Abstract
Myc-associated Zinc-finger Protein (MAZ) has been implicated in the malignant progression of various tumors. However, its expression and functional relationship of MAZ in melanoma have not been previously investigated. This study confirms elevated expression of MAZ in melanoma, correlating with poor patient prognosis. Furthermore, our findings demonstrate that MAZ enhances melanoma progression by promoting proliferation, migration and invasion. It is worth noting that we found that MAZ can target and regulate the transcription of NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 (NDUFS3), a core subunit of mitochondrial complex I, to enhance mitochondrial metabolism and thus promote malignant progression of melanoma. Predictive modeling indicates that the co-expression of MAZ and NDUFS3 could serve as a potential prognostic marker for melanoma patients.
Similar content being viewed by others
Introduction
Malignant melanoma (MM) arises from the gradual accumulation of genetic mutations in melanocytes, disrupting cell proliferation, differentiation, and apoptosis, primarily manifesting in the skin1. MM represents 90% of skin cancer related deaths2 and exhibits a high propensity for early metastasis3, emphasizing the critical need for in-depth investigations into its molecular mechanisms. Understanding these mechanisms is pivotal for developing new treatments to improve melanoma patient survival. Transcription factors (TFs) play pivotal roles in regulating material metabolic pathways during tumor development, migration, and invasion by modulating gene expression4. Zinc finger proteins (ZFPs) constitute the largest family of transcription factor in the human genome5, exerting influence on the development and metastasis of malignant tumors by directly recognizing or binding to specific DNA sequences and regulating gene transcription and translation processes6,7,8. Myc-associated zinc finger protein (MAZ) is frequently overexpressed in various malignant tumors and is associated with increased patient mortality9.
In humans, the MAZ gene resides on autosomal chromosome 16p11.2, comprising a promoter, five exons, four introns, and the 3’-UTR. Its transcription product of MAZ yields a 2.7 kb mRNA encoding a 60 kDa MAZ protein consisting of 477 amino acids10,11. Functionally, MAZ acts transcriptionally by binding to GC-rich portions within the promoter regions of target genes through its six C2H2-type zinc fingers located at the carboxyl terminus12,13. MAZ controls gene transcription initiation and termination. MAZ regulates binding to promoters at the initiation of DNA transcription14, and regulates RNA polymerase II activity during transcription elongation15. MAZ also controls selective splicing14,16 and activation of transcription termination induced by polyadenylation17.
The regulatory role of MAZ in various malignant tumors has been extensively reported. For instance, MAZ targets and binds to the promoter region of mitogen-activated protein kinase kinase 2 (MAP2K2), activating its transcription and promoting the growth of clear cell renal cell carcinomas (ccRCC)18. MAZ can also be recruited by G-quadruplexes (G4s) to the promoter of Cyclin D1 (CCND1), forming a MAZ/CCND1-G4 condenser, thereby enhancing the transcription of CCND1 and facilitating the proliferation of hepatocellular carcinoma cells (HCC)15. Similar regulatory roles of MAZ have been reported in triple-negative breast cancer (TNBC) and thyroid cancer (THCA)19,20. Moreover, MAZ not only directly regulates target gene transcription but also interacts with other TFs. For example, MAZ promotes colon cancer (CRC) growth in vitro and in vivo by activating the transcription factor signal transducer and activator of transcription 3 (STAT3) and maintaining high STAT3 signaling activity21. Notably, MAZ can promote malignant tumor progression by regulating key molecules in metabolic pathways. By binding to the promoter of nei-like DNA glycosylase 3 (NEIL3), MAZ promotes NEIL3 transcription, leading to increased extracellular acidification, glucose uptake, and lactate production levels, and accelerated angiogenesis in HCC22. However, whether MAZ may have a regulatory function in the malignant progression of melanoma remains unclear.
In addition, metabolic heterogeneity can significantly affect the metastatic potential of melanoma23, with its metabolic state being mitochondria-dependent24. Therefore, targeting mitochondrial therapy represents a new therapeutic option for melanoma25. NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 (NDUFS3), a molecular weight 30 kDa protein encoded by a nuclear gene, serves as a core subunit of Mitochondrial complex I (MCI)26,27. Dysregulation of NDUFS3 expression or activity results in mitochondrial structural28 and functional abnormalities29, affecting the assembly of MCI30 and the production of reactive oxygen species (ROS)31. NDUFS3 can be used as a target for MCI inhibitors, and its inhibition leads to MCI component dissolution and proliferation blockade in ovarian and colorectal cancer cells32. Decreased NDUFS3 expression correlates with mitochondrial damage and abnormal oxidative metabolism in breast cancer33, serving as a biomarker for invasive breast cancer34. Decreased expression of NDUFS3 in renal cancer35 and human plasma ovarian adenocarcinoma36 leads to poor prognosis of patients. On the contrary, elevated NDUFS3 expression in gastric cancer37 and colorectal cancer38 potentially promotes tumor infiltration and metastasis, suggesting its therapeutic targeting potential37. The above studies show the tissue specificity of NDUFS3 expression in tumor tissues, suggesting the diversity of its effects and the complexity of its mechanism. However, no literature has yet reported the role of NDUFS3 in melanoma development.
To date, no studies have investigated the correlation between MAZ and NDUFS3. In this work, we showed that the expression of NDUFS3 was significantly upregulated in melanoma, which was associated with higher levels of MAZ. Melanoma development and metastasis are boosted by highly expressed MAZ. Furthermore, we identified MAZ as an upstream transcription factor regulating NDUFS3 expression. MAZ enhances mitochondrial metabolism and promotes melanoma cells proliferation and metastasis by binding to the NDUFS3 promoter region and activating NDUFS3 transcription. And the co-expression of MAZ and NDUFS3 can provide a reference for the prognostic diagnosis of clinical melanoma patients. Our findings provide new insights and experimental foundations for targeted intervention strategies against MAZ in melanoma patients.
