Abstract
Oral tumors are relatively common in dogs, and canine oral squamous cell carcinoma (COSCC) is the most prevalent oral malignancy of epithelial origin. COSCC is locally aggressive with up to 20% of patients showing regional or distant metastasis at the time of diagnosis. The treatment of choice most typically involves wide surgical excision. Although long-term remission is possible, treatments are associated with considerable morbidity and can negatively impact functionality and quality of life. OSCCs have substantial upregulation of the RAS-RAF-MEK-MAPK signaling axis, and we had previously hypothesized that small-molecule inhibitors that target RAS signaling might effectively inhibit tumor growth and progression. Here, we demonstrate that the MEK inhibitor trametinib, an FDA-approved drug for human cancers, substantially inhibits the growth of six COSCC cell lines established from current patient tumor samples. We further show preliminary clinical evidence that the drug is able to cause ~ 40% and ~ 80% tumor regression in two out of four patients with spontaneously occurring COSCC, a partial response according to commonly used RECIST criteria. Given the limited treatment options available and the number of dogs for which standard of care is not acceptable, these preliminary findings provide new hope that more suitable treatment options may soon enter the veterinary clinic.
Similar content being viewed by others
Introduction
Oral tumors account for approximately 6% of all neoplasms in dogs. Canine oral squamous cell carcinoma (COSCC) represents by far the most common oral malignancy of epithelial origin1, exhibiting relatively high proliferation activity based on Ki67 immunostaining, particularly when compared to another common and locally invasive oral neoplasm of epithelial origin, canine acanthomatous ameloblastoma (CAA)2,3. Importantly, up to 20% of dogs presenting with COSCC have regional or distant metastasis at the time of initial diagnosis, leading to difficulties in treatment1,4,5. Locally, the primary tumor rapidly invades multiple anatomical planes including underlying osseous structures, resulting in severe pain, loss of function, and in some cases pathological fracture6. In the absence of regional or distant metastasis, the treatment of choice for COSCC is en bloc excision of the affected area of the mandible or maxilla (i.e., mandibulectomy or maxillectomy)1,7,8. Complete excision is associated with a high remission rate for e.g. oral tumors, while oropharyngeal tumors, such as those arising from the tonsils, have worse prognosis9. Moreover, excision is technically complex and expensive, and can result in substantial disfigurement and dysfunction1,7,10,11. Potential surgical sequelae include oronasal fistula, chronic ptyalism, protrusion of the tongue, and occlusal trauma1,7,12. Complications have been reported in ~ 37% of dogs, with ~ 38% of those complications being considered serious13. Conversely, incomplete excision often results in tumor recurrence and disease progression1. In our experience, a typical year in our clinic has ~ 15% of dogs arriving with COSCC that are not eligible for surgery due to disease progression, while even for dogs who are eligible, ~ 25% of owners reject standard of care due to cost, predicted morbidity, or other factors. Palliative courses of radiotherapy and chemotherapy have been described but are usually reserved for cases considered inoperable due to tumor size or the anatomical structures involved, for incompletely excised tumors, or for patients with regional or distant metastasis9,14,15. Moreover, radiotherapy and chemotherapy are also associated with negative side effects and mixed outcomes16,17. Thus, improved treatment options with reduced side effects are needed. Modern oncological targeted interventions based on small-molecule inhibitors have the potential to replace or complement traditional therapeutic options and help reduce patient morbidity while improving outcomes18,19.
We have previously demonstrated that most canine oral tumors of epithelial origin exhibit substantial activation of the RAS-RAF-MEK-MAPK (hereafter: RAS) pathway20,21,22. This is similar to human head and neck cancers, which also frequently hyperactivate RAS pathway components23,24. The RAS pathway is of major interest in oncology because it is mutated in a large percentage of cancers in humans and dogs25,26,27,28. The RAS pathway has been recognized as a driver in many cancers, and hyperactivation of RAS and its signaling partners leads to increased and unrestricted tumor growth29,30,31. Targeting the RAS pathway has therefore been a major objective globally. While RAS itself is traditionally difficult to inhibit directly, several of its downstream effectors have been targeted successfully with various inhibitors. One such target is MEK, a serine/threonine kinase that is activated by RAS and its partner RAF, and in turn activates MAPK and ERK in order to promote gene transcription and cell proliferation. Trametinib is a well-studied small-molecule inhibitor of MEK32, has been approved for use in several indications in humans33,34, and is being studied in combination with various other inhibitors to further improve its efficacy33,35,36,37,38. Moreover, trametinib is specifically approved for use in human melanoma with a BRAF p.V600E or p.V600K mutation34. Analogous BRAF p.V595E mutations are highly prevalent in COSCC, particularly in the papillary histological subtype of the tumor, which tends to proliferate very rapidly22. Although less frequent, somatic HRAS p.Q61L mutations, which constitutively activate RAS and downstream signaling, have also been identified in COSCC2,20,21. Trametinib has been shown in pre-clinical studies to be effective in treating several canine cancers39,40,41,42, and in the clinical setting trametinib has reportedly been used “off-label” for canine histiocytic sarcoma43. Taken together, these results suggest that trametinib may serve as an effective targeted therapy option for future management of neoplasms in dogs. In this study, we pursue this hypothesis through three primary objectives: developing a series of COSCC cell lines, using the cell lines to test chemotherapeutic and targeted therapeutic agents including trametinib, and piloting a clinical trial in canine patients to show that trametinib can debulk COSCC tumors.
Results
Development of COSCC model systems
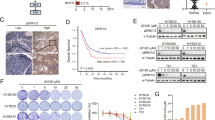
We began our study by generating patient-derived xenograft (PDX) models in NOD scid gamma (NSG) mice. We collected samples from seven dogs with spontaneously occurring OSCC tumors (Table 1). Minced tumor segments (average diameter 2–3 mm) were implanted into the left flank of 5–6 NSG mice within 6 h of collection. The mice exhibited no signs of distress, and tumors generally became palpable in ~ 24–28 weeks. Once the tumors reached a larger size at ~ 36–40 weeks, the mice were euthanized and the tumors were cryopreserved, with one tumor optionally being randomly selected and passaged into a new generation of mice. In each passage/generation, tumor sections were compared histologically to samples from the original tumor (Fig. 1). While one xenografted tumor (270,858, Fig. 1A) appeared to have noticeably altered phenotype by its second passage in mice, the other PDX models largely maintained the features of the parent tumor (Fig. 1B, C).
PDX Model Characterization. Representative 10X light photomicrograph images of formalin-fixed and paraffin-embedded samples taken from three primary COSCCs corresponding to the (A) basaloid (bOSCC; tumor 270,858,), (B) conventional (cOSCC; tumor 294,246) and (C) papillary (pOSCC, tumor 301,313) histological subtypes, and corresponding patient-derived xenograft (PDX) samples. Except for sample 270_PDX301 (P3) (panel A), which corresponded to a murine tumor that formed at the implantation site upon second passage, note the consistent neoplastic epithelial cell (arrows) invasion of the mesenchymal stroma (asterisks) in the primary tumor and PDX samples observed on hematoxylin and eosin (H&E) and pan-cytokeratin stains. Also, scattered Ki67 nuclear immunoreactivity of proliferating neoplastic epithelial cells (arrows) in the primary tumor samples is similar to the corresponding PDX samples.
We next turned to developing stable 2D cell culture models of COSCC. Tumor samples were harvested from either our PDX-bearing mice (n = 5), or directly from patients (n = 2, Table 1). Two sequential passages of PDX tumor 270,858 were used to generate cell lines Co-B-658 and Co-C-958. Two sequential passages of PDX tumor 294,236 were used to generate cell lines Co-F-1236 and Co-L-1236. A single cell line was derived from each other tumor sample. The tumor tissue in each case was digested in collagenase II and trypsin, and epithelial cells were enriched gravimetrically44. The cells were placed in tissue culture-treated plates, and the plastic-adherent cells were then cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). Figure 2 shows that the cells adopted a variety of different morphologies, but that different collections from genetically identical PDX models looked very similar, as expected.
Micrographs of COSCC cell lines. Photographs of cells (A–I) were taken on a phase contrast microscope. Cells did not noticeably change morphology over ~ 10 passages/2–3 months. A different cell line is shown in each panel A-I, and the cell line name is labeled immediately underneath the photograph. Scale bar = 200 µM.
