Abstract
Membrane proteins are considered the major source of drug targets and are indispensable for drug design and disease prevention. However, traditional biomechanical experiments are costly and time-consuming; thus, many computational methods for predicting membrane protein types are gaining popularity. The position-specific scoring matrix (PSSM) method is an excellent method for describing the evolutionary information of protein sequences. In this study, we propose an improved capsule neural network (ICNN) model based on a capsule neural network to acquire sufficient relevant information from the PSSM. Furthermore, accounting for the complementarity between traditional machine learning and deep learning, we propose a hybrid framework that combines both approaches to predict protein types. This framework trains 41 baseline models based on the PSSM. The optimal subset features, selected after traversal, are fused using a two-level decision-level feature fusion approach. Subsequently, comparisons are made using three combined strategies within an ensemble learning framework. The experimental results demonstrate that solely relying on PSSM input, the proposed method not only surpasses the optimal methods by 1.52\(\%\), 2.26\(\%\) and 2.67\(\%\) on Dataset1, Dataset2, and Datasets3, respectively, but also exhibits superior generalizability. Furthermore, the code and dataset can be free download at https://github.com/ruanxiaoli/membrane-protein-types.
Similar content being viewed by others
Introduction
Proteins on the cell or organelle membrane are called membrane proteins. These proteins account for approximately 30\(\%\) of the proteins in a cell1. More than 60\(\%\) of drug targets are derived from membrane proteins owing to their role in mediating various interactions, such as those between cells and the extracellular environment, as well as interactions within membranes and organelles2. In addition, these proteins participate in multiple processes, such as material transportation, information transduction, energy conversion, and immune defense, and are indispensable in maintaining normal physiological functions in the human body. Different binding strengths and positions between proteins and membranes cause membrane proteins to have different functions; therefore, the identification of membrane protein types is essential for understanding cell function and drug design3.
Feature extraction is a key step in membrane protein function prediction, and multifeature fusion not only enhances their complementary but also improves the model prediction performance. Siva et al.4 used pseudoamino acid composition (PseAAC), amino acid composition (AAC), and physical and chemical properties to describe protein sequence features and employed a variety of tree-based classifiers to predict the fusion features. The results indicated that, based on the above feature description method, the random forest (RF) algorithm outperformed other classifier algorithms. Zhen et al.5 learned the sequence (sequence information, secondary structure and solubility) and mutual information features (paired contact potential) of nonmembrane proteins based on a deep migration learning method. This method differs from the traditional method, which does not directly use any features related to membrane proteins for the prediction of their types. Clares et al.6 regarded the evolutionary features and structural features of protein sequences as image features and then used image texture description methods (local phase quantization, local binary pattern, Fourier histogram features and rotation invariant binary Gabor patterns) to extract feature vectors. Anak Kaya7 used the minimum redundancy maximum relevance and correlation to select the best subfeatures from PseACC, and then the support vector machine (SVM) and k-nearest neighbors (KNN) were applied to predict membrane protein types. The results showed that the prediction performance of mRMR-SVM and CFS-SVM were better than those of mRMR-KNN and CFS-KNN based on the benchmark. Muhammad Arif et al.8 proposed the IMEM-2LSAAC algorithm to predict membrane protein types with two layers, and feature description methods, including the AAC, PseAAC and new split amino acid composition (SAAC) methods, were compared based on KNN, SVM, probabilistic neural network (PNN), generalized regression neural network (GRNN) and random forest (RF). The results indicated that the SAAC based on the SVM achieved better prediction performance. Zhang9 used a hybrid model to predict membrane protein types, and two strategies were used to construct a variety of protein interaction information for fusion: one was direct fusion, and the other was input into the node2vec network model for feature relearning and subsequent fusion. In addition, for each fusion feature, the synthetic minority oversampling technique (SMOTE) was used to solve the class imbalance problem. Ref.10 trained different deep learning models using protein sequence and protein evolution features to predict membrane protein types; for other related studies, see Refs.11,12. The above feature fusion approaches mostly adopt a concatenation method, resulting in high dimensionality after fusion and thus increasing the complexity of the classifier.
Evolutionary features provide significant guidance for predicting protein function and can help us better understand the structure, function and mechanism of proteins. Ahmad et al.13 employed the position relative incident matrix to extract features from protein sequences and then fed them into a multilayer neural network for feature relearning and classification. Muhammad Kabir et al.14 segmented the position-specific scoring matrix (PSSM), and then each subregion was described by the PsePSSM. The results confirmed that in many classifiers, the SVM achieve superior prediction performance. Li et al.15 used a recurrent neural network to relearn and classify the PSSM features of membrane proteins. The model achieved 94.1\(\%\) and 89.48\(\%\) prediction accuracy based on D1 and D2, respectively, because the model used the amino acids in the protein sequence as the time step of the neural network and made full use of the information provided by the PSSM to improve the prediction performance. Wang16 adopted average blocks, discrete wavelet transforms (DWTs), discrete cosine transforms and histograms of oriented gradient methods to extract features based on PSSMs; these fusion features use multikernel SVMs for prediction. Although the DWT can remove the singularity of feature points, it is likely to miss some effective discriminant features by smoothing the singularity points directly.
