Abstract
Despite advances in research, studies on predictive models for Non-Alcoholic Fatty Liver Disease (NAFLD)-related fibrosis remain limited. Identifying new biomarkers to distinguish Non-Alcoholic Steatohepatitis (NASH) from NAFLD would aid in the treatment of NASH. Gene expression and clinical profiles of NAFL and NASH patients were collected from databases. Differentially expressed genes with prognostic value were used to construct predictive model. Validation of fibrosis stage-related pyroptosis-related genes (PRGs) was performed using Sprague-Dawley rats liver fibrosis models induced by CCl4 or PS. Immune cell infiltration assessment demonstrated that stromal score, immune score, and ESTIMATE score were higher in patients with NASH compared to those with NAFL. BAX, BAK1, PYCARD, and NLRP3 were identified as hub genes that exhibit a strong correlation with fibrosis stage. Additionally, the expression of these genes was increased in fibrotic liver tissues induced by CCl4 and PS. The pyroptosis-associated gene signature effectively predicts the degree of liver fibrosis in NASH patients. Our study indicates that BAX, BAK1, PYCARD, and NLRP3 might serve as biomarkers for NASH-associated fibrosis.
Similar content being viewed by others
Introduction
Non-alcoholic fatty liver disease (NAFLD) is characterized by the excessive accumulation of triglycerides in the liver without any secondary causes. Epidemiologically, it has been closely linked to metabolic disorders such as obesity, hyperlipidemia, and type 2 diabetes1,2. Over the past decades, profound shifts in lifestyle patterns have positioned NAFLD as the most prevalent liver disorder in many regions worldwide3,4. NAFLD spans a spectrum from benign steatosis (NAFL) to inflammatory non-alcoholic steatohepatitis (NASH), which may advance to fibrosis, cirrhosis, or hepatocellular carcinoma5,6. NAFLD-related liver transplantation is experiencing a significant rise and is projected to become a major indication in the foreseeable future7, exerting a significant societal and patient-specific burden8.
Non-alcoholic steatohepatitis (NASH), an inflammatory subtype of NAFLD, is defined by hepatic steatosis with lobular inflammation, hepatocyte injury, and potential fibrosis9. Patients with NASH are at a markedly elevated risk of fibrosis development, which is strongly associated with disease outcomes10 and 15–20% of these patients ultimately progress to cirrhosis11,12. The Brunt fibrosis staging system stratifies NASH patients into five distinct stages based on the histological assessment of liver biopsies, with a focus on the degree of perisinusoidal fibrosis in zone 3, a hallmark feature of steatohepatitis. Stage 0 indicates no fibrosis, while stage 4 signifies cirrhosis13.
At present, NASH presents a significant diagnostic and therapeutic challenge for clinicians. Liver biopsy continues to be the gold standard for diagnosing NASH14, and no approved therapeutic interventions are currently available, making lifestyle modification the cornerstone of management1,15. Despite extensive efforts to elucidate relevant pathophysiologic factors, the exact mechanisms underlying steatosis and fibrosis progression remain inadequately understood16. A comprehensive understanding of the underlying pathology is crucial for identifying novel targets for intervention.
The “two-hit” theory, which attributes the pathogenesis of NASH to steatosis and additional factors such as lipotoxicity, oxidative stress, and inflammation, has been widely embraced17. These factors, individually or in combination, activate cellular stress pathways leading to hepatocyte death, liver injury, inflammation, and fibrosis. The central role of inflammation in these processes suggests that various immune cells are involved in modulating the development and severity of NASH18. Recent advances have shed light on the ways in which the immune cell repertoire is reshaped during NASH progression. Neutrophils and monocytes are rapidly recruited to the liver in the early stages of the disease. Dendritic cells interact with T cells to promote hepatic inflammation by secreting cytokines, while B cells, natural killer T (NKT) cells, and innate lymphoid cells also contribute to liver fibrosis18. Despite these insights, the current understanding of the inflammatory cues driving NASH remains fragmented, and the specific contributions of various immune cell subsets to the disease are still not fully understood.
Pyroptosis represents a unique form of programmed cell death dependent on inflammatory caspases, specifically caspase-1 and caspase-4/5/1119,20. Research has shown that excessive pyroptotic cell death significantly promotes cardiovascular, autoimmune, and other diseases, as well as various tumors21,22,23. Recent studies have revealed a connection between pyroptosis and the progression from NAFL to NASH. Activation of the NLRP3 inflammasome induces the cleavage of caspase-1 and subsequent pyroptosis in NAFLD, which amplifies and perpetuates inflammasome-driven fibrogenesis in NASH22,24. Gasdermin D, the primary executor of pyroptosis, plays a critical role in the steatohepatitis associated with NASH by regulating NF-κB pathway activation, cytokine secretion, and lipogenesis25. However, the precise molecular mechanisms of pyroptosis in liver fibrosis, as well as the diagnostic value of pyroptosis-related genes (PRGs) in NASH, remain to be further elucidated.
In the present study, we conducted a comprehensive analysis to investigate immune cell infiltration levels and the expression of pyroptosis-related genes in patients with NAFL and NASH. We then constructed a logistic-LASSO regression model based on key features and developed a nomogram to predict the diagnosis of NASH. Additionally, we examined the pyroptosis-related genes corresponding to the Brunt fibrosis stages of NASH. Independent external cohorts and liver fibrosis rat models were utilized for validation. Our findings suggest suggest that pyroptosis-associated genes, fibrosis progression, and immune cell infiltration, characterized by distinct immune cell compositions and their interactions, are interconnected during the development of NASH, and may inform the development of novel therapeutic targets to mitigate or reverse disease progression.