Results
MAZ expression is elevated in melanoma and regulates mitochondrial function
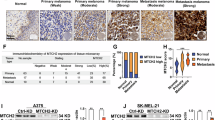
To explore the MAZ expression in melanoma, we analyzed mRNA expression of melanoma tissue and normal tissue samples differentially using the cancer genome atlas (TCGA, https://portal.gdc.cancer.gov/) database and gene expression omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database. The results showed elevated expression of MAZ in melanoma (Fig. 1a, b). We then examined MAZ protein and mRNA expression in the melanoma cell lines A375, A875, MeWo, SK-MEL-21, SK-MEL-28 and human epidermal melanocytes (HEM) by Western Blot and RT-qPCR, respectively. The results showed that MAZ was expressed at elevated protein and transcript levels within melanoma cell lines (Fig. 1c, d). The above experimental results indicate increased MAZ expression in melanoma cells.
a, b Analysis of MAZ expression in normal and melanoma tissues based on TCGA and GEO databases. The student’s t-test was used to analyse the differences in MAZ expression in normal and melanoma tissues. (**p < 2e-3, *p < 0.05). c, d Protein and mRNA expression of MAZ in HEM cells and melanoma cells A375, A875, MeWo, SK-MEL-21, SK-MEL-28 were analyzed by Western Blot (c) and RT-qPCR (d). The experimental data were normalized. Student’s t-test was used to analyse the differences in MAZ expression in HEM cells and melanoma cells. (mean ± SEM, n = 3). (****p < 1e-4, ***p < 2e-4, **p < 2e-3, *p < 0.05). e Prognostic survival analysis of MAZ high/low expression groups. (Shaded areas represent 95% confidence intervals for survival). f Time dependent ROC assessment of time prediction models. g Heat maps of different MAZ expressions corresponding to different pathologic features.
Next, to investigate the biological function of MAZ in melanoma progression, this study screened the most relevant genes for MAZ by spearman correlation analysis and performed gene ontology (GO) analysis based on the above genes. The findings suggest the composition and function of mitochondria and ribosomes were strongly interrelated, and MAZ primarily involved in DNA replication, transcription, and translation processes, and regulation of melanoma mitochondria-related gene expression (Supplementary Fig. 1a). Further analysis of differentially expressed genes between MAZ high- and low-expression groups (Supplementary Fig. 1b), along with gene set enrichment analysis (GSEA), indicated functional associations of MAZ with mitochondrial complexes and regulation of oxidative phosphorylation and assembly of respiratory chain complexes in mitochondria (Supplementary Fig. 1c).
High MAZ expression indicates poor prognosis in melanoma patients
To investigate the relationship between MAZ expression and survival prognosis for patients with melanoma, we performed a prognostic analysis comparing MAZ expression with the melanoma patients’ survival from the TCGA database and showed that elevated MAZ expression correlates with poor patient prognosis (Fig. 1e). Time-dependent receiver operating characteristic curve (ROC) analysis demonstrated predictive accuracy of MAZ expression for 1-, 3-, and 5-year survivals, with an area under curve (AUC) exceeding 0.5, indicating diagnostic significance (Fig. 1f). In conclusion, MAZ emerges as a risk gene and a robust predictor of survival status in melanoma patients.
Univariate COX regression analysis identified several pathologic characteristics as prognostic factors in melanoma patients, including high Clark grading, Age > 60, and advanced T and N stages (Supplementary Fig. 2a). Risk factors were then analyzed for prognostic survival, and the outcomes demonstrated that melanoma patients with high Clark grading and T staging, Age > 60, TNM staging III-IV, and staging N1-N3 would have a poorer prognosis (Supplementary Fig. 2b). And patients with different MAZ expression levels showed different clinical and pathological expression characteristics (Fig. 1g), showing significant differences in age and TNM staging, and a positive correlation with T staging malignancy in older patients (Supplementary Fig. 3a). And patients with higher MAZ expression levels experienced worse prognosis during the same period of time (Supplementary Fig. 3b), validating the diagnostic significance of all three predictive models (Supplementary Fig. 3c).
Next, we stained the tissue microarrays using immunohistochemical techniques to detect MAZ gene expression in paracancerous and melanoma tissues. The tissue microarrays contained 80 cases of paraneoplastic tissue, 66 cases of primary melanoma, and 14 patients with metastatic melanoma. Representative images of HE staining and immunohistochemical staining were examined (Fig. 2a). The positivity rates of MAZ in normal tissue, primary melanoma, and metastatic melanoma were 11.25% (9/80), 33.33% (22/66), and 57.14% (8/14), respectively (Fig. 2b). Moreover, the MAZ score in melanoma tissues was higher than that of paracancerous tissues (Fig. 2c). Subsequently, we counted the rate of MAZ positivity in melanoma tissues according to TNM, N and T stages (Fig. 2d). Notably, MAZ expression significantly varied among TNM, N and T stages (Fig. 2e). Elevated MAZ expression was associated with malignant pathologic progression of melanoma, consistent with previous bioinformatics analysis.
a HE staining and immunohistochemical morphological features of normal and melanoma tissues, scale bars: 50 μm. b Percentage of MAZ negative and positive tumors in normal tissue, primary melanoma, and metastatic melanoma, scale bars: 50 μm. c MAZ scores in normal tissue, primary melanoma and metastatic melanoma. d Number of negative and positive MAZ expressions in different pathological stages. e Differences in MAZ scores at different pathological stages. T stage groupings was analyzed using one-way anova. The grouping of TNM stage and N stage were analyzed by student’s t-test.