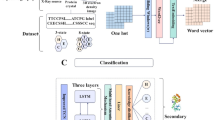
The key factors in solving this problem are the high dimensionality and heterogeneity among features after multifeature fusion. In summary, considering the superiority of the PSSM and the difference between traditional machine learning (TML) and deep learning (DL), a hybrid learning framework based on TML and DL is proposed. The classical TML features include Dpc-PSSM, Eedp-PSSM, DF-PSSM, Pse-PSSM, Kse-PSSM, Ab-PSSM, Rpm-PSSM and TG-PSSM to represent membrane proteins. Accordingly, five commonly used classical ML algorithms, the RF, SVM, extreme gradient boosting (XGBoost), KNN and gradient boosting tree (GBT) algorithms, were utilized to construct such TML baseline models. For DL, improved capsule neural network (ICNN) models were employed for feature reextraction. Furthermore, the prediction probabilities of the 41 baseline models (including the TML and DL baseline models) were investigated and selected as the final meta-features. We showed that the final meta model based on the stacking strategy achieves better overall performance than that of the other combination strategies on independent tests. The model framework is shown in Fig. 1.
Results and discussion
Performance evaluation of deep learning
The parameters and structure of CapNet were optimized, namely, the OCNN (optimized capsule neural network) and the ICNN (improved capsule neural network). To further compare the performance of CapNet, the OCNN, and the ICNN in predicting membrane protein types, we evaluated the test datasets of Dataset 1, Dataset 2, and Dataset 3. Pre, Se, F-m and Mcc were used as the evaluation indices, and the results are shown in Tables 1, 2 and 3. Moreover, Fig. 2 displays the variations in prediction accuracy and loss based on Dataset3 for CapNet, the OCNN and the ICNN. The results indicate that, compared to those of CapNet, the OCNN model achieved prediction accuracy improvements of 4.58\(\%\) and 2.08\(\%\) on the validation and test sets, respectively. Furthermore, the ICNN model achieved prediction accuracy improvements of 6.67\(\%\) and 2.78\(\%\) on the validation and test sets, respectively. The result of the ICNN model increased, demonstrating that increasing the model depth can effectively enhance its generalizability. For Dataset 1, the greater the Se value is, the better the predictive performance of the model for positive samples. For different membrane protein categories, the fluctuation ranges of the Se evaluation metric for the CapNet, OCNN, and ICNN deep models were 0–96\(\%\), 33–96\(\%\), and 50–97\(\%\), respectively. The ICNN model exhibited relatively smaller fluctuations, indicating its overall better predictive performance for positive samples. The MCC can measure the performance of classifiers, and based on different models, the multispanning membrane protein class achieved the highest predicted values. Furthermore, the macroaverage and weighted average of the evaluation metrics for the ICNN model were greater than those for the CapNet and OCNN models.
For Dataset 2 and Dataset 3, F-m is a comprehensive performance evaluation metric for addressing sample class imbalance issues. The fluctuation ranges on Dataset 2 were 15–96\(\%\), 9–96\(\%\), and 14–97\(\%\) for the different models. Compared to that of CapNet, F-m of the ICNN improved by 2\(\%\), but there is still significant room for improvement regarding sample imbalance issues. The fluctuation ranges on Dataset 3 were 11–96\(\%\), 36–96\(\%\), and 37–96\(\%\). In all three datasets, single-spanning membrane protein Class III had the fewest categories compared to those of the others, resulting in lower performance metrics. Conversely, the multispanning membrane protein class has a greater number of samples, leading to better predictive accuracy and performance metrics. In conclusion, the ICNN demonstrates significant performance improvements across various categories, such as Class I single-spanning membrane proteins and multispanning membrane proteins, in Dataset 1, Dataset 2, and Dataset 3. Additionally, the OCNN achieved minor improvements over CapNet, while the ICNN exhibited improvements over both the OCNN and CapNet. Finally, there is still considerable room for improvement in the performance of lipid chain-anchored membrane proteins and GPI-anchored membrane proteins in Dataset 2.
Figure 3 shows the mean and variance of Se, Sp, Mcc, F-m, OA and G-m for each membrane protein type under the CapNet, OCNN and ICNN models. The OA of the ICNN was higher than that of the other models, and the prediction performance for each type exhibited a smaller variance in different datasets. Second, the overall prediction performance of each type had an impact, the Mcc was lower, and the variance fluctuated greatly for each DL technique. Third, compared with that of the traditional feature description method based on local PSSMs, the OCNN model improved the prediction performance and alleviated the differences among the evaluation indices. In summary, the optimal results for the ICNN could be attributed to the following factors: (i) The model training time is increased after the batch_size is reduced and the convolution kernel size of the first convolution layer is increased, but the subsequent convolution operation achieves more effective features. (ii) By increasing the margin loss weight, the prediction accuracy of verification is improved effectively, while the loss value is reduced. (iii) A reduced dropout value not only avoids overfitting in the training process but also decreases the number of neural network parameters discarded and retains more valid feature information. (iv) The channel number of convolution layer three is decreased to better transmit information with the capsule layer.