Materials and methods
Datasets and preprocessing
Raw RNA sequencing (RNA-seq) data and clinical information for 241 NAFLD patients were obtained from the GEO datasets GSE162694 and GSE167523, both based on the GPL21290 platform (Illumina HiSeq 3000), and were combined to form a derivation cohort. For external validation, RNA-seq data and clinical information for 206 NAFLD patients were sourced from GEO dataset GSE135251 and data for an additional 31 NAFLD patients were obtained from GEO dataset GSE126848, which is based on the GPL18573 platform (Illumina NextSeq 500). After integrating the datasets using raw counts files, the derivation cohort was normalized with package “sva” and its “ComBat_seq” function26. Box plot and principal component analysis (PCA) plot were used to visualize general distribution of both datasets. As shown in Supplementary Fig. 1, batch effect was successfully removed after correction.
Identification of differentially expressed genes (DEGs)
Differentially expressed genes (DEGs) were identified using the DESeq2 R package, and Wald test were employed by default to assess statistical significance27. Genes whose |log2 fold change |>1 and adjust P value < 0.05 were considered as DEGs. All p-values were adjusted for multiple testing using the Benjamini-Hochberg procedure, as implemented in the DESeq2 package, to control the false discovery rate (FDR).
Functional enrichment analysis
Gene Ontology (GO) functional enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and Gene Set Enrichment Analysis (GSEA) were conducted using the “ClusterProfiler” R package28. GO analysis was performed to categorize DEGs into three primary domains: Biological Process (BP), Molecular Function (MF), and Cellular Component (CC). The analysis identified significantly enriched GO terms associated with DEGs, providing insight into their biological roles. Adjusted p-values were calculated using the Benjamini-Hochberg method. KEGG pathway analysis was performed to identify significantly enriched metabolic pathways and signaling pathways associated with DEGs29,30. The analysis mapped genes to predefined pathways in the KEGG database, and statistical significance was determined using hypergeometric testing. GSEA was conducted to assess the enrichment of predefined gene sets across the entire ranked list of genes, rather than focusing solely on DEGs. The ranking metric was based on the log2 fold change values of all genes. Enrichment scores were calculated for each gene set, and statistical significance was determined through permutation testing. Adjusted P < 0.05 were considered significant.
PPI network construction
STRING website (https://cn.string-db.org/) was used to analyze the potential protein–protein interaction by DEGs. Cytoscape software was used to visualize and embellish PPI network31. The Cytoscape plug-in app CytoHubba, utilizing the Maximal Clique Centrality (MCC) method, was used to calculate the interaction degree of each gene in the PPI network32. The results obtained reflect predicted and potential protein–protein interactions based on existing evidence.
Immune cell infiltration analysis
Immune cell infiltration was evaluated (based on transcriptomic data) using the single-sample Gene Set Enrichment Analysis (ssGSEA) method with the GSVA R package33. Representative immune cell gene sets were used to calculate ssGSEA scores, reflecting the relative abundance of immune cell types. Correlations between ssGSEA scores were assessed with Pearson’s correlation coefficient and visualized using the “corrplot” R package. Additionally, Stromal Score, Immune Score, and ESTIMATE Score were calculated using the “estimate” R package. Gene expression data were normalized to Transcripts Per Kilobase of exon model per Million mapped reads (TPM) before analysis34.
Identification of differentially expressed PRGs
A total of 45 pyroptosis-related genes (PRGs) (listed in Supplementary Table 1) were identified from previous literature35,36,37. Differentially expressed PRGs between NASH and NAFL tissues were analyzed using the “DESeq2” R package, with adjusted P < 0.01 as the significance threshold. A protein-protein interaction (PPI) network of these PRGs was constructed using the STRING database and visualized with Cytoscape software.
Construction and validation of predictive models
To identify key PRGs and immune-related scores associated with NASH progression, univariate logistic regression was performed on differentially expressed PRGs and immune-related scores, with P < 0.05 as the cut-off. Significant variables were refined using LASSO regression and then included in multivariate logistic regression, retaining only features with p < 0.05 for building the diagnostic model. A nomogram was constructed to visualize the model, and its performance was evaluated using ROC and calibration curves. External validation was performed using the GEO dataset GSE135251, with ROC and calibration curves generated to assess predictive accuracy and reliability.
Validation of the fibrosis stage related PRGs with GEO dataset
The GSE126848 dataset was used to validate fibrosis stage-related PRGs. The correlation between the expression levels of PRGs and fibrosis stages was analyzed using Spearman correlation.
Experimental validation in animal models
Male Sprague-Dawley rats, aged 6–8 weeks and weighing 120–150 g, were purchased from Slac Laboratory Animal (Changsha, Hunan, China). The rats were provided with standard rat chow and water ad libitum and housed under a 12-hour light/dark cycle in a pathogen-free environment. All animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Guangdong Provincial People’s Hospital. Animal care and experimental procedures complied with the hospital’s guidelines. The euthanasia process was conducted in accordance with ethical guidelines to minimize animal suffering. Sodium pentobarbital (150 mg/kg) was administered via intraperitoneal injection, followed by cervical dislocation to ensure death.
Two well-established experimental models of hepatic fibrosis, induced by either CCl4 (Aladdin Industrial, Shanghai, China) or porcine serum (PS) (Beijing YuanHeng ShengMa, Beijing, China), were used in this study. The rats were randomly assigned to a normal group or a treatment group receiving CCl4 or PS. Hepatic fibrosis was induced via intraperitoneal injection of CCl4 (2 mL/kg body weight, 1:1 in olive oil) or porcine serum (0.5 mL/kg body weight) twice per week for 8 weeks, as previously reported. The control group rats received an equal volume of vehicle with the same route of administration and schedule. All rats were sacrificed in the ninth week, and liver tissue was stored in liquid nitrogen for future use.