MAZ promotes melanoma cell proliferation
To investigate the influence of MAZ expression on melanoma cell function, we generated stable melanoma cell lines with MAZ overexpressing and knockdown using lentivirus-infected in A875 and SK-MEL-21 cells. Protein and transcript levels of MAZ in these cell lines were detected by Western Blot and RT-qPCR (Fig. 3a, b), respectively. Synthesis of MAZ expression in protein level and transcriptional level in both cell lines was analyzed, knockdown sequence 1 (KD1) was finally selected as the MAZ knockdown cell line.
a, b Stable MAZ overexpression and knockdown were established in A875 and SK-MEL-21 cells. Western Blot (a) and RT-qPCR (b) were used to detect MAZ expression. c GSEA analysis of MAZ positive regulation of cell proliferation. d–f Proliferation rates of MAZ overexpressing or knockdown A875 and SK-MEL-21 cells and corresponding control cells were detected using MTS assay (d), EdU assay (e) and plate cloning assay (f), scale bars: 100 μm. The experimental data were normalized. The coordinates on the Y-axis indicate the multiplicity of MAZ-OE and MAZ-KD proliferation relative to control. Student’s t-test was used to analyse the experimental results of the MAZ-OE and MAZ-KD groups with their control groups. (mean ± SEM, n = 3). (****p < 1e-4, ***p < 2e-4, **p < 2e-3, *p < 0.05, Ns means that the results are not statistically significant).
The relationship between MAZ and cell proliferation was predicted by GSEA in this study, and it was revealed that MAZ positively regulated cell proliferation (Fig. 3c). The MTS assay, EdU assay, and plate cloning assay verified the influence of MAZ on melanoma cell proliferation. MTS assay results showed that MAZ overexpression in A875 and SK-MEL-21 cell lines promoted cell proliferation compared to control cells (Fig. 3d). EdU and plate cloning experiments showed that MAZ overexpression increased the proportion of EdU-positive cells and the number of colony-forming cells, while knockdown of MAZ led to a significant reduction in both parameters (Fig. 3e, f). These findings collectively support the role of MAZ in promoting melanoma cell proliferation and tumor progression.
MAZ promotes melanoma cell proliferation by increasing S-phase and G2/M-phase cell numbers
To explore further how MAZ regulates the proliferation process of melanoma cells, GSEA analysis was utilized to predict the effect of MAZ on the cell cycle, and the findings indicated that MAZ had a positive regulatory effect on the cell cycle. MAZ might promote melanoma cell proliferation by enhancing S phase and G2/M phase cell counts (Fig. 4a). To validate this prediction, we examined changes in cell cycle distribution in A875 and SK-MEL-21 cell lines following MAZ overexpression/knockdown by flow cytometry. Results have shown that cells overexpressing MAZ exhibited reduced G0/G1-phase cell numbers and increased S-phase and G2/M-phase cell numbers compared to controls, while knockdown of MAZ yielded opposite effects (Fig. 4b, Supplementary Fig. 4).
a GSEA analyses MAZ in relation to the cell cycle. b Flow cytometry analysis of the effect of MAZ on melanoma cell cycle phase distribution. c Correlation of MAZ with cell cycle regulators analyzed by GEO and TCGA databases. d Western Blot detection of cell cycle regulatory factors in MAZ overexpression/knockdown cells at protein levels. The experimental data were normalized. Student’s t-test was used to analyse the experimental results of the MAZ-OE and MAZ-KD groups with their control groups. (mean ± SEM, n = 3). (***p < 2e-4, **p < 2e-3, *p < 0.05).
To elucidate the underlying mechanisms by which MAZ regulates the cell cycle, correlation analysis between MAZ and cell cycle regulators was performed using TCGA and GEO databases. Results suggested that MAZ may up-regulate G0/G1 phase regulatory factors CDK2, CDK4, and CCNE1 (Cyclin E1); S phase regulatory factors MYC (c-myc), MCM3, MCM4, and MCM5, as well as G2/M phase regulatory proteins CCNB1 (Cyclin B1) and CCNB2 (Cyclin B2) (Fig. 4c). We chose the S-phase regulator c-myc and the G2/M-phase regulator Cyclin B1 for validation, and Western Blot showed that c-myc and Cyclin B1 protein expression levels were elevated in cell lines overexpressing MAZ, whereas knockdown of MAZ decreased their expression levels (Fig. 4d). In summary, MAZ reduces the G0/G1 phase of cells and increases S and G2/M phase cells by up-regulating the expression of the cell cycle factors c-myc and Cyclin B1, thereby positively regulating the cell cycle.
MAZ overexpression enhances migration and invasion of melanoma cells
To investigate the potential regulatory role of MAZ in melanoma distal metastasis, we predicted its association with metastatic potential using GSEA analysis, which suggested a positive regulation of cell proliferation by MAZ (Fig. 5a). Meanwhile, the results of wound healing assay and transwell assay showed that in both A875 and SK-MEL-21 cell lines, cells overexpressing MAZ had higher migration and invasion rates than controls; on the contrary, knockdown of MAZ resulted in cells with significantly lower migration and invasion rates compared to controls (Fig. 5b–d).
a GSEA analysis of MAZ positively regulates cell migration. b Scratch assay to detect the migration rate of MAZ overexpressing and knockdown cells in 0 h and 24 h melanoma A875 and SK-MEL-21 cells, scale bars: 100 μm. c, d Transwell migration (c) and invasion (d) assays were performed to detect changes in cell migration and invasion rates after MAZ overexpression or knockdown, scale bars: 50 μm. The experimental data were normalized. The coordinates on the Y-axis indicate the multiplicity of MAZ-OE and MAZ-KD migration and invasion relative to control. e Correlation of MAZ with migratory invasion regulators analyzed in GEO and TCGA databases. f Western Blot detection of MAZ overexpression or knockdown of protein level expression of migration and invasion regulators in cell lines. The experimental data were normalized. Student’s t-test was used to analyse the experimental results of the MAZ-OE and MAZ-KD groups with their control groups. (mean ± SEM, n = 3). (****p < 1e-4, ***p < 2e-4, **p < 2e-3, *p < 0.05).
