Abstract
Osteoarthritis (OA) is the most common degenerative joint disease, leading to severe pain and functional disability for nearly 530 million people worldwide. OA is characterized by progressive loss of cartilage and synovial hyperplasia from the articulating surfaces of any diarthrodial joints, however, the majority of cases account for the hip and knee. Currently, regenerative therapy based on stem cells has emerged as one of the most promising and rapidly evolving strategies in OA. Although progression and potential regeneration can be monitored by magnetic resonance imaging (MRI), we lack proper molecular markers of joint regeneration for diagnostic and in vitro studies. Gene expression profiles of articular cartilage (chondrocytes) and synovium from OA-affected patients’ were downloaded from The Gene Expression Omnibus database (GSE179716, GSE206848, GSE239343, GSE48556) and analyzed using various bioinformatic tools and platforms: GEO2R, FunRich, C-Big, The Human Protein Atlas, STRING, Orange data mining, Jasp, Gene Ontology and Reactome. OA-affected synovium and chondrocytes present differences between aurora B and A signaling. However, major biological pathways are similarly enriched with SRI, SMC2, PSIP1, TLE4, and MSX1 genes identified as prominent molecular biomarkers of OA progression and mesenchymal stem cell-based OA regeneration. Additionally, in peripheral blood mononuclear cells (PBMCs) from OA patients PSIP1 and TLE4 present (respectively) down and up-regulated mRNA levels. mRNA expression levels of chosen genes can indicate OA progression mainly in the in vitro studies, whereas the mRNA level ratio of PSIP1:TLE4 from PBMCs derived from OA patients can help monitor OA progression in clinical practice.
Similar content being viewed by others
Introduction
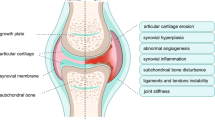
Osteoarthritis (OA) - a complex progressive degenerative disease of the whole joint- is one of the most common types of arthritis and the leading form of disability, affecting nearly 530 million patients worldwide (approximately 7% of the total human population)1. Globally, in the past 30 years, the incidence rate of OA increased by 113.25%, from 247.51 million OA-affected patients in 1990 to 527.81 million in 20192. Clinical data showed that OA is characterized by cartilage degeneration, synovial inflammation, subchondral bone thickening, and osteophyte formation, leading to severe structural alterations of the whole joint, causing pain and decreased joint flexibility3. Its etiology and major risk factors can be categorized into two subgroups according to the type of stimuli: (1) mechanical, such as joint injury and obesity that significantly increases wear and tear, and (2) non-mechanical, such as excessive and prolonged inflammation resulting in the generation of reactive oxygen species and chemokine secretion, dysregulation of signaling pathways, and mutations in key genes involved in the development of OA3,4,5. On the molecular basis, OA is characterized by dysregulation of Wnt/β-catenin, NF-κB, TGFβ/ΒΜP and FGF signaling pathways, resulting in increased expression of proteolytic enzymes and pro-inflammatory factors: ADAMTS-5, matrix metalloproteinases (MMP1, MMP2, MMP3, MMP9, MMP13), tumor necrosis factor-alpha (TNF-α), interleukin 1 beta (IL-1β), interleukin 6 (IL-6), as well as the deposition of collagen X, that results in imbalance in the delicate equilibrium of ECM synthesis and degradation in cartilage and synovium as well as altering the subchondral bone quality4,6,7.
Due to its physiology, cartilage has little self-renewal ability8. Currently, no drugs can reverse the progression of OA and prevent long-term disability9. The recommended treatments, such as non-steroidal anti-inflammatory drugs (NSAIDs), glucocorticoids, opioids, chondroprotective agents, and cytokine inhibitors, reduce the pain and improve joint functionality10,11. Recently, regenerative therapy based on mesenchymal stem cells (MSC) has emerged as one of the most promising and rapidly evolving strategies to treat OA12. In the case of OA patients, the course of the disease can be monitored by magnetic resonance imaging (MRI) and ultrasonography (USG)13. However, standard MRI analyses can predominantly detect alterations seen in the advanced stages of OA and might possess low sensitivity for early, subtle, and potentially reversible changes14. OA affecting the whole joint, dysregulates molecular, structural, and physiological homeostasis of different cell/tissue types, that create joints and significantly change their biochemical properties. Therefore, the identification of precise molecular markers that could be utilized to monitor OA progression and potential incremental regeneration and therapeutic outcomes is an urgent matter.
Recently, bioinformatic analyses of genomic, transcriptomic, and proteomic data, often supported by AI-driven engines, by simultaneous analysis of hundreds of records, proved to be useful in the selection of the new markers or important, dysregulated signaling pathways in many different disorders such as cancer or acute myocardial infarction15,16,17,18. Herein, we present the application of integrated bioinformatic tools to analyze transcriptomic data of OA-affected patients deposited in the publicly available Gene Expression Omnibus database (GEO) to identify genes encoding proteins directly involved in osteoarthritis development that could be utilized as prognostic biomarkers.
Materials and methods
Microarray data processing and analysis
Gene expression profiles with accession numbers: GSE were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/ accessed on 30 December 2024) and analyzed similarly to our previous work using the GEO2R online tool and integrated bioinformatics16,17. The Benjamini and Hochberg adjusted p-value (Padj.) from the stats package in R, which is the transformation of the p-value using the false discovery rate (FDR) – [P_values, method="fdr”], was used to set statistically significant differently expressed genes (DEGs)19.
The GSE179716 dataset is composed of transcriptomic data from chondrocytes derived from OA-affected patients (n = 3 early stage and n = 3 late stage). Acquired by Biomedical Informatics, University of Pittsburgh, Pittsburg, PA, USA.
The GSE206848 dataset is composed of transcriptomic data from synovium isolated from n = 7 normal (non-OA-affected) and n = 9 late OA patients. Acquired by NYU GROSSMAN SCHOOL OF MEDICINE, New York, NY, USA.
The GSE239343 dataset is composed of transcriptomic data (from n = 3 independent experiments) from chondrocytes (HC #P10970 https://innoprot.com/), inflamed chondrocytes (TNF-inflamed chondrocytes: cultured in a mix of serum-free DMEM and 1% penicillin/streptomycin with 25 ng/mL TNF), and inflamed chondrocytes treated with conditioned medium and conditioned medium (50 µg/mL) from adipose tissue-derived mesenchymal stem cells (AD-MSCs). Acquired by CIC bioGUNE, Genome Analysis Platform, Bizkaia, Spain as described in20.
The GSE48556 dataset is composed of transcriptomic data obtained from peripheral blood mononuclear cells (PBMCs) of n = 33 healthy controls and n = 106 OA patients, Acquired by Leiden University Medical Center, Department of Molecular Epidemiology, Leiden, Netherlands21.
Bioethical committee approval and patient consent statements are presented with the original publications20,21 and datasets, available in The Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/geo/ reference numbers: GSE239343, GSE179716, GSE206848, and GSE48556.