Real-time PCR
Total RNA was isolated from liver tissue using Trizol reagent, following the manufacturer’s instructions. A total of 1 µg of RNA was used to synthesize first-strand cDNAs using the RevertAid First Strand cDNA Synthesis Kit. Specific primers for BAX, BAK1, PYCARD, NLRP3, and GAPDH were designed based on their gene sequences and synthesized by Generay Biotech (Shanghai, China). The BAX primer pair consisted of forward primer 5’-GTGGGATGGGCTTCAGGAACAAC-3’ and reverse primer 5’-TCCAAGGTCAGCTCAGGTGTCTC-3’ ; BAK1 primer pair consisted of forward primer 5’-CAGAGTGAGGGCAGAGGTGAGAG-3’ and reverse primer 5’-ACTAGGTTGGTAGCAGTGGTCAGG-3’; PYCARD primer pair consisted of forward primer 5’-TTGCTGGATGCTCTGTATGG-3’ and reverse primer 5’-CCAAGTAGGGCTGTGTTTGC-3’; NLRP3 primer pair consisted of forward primer 5’-ACCCAAGGAGGAAGAAGAAGAG-3’ and reverse primer 5’-AGTCTGGAAGAACAGGCAACAT-3’; and GAPDH primers were forward primer 5’-CAGTGCCAGCCTCGTCTCAT-3’ and reverse primer 5’-AGGGGCCATCCACAGTCTTC − 3’. The protocol used for quantitative real-time PCR analysis was the same as previously described. Genes were normalized to GAPDH used as the internal control, and the amount of specific mRNA in each sample was calculated using the comparative 2−ΔΔCT method38.
Statistical analysis
All statistical analyses were performed using R software (version 4.1.2) or GraphPad Prism (version 9.0). Immune infiltration scores and group comparisons were analyzed using the Mann-Whitney U test, while correlations were assessed with Spearman’s or Pearson’s correlation as appropriate. For fibrosis-stage-related PRGs, the Kruskal-Wallis test was used for overall group comparisons, followed by Dunn’s Test with Benjamini-Hochberg correction for pairwise comparisons. For animal data, Student’s t-test was used to compare PRG expression between groups. A p-value < 0.05 was considered statistically significant.
Results
Identification of DEGs in liver sample between NAFL and NASH patients
A workflow overview of the present study is illustrated in Fig. 1.
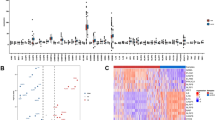
A total of 266 DEGs were identified between NAFL and NASH patients according to the predefined cut-off criteria, comprising 264 upregulated and 2 downregulated genes (Fig. 2A).
Differentially expressed genes (DEGs) in NASH vs. NAFL liver tissues, PPI network and functional enrichment analysis of DEGs. (A) volcano plot of all DEGs. (B) Functional PPI network, colors represent node correlativity degree evaluate by Cytoscape software. Redder colors represent higher interaction scores, while bluer colors indicate lower interaction scores. (C) Gene Ontology enrichment analysis, categorized into three domains: BP (Biological Process), MF (Molecular Function), and CC (Cellular Component). (D) KEGG enrichment analysis. (E) Gene Set Enrichment Analysis. Pathways enriched in the NAFL group are explicitly marked with an asterisk (*) on the top-right corner.
Functional enrichment analysis and PPI network construction of DEGs
Protein-protein interactions (PPIs) among DEGs were visualized using a network constructed with Cytoscape (Fig. 2B). Hub genes with the highest interaction scores, determined using the CytoHubba plug-in, were highlighted with red colors, while genes with fewer interactions were marked with light blue. Genes with Maximal Clique Centrality (MCC) < 10 was excluded from the visualization to simplify the network. Among the identified genes, CXCL8, CCL2, and MMP9 exhibited the highest degrees of interaction.
GO enrichment analysis revealed that the DEGs were primarily involved in fibrosis-related functions, including extracellular matrix (ECM) organization, collagen-containing ECM as a cellular component, and ECM structural constituents as molecular functions (Fig. 2C). KEGG pathway analysis indicated that the DEGs were enriched in ECM-receptor interaction and focal adhesion pathways, which are critical for tissue morphogenesis and structural integrity (Fig. 2D).
GSEA results showed that 20 pathways were enriched in the NASH group and 6 pathways were enriched in the NAFL group (Fig. 2E). Consistent with the previous GO and KEGG analyses, pathways related to cell and ECM interactions were prominently enriched.
Assessment of immune infiltration in NASH and NAFL liver tissues
The infiltration of 28 immune cell subpopulations (estimated values derived from the ssGSEA algorithmin) were assessed in NAFL and NASH patients by analyzing the derivation cohort,. The immune cell landscape in NASH and NAFL liver tissues is illustrated by a heatmap (Fig. 3A) and a bar plot (Fig. 3B). These visualizations indicate that activated T cells, activated B cells, Th1 cells, dendritic cells, NK cells, macrophages, mast cells, and monocytes were significantly elevated in NASH patients (P < 0.05), whereas Th2 cells, Th17 cells, eosinophils, and neutrophils did not exhibit significant differences between NAFL and NASH patients.
Assessment of immune infiltration in NASH vs. NAFL liver tissues. (A) Heatmap of the normalized abundance for 28 immune cells in individual samples. (B) Bar plot of 28 immune cells normalized abundance between NASH vs. NAFL liver tissues. P-values were adjusted for multiple testing using the Benjamini-Hochberg procedure. (C) Correlation matrix of 28 immune cells. The pie chart areas represent the correlation coefficients, with a full circle indicating a coefficient of 1. Red represents negative correlation; blue represents positive correlation. (D) Stromal Score, Immune Score and ESTIMATE Score in NASH vs. NAFL liver tissues. P values were calculated by Mann-Whitney test, *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we analyzed the correlations between immune cell ssGSEA scores, which revealed predominantly positive associations between different immune cell types. This is consistent with the complex immune cell interactions described in NASH pathogenesis, where diverse immune cell populations contribute to inflammation and liver injury18. Notably, we observed significant positive correlations between innate immune cells, particularly dendritic cells, macrophages, and mast cells (Fig. 3C).