Correlation analyses between MAZ and regulators of migration and invasion were conducted, revealing potential up-regulation of MMP14, MMP15, and ADAM15 in the extracellular matrix pathway, as well as VEGF and SNAI2 (Snail2), and downregulation of TP63 (P63) (Fig. 5e). We detected protein expression of MMP14 and ADAM15 in MAZ overexpression and knockdown cell lines by Western Blot. As shown, MAZ promoted the expression of MMP14 and ADAM15 in melanoma cells (Fig. 5f). These findings suggest that MAZ may promote melanoma cell migration and invasion by enhancing the expression of MMP14 and ADAM15.
NDUFS3 is highly expressed in melanoma and closely correlates with MAZ expression
We initially confirmed a positive correlation between MAZ and NDUFS3 expression in melanoma using two datasets from GEO database (Fig. 6a). Subsequently, we observed elevated expression of NDUFS3 in melanoma compared to normal skin and nevi (Fig. 6b), as validated by Western Blot analysis showing higher levels of NDUFS3 protein in melanoma cells lines A375, A875, MeWo, SK-MEL-21 and SK-MEL-28 compared to melanocyte cell line HEM (Fig. 6c). We then predicted the effect of high levels of NDUFS3 expression in survival status of melanoma patients, and the results suggested that high expression of NDUFS3 led to poor prognosis of patients (Fig. 6d).
a GEO database analysis of MAZ correlation with NDUFS3. b GEO database analysis of NDUFS3 expression in skin, nevi, dysplastic nevi and melanoma. Differential expression of NDUFS3 between different groups was analyzed by one-way anova. c Western Blot was performed to detect the protein level expression of NDUFS3 in HEM cells and melanoma cells A375, A875, MeWo, SK-MEL-21, SK-MEL-28. The experimental data were normalized. Student’s t-test was used to analyse the differences in NDUFS3 expression in melanocyte lines and melanoma cell lines. (mean ± SEM, n = 3). (**p < 2e-3, *p < 0.05). d Survival analysis of the NDUFS3 high and low expression groups. (Shaded areas represent 95% confidence intervals for survival). e Analysis of MAZ and NDUFS3 promoter region binding sites based on JASPAR database. f ChIP assay was used to detect MAZ in the NDUFS3 promoter region in A875 and SK-MEL-21 cells. Student’s t-test was used to analyse the experimental results of the MAZ and IgG group. (mean ± SEM, n = 3). (***p < 2e-4, ****p < 1e-4). g Dual-luciferase reporter assays to validate the binding of MAZ to the NDUFS3 promoter region. Student’s t-test was used to analyse the experimental results of the MAZ and vector groups. (mean ± SEM, n = 4). (****p < 1e-4, Ns means that the results are not statistically significant). h, i Western Blot (h) and RT-qPCR (i) were used to detect NDUFS3 protein level and transcript level expression in MAZ overexpression or knockdown cell lines. The experimental data were normalized. Student’s t-test was used to analyse the experimental results of the MAZ-OE and MAZ-KD groups with their control groups. (mean ± SEM, n = 3). (****p < 1e-4, ***p < 2e-4, **p < 2e-3).
KEGG/GO analysis predicted the functional role of NDUFS3 in melanoma, revealing its involvement in mitochondrial function, assembly of mitochondrial complex I, and various metabolic processes in vivo such as oxidative phosphorylation. NDUFS3 was also found to be associated with mitochondrial gene expression and positively correlated with cell cycle regulation and transcriptional regulatory processes, suggesting its potential role in promoting melanoma proliferation and transcriptional regulation (Supplementary Fig. 5a, b) (Supplementary Fig. 6a, b).
These experimental results suggest that NDUFS3 expression is elevated in melanoma and may serve as a key molecule in the regulation of multiple metabolic pathways, potentially contributing to poor patient prognosis.
MAZ targeted upregulation of NDUFS3 transcription promotes malignant phenotype in melanoma cells
The mechanisms underlying the high expression of NDUFS3 in melanoma have not been elucidated in previous studies. The outcomes of our enrichment analyses revealed associations of NDUFS3 with transcriptional processes and assembly of mitochondrial respiratory chain complex I in melanoma. This is consistent with the function of MAZ and the positive correlation between MAZ and NDUFS3 expression in melanoma; therefore, we hypothesized MAZ as an upstream transcription factor regulating NDUFS3 expression.