COSCC is a disease of epithelial origin, and our goal was to enrich for epithelial cells while discarding infiltrating fibroblasts. However, differentiating between epithelial and fibroblastic cells in this situation is difficult. Gene set enrichment analysis previously demonstrated that COSCC tumors highly express genes supporting epithelial-to-mesenchymal transition (EMT)20,23, which can cause epithelial cells to adopt a fibroblast-like morphology and to express many of the same protein markers which might traditionally be used to identify fibroblasts (e.g. SNAIL or vimentin45,46,47,48). Examination of our cell morphologies (Fig. 2) bears this out. Cell lines Co-G-1114, Co-I-913, and Co-O-172 (Fig. 2E, G, I) have block-like morphologies largely similar to other known epithelial cells. Co-L-1236 and Co-F-1236 (from genetically identical PDX model samples, Fig. 2C, D) are spindly, triangular cells appearing to be partway between epithelial and fibroblastic cell shapes and resemble some highly aggressive epithelial cell cancers such as the MDA-MB-231 human breast cancer cell line. Co-B-658 and Co-C-958 (from genetically identical PDX model samples, Fig. 2A, B), Co-N-1242 (Fig. 2F), and Co-J-1220 (Fig. 2H), meanwhile, are long, thin cells which look predominantly fibroblastic. Thus, more advanced characterization techniques than morphology analysis were required to help define these cell culture models.
Model system characterization
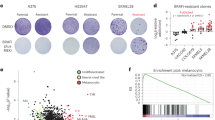
As we turned our attention to characterization of each model system, our major concern was determining whether each model was canine or murine in origin. We began by conducting Western blots for each cell line against a cyclophilin A (CyPA, an abundant and ubiquitous cytosolic protein) antibody known to bind murine protein specifically. This antibody had previously been demonstrated to selectively stain murine cells which invaded xenograft models of human cells in mice49. The canine CyPA has an even lower sequence identity to murine CyPA than does the human protein (Supplemental Figure S1A). Figure 3A shows that of the cell lines we isolated, three (Co-B-658, Co-C-958, and Co-J-1220) had detectable mouse protein, while the others did not (representative uncropped membranes are presented in Supplemental Figure S1B). Two murine cancers (4T1 and EO771 breast cancer cells), and one human cancer (U87 MG glioma cells), are included on the blot for comparison. We had previously found that canine oral cancers have broad upregulation of RAS-RAF-MEK-ERK signaling20,21,22, and so examined ERK phosphorylation as a marker of RAS activation. While phospho-ERK staining was different in each cell line (Fig. 3A), it was detectable in all cells to a similar extent to U87 MG cells, which are generally considered to have activated RAS signaling50, thus demonstrating that RAS signaling was intact in our cell lines.
Characterization of COSCC cell lines. (A) Cell lysates were resolved via SDS-Page, and blotted for murine cyclophilin A (CyPA), phospho-ERK (p.ERK), or vinculin (loading control). 4T1 and E0741 are murine breast cancer cell lines, which show a signal for the CyPA protein, while U87 is a human glioblastoma cell line which is negative for the protein. Three of the novel cell lines show signal for murine protein. All of the novel cell lines have some phospho-ERK signal, suggesting intact RAS signaling. (B) For each cell line, 1000 cells were plated into a 10 cm plate, and allowed to grow for 10–20 days. The plates were stained with crystal violet to visualize colony formation.
Following this, we addressed the issue of whether cells were derived from healthy or tumor tissue. Each cell line was cultured for a minimum of 10 weeks/15–20 passages to ensure some degree of immortality, with several cell lines being cultured for at least 50 passages/6 months. As healthy epithelial cells tend to senesce within 3–4 passages51, this demonstrated a considerable degree of immortalization in our cells, strongly suggesting that they were derived from tumor tissue. Notably, while we were able to passage cell lines Co-I-913 and Co-N-1242 for the 10-week period, both proved difficult to culture, and required that relatively high confluence be maintained at all times or the cells ceased to divide. The other cell lines were generally split from 70–80% confluence 1:10, and regained confluence within 3–5 days.
We next examined the ability of the cells to evade contact inhibition of growth, as most (but not all) cancer cells are able to sustain growth even following contact inhibition. Cells were plated at low density and allowed to form colonies (Fig. 3B). Most of the cell lines formed dense colonies under these conditions, strongly supporting a cancer phenotype. Cell lines Co-I-913 and Co-N-1242 did not form detectable colonies, but this was unsurprising given the previous observation that they must be kept at relatively high density to proliferate.
At this point, we wanted a simple mechanism by which to positively identify our cells as being related to the original tumors from which they were derived, and we turned to genotype analysis. The CanineHD beadchip (Illumina Inc.) contains approximately 220,000 single nucleotide polymorphisms (SNPs) across the canine genome52. We collected DNA from tumor samples, PDX-samples, and cell line samples, and submitted the DNA for genotyping. While multiple generations/passages of PDX model were available for some tumors, we conducted genotyping only of PDX passages leading up to cell line derivation. Cell lines Co-B-658, Co-C-958, and Co-J-1220 failed due to high levels of SNP missingness. This strongly suggests that they are not canine cells and are instead immortalized mouse cells. Interestingly, we examined three different generations of the PDX tumor (270,858) that led to the Co-B-658 and Co-C-958 cell lines. The first generation of the tumor was genotyped successfully, but the second and third generations (from which the cell lines were isolated) also had a high rate of SNP missingness. Combined with our histological analysis, which suggested a dramatic shift in tumor morphology from the original PDX tumor to later passages, this strongly supports the hypothesis that a murine tumor was growing from the second passage onward. The tumor and PDX samples leading to the Co-J-1220 cell line (tumor 303,520) did not fail, suggesting that cell isolation may simply have captured spontaneously immortalizing murine fibroblasts.
We proceeded to analyze the genotype data from the samples which were canine in nature. We began by pruning the dataset—we removed SNPs that were missing in some data, or which were highly correlated, leaving 13,178 SNPs in the dataset. The data for each canine tumor, PDX model (1–3 per tumor), and cell line (1–2 per tumor) were then compared by calculating ^π (pi-hat), an identity-by-descent metric which takes into account the probabilities that 0, 1 or 2 alleles are shared between two samples53. ^π values are ~ 1 if two samples are genetically identical. Table 2 shows that most tumor, PDX, or cell-line pairs from the same sample had ^π values over 0.97, suggesting near identity with one another. A notable outlier is Tumor 301,814, which showed ^π of 0.82 compared to the derived PDX-model, and 0.76 compared to derived cell line Co-G-1114. The PDX-model and cell line were more closely related to one another, with ^π of 0.94, which suggests that while the cell line is less similar to the original tumor than the PDX model is, a greater degree of genetic mutation might have occurred while establishing the PDX model from the tumor than while establishing the cell culture model from the PDX sample.
Following this analysis, we selected 168 SNPs to form a ‘fingerprint’ for each cell line (Fig. 4A, Supplemental Table S1), which could be used as authentication in the future to determine the extent of genetic drift due to passaging, or the presence of contaminating cells. Pairwise identity by descent (IBD) measures using the 168 fingerprint SNPs calculated between samples in the present study and previously genotyped dogs (n = 880) shows that no other sample is closely related (all ^π values < 0.25) (Supplemental Table S2). Of note, cell line Co-L-1236 was genotyped twice: at passage 14 and at passage 42, and these had a pairwise ^π value of 0.979 using 13,178 SNPs and were identical using our 168 SNP fingerprint. As a final test, we wanted to determine if our cell culture models could reconstitute tumors in mice. NSG mice were implanted with either the Co-F-1236 or Co-G-1114 cell lines (2 X 106 cells suspended in Matrigel) and tumors were allowed to grow. Growth for either line was initially slow, with Co-F-1236 taking ~ 4 months and Co-G-1114 taking ~ 12 months to form palpable tumors. However, when we transplanted a Co-F-1236 tumor into new mice as though it were a PDX model, we found that the host-adapted tumor grew rapidly, reaching a size mandating euthanasia (~ 2000 mm3) in ~ 2–3 months (Fig. 4B).
Further characterization of COSCC cell lines. (A) Select SNPs were chosen from genotyping data in order to develop a ‘fingerprint’ for each COSCC cell line. Colors represent genotypes (0 = blue, 1 = yellow, 2 = red). (B) Co-F-1236 cells were suspended in Matrigel and implanted into the left flank of NSG mice. The mice grew large tumors (indicated by red arrow) at the injection site.
New approaches to inhibiting COSCC growth
With our model systems in hand, we turned our attention to testing novel therapeutic approaches. Since comparatively few approaches to COSCC have been demonstrated, we began by testing approaches that work against some head and neck cancers in humans. The current chemical standard of care for OSCC in people is based around three chemotherapeutics: docetaxel, cisplatin, and 5-fluorouracil (5-FU)19. Cisplatin is comparatively difficult to handle due to chemical instability in solvating agents such as DMSO and the need to be prepared freshly before use, and so we began by studying docetaxel and 5-FU. Each cell line was dispensed at low density into 96 well plates and treated with different concentrations of each drug. Despite having been identified as murine cells, we conducted experiments on cell lines Co-B-658, Co-C-958, and Co-J-1220 as non-canine controls. Figure 5 shows the dose curves for each drug determined for cell line Co-F-1236, and IC50 data for all cell lines is compiled in Supplemental Table S3. These dose responses were compared to the average IC50 for either drug from the NCI-60, a set of 60 human cancer cell lines (Fig. 6). The doses for either drug were comparable to the NCI-60 averages, with no comprehensive difference between the canine cancer and murine cells. We proceeded to examine the EGF-receptor targeting antibody cetuximab, which has seen some use in human oral cancers54,55,56. This showed very little potency in our COSCC cell lines compared to the NCI-60 average, although it is unclear if this is due to resistance to EGF-receptor inhibition, species specificity of the antibody, or some other factor. We then examined two other chemotherapeutic agents commonly used in some non-oral human cancers, carmustine and temozolomide. While the cell lines were generally sensitive to carmustine, they were comparatively resistant to temozolomide. And again, in either case, there was no clear difference between the sensitivity of canine and murine cell lines.