Stromal signature genes and immune signature genes were used to estimate the levels of stromal and immune cell infiltration. The ESTIMATE score, which is the summation of the stromal and immune scores, was generated for each case. The Mann-Whitney test was employed to evaluate differences in stromal score, immune score, and ESTIMATE score between NAFL and NASH patients. As shown in Fig. 3D, NASH patients had significantly higher scores in all three metrics.
Identification of differentially expressed pyroptosis-related genes and fibrosis stage related PRGs
The expression of 45 pyroptosis-related genes (PRGs) was compared between NAFL and NASH patients, identifying 23 differentially expressed PRGs, including 18 upregulated and 5 downregulated genes. These differentially expressed PRGs were visualized using a heatmap (Fig. 4A). Interactions between these PRGs were depicted in a PPI network constructed using the same method as for DEGs (Fig. 4B), highlighting genes such as CASP1, NLRP3, and PYCARD as hub genes.
Identification of differentially expressed PRGs and fibrosis stage related PRGs (A) Heatmap of 23 differentially expressed PRGs in individual samples. The colorbar indicates normalized gene expression level. Red represents higher values, and blue represents lower values. (B) PPI network of differentially expressed PRGs, top 10 Hub genes of interaction were identified using Cytohubba. Node colors represent the interaction degree, redder colors indicate higher interaction scores, bluer colors indicate lower interaction scores. (C–G) Hepatic transcriptomic profiling of PRGs and liver fibrosis (C) BAX, (D) BAK1, (E) PYCARD, (F) NLRP3, (G) NLRP6. Kruskal–Wallis test were conducted for overall differences and Dunn’s Test with Benjamini–Hochberg correction for pairwise comparisons, *P < 0.05, **P < 0.01, ***P < 0.001.
Considering that fibrosis is a crucial predictor of prognosis in patients with NAFLD, we further examined the association of the correlation between differentially expressed PRGs and fibrosis stage. Our results showed that 10 PRGs were upregulated in the F0 fibrosis stage compared to patients without fibrosis, while another 9 PRGs were upregulated in the F4 fibrosis stage compared to F0 fibrosis stage patients (Supplementary Fig. 2). Four PRGs, including BAX, BAK1, PYCARD, and NLRP3, exhibited a consistent pattern of upregulation at both the F0 fibrosis stage compared to patients without fibrosis and the F4 fibrosis stage compared to the F0 fibrosis stage (Fig. 4C-G) and were recognized as fibrosis stage-related PRGs.
Construction of NASH diagnose prediction model
Twenty-three differentially expressed PRGs and three scores (stromal score, immune score, and ESTIMATE score) were included in the univariate logistic regression analysis for the primary screening of the NASH diagnostic prediction model. Variables with a P value less than 0.05 were retained for further analysis, resulting in 22 features being integrated into the least absolute shrinkage and selection operator (LASSO) regression model. Based on the optimal λ value (determined using the “cv.glmnet” function with cross-validation), 5 features were selected for multivariate logistic regression analysis. These features were further filtered based on their statistical significance, using p-value < 0.05 as the cutoff. (Fig. 5A-B). BAX, TP63, and stromal score were chosen to build the diagnostic prediction model for NASH and were presented in the nomogram (Fig. 5C).
Construction of NASH diagnose prediction model with Logistic- LASSO regression. (A, B) LASSO regression analysis and cross validation identified 5 factors to prevent overfitting. (C) Nomogram for prediction model. (D) ROC curve for derivation cohort (E) calibration curve for derivation cohort. P values were calculated by Mann-Whitney test, *P < 0.05, **P < 0.01, ***P < 0.001.
The ROC curve and calibration curve indicated robust predictive performance for the prediction model. The area under the ROC curve (AUC) was 0.870, and the calibration curve showed that the actual and predicted probabilities of NASH occurrence agreed well with each other (Fig. 5D-E).
Validation of the prediction model and fibrosis stage related PRGs
Validation of the NASH diagnostic prediction model was performed using the GSE126848 dataset, which includes 15 NAFL and 16 NASH patients. The processing procedure was identical to that of the derivation cohort, and the stromal score was generated using the same methodology. The area under the ROC curve (AUC) was 0.833, and the calibration curve showed that the validation dataset’s prediction model also demonstrated good agreement between the actual and predicted probabilities of NASH diagnosis (Fig. 6A-B).
Validation of the prediction model and fibrosis stage related PRGs. (A) ROC curve for external validation cohort GSE126848. (B) calibration curve for external validation cohort GSE126848. (C–F) Validation of the fibrosis stage related PRGs with GSE135251. (C) BAX, (D) BAK1, (E) PYCARD, (F) NLRP3. Spearman’s correlation coefficient ρ and P values were calculated by spearman correlation analysis.
Additionally, validation of fibrosis stage-related PRGs was performed using the GSE135251 dataset, which contains 206 NAFLD cases with different fibrosis stages. The correlation between the expression levels of the previously identified PRGs and fibrosis stages was analyzed using Spearman correlation. The results revealed that BAX, BAK1, PYCARD, and NLRP3 exhibited significant positive correlations with fibrosis stages, suggesting their potential as fibrosis markers (Fig. 6C-F).
