Abstract
Neurodegenerative diseases and serious mental illnesses often exhibit overlapping characteristics, highlighting the potential for shared underlying mechanisms. To facilitate a deeper understanding of these diseases and pave the way for more effective treatments, we have generated a population-scale multi-omics dataset consisting of genotype and single-nucleus transcriptome data from the prefrontal cortex of frozen human brain specimens. Encompassing over 6.3 million nuclei from 1,494 donors, our dataset represents a diverse range of neurodegenerative and serious mental illnesses, including Alzheimer’s and Parkinson’s diseases, schizophrenia, bipolar disorder and diffuse Lewy body dementia, as well as neurotypical controls. Our dataset offers a unique opportunity to study disease interactions, as 21% of donors had comorbid diagnoses of two or more major brain disorders. Additionally, it includes detailed phenotypic information on neuropsychiatric symptoms, such as apathy and weight loss, which commonly accompany Alzheimer’s disease and related dementias. We have performed stringent preprocessing and quality controls, ensuring the reliability and usability of the data. As a commitment to fostering collaborative research, we provide this valuable resource as an online repository, enabling widespread analyses across the scientific community.
Similar content being viewed by others

Background & Summary
Alterations in gene expression and changes in cell type abundances are commonly observed in various brain-related disorders, ranging from mental illnesses like schizophrenia (SCZ)1,2 to neurodegenerative diseases such as Alzheimer’s (AD)3,4,5 and Parkinson’s disease (PD)6,7. Traditional methods, using bulk tissue or broad populations of sorted cells, fail to fully capture the intricate, often highly cell type specific, molecular changes associated with these diseases. Recent advances in single-cell expression profiling address these limitations and have facilitated the generation of larger datasets, most notably for AD where the combined data now nears 1,000 cases3,8,9,10,11. However, single cell resolution datasets for other diseases are considerably smaller. For instance, the latest release from PsychENCODE, which consolidates all major sources of single-cell data on serious mental illnesses, reported only 77 cases of SCZ, 52 cases of autism, 34 cases of bipolar disorder (BD), and 10 cases of post-traumatic stress disorder2,12. Although the existing datasets offer valuable insights into each disorder separately, the potential for conducting complex analyses across different disorders to identify shared or distinct molecular pathways is still limited. This is mainly due to the small sample sizes and challenges caused by merging studies generated by different research groups, introducing an additional layer of systematic bias.
To enhance our ability to identify shared and distinct molecular pathways, causal variants, and genes involved in various brain-related disorders, we generated the largest collection, to date, of single-nucleus gene expression data in the human brain (Fig. 1). This collection comprises over 6.3 million individual nuclei, isolated from 1,494 frozen postmortem samples of the dorsolateral prefrontal cortex (DLPFC). We selected the DLPFC for our study due to its critical role in higher-level brain processes, including cognitive function, decision making, and emotional regulation, all of which are often impaired in the diseases under investigation13,14. Importantly, the DLPFC has been implicated in numerous neuroimaging and neuropathological studies as a region that undergoes significant pathological changes in both neurodegenerative diseases and serious mental illnesses15,16,17.
The sample cohort consists of neurotypical controls, as well as donors affected by more than 30 different disorders, including three represented by more than 100 cases (AD (n = 519), SCZ (n = 177), and diffuse Lewy Body disease (DLBD; n = 112)) and three by more than 40 (vascular dementia (n = 85), BD (n = 72) and PD (n = 48)). In addition to providing a more detailed characterization of these somewhat well-studied diseases, our dataset also includes a subset of cases with relatively understudied conditions, such as obsessive-compulsive disorder (n = 6), amyotrophic lateral sclerosis (n = 5), progressive supranuclear palsy (n = 5), argyrophilic grain disease (n = 3) or normal pressure hydrocephalus (n = 3). To our knowledge, many of these disorders have not yet been analyzed at the single-cell level. Despite the small number of cases, analyzing them alongside well-matched controls for sex, age, and race could offer valuable preliminary insights into these conditions. Lastly, we want to highlight the availability in our cohort of phenotypic information on neuropsychiatric symptoms (NPS), which frequently accompany AD and related dementias18. Throughout the course of the disease, more than 80% of individuals with AD are estimated to exhibit at least one NPS that significantly impacts their clinical outcomes19. So far, various studies have examined population data to characterize NPS along the AD continuum20,21,22. For example, depression and apathy are often the most observed symptoms in the early stages of AD, with delusions, hallucinations, and aggression becoming more prevalent as the disease advances20. Yet, beyond broad population-level observations, research into the mechanistic basis of these NPS remains scarce. We believe that our dataset provides a unique opportunity to explain NPS in AD at a more granular level, potentially leading to a better understanding of the disease and the identification of novel therapeutic targets.
The release of this dataset by the PsychAD consortium is accompanied by a series of manuscripts describing the cross-disorder analysis of transcriptomic vulnerability23, genetic regulation of gene expression24 and transcriptome-wide association studies25. The consortium has also leveraged neurotypical controls to assemble a map of transcriptomic changes across the lifespan26. Lastly, the computational scale and diversity of the generated data led to the development of analytical tools and databases, including dreamlet for differential gene expression27, PASSCODE for detecting phenotype-associated cells28 and iBrainMap for personalized functional genomics analysis, enabling the identification of cell-type-specific regulatory networks and phenotypic prioritization29.