Performance evaluation of traditional machine learning methods
To evaluate the impact of different classifiers on feature description methods, RF, KNN, light gradient boosting machine (LightGBM)17, SVM and XGBoost18 were compared based on independent test methods, and the parameters of each classifier were optimized via the grid search method. Each classifier successively predicts several classical feature description methods based on the PSSM: Dpc-PSSM, Rpm-PSSM19, Eedp-PSSM20, DF-PSSM21, Ab-PSSM, Smooth-PSSM, and TG-PSSM, and the results are shown in the blue histogram in Fig. 4.
The results of the blue histogram in Fig. 4 demonstrate that the average prediction accuracies of LightGBM and KNN for the above feature extraction methods were the best (92.59\(\%\), 87.83\(\%\) and 88.61\(\%\)) and the worst (90.41\(\%\), 81.71\(\%\) and 84.51\(\%\)), respectively. In addition, the Eedp-PSSM, Rpm-PSSM and TG-PSSM correspond to the Eedp-PSSM, PRSSM and TriPSSM description methods of Ref.22, respectively, but SVM rather than RF was adopted for prediction in this work. The results showed that the SVM was better than the RF based on the same feature description and parameter optimization of the classifier. The receiver operating characteristic (ROC) curve corresponding to each classifier is shown in Fig. 5. As shown in this figure, the same feature description method achieved different prediction performance under different classifiers, and the evaluation indices were positively correlated with the results in Fig. 4. Second, the orange histograms in Fig. 4 represent the prediction accuracies of XGBoost, KNN, LightGBM, SVM and RF for the corresponding meta-features. Although LightGBM was superior for each subfeature, it exhibited poor prediction accuracy in terms of its meta features. Based on the meta features, the prediction performance of LightGBM was lower than that of SVM, indicating that the generalizability of the model was weak. Moreover, for Dataset1, Dataset2 and Dataset3, SVM and RF, SVM and RF, and SVM and XGBoost achieved the best and worst performance, respectively, among the meta classifiers. Third, based on different classifiers, the prediction accuracy of meta features was generally greater than the average of the corresponding subfeature predictions for each classifier (the SVM obtained the highest increase of 4.03\(\%\), 5.76\(\%\) and 5.29\(\%\), respectively), indicating that meta features had a positive effect on improving the model prediction performance. Fourth, the same feature description methods yielded different prediction accuracies for different classifiers with the optimal parameters, indicating that different classifiers have complementary performance.
Comparison between DL and ML
Tables 4, 5 and 6 present the performance differences between the TML and DL methods. These tables show that the prediction accuracies of the ICNN were 3.05\(\%\), 3.86\(\%\), 12.83\(\%\), 3.35\(\%\), 2.93\(\%\), 4.2\(\%\), and 1.92\(\%\) greater than those of the Dpc-PSSM, Eedp-PSSM, DF-PSSM, Kse-PSSM, Ab-PSSM, Rpm-PSSM, and TG-PSSM for Dataset1, respectively. For Dataset2, the ICNN model results were 3.61\(\%\), 3.91\(\%\), 12.18\(\%\), 2.78\(\%\), 4.21\(\%\), 5.14\(\%\) and 1.66\(\%\) greater than those of Dpc-PSSM, Eedp-PSSM, DF-PSSM, Kse-PSSM, Ab-PSSM, Rpm-PSSM, and TG-PSSM, respectively. The ICNN model outperformed the Dpc-PSSM, Eedp-PSSM, DF-PSSM, Kse-PSSM, Ab-PSSM, Rpm-PSSM, and TG-PSSM models by 4.21\(\%\), 5.51\(\%\), 12.28\(\%\), 4.21\(\%\), 4.39\(\%\), 5.34\(\%\) and 2.69\(\%\), respectively, for Dataset3. In summary, based on the PSSM, the results indicate that DL is better than TML. In addition, the ICNN can better extract important features in the PSSM to improve membrane protein type prediction.
Among the many feature description methods based on the PSSM, the DF-PSSM and TG-PSSM achieved the worst and best prediction accuracies, respectively. However, the TG-PSSM increased the classifier complexity because it produced the highest dimensional eigenvectors (8000-D). Furthermore, to further compare the performance of the models, the Se, Mcc, F-m, OA, area under the receiver operating characteristic (AUROC) curve and area under the precision-recall (AUPR) curve were also used to compare and analyze the TG-PSSM and ICNN, and the detailed results are shown in Table 7.