Liver fibrosis models induced by CCl4 or PS were established to further verify fibrosis stage-related PRGs. Masson staining was used to measure hepatic fibrosis. As shown in Fig. 7A, the model group displayed increased liver lobule damage and collagen accumulation following administration of CCl4 or PS. After confirming the success of the liver fibrosis model, the expression of fibrosis stage-related PRGs was detected by real-time PCR. The results showed that the transcriptional levels of BAX (Fig. 7B), BAK1 (Fig. 7C), PYCARD (Fig. 7D), and NLRP3 (Fig. 7E) were significantly increased in the fibrotic liver of the CCl4 or PS model compared to the normal group, confirming the potential role of these PRGs in predicting liver fibrosis.
Validation of fibrosis stage related PRGs in liver fibrosis models. (A) Masson staining of liver tissue (100×). (B–E) Expression of BAX, BAK1, PYCARD, NLRP3 measured by real-time PCR in liver fibrosis models. (B) BAX, (C) BAK1, (D) PYCARD, (E) NLRP3. P values were calculated by unpaired t test analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
NAFLD has become the leading cause of cirrhosis and hepatocellular carcinoma worldwide, affecting approximately 25% of the global population39,40. The disease spectrum ranges from steatosis (NAFL) to non-alcoholic steatohepatitis (NASH), with approximately 10% of advanced NAFLD patients progressing to cirrhotic complications and hepatocellular carcinoma41. Higher degrees of liver fibrosis are associated with poorer prognoses and increased disease-specific mortality42. Consequently, there is an increasing demand for alternative biomarkers to differentiate NASH from simple steatosis and identify advanced hepatic fibrosis in NAFLD patients. In this study, we employed bioinformatics analyses to estimate immune cell infiltration and PRG expression patterns in NASH patients, as well as fibrosis-related PRGs, to explore the association between PRG upregulation and fibrosis in NASH patients.
We performed functional enrichment analysis to investigate the underlying mechanisms involved in the development and progression of NASH. Our findings indicated a significant enrichment of biological processes associated with ECM organization and structural components in NASH patients. This aligns with the hallmark features of liver fibrosis, which is driven by excessive ECM deposition, resulting in abnormal cell proliferation, angiogenic changes, and the eventual formation of scar tissue43,44.
The PPI network identified CXCL8, CCL2, and MMP9 as hub genes for NAFL-to-NASH progression. CXCL8, a CXC chemokine with pro-inflammatory and pro-angiogenic functions, has been reported at significantly higher concentrations in NAFL and NASH, suggesting its potential role in liver inflammation and disease progression45. CCL2, mainly produced by hepatocytes in NASH, drives monocyte-derived macrophage infiltration and liver fibrosis via Notch signaling, exacerbating liver inflammation and fibrosis46. MMP-9 is a zinc-dependent endopeptidase and a major protease involved in ECM degradation. Although our results showed increased levels of MMP9 in patients with NASH, previous studies have reported lower serum concentrations of MMP9 in patients with advanced fibrosis47. The specific mechanisms underlying this dynamic change require further exploration.
NASH represents a progressive stage of NAFLD characterized by significant immunoinflammatory alterations in patients with hepatic inflammation. A vast network of immune cells is activated during the onset of NASH. By analyzing transcriptome data of NAFL and NASH patients using the ssGSEA algorithm, our study offers deeper insights into the regulatory interactions between NASH progression and immune cell infiltration. Although innate immunity has traditionally been seen as a key element in supporting hepatic inflammation in NASH, current evidence increasingly highlights the role of adaptive immunity48. The aggregation of B and T cells in the liver positively correlates with lobular inflammation and fibrosis stages, consistent with our findings that highlight their role in NASH progression. Specifically, CD4 + T cell polarization into IFNγ-producing Th1 cells and the activation of cytotoxic CD8 + T cells exacerbate inflammation and insulin resistance. B cells further contribute to NASH by producing pro-inflammatory mediators and presenting antigens49. Dendritic cells (DCs) exhibit dual roles in NASH, with some studies suggesting a proinflammatory function50,51, while others indicate they mitigate inflammation by clearing apoptotic cells and regulating cytokine production52. Macrophages are involved in tissue remodeling during the initiation and progression of NASH, and monocytes can be recruited in response, though the underlying pathways remain unclear53. Mast cells have been shown to promote liver damage and microvesicular steatosis in a mouse model of NASH54 and correlate directly with the stage of fibrosis in NASH patients55. Neutrophils, as pivotal effectors of the innate immune response, are intricately involved in the onset and progression of NAFLD through inflammation, protease release, and neutrophil extracellular trap (NET) formation56. However, their infiltration gradually declines in the later stages of fibrosis, which might provide a plausible explanation for the observed lack of significant differences in neutrophil-related gene expression between the two groups in our study.
Pyroptosis is a newly recognized form of programmed cell death that triggers inflammasome activation. Emerging evidence indicates a crosstalk between pyroptosis and the etiopathology of NASH24,57,58. Our study sought to clarify the involvement of pyroptosis-related genes (PRGs) in NASH and liver fibrosis. We first examined the expression levels of all PRGs in patients with NAFL and NASH. Our findings revealed that several PRGs were differentially expressed in NASH tissue, with PLCG1, BAK1, IL1B, BAX, NLRP3, CASP1, TNF, IRF1, GZMA, CASP4, IL18, NOD2, CASP5, GSDMC, NLRP1, PYCARD, NLRP6, and TP63 upregulated, and HMGB1, GPX4, GSDMB, CASP6, and CHMP2A downregulated. These findings are consistent with previous studies59,60,61,62,63. We performed Logistic-LASSO regression analysis to develop a NASH diagnostic prediction model, selecting BAX, TP63, and stromal scores as the model’s components. Clustering of transcriptome data accurately distinguished NASH from NAFL in the validation cohort. BAX plays a critical role in maintaining tissue homeostasis and acts as a key executioner of mitochondrial-regulated cell death64. TP63, a member of the p53 family, is involved in epidermal development and squamous cell carcinoma tumorigenesis65,66. However, studies on the roles of BAX and TP63 in the development of NASH are limited. Further research is needed to uncover the detailed mechanisms of pyroptosis in NASH development.