Enrichment analysis
Functional enrichment software tool FunRich (v3.1.3) (http://www.funrich.org/) supported by Gene Ontology (GO) (http://geneontology.org/) (accessed on 30 December 2024) database was used to compare, analyze, and visualize the Biological Process (BP) and Cellular Components (CC) differences associated with the proteins encoded by differently expressed genes (DEGs) in OA-affected and normal patient samples. The cut of was set using p-value < 0.05.
Hierarchical clustering analysis
After extracting the expression values from the gene expression profiles, a bidirectional hierarchical clustering heatmap of differently expressed genes (DEGs) was calculated and visualized using the Orange open-source machine learning and data visualization platform as previously described by us16,17.
Pearson correlation analysis
Pearson correlation analysis of chosen DEGs was calculated and visualized using JASP 0.14.1.0 software (https://jasp-stats.org/ accessed on 30 December 2024) with a color depiction of “r” value and significance * p < 0.05, ** p < 0.005, and *** p < 0.001 as previously described by us16,17.
Radial data visualization
Radial data visualization was performed using Orange data mining 3.31.1 software as described by us using a modified Radial Visualization (RadViz) algorithm17,22,23. Briefly: data were subjected to a data-driven linear transformation, using the RadViz algorithm, which focuses on visualizing multivariate data in a two-dimensional (2D) space, optimally preserving the data structure. This method visualizes data represented by points inside a circle, using data attributes (mRNA expression level) of N number variables (genes) equidistantly distributed along the circle’s circumference. Next, data are placed as points in a location set by the mutual interaction of attributes of N variables, assuming that the greater the attribute value, the closer the data point is drawn. Thus, finally, the data point is placed where the sum of all forces generated by the data value equals 022,23. The homogeneity of formed data clusters was verified using Inner Cluster Deviation (wc).
ri center of Ci cluster.
Protein-protein interaction (PPI) network
A PPI network of proteins encoded by chosen genes was created using STRING version 12.0 online software (https://string-db.org/). Network analysis was performed using Cytoscape software version 3.9.1 (https://cytoscape.org/), the Gene Ontology (GO) database (https://amigo.geneontology.org/ ) and the Reactome database (https://reactome.org/ ) as previously reported in16,24. Active interaction source: “Text-mining”, “Experiments”, “Databases”, “Co-expression”, “Neighborhood”, “Gene Fusion”, “Co-occurrence”. Minimum required interaction score: 0.4. Data were additionally verified using the Reactome 2015 database and the Pathway-to-Gene option of the Pathway Connector bioinformatics web tool (https://bioinformatics.cing.ac.cy/ ) run by the Bioinformatics Department (C-BIG) at the Cyprus Institute of Neurology & Genetics (CING) and the High-Performance Computing Facility (HPCF) of the Cyprus Institute (CyI).
Protein visualization
The tissue sections (immunohistochemistry) and immunofluorescence (confocal microscopy) images of cellular localization of proteins encoded by chosen genes were downloaded from The Human Protein Atlas (HPA) (https://www.proteinatlas.org/)25. Immunohistochemistry imaging was performed using antibodies: anti-Sorcin (HPA073666), anti-Structural Maintenance Of Chromosomes 2 (HPA071309), anti-PC4 And SRSF1 Interacting Protein 1 (HPA019697), and anti-Msh Homeobox 1 (CAB026198). Transducin-like enhancer of split protein 4 tissue immunohistochemistry data are not available (NA) in the HPA database. Immunofluorescence imaging was performed using U-2 OS human osteosarcoma cell line and antibodies: anti-Sorcin (HPA019004), anti-Structural Maintenance Of Chromosomes 2 (HPA071309), anti-PC4 And SRSF1 Interacting Protein 1 (HPA019697), anti-Transducin-like enhancer of split protein 4 (HPA065357), and anti-Msh Homeobox 1 (HPA063895).
Construction of the receiver operating characteristic curve (ROC)
To evaluate the predictive capabilities of the chosen gene, the Area Under the Receiver Operating Characteristic curve (AUC-ROC) curve was calculated and visualized using the JASP 0.16.0.0 software.
Statistics
Statistical evaluation was performed using the normality test (Shapiro–Wilk) followed by the T-Student test (for normally distributed data) or Mann-Whitney U test (for not normally distributed data). Multifactorial statistical evaluation was performed using the ANOVA (one-way) test with Tukey post hoc correction. Calculations and graphs were performed using Orange data mining 3.31.1 software and JASP 0.16.0.0 software; p values < 0.05 were considered statistically significant for all analyses: * p < 0.05; ** p < 0.005; *** p < 0.001, NS-not statistically significant.
Results.
Identification of major OA-related dysregulated genes and biological processes enrichment
OA, as a complex joint degenerative condition, affects several tissue and cell types, resulting in different molecular responses. Thus, to select useful molecular markers to monitor OA progression and joint regeneration upon given therapy, we decided to analyze chondrocytes and synoviocytes from normal and OA-affected knees to find dysregulated genes and molecular pathways (workflow schematic depiction is presented in Fig. 1). We downloaded patients’ transcriptomic data deposited in the Gene Expression Omnibus database: the GSE179716 data set is composed of transcriptomic data from chondrocytes derived from OA-affected patients, and the GSE206848 data set is composed of transcriptomic data from synovium. Next, using the GEO2R tool, statistically significant (Padj.<0.05), differently expressed genes (DEGs) were selected for further analysis, receiving 1683 up regulated and 1641 down regulated genes for OA-affected synoviocytes and respectively 3038 and 1513 for OA-affected chondrocytes. Using the Gene Ontology database, and functional enrichment analysis platform FunRich, we noticed that many biological pathways were similarly affected by ongoing OA in both chondrocytes and synovium (Tables 1 and 2). However, significant differences were also noticed, especially once including Aurora B and A signaling that may cause senescence and heterochromatin instability, EMT, and overall mechanisms of gene regulation (Tables 1 and 2). Representative Biological Pathways were presented in Fig. 2A, B. Nevertheless, to find potential molecular biomarkers of OA progression, we have selected genes that were up or downregulated in both OA-affected chondrocytes and synoviocytes, receiving respectively 142 and 319 DEGs (Fig. 3A). Obtained PPI network was further analyzed using Cytoscape 3.9.1. Average metrics, for PPI network of up-regulated genes and for PPI network of down-regulated genes, of how often a node appears on the shortest paths between other nodes – BetweennessCentrality are respectively: 0.036 and 0.019, average shortest path from a node to all other nodes – ClosenessCentrality are: 0.389 and 0.267, numerical depiction of how interconnected a node’s neighbors are – ClusteringCoefficient are: 0.234 and 0.265 and the average number of direct interactions (edges) a node has (Degree) 3.037 and 4.172. The highest Degree in the PPI network of up-regulated genes presents AKT1 (22) and for PPI network of down-regulated genes EGFR (44). Analysis of Fold change enrichment of Biological Processes and cellular components proved that the major impact is related to apoptosis, immune response, cell adhesion, migration, cell-matrix degradation, and ion transport (Fig. 3B).