Table 7 shows that the prediction accuracies of the ICNN were 1.91\(\%\), 1.55\(\%\) and 2.69\(\%\) higher than those of the TG-PSSM for Dataset1, Dataset2 and Dataset3, respectively. Second, for Dataset1, Dataset2 and Dataset3, the AUROC and AUPR of the ICNN were 1.5\(\%\) and 1.46\(\%\), 2.04\(\%\) and 1.61\(\%\), and 2.17\(\%\) and 2.32\(\%\) lower than those of the TG-PSSM, respectively. Third, the Se, Mcc and F-m of the ICNN were higher than those of the TG-PSSM, indicating that the TG-PSSM can better relieve sample imbalance and that they have some complementary effects. To intuitively validate the effectiveness of the model, t-distributed Stochastic Neighbor Embedding (t-SNE) was employed to visualize the reduced dimensions of the meta-features of the ICNN model. The ICNN model takes PSSM as input and is designed with an 8-layer deep neural network structure, comprising 3 convolutional layers, 3 pooling layers, and 2 capsule layers for feature enrichment. Specifically, to preserve more effective features and leverage the feature learning capability of capsule layers, the primary capsule layer is set with 8 capsules. The final prediction probabilities are generated through the output layer. t-SNE primarily utilizes the Kullback-Leibler divergence (KL divergence) to measure the difference between the conditional probability distributions in high-dimensional space and the Student-t distribution in low-dimensional space. It employs gradient descent to minimize the sum of KL divergences across all data points. After optimization, t-SNE outputs the positions of each data point in three-dimensional space, as illustrated in Fig. 6.
Performance of the hybrid meta model
In view of the diversity of the same feature extraction method for different classifiers in Sect. "Architecture of the proposed ICNN", the meta feature adds different classifier outputs. Therefore, the feature dimensions were increased from 64-D to 328-D, and the results are shown in Table 8. Compared with that of the other classifiers, the SVM achieved the best predictive performance except for Dataset1. Combined with the results in Sect. "Evaluation measurements", these results indicate that the SVM achieved better performance for processing meta features. Second, on account of the dataset, the F-m corresponding to KNN was highest. The metrics were 0.49\(\%\), 7.29\(\%\), 9.41\(\%\) and 9.45\(\%\); 9.55\(\%\), 12.06\(\%\), 15.89\(\%\) and 23.7\(\%\); and 6.26\(\%\), 4.39\(\%\), 16.69\(\%\) and 19.24\(\%\) greater than those of SVM, RF, LightGBM and XGBoost, respectively. The higher the value is, the less the presence of class imbalance. Third, the Se of the different models were lower than the Sp, indicating that the prediction accuracy of the hybrid model was lower than the error detection performance. Fourth, the meta features of the hybrid classifier improved the prediction accuracy compared with that of the meta features of a single classifier, and their average increased by 1.01\(\%\), 1.22\(\%\) and 0.12\(\%\). However, the highest prediction performance of the mixed classifier was still lower than that of the meta features of a single classifier.
Comparison of different ensemble strategies
To improve the prediction capability of the model, we constructed a new hybrid learning framework based on TML and DL. In this framework, we selected the top 1 to top 9 new feature vectors (8-72 D, where D represent dimensions) in descending order based on the Acc evaluation metric of each feature description method. While combination strategies can improve the stability of the model and avoid local optima, different combination strategies may result in different performance improvements. Therefore, we compared and analyzed the predictive performance of three combination strategies (majority voting, averaging, and stacking). The detailed comparison results are shown in Fig. 7, where the horizontal axis represents the nine selected new feature vectors. For Dataset1, the horizontal axes from 1 to 9 represent the meta features corresponding to the ICNN, TG-PSSM, Ab-PSSM, Dpc-PSSM, Kse-PSSM, Pse-PSSM, Eedp-PSSM, Rpm-PSSM, and DF-PSSM, respectively. For Dataset2, the horizontal axes from 1 to 9 represent the meta features corresponding to the ICNN, TG-PSSM, Kse-PSSM, Pse-PSSM, Dpc-PSSM, Eedp-PSSM, Ab-PSSM, Rpm-PSSM, and DF-PSSM, respectively. For Dataset3, the horizontal axes from 1 to 9 represent the meta features corresponding to the ICNN, TG-PSSM, Dpc-PSSM, Kse-PSSM, Ab-PSSM, Pse-PSSM, Rpm-PSSM, Eedp-PSSM, and DF-PSSM, respectively.
The results indicate that the performance of the average stacking method surpassed that of majority voting and averaging. First, for Dataset1, based on Dataset1, when the new feature vectors were selected as 4, 4, and 1, the stacking, majority voting, and average scoring combination strategies achieved the best prediction results, with accuracies of 95.85\(\%\), 95.66\(\%\), and 95.92\(\%\), respectively. Second, for Dataset2, when the new feature vectors were selected as 2, 1, and 1, the stacking, majority voting, and average scoring combination strategies achieved the best prediction results, with accuracies of 92.04\(\%\), 90.64\(\%\), and 90.51\(\%\), respectively. Finally, for Dataset3, when the new feature vectors were selected as 5, 1, and 2, the stacking, majority voting, and average scoring combination strategies achieved the best prediction results, with accuracies of 93.15\(\%\), 92.76\(\%\), and 93.37\(\%\), respectively.