Our data demonstrated that the expression levels of BAX, BAK1, PYCARD, and NLRP3 were among the PRGs upregulated in both the F0 fibrosis stage compared to patients without fibrosis and the F4 fibrosis stage compared to the F0 fibrosis stage, indicating their association with fibrosis progression. Previous research has suggested that NLRP3 plays a role in liver fibrosis, with its blockade reducing liver inflammation and fibrosis59,67. Other genes may be involved in the extracellular release of inflammasomes from hepatocytes, mediating inflammatory and pro-fibrogenic signals. The identification of upregulated pyroptosis-related genes associated with fibrosis progression presents potential avenues for developing novel therapies for hepatic fibrosis and cirrhosis.
Our study has several limitations. Due to the limited number of patients who underwent liver biopsy, the sample size was relatively small, particularly for stratified analyses across different fibrosis stages. The reliance on liver biopsy to measure the expression of pyroptosis-associated genes also limits their current clinical utility as biomarkers. MicroRNAs and extracellular vesicles have demonstrated potential as regulators, biomarkers, and therapeutic targets in liver diseases68,69. Future studies could explore non-invasive markers, such as extracellular vesicles or circulating RNAs, associated with these genes. Additionally, the prevalence of NASH and fibrosis in our NAFLD cohorts was higher than what is typically observed in the broader NAFLD population, likely due to the specific selection criteria for liver biopsy, which may introduce selection bias. The immune cell infiltration levels and protein-protein interactions analyzed in this study are computationally inferred rather than experimentally quantified, and do not encompass all immune cell types present in the liver. Further validation using experimental approaches, such as single-cell cytometry, is necessary to confirm these findings.
Conclusion
Our study identified BAX, BAK1, PYCARD, and NLRP3 as key pyroptosis-associated genes upregulated with fibrosis progression in NASH. Validation in external datasets and animal models confirmed their strong correlation with fibrosis stages. Immune infiltration analysis revealed distinct immune cell compositions, further linking pyroptosis to immune-mediated fibrosis. These findings highlight pyroptosis-associated genes could serve as potential targets for investigating NASH-related fibrosis.
Data availability
The data that endorse the results of this research are available upon reasonable request from the authors (S.T. or X.J.) Besides, original data for bioinformatics analysis could be download from Gene Expression Omnibus(GEO) datasets GSE162694, GSE135251 and GSE126848.
References
Sheka, A. C. et al. Nonalcoholic steatohepatitis: A review. JAMA 323, 1175–1183. https://doi.org/10.1001/jama.2020.2298 (2020).
Chen, H. et al. PPAR-gamma signaling in nonalcoholic fatty liver disease: Pathogenesis and therapeutic targets. Pharmacol. Ther. 245, 108391. https://doi.org/10.1016/j.pharmthera.2023.108391 (2023).
Zhou, J. et al. Epidemiological features of NAFLD from 1999 to 2018 in China. Hepatology 71, 1851–1864. https://doi.org/10.1002/hep.31150 (2020).
Younossi, Z. et al. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15, 11–20. https://doi.org/10.1038/nrgastro.2017.109 (2018).
Paternostro, R. & Trauner, M. Current treatment of non-alcoholic fatty liver disease. J. Intern. Med. 292, 190–204. https://doi.org/10.1111/joim.13531 (2022).
Meroni, M., Longo, M., Rustichelli, A. & Dongiovanni, P. Nutrition and genetics in NAFLD: The perfect binomium. Int. J. Mol. Sci. 21 https://doi.org/10.3390/ijms21082986 (2020).
Terrault, N. A., Francoz, C., Berenguer, M., Charlton, M. & Heimbach, J. L. T. Status report, current and future challenges. Clin Gastroenterol Hepatol 21, 2150–2166, (2023). https://doi.org/10.1016/j.cgh.2023.04.005 (2023).
Huang, D. Q. et al. Global epidemiology of cirrhosis—aetiology, trends and predictions. Nat. Rev. Gastroenterol. Hepatol. 20, 388–398. https://doi.org/10.1038/s41575-023-00759-2 (2023).
Tincopa, M. A. & Loomba, R. Non-invasive diagnosis and monitoring of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Lancet Gastroenterol. Hepatol. 8, 660–670. https://doi.org/10.1016/S2468-1253(23)00066-3 (2023).
Parola, M. & Pinzani, M. Liver fibrosis in NAFLD/NASH: From pathophysiology towards diagnostic and therapeutic strategies. Mol. Aspects Med. 95, 101231. https://doi.org/10.1016/j.mam.2023.101231 (2024).
Loomba, R. & Adams, L. A. The 20% rule of NASH progression: The natural history of advanced fibrosis and cirrhosis caused by NASH. Hepatology 70, 1885–1888. https://doi.org/10.1002/hep.30946 (2019).
Samy, A. M., Kandeil, M. A., Sabry, D., Abdel-Ghany, A. A. & Mahmoud, M. O. From NAFLD to NASH: Understanding the spectrum of non-alcoholic liver diseases and their consequences. Heliyon 10, e30387. https://doi.org/10.1016/j.heliyon.2024.e30387 (2024).
Brunt, E. M., Janney, C. G., Di Bisceglie, A. M., Neuschwander-Tetri, B. A. & Bacon, B. R. Nonalcoholic steatohepatitis: A proposal for grading and staging the histological lesions. Am. J. Gastroenterol. 94, 2467–2474. https://doi.org/10.1111/j.1572-0241.1999.01377.x (1999).
Chalasani, N. et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American association for the study of liver diseases. Hepatology 67, 328–357. https://doi.org/10.1002/hep.29367 (2018).