Schematic presentation of research design. GSE179716 and GSE206848 datasets were analyzed to identify OA-related DEGs (142 upregulated and 319 downregulated) common for chondrocytes and synovium, supported by enrichment analysis. Next, DEGs from control chondrocytes and OA chondrocytes (obtained by TNF-related inflammation) from the GSE239343 dataset were compared to the obtained OA-related DEGs. Common DEGs patterns were then analyzed, and only genes that present dysregulated expression upon OA and regenerated level upon AD-MSC conditioned medium treatment were selected: SRI, SMC2, PSIP1, TLE4, and MSX1. Biological pathways enrichment of proteins encoded by chosen genes was analyzed using various bioinformatical tools and platforms. Finally, selected genes expression levels were validated in PBMCs using the GSE48556 dataset with PSIP1 and TLE4, presenting significant OA-predictive value. Image provided by Servier Medical Art (https://smart.servier.com/), licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Functional enrichment analysis of up and down-regulated genes for OA-affected chondrocytes and synoviocytes. Data were downloaded from the GEO database GSE179716 and GSE206848. Major similarly (A) and differently (B) enriched biological pathways were analyzed using FunRich (v3.1.3) (http://www.funrich.org/) software supported by Gene Ontology (GO) (http://geneontology.org/).
Selection and functional analysis of OA-related genes. Data were downloaded from the GEO database GSE179716 and GSE206848. 142 upregulated and 319 down-regulated genes commonly dysregulated in OA-affected chondrocytes (GSE179716) and synoviocytes (GSE206848) were selected and visualized by Venn diagram using FunRich (v3.1.3) (http://www.funrich.org/) software (A). Protein-protein interaction network visualized using STRING platform. Active interaction source: “Text-mining”, “Experiments”, “Databases”, “Co-expression”, “Neighborhood”, “Gene Fusion”, “Co-occurrence”. Minimum required interaction score: 0.4. Network analysis performed via Cytoscape 3.9.1. presenting average value of BetweennessCentrality, ClosenessCentrality, ClusteringCoefficient, and Degree calculated for each obtained network. Analysis of Fold change enrichment of Biological Processes and cellular components performed and visualized using FunRich (v3.1.3) (http://www.funrich.org/) software supported by Gene Ontology (GO) (http://geneontology.org/) (B).
Selection of potential markers for OA regeneration based on AD-MSCs therapy
In the next step of our analysis, we decided to use patient-derived data as reference data to select genes that, in the case of chondrocytes, were dysregulated upon inflammation and restored following the treatment with adipose tissue-derived mesenchymal stem cells (AD-MSCs). Thus, having established common OA-related differently expressed genes (DEGs) for chondrocytes and synoviocytes, we tried to select genes which the expression level would be restored by mesenchymal stem cell-based therapy. Hence, we downloaded data from the GSE239343 data set, composed of transcriptomic data from chondrocytes (named Control), TNF-α treated chondrocytes (named Inflammation), that to some extent mimics OA-related joint inflammation and TNF-α inflamed chondrocytes treated with conditioned medium from adipose tissue-derived mesenchymal stem cells (named MSC CM) as described in method section and in20. The selection of potential OA markers was performed in several stages. In the first stage, genes that present significantly different (up or down-regulated) expression levels in TNF-α treated chondrocytes vs. control chondrocytes were identified. Next, the obtained set of genes was verified using our previously established OA-related 142 upregulated and 319 down-regulated genes for both chondrocytes and synoviocytes (Fig. 1). Finally, genes, which expression levels were not affected by MSCs secretome (no significant changes between Inflammation vs. MSC CM groups) were excluded. This approach resulted in the selection of 5 genes: SRI, SMC2, PSIP1, TLE4, and MSX1 (Table 3). Well established molecular markers of OA such as IL-1B, IL6, MMP1, MMP2, MMP3, MMP9, MMP13 and ADAMTS5 were additionally tested. As expected, all tested markers presented upregulated mRNA level in TNF-α treated chondrocytes, proving that this in vitro model molecularly resembles OA (data not shown). Interestingly, only MMP9 level was partially downregulated upon adipose tissue-derived mesenchymal stem cells medium treatment.
SRI, SMC2, PSIP1, TLE4, and MSX1 mRNA expression levels were analyzed in all chondrocyte variants of the GSE239343 data set, and differences were analyzed using the ANOVA test with Tukey post hoc correction (Fig. 4A). Treatment of inflammation-based damaged chondrocytes with conditioned medium from adipose tissue-derived mesenchymal stem cells leads to full (SRI, SMC2, PSIP1) or partial (TLE4, MSX1) restoration of normal mRNA expression level observed in control, untreated chondrocytes. Next, to analyze the mutual interaction of chosen genes and verify whether changes in their expression levels correlate to one another during OA progression and regeneration processes, the Pearson correlation matrix was created using combined data of the GSE239343 data set and Orange data mining software as previously described by us (Fig. 4B). Our analysis proves that mRNA expression of MSX1, TLE4, PSIP1, and SMC2 present very strong, statistically significant mutual correlation efficiency (r > ± 0.8). However, mRNA expression of SRI, shows a moderate correlation efficiency (r > ± 0.4) with other analyzed genes, but this correlation is statistically insignificant. Hierarchical clustering analysis allows for the construction of a bidirectional tree diagram (dendrogram), where the most similar subjects are placed on branches that are close together visualizing their mutual relations. Thus, using this technique, we have verified if chosen up and down-regulated genes are located on separate arms of the hierarchical dendrogram (Fig. 4C). This technique additionally visualized mRNA expression as shades of an “inferno” color scheme (Orange data mining). Our analysis proved the possible discriminative ability of chosen genes which was further verified using radial data visualization via the RadViz algorithm and Orange data mining software (3.31.1) (Fig. 4D). This method was recently described by us in17, briefly: the greater the attribute value, the closer the data point is drawn, and as a result, the data point is placed where the sum of all forces generated by the data value equals 0. Calculation of inner cluster deviation (wc) for data of each chondrocyte variant, revealed that data points from all form three separate clusters, with the Control and MSC CM one, being only slightly separated, thus proving high similarity between mRNA expression of all analyzed genes in these two data groups.
Analysis of selected genes as potential OA markers. The expression levels of selected SRI, SMC2, PSIP1, TLE4, and MSX1 genes were analyzed using the GSE239343 dataset in the Control (chondrocytes), Inflammation (chondrocytes cultured in a mix of serum-free DMEM and 1% penicillin/streptomycin with 25 ng/mL TNF), MSC CM (chondrocytes cultured in a mix of serum-free DMEM and 1% penicillin/streptomycin with 25 ng/mL TNF and treated with 50 µg/mL of conditioned medium from adipose tissue-derived mesenchymal stem cells). Data were calculated using ANOVA with Tukey post hoc correction and visualized using JASP software (A). The Pearson correlation matrix was calculated and visualized using JASP software. The color intensity and presented value represent correlation efficiency whereas *p < 0.05, **p < 0.005, and ***p < 0.001 denote the statistical significance of the correlation (B). Hierarchical clustering analysis and hierarchical dendrogram were calculated and visualized using Orange data mining software (C). 2D depiction of mRNA expression level ability of data discrimination using Radial data visualization algorithm RadViz and SRI, SMC2, PSIP1, TLE4, and MSX1 genes mRNA expression as attributes, calculated and visualized using Orange data mining software (D).
