Abstract
Increasing evidence suggests that the synaptotagmin (SYT) family is associated with the development of multiple types of cancer, yet research on SYT15 remains limited. The aim of this study was to elucidate the clinical significance and possible molecular mechanisms of SYT15 expression in lung adenocarcinoma (LUAD). The expression, clinical value, cellular function and possible molecular mechanisms of SYT15 in LUAD were evaluated by using bioinformatics analysis combined with cellular experiments. In LUAD, SYT15 expression was downregulated. Notably, this reduced expression was negatively associated with patient survival but positively correlated with immune cell infiltration in the tumor microenvironment. Furthermore, SYT15 expression correlated with three key aspects: sensitivity to 20 distinct drugs, the expression of immune checkpoint genes (ICGs), and the efficacy of immunotherapy. SYT15 was most highly expressed in Mono/Macro, CD8Tex, and CD8T cells within LUAD tissues. Cellular experiments confirmed that SYT15 overexpression reduced the malignant phenotype of A549 and H1975 cells. RNA sequencing results showed that the PI3K/AKT signaling pathway was significantly enriched after overexpression of SYT15 in A549 cells, and qPCR and WB experiments confirmed significant downregulation of genes related to this pathway.. Further experiments confirmed that PI3K agonist 740Y-P reversed the regulatory effects of SYT15 overexpression on A549 cell malignant phenotype and the PI3K/Akt signaling pathway. In LUAD, SYT15 acts as a tumor suppressor by potentially regulating immune cell infiltration in the tumor microenvironment and inhibiting the expression of key PI3K/AKT pathway genes in A549 cells. Consequently, it holds promise as a novel prognostic marker and potential therapeutic target..
Similar content being viewed by others
Introduction
Lung cancer is the tumor with the highest incidence and mortality rate among malignant tumors1. Non-small cell lung cancer (NSCLC) is the major type of lung cancer, accounting for about 85% of all lung cancers, of which about 40–55% are LUAD2. Despite significant advancements in NSCLC treatment—such as the development of a multidisciplinary strategy integrating surgery, radiotherapy, chemotherapy, molecular targeted therapy, and traditional Chinese medicine—many patients are still diagnosed with advanced tumors and extensive metastases. This contributes to a poor 5-year survival rate. Consequently, investigating the molecular mechanisms of this disease is of considerable scientific importance and potential clinical value.
Synaptotagmins (SYT) are transmembrane (TM) proteins that traverse vesicle membranes, with their N-terminus positioned within the vesicle lumen and the C-terminus in the cytoplasm3. These proteins primarily function as calcium ion sensors, and they regulate vesicle trafficking and exocytosis—two processes essential for cellular secretion4,5. As membrane transport proteins, Synaptotagmins play a key role in membrane fusion. This crucial process participates in autophagy, exosome release, and other cellular events, all of which are closely associated with the progression and metastasis of malignant tumors6,7. As a result, Synaptotagmins have attracted increasing attention in cancer research. Distinct from other Synaptotagmin family members, SYT15 encodes a protein that is may be involved in membrane transport in non-neuronal tissues. SYT15 mRNA is primarily expressed in non-neural tissues such as the heart, lungs, skeletal muscle, and testes of mice8. However, research on SYT15 in lung cancer, and in malignant tumors in general, remains limited.
In this study, we combined bioinformatics analysis with cellular experiments to explore the clinical significance and possible molecular mechanisms of SYT15 expression in LUAD.
Results
Expression of the SYT15 gene in LUAD
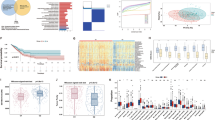
In TCGA and GEO cohorts, as well as in clinical samples, SYT15 mRNA expression was notably reduced in tumor compared to normal (Fig. 1A–C). This reduction was further corroborated by data from the CPTAC platform, which showed a significant decrease in SYT15 protein levels in tumor (Fig. 1D). To investigate SYT15 expression patterns in LUAD, we categorized patients by clinical factors (age, gender, smoking status, T classification, pathological stage) and subsequently compared SYT15 expression across these subgroups. In the TCGA cohort, lower SYT15 mRNA expression was observed in males, those with higher T classification, advanced-stage disease, and (Fig. 1E). Similarly, in the GEO cohort, The expression of SYT15 was significant reduction in males and smokers compared to females and non-smokers(Fig. 1F).