Methods
Cohort data collection
The “PsychAD cohort” comprises 1,494 donors, all of whom have undergone single nucleus RNA-seq (snRNA-seq) analysis. Among these donors, genotype data is available for 1,381 (92%) of them. Specimens came from multiple sources, the Mount Sinai NIH Brain Bank and Tissue Repository (MSSM; 1,042 samples), the NIMH-IRP Human Brain Collection Core (HBCC; 300 samples), and five prospective cohort studies at the Rush Alzheimer’s Disease Center (RADC; 152 samples)30,31 (Fig. 2a). All five RADC cohorts were approved by an Institutional Review Board of Rush University Medical Center and participants signed informed and repository consents and an Anatomic Gift Act for organ donation. Importantly, 60% of the PsychAD cohort, totaling 896 donors, had previously been included in MSSM AMP-AD32, CommonMind33 and/or RADC studies34, which had already generated a wealth of omics data for these individuals, including SNP-array33, whole-genome sequencing (WGS)32, RNA sequencing (RNA-seq)35,36,37, assay for transposase-accessible chromatin (ATAC-seq)4,7,35,38, DNA methylation37, proteomics35, chromatin immunoprecipitation sequencing for histone 3 lysine 27 acetylation (ChIP-seq H3K27ac)4,39 and for histone 3 lysine 4 trimethylation (ChIP-seq H3K4me3). However, it is important to emphasize that, prior to this study, no snRNA-seq data was available for the PsychAD cohort, with the exception of 53 donors from MSSM2 and 7 from RADC3,11.
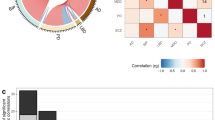
Summary of demographics and clinical data of the PsychAD cohort. (a) Overlap of the PsychAD cohort with MSSM AMP-AD, CommonMind and RADC cohorts. (b) Correlations among AD-related phenotypes. This analysis includes donors with either sole AD diagnosis (without comorbidities) or control samples (free of any diagnosis). For the “cognitive impairment” phenotype, untransformed CDR values are used for MSSM donors. RADC donors are numerically scaled as follows: Mild Cognitive Impairment (MCI) = 0.75, clinical dementia = 3. (c) Distribution of the number of diagnoses per donor. Note that “Dementia” and “MCI” are not counted as separate diagnoses if the donor already has a neurodegenerative or neurological disease. Also, NPS are excluded from this comparison. (d) Analysis of the counts and intersections among the top 10 most frequently represented diagnoses plus controls, with a minimum intersection size for plotting set to 10. FTD: Frontotemporal dementia; ASCVD: Atherosclerotic cardiovascular disease; PD: Parkinson’s disease; BD: Bipolar disorder; Diabetes: Diabetes mellitus Type 1/2/unspecified; Vascular: Vascular dementia; DLBD: Diffuse Lewy body disease; SCZ: Schizophrenia. (e–h) Exploration of demographic and clinical variables within subcohorts of samples from the three brain tissue sources, encompassing sex (e), genetically inferred ancestry (f), age (g), and disease status (h). NPS are not included in the disease count in (h). (i) Dendrogram of NPS based on their co-occurrence with three highlighted clusters.
Because the institutions provided the donor’s clinical records in different formats, our first imperative was to harmonize the diagnosis status prior to downstream analyses. The full sample cohort captures 20 neurodegenerative/neurologic diseases (e.g. AD, PD, DLBD), 13 serious mental illnesses (e.g. SCZ, BD), 19 NPS (e.g. insomnia, weight loss) and 4 metabolic diseases (e.g. Type 1/2 diabetes) (Table 1). The presence of these diseases is typically encoded in a binary format, except for AD, for which we have: (1) case-control status defined using the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) criteria40; (2) Braak AD-staging score for progression of neurofibrillary neuropathology15,41; (3) mean density of neuritic plaques (plaque mean); and (4) assessment of dementia and cognitive status based on clinical dementia rating scale (CDR) for MSSM samples42, or final summary clinical diagnosis (cogdx) for RADC43. For binary diagnosis classification within the PsychAD dataset, we define the AD category as follows: CERAD ≥ 2, Braak ≥ 3 and CDR ≥ 1/cogdx ≥ 4 (MSSM/RADC). Donors with a neuropathological burden but no cognitive loss are categorized as “Tauopathy” (CERAD = 1, Braak ≥ 3 for both MSSM and RADC and, additionally, CDR = 0 for MSSM/no cognitive impairment for RADC) (Tables 1, 2). Depending on the severity of cognitive loss, the donors are classified as Mild Cognitive Impairment (MCI; CDR = 0.5 for MSSM; MCI for RADC) or Dementia (CDR ≥ 1 for MSSM; clinical dementia for RADC) (Tables 1, 2). The AD-related neuropathological and clinical phenotypes are moderately correlated (Fig. 2b), indicating shared and distinct disease processes4. While the PsychAD cohort contains 420 donors with no diagnosis (28% of the dataset, referred to as “control” samples) and 754 donors with exactly 1 diagnosis (51% of the dataset), the remaining 320 donors (21%) are associated with 2 or more diagnoses (Fig. 2c,d). The demographics and clinical characteristics of donors varied significantly among the sources (Fig. 2e–h), with donors of European ancestry constituting over 79% of the MSSM subcohort, compared to 50% in HBCC, and 18% in RADC (Fig. 2f). This variation enables the exploration of ancestry-specific disease signatures. Regarding age distribution, 89% and 99% of individuals in the MSSM and RADC subcohorts are over 60 years old, respectively, while only 10% of HBCC are among this older age category, with 49% of donors under the age of 40 (Fig. 2g). These age distribution patterns partly align with disease distribution, as MSSM and RADC primarily consist of donors with or at risk for neurodegenerative/neurological diseases, for which age is a major risk factor (Fig. 2h). In contrast, in addition to controls, HBCC exclusively includes serious mental illnesses, which typically manifest during childhood or adolescence44,45,46,47. Due to HBCC’s different disease focus, we lack certain AD-related metrics (CERAD, BRAAK, CDR, Plaque Mean) that are available for MSSM and RADC. However, HBCC’s sample selection process involved reviewing neuropathology reports to ensure the absence of significant plaque and/or tangle pathology. As a result, HBCC donors without brain-related diagnoses can reliably be used as controls for comparison with neurodegenerative diseases, even in the absence of additional neuropathological data.