Cellular localization of proteins encoded by SRI, SMC2, PSIP1, TLE4, and MSX1 genes
To validate the cellular localization of proteins encoded by SRI, SMC2, PSIP1, TLE4, and MSX1 genes, we used the immunohistochemistry images of and immunofluorescence bone marrow and cartilage (chondrocytes from soft tissue) tissue sections stained with respective antibodies (method section), obtained from The Human Protein Atlas (Fig. 5.)25. Protein detection and staining intensity are presented in Table 4. Unfortunately, in the case of cartilage (chondrocytes), only Sorcin data were available (presenting strong staining intensity). Additionally, as data for Transducin-like enhancer of split protein 4 are not available for both bone marrow and cartilage, to further analyze cellular localization of all proteins, high-resolution immunofluorescence (confocal microscopy) images of U-2 OS human osteosarcoma cell line staining was presented. Even though U-2OS is not a cellular model of OA, it is often used to verify the subcellular localization of various proteins26,27. Structural Maintenance Of Chromosomes 2, PC4 and SRSF1 Interacting Protein 1, Transducin-like enhancer of split protein 4, and anti-Msh Homeobox 1 presents predominantly nucleoplasm localization, whereas Sorcin is present in both nucleoplasm and cytoplasm (Fig. 5.).
Immunochemistry and immunofluorescence staining of proteins encoded by SRI, SMC2, PSIP1, TLE4, and MSX1 genes. Data were obtained from the Human Protein Atlas database (https://www.proteinatlas.org/). NA – data not available. Detection and staining intensity are resented in Table 4. In the immunofluorescence staining, proteins encoded by SRI, SMC2, PSIP1, TLE4, and MSX1 genes were shown as green, and microtubules as red. https://www.proteinatlas.org/ENSG00000075142-SRI/tissue/soft+tissue#imghttps://www.proteinatlas.org/ENSG00000075142-SRI/tissue/bone+marrow#imghttps://www.proteinatlas.org/ENSG00000136824-SMC2/tissue/bone+marrow#imghttps://www.proteinatlas.org/ENSG00000164985-PSIP1/tissue/bone+marrow#imghttps://www.proteinatlas.org/ENSG00000163132-MSX1/tissue/bone+marrow#imghttps://www.proteinatlas.org/ENSG00000136824-SMC2/subcellular#imghttps://www.proteinatlas.org/ENSG00000163132-MSX1/subcellular#imghttps://www.proteinatlas.org/ENSG00000075142-SRI/subcellular#imghttps://www.proteinatlas.org/ENSG00000106829-TLE4/subcellular#imghttps://www.proteinatlas.org/ENSG00000164985-PSIP1/subcellular#img.
Molecular functions and biological pathways enrichment of proteins encoded by selected genes
To further elucidate the molecular functions of proteins encoded by SRI, SMC2, PSIP1, TLE4, and MSX1 PPI network was composed using STRING version 12.0 online software (https://string-db.org/). Genes that were down-regulated and up-regulated in OA, were analyzed separately, using a minimal number of additional known proteins to fill missing connections and interactions (Fig. 6). The network created by SRI, SMC2, and PSIP separates into two distinct clusters with SMC2, and PSIP involved in the regulation of DNA condensation (GO:0000796 Condensin complex) and SRI in the regulation of lead ion binding and SOUL hem-binding protein (Fig. 6A). PPI network created using proteins encoded by TLE4, and MSX1 proved involvement in the repression of Wingless and Int-1 (Wnt) target genes (HSA-4641265) (Fig. 6B). Finally, using the Pathway-to-Gene mode of the Pathway Connector tool we have analyzed additional biological pathways enriched and regulated by proteins encoded by selected genes (Fig. 6C). PSIP1 enriches processes involved in the virus (including HIV) infection such as 2-LTR circle junctions of Viral DNA and integration of provirus. SMC2 is involved in the condensation of chromatin and SRI in the transport of Ca2 + ions. The major pathway regulated by TLE4 is related to the repression of Wnt target genes. Unfortunately, the analyzed system version lacks MSX1 interaction with known biological pathways.
Analysis of protein-protein interaction of SRI, SMC2, PSIP1, TLE4, and MSX1 genes. PPI network of downregulated (A) and upregulated (B) genes in OA. Data visualized using the STRING platform. (C) Biological pathways enriched and regulated by proteins encoded by PSIP1, SMC2, SRI, and TLE4 genes. Data analyzed and visualized using pathway-to-Gene analysis of the Pathway Connector tool and Reactome 2015 database.
Verification of chosen genes’ feasibility as blood-derived OA markers
SRI, SMC2, PSIP1, TLE4, and MSX1 were proven to be capable molecular markers (in cartilage and synovium) of knee OA and potential markers of OA regeneration upon MSCs treatment, that could be easily isolated in vitro. However, in vivo, knee biopsy is required, which might bring additional complications for patients. OA progression triggers acute inflammation of the joint and its surrounding tissues leading to an acute systemic inflammatory response with increased cytokine and peripheral blood immune cell levels28. This prolonged inflammation may impact actual PBMC gene expression patterns, and several PBMC OA markers were previously identified21,29. Thus, we analyzed the potential role of our chosen genes as OA markers that could be potentially detected from the patient’s blood29. The GSE48556 dataset, composed of transcriptomic data obtained from peripheral blood mononuclear cells of 33 healthy controls and 106 OA patients was downloaded21. Analysis of mRNA expression levels of chosen genes showed that only PSIP1 and TLE4 present statistically significant differences (p < 0.001) between PBMCs isolated from OA patients and healthy control (Fig. 7A). ROC analysis showed that AUC values of PSIP1 and TLE4 present lower values than 0.75, respectively 0.692 and 0.742, whereas, calculated precision equals 0.774 and 0.814 respectively (Fig. 7B). However, knowing that PSIP1 mRNA expression is lower in PBMCs isolated from OA patients and TLE4 is higher, we reasoned that PSIP1:TLE4 mRNA expression ratio could be far more effective as a prognostic biomarker. Obtained values showed significant discriminating abilities of OA/control, with ROC analysis proving that the AUC value of PSIP1:TLE4 equals 0.786 and a precision of 0.823 (Fig. 7C). Additionally due to sample size imbalance, sensitivity analysis was performed, down-sampling of the OA group, randomly assigning n = 33 OA samples using “data sampler” widget of orange data mining software. This analysis proved consistent results, supporting the robustness of our conclusions (data not shown).
Analysis of PSIP1 and TLE4 mRNA expression level in PBMCs as potential prognostic markers of OA development. mRNA expression levels of PSIP1 and TLE4 in PBMCs isolated from healthy (Control) and OA-affected (OA) patients (A). ROC analysis of prognostic properties of PBMCs mRNA levels of PSIP1 and TLE4 in OA development (B). Analysis of PBMCs isolated PSIP1:TLE4 mRNA expression levels ratio as a prognostic tool of OA C). All calculations and visualizations were performed using Jasp software. Samples were tested with the Shapiro–Wilk test, presenting a not-normal distribution, thus followed by the Mann-Whitney U test *** p < 0.001.