SYT15 expression difference between LUAD and normal tissues and its association with clinical stages. (A–B) SYT15 mRNA levels in LUAD as analyzed by the TCGA and GEO databases. (C) SYT15 mRNA expression in 40 clinical LUAD samples. (D) SYT15 protein expression in LUAD according to the CPTAC database. (E) SYT15 mRNA expression in different patient subgroups of the TCGA population. (F) SYT15 mRNA expression across different patient subgroups in the GEO cohort. n.s. p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Correlation analysis between SYT15 expression and prognostic indicators of LUAD
As shown in Fig. 2A, the OS, DFS, and PFS of the SYT15 low-expression group were shorter than those of the SYT15 high-expression group. Using TCGA cohort data, we further analyzed overall survival (OS) across several patient subgroups: age (low/high), gender (male/female), disease stage (early/mid-late), and metastasis status (metastasis-free/metastatic). Notably, SYT15 expression showed stronger predictive value for OS in the high-age, mid-late stage, and metastatic subgroups than in the low-age, early stage, and non-metastatic subgroups. (Fig. 2B). To determine whether SYT15 was an independent risk factor for prognosis in LUAD patients, we performed univariable and multivariable Cox regression analyses. In the TCGA cohort, low expression of SYT15 was an independent risk factor for OS along with pathologic stage (Fig. 2C).
Association of SYT15 expression with overall survival and key clinical prognostic factors in LUAD patients. (A) Prognostic impact of SYT15 expression on patient outcomes in the TCGA cohort. (B) OS analysis by SYT15 expression in various LUAD subgroups in the TCGA cohort. (C) Univariable and multivariable Cox regression analyses of OS in the TCGA cohort.
Evaluation of SYT15 expression and drug responsiveness in LUAD patients
In order to assess the impact of SYT15 expression on drug sensitivity in LUAD patients. We observed a significant difference in drug sensitivity between the SYT15 low/high expression groups (Supplementary Fig. S1). Patients with elevated SYT15 expression showed increased sensitivity to a range of drugs, including Eriotinib, Rapamycin, Sunitinib, PHA-665752, MG-132, Cyclopamine, Sorafenib, VX-680, Crizotinib, Dasatinib, Parthenolide, Roscovitine, Zibotentan, Bosutinib, YM201636, Phenformin, and Tamoxifen. Conversely, these patients exhibited decreased sensitivity to Thapsigargin, Obatoclax Mesylate, and Lisitinib.
Association between SYT15 expression and tumor microenvironment in LUAD
We used CIBERSORT to estimate immune cell populations in the TCGA cohort. Specifically, we analyzed immune cell infiltration based on a signature matrix covering 22 immune cell types and performed deconvolution of normalized gene expression data to quantify the proportion of each immune cell type in individual samples (Fig. 3A). When comparing high/low SYT15 expression groups, significant differences were observed in immune cell abundance, with higher SYT15 expression being positively associated with several immune cell types (Fig. 3B). Single-cell analysis further revealed differential expression of SYT15 across various cell types within LUAD tumors, with the highest expression observed in Mono/Macro cells, followed by CD8Tex, CD8T, CD4Tconv, Treg, and Tprolif cells (Fig. 3C–E).
To investigate the link between SYT15 expression and immunotherapy outcomes, we analyzed the expression of 10 ICGs in the high/low SYT15 expression groups. The high SYT15 expression group showed significantly elevated levels of HAVCR2,CD274, PDCD1,CTLA4 , ITPRIPL1, PDCD1LG2, TIGIT, and SIGLEC15 compared to the low expression group (Supplementary Fig. S2A). Furthermore, patients with high SYT15 expression had lower TIDE scores as predicted by TIDE analysis, indicating a better potential response to immunotherapy (Supplementary Fig. S2B-C).
Effect of SYT15 overexpression on the malignant phenotype of LUAD cells
Given the strong association between SYT15 and LUAD progression, we performed cell-based experiments to explore SYT15’s role in LUAD. The results showed that SYT15 expression levels were the lowest in A549 and H1975 cells. (Fig. 4A). After transducing A549 cells with Lenti-SYT15, SYT15 expression was significantly elevated (Fig. 4B–C). We then examined proliferation-related phenotypes including CCK8, colony formation, and KI67 immunofluorescence and found that A549 cells overexpressing SYT15 showed significantly reduced proliferation (Fig. 4D–F). Additionally, Transwell migration and wound healing assays showed that there was a significant decrease in the migratory capacity of A549 cells following SYT15 overexpression (Fig. 4G–H). The Transwell invasion assay further confirmed that SYT15 overexpression significantly impaired the invasive capacity of A549 cells (Fig. 4I). Consistent with A549 cells, H1975 cells also presented significantly reduced proliferation, migratory capacity, and invasive capacity upon SYT15 overexpression.(Fig. 4D–I , right panels).