In addition to offering disease-related phenotypes, we included a set of 19 NPS, each of which affected between 23 and 438 individuals from the 1,042 MSSM donors. These symptoms constitute commonly associated features of AD and related dementias, and are linked to significant adverse effects on daily function and quality of life48. Utilizing hierarchical clustering analysis, we observed that these NPS tend to group into three distinct clusters, broadly aligning with established associations (Fig. 2h). Therefore, as an alternative to analyzing the 19 individual classes, we also offer a categorization of donors into the three aggregated NPS classes (Table 3).
snRNA-seq data generation and analysis
Nuclei isolation and snRNA-seq library preparation
All buffers were supplemented with RNAse inhibitors (Takara). 6 samples were processed in parallel. 25 mg of frozen postmortem human brain tissue from each specimen was homogenized in cold lysis buffer (0.32 M Sucrose, 5 mM CaCl2, 3 mM Magnesium acetate, 0.1 mM, EDTA, 10 mM Tris-HCl, pH8, 1 mM DTT, 0.1% Triton X-100) and filtered through a 40 µm cell strainer. The flow-through was underlaid with sucrose solution (1.8 M Sucrose, 3 mM Magnesium acetate, 1 mM DTT, 10 mM Tris-HCl, pH8) and centrifuged at 107,000 xg for 1 hour at 4 °C. Pellets were resuspended in PBS and quantified (Countess II, Life Technologies). 2 M nuclei from each sample were then pelleted at 500 xg for 10 minutes at 4 °C and re-suspended in 100 µl staining buffer (2% BSA, 0.02% Tween-20, 10 mM Tris, 146 mM NaCl, 1 mM CaCl2 and 21 mM MgCl). Each sample was incubated with 1 µg of a distinct TotalSeq-A nuclear hashing antibody (Biolegend) for 30 min at 4 °C. Prior to fluorescence activated nuclei sorting (FANS), volumes were brought up to 250 µl with staining buffer and 7-AAD (Invitrogen) added to facilitate the detection of nuclei. 7-AAD positive nuclei were sorted into tubes pre-coated with 5% BSA using a FACSAria flow cytometer (BD Biosciences).
Following FANS, nuclei were washed in staining buffer before being re-suspended in 22 µl PBS and quantified. Nuclei concentrations were normalized and equal amounts from each sample were pooled together. 2 aliquots of 60,000 pooled nuclei (i.e. 10,000 per sample) were processed in parallel using 3′ v3.1 reagents (10x Genomics). At the cDNA amplification step (step 2.2), reactions were supplemented with a hash-tag oligo (HTO) cDNA “additive” primer (GTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T; *Phosphorothioate bond). Following cDNA amplification, supernatants from the 0.6x SPRI selection step were retained for HTO library generation. Otherwise, cDNA libraries were prepared according to the manufacturer’s instructions (10x Genomics). HTO libraries were prepared as described previously49
Computational processing
Sequencing reads from all pools of multiplexed samples were aligned to the hg38 reference genome using STARsolo50,51. To assign the nuclei from each pool to their respective donors, we applied a genotype-based demultiplexing approach followed by a genotype concordance check. First, cellSNP52 was used to pile up the alleles from polymorphic sites overlapping snRNA-seq reads within expressed genes (for inclusion, a gene needed to be expressed by at least 10 cells). Polymorphic sites had to show a minimum minor allele frequency of 0.1 and a minimum aggregated UMI count of 20. Then, vireo53 utilized those pile-ups to split cells into clusters corresponding to six distinct donors per pool. The assignment of the identity of each cluster of cells to a particular donor was derived from genotype concordance analysis that compared the clusters of cells against reference genotyping data using QTLtools-mbv54. This analysis could be accurately performed only for cells exceeding baseline quality control (QC) metrics, i.e. minimum number of expressed genes (n ≥ 1,000) and maximum fraction of mitochondrial reads (less than 5%). Cells that didn’t meet these criteria were excluded. While the majority of pools contained the cells from the expected sets of donors, we leveraged the genotype concordance results to detect and correct occasional sample swaps and mislabeling.
