Abstract
Our study aimed to identify PANoptosis in Aβ1-40-induced AMD, both in vivo and in vitro, and to determine if AIM2-PANoptosome mediates this process. We used transcriptomics to explore the signaling pathways and target genes linked to PANoptosis within a mouse model of AMD triggered by Aβ1-40. Optical coherence tomography (OCT), hematoxylin and eosin (H&E) staining, and electroretinography (ERG) were employed to assess retinal damage in terms of morphology and function. Morphological changes in ARPE-19 cells were observed using optical microscopy and scanning electron microscopy. Enzyme-linked immunosorbent assay (ELISA) was used to detect the levels of cytokines in cell supernatants, mouse orbital serum, and human plasma to evaluate the severity of inflammation. CO-immunoprecipitation(CoIP) and molecular docking were performed to assess the impact and expression of proteins associated with the AIM2-PANoptosome. Quantitative polymerase chain reaction (qPCR), Western blot (WB), immunofluorescence, and apoptosis detection kits were used to evaluate the expression levels of genes and proteins related to PANoptosis-like cell death. Our results showed that the Aβ1-40-induced AMD model had increased expression of apoptosis, necroptosis, and pyroptosis pathways, and AIM2-PANoptosome components. CoIP and docking confirmed increased AIM2, ZBP1, and PYRIN levels under Aβ1-40 treatment. WB and immunofluorescence showed upregulation of PANoptosis-related proteins. Inhibitors reduced Aβ-induced protein expression. ELISA showed increased inflammatory cytokines. Apoptosis assays and microscopy revealed Aβ1-40-induced ARPE-19 cell loss and morphological changes. In conclusion, the Aβ1-40-induced AMD model displayed PANoptosis-like cell death, offering insights into disease pathogenesis.
Similar content being viewed by others
Introduction
AMD is a progressive retinal condition that predominantly impacts the macula, which is the central, high-acuity portion of the retina tasked with sharp and intricate vision1. This condition is a major cause of vision impairment in individuals over the age of 55, constituting 6–9% of global cases of legal blindness2,3. AMD affects individuals from various economic backgrounds, highlighting its widespread impact on global health. Current global estimates anticipate a significant increase in the prevalence of AMD, with the number of affected individuals expected to rise from approximately 196 million in 2020 to an estimated 288 million by 2040 3.
Amyloid-beta (Aβ), a peptide closely associated with Alzheimer’s disease (AD), is also a significant pathological hallmark in AMD4. Aβ is a component of drusen, which are extracellular, yellowish deposits that accumulate below the retinal pigment epithelium (RPE) and are associated with AMD5,6. This peptide plays an important role in early AMD development and signals disease onset7. Macular degeneration is a progressive disease involving the dysfunction of the RPE, leading to cellular damage and ultimately cell death8. Initially, early disease is characterized by few drusen or mild RPE abnormalities. As the condition progresses, drusen enlarge and RPE hypopigmentation and hyperpigmentation occur. Late AMD is divided into dry AMD with geographic atrophy and wet AMD with neovascularization9. Understanding this condition is critical, as it can provide insights into the key components that influence retinal health and are central to the evolution and progression of AMD. Comprehending these processes will be essential for designing effective interventions and treatments to preserve vision in affected individuals.
Organismal development is dependent on the phenomenon of Programmed cell death (PCD). The defense against pathogens and the maintenance of tissue stability are essential, and PCD, with its multiple distinct mechanisms, aids in the regulated cell death. These include apoptosis, necroptosis, pyroptosis, ferroptosis, and PANoptosis10,11. Each of these cell death forms is closely linked to the body’s innate immune response, highlighting their significance in maintaining cellular health and fighting disease12,13,14. PANoptosis, a term introduced by Malireddi in 2019, represents a novel mode of PCD that integrates features of pyroptosis, apoptosis, and necroptosis, yet is not fully defined by any single processs15. This process is tightly regulated by a complex network of upstream receptors and molecular signals, which converge to form a composite structure known as the PANoptosome, essential for executing this type of cell death16. Recent studies have emphasized the significance of PANoptosis in various pathological conditions. Wan et al.17 demonstrated its role in retinal ischemia-reperfusion injury, while Lan et al.18 showed its importance in cerebral ischemia-reperfusion injury. Wang et al.19 identified a noncanonical feedback loop between the RIP3–MLKL and 4EBP1–eIF4E pathways, highlighting PANoptosis’s unique integration of multiple cell death pathways and its therapeutic potential for AMD and stroke. While the influence of apoptosis, necroptosis, and pyroptosis on RPE cells is well-documented20,21,22, the specific impact of PANoptosis on these cells in pathological conditions such as AMD is not well understood. Further research into how PANoptosis affects the disease process in AMD could deepen our comprehension of this PCD pathway and potentially inform the development of therapeutic strategies.
Our research aimed to determine whether the Aβ1-40-induced AMD model exhibits characteristics of PANoptosis-like cell death. Our findings revealed the coexistence of apoptosis, necroptosis, and pyroptosis in both in vivo and in vitro environments following Aβ1-40 exposure, as well as increased expression of proteins associated with PANoptosis and activation of the AIM2-PANoptosome.
Materials and techniques
Reagents and materials
Relevant antibody information (Table 1) and reagents information (Table 2) are provided below.
Cell culture
We obtained the ARPE-19 cells from the American Type Culture Collection (ATCC, Manassas, VA). For culturing ARPE-19 cells, complete culture medium (100 µg/ml penicillin and streptomycin: FBS: DMEM = 1:10:89) was used. The cells were incubated at 37℃ in a 5% CO2 incubator (Thermo Fisher, USA). Cells sourced from passages 9 to 15 of the ARPE-19 line were plated into distinct well plates and incubated for 48 h at 37℃ in DMEM/F12 medium under 5% CO2. The cells were then prepared for various analyses.
Human blood samples collection
This research was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University and received approval under the number 2023-29. All participants provided their informed consent prior to their involvement in the study. Blood collection was conducted following the principles of the Declaration of Helsinki and the ARVO Statement on Human Subjects in Research. Peripheral blood was collected from 12 age-matched subjects with cataracts and 12 subjects with untreated age-related macular degeneration (AMD) (Supplementary Table 1). The diagnosis of AMD was confirmed by experienced ophthalmologists using OCT and optical coherence angiography(OCT-A) according to revised criteria. Participants with cardiovascular disease, autoimmune conditions, hematological disorders, active infections, or elevated neutrophil counts were excluded to minimize the risk of background activation that could confound the results. The control group consisted of age- and gender-matched individuals without AMD, ensuring a relevant comparison with the patient group.
