S. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun. 61, 4654–4661 (1993).
Fachi, J. L. et al. Hyperbaric oxygen augments susceptibility to C. difficile infection by impairing gut microbiota ability to stimulate the HIF-1α–IL-22 axis in ILC3. Gut Microbes 16, 2297872 (2024).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet 53, 403–411 (2021).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Acknowledgements
We thank S. Gilfillan, M. Cella, V. Peng and members of the Colonna lab for helpful discussions; J. M. Dimitry in Musiek lab for sharing Nr1d1fl/flNr1d2fl/fl mice and S. Panda in Colonna lab for sharing CD45.1 mice; W. Wang and P. Rodrigues for technical assistance; E. Lantelme and D. Brinja for cell sorting; and the Genome Technology Access Center at the McDonnell Genome Institute for scRNA-seq and scATAC–seq. The Center is partially supported by NCI Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center from the National Center for Research Resources, a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This study was supported by the NIH (1R01DK126969, R01DK132327 and R01DK30292 to M.C.; R01AI134035 to M.C. and E.M.O.; R01DK45586 to M.A.L.; T32 AI165391 to L.S.H.). A.S. was supported by Pelotonia Foundation Postdoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
B.B. and M.C. conceived the project. B.B. designed the experiments and analyzed the data. B.B. and J.L.F. performed the experiments. A.U.A. performed multiome analysis. W.L.B. performed electron microscopy analysis. S.d.O. analyzed 16S sequencing data. L.S.H., M.V.D.M. and A.S. generated the MNK3i cell lines. E.S.M., V.K.K., M.A.L. and E.M.O. provided reagents. B.B. and M.C. wrote the paper with input from all other authors.
Corresponding author
Ethics declarations
Competing interests
M.C. is a member of the scientific advisory board of Vigil, Cell Signaling Technologies, Cartesian and Halyard. M.A.L. is a member of a scientific advisory board for Pfizer and is a cofounder and scientific advisory board member for Flare Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Ioana Staicu, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Selective impact of REV-ERBαβ deficiency on gut ILC3s.
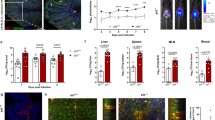
a, Representative gating strategy for identification of siLP ILCs. b, Representative flow cytometry plot and quantification of frequency of siLP ILC3s out of CD45+Lin− cells, absolute number of siLP ILC3s, absolute number of mesenteric lymph node (mLN) ILC3s of WT (n = 9) and DKO (n = 8) mice. c, Representative flow cytometry plot and quantification of frequency of IL-17-producing cells and amount of IL-17 per cell (gMFI) in DN and CCR6+ ILC3s from siLP of WT (n = 7) and DKO (n = 5) mice, in response to ex vivo stimulation with IL-23 + IL-1β for 3.5 hrs. d, Representative flow cytometry plots and quantification of frequency of GM-CSF-producing cells and amount of GM-CSF per cell (gMFI) in NKp46+, DN, and CCR6+ ILC3s from siLP of WT (n = 5) and DKO (n = 5) mice, in response to ex vivo stimulation with PMA+ionomycin for 3.5 hrs. e, Quantification of IL-17+ CD4+ T cells from siLP of WT (n = 4) and DKO (n = 4) mice 6hrs after ex vivo stimulation with PMA+ionomycin. f, Quantification of absolute numbers of siLP ILC2s and NK cells from WT and DKO mice (n = 7 or 8 per group). g, Quantification of gMFI of GATA3 in siLP ILC2s of WT and DKO mice (n = 5 per group). h-i, Representative flow cytometry plot and quantification of liver ILC1 and NK cells, gated on CD45+Lin(CD3/CD5/CD19)−NK1.1+NKp46+ cells from WT (n = 5) and DKO mice (n = 5). j, Representative flow cytometry plot and quantification of liver IFNγ+ ILC1s, gated on CD45+Lin(CD3/CD5/CD19)−NK1.1+NKp46+CD49a+ cells as in h-i. k, Quantification of IFNγ+ NK cells [gated on CD45+Lin(CD3/CD5/CD19)−NK1.1+NKp46+CD49b+ cells] of WT (n = 5) and DKO (n = 5) mice. Data pooled from two independent experiments (b, f) and representative of 2-3 independent experiments (c-e, g-k). Statistical analysis was performed using unpaired two-tailed Student’s t-test (b, c, e-k); two-way ANOVA with Sidak’s multiple comparison test for d. Bars indicate means ( ± SEM).