After genome alignment and demultiplexing, the downstream processing was performed using Pegasus and scanpy55. We applied rigorous three-step QC to remove ambient RNA and retain viable cells for downstream analysis. First, we implemented a more stringent QC for individual cells, in addition to the initial QC carried out during the demultiplexing stage. Cells falling outside the defined ranges for UMI counts (1,500 ≤ n_UMIs ≤ 110,000), gene counts (1,100 ≤ n_genes ≤ 12,500), and mitochondrial content (less than 5%) were removed. We also checked for possible contamination from ambient RNA using CellBender56. Further filtering was carried out by removing doublets using the Scrublet method57. Second, the QC was applied at the feature level. We removed features (genes) that were not robustly expressed by at least 0.05% of the nuclei. Lastly, the QC was applied at the donor level and, because they could introduce noise in downstream analysis, those with less than 50 nuclei were removed. Then, to correct for unwanted (non-biological) variance, such as dissection biases arising from differing tissue source protocols across source brain banks, we employed Canonical Correlation Analysis using the Harmony method58. Highly variable features were selected from mean and variance trends, and we used the k-nearest-neighbor (kNN) graph calculated on the basis of harmony-corrected PCA embedding space to cluster cells in the same cell type using Leiden59 clustering algorithms. We used UMAP (Uniform Manifold Approximation and Projection)60 for the visualization of the resulting clusters.
Defining cellular taxonomy using iterative clustering
Cellular taxonomy was defined using a divide-and-conquer strategy. From the full dataset containing over 6 million nuclei, 8 major cell classes were defined as described above. After subsetting the data by each class, we re-calculated highly variable genes (HVGs) among cells in the same class. This allowed us to re-focus on feature space that is more relevant for the same class of cells. We then calculated kNN graph on the basis of the harmony-corrected PCA of the selected HVGs. Leiden-clustering was used to annotate subclass-level annotation. We iterated to the second level of taxonomy which returned us with 67 subtypes of human brain cell types.
Genotyping
Overall strategy
The majority of donors from the HBCC (98%) and RADC (87%) had previously undergone genotyping32,33,61 and we opted to utilize this existing data. For the MSSM donors, however, the coverage from previous genotyping efforts was notably lower, with only 57% (598 donors) having been genotyped. Therefore, we conducted genotyping for all MSSM donors for whom we had a sufficient amount of material, as detailed in the “Library preparation for MSSM donors” and “Computational processing for MSSM donors” sections. We made use of the existing genotyping data from MSSM to assess genotype concordance with the newly generated SNP array. This allowed us to identify sample swaps and unintended duplicates. Lastly, we implemented a multi-step procedure to merge genotypes from the existing and newly generated WGS and SNP array data, resulting in the creation of a comprehensive genotype dataset for the PsychAD cohort, as outlined in the “Integration of multi-source genotype data” section.
SNP arrays
Genomic DNA was isolated using the QIAamp DNA mini kit (Qiagen), according to the manufacturer’s instructions, and quantified via Qubit (Life Technologies). 400 ng of DNA was then whole-genome amplified, fragmented, precipitated and resuspended in the appropriate hybridization buffer. Genotyping was performed using the Infinium Global Screening Array-24 Kit (Illumina) according to the manufacturer’s protocol. Briefly, denatured samples were hybridized on prepared Illumina Bead Chips. After hybridization, the Bead Chip oligonucleotides were extended by a single fluorescent labeled base, which was detected by fluorescence imaging with an Illumina Bead Array Reader, iScan.
Computational processing for MSSM donors
Pre-imputation processing of the PsychAD MSSM subcohort genotype data consisted of running the quality control script HRC-1000G-check-bim.pl from the McCarthy Lab Group (https://www.well.ox.ac.uk/~wrayner/tools/). Genotypes were then phased and imputed on the Trans-Omics for Precision Medicine (TOPMed) Imputation Server62. Only variants with an imputation R2 > 0.3 were retained. Biallelic variants were additionally annotated with ancestry-specific MAF values from the National Center for Biotechnology Information’s Allele Frequency Aggregator (ALFA) (https://ftp.ncbi.nih.gov/snp/population_frequency/latest_release/). The populations included in the ALFA database are described at https://www.ncbi.nlm.nih.gov/snp/docs/gsr/data_inclusion/#population.
Integration of multi-source genotype data
To compile a combined dataset covering the largest possible fraction of the PsychAD cohort, we leveraged the following datasets:
-
PsychAD-MSSM SNP array: Genotyping data for 882 donors that cover the majority of MSSM donors were generated and described in this manuscript.
-
CommonMind SNP array: Genotyping data for 513 samples overlapping the PsychAD cohort were obtained from previously generated SNP array on Illumina Infinium HumanOmniExpressExome 8 v 1.1b chip33. These data are accessible to all registered users of the NIMH Data Archive (RRID:SCR_004434) under the collection identifier C5063.
-
RADC WGS: Whole-genome sequencing data for 131 samples overlapping the PsychAD cohort were obtained from a Diverse study61 available to all registered users of the AD Knowledge Portal (RRID:SCR_006307) under the release number 20.6, accession identifier syn51757644.