Aβ oligomerization
Initially, Aβ1-40 was dissolved in HFIP (222µL/mg), aliquoted into sterile tubes, and dried under nitrogen gas to form an Aβ peptide film. Following evaporation, the film was dissolved in DMSO (5µL). The solution was then diluted to 100 µM (0.45 µg/µL) with phosphate-buffered saline (PBS, pH 7.4, 545 µL). To form oligomeric Aβ, the diluted solution was incubated at 37 °C for 6 h, after which it was stored at -80 °C for later use.
Drug preparation
DSF was dissolved in DMSO and diluted in PBS to a final concentration of 8 µM. Z-VAD-FMK was dissolved in DMSO and diluted in PBS to a final concentration of 10 µM. Nec-1 was diluted in DMSO and diluted in PBS to a final concentration of 20 µM. Before exposure to Aβ1-40, ARPE-19 cells were pretreated with each inhibitor for 1 h. When used in combination with Aβ1-40 in ARPE-19 cells, the inhibitors were maintained at their individual concentrations.
Animals and treatments
Male C57BL/6 mice (6- to 8-week-old) were selected based on previous studies23,24 and were obtained from the Animal Health Committee of Chongqing Medical University. The mouse studies in this manuscript were reported following the ARRIVE guidelines (https://arriveguidelines.org/). The mice were euthanized according to the American Veterinary Medical Association (AVMA) Guidelines for the Euthanasia of Animals (2020). Intraperitoneal injection with sodium pentobarbital solution at 100 mg/kg was used to euthanize. The animals were housed in a sterile environment with controlled conditions: a room temperature ranging from 20 °C to 24 °C, relative humidity between 40% and 60%, and alternating 12-hour periods of light and darkness. Food and water were available to the animals without restriction. The project’s animal protocols were approved by the Institutional Animal Care and Use Committee at Chongqing Medical University (IACUC-CQMU-2023-0355). Intraperitoneal injection of 2.0% tribromoethanol anesthesia (at 6–8 µl/g). 2 µl of Aβ1-40 (350 µM) was injected into mice vitreous using a 10 µl syringe under a stereomicroscope. On day 4, the mice were euthanized for experiments25 (e.g., RNA-seq, H&E, OCT, qPCR, WB, and ELISA). The control group (PBS + 1% DMSO) and the Aβ1-40 group were both injected in the ipsilateral eye (n = 27, Power = 1.0). The ocular area was disinfected before the invasive procedure and antibiotic ointment was applied afterward. After injection, the animals showed no changes in body weight, abnormal activity, or decreased appetite.
Cell counting Kit8
1 × 104 ARPE-19 cells per well was used to seed the cells into 96-well plates, following the manufacturer’s protocol. The cells were exposed to varying concentrations of Aβ1-40 (NC, vehicle-treated, 2.5, 5, 10, 20µM), and their viability was assessed at 24 and 48 h post-exposure. Cell viability was then conducted using the CCK-8 (Beyotime, Shanghai, China). Absorbance values were analyzed applying a microplate reader Varioskan Flash (Thermo Scientific, USA), with the wavelength set at 450 nm for analysis.
Western blotting
RIPA was added to lyse cells and tissues, and the supernatant was collected after centrifugation. Protein content was quantified using a BCA assay kit. Proteins extracted from cellular samples (30 µg) and animal tissues (60 µg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently blotted onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA) through a Tank blot carried out at 375 mA for 35–60 min at 4℃. The membranes were treated with a non-protein rapid blocking reagent at ambient temperature for 60 min, then exposed to the primary antibody at 4 °C for an extended period. After extensive rinsing with Tris-buffered saline (TBS), they were further incubated with horseradish peroxidase-tagged secondary antibodies at 37 °C for 120 min. The protein bands were detected on the membranes with the help of a gel imager (Thermo Fisher, USA), and image analysis was conducted using ImageJ software (National Institutes of Health, USA).
Co-Immunoprecipitation assay
Co-immunoprecipitation assay was conducted on ARPE-19 cell proteins using antibodies at 4℃ overnight. The immune complexes were precipitated following incubation, employing protein A/G-agarose beads (HY-K0202, MedChemExpress) for 4 h at 4 °C. The beads were subsequently rinsed five times with lysis buffer, and after each rinse, they were centrifuged at 1000×g for 5 min at 4℃ to collect the beads. The immunoprecipitated proteins were resolved by SDS-PAGE and subsequently transported to a membrane for WB with the corresponding primary antibodies.
Immunofluorescence assay
Cells were plated at a concentration of 5 × 104 ARPE-19 cells/6-well plates and cultured overnight to allow for attachment. Cells were exposed to Aβ1-40 oligomers (5, 10, 20µM) for 48 h, then fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and subsequently blocked with 5% goat serum. Subsequently, the cells were cultured with the primary antibody for an extended period at 4°C, rinsed with PBS, and then further incubated with the appropriate secondary antibody for 2 h at ambient temperature. Then washing with PBS, and an anti-fade solution containing DAPI was added. Fluorescence imaging was conducted using the Leica TE2000 inverted microscope as the imaging instrument (Leica Microsystems GmbH, Germany).
Histological analysis
The entire mouse eyeballs were extracted, washed with PBS, and subsequently immersed in FAS Ocular Fixation Solution for a 24-hour fixation period. The tissues underwent dehydration through a sequence of ethanol concentrations and were enbedded in paraffin. Sections of the eye tissues, 5 μm thick, were cut and subjected to H&E staining for routine histological analysis. Ultimately, the morphological structures of the tissues were observed and documented with an upright fluorescence microscope (Leica Microsystems GmbH, Germany).
Quantitative real-time PCR (qPCR)
Quantitative PCR evaluation (qPCR) was conducted utilizing Real-Time PCR System (Thermo Fisher Scientific, USA). The data were adjusted to β-actin levels in the control (sham) cohort, using this as an internal standard for ensuring the comparability of results across all samples. The specifications of the primers are delineated in Table 3.
ERG assessment
The mice were kept in the dark for an entire night. The mice were fully anesthetized under dim red lighting. Mydriatics are used to dilate the pupils(1% tropicamide), and local anesthetics are used to numb the eyes in preparation for further examination. Insert the ground needle electrode caudally and place the reference needle electrode subcutaneously under the eye. A drop of 2.5% methylcellulose was placed into both eyes and the contact lens electrode was placed at the cornea’s center. Scotopic (dark-adapted) flash electroretinograms (ERGs) were performed using a visual electrophysiological system, which included a Ganzfeld Bowl and a Q450SC device from Roland Consult. The system was utilized to record the ERG responses under a range of flash intensities: 0.1, 1.0, oscillatory potential (OP), 3.0, and 10 cd·s/m.