Table 9 presents a performance comparison of the best new eigenvectors under different combination strategies. First, Sp was greater than Se, indicating that our model’s diagnostic performance for negative samples was greater than that for positive samples. Higher values of both indicate better model performance. Second, under the optimal combination strategy, the hybrid model outperformed the best classical feature, TG-PSSM, by 2.18\(\%\), 3.10\(\%\), and 3.30\(\%\) across the three datasets and outperformed the best deep learning model, ICNN, by 0.26\(\%\), 1.55\(\%\), and 0.61\(\%\). Again, this finding demonstrates that the hybrid model can enhance the prediction performance. Third, across Dataset1, Dataset2, and Dataset3, majority voting exhibited the lowest prediction accuracy. These values were lower than those of the stacking method by 0.19\(\%\), 1.42\(\%\), and 0.39\(\%\) and lower than those of the averaging method by 0.26\(\%\), 0.13\(\%\), and 0.61\(\%\), respectively. This finding is attributed to the fact that stacking and averaging methods employ different strategies to relearn meta features. Although majority voting is the simplest approach, it may overlook discriminative minority outputs.
In Fig. 8, subplots A, B, C, and D represent the comparison of prediction accuracies for different feature description methods and the different evaluation metrics corresponding to each membrane protein type under the best hybrid models based on Dataset1, Dataset2, and Dataset3. Under the optimal combination strategy, the hybrid model outperformed the best feature description methods in traditional machine learning methods by 2.18\(\%\), 3.1\(\%\), and 3.3\(\%\) and surpassed the worst feature description methods in traditional machine learning methods by 13.09\(\%\), 13.62\(\%\), and 12.89\(\%\), respectively. These data once again validate the effectiveness of our model. In the box plot, a significant fluctuation in the F-m score was observed compared to the other evaluation metrics, which is attributed to the relatively low sample count of the single-span type III evaluation metric compared to that of the others. Compared to the F-m scores of the DL models, the optimal hybrid model improved the F-m score by 4.84\(\%\), 6.5\(\%\), and 3.12\(\%\), respectively. Overall, the results once again demonstrate that the proposed hybrid model not only exhibits better predictive performance but also improves prediction accuracy, addressing the issue of sample imbalance.
Comparison and analysis with other models
To verify the effectiveness of the proposed model, we compared it with other existing methods. To ensure fairness, the methods used for comparison were all based on the same data and validation methods. These methods include PsePSSM_Ensemble23, Physicochemical-Ensemble24, PsePSSM-LLDA25, PsePSSM-DC26, PsePSSM-PCA25, FEA-Fusion27, the sequence information model (SIM)22, CapNet22, recurrent neural network (RNN)15, Ave-WT16, and MKSVM-HSIC16. Most of these methods are based on the PSSM for feature extraction. The results are reported in Table 10.
On Dataset1, the performance values of the OCNN were 0.2\(\%\) and 13.9\(\%\) greater than those of the traditional classical algorithm (FEA-Fusion) and the worst algorithm (PsePSSM-PCA), respectively, and 0.5\(\%\) and 3\(\%\) greater than those of the RNN and SIM, respectively. The performance values of the ICNN method were 1.3\(\%\) and 15\(\%\) better than those of the best (FEA-Fusion) and worst (PsePSSM-PCA) TML methods and 1.6\(\%\) and 4.1\(\%\) greater than those of the RNN and SIM, respectively. Second, the gaps between the validation set and the CapNet, SIM, and ICNN test sets were 3.5\(\%\), 6.1\(\%\) and 0.9\(\%\), respectively. Compared with those of the CapNet and SIM models, those of the ICNN were lower by 2.6\(\%\) and 5.2\(\%\), respectively, indicating that the model has good generalizability. Third, based on Dataset2, the performance values of the OCNN were 0.7\(\%\) and 12.2\(\%\) greater than those of the best model (MKSVM-HSIC) and the worst model (PsePSSM_Ensemble), respectively, and 1.1\(\%\) and 1.4\(\%\) greater than those of the RNN and SIM of DL, respectively. Fourth, compared with the performance of the other algorithms, the prediction performance of the hybrid model was the best. Although the calculations of the hybrid model are relatively complicated, the meta feature dimension of the final construction is relatively low, reducing the complexity of the calculation of the classifier. Furthermore, based on Dataset2, the prediction accuracy of the hybrid model was 2.5\(\%\) higher than that of the ICNN.