Discussion
Annually, millions of patients around the world, suffer from OA-related mild to severe joint pain and movement impediments leading to long-term disabilities. OA’s economic and social impact can not be dismissed, especially considering the aging society and increasing obesity rate1,2,5. Thus, since currently, no drugs can alter the progression of OA, the development of new therapeutic strategies such as regenerative therapy is extremely important. The first concept of regenerative therapy in OA assumed that administered cells may engraft to a lesion site and differentiate into chondrocytes with autologous chondrocytes being the first cell candidates30. Unfortunately, important shortcomings of such treatment modality, including loss of chondrocyte phenotypes and additional problems associated with the process of chondrocyte acquisition, led scientists to search for other sources of therapeutic cells, predominantly mesenchymal stem cells (derived from either: bone marrow, umbilical cord, or adipose tissue)12,30. However, we lack well-established molecular indicators of OA advancement. Currently, serious efforts are focused on the molecular understanding of the disease, that could be utilized to address disease prevention, diagnosis, and treatment leading to the qualification of biomarkers, intended for use as companion diagnostics for approval of a new drug or therapeutic scheme31. Therefore, we have focused on establishing potential molecular markers, that, on the mRNA level, would allow us to monitor OA development, progression, and potential MSC-based regeneration of OA-affected cells (in vitro) and tissue (in vivo).
In our study, we have focused on the knee joint as one of the most prevalent sites3. Our analysis proved that aurora B and A signaling pathways are significantly enriched in OA-affected chondrocytes but not in synovium. Aurora kinase B (AURKB) is a key component of the chromosomal passenger complex involved in heterochromatin destabilization and mitosis progression, which in the case of OA-affected chondrocytes may induce senescence, leading to MMPs overexpression and OA progression32. Nevertheless, both OA-affected knee synovium and knee chondrocytes present similar enrichment of major biological pathways that are dysregulated during OA progression (such as PDGF, IGF1, VEGF, S1P1, glypican or TRAIL signaling pathways). Thus, in the first step, we have identified dysregulated genes for both (patient-derived) synovium and chondrocytes, focusing on those presented in both OA-affected cell types. This approach allowed us to select up and down-regulated genes that are generally relevant to OA development. Enrichment analysis proved that selected up- and down-regulated genes are involved mainly in the process such as apoptosis, immune response, cell proliferation, cell adhesion, migration, cell-matrix degradation, and ion transport. Apoptosis as an etiological factor in osteoarthritis onset, combined with dysregulated immune response and impaired cell-matrix interactions, often leads to increased and prolonged inflammation and cartilage or tendon tissue degradation, contributing to the significant deterioration of the patient’s condition4,21. PPI networks created by up and down-regulated genes that are generally relevant to OA development present a similar value of average ClusteringCoefficient parameter, which is a numerical depiction of how interconnected a node’s neighbors are, enabling quantification of the local cohesiveness of interactions. Low average BetweennessCentrality and high average Degree parameter in both networks suggests that the majority of the nodes are less involved in connecting distant parts of the network, and few proteins, such as AKT1 (Degree 22) and EGFR (Degree 44), act as key bottleneck hubs, regulating signaling pathways related to the development of OA6,33,34. Additionally, higher average ClosenessCentrality in the PPI network of up-regulated genes suggests higher signaling efficiency of hub genes.
Next, we have shifted our interest to an in vitro model of OA caused by TNF-derived inflammation of chondrocytes to identify potential new markers of OA reversion via the regenerative properties of AD-MSCs secretome20. TNF-treated chondrocytes, presents molecular characteristic of in vivo OA affected cartilage, especially regarding expression of well know OA markers such as: IL-1B, IL6, MMP1, MMP2, MMP3, MMP9, MMP13 and ADAMTS5. Analyzed data were verified using previously obtained DEGs (patient-derived) that are generally relevant to OA development. Selected genes SRI, SMC2, PSIP1, TLE4, and MSX1 presented dysregulated expression levels in the TNF-inflammation-derived in vitro model of OA, as well as in patient samples – compared to respective non-OA controls, and their expression levels were reversed to control level upon AD-MSCs secretome treatment.
All proteins encoded by chosen genes present nucleoplasm localization, with only Sorcin (soluble resistance-related calcium-binding protein) encoded by SRI being detected in both nucleoplasm and cytoplasm (predominantly). Cell nuclei, with well-defined chromatin compartmentalization and ribosome biogenesis, is a key player that regulates many aspects of cell biology. Therefore, nucleoplasm proteins are extremely sensitive even to incremental changes in cell physiology, which can be exploited to monitor cores of the disease35. Sorcin is a key protein in the endoplasmic reticulum (ER) calcium-dependent cascades and signaling that regulates cellular homeostasis via important functions, such as gene expression, cell differentiation, proliferation, survival, apoptosis, and xenobiotics (drugs) resistance36,37. Interestingly, Gong et al. observed different levels of extracellular calcium-dependent spontaneous signaling between different zones in healthy cartilage, whereas this zonal difference was not observed in OA-affected cartilage, proving that an imbalance in calcium signaling relates to OA38. The protein encoded by SMC2 is involved in the regulation of chromosome condensation during mitosis39. There are no known alterations regarding SMC2 and OA development, however, it is a known fact that SMC2 downregulation and/or knock-out impairs bovine embryo development40. Interestingly, SMC2 was proven to be one of the top hub genes involved in dynamic compression-enhanced chondrogenesis leading to cartilage regeneration8. PC4 And SRSF1 Interacting Protein 1 also known as Lens epithelium-derived growth factor is a transcriptional co-activator and cellular stress survival factor regulating processes such as DNA repair and RNA transcription essential for the cell/tissue development. PSIP1 plays an important role in the course of several human diseases, including acute leukemia, virus infection, and autoimmune disorders41. Proteins encoded by SMC2 and PSIP1 genes, via regulation of chromatin condensation, influence the expression of many, potentially OA-related genes. Transducin-like enhancers of split protein 4 (TLE4), share identical structures and molecular properties as the Drosophila protein Groucho which is involved in the regulation of cell-determination events during insect neurogenesis and segmentation. Its expression pattern correlates to TLE242. TLE4 and TLE2 transcriptionally repress Wnt-β-catenin signaling, repressing Wnt-mediated inflammation43. In the case of OA, a positive correlation between disease severity and Wnt-β-catenin signaling was observed, thus upregulation of its repressor in the OA-affected cells, at first sight, is rather surprising and difficult to explain44. However, canonical (Wnt/β-catenin, Wnt3a) and non-canonical (β-catenin independent, Wnt5a) Wnt signaling pathways substantially differ, influencing transcription of target genes via (respectively) β-catenin or Jun/ATF2 and NAFT transcription factors45. Furthermore, there is substantial heterogeneity within OA patients and caused by the multifactorial nature of this disease. Canonical Wnt signaling plays a major role in injury-induced osteoarthritis mouse models but is not observed in the STR/ort mouse OA model46. Treatment of OA-affected sheep cartlidge with Wnt signaling antagonist - sclerostin, results in inhibition of Wnt/β-catenin signaling and predominately an anti-catabolic effect by reducing Mmp, Adamts, Acan, and Col2a1 gene expression, as well as IL-1a-mediated aggrecanolysis47. The complex nature of the Wnt regulatory network in skeletal homeostasis proves that both excessive activation and inactivation of Wnt signaling can cause skeletal malformation, bone diseases, and cartilage loss. Recent reports suggest that Wnt16 presents a strong association with bone mineral density and strength, overall cortical bone thickness, additionally playing a chondroprotective role, limiting cartilage destruction during OA progression48. Associated with bone formation, the MSX1 gene was previously shown by Karlsson et al. to be significantly upregulated in OA-affected cartilage compared to normal (ON-not-affected) control cartilage49. On the other hand, recent studies have shown that MSX1+ mesenchymal progenitor cells exhibit strong chondrogenic potential, that could be utilized for cartilage regeneration. In the rat models of acute rotator cuff injury, overexpression of the Msx1 inhibits osteogenic and chondrogenic differentiation of the engrafted bone marrow mesenchymal stem cells simultaneously increasing their differentiation and migratory potential50. Thus, the role of MSX1 in OA development and cartilage damage and regeneration requires further analysis.