Effect of SYT15 overexpression on proliferation, migration and invasion of A549 and H1975 cells (A) SYT15 mRNA levels in different lung cancer cell lines. (B–C) Efficiency of SYT15 overexpression in A549 cells. (D–F) Results of CCK8, colony formation and immunofluorescence assays. Scale bar = 50 µm. (G–I) Results and statistical analysis of wound healing, migration, and invasion assays. Scale bar = 50 µm. n.s. p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Gene expression changes in SYT15 overexpressing cells
To explore the gene expression changes associated with SYT15 overexpression, RNA sequencing was conducted (Fig. 5A–C). Differential gene expression analysis was performed with a threshold of padj < 0.05 and |FoldChange|> 2. This analysis identified 307 differentially expressed genes, of which 223 were upregulated and 84 were downregulated relative to the control group. The top 10 pathways identified through GO Biological Process (GO BP) analysis are shown in Fig. 5D, while Fig. 5E9,10,11 highlights the top 10 pathways from the KEGG enrichment analysis. Both analyses pointed to significant enrichment of the PI3K/AKT pathway. Specifically, genes such as PIK3CA, PIK3R1, PIK3R3, PIK3C2A, AKT1S1, and AKTIP were significantly downregulated in SYT15 overexpressing cells. We further validated these RNA sequencing results through quantitative real-time PCR (qPCR) and Western blot (WB) assays, confirming that the expression patterns of the aforementioned genes were consistent with the sequencing results (Fig. 5F–H).
Expression changes of PI3K/AKT pathway-related genes in A549 cells after SYT15 overexpression. (A) Sample correlation analysis. (B) Valcano map (C) Heatmap. (D–E) KEGG and GO BP pathway enrichment analyses. (F–H) Validation of RNA sequencing results by qPCR and WB: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
PI3K agonist 740Y-P reverses the regulatory effects of SYT15 overexpression on A549 cells and the PI3K/Akt signaling pathway
To clarify the specific mechanism of action of SYT15 in A549 cells, the PI3K agonist 740Y-P was used for intervention experiments. Three experimental groups were established: (1) SYT15-negative control (NC) A549 cells; (2) SYT15-overexpressing A549 cells; and (3) SYT15-overexpressing A549 cells treated with 20 μM 740Y-P. Detection via CCK-8 proliferation assay, wound healing assay, and Transwell invasion assay revealed that 740Y-P treatment could significantly reverse the inhibitory effects of SYT15 overexpression on the proliferation, migration, and invasion capacities of A549 cells (Fig. 6A–C). Further verification by WB assay demonstrated that 740Y-P was able to reverse the regulatory effect of SYT15 on the PI3K/Akt signaling pathway, which was specifically manifested by the significant upregulation of the protein expression levels of key molecules in this pathway, including PIK3CA, PIK3R1, PIK3R3, PIK3C2A, AKT1S1, and AKTIP (Fig. 6D).
PI3K agonist 740Y-P reverses SYT15 overexpression-mediated regulation of A549 cells and PI3K/Akt pathway. (A–C):Results of CCK-8 proliferation assay, wound healing assay, and Transwell invasion assay. Scale bar = 50 µm. (D) WB detection of protein expression levels of PI3K/AKT pathway-related genes. n.s. p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001.
Discussion
The SYT15 protein exists in two isoforms, SYT15-a and SYT15-b. Biochemical and cellular studies demonstrate that SYT15-a contains a short hydrophobic region at its N-terminus (amino acids 5–22). This segment alone is sufficient to establish a type I membrane topology in cultured cells, analogous to other members of the synaptotagmin (SYT) family. On the other hand, SYT15-b lacks the C-terminal portion of the C2B peptide.The mRNA expression of both SYT15-a and SYT15-b is predominantly observed in non-neuronal tissues, particularly in the lungs and testes12.