High-resolution SD‐OCT
Mice were anesthetized, and fundus images were captured using the Bioptigen R-Class System (Envisu R4310; Bioptigen Inc.). Pupil dilation was achieved by administering eye drops containing 0.5% tropicamide and 0.5% noradrenaline hydrochloride, while sodium hyaluronate eye drops were used to maintain ocular moisture.
Scanning electron microscopy
Cells after their exposure to Aβ1-40 oligomers (10µM) for 48 h to observe morphological changes. After the treatment was finished, the cells were fixed with 4% paraformaldehyde for 40 min and then washed four times with 1×PBS. Subsequently, the specimens underwent dehydration through a sequential ethanol gradient, followed by critical point drying to eliminate residual solvent. To facilitate observation with a scanning electron microscope(model SU8020, Hitachi, Japan), the sample surfaces were first coated with a gold-palladium layer to boost conductivity and mitigate charging.
ELISA assay
ARPE-19 cell supernatant, mouse orbital serum, and human plasma were prepared. Commercial assay kits compatible with both human and mouse samples were used to measure levels of IFN-γ, TNF-α, IL-1β, and IL-18. Absorbance readings were taken at 450 nm, and results from three independent experiments are reported in pg/ml.
Apoptosis assay
Cells were seeded at 8 × 10^4 cells per well in a 12-well plate and cultured overnight. They were then treated with 10 µM oligomeric Aβ1-40 for 48 h. Following the treatment, cells were harvested and analyzed for apoptosis using the Annexin V-FITC/PI Apoptosis Detection Kit according to the manufacturer’s guidelines. Apoptosis analysis was performed using a NovoCyte Advanteon flow cytometer (Agilent Technologies, USA), adhering to the protocol provided by the manufacturer.
RNA sequencing (RNA-seq) data
The cornea, lens, and vitreous of the mouse eyeball were excised under a microscope. By tiling the eye cup, the retina and sclera were separated, leaving only the RPE-Bruch’s membrane complex layer. Retinal choroidal complex tissues were collected from successfully modeled mice for sequencing. The integrity of the sequencing data was assessed and confirmed using the FastQC program (version 0.23.0). They were mapped to the mouse reference genome (ftp://ftp.ensembl.org/pub/release-101/fasta/mus_musculus/dna/) using STAR software (version 2.5.3a) with default parameters. The sequencing libraries were constructed using the kcUID software from Seqhealth. RPKM (Reads per Kilobase per Million Reads) was employed as an indicator of gene expression. Subsequently, the absolute value of logFC with a cutoff of 0.58 and a p-value set at 0.05 were used as thresholds for identifying differentially expressed genes. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the ClusterProfiler package26.
Molecular docking
In this study, the 3D protein frameworks of Amyloid (PDB ID: 1AML), AIM2 (PDB ID: 3RN2), ZBP1 (PDB ID: 2LNB), and PYRIN (PDB ID: 2MPC) were retrieved from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB). (https://www.rcsb.org/). The molecular docking program employed is HADDOCK (Version 2.4)27, an information-driven flexible docking method for modeling biomolecular complexes. HADDOCK integrates various experimental and/or bioinformatics data to guide modeling, enabling the use of more sophisticated conformational flexibility treatments to focus the search on relevant areas of the interaction space. The HADDOCK program was utilized to dock the proteins and calculate the complex model. The PyMOL software (Version 3.0.3) was utilized for result analysis and visualization.
Statistical analysis
Data analysis was conducted using GraphPad Prism software (version 10.0). The data conforms to normal distribution via the K-S test. A one-way ANOVA and Student’s t-test were employed to assess differences among groups. Results are expressed as mean ± SEM, with statistical significance defined as a p-value below 0.05.
Results
PANoptosis-like cell death in Aβ1-40-induced AMD
To explore the effects of pathways and target genes involved in PANoptosis-like cell death in the Aβ-induced AMD model, we obtained retinal choroidal complex tissues from mouse eyes after confirming the success of the model using in vivo OCT and ERG examinations for transcriptome sequencing. Transcriptome differential analysis identified 695 differentially expressed genes (DEGs) with a threshold of |log2FoldChange| < 0.5& Pvalue < 0.05. This data comprises 515 genes that were up-regulated. Based on the literature related to PANoptosis28, a Venn diagram was created to show the intersection of transcriptomic DEGs and PANoptosis-related genes, which includes 2 genes for Necroptosis, 21 genes for Pyroptosis, and 307 genes for Apoptosis(Fig. 1a).
A total of 14 differentially expressed genes were identified in the Venn diagram, with 13 being related to apoptosis and 1 to pyroptosis (Fig. 1a). Protein-protein interaction (PPI) analysis was conducted utilizing the STRING database (https://cn.string-db.org/) to explore the interactions among the 14 genes (Fig. 1b). GO analysis revealed the occurrence of pyroptosis, apoptosis, and inflammation in Aβ1-40-induced AMD model (Fig. 1c). KEGG29 analysis revealed activation of JAK-STAT signaling pathway, NOD-like receptor pathway, natural killer cell-mediated cytotoxicity, cytokine-cytokine receptor interaction, and autoimmune thyroid disease associated with necroptosis, pyroptosis, and apoptosis in Aβ1-40-treated mice (Fig. 1d). Genes related to the AIM2-PANoptosome, including Aim2, Zbp1, Mefv, Mlkl, Casp8, Nlrp1b, Casp1, and Gsdmd, were all upregulated (Fig. 1e). This suggests that PANoptosis-like cell death may exist in the Aβ1-40-induced AMD model, and Aβ1-40 oligomers may trigger this form of cell death by activating the AIM2 PANoptosome.
Gene expression data from RNA sequencing in Aβ1-40-injected mice. (a) A Venn diagram constructs the intersection of genes associated with apoptosis, necroptosis, and pyroptosis, revealing 14 DEGs implicated in these processes. (b) PPI network analysis demonstrates the enrichment of differentially expressed genes in mice injected with Aβ1-40 as opposed to the control cohort. (c) GO analysis discloses the enrichment of differentially expressed genes in mice treated with Aβ1-40 relative to the control group. (d) A bubble chart illustrates the KEGG pathway enrichment analysis. (e) A heatmap is generated for the cluster analysis of genes involved in necroptosis, pyroptosis, and apoptosis.