Tacke, F., Puengel, T., Loomba, R. & Friedman, S. L. An integrated view of anti-inflammatory and antifibrotic targets for the treatment of NASH. J. Hepatol. 79, 552–566. https://doi.org/10.1016/j.jhep.2023.03.038 (2023).
Wei, S., Wang, L., Evans, P. C. & Xu, S. NAFLD and NASH: Etiology, targets and emerging therapies. Drug Discov. Today. 29, 103910. https://doi.org/10.1016/j.drudis.2024.103910 (2024).
Khairnar, R., Islam, M. A., Fleishman, J. & Kumar, S. Shedding light on non-alcoholic fatty liver disease: Pathogenesis, molecular mechanisms, models, and emerging therapeutics. Life Sci. 312, 121185. https://doi.org/10.1016/j.lfs.2022.121185 (2023).
Huby, T. & Gautier, E. L. Immune cell-mediated features of non-alcoholic steatohepatitis. Nat. Rev. Immunol. 22, 429–443. https://doi.org/10.1038/s41577-021-00639-3 (2022).
Al Mamun, A. et al. Role of pyroptosis in liver diseases. Int. Immunopharmacol. 84, 106489. https://doi.org/10.1016/j.intimp.2020.106489 (2020).
Hammerich, L. & Tacke, F. Hepatic inflammatory responses in liver fibrosis. Nat. Rev. Gastroenterol. Hepatol. 20, 633–646. https://doi.org/10.1038/s41575-023-00807-x (2023).
Erkes, D. A. et al. Mutant BRAF and MEK inhibitors regulate the tumor immune microenvironment via pyroptosis. Cancer Discov. 10, 254–269. https://doi.org/10.1158/2159-8290.CD-19-0672 (2020).
Rao, Z. et al. Pyroptosis in inflammatory diseases and cancer. Theranostics 12, 4310–4329. https://doi.org/10.7150/thno.71086 (2022).
Yu, P. et al. Pyroptosis: Mechanisms and diseases. Signal. Transduct. Target. Ther. 6, 128. https://doi.org/10.1038/s41392-021-00507-5 (2021).
Gaul, S. et al. Hepatocyte pyroptosis and release of inflammasome particles induce stellate cell activation and liver fibrosis. J. Hepatol. 74, 156–167. https://doi.org/10.1016/j.jhep.2020.07.041 (2021).
Xu, B. et al. Gasdermin D plays a key role as a pyroptosis executor of non-alcoholic steatohepatitis in humans and mice. J. Hepatol. 68, 773–782. https://doi.org/10.1016/j.jhep.2017.11.040 (2018).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The Sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. https://doi.org/10.1093/bioinformatics/bts034 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. A database for post-genome analysis. Trends Genet. 13, 375–376. https://doi.org/10.1016/s0168-9525(97)01223-7 (1997).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Chin, C. H. et al. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8 (Suppl 4). https://doi.org/10.1186/1752-0509-8-S4-S11 (2014).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14 https://doi.org/10.1186/1471-2105-14-7 (2013).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612. https://doi.org/10.1038/ncomms3612 (2013).
Karki, R. & Kanneganti, T. D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer. 19, 197–214. https://doi.org/10.1038/s41568-019-0123-y (2019).
Wang, X. et al. Cross-talk of pyroptosis and tumor immune landscape in lung adenocarcinoma. Transl Lung Cancer Res. 10, 4423–4444. https://doi.org/10.21037/tlcr-21-715 (2021).
Kesavardhana, S., Malireddi, R. K. S. & Kanneganti, T. D. Caspases in cell death, inflammation, and pyroptosis. Annu. Rev. Immunol. 38, 567–595. https://doi.org/10.1146/annurev-immunol-073119-095439 (2020).
Tu, S. et al. Fluorofenidone protects liver against inflammation and fibrosis by blocking the activation of NF-kappaB pathway. FASEB J. 35, e21497. https://doi.org/10.1096/fj.202002402R (2021).
Powell, E. E., Wong, V. W. & Rinella, M. Non-alcoholic fatty liver disease. Lancet 397, 2212–2224. https://doi.org/10.1016/S0140-6736(20)32511-3 (2021).
Riazi, K. et al. The prevalence and incidence of NAFLD worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 7, 851–861. https://doi.org/10.1016/S2468-1253(22)00165-0 (2022).
Shah, P. A., Patil, R. & Harrison, S. A. NAFLD-related hepatocellular carcinoma: The growing challenge. Hepatology 77, 323–338. https://doi.org/10.1002/hep.32542 (2023).
Llovet, J. M. et al. Nonalcoholic steatohepatitis-related hepatocellular carcinoma: Pathogenesis and treatment. Nat. Rev. Gastroenterol. Hepatol. 20, 487–503. https://doi.org/10.1038/s41575-023-00754-7 (2023).
Tong, G. et al. Fibroblast growth factor 18 attenuates liver fibrosis and HSCs activation via the SMO-LATS1-YAP pathway. Pharmacol. Res. 178, 106139. https://doi.org/10.1016/j.phrs.2022.106139 (2022).
Yuan, S. et al. Sorafenib attenuates liver fibrosis by triggering hepatic stellate cell ferroptosis via HIF-1alpha/SLC7A11 pathway. Cell. Prolif. 55, e13158. https://doi.org/10.1111/cpr.13158 (2022).
Pan, X. et al. Chemokines in Non-alcoholic fatty liver disease: A systematic review and network meta-analysis. Front. Immunol. 11, 1802. https://doi.org/10.3389/fimmu.2020.01802 (2020).
Kang, J. et al. Notch-mediated hepatocyte MCP-1 secretion causes liver fibrosis. JCI Insight. 8 https://doi.org/10.1172/jci.insight.165369 (2023).