Materials and methods
Dataset
To evaluate the performance of the model, we utilized datasets similar to those used in cutting-edge research in the field; these datasets were constructed by Chou23, Chen28 and Li28. The specific distribution of these datasets is presented in Table 11. The protein sequence length and probability density distribution for each membrane protein sequence are depicted in Fig. 9. Dataset 1 was obtained from the Swiss-Prot database. To ensure a balanced number of sequences between the training and testing datasets, Chou23 employed a percentage-based random allocation method. Dataset 2 was derived by removing redundant protein sequences from Dataset 1, as performed by Chen28 et al. The filtration of nonredundant sequences primarily involves two steps: utilizing CD-hit to filter redundant sequences and ensuring that the remaining sequences have a sequence identity of no more than 40\(\%\) with any other sequence. To obtain updated data, Chen28 and Li28 constructed a new dataset from the Swiss-Prot database, applying the following criteria for sequence selection: (1) removal of protein sequences with lengths less than 50 amino acid residues, (2) exclusion of proteins annotated with multiple topologies or nonexperimental qualifiers in topology, and (3) elimination of protein sequences with a sequence identity exceeding 40\(\%\) using CD-hit. All three datasets include eight types of membrane proteins, and 20\(\%\) of each training set was randomly selected as the validation set to optimize the model parameters.
Architecture of the proposed ICNN
PSSM description
As organisms gradually evolve, some residues in protein sequences may be replaced, inserted, or deleted. Mutation of residues in the primary protein sequence may cause changes in the high-level structure and function of the protein, which may lead to some diseases. The PSSM method is a method that calculates the possibility of amino acid peptide mutation at a certain position29, which not only contains the order information of the protein sequence but also expresses the implicit information in the process of protein evolution via numerical information. By searching and comparing each protein sequence with the homologous sequences in the NCBI NR database (ftp://ftp.ncbi, nih.gov/blast/db/), a PSSM can be obtained. For any protein sequence P with length L, the PSSM can be expressed as follows:
where W(i,k) represents the ratio of the k-th benchmark amino acid residues that appear at the i-th position in the target protein sequence in a similar protein group. Y(j,k) denotes Dayhoff’s mutation matrix30 between the j-th and k-th amino acid peptides, which represents the score of amino acid mutation from the i-th original position to the j-th along the protein sequence, and finally, a score matrix of 20\(\times \)20 is obtained. When \({M}_{i,j}\) is positive, the value becomes larger, which indicates that the residues are more likely to be replaced at this position.
ICNN
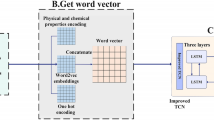
To improve the prediction performance of membrane protein types, the parameters and structure of the capsule neural network (CapNet) were optimized. The architecture of the ICNN is shown in Fig. 10. The ICNN contains three 2D convolutional layers, three average pooling layers, two primary capsule layers and one fully connected layer. Each 2D convolutional layer includes a dropout that used to obtain more valid features while preventing model overfitting. The length of the PSSM was normalized and then fed into the first 2D convolutional layers (number of filters: 256; kernel size: (\(7\times {7}\)); activation function: rectified linear unit (ReLU); and stride: 1). Fig. 9 shows that the sequence length range is 50–5000 amino acids, and more than 98\(\%\) of protein sequence lengths are shorter than 1500 amino acids. Therefore, the input length of the model is fixed to 1500 amino acids. To improve the model generalizability and prevent overfitting, the batch size, margin loss and dropout were set to 8, 0.25 and 0.3, respectively. The learning rate and optimizer were set to 0.0001 and 0.0001, respectively.
Dynamic routing algorithm and loss function
CapNet was proposed by Geoffrey Hinton31 and can better express the relationships between model hierarchies; additionally, it is more suitable for small sample data. The capsule network layer uses vectors rather than scalars as the inputs and outputs of the model. Moreover, instead of traditional back propagation, the dynamic routing algorithm is used to update the weights of the capsule neural network, alleviating the loss of important information in the max pooling layer using most DL approaches. The propagation process between capsule units is shown in Fig. 11.
The bottom layer input vector of the capsule network model is \({{u}_{i}}\), and the mapping matrix \({{w}_{ij}}\) is multiplied to obtain the feature vector \({{\hat{u}}_{j|i}}\) of the high-level capsule. The output \({{v}_{j}}\) of each capsule is obtained by the sum of the weighted \({{u}_{i}}\) input to the nonlinear compression function.
The sum of all coupling coefficients between the capsule and the upper layer is 1, and the coupling coefficient \({{c}_{ij}}\) is updated by dynamic routing every iteration32. The similarity is determined by the direction between the input vector \({{\hat{u}}_{j|i}} \)and the output vector \({{v}_{1}}\) at a high-level, and the updating of the dynamic routing coupling coefficient will be reduced when the dot product is negative, otherwise.