Molecular interactions of Aβ with key proteins in PANoptosomes
The PANoptosome complex is integral to the PANoptosis process, connecting key components such as AIM2, CASP8, and RIPK3, which are sourced from three major cell death pathways: apoptosis, necroptosis, and pyroptosis16,30. The results of the Co-IP experiment revealed the interaction between PYRIN and AIM2, ZBP1 and AIM2 before and after drug treatment. There is a certain interaction between PYRIN and AIM2, ZBP1 and AIM2 after drug treatment (Fig. 2a).
Furthermore, we performed molecular docking simulations to investigate the interactions between Aβ and AIM2 (Fig. 2b), as well as the relationships between the molecular conformations of AIM2, ZBP1, and PYRIN, with these proteins being connected via hydrogen bonds (Fig. 2c). The spatial conformations of the proteins and their interactions reveal the formation of the AIM2-PANoptosome in the Aβ-induced AMD model.
CoIP and protein molecular docking reveal AIM2-PANoptosome interaction. (a) CoIP analysis of PYRIN and ZBP1 bound to AIM2 with or without Aβ1-40 in ARPE cell. Western blot results were quantified (n = 3). (b) Structural analysis of the protein interaction interface between Aβ1-40 and AIM2. Illustrates protein-protein interactions, highlighting the amino acid residues involved in these interactions. The cyan section represents the Amyloid protein, while the green section denotes the AIM2 protein. (c) Structural examination of the protein interaction interfaces among AIM2, ZBP1, and PYRIN. The diagram illustrates protein-protein interactions, highlighting the amino acid residues involved in hydrogen bond interactions between the respective proteins. The cyan section represents the AIM2 protein, the blue section denotes the ZBP1 protein, and the orange section corresponds to the Pyrin protein. Values are expressed as mean ± SEM. *P < 0.05, vs. the Aβ1-40 group.
PANopotosis-like cell death was induced by Aβ1-40 in vivo
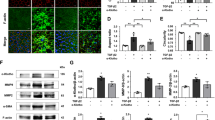
In this study, we injected Aβ1-40 into the vitreous of C57BL/6 mice. On day 4 post-injection, OCT revealed disruptions in the structural continuity and integrity of the RPE layer in the Aβ1-40-injected group (Fig. 3a). Additionally, we performed H&E staining on retinal sections to assess structural changes in the retina and RPE areas. We observed discontinuities and elevations in the RPE layer (Fig. 3b), indicating damage to the RPE in the Aβ1-40-induced AMD model.
To assess the effects of Aβ1-40 on visual function in mice, we conducted ERG examinations. To elicit the dark-adapted ERG response, various light stimulation intensities were applied with 20-second intervals between each stimulus. We induced dark adaptation using different light intensities (0.1, 1.0, 3.0, 10.0) (Supplementary Fig. S1)and observed a notable reduction in the b-wave amplitudes in the Aβ1-40 injection group at a light intensity of 1.0 (Fig. 3c). The a-wave of the ERG represents the function of photoreceptors and the RPE, while the b-wave is produced by the depolarization of bipolar cells. The b-wave is also dependent on the functional integrity of the outer retina31,32. This decline suggests that the presence of Aβ1-40 may cause functional deficits in the retina.
Aβ1-40-induced retinal damage in vivo. (a) OCT scans of control and Aβ1-40 injected mice. (b) Retinal paraffin sections were stained with H&E. (c) The impact on retinal function in mice injected with Aβ1-40 is demonstrated through exemplary 1.0 scotopic ERG waveforms for each group, along with the quantification of b-wave amplitudes (n = 3).
As shown in Fig. 4, WB analysis of retinal choroidal complex tissues extracted from mice revealed upregulation of proteins associated with the AIM2-PANoptosome, including AIM2, PYRIN, ZBP1, ASC, and NLRP3 (Fig. 4a). Additionally, the Aβ1-40-injected group exhibited activation of key proteins related to PANoptosis across three signaling pathways: pyroptosis (ASC, GSDMD-N, CASP-1, Cleaved CASP-1), apoptosis (FADD, BAX, BCL-2, CASP-7, CASP-3, CASP-8), and necroptosis (p-MLKL, MLKL, RIPK1, RIPK3) (Fig. 4a-c). Furthermore, qPCR analysis of the retinal choroidal complex demonstrated significant increases in mRNA levels of Aim2, Zbp1, and Mefv in the Aβ1-40-exposed group (Fig. 4d). These results indicate that the initiation of AIM2-PANoptosome and PANoptosis-like cell demise are present in the Aβ1-40-mediated AMD model.
WB and qPCR analysis of proteins and genes. (a) WB analysis of AIM2, PYRIN, ZBP1, ASC, GSDMD-N, CASP-1, Cle-CASP1 and NLRP3 protein levels in the retina-choroid complex (n = 3). (b) WB analysis of FADD, BAX, BCL-2, CASP-7, CASP-3, CASP-8 and protein levels in the retina-choroid complex (n = 3). (c) WB analysis of p-MLKL; MLKL; RIPK1; RIPK3 protein levels in retina-choroid complex (n = 3). (d) qPCR detection of Aim2, Zbp1 and Mefv in retina-choroid complex. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the Aβ1-40 group. NFL: Nerve fiber layer; INL: Inner nuclear layer; ONL: Outer nuclear layer.
Induction of PANopotosis-like cell death by Aβ1-40 was investigated in vitro
After a 24-hour exposure to Aβ1-40, microscopic examination and CCK8 assays did not detect any significant alterations in cellular morphology or a reduction in cell viability (Fig. 5a-b). However, following a 48-hour exposure to 10 µM Aβ1-40, alterations in cell shape and a substantial decline in cell viability were observed (Fig. 5b). At lower concentrations (2.5 and 5 µM), Aβ1-40 had minimally affected cell morphology and viability over 24–48 h. In contrast, higher concentrations (10 and 20 µM) induced significant morphological changes and dose-dependent cell death, with stronger effects at 48 h. Exposure to 20 µM Aβ1-40 for 48 h caused significant damage in ARPE-19 cells. Subsequent experiments were used for study using 10µM. SEM imaging revealed that Aβ1-40 exposure led to cell enlargement, cell membrane rupture, and the appearance of membrane fragments (Fig. 5c). Previous studies have confirmed that the morphological changes observed after exposure to Aβ1-40 oligomers are indicative of pyroptosis pathway21,32. Additionally, immunofluorescence results showed increased expression levels of AIM2, GSDMD-N, and CASP-1 in the Aβ1-40 group (Supplementary Fig. S2). These findings reveal the involvement of pyroptosis in Aβ-induced RPE damage. Apoptosis assay kit detection revealed that as the concentration of Aβ1-40 increased, the apoptosis rate in ARPE-19 cells progressively rose (Fig. 5d).