Extended Data Fig. 2 An altered ILC3/ILC1 ratio increases susceptibility to C. rodentium infection.
a, Representation of relative abundances of specific bacteria at the phylum level between WT and DKO mice (n = 4 per group). b, Representation of relative abundance of specific bacteria at the genus level as in a. c, Boxplot representation of richness (Shannon’s alpha diversity index) for microbiome composition as in a. Boxes extend from the 25th to 75th percentiles, whiskers extend to 1.5 times the Interquartile Range (IQR), and the center line is the median. d, Principal coordinate analysis (PCoA) plot of bacterial beta diversity of 2314 unique taxa at the species level using Bray-Curtis dissimilarities. e, Representative hematoxylin and eosin (H&E) staining of gut sections from WT (n = 2) and DKO mice (n = 2) at steady state: scale bar, 100μm. f, Fecal lipocalin-2 (Lcn2) in WT (n = 4) and DKO (n = 3) mice at steady state measured by enzyme-linked immunosorbent assay (ELISA). g, Intestinal permeability measured by serum FITC-Dextran level and FITC-Dextran gMFI in DKO and WT littermates (n = 8 per group) at steady state. h, Body weight variation over time in WT (n = 5) and DKO mice (n = 4) infected with C. rodentium (2 × 109). i, C. rodentium fecal colony-forming units (c.f.u.) at indicated time point between 0 to 20 days post infection (dpi) as in h. Results are representative of two independent experiment (f, h, i) or pooled from two independent experiments (g). Statistics was performed using Wilcoxon rank sum exact test (c); unpaired two-tailed Student’s t-test (f, g); two-way ANOVA with Bonferroni’s multiple comparison test (h); Sidak’s multiple comparison test (i). Bars indicate means ± SEM.
Extended Data Fig. 3 REV-ERBαβ deficiency alters ILC3 metabolism and impairs antigen presentation in CCR6⁺ ILC3s.
a, Volcano plot showing top 50 DEGs of NKp46 + ILC3s from WT and DKO mice (P value adjusted<0.05 and |Log2 Fold Change | > 1). Gene Ontology (GO) analysis of upregulated and downregulated DEGs in DKO NKp46 + ILC3s compared to WT cells. b, Volcano plot showing top 50 DEGs of DN ILC3s from WT and DKO mice (P value adjusted<0.05 and |Log2 Fold Change | > 1. GO analysis of upregulated and downregulated DEGs in DKO DN ILC3s compared to WT cells. c, Volcano plot showing top 50 DEGs of CCR6 + ILC3s from WT and DKO mice (P value adjusted<0.05 and |Log2 Fold Change | > 1). GO analysis of upregulated and downregulated DEGs in DKO CCR6 + ILC3s compared to WT cells. d, Heatmap of selected genes related to antigen presentation and MHC-II processing in bulk-RNA seq samples from WT and DKO CCR6 + ILC3s. e, Representative flow cytometry plot and quantification of CCR6 + MHCII+ ILC3s from WT and DKO mice (n = 4 per group). f, Quantification of pTregs and RORγt–Tregs in the siLP of WT and DKO mice (n = 5 per group). Results are representative of two independent experiments (e,f). FDR was determined by two-sided Wald test (DESeq2) for bulk RNA-seq analysis. Statistical analysis was performed using unpaired two-tailed Student’s t-test. Bars indicate means ( ± SEM).
Extended Data Fig. 4 REV-ERBαβ deletion disrupts mitochondrial homeostasis in ILC3s.
a, Heat map showing normalized expression of selected genes involved in lipid metabolism (highlighted in orange) and glycolysis. b, Transmission electron microscopy images of mitochondria from sorted WT and DKO ILC3s (Scale bars, 500 nm; Left-10,000X, right-25,000X) and quantification of mitochondrial matrix (upper panel) and mitochondrial length (lower panel). c, Transmission electron microscopy showing images of autophagosome structures (Scale bars, 500 nm, 25,000X magnification) and quantification of number of autophagosomes per 40 cells. d, GSEA analysis (top) and heatmap (bottom) showing normalized expression of genes related to amino acid metabolic process in DKO ILC1s. e, GSEA analysis and heatmap showing key genes in sphingolipid biosynthetic process in DKO ILC1s. Data is representative of pooled experiment (n = 3 per group) (b, c). Statistical analysis was performed using unpaired two-tailed Student’s t-test. No statistical analysis was performed on c.