We initially searched the NDUFS3 gene promoter sequences through national center of biotechnology information (NCBI, https://www.ncbi.nlm.nih.gov/) and university of california santa cruz (UCSC, https://genome.ucsc.edu/) genomic browser database and predicted that MAZ could bind to the NDUFS3 promoter region on the JASPAR database (https://jaspar.elixir.no/) (Fig. 6e). To make clear whether MAZ can be targeted to bind in the NDUFS3 promoter region, this study designed primers based on MAZ and NDUFS3 promoter binding sites analyzed by the JASPAR database and performed ChIP-qPCR experiments and agarose gel electrophoresis assays. The results of the ChIP-qPCR experiments showed that, in A875 and SK-MEL-21 melanoma cells, the expression of MAZ in the NDUFS3 promoter region at the binding site 1 (1147-1157) and binding site 2 (1735-1745) were increased, respectively (Fig. 6f, Supplementary Fig. 7). In addition, the results of the dual luciferase assay showed that the fluorescence activity of MAZ was reduced after we mutated the binding site of MAZ to the NDUFS3 promoter region (Fig. 6g). Then, this study was applied to detect NDUFS3 protein level and mRNA level in A875 and SK-MEL-21 cells after MAZ overexpression/knockdown by Western Blot and RT-qPCR. Compared with controls, both protein and transcript levels of NDUFS3 were upregulated in MAZ overexpressed A875 and SK-MEL-21 cell lines, whereas knockdown of MAZ leads to decreased levels of NDUFS3 protein and transcripts (Fig. 6h, i). These results indicate that MAZ may be an upstream transcription factor of NDUFS3.
To assess whether MAZ affects melanoma malignant phenotype via NDUFS3 regulation, we transfected MAZ knockdown cell lines A875 and SK-MEL-21 with NDUFS3 overexpression plasmid. Western Blot analysis confirmed increased NDUFS3 protein levels in these cells (Fig. 7a). Functional rescues experiments demonstrated that NDUFS3 overexpression reversed the reduce proliferation and migration capacity induced by MAZ knockdown (Fig. 7b–d). Meanwhile, the expression of Cyclin B1, c-myc, ADAM15 and MMP14 was rebounded after NDUFS3 overexpression (Fig. 7e).
a Western Blot detection of NDUFS3 expression in MAZ knockdown cell lines after transfection with NDUFS3 overexpression plasmid. b, c MTS (b) assay and plate cloning (c) assay were used to detect the effect of NDUFS3 overexpression on cell proliferation in MAZ knockdown cell lines. d Transwell migration assay was used to detect the effect of NDUFS3 overexpression on cell migration in MAZ knockdown cell lines, scale bars: 50 μm. e Western Blot detected the protein expression levels of Cyclin B1, c-myc, ADAM15 and MMP14 after NDUFS3 overexpression. The experimental data were normalized. Student’s t-test was used to analyze the experimental results of the MAZ-NC and MAZ-KD groups, the MAZ-KD-NDUFS3-NC group and the MAZ-KD-NDUFS3-OE group. (mean ± SEM, n = 3). (* represents statistical significance between MAZ-NC and MAZ-KD groups. ****p < 1e-4, ***p < 2e-4, **p < 2e-3; # represents statistical significance between MAZ-KD-NDUFS3-NC and MAZ-KD-NDUFS3-OE groups. ####p < 1e-4, ###p < 2e-4, ##p < 2e-3, #p < 0.05).
These findings indicate that MAZ can target the NDUFS3 promoter region, promoting its transcription and upregulating NDUFS3 expression, thereby promoting the malignant progression of melanoma.
MAZ promotes melanoma growth in vivo
We injected the MAZ over-expressed/knocked down stable A875 melanoma cell line as well as its control cells into the axilla of BALB/c nude mice to explore further whether MAZ promotes melanoma growth in vivo and constructed a subcutaneous implantation tumor model in nude mice. As shown, the tumor volume in MAZ overexpression group was significantly larger than in controls, whereas knockdown of MAZ significantly reduced the melanoma formation ability (Fig. 8a, b). It is also worth noting that the growth rate as well as the weight of the tumor was higher after MAZ overexpression than controls, whereas after knockdown of MAZ, tumor showed reduced growth (Fig. 8c, d).
a, b BALB/c nude mice injected with stably transfected MAZ overexpressing/knockdown and control A875 cells. c Mice tumor volume growth curves were plotted. d Measurement of mice tumor weight. Student’s t-test was used to analyse the experimental results of the MAZ-OE and MAZ-KD groups with their control groups. (mean ± SD, n = 4). (**p < 2e-3, *p < 0.05) e, f Western Blot (e) and RT-qPCR (f) were used to detect the protein expression level and mRNA level expression of MAZ, NDUFS3 in tumor tissues of each group. g The protein expression levels of c-myc and Cyclin B1 were expressed in the tumor tissues of each group using Western Blot. The experimental data were normalized. Student’s t-test was used to analyse the experimental results of the MAZ-OE and MAZ-KD groups with their control groups. (mean ± SEM, n = 3). (****p < 1e-4, ***p < 2e-4, **p < 2e-3, *p < 0.05).
In this work we analyzed MAZ and NDUFS3 protein and mRNA expression in tumor tissues using Western Blot and RT-qPCR, which showed a positive correlation between tumor volume and the expression of MAZ and NDUFS3 (Fig. 8e, f). We further examined the expression of cell cycle regulators c-myc and Cyclin B1 in tumor tissues. The results reveal that these proteins were upregulated in MAZ overexpressing tumor tissues and significantly reduced in MAZ knockdown tissues (Fig. 8g). These results demonstrate that MAZ can regulate downstream NDUFS3 expression, affect the melanoma cell cycle through c-myc and Cyclin B1, and promote melanoma growth in vivo.