In the study, we observed a significant downregulation of SYT15 expression in LUAD tumor, as demonstrated by data from both the TCGA and GEO, also in clinical samples. This reduced expression of SYT15 was inversely associated with key clinical indicators, including T classification, tumor stage, and metastasis status. Moreover, patients with lower SYT15 expression exhibited shorter OS, DSS, and PFS. Importantly, low SYT15 expression emerged as an independent predictor of poorer OS. Above results indicate that SYT15 could play a critical role in LUAD pathogenesis, potentially functioning as a tumor suppressor.
The TME, composed primarily of immune and stromal cells, has become a key area of interest in cancer prognosis research. The genetic, immune, and metabolic heterogeneity of the tumor microenvironment (TME) significantly impacts cancer treatment outcomes. Moreover, this heterogeneity is a major driver of treatment resistance13,14. Recent studies have reported the important role of immune infiltration in TME, which facilitates interactions between the host and the tumor. Immune cells are critical in cancer progression and treatment response, making them essential prognostic markers in oncology15,16,17. In this study, we utilized various bioinformatics tools to explore the relationship between SYT15 expression and TME components. Our results showed a positive correlation between SYT15 expression and the presence of multiple immune cell types within tumor tissues. Specifically, the SYT15 high-expression group exhibited higher proportions of B.cells.memory, Macrophages.M1, Mast.cells.resting, and T.cells.CD4.memory.resting, suggesting that elevated SYT15 expression is linked to increased immune cell infiltration. As well, single-cell analysis demonstrated that SYT15 expression varied across multiple cell types in LUAD, with the highest expression observed in Mono/Macro, CD8Tex, and CD8T cells, which are pivotal in tumor immunity18,19.
Immune checkpoints, such as programmed death receptors and their ligands, play a pivotal role in cancer immunotherapy by regulating the immune system’s response to tumors. Blocking these checkpoints, the interaction between PD-1 on immune cells and the ligand PD-L1/PD-L2 on tumor cells enhances the ability of the immune system to target and kill cancer cells20. CTLA-4 is typically expressed on T cells21, while LAG3 and TIGIT are found on T cells, B cells and Tregs, playing crucial roles in T-cell regulation and immune homeostasis22,23,24. HAVCR2, a more recently identified immune checkpoint protein, is present on T cells, Tregs, dendritic cells, and macrophages25,26,27. In our study, We observed expression of a number of ICGs, including, PDCD1LG2, CTLA4, ITPRIPL1, PDCD1, CD274, SIGLEC15, TIGIT and HAVCR2, were significantly elevated in the high SYT15 expression group. The TIDE scores further indicated that patients with high SYT15 expression might respond more favorably to immunotherapy. These discoveries imply that SYT15 may be play a key role in modulating the TME, particularly in terms of immune cell infiltration and tumor immunotherapy, thus affecting the development of LUAD.
To explore the role of SYT15 in LUAD further, we performed a series of cellular experiments. We found that SYT15 expression was significantly lower in A549 and H1975 cells compared to other lung cancer cell lines. Overexpressing SYT15 in A549 and H1975 cells significantly reduced their proliferation, colony formation, migration, and invasion capabilities. These experimental outcomes are consistent with our bioinformatics analyses, supporting the hypothesis that SYT15 acts as a tumor suppressor in LUAD.
The PI3K/AKT pathway is crucial for maintaining cell survival within the tumor microenvironment and is involved in cellular stress responses28. This pathway is typically activated by growth factors such as fibroblast growth factor (FGF) and epidermal growth factor (EGF), which bind to enzyme-coupled tyrosine kinases (RTKs). Phosphatidylinositol 3-kinase (PI3K), a lipid kinase, phosphorylates the 3′-OH group of phosphatidylinositol phosphates29. PI3K is classified into three classes, among which class I PI3K has been most extensively studied. This class forms a heterodimer composed of p110 (encoded by PIK3CA) and p85 (encoded by PIK3R1)30. PI3K activation catalyzes the phosphorylation of hydroxyl groups at position 3 of PIP2 to generate PIP3, which activates AKT. Subsequently, AKT exerts two key effects: it regulates cell proliferation and division by modulating GSK3β, and it recruits mTOR. mTOR then influences transcription factors such as S6K1 and 4EBP, which in turn promotes protein synthesis, accelerates the cell cycle, and inhibits apoptosis31. During cancer progression, the hyperactivation of the PI3K/AKT pathway supports the tumor’s metabolic and nutritional needs, driving excessive cell division, angiogenesis, and growth while suppressing apoptosis and autophagy, thereby facilitating tumor progression32. Our study found that overexpression of SYT15 in A549 cells resulted downregulation of gene expression associated with the PI3K/AKT pathway, as revealed by RNA sequencing. These findings were confirmed by qPCR and WB, indicating that overexpression of SYT15 inhibits proliferation, migration, and invasion of A549 cells through down-regulation of the PI3K/AKT pathway. To further validate this regulatory mechanism, we treated SYT15-overexpressing A549 cells with the PI3K agonist 740Y-P. The results showed that 740Y-P could significantly reverse the inhibitory effects of SYT15 overexpression on the proliferation, migration, and invasion capacities of A549 cells. Meanwhile, WB assay confirmed that 740Y-P was able to abrogate the regulatory effect of SYT15 on the PI3K/Akt signaling pathway.