Dose curves for assorted drugs vs. COSCC cell line Co-F-1236. Cells were plated at low density (2000 cells per well) in 96 well plates and treated for 6 days with the indicated amounts of each inhibitor (A) docetaxel, (B) 5-fluorouracil, (C) cetuximab, (D) carmustine, (E) temozolomide, (F) trametinib, (G) dabrafenib). The cells were then treated with PrestoBlue reagent, and cell viability was determined. %Inhibition was determined by comparison to no-drug controls (representing max growth) and cell-free lanes (representing 100% cell death). Each data point represents the average measurement from three samples, and the error bars represent the standard deviation from the mean. The lowest-concentration data point for each graph is a no-drug control and is given a non-zero value to be visible on the semi-log plot.
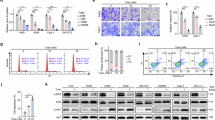
Sensitivity of COSCC to assorted drug approaches. (A–G) The determined IC50 value for each drug (A) docetaxel, (B) 5-fluorouracil, (C) cetuximab, (D) carmustine, (E) temozolomide, (F) trametinib, (G) dabrafenib) for each COSCC cell line was plotted against data from the NCI-60. For each graph, the canine cell lines are highlighted as red bars, and the y-axis crosses the x-axis at the average IC50 value across all NCI-60 cell lines. Notably, the data for carmustine included an extended set of cell lines, so more bars are present than for the other drugs. (H) Co-F-1236 or Co-O-172 cells were treated with trametinib (125 nM or 315 nM respectively) and the indicated amounts of dabrafenib (Dab: 0, 2.5, 5, or 10 µM) for 6 days, and %inhibition of cell growth was determined, as scaled to wells containing zero cells (100% inhibition) and wells containing cells not exposed to any drug (100% growth). Inhibition was only minimally additive for Co-F-1236 cells, while combination treatment actively reduced inhibition rate in Co-O-172 cells. (I) As (H), except the data for each bar is scaled to wells containing dabrafenib alone (100% growth for that condition) to examine the effects of trametinib alone. Here, as the concentration of dabrafenib is increased, the effect due to trametinib appears to decrease.
Each of the chemotherapeutics we examined is known for use-limiting side effects. We set out to determine if a more targeted agent might have similar effects in reducing COSCC growth. For this we chose trametinib, an FDA-approved drug which inhibits MEK1 and MEK2, downstream of RAS and RAF, and upstream of MAPK. Cells were exposed to trametinib (Fig. 5F), and the IC50 values compared to the average from the NCI-60. Interestingly, the cell lines were generally more sensitive to trametinib when compared to other susceptible cancer cell lines (Fig. 6F). Notably, trametinib was the only tested inhibitor that had a strong effect on Co-I-913 cells, and it had the least effect on the three murine cell lines.
Trametinib is often given alongside the BRAF inhibitor dabrafenib, specifically in BRAF p.V600E mutant cancers32,33,38,57. Two of our cell lines were derived from BRAF p.V595E papillary COSCC tumors, and we set out to determine if the two drugs could combine in these cell lines to provide a synergistic effect. We began by treating each cell line with dabrafenib alone and found mixed sensitivity to the drug as a monotherapy (Figs. 5G, 6G), with only one line (the BRAF p.V595E positive Co-O-172) being substantially more sensitive than the NCI-60 average, and four cell lines having very low sensitivity, with less than 50% inhibition of growth at 20 µM dabrafenib. We then treated two cell lines, the BRAF-mutant cell line Co-O-172 and the BRAF-wildtype cell line Co-F-1236, with trametinib and dabrafenib simultaneously. When compared to control DMSO-treated cells, the combination of trametinib and dabrafenib appeared to have a very mildly additive effect in Co-F-1236, but unexpectedly had a subtractive effect in Co-O-172 cells, where lesser inhibition of cell growth was observed with the combined drugs than with trametinib alone (Fig. 6H). However, when dabrafenib-treated cells were used as full-growth controls, and the effect of trametinib alone was calculated, it became apparent that the combination was antagonistic in both cell lines, with the effect due to trametinib decreasing as the concentration of dabrafenib increased (Fig. 6I).
Trametinib treatment of COSCC in vivo
Trametinib as a single treatment had a substantial effect on nearly all of our COSCC cell culture models, while combining it with dabrafenib resulted in only minimal improvement. Thus, we were interested to see if trametinib alone could reduce the growth rate of COSCC tumors in mice. Since our PDX-tumors were very slow growing, we chose to conduct testing on the aggressive and rapidly growing Co-F-1236 xenograft model. Cells (2 × 106) were suspended in Matrigel, and then injected into the left flank of NSG mice. Once tumors became palpable, mice were randomly sorted into two cohorts (N = 6 per group). One cohort was treated with trametinib (0.5 mg/kg daily by oral gavage, 0.125 mg/mL drug stock concentration, Figure S2), and one was treated with an equivalent volume of vehicle alone (10% Cremophor EL, 10% PEG400, 80% PBS), similar to previously described protocols57,58. The amount of drug administered was increased to 0.75 mg/kg on day 12, and mice were sacrificed due to tumor burden on day 25. Tumors were measured 3 times per week with calipers. Representative mice were subjected to MRI before drug treatment began, and immediately prior to sacrifice. Of note, a single mouse died the evening before mice were sacrificed, presumably due to aspiration of the liquid medication. It is included in all of our data due to the brief duration between its death and final tumor measurement.
MRI imaging conducted on representative mice before treatment, and immediately before sacrifice, show that trametinib treatment almost entirely halted tumor growth (single slice images are shown in Fig. 7A–D). Measurement of tumor length and width with calipers and estimation of tumor volume (0.5 * width * width * length) showed a statistically significant difference between control and treated tumors by the 5th day of treatment, with drug-treated tumors showing little to no growth in that timeframe. However, even increasing the dose of drug on day 12 did not cause the tumors to shrink (Fig. 7E). Individual tumor sizes also clustered relatively well, with the exception of a single vehicle-control mouse with a slower growing tumor (Fig. 7F, red arrow). MRI and caliper measurements were mirrored by tumor appearance and weight (Fig. 7G, H). The mice did not show any signs of distress due to drug treatment, and no change in body weight was apparent (Fig. 7I–K), with the exception of the one mouse which died prematurely (Fig. 7J, red arrow). Overall, these results show that trametinib was able to substantially inhibit the growth of an aggressive COSCC tumor without having any acute, adverse effects on the mice hosting the tumor.
In vivo potency of trametinib against COSCC. (A, B) MRI images of tumor-bearing mice before and after treatment with trametinib. Tumors are shown in red. (C, D) MRI images of tumor-bearing mice before and after treatment with carrier control. Tumors are shown in red. (E) Mouse tumors were measured (length and width) with calipers twice a week. Tumor volume was estimated as 0.5 * width (shorter measurement) * width * length (longer measurement). The drug treatment began at 0.5 mg/kg but was increased to 0.75 mg/kg on day 12 (marked with ¥). Datapoints represent the mean value from all six mice in each group, with error bars representing the standard deviation from the mean. The * represents data for which the p-value between treated and untreated samples is < 0.05. The exact p-value for the first statistically significant datapoint is shown. (F) Individual growth curves for each mouse tumor. Curves are grouped into ‘vehicle’ (V) or ‘treated’ (T) groups. One mouse (V1, indicated with a red arrow) was treated only with carrier control, but had a comparatively slow growing tumor. (G) Individual tumors removed from untreated and trametinib treated mice. Tumors from untreated mice are substantially larger than those from treated mice. (H) The weights of tumors in (G) were determined. (I) The body weight of each mouse was determined immediately before drug was injected. Drug treatment had no noticeable impact on body weight, and untreated mice grew slightly heavier throughout the course of the experiment. (J) Individual body weight plots for each mouse. One mouse in the drug treated group (T5, indicated with red arrow) was consistently lighter than all other mice, and died of treatment complications on the final day of the experiment, resulting in desiccation and body weight loss. (K) The weight of each mouse’s tumor was subtracted from its total weight, and then the weights of the treated and untreated mice were compared. There is no statistical difference between their body weights on the final day of the experiment once tumor weights are controlled for.