The effect of amyloid-beta 1-40 (Aβ1-40) on PANoptosis-associated factor expression in cells. (a) Optical microscopy was employed to examine ARPE-19 cells treated with Aβ1-40 at concentrations varying from 2.5 µM to 20 µM for durations of either 24–48 h. (b) The CCK-8 assay was applied to evaluate the viability of ARPE-19 cells after incubation periods of 24–48 h (n = 3). (c) Scanning electron microscopy of Aβ1-40-treated ARPE-19 cells (n = 3). (d) Apoptosis rates in Aβ1-40-treated ARPE-19 cells were assessed using an apoptosis assay kit (n = 3).
As shown in Fig. 6, WB analysis showed upregulation of proteins associated with the AIM2-PANoptosome, including AIM2, PYRIN, ZBP1, ASC, and NLRP3, which is consistent with our in vivo experimental results(Fig. 6a). Furthermore, WB analysis following Aβ1-40-induced RPE injury revealed the expression of proteins associated with PANoptosis-related death pathways, including apoptosis (FADD, BAX, BCL-2, CASP-7, CASP-3, CASP-8), pyroptosis (ASC, GSDMD-N, CASP-1, Cleaved CASP-1), and necroptosis (p-MLKL, MLKL, RIPK1, RIPK3) (Fig. 6a-c). These in vitro experiments further validate that RPE damage triggered by Aβ1-40 is marked by signs of cell death that are indicative of apoptosis, pyroptosis, and necroptosis occurring concurrently.
WB analysis of proteins in vitro. (a) WB analysis of AIM2, PYRIN, ZBP1, ASC, GSDMD-N, CASP-1, Cle-CASP1 and NLRP3 protein levels in vitro (n = 3). (b) WB analysis of FADD, BAX, BCL-2, CASP-7, CASP-3, CASP-8 and protein levels in vitro (n = 3). (c) WB analysis of p-MLKL; MLKL; RIPK1; RIPK3 protein levels in vitro (n = 3). Values are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar = 100 μm.
Triple RCD inhibitors shield ARPE-19 cells from Aβ1-40 damage
WB analysis showed that the expression of apoptosis-associated proteins Caspase3 and Bax was upregulated, while the expression of BCL-2 was downregulated in the Aβ1-40 group. The addition of Z-VAD-FMK led to a downregulation of Caspase3 and BAX, and an upregulation of BCL-2 expression (Fig. 7a). Aβ1-40 also elevated the levels of cleaved caspase-1 and GSDMD-N, which are pivotal in pyroptosis, an effect that was attenuated by DSF (Fig. 7b). Additionally, Aβ1-40 increased p-MLKL, a necroptosis marker, which was inhibited by necroptosis inhibitors (Fig. 7c).
When three inhibitors are used in combination, proteins related to apoptosis, pyroptosis, and necroptosis were all affected (Fig. 7d). These findings suggest that Aβ1-40 induces simultaneous activation of pyroptosis, apoptosis, and necroptosis in RPE cells, and the combined use of these three inhibitors can ameliorate Aβ1-40-induced damage in RPE cells.
Three Inhibitors of Regulated Cell Death Protect ARPE-19 Cells from Injury Induced by Aβ1-40. (a) Post-Aβ1-40 and Z-VAD treatment, WB of BAX, BCL-2, CASP-3 in ARPE-19 cells. (b) Post-Aβ1-40 and DSF treatment, Western blots of Cle-CASP1, GSDMD-N in ARPE-19 cells. (c) Post-Aβ1-40 and Nec-1 treatment, WB of p-MLKL, MLKL in ARPE-19 cells. (d) After RCD inhibitor and Aβ1-40 mix treatment, WB of BAX, caspase-3, Bcl-2, GSDMD-N, Cle-cas1, p-MLKL, MLKL in Vitro. Values are expressed as the mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 vs. Aβ1-40. DSF: disulfiram; Nec1: necrostatin-1; Z-VAD: Z-VAD-FMK.
Secretion of pro-inflammatory cytokines
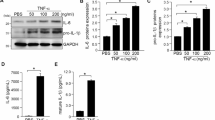
In this study, we conducted an ELISA assay kit to evaluate the inflammatory cytokine. In our analysis, Aβ1-40-treated cell supernatants showed significantly elevated concentrations of various inflammatory biomarkers, such as IL-18, IL-1β, IFN-γ, and TNF-α, compared to the control group (Fig. 8a). In vivo, Aβ1-40-treated mouse orbit serum had markedly higher inflammatory marker levels than controls (Fig. 8b). The analysis of cytokines in the plasma from peripheral blood of 12 patients with cataracts and 12 patients with AMD showed increased expression of cytokines (Fig. 8c). The study reveals the involvement of inflammation in the Aβ1-40-induced AMD model and in human patients.
Cytokine secretion in response to various sample detections. (a) ELISA quantify the levels of IL-1β,IL-18 IFN-γ and TNF-α in cell supernatant (n = 4). (b) Quantify the level in mouse orbit serum (n = 9). (c) Quantify the level of IL-1β, IL-18, IFN-γ, and TNF-α in plasma from cataract and AMD patients (n = 12). Values are expressed as mean ± SEM. ***P < 0.001.
Discussion
Numerous prior investigations have demonstrated that the activation of inflammatory processes is a pivotal factor in the pathogenesis of AMD21,33,34. β-Amyloid, the component of drusen, activates Caspase-3 and triggers apoptosis in RPE via the glycosylation end product receptor/NF-κB signaling pathway35,36. Furthermore, β-Amyloid can induce RPE cells to exhibit morphological features of pyroptosis, including swelling, blistering, and membrane rupture, along with an increase in pyroptosis phenotype indicators such as IL-18 and IL-1β. This mechanism may involve the NLRP3/Caspase-1/GSDMD-N axis, leading to pyroptosis37. Lycium barbarum polysaccharide has been shown to alleviate RPE cell dysfunction by inhibiting β-amyloid-induced pyroptosis38. In various AMD animal models, RIPK1 inhibitors have been found to protect RPE cells from necroptosis and preserve retinal visual function39. Nevertheless, the specific molecular processes that cause AMD are still not well understood. Previous studies have confirmed that the Aβ-induced AMD model can undergo single cell death pathways. Building on these previous studies, this research reveals that RNA sequencing data during the Aβ-induced AMD model show concurrent cell death through apoptosis, pyroptosis, and necroptosis. Additionally, the study demonstrates enrichment of the NOD-like receptor signaling pathway, JAK-STAT signaling pathway, natural killer cell-mediated cytotoxicity, cytokine-cytokine receptor interaction, and autoimmune thyroid disease pathways. The main genes associated with the AIM2-panoptosome show upregulated expression. Molecular docking analysis indicates potential interactions between Aβ and AIM2, as well as between AIM2, ZBP1, and PYRIN, which may be associated with the activation of PANoptosis-like cell death mechanisms in Aβ1-40-induced AMD.