Codependent expression of MAZ and NDUFS3 may serve as prognostic marker for melanoma patients
The study employed the LASSO Cox regression algorithm to identify MAZ and NDUFS3 as prognostic risk genes for melanoma patients (Fig. 9a). A bi-genetic risk factor was established, and a risk score was calculated for each patient. Subsequently, patients were categorized into high and low risk scoring groups based on the median characteristic risk score. The two groups were analyzed for prognostic survival, which showed that melanoma patients who scored a higher risk had a poorer prognosis compared to those with lower risk score (Fig. 9b).
a Progression-free survival (PFS) screening parameters in LASSO regression models. The left dashed line is “λ.min” and the right dashed line is “λ.1-s.e.”. The dashed line on the left is λ min, meaning the λ with the smallest deviation, representing the highest model fit at that lambda value. Therefore, the λ1-s.e on the right side is usually chosen as the final equation screening criterion in clinical practice. b Survival analysis of patients in the high-risk group in the TCGA database based on risk scores. (Shaded areas represent 95% confidence intervals for survival). c Nomogram combining risk scores with clinicopathologic characteristics. The scale is developed based on the magnitude of the risk factor regression coefficients, giving each risk factor a score for each taken level; for each patient, a total score can then be calculated, and the probability that the outcome will occur at the time of the outcome is calculated for each patient by means of a conversion function between the score and the probability of the outcome occurring. d Nomogram correction curves, which assess how close the estimated risk from the column line graph is to the actual risk. e Heatmap for different risk scores corresponding to different pathologic features.
To investigate the combined effects of MAZ and NDUFS3 on the clinicopathological progression of melanoma patients, independent prognostic parameters were integrated into a Nomogram model (Fig. 9c). This model predicted patient survival at 1-, 3-, 5-, and 10-year intervals based on risk scores and pathological characteristics. The calibrated plot confirmed the accuracy of the risk prediction model (Fig. 9d). The analysis of risk scores in relation to pathologic characteristics revealed significant differences in expression among melanoma patients at different TNM stages, particularly in stages III to IV. Within these stages, higher risk scores were associated with malignant progression in both T and N stages (Fig. 9e, Supplementary Fig. 8a). Subsequent survival analyses for patients at TNM stage III to IV, T1 to T4, and N1 to N3 demonstrated a poorer prognosis for those with high risk scores, with results deemed diagnostically significant (Supplementary Fig. 8b), with results deemed diagnostically significant (Supplementary Fig. 8c).
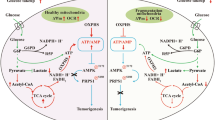
Further exploration focused on the effects of high co-expression of MAZ and NDUFS3 on melanoma biological functions. GO analysis of genes associated with melanoma risk scores indicated that MAZ’s regulation of NDUFS3 expression influenced the function of mitochondrial complex I and controlled the mitochondrial oxidative phosphorylation process by regulating NADH electron transfer through the respiratory chain. This regulation also impacted mitochondrial gene expression (Supplementary Fig. 9a). Differential gene expression analysis between high and low risk scoring groups (Supplementary Fig. 9b), followed by GSEA analysis on the differential genes, revealed that MAZ’s regulation of NDUFS3 was closely related to mitochondrial metabolic processes. Furthermore, it positively regulated DNA replication processes and the cell cycle, which is consistent with the results of previous experiments (Supplementary Fig. 9c). To confirm the effect of MAZ-regulated NDUFS3 on mitochondrial metabolic processes, we detected changes in mitochondrial function by seahorse experiments. It was confirmed that basal respiration, maximal respiration and real-time ATP production were all affected by MAZ-regulated NDUFS3 expression, and MAZ and NDUFS3 expression were positively correlated with mitochondrial energy metabolism (Fig. 10a). We subsequently determined the activity and total ATP content of mitochondrial complex I (Fig. 10b, c), which also confirmed this conclusion. In brief, MAZ promotes the malignant phenotype of melanoma by regulating NDUFS3 expression and promoting mitochondrial energy metabolism.
a For mitochondrial respiration, the oxygen consumption rate (OCR) of melanoma cells with MAZ overexpression and knockdown was determined using a Seahorse XF24 Analyzer. Maximum respiration was determined by sequential addition of oligomycin, FCCP, Rot/AA, respectively. b Changes in mitochondrial complex I activity in MAZ overexpression knockdown melanoma cell lines detected by UV spectrophotometry. c Microplate reader detects changes in ATP content in MAZ overexpression or knockdown melanoma cell lines. d Mechanistic modeling MAZ-targeted regulation of NDUFS3 in melanoma. The experimental data were normalized. Student’s t-test was used to analyse the experimental results of the MAZ-OE and MAZ-KD groups with their control groups. (mean ± SEM, n = 3). (****p < 1e-4, ***p < 2e-4, **p < 2e-3, *p < 0.05).
The analysis of risk scores in correlation with cell cycle regulators and migration and invasion regulators revealed significant effects of MAZ regulation of NDUFS3 expression on various aspects of melanoma biology. Specifically, MAZ’s regulation of NDUFS3 expression upregulated CDK2/CKD4, promoting the transition of the cell cycle from G0/G1 to S phase. It may upregulate MCM3, MCM4, MCM5, and MYC (c-myc), increasing the number of cells in S phase, as well as up-regulate CCNB1/CCNB2 (Cyclin B1/B2) expression to increase the number of cells in the G2/M phase (Supplementary Fig. 9d). Furthermore, MAZ’s regulation of NDUFS3 expression ultimately up-regulated the expression of MMP14, MMP15 and ADAM15 in the MMPs family and ADAM family, promoting the migration and invasion process of cells. It may also enhance the migration and invasion of melanoma cells by up-regulating both VEGF and SNAI2 (Snail2) expression and down-regulating TP63 (P63) expression (Supplementary Fig. 9e).