Preliminary clinical results
Trametinib showed efficacy in our mouse CDX model and is an FDA-approved drug for which substantial safety testing has already been conducted33. We were thus eager to determine if it could help dogs with spontaneously occurring oral tumors. While we cannot yet present the full results of our ongoing clinical trial, our preliminary results are encouraging and are presented here in full. For this, client-owned dogs diagnosed with COSCC that are older than 1 year, are not pregnant or lactating, have not previously undergone chemotherapy or radiation, and do not have signs of metastatic disease or other chronic disease, are enrolled. Qualifying dogs are prescribed 0.015 mg/kg or 0.020 mg/kg trametinib to be administered orally once per day. Dogs exit the study at day ~ 56, or if disease progression occurs based on biweekly medical examination including full tumor staging at week ~ 4. This trial is predominantly aimed at debulking the tumor, and the anticipated endpoint for both responsive and non-responsive dogs is standard-of-care surgical intervention, although if tumor regression occurs owners are offered the option to maintain their dog on the drug. To date, we have treated four patients (Supplemental Table S4 and Figure S3), two of which exhibited a partial response to the drug based on RECIST criteria59, with tumors shrinking markedly (Fig. 8, Supplemental Table S3 and Figure S4), while the other two experienced progressive disease and exited the study. It must be reinforced however that these preliminary results apply to just four dogs and should be considered not as statistically significant evidence of a reliable drug response, but as exciting early data.
Preliminary clinical trial results. (A) Chart showing initial tumor volumes when patients arrived for diagnosis (black bars), and at terminal point of trial (white bars). Disease progress is labeled via RECIST criteria as progressive disease (PD, > 20% increase in tumor size) or partial response (PR, > 30% reduction in tumor size, but not complete disappearance). (B) Photograph of patient 4 (see Supplemental Tables S4-S5 and Figures S3-S4) at the time of pre-enrollment, (C) photograph of the same patient following 14 days of treatment with trametinib, (D) photograph of the same patient at 56 days, upon exiting the trial. (E) CT bone window slice of patient 3 showing osseous tumor invasion at initial diagnosis, and (F) corresponding CT bone window slice of following 4 weeks of treatment with trametinib, exhibiting tumor size reduction and osseous regeneration.
Methods
Culture medium, FBS, and Tryp-LE were from Invitrogen. Collagenase type II and trypsin were from Worthington Biochemical Corporation. Trametinib was from Cayman Chemicals. All other chemicals were from Fisher unless otherwise noted. All animal procedures were approved by Cornell University’s Institutional Animal Care and Use Committee (IACUC), protocols 2005-0151 (clinical sample collection), 2017-0035 (experimental mouse work), and 2023-0034 (clinical trial). Animal experimentation was consistent with the ARRIVE guidelines’ best practices except as noted, specifically in that no placebo control group of dogs was used for our veterinary clinical trial, and so researchers were not blinded when conducting tumor measurements.
Tumor sample collection
After obtaining informed consent from owners for sample collection, dogs were placed under general anesthesia. Two tumor samples were collected, with a 4 mm punch device for diagnostic use, and with a 2 mm punch device for deposit with the Cornell Veterinary Biobank (CVB). When the tumor was later surgically excised as part of the dog’s treatment, it was delivered to the Cornell University Progressive Development of Therapeutics (PATh) facility to generate PDX models. All procedures conducted on dogs were in accordance with accepted best practices and AVMA guidelines, and in accordance with approved Cornell University’s IACUC (protocol 2005–0151), and no extraordinary steps were conducted to obtain samples for use in these experiments.
Histology and immunohistochemistry
Tumor samples were embedded in formalin, and 4 µm slices were taken. These slices were mounted to charged slides, and then samples were processed with an automated IHC stainer largely as previously described60. Paraffin was removed with Bond dewax solution (Leica), and then the samples were exposed to Bond epitope retrieval solution (Leica) for 30–40 min. Samples were then incubated with anti AE1/AE3 (DakoCytomation, #M3515, to detect cytokeratin2) or anti-MIB-1 (DakoCytomation, #M7240, to detect Ki67), followed by alkaline phosphatase conjugated anti-mouse IgG (Leica, #PV6110) and RedDetection CM (Leica, #DS9390). Samples were alternately stained with hematoxylin and eosin stain. Slides were scanned with an Aperio CS2 ScanScope (Vista). All histological analyses were conducted by board-certified pathologists blinded to the condition being examined.
Patient-derived xenograft development
NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were bred in-house by the Cornell PATh facility and were thus fully acclimated before experiments began. Tumor samples were collected from dogs as described above. Samples ~ 2–3 cm3 were placed in DMEM with penicillin and streptomycin for transport. Within 2–3 h, the samples were minced into 2–3 mm3 pieces. Several samples were cryopreserved, and the remaining samples were divided evenly and implanted into five NSG mice (female, 6–8 weeks old). Mice were anesthetized with 2.5% isofluorane, and pieces were implanted into the left flank. Mice were administered analgesics as needed for pain and monitored carefully for two hours following surgery. Once tumors reached ~ 100 mm3, approximately 36 weeks after implantation, the mouse was sacrificed using carbon dioxide euthanasia (3.5 L/min), the tumor was harvested, minced into segments, and the segments were implanted into new mice (male or female as available, no more than 10 weeks old). Passaging of the tumor from one mouse to another cohort of mice was continued as necessary to conduct downstream experiments. Samples were also cryopreserved, and other samples were used to develop cell culture models as described below.
Cell line development
Cell lines were isolated from tumors by adapting a procedure from a study on breast cancers44. Briefly, tumor samples were collected from PDX-carrying mice, or directly from dogs. Tumors were soaked in 70% ethanol for ~ 30 s, and then minced with sterile blades. The tumors were washed twice with PBS to remove blood, and then transferred to a sterile conical tube. 5 mL of digestion buffer (2.5 mg/mL trypsin, 5 mg/mL albumin, 850 units/mL collagenase type II in PBS) was added to the tumor samples. The tube was capped and placed in a 37°C shaker and shaken at 550 RPM for 20 min. The mixture was then filtered through a cell strainer, and the tumor pieces captured by the strainer were transferred to a clean tube, suspended in 5 mL of fresh digestion buffer, and shaken for another 20 min. To this solution, 10 mL of wash medium (F12 medium supplemented with 5% FBS and 250 ng/mL gentamicin) was added. The tube was spun down (250 × g for 10 min), and 10 mL of medium (expected to contain primarily fibroblasts) was removed. The remaining 5 mL of medium, and tumor pieces, were expected to contain primarily epithelial cells.
Epithelial cells were collected via gravimetric enrichment. To the tube containing 5 mL of medium and tumor pieces, 10 mL of wash medium was added. The tube was inverted several times to suspend cells, and the cells were allowed to settle for 20 min. 10 mL of medium was removed and spun down to collect a cell pellet, which was suspended in RPMI-1640 supplemented with 10% FBS and dispensed into a single well of a 6-well plate. This ‘wash + collect + spin’ procedure was repeated 12 more times on the tube with tumor pieces, to create 13 culture samples. The tumor pieces were then suspended in RPMI 1640 + 10% FBS and dispensed into a well plate as well. Cells were kept highly confluent for 1–2 weeks, after which we began passaging them with greater dilution. In general, we found that each subsequent wash resulted in more homogeneous looking cells, but that cells in the high-wash wells were sometimes not confluent enough to grow successfully. We made use of the highest wash-number sample which showed robust growth to perform further experiments.
Clonogenic colony formation assay
Cells were grown to ~ 70% confluence, and then passaged using Tryp LE. Cells in suspension were counted, and 1000 cells were dispensed into a 10 cm dish. The cells were grown for 4 weeks, or until colonies were readily visible on inspection, whichever occurred first. The cells were then washed three times with PBS, fixed to the plates with 3.7% formaldehyde in PBS, and stained with 4% crystal violet in methanol for 5 min. The crystal violet was removed, and the cells were washed three times with water (5 min each wash) then one final water wash (2 h). The plated colonies were then photographed.
Cell drug dose studies
Cells at ~ 70% confluence were washed with PBS and exposed to Tryp-LE solution for ~ 5 min. The cells were then suspended in RPMI-1640 supplemented with 10% FBS and transferred to a sterile conical tube. The cells were counted, then dispensed into 96-well plates at a density of 1000 cells per well. The cells were allowed to settle overnight, and the following day the medium was removed and replaced with medium containing the indicated DMSO-solvated drugs at assorted concentrations, or with DMSO alone as a control. The medium was again replaced on the fourth day of culture. On the sixth day of culture, the medium was removed, replaced with fresh medium, and cell viability was determined with Presto Blue reagent (Thermo). Viability was determined colorimetrically, following ~ 1–2 h incubation with the reagent, on a Tecan Spark microplate reader. IC50 values were determined in GraphPad Prism using a two-parameter, variable slope logistic curve. Drugs were generally considered ‘effective’ if the IC50 for the majority of COSCC cell lines was lower than the average IC50 for cell lines in the NCI-60.