Research shows the composition of the PANoptosome, with components like AIM2, ZBP-1, Pyrin, NLRP3, and caspases, is vital for PANoptosis16,40,41,42. Understanding this is essential for researching inhibitors and regulators of inflammatory cell death in infections10,11,15,43. To demonstrate the occurrence of PANoptosis, it is necessary to promote the formation of the PANoptosome. In this study, co-immunoprecipitation proved the enhanced interaction between AIM2, Pyrin, and ZBP1 in the Aβ-induced AMD model. qPCR showed a significant increase in the mRNA levels of AIM2, Pyrin, and ZBP1 in the retinal choroidal complex exposed to Aβ1-40. In our previous work24, we have confirmed that Aβ1-40 induces pyroptosis in ARPE-19 cells. Then we assessed the expression levels of various proteins associated with apoptosis (BCL-2, BAX, Caspase-3, FADD), pyroptosis (GSDMD-N, Caspase-1, Cleaved Caspase-1), and necroptosis (p-MLKL, Caspase-8, MLKL, RIPK1, RIPK3), and observed an upregulation in their expression. Combining the above findings, it appears that the activation of the AIM2 PANoptosome may occur in the Aβ1-40-induced AMD, leading to the occurrence of three distinct forms of cell death. A combination of three RCD inhibitors, disulfiram, inhibits GSDMD pores and reduces pyroptosis and IL-1β release in cells44. Z-VAD-FMK blocks Fas-mediated apoptosis45. Necrostatin-1 (Nec-1) inhibits necroptosis and has research and therapeutic potential in necroptosis-related diseases46. Aβ1-40 simultaneously induces pyroptosis, apoptosis, and necrosis in ARPE-19 cells, and the combination of three inhibitors ameliorates Aβ1-40-induced damage to ARPE-19 cells by modulating the corresponding pathway proteins. Taken together, these results indicate that Aβ1-40 exposure in ARPE-19 cells induces simultaneous activation of pyroptosis, apoptosis, and necroptosis at the protein level, suggesting that Aβ1-40 may induce a PANoptosis-like form of cell death.
Similarly, studies on RPE injury associated with retinal cell death (RCD) have reported a close relationship between the occurrence of pyroptosis and inflammatory factors38,47,48,49,50. Moreover, the principal molecules involved in apoptosis can be modulated via inflammatory responses35,51,52. Necroptosis can similarly be triggered by pro-inflammatory cytokines53,54. Previous studies have constructed a framework outlining the mechanism behind a cytokine storm, which is characterized as systemic inflammation resulting from cytokine-driven cell death of inflammatory cells, termed PANoptosis55. PANoptosis plays a significant role in the release of factors and inflammatory mediators, potentially worsening the progression of inflammatory diseases by triggering a cytokine storm. ELISA tests were performed on cell supernatant, mouse orbital serum, and human plasma. The concentrations of IL-18, IL-1β, IFN-γ, and TNF-α were all observed to increase. Besides, TNF-α and IFN-γ are associated with apoptosis, necroptosis, and pyroptosis, while IL-1β and IL-18 are closely related to the process of pyroptosis. The release of these inflammatory cytokines can promote the further development of PANoptosis-like cell death55.
Aβ1-40 is a component of drusen and has been implicated in various physiological and pathological processes in AMD. Our study shows that Aβ1-40 not only induces pyroptosis via the AIM2/Caspase-1/GSDMD-N axis24 but also activates apoptosis and necroptosis pathways. The concurrent activation of these cell death pathways by Aβ1-40 suggests a PANoptosis-like mechanism, which may exacerbate RPE cell dysfunction and contribute to AMD progression. Additionally, Aβ1-40-induced cell death is associated with the release of inflammatory cytokines such as IL-1β, IL-18, IFN-γ, and TNF-α, which can further drive PANoptosis and worsen AMD progression.
This study represents the initial exploration of AMD PANoptosis. Anticipated limitations of the experiments may impact the thoroughness and precision of the conclusions. During the collection of clinical specimens, patients with AMD were not divided into clinical types (non-exudative or exudative) or clinical stages (no AMD, early stage, intermediate stage, or late stage). In the initial phase of AMD, the formation of drusen was not precisely depicted using animal models. The study aims to conduct a phenotypic analysis of PANoptosis to identify and understand its occurrence. Upcoming experimental research should concentrate on confirming the role of the PANoptosis pathway in AMD and on discovering the crucial molecules that govern PANoptosis.
Conclusion
In summary, our study has demonstrated that following Aβ induction in AMD, three distinct cell death pathways are activated. We have unveiled the link between AIM2 PANoptosome activation and PANoptosis, thereby establishing a molecular foundation for PANoptosis within our model. Our findings validate the dual aspects of PANoptosis cascade characteristics: firstly, the engagement of AIM2-PANoptosome in Aβ-induced models, and secondly, the triggering of three distinct cell death pathways. This research represents an initial foray into PANoptosis, with the goal of delineating the cellular death features associated with it. Future experimental studies should prioritize elucidating the role of PANoptosomes in RPE damage within the retina and identifying the pivotal molecules and pathways that regulate PANoptosis.
Data availability
The raw data provided in the study can be downloaded from GEO database (https://dataview.ncbi.nlm.nih.gov/object/PRJNA1242203?reviewer=3curk3dmo84ivgmv799mldo936). For further clarification, contact the corresponding author.
References
Fleckenstein, M. et al. Age-related macular degeneration. Nat. Rev. Dis. Prim. 7, 31. https://doi.org/10.1038/s41572-021-00265-2 (2021).
Jonas, J. B., Cheung, C. M. G. & Panda-Jonas, S. Updates on the epidemiology of age-related macular degeneration. Asia-Pacific J. Ophthalmol. 6, 493–497. https://doi.org/10.22608/apo.2017251 (2017).