Conclusion
We investigated the pathological role and intrinsic mechanism of SYT15 in LUAD using bioinformatics and experimental methods. The results indicate that SYT15 plays a tumor-suppressive role in LUAD pathogenesis. Due to its strong association with disease progression, SYT15 is expected to be a novel prognostic biomarker and a new target for the treatment of LUAD.
Materials and methods
Patients and data sources
We collected postoperative cancer tissue and adjacent non-cancerous tissue samples from 40 LUAD patients treated at the First Affiliated Hospital of Bengbu Medical College between November 2023 and April 2025. The TCGA cohort comprised 516 LUAD patients, while the GEO cohort included 226 LUAD patients from the GSE31210 dataset. Clinical information and SYT15 expression data were obtained from the TCGA and GEO cohorts, and single-cell data for LUAD were sourced from the GSE99254 dataset. All patients were fully informed about the study and provided written consent. The study was approved by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical College (Ethics Approval Number: 2023–265).
Prognostic analysis
Patients in the TCGA cohort were categorized into high/ low SYT15 expression groups based on the median SYT15 expression level. Kaplan–Meier survival curves were generated using the R software package survival and assessed for significance by Log-rank test. Univariate-Cox and multivariate-Cox regression analyses were performed and the results of the analyses were presented using forest plots drawn by the forestplot software package, which showed the p-values.
Drug sensitivity analysis
Patients in the TCGA cohort were classified into high-SYT15 and low-SYT15 expression groups based on the median SYT15 expression level. To predict chemotherapy response in both groups, we utilized the Genomics of Drug Sensitivity in Cancer (GDSC) database, the largest publicly available pharmacogenomic resource. We used the R package pRRophetic to predict drug sensitivity by estimating the half-maximal inhibitory concentration (IC50) of each sample via ridge regression. For this analysis, we applied default settings, corrected batch effects using ComBat, included all tissue types, and averaged repeated gene expression values.
Tumor microenvironment (TME) analysis
The CIBERSORT algorithm was employed to estimate immune cell abundance in the TCGA cohort. Immune cell infiltration for each sample was evaluated using a matrix of expression profiles for 22 immune cell types provided by CIBERSORT. The standardized gene expression data were deconvoluted with the CIBERSORT package using default settings, yielding the proportions of various immune cells in each sample. For single-cell data from the GSE99254 dataset, the R software packages MAESTRO and Seurat were used for processing and analysis, with t-SNE applied for cell re-clustering. Patients in the TCGA cohort were classified into high-SYT15 and low-SYT15 expression groups based on the median SYT15 expression level. The expression values of 10 immune checkpoint-related genes were extracted and analyzed in both groups using the R packages ggplot2 and pheatmap. Additionally, the TIDE algorithm was used to predict potential responses to immunotherapy.
Cell culture and transfection
The two cell lines (A549 and H1975) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and the cells were cultured with high-saccharide DMEM (gibco, USA) containing a mixture of 10% FBS (Procell, China) and 1% mass fraction penicillin–streptomycin (gibco, USA), and incubated at 37 °C in a 5% CO2 incubator by volume. The SYT15 coding sequence was inserted into the lentiviral vector CMV-MCS-EF1α-ZsGreen1-PGK-Puro, which was then packaged into lentivirus and used to transduce the cells. Stable SYT15-overexpressing cell lines were selected with puromycin.