Accurate molecular markers for monitoring OA progression and regeneration, which are valid in patients, are essential for in vitro studies to develop new therapies. However, in the case of OA patients, analysis of their expression (thus monitoring of OA regeneration) would require constant joint biopsy. Therefore, less invasive methods, of OA marker isolation, such as blood liquid biopsy are being developed29,51. Progression of OA is related to the inflammation and recruitment of immunological cells which can impact actual PBMC gene expression, leading to the identification of PBMC-derived OA markers21,29,52. Interestingly, our data proves the expression level of PSIP1 and TLE4 in the PMBCs correlates with OA development, however, when used separately, as a single prognostic marker, each of them presents less than 0.75 ROC AUC, thus poor discrimination. On the other hand, the PSIP1:TLE4 mRNA expression levels ratio used as a prognostic marker presents AUC for ROC 0.786, which could be regarded as more than acceptable.
The major limitation of this study is that data, are obtained mainly from the European descent population of Western society (mainly due to the higher OA incident rate). Consequently, our findings may not account for racial-specific factors, especially in the light of the evidence that in the case of knee OA, pain and functional limitations are greater for African and African-Americans than European descent4,53. Additionally, we have based our analysis on available clinical descriptions of patients’ OA status. However, standard MRI are focused mostly on alterations seen in advanced OA stages possibly incorrectly identifying early, potentially reversible stages14. Furthermore, GSE48556 dataset consists of imbalanced sample size (33 healthy subjects and 106 OA patients) and highlights one of the biggest problems with clinical data, that disease cohorts are usually better characterized than healthy controls. Additionally, we have based our study on microarray data that are less sensitive than RNA sequencing (RNA-seq) especially for low-abundance RNAs (Spearman correlations coefficients of ∼0.8 between expression data obtained from the Affymetrix one-channel microarray and RNAseq for high-abundance genes and ∼0.2 for low-abundance genes), but are more reliable and easier to use regarding analysis of gene expression profiling of well-know sequences. Furthermore, results derived from microarrays accumulated over the last 20 years, and stored in repositories such as GEO, can still serve as excellent data mining resources54,55. Finally, despite the fact that recent transcriptomic analyses of OA patient samples deposited in the GEO database, have indeed yielded clinically valuable insights, particularly for potential new biomarker discovery and therapeutic targets, translating these findings into real-world clinical practice requires additional experimental confirmation and preferably prospective or controlled clinical studies56,57.
Conclusion
Our analysis proved that mRNA expression levels of SRI, SMC2, PSIP1, TLE4, and MSX1 are dysregulated in osteoarthritis (SRI, SMC2, PSIP1 downregulated in OA, TLE4, and MSX1 upregulated in OA ) and potentially could be utilized to monitor its development and potential regeneration upon AD-MSCs-based regenerative therapy in both chondrocytes and synovium. Additionally, expression levels of PSIP1 and TLE4 in the PMBCs may be considered as less invasive OA markers. These genes have potential as diagnostic and prognostic biomarkers not only in osteoarthritis but also in other chronic joint diseases, traumatic injuries, and overuse syndromes. Further research is needed to validate their clinical utility across these diverse conditions.
Data availability
The datasets analyzed during the current study are available in the Gene Expression Omnibus repository at http://www.ncbi.nlm.nih.gov/geo/ reference numbers: GSE239343, GSE179716, GSE206848 and GSE48556. URLs: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239343, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179716, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE206848, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48556.
Abbreviations
- ADAMTS-5:
-
A disintegrin and metalloproteinase with thrombospondin motifs protein 5
- AD-MSCs:
-
Adipose tissue-derived mesenchymal stem cells
- AUC-ROC:
-
The area under the receiver operating characteristic curve
- BP:
-
Biological process
- ΒΜP:
-
Bone morphogenic protein
- CC:
-
Cellular components
- CING:
-
Cyprus Institute of Neurology & Genetics
- DEGs:
-
Differently expressed genes
- DNA:
-
Deoxyribonucleic acid
- EMT:
-
Epithelial to mesenchymal transition
- FGF:
-
Fibroblast growth factor
- GEO:
-
The Gene Expression Omnibus database
- GO:
-
The Gene Ontology
- HPA:
-
The Human Protein Atlas
- HPCF:
-
The High-Performance Computing Facility
- IGF1:
-
Insulin-like growth factor
- IL-1β:
-
Interleukin 1 beta
- IL-6:
-
Interleukin 6
- MMPs:
-
Matrix metalloproteinases
- MRI:
-
Magnetic resonance imaging
- mRNA:
-
Messenger ribonucleic acid
- MSC:
-
Mesenchymal stem cells
- NF-κB:
-
Nuclear factor-kappa B
- NSAIDs:
-
Non-steroidal anti-inflammatory drugs
- OA:
-
Osteoarthritis
- PBMCs:
-
Peripheral blood mononuclear cells
- PDGF:
-
Platelet-derived growth factor
- PPI:
-
Protein-protein interaction
- S1P1:
-
Sphingosine-1-phosphate receptor 1
- TGFβ:
-
Transforming growth factor beta
- TNF-α:
-
Tumor necrosis factor alpha
- USG:
-
Ultrasonography
- VEGF:
-
Vascular endothelial growth factor
- Wnt:
-
The Wingless and Int-1
References
Leifer, V. P., Katz, J. N. & Losina, E. Chap. 1: the burden of OA-health services and economics. Osteoarthr. Cartil. 30, 10–16. https://doi.org/10.1016/j.joca.2021.05.007 (2022).
Long, H. et al. Prevalence Trends of Site-Specific Osteoarthritis From 1990 to 2019: Findings From the Global Burden of Disease Study 2019, Arthritis Rheumatol. 74 1172–1183. https://doi.org/10.1002/art.42089. (2022).