-
ADSP WGS: Whole-genome sequencing data for 284 samples overlapping the PsychAD cohort were obtained from the seventh release of WGS data under the ADSP Umbrella Study (NG00067.v7) from the National Institute of Aging Genetics of Alzheimer’s Disease Data Storage Service63.
These datasets exhibited a significant overlap, resulting in 377 donors with one or more duplicates, as identified by KING64 (kinship score ≥ 0.4; Fig. 3a). To determine which of the duplicated samples were retained for use as the final genotype file, the following criteria were applied: First, if one sample was obtained through WGS and the other through SNP array genotyping, we retained the WGS sample. This prioritization criterion was applied to 249 donors. Next, if one sample displayed a heterozygosity value falling within ± 2 standard deviations from the mean, we retained the other sample, thus excluding an additional 8 pairs. All of the remaining 120 donors had samples in both the PsychAD-MSSM and CommonMind SNP arrays. We opted to prioritize the PsychAD-MSSM SNP array due to inherent imperfections of the CommonMind SNP array, which was initially provided in hg19 coordinates and necessitated conversion to hg38 coordinates. As a result, the final combined genotype file encompasses 92% (1,381) of the PsychAD donors.
Analysis of genotyping data. (a) Counts and intersections among sources of genotyping data available for donors from the PsychAD cohort. (b) Distribution of genetic similarities estimated between combined genotype dataset and genotypes called from snRNA-seq data. (c) F-statistic from plink’s “check-sex” function plotted by reported sex (samples with known sex chromosome aneuploidies not shown). (d,e) The first two PCs of genetic ancestry were calculated separately for the PsychAD-MSSM genotype dataset of 882 samples (d) and for the combined dataset of 1,381 samples (e).
Whole-genome sequencing (WGS) variant calling for the RADC and ADSP samples was carried out according to the best practice guidelines of the Genome Analysis Toolkit (GATK)65. In summary, the identification of single nucleotide variants (SNVs) and insertions/deletions (indels) was performed jointly using GATK’s HaplotypeCaller and GenotypeGVCFs tools. The refinement and annotation of variants were achieved through Variant Quality Score Recalibration (VQSR) within the GATK environment. Quality control (QC) steps followed previously established pipelines66,67,68. For sample-level QC, relatedness, DNA contamination, and sample-level missingness (samples excluded if > 0.05) were evaluated, as well as overall coverage (samples excluded if < 25x). Outlier samples were identified and excluded based on several criteria, including the number of called SNVs and indels, insert size length, alignment mapping quality score (MQ), CRAM file size, transition/transversion ratio (Ti/Tv), the ratio of novel variants to all variants, and the ratio of mapped reads to paired reads, as detailed in previous studies66,67,68. At the variant level, filtering removed variants with more than 10% missingness and high levels of heterozygosity (InbreedingCoeff <−0.8). Individual genotype calls with a depth (DP) of less than 10 or a genotype quality (GQ) of less than 20 were marked as missing. Analyses were limited to biallelic variants only.
Ancestry estimation
Based on the success of Mahalanobis distance techniques in ancestry assignment69,70, we leveraged quadratic discriminant analysis (QDA) to assign ancestry using scikit-learn71. We determined the genetic ancestry of our samples based on the five superpopulations defined by the 1000 Genomes Project. Initially, we merged unimputed genotypes with the 1000 Genomes Project data on the GRCh38 v2a reference using BCFtools version 1.9. We then computed the principal components (PCs) of the merged genotypes using PLINK PCA. The merged genotype used for this calculation was variant-level filtered to keep only single nucleotide variants (SNVs) with a minor allele frequency (MAF) of at least 0.01, a Hardy-Weinberg equilibrium (HWE) P-value of at least 10−10, and a variant-level missingness of no more than 0.01. We also performed linkage disequilibrium (LD) pruning with a window size of 1,000 kb, a step size of 10, and an R2 threshold of 0.2. Finally, we used forward selection to choose PC1 through PC6 for training the QDA models, applying a regularization parameter of 5−7.
Data Records
Raw and processed data described herein are available for use by the research community and have been deposited in the AMP-AD Knowledge Portal in the study-specific folder72. These include sample metadata, as well as raw and processed sequencing data for snRNA-seq and genotyping. Single nuclei data can be inspected at the CELLxGENE (RRID:SCR_021059) portal at https://cellxgene.cziscience.com/collections/84ce6837-548d-4a1f-919f-0bc0d9a3952f.
Technical Validation
Genotype data quality control
Out of four genotype datasets used in this study, three external datasets already underwent QC before they were released so we performed only a limited check. For the newly generated PsychAD-MSSM SNP array data, we started by removing samples with missingness over 0.05 (calculated within a subset of high-quality variants with variant-level missingness ≤ 0.02). All SNP-array samples were compared against genotypes called from snRNA-seq samples to check the across-assay concordance for samples originating from the same donor. This comparison allowed us to resolve sample swaps in both assays, as well as to detect and remove duplicated and contaminated samples (Fig. 3b). Next, the samples with a mismatch between the self-reported and genetically inferred sex were removed, as well as those with outlier heterozygosity defined by ± 3 standard deviations from the mean (samples with known sex chromosome aneuploidies were not subjected to this check). After performing all QC steps, we observed unambiguous separation of male and female samples and good concordance of inferred and reported ancestry for all remaining 882 samples (Fig. 3c,d). Comparable results were obtained for the merged genotype dataset, which included 1,381 samples (Fig. 3e).