Quantitative PCR
Tissues or cells were lysed with TRIzol reagent (Tiangen, Beijing). After sequential extraction with chloroform, isopropanol, and 70% ethanol, mRNA was isolated. This mRNA was then reverse-transcribed into cDNA using the HiScript III 1st Strand cDNA Synthesis Kit (with gDNA wiper; Vazyme, China). cDNA was quantified using TB Green (Takara, Japan), and the relative expression of the target genes was calculated by 2-ΔΔCt with β-ACTIN as the internal reference gene. The primers were synthesized by Shanghai Ltd. and are shown in Supplementary Table S1.
Western blot
Cells were lysed and total proteins were extracted using RIPA lysing solution containing 1% protease inhibitor (Beyotime, Shanghai), and the concentration of extracted proteins was determined using the BCA method (Beyotime, Shanghai). After electrophoresis, proteins were transferred to a membrane. The membrane was then blocked with 5% bovine serum albumin (BSA), followed by incubation with the primary antibody at 4 °C overnight. On the next day, the secondary antibody was added and incubated for 1 h, and protein signals were detected using ECL luminescent solution (Millipore, USA). Primary antibody: mouse GAPDH antibody (Sanying, Wuhan), rabbit SYT15 antibody (Biorbyt, UK), rabbit PIK3CA antibody (Sanying, Wuhan), rabbit PIK3R1 antibody (Sanying, Wuhan), rabbit PIK3C2A antibody (Sanying, Wuhan), rabbit PIK3R3 antibody (Sanying, Wuhan), rabbit AKT1S1 antibody (Sanying, Wuhan) rabbit AKTIP antibody (Sanying, Wuhan). Secondary antibody: goat anti-rabbit lgG (Sanying, Wuhan), goat anti-mouse lgG (Sanying, Wuhan).
CCK-8 cell viability assay
After cell counting, the cells were inoculated in 96-well plates at a density of 2000 cells/well, and the absorbance values at the time points of 0 h,24 h,48 h and 72 h were detected after the cells were attached to the wall. Before detection, 10 μL of CCK-8 reagent (UElandy, Shanghai) was added to each well, and then incubated in a CO2 incubator for 30 min, and the absorbance at 450 nm was measured using an enzyme counter.
Colony formation assay
After cell counting, cells were inoculated in 6-well plates at a density of 1000 cells/well. after two weeks of incubation in a CO2 incubator, cells were fixed with 4% paraformaldehyde for 15 min, washed with PBS, stained with 1% crystal violet for 10 min, and washed with PBS for three times and then counted under a microscope (≥ 50 cell colony numbers).
Wound healing assay
Prior to cell seeding, a horizontal line was drawn on the bottom of each 6-well plate using a marker pen. An appropriate number of cells were then seeded into the plates. Once the cells reached confluence, they were scratched longitudinally with a sterile 10 μL pipette tip. The medium was replaced with serum-free medium, and the plates were placed in a CO2 incubator; this time point was recorded as 0 h. Photographs were taken with the use of a microscope at 0 h, 24 h, 48 h, and 72 h, respectively.
Cell migration assay
After cell counting, 50,000 cells were taken, resuspended in 200ul of serum-free medium and then inoculate into Transwell chambers. In the lower chamber, 500 μL of complete medium was added. After 36 h of cell culture, the cells were fixed with 4% PFA for 20 min, washed with PBS, stained with 1% violet crystal for 15 min, the cells were wiped off from the inner surface of the Transwell membrane with a cotton swab, photographed and counted under an inverted microscope.
Cell invasion assay
Spread 60 μL of 1:8 diluted matrix gel (Corning USA) into the upper chamber of the Transwell in advance, and place it in a CO2 incubator for 3 h. After cell counting, 100,000 cells were taken and resuspended in 200ul of serum-free medium and then inoculate into Transwell chambers. In the lower chamber, 500 μL of complete medium was added. After 48 h of cell culture, the cells were fixed with 4% PFA for 20 min, washed with PBS, stained with 1% violet crystal for 15 min, the cells were wiped off from the inner surface of the Transwell membrane with a cotton swab, photographed and counted under an inverted microscope.