Extended Data Fig. 5 Deletion of REV-ERBαβ has no impact on liver ILC1s and NK cells.
a, Representative flow cytometry plots and quantification of the chimerism for liver CD45.1/2 WT and CD45.2 DKO ILC1s compared with chimerism of splenic CD45+ cells. b, Representative flow cytometry plots and quantification of liver NK cells chimerism compared with chimerism of splenic CD45+ cells. c, Schematic of BrdU administration. d, Quantification of Ki-67 + ILC1s from siLP of WT and DKO mice (n = 6 or 7 mice per group). e, Quantification of Ki-67 + NKp46 + ILC3s from siLP of WT and DKO mice (n = 4 mice per group). f, Representative flow cytometry plots showing frequency of Lin–CD45.2+ cells in salivary gland of NSG recipient mice 8 weeks post transfer of NKp46 + ILC3 from WT (n = 2) and DKO (n = 2) mice. Data is representative of two or three independent experiments. Statistical analysis was performed using Student’s t-test. Bars indicate means ± SEM.
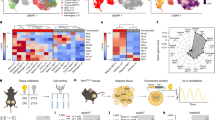
Extended Data Fig. 6 Multiome analysis reveals altered gene expression and regulatory element accessibility in DKO mice.
a, Bubble plot shows the expression of key genes by pseudo-bulk RNA clusters (related to Fig. 5a). Bubble size is proportional to percentage of cells expressing a gene, and color intensity is proportional to average scaled gene expression within a cluster. b, Number of peaks called for each pseudo-bulk ATAC clusters (related to Fig. 6a).
Extended Data Fig. 7 Pseudo-time trajectory analysis of scATAC-seq data reveals potential ILC3-to-ILC1 differentiation paths.
a-b, Heat maps showing transcription factor motif deviation scores and integrated scRNA-seq expression values along pseudo-time trajectories from WT ILC3s to ILC1s (a) and from DKO ILC3s to ILC1s (b).
Extended Data Fig. 8 Generation of MNK3i cell lines targeting NFIL3 and the Rorc -2 kb cis-regulatory element.
a, Schematic of the constructs used for generating MNK3i_Scrambled and MNK3i_NFIL3 cell lines. b, Schematic representation of the Rorc locus as well as the -2 kb cis-regulatory element that was targeted to generate the MNK3i-Rorc−2kbΔ/Δ cell line.
Extended Data Fig. 9 REV-ERBαβ deficiency alters the chromatin landscape of key ILC3 program genes.
a-d, Washington University Epigenome Browser display of pseudobulk sc-ATAC clusters [Zfp683+ ILC1, Gzma+ ILC1, tILC3, WT (NKp46+, DN, CCR6+) ILC3s and DKO (NKp46+, DN, and CCR6+) ILC3s] of Il23r and Il12rb2 (a), IL17a and IL17f (b), Ifnγ (c), and Ccr9 loci (d), alongside REV-ERBα, NFIL3, T-bet and RORγt ChIP-seq. Differentially accessible regions are marked with discontinuous boxes.
Extended Data Fig. 10 REV-ERBαβ deficiency reshapes the chromatin landscape of metabolism-related genes in ILC3s.
a, Heatmap showing key metabolism related genes from scRNA-seq in Fig. 5a. b-f, Washington University Epigenome Browser display of pseudobulk scATAC clusters [Zfp683+ ILC1, Gzma+ ILC1, tILC3, WT (NKp46+, DN, CCR6+) ILC3s and DKO (NKp46+, DN, and CCR6+) ILC3s] of Ucp2 (b), Lpl (c), Pkm (d), Ass1 (e), and Acat1 (f), alongside REV-ERBα and NFIL3 ChIP-seq. Differentially accessible regions are marked with discontinuous boxes.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhattarai, B., Antonova, A.U., Fachi, J.L. et al. Circadian circuits control plasticity of group 3 innate lymphoid cells by sustaining epigenetic configuration of RORγt. Nat Immunol 26, 1527–1539 (2025). https://doi.org/10.1038/s41590-025-02240-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-025-02240-5
This article is cited by
-
Clock genes tune plasticity of group 3 innate lymphoid cells
Nature Immunology (2025)
-
Clock genes keep intestinal ILC3s ticking
Nature Reviews Immunology (2025)