It was indicated by these results that co-expression between MAZ and NDUFS3 could serve as a prognostic marker for melanoma patients, and MAZ represents a potential new therapeutic target for melanoma treatment.
Discussion
Metabolic reprogramming is not only a hallmark of tumors, but key molecules in metabolic pathways are also potential targets for targeted therapies to prevent tumor progression39. Emerging studies have highlighted the regulatory role of transcription factors in metabolic pathways40. By regulating the expression of genes central to tumor metabolism, transcription factors can significantly affect metabolic reprogramming and signal transduction41, making them promising targets for tumor therapy42. MAZ regulates the transcription of nucleolar protein 2 (NOP2) and promotes glycolysis in HCC cells43. Previous studies have shown that MAZ is aberrantly expressed in various malignant tumors, with complex and diverse regulatory mechanisms44. However, no relevant study has elucidated the expression and functional mechanisms of MAZ in melanoma. Our data suggest that MAZ is highly expressed in melanoma and promotes tumor proliferation and metastasis, correlating with poor patient prognosis. High MAZ expression is particularly associated with older age, advanced TNM stage III-IV, and N-N3 stages, leading to increased patient mortality. Inhibition of MAZ expression could potentially offers a new therapeutic option for melanoma patients.
Mitochondria play a central role in energy synthesis of organisms45, providing ATP for biomolecule synthesis to sustain hypermetabolic processes in tumors46. Mitochondrial dysfunction strongly affects melanoma cell viability47. Therefore, targeting mitochondria for melanoma treatment is expected to improve patient prognosis. MCI, the largest complex in the respiratory chain, is crucial for electron transfer in the NADH oxidative respiratory chain and is considered a potential therapeutic target for cancer48,49. NDUFS3, a core subunit of MCI, is essential for its function32, and its aberrant NDUFS3 expression is usually connected tumor malignancy. However, there has been no report on the expression and mechanism of NDUFS3 in melanoma. Our study demonstrated high expression of NDUFS3 in melanoma, correlating with poor patient prognosis.
No study has yet revealed the upstream transcription factors that regulate NDUFS3 expression. Our findings suggested that MAZ regulates the expression of NDUFS3 in melanoma. Mechanistically, MAZ can bind to the 1147-1157 and 1735-1745 regions of the NDUFS3 promoter, promoting the transcription of NDUFS3, which accelerates the process of melanoma proliferation and migration.
Previously identified as a core gene involved in mitochondrial dysfunction in breast cancer, MAZ serve as a biomarker and therapeutic target for breast cancer diagnosis and prognosis50. However, the connection between MAZ and mitochondrial metabolism remains unclear and has not been experimentally confirmed. Our GO and GSEA analyses revealed that MAZ also regulates the expression of mitochondrial genes in melanoma and participates in the assembly of respiratory chain complexes, suggesting that MAZ could be targeted to molecular mitochondrial function in melanoma treatment. Functionally, NDUFS3 is closely associated with mitochondrial function31,51. This suggests that MAZ may influence melanoma’s malignant progression by regulating the expression of the core mitochondrial gene NDUFS3 and controlling mitochondrial metabolic processes.
Risk score prediction models, which predict the probability of a particular disease or adverse outcome based on a patient’s pathological characteristics, are now widely used in tumor research52,53,54. In this work, we constructed a personalized melanoma patient risk prediction model, combining MAZ and NDUFS3 as risk factors. Our model revealed that MAZ’s regulation of NDUFS3 expression affects the pathological process of melanoma patients in T staging, N staging and TNM staging. Increased expression of MAZ with NDUFS3 leads to higher mortality in melanoma patients with T1 to T4, N1 to N3 stage and TNM stage III to IV, thereby impacting overall patient survival. Therefore, the co-expression of MAZ and NDUFS3 could serve as a prognostic marker for assessing the survival status of patients with melanoma.
Next, the biological function of MAZ in regulating NDUFS3 transcription was predicted by enrichment analysis of risk scores. High expression of MAZ and NDUFS3 modulates metabolic processes in melanoma cells and promotes malignant progression by affecting the function of mitochondrial complex I and the respiratory chain. This suggests that MAZ and NDUFS3 could be targeted for melanoma mitochondrial therapy. We also confirmed this by measuring the mitochondrial energy metabolism and mitochondrial complex I activity. This study contributes valuable insights to melanoma diagnosis and treatment strategies.
In conclusion, our study reveals that MAZ is highly expressed in melanoma and regulates the transcription of NDUFS3, which enhances mitochondrial function in melanoma and thereby promoting melanoma growth and invasive migration. We propose that MAZ and NDUFS3 could serve as new tumor biomarkers for melanoma, as evidenced by the construction of a clinical risk prediction model (Fig. 10d). This work provides a new target for therapeutic interventions aimed at targeting the mitochondrial energy metabolism pathway in melanoma treatment.
Materials and methods
Bioinformatics Analysis
RNA-seq data and clinical information from skin cutaneous melanoma (SKCM) patients were obtained from TCGA database (https://portal.gdc.cancer.gov/). The GSE15605 and GSE114445 datasets were retrieved from the GEO database55,56 (https://www.ncbi.nlm.nih.gov/geo/). The GSE15605 dataset contains 16 normal skin samples and 58 melanoma samples. Samples in the GSE114445 dataset include 5 normal pigmented nevi, 7 dysplastic nevi, 6 normal skin and 16 melanomas. The 500 genes with the highest correlation with MAZ in melanoma were screened by Spearman’s correlation coefficient and subsequently analyzed by KEGG/GO. High-expression and low-expression groups were divided according to the median MAZ expression, and differential genes between the two groups were analyzed by the R language package Limma. The obtained differential genes were subjected to GSEA analysis.