Western blotting
Cells were grown to ~ 70% confluence, washed 3 times with PBS, and then lysed in lysis buffer (20 mM HEPES pH 7.6, 150 mM NaCl, 1 mM EDTA, and 1% NP-40). Protein concentration in the lysate was determined with BioRad Protein Assay Dye per the manufacturer’s instructions. 30 µg of protein in Laemmli buffer was then loaded onto 4–20% Tris–glycine gels and resolved via SDS-PAGE. Protein was transferred to PVDF membrane, and the membrane was blocked overnight in milk. The membrane was rinsed with TBST and stained for one hour with CypA antibody (1:1000 dilution in TBST, Cell Signaling #51418), phospho-ERK antibody (1:2000 dilution, Cell Signaling #9106), or vinculin antibody (1:2000 dilution, Cell Signaling #13901). The membrane was washed 4X with TBST, and then exposed for two hours to HRP-linked anti-rabbit IgG (Cell Signaling #7074, 1:5000 dilution in TBST) or anti-mouse IgG (Cell Signaling #7076, 1:5000 dilution). The membrane was washed 4X with TBST, exposed to Western Lighting Plus Chemiluminescence reagent (Perkin Elmer), and imaged on a Bio-Rad ChemiDoc. Band density was quantitated using ImageJ61.
Genotyping analysis
DNA was collected from dog tumor, PDX tumor, and cell line samples using the Zymo DNA micro-prep kit per the manufacturer’s instructions. DNA was then submitted to Illumina for genotyping on the EMBARK version of the CanineHD 220k bead array. Quality control on the genotype data was performed in PLINK 1.9 (www.cog-genomics.org/plink/1.9/)53. Samples with missingness values of > 3% were considered to have failed and were removed. For the remaining samples, SNPs with any missingness were removed, and then linked SNPs were removed, resulting in 13,178 informative SNPs being maintained. For each sample pair, the IBD was calculated using a metric called pi-hat (^π), which is a measure of the probability that the two samples share 0, 1 or 2 alleles. To generate a ‘fingerprint’ for each cell line, we selected 168 SNPs across the genome to differentiate the cell-lines from each other. Criteria for SNP selection included: no missingness, located > 10Mb apart, minor allele frequency (MAF) > 30% in the samples for the present study, and MAF > 30% in a group of 880 dogs genotyped on the same 220k array for other studies. Further manual selection was done to maximize genotype differences between the cell-lines. To show the relationship between each of the samples for these 168 SNPs, a heatmap was generated using the package pheatmap v 1.0.1262 in R63. To determine the uniqueness of the SNPs chosen for fingerprinting, pairwise IBD was calculated using only the 168 SNPs for all samples in this study and the 880 dogs that have been previously genotyped.
BRAF mutation analysis
DNA from each cell line, PDX, or original tumor sample was collected using the Zymo DNA kit as described above. DNA concentration and purity were determined with a Nanodrop ND1000 (Thermo Scientific). BRAF was amplified via PCR using AmpliTaq Gold (Thermo Scientific), with forward primer 5’-GGAAAGCATCTCACCTCATCC-3’, and reverse primer TTTCTAGTCACTCAGTAGCACCTC-3’, to generate a product with an expected size of 458 bp. The PCR cycles were as follows: 95 °C for 10 min initial; then 10 cycles of 95 °C for 30 s, 65 °C for 30 s, and 65* °C for 45 s [decreasing by 1 °C every cycle for a final cycle temperature of 55 °C]; 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s; and then finally 72 °C for 5 min. The product was cleaned up with ExoSAP-IT (Applied Biosystems), and Sanger sequencing was performed by the Biotechnology Resource Center (BRC) Genomics Facility at the Cornell Institute of Biotechnology, using either primer 5’-TGGCCTCAATTCTTACCATCCAC-3’ or primer 5’-TTTCTAGTCACTCAGTAGCACCTC-3’).
In vivo drug study
Trametinib was dissolved in vehicle (10% Cremophor EL, 10% PEG400, 80% PBS). Co-F-1236 cells (2 × 106) were suspended in Matrigel, and then injected into the left flank of male NSG mice (8–10 weeks old). Drug treatments began once the majority of mice had palpable tumors. Mice were marked by ear punching, and then randomly sorted into drug-treatment or vehicle-treatment groups using Excel’s randomization function. Mice were weighed daily to determine proper treatment volume; mouse handling was conducted by grasping a mouse by the tail, lifting it from the cage, then scruffing the neck and securing the tail in one hand. Six mice were treated with the indicated amount of trametinib daily by oral gavage, and six were treated with an equivalent volume of vehicle alone. Tumors were measured 3 times per week with calipers. Tumor volume was estimated as 0.5 * length * width * width. Representative mice were subjected to MRI before drug treatment began, and immediately prior to sacrifice. MRI images were analyzed using VivoQuant Imaging Software v3.5 by Invicro. At the experimental endpoint, mice were euthanized with carbon dioxide (3.5 L/min), then tumors were harvested, photographed, and immediately weighed before desiccation could occur. While animal drug treatment was not masked, and blinding was not possible during outcome assessment, different endpoint assessments were conducted by different investigators, and analysis of data was conducted by another investigator who was blinded to the condition with which each mouse was treated. Data for tumor growth were analyzed for statistical significance using a two-tailed Student’s t-test in Microsoft Excel.
Ongoing clinical study
Client-owned dogs with a confirmed diagnosis of COSCC arising from an oral mucosal surface are enrolled in an 8-week study to evaluate the response and tolerability of trametinib. Eligibility requirements include completely staged patients that have no evidence of regional or distant metastasis, have not received previous treatment of the oral tumor (i.e., surgical excision, chemotherapy or radiation therapy), are considered suitable candidates to undergo general anesthesia, are not diagnosed with other debilitating or chronic systemic diseases, are not pregnant or lactating, and the owners can comply with the required follow up appointments during the study. All enrolled canine patients receive an oral dose of trametinib (0.015–0.02 mg/kg once daily) during the study. Response to therapy is monitored via a weekly phone call to document potential side effects observed at home by the owner, biweekly physical and oral examination including caliper measurements of visible oral mass and routine bloodwork (i.e., complete blood cell count and serum biochemistry panel), and full oral tumor staging under general anesthesia at days ~ 28 and ~ 56, including a contrast-enhanced (2ml/kg Iohexol 350mg iodine/ml) computed tomography (CT) head exam (16-slice Aquilion LB; Canon Medical Systems USA, Inc, Tustin, CA), diagnostic imaging of the abdominal cavity (CT or ultrasound) and thorax (CT or 3-view radiographs), and cytological or histological assessment of regional lymph nodes. The head CT exams are acquired in continuous helical axial slices, with slice thickness ranging from 0.5mm to 2.0 mm and reconstructed into volumes (effective slice thickness 0.3mm to 1.0 mm). The CT studies are reviewed by a board-certified veterinary radiologist as DICOM studies in the hospital’s Picture Archiving and Communication system [PACS] (Carestream VuePACS; Rochester, NY, USA) on medical diagnostic-quality monitors (Dell U3219Q; Dell Technologies, Round Rock, TX, USA). Tumor volume (cm3) is calculated as the sum of the hand-drawn cross-sectional area (cm2) of the abnormal [tumor] tissue on each axial slice, multiplied by the effective slice thickness. Tumor volume is determined on each head CT exam (timepoints: enrollment, d. 28, and d.56) and compared to document changes between each timepoint. All clinical procedures are done in accordance with a protocol (2023-0034) reviewed and approved Cornell University’s IACUC. Because there was no control group in this preliminary study, and because animals were seen so frequently for follow up visits to monitor tumor growth, it was impossible to meaningfully blind the clinical staff who collected outcome measurements, or to blind the investigators analyzing the data.
Discussion
A major challenge in determining new treatments for veterinary patients is a relative lack of access to animal-derived cell culture model systems. In the case of COSCC, popular repositories such as the ATCC are extremely limited, with only 19 cell culture models available from Canis familiaris (domestic dog). Of these, only 1 cell line is derived from the head (the trachea, CF52.Tr cells), and it is not a cancer cell line. Of the dog cells, six are described as ‘normal’, eight as some form of cancer, and the rest lack a disease designation64. This is a particular problem as drug responses are not identical even in samples of the same cancer type, and so ideally a large number of model systems will be used to determine if any given drug treatment is appropriate for a certain disease. To address this problem, we have developed several new model systems representing different COSCCs. We developed both cell culture models, which are easy to work with and conduct multiple drug tests on, and PDX models, which are widely considered to be among the most accurate models of a tumor, as they contain all of the diverse cell types a tumor is comprised by, and also maintain a tumor’s extracellular environment65,66,67,68. Our PDX models largely replicate the phenology of their originating tumors, and our cell lines generally have morphology matching epithelial cells with high expression of EMT signals, which cause epithelial cells to develop fibroblast-like characteristics, and which had been expected following gene-set enrichment analyses of previous RNAseq datasets20. Indeed, some of our cell lines look similar to aggressive epithelial breast cancers such as MDA-MB-231 cells69, or ovarian epithelial cells70, each of which are generally considered to express EMT signals.