Boer, C. G. et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell. 184, 4784–4818.e17. https://doi.org/10.1016/j.cell.2021.07.038 (2021).
Yao, Q. et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Sig Transduct. Target. Ther. 8, 1–31. https://doi.org/10.1038/s41392-023-01330-w (2023).
Xiang, Q. et al. Cellular and molecular mechanisms underlying obesity in degenerative spine and joint diseases. Bone Res. 12, 71. https://doi.org/10.1038/s41413-024-00388-8 (2024).
Li, H. et al. TNF-α increases the expression of inflammatory factors in synovial fibroblasts by inhibiting the PI3K/AKT pathway in a rat model of monosodium iodoacetate-induced osteoarthritis. Exp. Ther. Med. 16, 4737–4744. https://doi.org/10.3892/etm.2018.6770 (2018).
Chisari, E., Yaghmour, K. M. & Khan, W. S. The effects of TNF-alpha inhibition on cartilage: a systematic review of preclinical studies. Osteoarthr. Cartil. 28, 708–718. https://doi.org/10.1016/j.joca.2019.09.008 (2020).
Chen, J., Chen, L., Hua, J. & Song, W. Long-term dynamic compression enhancement TGF-β3-induced chondrogenesis in bovine stem cells: a gene expression analysis. BMC Genom Data. 22, 13. https://doi.org/10.1186/s12863-021-00967-2 (2021).
Lohmander, L. S. & Roos, E. M. Disease modification in OA — will we ever get there? Nat. Rev. Rheumatol. 15, 133–135. https://doi.org/10.1038/s41584-019-0174-1 (2019).
Richard, M. J., Driban, J. B. & McAlindon, T. E. Pharmaceutical treatment of osteoarthritis. Osteoarthr. Cartil. 31, 458–466. https://doi.org/10.1016/j.joca.2022.11.005 (2023).
Zhang, W., Robertson, W. B., Zhao, J., Chen, W. & Xu, J. Emerging trend in the pharmacotherapy of osteoarthritis. Front. Endocrinol. 10 https://doi.org/10.3389/fendo.2019.00431 (2019).
Zhu, C., Wu, W. & Qu, X. Mesenchymal stem cells in osteoarthritis therapy: a review. Am. J. Transl Res. 13, 448–461 (2021).
Tortorella, F. et al. Meniscal extrusion correlates with symptom severity in knee osteoarthritis: an ultrasound and magnetic resonance imaging analysis of 100 patients. J. Clin. Med. 13, 7716. https://doi.org/10.3390/jcm13247716 (2024).
Mallio, C. A. et al. Advanced MR imaging for knee osteoarthritis: A review on local and brain effects. Diagnostics (Basel). 13, 54. https://doi.org/10.3390/diagnostics13010054 (2022).
Wei, Q. et al. Plasma proteomics implicate glutamic oxaloacetic transaminases as potential markers for acute myocardial infarction. J. Proteom. 308, 105286. https://doi.org/10.1016/j.jprot.2024.105286 (2024).
Kryczka, J. & Boncela, J. Integrated bioinformatics analysis of the hub genes involved in Irinotecan resistance in colorectal cancer. Biomedicines 10, 1720. https://doi.org/10.3390/biomedicines10071720 (2022).
Kryczka, J., Bachorz, R. A., Kryczka, J. & Boncela, J. Radial data visualization-based step-by-step eliminative algorithm to predict colorectal cancer patients’ response to FOLFOX therapy. Int. J. Mol. Sci. 25, 12149. https://doi.org/10.3390/ijms252212149 (2024).
Tahir, M. et al. Artificial intelligence and deep learning algorithms for epigenetic sequence analysis: A review for epigeneticists and AI experts. Comput. Biol. Med. 183, 109302. https://doi.org/10.1016/j.compbiomed.2024.109302 (2024).
Jafari, M. & Ansari-Pour, N. Why, when and how to adjust your P values?? Cell. J. 20, 604–607. https://doi.org/10.22074/cellj.2019.5992 (2019).
González-Cubero, E. et al. The therapeutic potential of adipose-derived mesenchymal stem cell secretome in osteoarthritis: A comprehensive study. Int. J. Mol. Sci. 25, 11287. https://doi.org/10.3390/ijms252011287 (2024).
Ramos, Y. F. M. et al. Genes expressed in blood link osteoarthritis with apoptotic pathways. Ann. Rheum. Dis. 73, 1844–1853. https://doi.org/10.1136/annrheumdis-2013-203405 (2014).
Koren, Y. & Carmel, L. Visualization of labeled data using linear transformations. In IEEE Symposium on Information Visualization 2003 (IEEE Cat. No.03TH8714), 121–128. https://doi.org/10.1109/INFVIS.2003.1249017 (2003).
Leban, G., Zupan, B., Vidmar, G. & Bratko, I. VizRank: Data Visualization Guided by Machine Learning, Data Min. Knowl. Disc 13 119–136. https://doi.org/10.1007/s10618-005-0031-5. (2006).
Kryczka, J. & Boncela, J. Characteristics of ABCC4 and ABCG2 high expression subpopulations in CRC-A new opportunity to predict therapy response. Cancers (Basel). 15, 5623. https://doi.org/10.3390/cancers15235623 (2023).
Uhlén, M. et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 4, 1920–1932. https://doi.org/10.1074/mcp.M500279-MCP200 (2005).
Beijer, D. et al. Biallelic ADPRHL2 mutations in complex neuropathy affect ADP ribosylation and DNA damage response. Life Sci. Alliance. 4 https://doi.org/10.26508/lsa.202101057 (2021).
Nusinow, D. P. et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell. 180 387–402.e16. https://doi.org/10.1016/j.cell.2019.12.023. (2020).
Weber, P. et al. Zenobi-Wong, the collagenase-induced osteoarthritis (CIOA) model: where mechanical damage meets inflammation. Osteoarthr. Cartil. Open. 6, 100539. https://doi.org/10.1016/j.ocarto.2024.100539 (2024).
Li, Y. & Dong, B. Exploring liquid-liquid phase separation-related diagnostic biomarkers in osteoarthritis based on machine learning algorithms and experiment. Immunobiology 229, 152825. https://doi.org/10.1016/j.imbio.2024.152825 (2024).
Im, G. I. & Kim, T. K. Regenerative therapy for osteoarthritis: A perspective. Int. J. Stem Cells. 13, 177–181. https://doi.org/10.15283/ijsc20069 (2020).
Kraus, V. B. Biomarkers as drug development tools: discovery, validation, qualification and use. Nat. Rev. Rheumatol. 14, 354–362. https://doi.org/10.1038/s41584-018-0005-9 (2018).
Ren, X., Zhuang, H., Jiang, F., Zhang, Y. & Zhou, P. Barasertib impedes chondrocyte senescence and alleviates osteoarthritis by mitigating the destabilization of heterochromatin induced by AURKB. Biomed. Pharmacother. 166, 115343. https://doi.org/10.1016/j.biopha.2023.115343 (2023).