Immunofluorescence
Take the appropriate amount of cells inoculated in the chamber slides, when the cell confluence to about 80%, discard the medium, PBS wash and add 200ul 4% PFA room temperature fixed 20 min, PBS wash 3 times. Add the blocking punch solution (PBS containing 3% BSA + 0.4% Triton X-100) for 1 h. Add primary antibody and incubate at 4 °C overnight, then PTW (PBS containing 1/1000 Tween 20) for 3 times on the next day, and incubate the secondary antibody for 1 h. After PTW washed for 3 times, add DAPI (Beyotime, Shanghai), and then take pictures under a fluorescence microscope and count them. Primary antibody: rabbit KI67 antibody (Sanying, Wuhan), secondary antibody: sheep anti-rabbit CY3 (Sanying, Wuhan).
Statistical analysis
Image analysis using Image J software, and plotting using GraphPad Prism 9.0 software. Other differences between the two groups were compared using the independent samples t-test. p < 0.05 was considered significant.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74(3), 229–263. https://doi.org/10.3322/caac.21834 (2024).
Thai, A. A., Solomon, B. J., Sequist, L. V., Gainor, J. F. & Heist, R. S. Lung cancer. Lancet 398(10299), 535–554. https://doi.org/10.1016/s0140-6736(21)00312-3 (2021).
Moghadam, P. K. & Jackson, M. B. The functional significance of synaptotagmin diversity in neuroendocrine secretion. Front. Endocrinol. 4, 124. https://doi.org/10.3389/fendo.2013.00124 (2013).
Henry, D., Joselevitch, C., Matthews, G. G. & Wollmuth, L. P. Expression and distribution of synaptotagmin family members in the zebrafish retina. J. Comp. Neurol. 530(4), 705–728. https://doi.org/10.1002/cne.25238 (2022).
Wolfes, A. C. & Dean, C. The diversity of synaptotagmin isoforms. Curr. Opin. Neurobiol. 63, 198–209. https://doi.org/10.1016/j.conb.2020.04.006 (2020).
Tomes, C. N. et al. The proteins of exocytosis: Lessons from the sperm model. Biochem. J. 465(3), 359–370. https://doi.org/10.1042/bj20141169 (2015).
Debnath, J., Gammoh, N. & Ryan, K. M. Autophagy and autophagy-related pathways in cancer. Nat. Rev. Mol. Cell Biol. 24(8), 560–575. https://doi.org/10.1038/s41580-023-00585-z (2023).
Stefańska, K. et al. The role of exosomes in human carcinogenesis and cancer therapy-recent findings from molecular and clinical research. Cells 12(3), 356. https://doi.org/10.3390/cells12030356 (2023).
Minoru, K. et al. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/27.1.29 (2000).
Fukuda, M. et al. Moleclar cloning and characterization of human, rat, and mouse synaptotagmin XV. Biochem. Biophys. Res. Commun. 306(1), 64–71. https://doi.org/10.1016/s0006-291x(03)00911-2 (2003).
Junttila, M. R. & de Sauvage, F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501(7467), 346–354. https://doi.org/10.1038/nature12626 (2013).
Ghoshdastider, U. et al. Pan-cancer analysis of ligand–receptor cross-talk in the tumor microenvironment. Cancer Res. 81, 1802–1812. https://doi.org/10.1158/0008-5472.can-20-2352 (2021).
Li, Z. X. et al. Comprehensive characterization of the alternative splicing landscape in head and neck squamous cell carcinoma reveals novel events associated with tumorigenesis and the immune microenvironment. Theranostics 9, 7648–7665. https://doi.org/10.7150/thno.36585 (2019).
Zhou, R. et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I–III colon cancer. Cancer Immunol. Immun. 68, 433–442. https://doi.org/10.1007/s00262-018-2289-7 (2019).
Xie, P. et al. Development of an immune-related prognostic signature in breast cancer. Front. Genet. 10, 1390. https://doi.org/10.3389/fgene.2019.01390 (2020).
Fendl, B., Berghoff, A. S., Preusser, M. & Maier, B. Macrophage and monocyte subsets as new therapeutic targets in cancer immunotherapy. ESMO Open 8(1), 100776. https://doi.org/10.1016/j.esmoop.2022.100776 (2023).
Yang, G. et al. Functional status and spatial architecture of tumor-infiltrating CD8+ T cells are associated with lymph node metastases in non-small cell lung cancer. J. Transl. Med. 21(1), 320. https://doi.org/10.1186/s12967-023-04154-y (2023).