Our study proceeded in several phases. Previous work involved collection of tumor or healthy gingival tissue, and RNA-seq experiments to understand genetic differences underlying the disease20. In this work, we generated novel PDX models from original tumor tissue, and cell lines from PDX or original tumor tissue. We then tested the cell lines against an assortment of drugs and tested the most promising drug (trametinib) against a murine cell-line xenograft model, and then conducted a pilot clinical study in dogs with spontaneously occurring COSCC. The key limitations of this study pertain primarily to sample numbers. We tested drugs against six OSCC cell lines (including one semi-duplicate derived from different passages of the same PDX tumor). While it is common in cancer drug testing to examine a drug in three different cell lines, and the NCI-60 cancer cell line panel uses an average of 6.7 cell lines per cancer type to test drugs with, the fact remains that every tumor in every patient is different, and that outliers will always exist if enough cell lines are studied. An important biological limitation of these, and all, cell lines is that we can study only the cells which grow well on plastic. Several attempts to generate cell lines from original tumor samples in particular failed; it is possible that had we developed cell lines from those tumors, we might have seen different results for one drug or another. Moreover, newly developed cell culture models in general select for rapidly growing cells, which dominate the culture plate, exaggerating the problems of homogeneity in cell culture when compared to the heterogeneous nature of a whole tumor. Finally, of course, our pilot clinical examination included just four dogs. While we consider the findings to be very encouraging, we must stress again that there is no statistical significance to these findings, and an expanded study (which we are currently conducting) will be necessary to determine if trametinib is broadly pertinent as a treatment for COSCC.
We had developed multiple cell lines in order to get a relatively clear understanding of how well various drugs would work in COSCC. This approach proved to be important, as no drug had identical efficacy in all cell lines. We also noticed some differences in drug efficacy between each member of the two paired cell lines (Co-B-658 and Co-C-958, and Co-F-1236 and Co-L-1236), but these IC50 values were still generally similar when compared to other cell lines, suggesting that there were only minor differences between the cells from either isolation. That the differences between unique tumor-derived cell lines were generally larger than the differences between cell lines derived from the same tumor also suggests that approaches such as this might eventually be used for personalized medicine.
We investigated three different groups of drugs in this study: current standard of care agents for oral cancers in humans, other common chemotherapeutic agents, and agents which specifically target the RAS pathway. While drugs do not always interact identically in humans and dogs, it remains the case that dogs are often used to model drug efficacy and safety in humans71,72,73. Correspondingly, we tested the various drugs at concentrations flanking the reported IC50 values from the NCI-60, on the basis that these were biologically meaningful concentrations, and that if substantially higher amounts of drug were needed to inhibit cell proliferation, whole tumors would likely require doses outside of established therapeutic safety windows. Encouragingly, our COSCC cell lines were generally highly sensitive to trametinib, an inhibitor of MEK1 and MEK2. Moreover, the COSCC cell lines were more sensitive than the three mouse cell lines we tested, and similarly more sensitive than the average human cancer cell line in the NCI-60. We did find that for most of the cell lines tested, the trametinib dose curve had a very broad slope, and that while a good effect could be obtained at relatively low concentrations of the drug, obtaining a more complete effect would require substantially more drug (Fig. 5F). That led us to attempt a combination treatment with dabrafenib, which is often co-administered with trametinib in humans, and has recently been tested in oral ameloblastomas38. Unfortunately, the improvement in treatment efficacy in our COSCC cell lines was marginal at best (Fig. 6H, I). We suspect that this may reflect how both inhibitors target the same signaling pathway, and since trametinib was already fairly potent in each cell line, that left little additional work for dabrafenib to do. More synergistic results may emerge in a whole tumor, where a heterogeneous cell population exists and it is more likely that single-drug resistant cell populations would be encountered. Alternatively, dabrafenib is used to target BRAF-mutant tumors. While one of our BRAF-mutant cell lines was more sensitive than any other cell line to dabrafenib, the other had no sensitivity at all to the drug, so it is possible that canine BRAF p.V595E is simply not strongly inhibited by dabrafenib. Happily, however, the trametinib response alone was substantial in our 3D murine tumor graft models. We were also able to increase the dose from 0.5 to 0.75 mg/kg in these in vivo experiments without the mice suffering any ill effect. While this increased dose also had no noticeably increased effect in reducing tumor growth, this does suggest a therapeutic window may be available for inhibiting COSCC growth without causing any negative side effects in the patient.
Trametinib is an FDA-approved drug in various indications and can be used off-label by veterinarians with little additional regulatory burden. The drug did not cause xenografted tumors to actually shrink, and instead only prevented growth. However, our xenograft model grew much faster than our PDX models do and was derived from a cell culture model that had already presumably selected for the fastest growing cells, suggesting that greater overall effects might be observed in a whole tumor. And indeed, our preliminary clinical trial results show two of four tumors shrinking after only a few weeks of trametinib treatment, with one large tumor almost entirely vanishing (Fig. 8B–D). While none of these four dogs showed any signs of drug-induced toxic effects, it should be noted that some side effects have been demonstrated in other studies, with the most common ones in dogs being lethargy, elevated blood pressure, nausea, vomiting, and diarrhea74. Further, it must be stressed that a single-agent therapy is unlikely to result in a full cure, and that treatment should currently be considered as an approach to debulking a tumor prior to surgery. While we will need to recruit more dogs to make strong statistical arguments and explore differences across clinical and molecular phenotypes including mutational profiles, the fact that the two patients with COSCC responded well and without any observed toxicity is encouraging. We are excited by the results to date, and hope that continuation of our study will result in important benefit to canine patients in the clinic.
Conclusion
We have developed several new cell lines derived from COSCC tumors, either taken directly from dogs with spontaneously developing tumors or first passaged as PDX models in mice. While the cells are generally sensitive to several chemotherapeutic agents, they also show a strong sensitivity to trametinib, a MEK inhibitor used in some RAS-dependent cancers, even in vivo. Indeed, to the best of our knowledge this study is the first to demonstrate the efficacy of trametinib in COSCC. Preliminary clinical results raise hope that the inhibitor may be of benefit to canine patients as well. Since targeted therapeutic agents generally have fewer side effects than traditional chemotherapeutics, we hope that these results will be a first step in deploying an effective, and well tolerated, treatment for COSCCs.
Data availability
Genotype data generated for this study and the 168 SNPs chosen for cell-line fingerprinting are publicly available as binary PLINK files in the datadryad.org repository, https://doi.org/https://doi.org/10.5061/dryad.zs7h44jh9. The 168 SNP fingerprinting data are also available in vcf format at the European Variation Archive (EVA) repository, accession number PRJEB77007.
References
Fulton, A. J., Nemec, A., Murphy, B. G., Kass, P. H. & Verstraete, F. J. M. Risk factors associated with survival in dogs with nontonsillar oral squamous cell carcinoma 31 cases (1990–2010). J. Am. Vet. Med. Assoc. 243, 696–702. https://doi.org/10.2460/javma.243.5.696 (2013).
Peralta, S., Grenier, J. K., McCleary-Wheeler, A. L. & Duhamel, G. E. Ki67 labelling index of neoplastic epithelial cells differentiates canine acanthomatous ameloblastoma from oral squamous cell carcinoma. J. Comput. Pathol. 171, 59–69. https://doi.org/10.1016/j.jcpa.2019.08.001 (2019).
Mestrinho, L. A. et al. Comparison of histological and proliferation features of canine oral squamous cell carcinoma based on intraoral location: 36 Cases. J. Vet. Dent. 34, 92–99. https://doi.org/10.1177/0898756417713979 (2017).
Grimes, J. A. et al. Histologic evaluation of mandibular and medial retropharyngeal lymph nodes during staging of oral malignant melanoma and squamous cell carcinoma in dogs. J. Am. Vet. Med. Assoc. 254, 938–943. https://doi.org/10.2460/javma.254.8.938 (2019).
Mestrinho, L. A. Current status and future perspectives in canine oral squamous cell carcinoma. Vet. Pathol. 55, 200–201. https://doi.org/10.1177/0300985817732114 (2018).
Amory, J. T. et al. Computed tomographic characteristics of odontogenic neoplasms in dogs. Vet. Radiol. Ultrasound 55, 147–158. https://doi.org/10.1111/vru.12101 (2014).
Verstraete, F. J. M. Mandibulectomy and maxillectomy. Vet. Clin. N Am: Small Anim Pract 35, 1009–1039. https://doi.org/10.1016/j.cvsm.2005.03.005 (2005).
Wright, A. L., Peralta, S. & Fiani, N. Case report: Spontaneous mandibular body regeneration following unilateral subtotal mandibulectomy in a 3-month-old French bulldog. Front. Vet. Sci. 10 (2023). https://doi.org/10.3389/fvets.2023.1281232
Brooks, M. B., Matus, R. E., Leifer, C. E., Alfieri, A. A. & Patnaik, A. K. Chemotherapy versus chemotherapy plus radiotherapy in the treatment of tonsillar squamous cell carcinoma in the dog. J. Vet. Internal Med. 2, 206–211. https://doi.org/10.1111/j.1939-1676.1988.tb00318.x (1988).