Wong, W. L. et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Global Health. 2, e106–e116. https://doi.org/10.1016/s2214-109x(13)70145-1 (2014).
Roos, T. T. et al. Neuronal spreading and plaque induction of intracellular Aβ and its disruption of Aβ homeostasis. Acta Neuropathol. 142, 669–687. https://doi.org/10.1007/s00401-021-02345-9 (2021).
Hogg, R. E., Stevenson, M. R., Chakravarthy, U., Beirne, R. O. & Anderson, R. S. Early features of AMD. Ophthalmology 114, 1028–1028e1023. https://doi.org/10.1016/j.ophtha.2006.11.013 (2007).
Isas, J. M. et al. Soluble and mature amyloid fibrils in Drusen deposits. Invest. Ophthalmol. Vis. Sci. 51, 1304–1310. https://doi.org/10.1167/iovs.09-4207 (2010).
Yoshida, T. The potential role of amyloid in the pathogenesis of age-related macular degeneration. J. Clin. Invest. 115, 2793–2800. https://doi.org/10.1172/jci24635 (2005).
Bhutto, I. & Lutty, G. Understanding age-related macular degeneration (AMD): Relationships between the photoreceptor/retinal pigment epithelium/bruch’s membrane/choriocapillaris complex. Mol. Aspects Med. 33, 295–317. https://doi.org/10.1016/j.mam.2012.04.005 (2012).
Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 85, 845–881. https://doi.org/10.1152/physrev.00021.2004 (2005).
Banoth, B. et al. ZBP1 promotes fungi-induced inflammasome activation and pyroptosis, apoptosis, and necroptosis (PANoptosis). J. Biol. Chem. 295, 18276–18283. https://doi.org/10.1074/jbc.RA120.015924 (2020).
Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol 1, aag2045 (2016) https://doi.org/10.1126/sciimmunol.aag2045
Boada-Romero, E., Martinez, J., Heckmann, B. L. & Green, D. R. The clearance of dead cells by efferocytosis. Nat. Rev. Mol. Cell. Biol. 21, 398–414. https://doi.org/10.1038/s41580-020-0232-1 (2020).
Koren, E. & Fuchs, Y. Modes of regulated cell death in cancer. Cancer Discov. 11, 245–265. https://doi.org/10.1158/2159-8290.Cd-20-0789 (2021).
Man, S. M., Karki, R. & Kanneganti, T. D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 277, 61–75. https://doi.org/10.1111/imr.12534 (2017).
Malireddi, R. K. S., Kesavardhana, S. & Kanneganti, T. D. ZBP1 and TAK1: Master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis). Front. Cell. Infect. Microbiol. 9, 406. https://doi.org/10.3389/fcimb.2019.00406 (2019).
Christgen, S. et al. Identification of the PANoptosome: A molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 237. https://doi.org/10.3389/fcimb.2020.00237 (2020).
Wan, H. et al. Voltage-dependent anion channel 1 oligomerization regulates PANoptosis in retinal ischemia–reperfusion injury. Neural Regen. Res. https://doi.org/10.4103/NRR (2025). NRR-D-4124-00674.
Lan, Z. et al. Curcumin-primed olfactory mucosa-derived mesenchymal stem cells mitigate cerebral ischemia/reperfusion injury-induced neuronal PANoptosis by modulating microglial polarization. Phytomedicine 129, 155635. https://doi.org/10.1016/j.phymed.2024.155635 (2024).
Wang, S. et al. Noncanonical feedback loop between RIP3–MLKL and 4EBP1–eIF4E promotes neuronal necroptosis. MedComm (2020) 6, e70107. https://doi.org/10.1002/mco2.70107 (2025).
Hanus, J., Anderson, C. & Wang, S. S. RPE necroptosis in response to oxidative stress and in AMD. Ageing Res. Rev. 24, 286–298. https://doi.org/10.1016/j.arr.2015.09.002 (2015).
Tarallo, V. et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149, 847–859. https://doi.org/10.1016/j.cell.2012.03.036 (2012).
Kaneko, H. et al. DICER1 deficit induces RNA toxicity in age-related macular degeneration. Nature 471, 325. https://doi.org/10.1038/nature09830 (2011).
Chen, J. et al. MicroRNA-191‐5p ameliorates amyloid‐β1‐40–mediated retinal pigment epithelium cell injury by suppressing the NLRP3 inflammasome pathway. FASEB J. 35, e21184. https://doi.org/10.1096/fj.202000645RR (2021).
Lu, J. et al. Atorvastatin alleviates age-related macular degeneration via AIM2-regulated pyroptosis. Inflammation https://doi.org/10.1007/s10753-10024-02179-z (2024).
Chen, J. et al. MicroRNA-191-5p ameliorates amyloid-β1-40 -mediated retinal pigment epithelium cell injury by suppressing the NLRP3 inflammasome pathway. FASEB J. 35, e21184. https://doi.org/10.1096/fj.202000645RR (2021).
Yu, G. Thirteen years of clusterprofiler. Innov. 5, 100722. https://doi.org/10.1016/j.xinn.2024.100722 (2024).
Honorato, R. V. et al. The HADDOCK2.4 web server for integrative modeling of biomolecular com plexes. Nat. Protoc. https://doi.org/10.1038/s41596-41024-01011-41590 (2024).
Song, F. et al. PANoptosis-based molecular subtyping and HPAN-index predicts therapeut Ic response and survival in hepatocellular carcinoma. Front. Immunol. 14, 1197152. https://doi.org/10.3389/fimmu.2023.1197152 (2023).
Kanehisa, M. K. E. G. G. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Lee, S. et al. AIM2 forms a complex with Pyrin and ZBP1 to drive PANoptosis and host defence. Nature 597, 415–419. https://doi.org/10.1038/s41586-021-03875-8 (2021).
Hood, D. C. & Birch, D. G. Assessing abnormal rod photoreceptor activity with the a-wave of the electroretinogram: Applications and methods. Doc. Ophthalmol. 92, 253–267. https://doi.org/10.1007/bf02584080 (1996).
Samuels, I. S., Bell, B. A., Pereira, A., Saxon, J. & Peachey, N. S. Early retinal pigment epithelium dysfunction is concomitant with hyper glycemia in mouse models of type 1 and type 2 diabetes. J. Neurophysiol. 113, 1085–1099. https://doi.org/10.1152/jn.00761.2014 (2015).