The similarity correlation score between capsule i and capsule j for each update iteration is calculated by \({{b}_{ij}}\):
Each capsule output vector uses the squash nonlinear function33, so that the modulus length is distributed between 0 and 1. This feature is more likely to exist when the modulus length is closer to 1. The output vector \({{v}_{j}}\) of capsule j is calculated by Eqs. (7) and (8), and the \({{s}_{j}}\) is all of its inputs.
where \(\frac{{{s}_{j}}}{||{{s}_{j}}||}\) represents a direction vector of length 1, and the \(\frac{||{{s}_{j}}|{{|}^{2}}}{1+||s|{{|}^{2}}}\) represents the length.
The reconstruction loss function can force the network to retain all the needed information for reconstructing the features, reducing the risk of overfitting and improving the generalizability of the model. The marginal loss function34 for class C is as follows:
The object exists for class C when Tc=1; otherwise, Tc=0. m+ and m- are the upper and lower bounds, respectively, and the initial values are usually set to 0.9 and 0.1, respectively. lambda \(\text {=}0.5\) is used to avoid weakening the activity vectors of all classes during initial network learning, and the reconstruction loss function generally uses the mean square error (MSE).
Evaluation measurements
To objectively compare the prediction performance with that of other works, several measurement metrics were applied to evaluate the model, including the overall accuracy (OA), sensitivity (Se), specificity (Sp), precision (Pre), G-mean (G-m), F-measure (F-m) and Mathew’s correlation coefficient (Mcc)35, which are defined as follows:
where \({{N}^{+}},{{N}^{-}}\) denote the total numbers of positive samples and negative samples, respectively. \(N_{+}^{-}\) represents the number of false positives, which indicates the number of true negative samples incorrectly predicted as positive samples, and \(N_{-}^{+} \)is the number of false negatives, which indicates the number of true positive samples incorrectly predicted as negative samples (Supplementary Information).
Conclusion
Considering that CapNet can better address small sample problems, it was applied to predict membrane protein types. First, the size of the convolution kernel, the weight of the margin loss function, the dropout and other parameters of the CapNet were optimized to alleviate overfitting on the training set. Second, to avoid effective feature loss, the model structure between the network layer and the capsule layer was optimized to boost the model generalizability. A hybrid framework was constructed based on DL and TML, and the subfeatures were compared and analyzed using three combination strategies for optimal traversal. The experimental results showed that the hybrid model not only improves the model prediction performance but also addresses the class imbalance problem. However, investigations could still be improved in the future; for example, the loss function of a new DL can be constructed to solve the problem of class imbalance in membrane protein type prediction. Moreover, in future research, to better utilize our method, we will establish a user-friendly web server for convenience, as we hope our method can make a certain contribution to the prediction of membrane protein types.
Data availability
The code and dataset can be free download at https://github.com/ruanxiaoli/membrane-protein-types, and separate data sets are provided in Supplementary Information for readers to download.
References
Levental, I. & Lyman, E. Regulation of membrane protein structure and function by their lipid nano-environment. Nat. Rev. Mol. Cell Biol. 24(2), 107–122 (2023).
Zhang, X. & Chen, L. Prediction of membrane protein types by fusing protein-protein interaction and protein sequence information. Biochim. Biophys. Acta (BBA)-Proteins Proteomics 1868(12), 140524 (2020).
Gulsevin, A. et al. Template-free prediction of a new monotopic membrane protein fold and oligomeric assembly by Alphafold2. Biophys. J . 122(3), 194a (2023).
Sankari, E. S. & Manimegalai, D. Predicting membrane protein types using various decision tree classifiers based on various modes of general PseAAC for imbalanced datasets. J. Theor. Biol. 435, 208–217 (2017).
Li, Z. et al. Predicting membrane protein contacts from non-membrane proteins by deep transfer learning. (2017). Preprint at http://arxiv.org/abs/1704.07207.
Clares, J. D. et al. Improved Image Based Protein Representations with Application to Membrane Protein Type Prediction (IEEE, 2017).
Kaya, G. A. & Kamal, N. A. M. Feature selection for human membrane protein type classification using filter methods. IAES Int. J. Artif. Intell. 8(4), 375 (2019).
Arif, M., Hayat, M. & Jan, Z. iMem-2LSAAC: A two-level model for discrimination of membrane proteins and their types by extending the notion of SAAC into Chou’s pseudo amino acid composition. J. Theor. Biol. 442, 11–21 (2018).
Zhang, X. et al. Identification of human membrane protein types by incorporating network embedding methods. IEEE Access 7, 140794–140805 (2019).
Guo, L. et al. Accurate classification of membrane protein types based on sequence and evolutionary information using deep learning. BMC Bioinform. 20, 1–17 (2019).
Sankari, E. S. & Manimegalai, D. Predicting membrane protein types by incorporating a novel feature set into Chou’s general PseAAC. J. Theor. Biol. 455, 319–328 (2018).
Alphonse, A. S., Mary, N. A. B. & Starvin, M. S. Classification of membrane protein using tetra peptide pattern. Anal. Biochem. 606, 113845 (2020).