Goyale, A. et al. Assessment of non-alcoholic fatty liver disease (NAFLD) severity with novel serum-based markers: A pilot study. PLoS ONE. 16, e0260313. https://doi.org/10.1371/journal.pone.0260313 (2021).
Sutti, S. & Albano, E. Adaptive immunity: An emerging player in the progression of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 17, 81–92. https://doi.org/10.1038/s41575-019-0210-2 (2020).
Barrow, F. et al. Microbiota-driven activation of intrahepatic B cells aggravates NASH through innate and adaptive signaling. Hepatology 74, 704–722. https://doi.org/10.1002/hep.31755 (2021).
Schuster, S., Cabrera, D., Arrese, M. & Feldstein, A. E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 15, 349–364. https://doi.org/10.1038/s41575-018-0009-6 (2018).
Wang, X. et al. Characteristic gene expression in the liver monocyte-macrophage-DC system is associated with the progression of fibrosis in NASH. Front. Immunol. 14, 1098056. https://doi.org/10.3389/fimmu.2023.1098056 (2023).
Henning, J. R. et al. Dendritic cells limit fibroinflammatory injury in nonalcoholic steatohepatitis in mice. Hepatology 58, 589–602. https://doi.org/10.1002/hep.26267 (2013).
Daemen, S. et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell. Rep. 34, 108626. https://doi.org/10.1016/j.celrep.2020.108626 (2021).
Kennedy, L. et al. Mast cells promote nonalcoholic fatty liver disease phenotypes and microvesicular steatosis in mice fed a Western diet. Hepatology 74, 164–182. https://doi.org/10.1002/hep.31713 (2021).
Lombardo, J. et al. Hepatic mast cell concentration directly correlates to stage of fibrosis in NASH. Hum. Pathol. 86, 129–135. https://doi.org/10.1016/j.humpath.2018.11.029 (2019).
Herrero-Cervera, A., Soehnlein, O. & Kenne, E. Neutrophils in chronic inflammatory diseases. Cell. Mol. Immunol. 19, 177–191. https://doi.org/10.1038/s41423-021-00832-3 (2022).
Koh, E. H. et al. Sphingomyelin synthase 1 mediates hepatocyte pyroptosis to trigger non-alcoholic steatohepatitis. Gut 70, 1954–1964. https://doi.org/10.1136/gutjnl-2020-322509 (2021).
Zhao, R. et al. Hepatocyte-specific NR5A2 deficiency induces pyroptosis and exacerbates non-alcoholic steatohepatitis by downregulating ALDH1B1 expression. Cell. Death Dis. 15, 770. https://doi.org/10.1038/s41419-024-07151-1 (2024).
Mridha, A. R. et al. NLRP3 inflammasome Blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 66, 1037–1046. https://doi.org/10.1016/j.jhep.2017.01.022 (2017).
Qi, J., Kim, J. W., Zhou, Z., Lim, C. W. & Kim, B. Ferroptosis affects the progression of nonalcoholic steatohepatitis via the modulation of lipid Peroxidation-Mediated cell death in mice. Am. J. Pathol. 190, 68–81. https://doi.org/10.1016/j.ajpath.2019.09.011 (2020).
Wree, A., Inzaugarat, M. E. & Feldstein, A. E. Transmembrane BAX inhibitor motif-containing 1, a novel anti-inflammatory approach for nonalcoholic steatohepatitis treatment. Hepatology 67, 438–441. https://doi.org/10.1002/hep.29495 (2018).
Chen, X., Wang, D., Wang, Z. & Huang, W. Identification of pyroptosis-related genes in NASH based on bioinformatic analysis. Biochem. Biophys. Res. Commun. 679, 90–97. https://doi.org/10.1016/j.bbrc.2023.08.067 (2023).
Mao, F. et al. Identification of pyroptosis-related gene signature in nonalcoholic steatohepatitis. Sci. Rep. 14, 3175. https://doi.org/10.1038/s41598-024-53599-8 (2024).
Spitz, A. Z. & Gavathiotis, E. Physiological and pharmacological modulation of BAX. Trends Pharmacol. Sci. 43, 206–220. https://doi.org/10.1016/j.tips.2021.11.001 (2022).
Jiang, Y. et al. Reciprocal Inhibition between TP63 and STAT1 regulates anti-tumor immune response through interferon-gamma signaling in squamous cancer. Nat. Commun. 15, 2484. https://doi.org/10.1038/s41467-024-46785-9 (2024).
Moses, M. A. et al. Molecular mechanisms of p63-Mediated squamous cancer pathogenesis. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20143590 (2019).
Charan, H. V., Dwivedi, D. K., Khan, S. & Jena, G. Mechanisms of NLRP3 inflammasome-mediated hepatic stellate cell activation: Therapeutic potential for liver fibrosis. Genes Dis. 10, 480–494. https://doi.org/10.1016/j.gendis.2021.12.006 (2023).
Wang, X., He, Y., Mackowiak, B. & Gao, B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut 70, 784–795. https://doi.org/10.1136/gutjnl-2020-322526 (2021).
Szabo, G. & Momen-Heravi, F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 14, 455–466. https://doi.org/10.1038/nrgastro.2017.71 (2017).
Author information
Authors and Affiliations
Contributions
The data were gathered, the analysis was carried out, the experiments were performed, and the manuscript was written by ST and XJ. Some of data analysis was assisted by XL B, QF Z, SY L, YG and XB Z. WH S revised the manuscript, designed the experiment.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The animal experimental protocol is in accordance with The Institutional Animal Care and Use Committee Guangdong Provincial People’s Hospital. The current study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tu, S., Jing, X., Bu, X. et al. Identification of pyroptosis-associated gene to predict fibrosis and reveal immune characterization in non-alcoholic fatty liver disease. Sci Rep 15, 14944 (2025). https://doi.org/10.1038/s41598-025-96158-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-96158-5