Xie, J. et al. Sustained Akt signaling in articular chondrocytes causes osteoarthritis via oxidative stress-induced senescence in mice. Bone Res. 7, 23. https://doi.org/10.1038/s41413-019-0062-y (2019).
Wei, Y. et al. Targeting cartilage EGFR pathway for osteoarthritis treatment. Sci. Transl. Med. 13, eabb3946. https://doi.org/10.1126/scitranslmed.abb3946 (2021).
Su, H., Kodiha, M., Lee, S. & Stochaj, U. Identification of novel markers that demarcate the nucleolus during severe stress and chemotherapeutic treatment. PLoS One. 8, e80237. https://doi.org/10.1371/journal.pone.0080237 (2013).
Ghosh, S., Sharma, A., Kumar, R. S. & Nasare, V. Sorcin: mechanisms of action in cancer hallmarks, drug resistance and opportunities in therapeutics. Med. Oncol. 42, 29. https://doi.org/10.1007/s12032-024-02580-6 (2024).
Genovese, I. et al. Sorcin is an early marker of neurodegeneration, Ca2 + dysregulation and endoplasmic reticulum stress associated to neurodegenerative diseases. Cell. Death Dis. 11, 861. https://doi.org/10.1038/s41419-020-03063-y (2020).
Gong, X. et al. Altered spontaneous calcium signaling of in situ chondrocytes in human osteoarthritic cartilage. Sci. Rep. 7, 17093. https://doi.org/10.1038/s41598-017-17172-w (2017).
Song, W. M. et al. Multiscale protein networks systematically identify aberrant protein interactions and oncogenic regulators in seven cancer types. J. Hematol. Oncol. 16, 120. https://doi.org/10.1186/s13045-023-01517-2 (2023).
Alba, P. G. et al. Pablo, SMC2 ablation impairs bovine embryo development shortly after blastocyst hatching. Reproduction 168, e240211. https://doi.org/10.1530/REP-24-0211 (2024).
Brouns, T. et al. The impact of lens epithelium-derived growth factor p75 dimerization on its tethering function. Cells 13, 227. https://doi.org/10.3390/cells13030227 (2024).
Yao, J. et al. Combinatorial expression patterns of individual TLE proteins during cell determination and differentiation suggest non-redundant functions for mammalian homologs of drosophila Groucho. Dev. Growth Differ. 40, 133–146. https://doi.org/10.1046/j.1440-169x.1998.00003.x (1998).
Shin, T. H., Brynczka, C., Dayyani, F., Rivera, M. N. & Sweetser, D. A. TLE4 regulation of wnt-mediated inflammation underlies its role as a tumor suppressor in myeloid leukemia. Leuk. Res. 48, 46–56. https://doi.org/10.1016/j.leukres.2016.07.002 (2016).
Zheng, H. et al. An injectable hydrogel loaded with Icariin attenuates cartilage damage in rabbit knee osteoarthritis via Wnt/β-catenin signaling pathway. Int. Immunopharmacol. 145, 113725. https://doi.org/10.1016/j.intimp.2024.113725 (2025).
Shi, D. L. Canonical and non-canonical Wnt signaling generates molecular and cellular asymmetries to establish embryonic axes. J. Dev. Biology. 12, 20. https://doi.org/10.3390/jdb12030020 (2024).
Poulsen, R. C., Jain, L. & Dalbeth, N. Re-thinking osteoarthritis pathogenesis: what can we learn (and what do we need to unlearn) from mouse models about the mechanisms involved in disease development. Arthritis Res. Therapy. 25, 59. https://doi.org/10.1186/s13075-023-03042-6 (2023).
Zhou, Y., Wang, T., Hamilton, J. L. & Chen, D. Wnt/β-catenin signaling in osteoarthritis and in other forms of arthritis. Curr. Rheumatol. Rep. 19, 53. https://doi.org/10.1007/s11926-017-0679-z (2017).
Ye, X. & Liu, X. Wnt16 signaling in bone homeostasis and osteoarthristis. Front. Endocrinol. (Lausanne). 13, 1095711. https://doi.org/10.3389/fendo.2022.1095711 (2022).
Karlsson, C. et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthr. Cartil. 18, 581–592. https://doi.org/10.1016/j.joca.2009.12.002 (2010).
Liu, K., Fu, X. W. & Wang, Z. M. Msx1-modified rat bone marrow mesenchymal stem cell therapy for rotator cuff repair: A comprehensive analysis of tendon-bone healing and cellular mechanisms. J. Orthop. Res. https://doi.org/10.1002/jor.26039 (2024).
Tchetina, E. et al. Upregulation measured in the peripheral blood mononuclear cells prior to surgery points to postoperative pain development in patients with hip osteoarthritis. Diagnostics 13, 1739. https://doi.org/10.3390/diagnostics13101739 (2023).
Hu, S., Tao, Y., Hu, F. & Liu, X. Diminished LAG3 + B cells correlate with exacerbated rheumatoid arthritis. Ann. Med. 55, 2208373. https://doi.org/10.1080/07853890.2023.2208373 (2023).
Allen, K. D. Racial and ethnic disparities in osteoarthritis phenotypes. Curr. Opin. Rheumatol. 22, 528–532. https://doi.org/10.1097/BOR.0b013e32833b1b6f (2010).
Guo, Y. et al. Large Scale Comparison of Gene Expression Levels by Microarrays and RNAseq Using TCGA Data, PLoS One 8 e71462. https://doi.org/10.1371/journal.pone.0071462. (2013).
Mantione, K. J. et al. Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Med. Sci. Monit. Basic. Res. 20, 138–141. https://doi.org/10.12659/MSMBR.892101 (2014).
He, M. et al. Screening and validation of key genes associated with osteoarthritis. BMC Musculoskelet. Disord. 25, 954. https://doi.org/10.1186/s12891-024-08015-7 (2024).
Zhu, Y. S., Yan, H., Mo, T. T., Zhang, J. N. & Jiang, C. Identification of diagnostic markers in synovial tissue of osteoarthritis by weighted gene coexpression network. Biochem. Genet. 61, 2056–2075. https://doi.org/10.1007/s10528-023-10359-z (2023).
Acknowledgements
The authors gratefully acknowledge the Polish Muscles Ligaments and Tendons Society (PolMuLTS) for the constructive feedback.The authors declare that they have not use AI-generated work in this manuscript.
Funding
This research was supported by the statutory fund of the Institute of Medical Biology of the Polish Academy of Sciences.
Author information
Authors and Affiliations
Contributions
J.K.: investigation, methodology, writing—original draft, formal analysis; validation.E.B-L.: formal analysis, writing—review and editing. M.P.: formal analysis; writing—review and editing. J.B.: funding acquisition; project administration; formal analysis; writing—review and editing. J.M.K.: conceptualization; data curation; formal analysis; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kryczka, J., Brzeziańska-Lasota, E., Pikuła, M. et al. A preliminary bioinformatic screen to identify SRI SMC2 PSIP1 TLE4 and MSX1 as potential diagnostic and prognostic markers of osteoarthritis. Sci Rep 15, 30583 (2025). https://doi.org/10.1038/s41598-025-13575-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13575-2