Butt, A. H. et al. A prediction model for membrane proteins using moments based features. BioMed. Res. Int. 2016, 8370132 (2016).
Kabir, M. et al. Prediction of membrane protein types by exploring local discriminative information from evolutionary profiles. Anal. Biochem. 564, 123–132 (2019).
Wang, S. et al. Efficient utilization on PSSM combining with recurrent neural network for membrane protein types prediction. Comput. Biol. Chem. 81, 9–15 (2019).
Wang, H. et al. Identification of membrane protein types via multivariate information fusion with Hilbert-Schmidt independence criterion. Neurocomputing 383, 257–269 (2020).
Li, L. et al. A LightGBM-based strategy to predict tunnel rockmass class from TBM construction data for building control. Adv. Eng. Inform. 58, 102130 (2023).
Ben Jabeur, S., Stef, N. & Carmona, P. Bankruptcy prediction using the XGBoost algorithm and variable importance feature engineering. Comput. Econ. 61(2), 715–741 (2023).
Zhou, S. et al. PredLLPS_PSSM: A novel predictor for liquid-liquid protein separation identification based on evolutionary information and a deep neural network. Brief. Bioinform. 24(5), d299 (2023).
Nadia, Gandotra E. & Kumar, N. Comparison of machine learning techniques for predicting NLR proteins. Biomed. Eng. Appl. Basis Commun. 35(02), 2250050 (2023).
Sofi, M. A. & Arifwani, M. Improving Prediction of Amyloid Proteins Using Secondary Structure Based Alignments and Segmented-PsSm (IEEE, 2021).
Guo, L. et al. Accurate classification of membrane protein types based on sequence and evolutionary information using deep learning. BMC Bioinform. 20(25), 1–17 (2019).
Chou, K. & Shen, H. MemType-2L: A web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM. Biochem. Biophys. Res. Commun. 360(2), 339–345 (2007).
Nanni, L. & Lumini, A. An ensemble of support vector machines for predicting the membrane protein type directly from the amino acid sequence. Amino Acids 35, 573–580 (2008).
Wang, T. et al. Predicting membrane protein types by the LLDA algorithm. Protein Pept. Lett. 15(9), 915–921 (2008).
Wang, T., Xia, T. & Hu, X. Geometry preserving projections algorithm for predicting membrane protein types. J. Theor. Biol. 262(2), 208–213 (2010).
Guo, L. et al. Prediction for membrane protein types based on effective fusion representation and MIC-GA feature selection. IEEE Access 6, 75669–75681 (2018).
Chen, Y. & Li, K. Predicting membrane protein types by incorporating protein topology, domains, signal peptides, and physicochemical properties into the general form of Chou’s pseudo amino acid composition. J. Theor. Biol. 318, 1–12 (2013).
Liu, S. et al. Integrating reduced amino acid composition into PSSM for improving copper ion-binding protein prediction. Int. J. Biol. Macromol. 244, 124993 (2023).
Duarte-González, M. E. et al. Cyclic concatenated genetic encoder: A mathematical proposal for biological inferences. Biosystems 163, 47–58 (2018).
Qin, Y., Guan, Y. L. & Yuen, C. Spatiotemporal capsule neural network for vehicle trajectory prediction. IEEE Trans. Veh. Technol. 72, 9746–9756 (2023).
Madhu, G. et al. Intelligent diagnostic model for malaria parasite detection and classification using imperative inception-based capsule neural networks. Sci. Rep. 13(1), 13377 (2023).
Weyori, B. A., Afriyie, Y. & Opoku, A. A. Analyzing the performances of squash functions in capsnets on complex images. Cogent Eng. 10(1), 2203890 (2023).
Li, J. et al. A hybrid neural network model based on optimized margin softmax loss function for music classification. Multimed. Tools Appl. 83, 1–36 (2023).
Li, Y. et al. DeepCellEss: Cell line-specific essential protein prediction with attention-based interpretable deep learning. Bioinformatics 39(1), c779 (2023).
Acknowledgements
This work was supported in part by the Guizhou Provincial Basic Research Program (Natural Science) under GrantZK[2023]YB054, in part by the National Natural Science Foundation of China under Grant 62166005, in part by the Guizhou University Talents Project under Grant [2022]29, and in part by the Developing Objects and Projects of Scientific and Technological Talents in Guiyang City under Grant ZKH[2023]48-8.
Author information
Authors and Affiliations
Contributions
X. R. wrote the main manuscript text, conducted all the simulations, and provided research ideas. S. X. revised the paper, prepared it, and assisted with manuscript writing. S. L. prepared all figures and tables presented within the manuscript. Z. S. played a significant role in data collection, analysis, and interpretation. J. Y. assisted with manuscript writing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ruan, X., Xia, S., Li, S. et al. Hybrid framework for membrane protein type prediction based on the PSSM. Sci Rep 14, 17156 (2024). https://doi.org/10.1038/s41598-024-68163-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-68163-7