Fiani, N. & Peralta, S. Extended subtotal mandibulectomy for the treatment of oral tumors invading the mandibular canal in dogs: A novel surgical technique. Front. Vet. Sci. 6, 339. https://doi.org/10.3389/fvets.2019.00339 (2019).
Warshaw, S. L., Carney, P. C., Peralta, S. & Fiani, N. Piezosurgical bone-cutting technology reduces risk of maxillectomy and mandibulectomy complications in dogs. J. Am. Vet. Med. Assoc. 261, 1–7. https://doi.org/10.2460/javma.23.03.0130 (2023).
Bar-Am, Y. & Verstraete, F. J. M. Elastic training for the prevention of mandibular drift following mandibulectomy in dogs: 18 cases (2005–2008). Vet. Surg. 39, 574–580. https://doi.org/10.1111/j.1532-950X.2010.00703.x (2010).
Cray, M. et al. Analysis of risk factors associated with complications following mandibulectomy and maxillectomy in dogs. J. Am. Vet. Med. Assoc. 259, 265–274. https://doi.org/10.2460/javma.259.3.265 (2021).
LaDue-Miller, T., Price, G. S., Page, R. L. & Thrall, D. E. Radiotherapy of canine non-tonsillar squamous cell cercinoma. Vet. Radiol. Ultrasound 37, 74–77. https://doi.org/10.1111/j.1740-8261.1996.tb00817.x (1996).
Evans, S. M. & Shofer, F. Canine oral nontonsillar squamous cell carcinoma: Prognostic factors for recurrence and survival following orthovoltage radiation therapy. Vet. Radiol. 29, 133–137. https://doi.org/10.1111/j.1740-8261.1988.tb01763.x (1988).
Nemec, A. et al. Osteonecrosis of the Jaws in dogs in previously irradiated fields: 13 Cases (1989–2014). Front. Vet. Sci. 2, 5–5. https://doi.org/10.3389/fvets.2015.00005 (2015).
Mayer, M. N. & Anthony, J. M. Radiation therapy for oral tumors: Canine acanthomatous ameloblastoma. Can. Vet. J. 48, 99–101 (2007).
Martin, D. et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget 5, 8906–8923. https://doi.org/10.18632/oncotarget.2417 (2014).
Devaraja, K. Current prospects of molecular therapeutics in head and neck squamous cell carcinoma. Pharm. Med. 33, 269–289. https://doi.org/10.1007/s40290-019-00288-x (2019).
Peralta, S. et al. Comparative transcriptional profiling of canine acanthomatous ameloblastoma and homology with human ameloblastoma. Sci. Rep. 11, 17792. https://doi.org/10.1038/s41598-021-97430-0 (2021).
Peralta, S., McCleary-Wheeler, A. L., Duhamel, G. E., Heikinheimo, K. & Grenier, J. K. Ultra-frequent HRAS p.Q61R somatic mutation in canine acanthomatous ameloblastoma reveals pathogenic similarities with human ameloblastoma. Vet. Comp. Oncol. 17, 439–445. https://doi.org/10.1111/vco.12487 (2019).
Peralta, S., Webb, S. M., Katt, W. P., Grenier, J. K. & Duhamel, G. E. Highly recurrent BRAF p.V595E mutation in canine papillary oral squamous cell carcinoma. Vet. Comp. Oncol. 21, 138–144. https://doi.org/10.1111/vco.12869 (2023).
Guscetti, F. et al. Molecular homology between canine spontaneous oral squamous cell carcinomas and human head-and-neck squamous cell carcinomas reveals disease drivers and therapeutic vulnerabilities. Neoplasia 22, 778–788. https://doi.org/10.1016/j.neo.2020.10.003 (2020).
Ngan, H.-L., Law, C.-H., Choi, Y. C. Y., Chan, J.Y.-S. & Lui, V. W. Y. Precision drugging of the MAPK pathway in head and neck cancer. npj Genomic Med. 7, 20. https://doi.org/10.1038/s41525-022-00293-1 (2022).
Barault, L., Lamba, S. & Di Nicolantonio, F. Ras mutations in cancer. eLS https://doi.org/10.1002/9780470015902.a0025010 (2013).
Mochizuki, H. & Breen, M. Sequence analysis of RAS and RAF mutation hot spots in canine carcinoma. Vet. Comp. Oncol. 15, 1598–1605. https://doi.org/10.1111/vco.12275 (2017).
Mochizuki, H., Kennedy, K., Shapiro, S. G. & Breen, M. BRAF mutations in canine cancers. PLOS ONE 10, e0129534–e0129534. https://doi.org/10.1371/journal.pone.0129534 (2015).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558. https://doi.org/10.1126/science.1235122 (2013).
Chang, W.-H. et al. KRAS-dependent cancer cells promote survival by producing exosomes enriched in Survivin. Cancer Lett. 517, 66–77. https://doi.org/10.1016/j.canlet.2021.05.031 (2021).
Drosten, M. & Barbacid, M. Targeting the MAPK pathway in KRAS-driven tumors. Cancer Cell 37, 543–550. https://doi.org/10.1016/j.ccell.2020.03.013 (2020).
Tan, A. C. et al. Management of glioblastoma: State of the art and future directions. CA: A Cancer J. Clin. 70, 299–312. https://doi.org/10.3322/caac.21613 (2020).
Lugowska, I., Kosela-Paterczyk, H., Kozak, K. & Rutkowski, P. Trametinib: a MEK inhibitor for management of metastatic melanoma. Onco Targets Ther 8, 2251–2259. https://doi.org/10.2147/Ott.S72951 (2015).
Odogwu, L. et al. FDA approval summary: Dabrafenib and trametinib for the treatment of metastatic non-small cell lung cancers harboring BRAF V600E mutations. Oncologist 23, 740–745. https://doi.org/10.1634/theoncologist.2017-0642 (2018).
Wright, C. J. M. & McCormack, P. L. Trametinib: First global approval. Drugs 73, 1245–1254. https://doi.org/10.1007/s40265-013-0096-1 (2013).
Wang, Z. Y., He, D., Chen, C., Liu, X. B. & Ke, N. W. Vemurafenib combined with trametinib significantly benefits the survival of a patient with stage IV pancreatic ductal adenocarcinoma with BRAF V600E mutation: A case report. Front. Oncol. 11, 801320. https://doi.org/10.3389/fonc.2021.801320 (2022).
Hanft, K. M. et al. Combinatorial effects of azacitidine and trametinib on NRAS-mutated melanoma. Pediatr Blood Cancer 69, e29468. https://doi.org/10.1002/pbc.29468 (2022).
Alshammari, K. et al. Phase II trial of trametinib and panitumumab in RAS/RAF wild type metastatic colorectal cancer. Clin Colorectal Canc 20, 334–341. https://doi.org/10.1016/j.clcc.2021.07.004 (2021).
Grynberg, S. et al. Neoadjuvant BRAF-targeted therapy for ameloblastoma of the mandible: an organ preservation approach. JNCI: J. Natl. Cancer Inst., djad232 (2023). https://doi.org/10.1093/jnci/djad232
Wei, B. R. et al. Efficacy, tolerability, and pharmacokinetics of combined targeted MEK and dual mTORC1/2 inhibition in a preclinical model of mucosal melanoma. Mol. Cancer Therap. 19, 2308–2318. https://doi.org/10.1158/1535-7163.Mct-19-0858 (2020).
Cronise, K. E., Hernandez, B. G., Gustafson, D. L. & Duval, D. L. Identifying the ErbB/MAPK signaling cascade as a therapeutic target in canine bladder cancer. Mol. Pharmacol. 96, 36–46. https://doi.org/10.1124/mol.119.115808 (2019).
Takada, M. et al. Targeting MEK in a translational model of histiocytic sarcoma. Mol. Cancer Therap. 17, 2439–2450. https://doi.org/10.1158/1535-7163.Mct-17-1273 (2018).
Wei, B. R. et al. Synergistic targeted inhibition of MEK and dual PI3K/mTOR diminishes viability and inhibits tumor growth of canine melanoma underscoring its utility as a preclinical model for human mucosal melanoma. Pigm Cell Melanoma R 29, 643–655. https://doi.org/10.1111/pcmr.12512 (2016).
Tufts Cummings Veterinary Medical Center. Pilot study of a MEK inhibitor in dogs with histiocytic sarcoma, https://trials.vet.tufts.edu/clinical_trials/pilot-study-of-a-mek-inhibitor-in-dogs-with-histiocytic-sarcoma/# (2022).