Kauppinen, A., Paterno, J. J., Blasiak, J., Salminen, A. & Kaarniranta, K. Inflammation and its role in age-related macular degeneration. Cell. Mol. Life Sci. 73, 1765–1786. https://doi.org/10.1007/s00018-016-2147-8 (2016).
Zhao, T. et al. Age-related increases in amyloid beta and membrane attack complex: evi dence of inflammasome activation in the rodent eye. J. Neuroinflam. 12, 121. https://doi.org/10.1186/s12974-015-0337-1 (2015).
Ye, Z., He, S. Z. & Li, Z. H. Effect of Aβ protein on inhibiting proliferation and promoting apoptosis of retinal pigment epithelial cells. Int. J. Ophthalmol. 11, 929–934. https://doi.org/10.18240/ijo.2018.06.06 (2018).
Liu, R. Z. T. et al. Vinpocetine inhibits amyloid-beta induced activation of NF-κB, NLRP3 inflammasome and cytokine production in retinal pigment epithelial cells. Exp. Eye Res. 127, 49–58. https://doi.org/10.1016/j.exer.2014.07.003 (2014).
Liu, X. et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158. https://doi.org/10.1038/nature18629 (2016).
Yang, M., So, K. F., Lo, A. C. Y. & Lam, W. C. The effect of Lycium barbarum polysaccharides on pyroptosis-associated amyloid β1-40 oligomers-induced adult retinal pigment epithelium 19 cell damage. Int. J. Mol. Sci. 21, 4658. https://doi.org/10.3390/ijms21134658 (2020).
Jang, K. H. et al. Protective effect of RIPK1-inhibitory compound in in vivo models for retinal degenerative disease. Exp. Eye Res. 180, 8–17. https://doi.org/10.1016/j.exer.2018.11.026 (2019).
Samir, P., Malireddi, R. K. S. & Kanneganti, T. D. The PANoptosome: A deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10, 238. https://doi.org/10.3389/fcimb.2020.00238 (2020).
Wang, Y. & Kanneganti, T. D. From pyroptosis, apoptosis and necroptosis to PANoptosis: A mechanistic compendium of programmed cell death pathways. Comput. Struct. Biotechnol. J. 19, 4641–4657. https://doi.org/10.1016/j.csbj.2021.07.038 (2021).
Chowdhary, A., Briard, B., Malireddi, R. K. S. & Kanneganti, T. D. Role of inflammasomes/pyroptosis and PANoptosis during fungal infection. PLoS Pathog. 17, e1009358. https://doi.org/10.1371/journal.ppat.1009358 (2021).
Liu, Y. et al. Pyroptosis in health and disease: Mechanisms, regulation and clinical perspective. Signal. Transduct. Target. Ther. 9, 245. https://doi.org/10.1038/s41392-024-01958-2 (2024).
Hu, J. J. et al. FDA-approved Disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat. Immunol. 21, 736–745. https://doi.org/10.1038/s41590-020-0669-6 (2020).
Slee, E. A. et al. Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem. J. 315, 21–24. https://doi.org/10.1042/bj3150021 (1996).
Cao, L. & Mu, W. Necrostatin-1 and necroptosis Inhibition: Pathophysiology and therapeutic implications. Pharmacol. Res. 163, 105297. https://doi.org/10.1016/j.phrs.2020.105297 (2021).
Varinthra, P., Huang, S. P., Chompoopong, S., Wen, Z. H. & Liu, I. Y. 4-(Phenylsulfanyl) Butan-2-one attenuates the inflammatory response in duced by amyloid-β oligomers in retinal pigment epithelium cells. Mar. Drugs 19, 1. https://doi.org/10.3390/md19010001 (2020).
Sun, H. J., Jin, X. M., Xu, J. & Xiao, Q. Baicalin alleviates age-related macular degeneration via miR-223/NLRP3-regulated pyroptosis Pharmacology 105, 28–38 https://doi.org/10.1159/000502614 (2020).
Kaarniranta, K., Tokarz, P., Koskela, A., Paterno, J. & Blasiak, J. Autophagy regulates death of retinal pigment epithelium cells in age-related macular degeneration. Cell. Biol. Toxicol. 33, 113–128. https://doi.org/10.1007/s10565-016-9371-8 (2016).
Sekar, R. et al. Impairing gasdermin D-mediated pyroptosis is protective against retinal degeneration. J. Neuroinflam. 20, 239. https://doi.org/10.1186/s12974-023-02927-2 (2023).
Masuda, N. et al. Effects of concentration of amyloid Β (Aβ) on viability of cultured re Tinal pigment epithelial cells. BMC Ophthalmol. 19, 70. https://doi.org/10.1186/s12886-019-1076-3 (2019).
Yang, P. et al. Caspase-8-mediated apoptosis in human RPE cells. Invest. Opthalmol. Vis. Sci. 48, 3341–3349. https://doi.org/10.1167/iovs.06-1340 (2007).
Duprez, L. et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 35, 908–918. https://doi.org/10.1016/j.immuni.2011.09.020 (2011).
Kaczmarek, A., Vandenabeele, P., Krysko, D. V. & Necroptosis The release of damage-associated molecular patterns and its physiological relevance. Immunity 38, 209–223. https://doi.org/10.1016/j.immuni.2013.02.003 (2013).
Karki, R. et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184, 149–. https://doi.org/10.1016/j.cell.2020.11.025 (2021).
Funding
This research was funded by the National Natural Science Foundation of China (Grant No.: 81670881) and the Natural Science Foundation of Chongqing, China (Grant No.: CSTB2024NSCQ-MSX0705).
Author information
Authors and Affiliations
Contributions
Conceptualization, Yuxia He; Data curation, Yuxia He, Jing Lu, Yong Du, Long Zhao, Lili Gong, Ping Wu, Qinxin Shu; Funding acquisition, Hui Peng; Visualization, Yuxia He; Investigation, Yuxia He, Jing Lu; Writing – original draft, Yuxia He; Writing – review & editing, Xing Wang, Hui Peng. All authors who contributed to the research have examined and agreed to the final version of the manuscript before it is released for publication. Yuxia He and Jing Lu contributed equally to this study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
This study was approved by the ethical standards set by the First Affiliated Hospital of Chongqing Medical University and the Institutional Animal Care and Use Committee at Chongqing Medical University. All experiments were conducted in line with relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, Y., Lu, J., Du, Y. et al. Investigation of PANoptosis pathway in age-related macular degeneration triggered by Aβ1-40. Sci Rep 15, 13514 (2025). https://doi.org/10.1038/s41598-025-98174-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98174-x