snRNA-seq data quality control
After the QC processes, our snRNA-seq dataset consisted of 6.32 million nuclei spread across 561 pools. Each pool contained six libraries, and each library underwent sequencing in duplicate on different sequencing lanes. The typical yield was about 21,238 nuclei per pool (Fig. 4a), which were anticipated to be distributed evenly across the libraries. Nevertheless, we observed considerable variability in nuclei counts, largely attributed to variances in tissue quality that impact cell viability and capture efficiency73. The largest replicate in a typical pool accounted for about 32% of the nuclei (3,367 nuclei), while the smallest only captured about 5% (721 nuclei) (Fig. 4b). Such fluctuations are not unusual and align with findings from other studies53,73,74. Despite these discrepancies in nuclei numbers, the replicates consistently showed a high correlation in gene expression signals (Spearman’s ρ = 0.82), underscoring the data’s robustness (Fig. 4c). Notably, samples discarded at the QC step had roughly 55% fewer nuclei than those that met the QC criteria (1,920 vs. 4,240 nuclei, Fig. 4d). The cellular taxonomy revealed eight major cell classes and 27 subclasses (Fig. 4e).
Analysis of the snRNA-seq dataset. (a) Distribution of the number of nuclei across sample pools. Dashed line indicates the mean. (b) Distribution of nuclei to libraries within pools, ordered by nuclei count (top) and fraction of nuclei (bottom). Each replicate is depicted using two boxplots representing the nuclei distribution before (blue) and after QC (green). The center line (black) indicates the median, the box shows the interquartile range, and the whiskers indicate the highest/lowest values within 1.5 × the interquartile range. (c) Comparison of QC-passed nuclei counts between pairs of replicates from the same sequencing pools (Spearman’s ρ = 0.84). (d) Distribution of nuclei counts in samples that passed or failed QC (vertical line indicates the mean values). (e) UMAP visualization of snRNA-seq data. IN: inhibitory/GABAergic neurons, EN: excitatory/glutamatergic neurons, SMC: smooth muscle cells, VLMC: vascular leptomeningeal cells, PVM: perivascular macrophages, OPC: oligodendrocyte progenitor cells, Astro: astrocytes, Oligo: oligodendrocytes, Micro: Microglia, Endo: endothelial, Adaptive: adaptive immune cells, PC: Pericytes.
Usage Notes
Having a dataset with such a large scope, including over 6 million nuclei, 1,494 donors, 33 diagnoses, and ages ranging from 0 to 108 years, presents many opportunities but also demands careful handling. A common use case involves a statistical comparison of two groups of donors, typically those from disease carriers and neurotypical controls. In such scenarios, it is important to ensure the careful selection of donors for the control group because controls have typically much lower age at the time of death in our dataset (Fig. 5). Due to the impact of normal aging on cell function11, a wide variation in the age distributions of the groups being compared can obscure the actual effects of the disease. While complex non-linear modeling can adjust for some of these effects in differential analysis, we still recommend setting a minimum age threshold. In disease-oriented papers published using this dataset23,24,25,28,29, we established a minimum age of 17 years for serious mental illnesses and 60 years for neurodegenerative disorders. While researchers can choose which effects they want to correct for in their analysis, it’s worth noting that our studies typically adjust for demographic factors such as sex, brain bank, and postmortem interval, each of which was modeled as having a linear effect.
All data are available via the AD Knowledge Portal (https://adknowledgeportal.org). The AD Knowledge Portal is a platform for accessing data, analyses, and tools generated by the Accelerating Medicines Partnership (AMP-AD) Target Discovery Program and other National Institute on Aging (NIA)-supported programs to enable open-science practices and accelerate translational learning. The data, analyses and tools are shared early in the research cycle without a publication embargo on secondary use. Data is available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.synapse.org/Data%20Access).
Code availability
The source code used to analyze the metadata and create figures for this manuscript can be found on GitHub at this location: https://github.com/DiseaseNeuroGenomics/psychAD_SciData.
References
Skene, N. G. et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018).
Ruzicka, W. B. et al. Single-cell multi-cohort dissection of the schizophrenia transcriptome. Science 384, eadg5136 (2024).
Mathys, H. et al. Single-cell atlas reveals correlates of high cognitive function, dementia, and resilience to Alzheimer’s disease pathology. Cell 186, 4365–4385.e27 (2023).
Bendl, J. et al. The three-dimensional landscape of cortical chromatin accessibility in Alzheimer’s disease. Nat. Neurosci. 25, 1366–1378 (2022).
Zeng, B. et al. Genetic regulation of cell type–specific chromatin accessibility shapes brain disease etiology. Science 384, eadh4265 (2024).
Kamath, T. et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat. Neurosci. 25, 588–595 (2022).
Bryois, J. et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet. 52, 482–493 (2020).
Gabitto, M. I. et al. Integrated multimodal cell atlas of Alzheimer’s disease. Nature Neurosci. 7, 2366–2383 (2024).