Zheng, H. et al. New insights into the important roles of tumor cell-intrinsic PD-1. Int. J. Biol. Sci. 17(10), 2537–2547. https://doi.org/10.7150/ijbs.60114 (2021).
Van Coillie, S., Wiernicki, B. & Xu, J. Molecular and cellular functions of CTLA-4. Adv. Exp. Med. Biol. 1248, 7–32. https://doi.org/10.1007/978-981-15-3266-5_2 (2020).
Durham, N. M. et al. Lymphocyte activation gene 3 (LAG-3) modulates the ability of CD4 T-cells to be suppressed in vivo. PLoS ONE 9(11), e109080. https://doi.org/10.1371/journal.pone.0109080 (2014).
Godfrey, J. et al. TIGIT is a key inhibitory checkpoint receptor in lymphoma. J. Immunother. Cancer 11(6), e006582. https://doi.org/10.1136/jitc-2022-006582 (2023).
Pan, C. et al. Poliovirus receptor-based chimeric antigen receptor T cells combined with NK-92 cells exert potent activity against glioblastoma. J. Natl. Cancer I. 116(3), 389–400. https://doi.org/10.1093/jnci/djad226 (2024).
Phong, B. L. et al. Tim-3 enhances FcεRI-proximal signaling to modulate mast cell activation. J. Exp. Med. 212(13), 2289–2304. https://doi.org/10.1084/jem.20150388 (2015).
Li, H., Yang, D., Hao, M. & Liu, H. Differential expression of HAVCR2 gene in pan-cancer: A potential biomarker for survival and immunotherapy. Front. Genet. 13, 972664. https://doi.org/10.3389/fgene.2022.972664 (2022).
Song, D. et al. Case Report: HAVCR2 mutation-associated Hemophagocytic lymphohistiocytosis. Front. Immunol. 14, 1271324. https://doi.org/10.3389/fimmu.2023.12713249 (2023).
Johnson, D. E. et al. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 6(1), 92. https://doi.org/10.1038/s41572-020-00224-3 (2020).
Fresno Vara, J. A. et al. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 30(2), 193–204. https://doi.org/10.1016/j.ctrv.2003.07.007 (2024).
Liu, Y. et al. The regulatory role of PI3K in ageing-related diseases. Ageing Res. Rev. 88, 101963. https://doi.org/10.1016/j.arr.2023.101963 (2023).
Ma, L., Zhang, R., Li, D., Qiao, T. & Guo, X. Fluoride regulates chondrocyte proliferation and autophagy via PI3K/AKT/mTOR signaling pathway. Chem. Biol. Interact. 349, 109659. https://doi.org/10.1016/j.cbi.2021.1096599 (2021).
Carnero, A., Blanco-Aparicio, C., Renner, O., Link, W. & Leal, J. F. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr. Cancer Drug tar. 8(3), 187–198. https://doi.org/10.2174/156800908784293659 (2008).
Acknowledgements
Thanks to the First Affiliated Hospital of Bengbu Medical College for his permission to publish this article. We would also like to extend special thanks to the technical staff of Anhui Clinical and Preclinical Key Laboratory of Respiratory Disease for their excellent laboratory assistance. Moreover, we thank all the clinicians of Thoracic Surgery Department who participated in the patient’s management.
Funding
This work was supported by Scientific Research Project of Colleges and Universities in Anhui Province(No. 2024AH051254), National Natural Science Foundation of China (82373329); Research Funds of Joint Research Center for Regional Diseases of Institute of Health and Medicine (IHM), Hefei Comprehensive National Science Center (2024bydjk007, 2024bydjk008); Anhui Provincial Major Science and Technology Project for Key Laboratory (S202305a12020038).
Author information
Authors and Affiliations
Contributions
M.T. , L.D. and N.W.conceived and designed the study. Y. L. and X L. conducted bioinformatics analysis. L. J. and H W. conducted cytological experiments. M.T. drafted the manuscript. X.Q., X.W. and N.W. reviewed the manuscript and provided valuable edits. All authors have read and approved the submitted version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
All patients were fully informed about the study and provided written consent. The study was approved by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical College (Ethics Approval Number: 2023–265). This study confirms that all experiments were conducted in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tian, M., Deng, L., Jin, L. et al. Clinical significance and potential molecular mechanisms of SYT15 expression in lung adenocarcinoma. Sci Rep 16, 2554 (2026). https://doi.org/10.1038/s41598-025-32512-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-32512-x