Kazerounian, S. Isolation of mammary epithelial cells and fibroblasts from mouse tumor. Bio-protocol 4, e1050–e1050 (2014). https://doi.org/10.21769/BioProtoc.1050
Cheng, F. et al. Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-β–Slug signaling. Proc. Natl. Acad. Sci. 113, E4320–E4327. https://doi.org/10.1073/pnas.1519197113 (2016).
Stanisavljevic, J. et al. Snail1-expressing fibroblasts in the tumor microenvironment display mechanical properties that support metastasis. Cancer Research 75, 284–295. https://doi.org/10.1158/0008-5472.CAN-14-1903 (2015).
Nishioka, R. et al. SNAIL induces epithelial-to-mesenchymal transition in a human pancreatic cancer cell line (BxPC3) and promotes distant metastasis and invasiveness in vivo. Exp. Mol. Pathol. 89, 149–157. https://doi.org/10.1016/j.yexmp.2010.05.008 (2010).
Medici, D., Hay, E. D. & Olsen, B. R. Snail and slug promote epithelial-mesenchymal transition through β-catenin–T-cell factor-4-dependent expression of transforming growth factor-β3. Mol. Biol. Cell 19, 4875–4887. https://doi.org/10.1091/mbc.e08-05-0506 (2008).
Hengstschlager, M. & Rosner, M. An antibody-based approach for the in situ evaluation of mouse contribution in human stem cell-derived xenografts. Protocol Exchange https://doi.org/10.1038/protex.2018.009 (2018).
Blum, R., Jacob-Hirsch, J., Amariglio, N., Rechavi, G. & Kloog, Y. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1α, causing glycolysis shutdown and cell death. Cancer Res. 65, 999–1006. https://doi.org/10.1158/0008-5472.999.65.3 (2005).
Dimri, G., Band, H. & Band, V. Mammary epithelial cell transformation: insights from cell culture and mouse models. Breast Cancer Res. 7, 171. https://doi.org/10.1186/bcr1275 (2005).
Kawakami, T. et al. Early onset adult deafness in the Rhodesian Ridgeback dog is associated with an in-frame deletion in the EPS8L2 gene. Plos One 17, e0264365. https://doi.org/10.1371/journal.pone.0264365 (2022).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. https://doi.org/10.1086/519795 (2007).
Banisadr, A., Safdari, Y., Kianmehr, A. & Pourafshar, M. Production of a germline-humanized cetuximab scFv and evaluation of its activity in recognizing EGFR- overexpressing cancer cells. Hum. Vaccines Immunotherap. 14, 856–863. https://doi.org/10.1080/21645515.2017.1407482 (2018).
Cai, W.-Q. et al. The latest battles between EGFR monoclonal antibodies and resistant tumor cells. Front. Oncol. https://doi.org/10.3389/fonc.2020.01249 (2020).
Kim, H. R. et al. Mouse–human co-clinical trials demonstrate superior anti-tumour effects of buparlisib (BKM120) and cetuximab combination in squamous cell carcinoma of head and neck. Br. J. Cancer 123, 1720–1729. https://doi.org/10.1038/s41416-020-01074-2 (2020).
Corcoran, R. B. et al. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600–mutant colorectal cancer. J. Clin. Oncol. 33, 4023–4031. https://doi.org/10.1200/JCO.2015.63.2471 (2015).
Kobayashi, M. et al. Establishment of a BRAF V595E-mutant canine prostate cancer cell line and the antitumor effects of MEK inhibitors against canine prostate cancer. Vet. Comp. Oncol. 21, 221–230. https://doi.org/10.1111/vco.12879 (2023).
Nguyen, S. M., Thamm, D. H., Vail, D. M. & London, C. A. Response evaluation criteria for solid tumours in dogs (v1.0): a Veterinary Cooperative Oncology Group (VCOG) consensus document. Vet. Compar. Oncol. 13, 176–183 (2015). https://doi.org/10.1111/vco.12032
Malatos, J. M., Kurpios, N. A. & Duhamel, G. E. Small intestinal lymphatic hypoplasia in three dogs with clinical signs of protein-losing enteropathy. J. Comp. Pathol. 160, 39–49. https://doi.org/10.1016/j.jcpa.2018.02.005 (2018).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nature Methods 9, 676–682. https://doi.org/10.1038/nmeth.2019 (2012).
pheatmap: Pretty Heatmaps (R package version 1.0.12, 2019).
R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2023).
American Type Culture Collection, https://www.atcc.org/ (2022).
Dobrolecki, L. E. et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 35, 547–573. https://doi.org/10.1007/s10555-016-9653-x (2016).
Hidalgo, M. et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discovery 4, 998–1013. https://doi.org/10.1158/2159-8290.CD-14-0001 (2014).
Jung, J., Seol, H. S. & Chang, S. The generation and application of patient-derived xenograft model for cancer research. Cancer Res. Treatment: Off. J. Korean Cancer Assoc. 50, 1–10. https://doi.org/10.4143/crt.2017.307 (2018).
Karamboulas, C. et al. Patient-derived xenografts for prognostication and personalized treatment for head and neck squamous cell carcinoma. Cell Rep. 25, 1318–1331. https://doi.org/10.1016/j.celrep.2018.10.004 (2018).
Paramanantham, A. et al. Activated ERK signaling is one of the major hub signals related to the acquisition of radiotherapy-resistant MDA-MB-231 breast cancer cells. Int. J. Mol. Sci. 22 (2021). https://doi.org/10.3390/ijms22094940
Li, N. F. et al. A modified medium that significantly improves the growth of human normal ovarian surface epithelial (OSE) cells in vitro. Lab. Investig. 84, 923–931. https://doi.org/10.1038/labinvest.3700093 (2004).
Pinho, S. S., Carvalho, S., Cabral, J., Reis, C. A. & Gärtner, F. Canine tumors: a spontaneous animal model of human carcinogenesis. Translat Res. 159, 165–172. https://doi.org/10.1016/j.trsl.2011.11.005 (2012).
Kwon, J. Y., Moskwa, N., Kang, W., Fan, T. M. & Lee, C. Canine as a comparative and translational model for human mammary tumor. J Breast Cancer 26, 1–13. https://doi.org/10.4048/jbc.2023.26.e4 (2023).
Nance, R. L., Sajib, A. M. & Smith, B. F. Chapter three-canine models of human cancer: Bridging the gap to improve precision medicine. Prog. Mol. Biol. Translat. Sci. 189, 67–99. https://doi.org/10.1016/bs.pmbts.2021.12.003 (2022).
Takada, M. et al. Population pharmacokinetics, pharmacodynamics and safety properties of trametinib in dogs with cancer: A phase I dose escalating clinical trial. Vet. Comp. Oncol. 22, 410–421. https://doi.org/10.1111/vco.12989 (2024).
Acknowledgements
We would like to acknowledge funding from the Cornell Richard P. Riney Canine Health Center to WPK and SP, from the National Cancer Institute (5R01CA201402-09) to RAC, and from the Maritza and Reino Salonen Foundation to KH. Biological material and associated data were provided by the Cornell Veterinary Biobank, a resource built with the support of NIH grant R24 GM082910 and the Cornell University College of Veterinary Medicine, and by the Cornell Animal Health Diagnostic Center. Experimental sample collection and processing, as well as client-owned dog enrollment in the clinical study were kindly facilitated by the Cornell University Hospital for Animals Clinical Trials group, led by Ms. Carol E. Frederick, LVT, VTS (ECC) and Ms. Andrea L. King, LVT; by Cornell’s Innovation laboratory, led by MB and Ms. Lisa M. Mitchell, LVT; and by the Dentistry and Oral Surgery, and Medical Oncology staff including Ms. Kalli Anderson, LVT, Ms. Laura M. Barlow, LVT, and Ms. Kaitlin Perez, LVT.
Author information
Authors and Affiliations
Contributions
SP directed all clinical work to originally harvest patient tumor samples. CEB, ABT, NF, KRH, and SRS assisted in clinical tasks. GED conducted all histological examinations. WPK isolated cell lines and conducted tests on cell culture models. RP and SB conducted all work in mouse models. SMW isolated DNA from tissue samples. JJH interpreted Embark data and conducted related calculations. PCC assisted in all statistical workup. JME, JKG, KH, ESM, ACW, and RAC provided crucial and valuable input that informed the design of various experiments. WPK and SP wrote the manuscript. All authors provided input into the final draft of the manuscript prior to submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Katt, W.P., Balkman, C.E., Butler, S.D. et al. The MEK inhibitor trametinib is effective in inhibiting the growth of canine oral squamous cell carcinoma. Sci Rep 15, 7069 (2025). https://doi.org/10.1038/s41598-025-90574-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-90574-3
Keywords
This article is cited by
-
Tyrosine kinase inhibitors in canine solid tumours: a systematic review of indications, response and safety
Veterinary Oncology (2025)
-
An editorial spotlight on recent progress in veterinary oncology
Veterinary Oncology (2025)
-
Clinical application of the MEK inhibitor trametinib in dogs with oral squamous cell carcinoma
Veterinary Oncology (2025)