Lee, D. et al. Plasticity of human microglia and brain perivascular macrophages in aging and Alzheimer’s disease. medRxiv https://doi.org/10.1101/2023.10.25.23297558 (2024).
Wang, Q. et al. Single cell transcriptomes and multiscale networks from persons with and without Alzheimer’s disease. Nat. Commun. 15, 5815 (2024).
Green, G. S. et al. Cellular communities reveal trajectories of brain ageing and Alzheimer’s disease. Nature 633, 634–645 (2024).
Emani, P. S. et al. Single-cell genomics and regulatory networks for 388 human brains. Science 384, eadi5199 (2024).
Huntley, J. D. & Howard, R. J. Working memory in early Alzheimer’s disease: a neuropsychological review. Int. J. Geriatr. Psychiatry 25, 121–132 (2010).
Howes, O. D., Bukala, B. R. & Beck, K. Schizophrenia: from neurochemistry to circuits, symptoms and treatments. Nat. Rev. Neurol. 20, 22–35 (2024).
Braak, H., Alafuzoff, I., Arzberger, T., Kretzschmar, H. & Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 112, 389–404 (2006).
Braak, H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211 (2003).
Zhao, B. et al. Common variants contribute to intrinsic human brain functional networks. Nat. Genet. 54, 508–517 (2022).
Argyriou, S. et al. Beyond memory impairment: the complex phenotypic landscape of Alzheimer’s disease. Trends Mol. Med. https://doi.org/10.1016/j.molmed.2024.04.016 (2024).
Lyketsos, C. G. et al. Prevalence of neuropsychiatric symptoms in dementia and mild cognitive impairment: results from the cardiovascular health study. JAMA 288, 1475–1483 (2002).
Lyketsos, C. G. et al. Neuropsychiatric symptoms in Alzheimer’s disease. Alzheimers. Dement. 7, 532–539 (2011).
Zhao, Q.-F. et al. The prevalence of neuropsychiatric symptoms in Alzheimer’s disease: Systematic review and meta-analysis. J. Affect. Disord. 190, 264–271 (2016).
Pless, A. et al. Understanding neuropsychiatric symptoms in Alzheimer’s disease: challenges and advances in diagnosis and treatment. Front. Neurosci. 17, 1263771 (2023).
Lee, D. et al. Single-cell atlas of transcriptomic vulnerability across multiple neurodegenerative and neuropsychiatric diseases. medRxiv 2024.10.31.24316513, https://doi.org/10.1101/2024.10.31.24316513 (2024).
Zeng, B. et al. Single-nucleus atlas of cell-type specific genetic regulation in the human brain. medRxiv 2024.11.02.24316590, https://doi.org/10.1101/2024.11.02.24316590 (2024).
Venkatesh, S. et al. Single-nucleus transcriptome-wide association study of human brain disorders. medRxiv 2024.11.04.24316495 https://doi.org/10.1101/2024.11.04.24316495 (2024).
Yang, H. et al. A single-cell transcriptomic atlas of the prefrontal cortex across the human lifespan. medRxiv 2024.11.06.24316592, https://doi.org/10.1101/2024.11.06.24316592 (2024).
Hoffman, G. E. et al. Efficient differential expression analysis of large-scale single cell transcriptomics data using dreamlet. bioRxiv https://doi.org/10.1101/2023.03.17.533005 (2023).
He, C. et al. Phenotype scoring of population scale single-cell data dissects Alzheimer’s disease complexity. medRxiv 2024.11.01.24316586, https://doi.org/10.1101/2024.11.01.24316586 (2024).
Chandrashekar, P. B. et al. Personalized single-cell transcriptomics reveals molecular diversity in Alzheimer’s disease. medRxiv 2024.11.01.24316589, https://doi.org/10.1101/2024.11.01.24316589 (2024).
Bennett, D. A. et al. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers. Dis. 64, S161–S189 (2018).
Marquez, D. X. et al. Representation of Older Latinxs in Cohort Studies at the Rush Alzheimer’s Disease Center. Neuroepidemiology 54, 404–418 (2020).
Wang, M. et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Scientific Data 5, 1–16 (2018).
Hoffman, G. E. et al. CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder. Sci Data 6, 180 (2019).
De Jager, P. L. et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci Data 5, 180142 (2018).
Coleman, C. et al. Multi-omic atlas of the parahippocampal gyrus in Alzheimer’s disease. Sci Data 10, 602 (2023).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
Wang, E. et al. Genome-wide methylomic regulation of multiscale gene networks in Alzheimer’s disease. Alzheimers. Dement. https://doi.org/10.1002/alz.12969 (2023).
Girdhar, K. et al. The neuronal chromatin landscape in adult schizophrenia brains is linked to early fetal development. medRxiv https://doi.org/10.1101/2023.10.02.23296067 (2023).
Girdhar, K. et al. Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat. Neurosci. 21, 1126–1136 (2018).
Fillenbaum, G. G. et al. Consortium to Establish a Registry for Alzheimer’s Disease (CERAD): The first twenty years. Alzheimers. Dement. 4, 96–109 (2008).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Morris, J. C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43, 2412–2414 (1993).
Schneider, J. A., Arvanitakis, Z., Bang, W. & Bennett, D. A. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology 69, 2197–2204 (2007).
Lewine, R. & Hart, M. Schizophrenia spectrum and other psychotic disorders. Handb. Clin. Neurol. 175, 315–333 (2020).
Lopez, O. L. & Kuller, L. H. Epidemiology of aging and associated cognitive disorders: Prevalence and incidence of Alzheimer’s disease and other dementias. Handb. Clin. Neurol. 167, 139–148 (2019).
Willis, A. W. et al. Incidence of Parkinson disease in North America. NPJ Parkinsons Dis 8, 170 (2022).
Szatmari, P. et al. Prospective Longitudinal Studies of Infant Siblings of Children With Autism: Lessons Learned and Future Directions. J. Am. Acad. Child Adolesc. Psychiatry 55, 179–187 (2016).
Chen, Y., Dang, M. & Zhang, Z. Brain mechanisms underlying neuropsychiatric symptoms in Alzheimer’s disease: a systematic review of symptom-general and -specific lesion patterns. Mol. Neurodegener. 16, 38 (2021).
Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Kaminow, B., Yunusov, D. & Dobin, A. STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. bioRxiv 2021.05.05.442755, https://doi.org/10.1101/2021.05.05.442755 (2021).
Huang, X. & Huang, Y. Cellsnp-lite: an efficient tool for genotyping single cells. Bioinformatics https://doi.org/10.1093/bioinformatics/btab358 (2021).
Huang, Y., McCarthy, D. J. & Stegle, O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 20, 273 (2019).
Fort, A. et al. MBV: a method to solve sample mislabeling and detect technical bias in large combined genotype and sequencing assay datasets. Bioinformatics 33, 1895–1897 (2017).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Fleming, S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods 20, 1323–1335 (2023).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291.e9 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3, 861 (2018).
Reddy, J. S. et al. Bridging the gap: Multi-omics profiling of brain tissue in Alzheimer’s disease and older controls in multi-ethnic populations. Alzheimers. Dement. 20, 7174–7192 (2024).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Leung, Y. Y. et al. Alzheimer’s Disease Sequencing Project Release 4 Whole Genome Sequencing Dataset. medRxiv. Preprint at https://doi.org/10.1101/2024.12.03.24317000 (2024).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178, https://doi.org/10.1101/201178 (2018).
Adelson, R. P. et al. Empirical design of a variant quality control pipeline for whole genome sequencing data using replicate discordance. Sci. Rep. 9, 16156 (2019).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Naj, A. C. et al. Quality control and integration of genotypes from two calling pipelines for whole genome sequence data in the Alzheimer’s disease sequencing project. Genomics 111, 808–818 (2019).
Peterson, R. E. et al. The utility of empirically assigning ancestry groups in cross-population genetic studies of addiction. Am. J. Addict. 26, 494–501 (2017).
Conomos, M. P. et al. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 98, 165–184 (2016).
Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
PsychAD Consortium. The PsychAD Dataset. Synapse https://doi.org/10.7303/syn60084804 (2024).
Brown, D. V. et al. A risk-reward examination of sample multiplexing reagents for single cell RNA-Seq. Genomics 116, 110793 (2024).
Cardiello, J. F. et al. Evaluation of genetic demultiplexing of single-cell sequencing data from model species. Life Sci Alliance 6, (2023).
Acknowledgements
We thank the patients and families who donated material for these studies. We thank members of the Roussos laboratory for thoughtful advice and critique. Human tissue was obtained from the NIH NeuroBioBank at the Mount Sinai and supported by NIMH-75N95019C00049. Human tissue was also obtained from the Rush Alzheimer’s Disease Center (RADC; funding: P30AG10161, P30AG72975, R01AG15819, R01AG17917, R01AG22018, U01AG46152, and U01AG61356), and NIMH-IRP Human Brain Collection Core (HBCC, project # ZIC MH002903). This study was supported by grants from the National Institute on Aging, the National Institutes of Health (NIH) grant R01AG067025 (to P.R., V.H., S.K. and D.W.), R01AG082185 (to P.R., V.H. and D.L.) and R01AG065582 (to P.R. and V.H.). This work was supported in part through the computational and data resources and staff expertise provided by Scientific Computing and Data at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences. Research reported in this publication was also supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD026880 and S10OD030463. The results published here are in whole or in part based on data obtained from the AD Knowledge Portal. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Consortia
Contributions
P.R. conceived and designed the study. P.K.A., L.L.B., D.A.B., S.M., V.H., contributed to the selection of donors, obtained informed consents, reviewed clinical examinations and/or pathological analysis and dissected tissue. J.F.F., C.C., A.H., M.A., Z.S. and S.A. processed tissue and generated snRNA-seq and/or SNP-arrays. J.F.F. supervised data generation. P.N.M., D.L., D.M., K.T., T.C., D.B., S.V. and J.B. conducted bioinformatics data analysis. D.L., G.E.H., G.V. and J.B. supervised bioinformatics data analysis. J.F.F., P.N.M., J.B. and P.R. wrote and edited the manuscript with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
Panos Roussos is an Editorial Board Member for Scientific Data.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fullard, J.F., NM, P., Lee, D. et al. Population-scale cross-disorder atlas of the human prefrontal cortex at single-cell resolution. Sci Data 12, 954 (2025). https://doi.org/10.1038/s41597-025-04687-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04687-5