Construction of personalized prediction models
To predict overall survival of patients, we developed a predictive model for the co-expression of MAZ and NUDFS3 by a combination of characteristic risk scores and clinicopathological features. We used the least absolute shrinkage and selection operator (LASSO) to identify the most useful predictive features to develop the optimal risk profile57. Among them, MAZ and NUDFS3 gene co-expression was determined to be associated with survival in melanoma patients. Patients were then categorized into high and low risk groups based on the median cutoff value of the risk score. Kaplan-Meier survival curves were used to estimate the survival distribution of patients in the high- and low-risk groups. Model accuracy and discrimination were evaluated using calibration curve.
Cell culture
Cells were cultured in a cell culture incubator at 37 °C with 90% humidity and 5% carbon dioxide concentration, using DMEM supplemented with 10% fetal bovine serum. Human epidermal melanocytes (HEM) and human melanoma cells (A375, A875, MeWo, SK-MEL-21 and SK-MEL-28) were obtained from the Kunming Institute of Animal Science, Chinese Academy of Sciences.
Cell transfection
A875 and SK-MEL-21 cells were transfected with MAZ overexpression (using overexpression vector LV-MAZ) or MAZ knockdown (using shRNA vector LV-MAZ - RNAi), along with corresponding controls (CON335 and CON313), for at least 48 h. The 6-well plate was 1×105 cells per well, and the lentiviral solution was added by mixing well with the transfection reagent HiTransG A/P (REVG004/ REVG005, Jikai Gene, Shanghai, China). Subsequently, the culture was replaced with medium containing 10% FBS to continue the culture. Cell constructs transfected with puromycin-containing puromycin-resistant viral vectors were screened by adding 5 μg/ml of puromycin to each well of transfected cells in six-well plates. Cells were passaged based on growth and screened using successive puromycin additions until fluorescence microscopy revealed more than 95% of cells exhibiting a positive green fluorescent protein signal. Finally, MAZ expression was detected by Western Blot and RT-qPCR.
MAZ knockdown A875 and SK-MEL-21 cells were transfected with NDUFS3 overexpression plasmid and its control (CON520). Plasmids and transfection vectors were purchased from Shanghai Jikai Gene Chemical Technology Company Limited, Shanghai, China. Transfection was performed using 5 μl Lipofectamine 2000 Transfection Reagent (11668500, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. After 6–8 h the medium was replaced with medium containing 10% FBS and incubation was continued until 24 h. Cells after plasmid transfection were collected for protein extraction. Western Blot detection of NDUFS3 protein expression after cell transfection. Cells successfully identified were used for subsequent experiments
Western Blot
Cells were lysed in protease and phosphatase (1:100) inhibitor (P0100, Solarbio Life Sciences, Beijing, China) and RIPA buffer (R0020, Solarbio Life Sciences, Beijing, China). Concentrations of proteins were determined using the BCA™ Protein Assay Kit (P1511, Applygen, Beijing, China). Protein samples (40 mg/sample) were separated by 10% polyacrylamide gel electrophoresis and transferred onto a PVDF membrane (IPVH00010, Millipore, Phillipsburg, NJ, USA). Membranes were incubated overnight at 4 °C in the refrigerator with the appropriate primary antibodies, followed by incubation with secondary antibodies for 1 h at room temperature. Protein bands were detected using a chemiluminescent reagent ECL kit (K-12045-D50, Bioship, Shanghai, Beijing, China) on a Bio-Rad ChemiDoc XRS system (Bio-Rad, Hercules, CA, USA). All experiments were conducted with three independent replicates.
The following primary antibodies were used: anti-MAZ (1:1000, #21068-1-AP, Proteintech, Wuhan, China); anti-NDUFS3 (1:3000, #R382058, Zenbio, Chengdu, China); anti-c-Myc (1:1000, #343250, Zenbio, Chengdu, China); anti-Cyclin B1 (1:1000, #R23324, Zenbio, Chengdu, China); anti-ADAM15 (1:1000, #R389200, Zenbio, Chengdu, China); anti-MMP14 (1:1000, #R22533, Zenbio, Chengdu, China); and anti-GAPDH (1:5000, #60004-1-IG, Proteintech, Wuhan, China). Fluorescent secondary antibodies used were anti-rabbit IgG (1:10000, #ab6721, Abcam, Cambridge, USA) and anti-mouse IgG (1:10000, #115-035-003, Jackson Immuno Research Laboratories, Maine, USA).
Quantitative real-time PCR
Trypsinized cells were collected into sterile EP tubes and centrifuged. Subsequently, 1 ml of Trizol and 200 µl of chloroform were added and mixed. The supernatant was then extracted by centrifugation and added to an equal volume of isopropanol. The supernatant is removed by centrifugation for 10 min and DEPC water is added to the precipitate to dissolve the RNA. The finally extracted RNA is stored in a refrigerator at -80 °C.
The extracted RNA was reverse-transcribed using a cDNA reverse transcription kit (K1622, Thermo Fisher Scientific, MA, USA). Expression of gene transcript levels was detected by real-time fluorescence quantitative PCR (RT-qPCR). The synthesis of real-time fluorescence quantitative PCR (Applied Biosystems, USA) and SYBR Green (06924204001, Roche, Switzerland) was performed by following the manufacturer’s guidelines. Table 1 shows the PCR primer pairs used for amplifying the target genes.
Chromatin immunoprecipitation
At least 3 × 106 cells were counted and collected. Configure buffer A/formaldehyde to cross-link for 10 min at room temperature, followed by the addition of glycine to terminate the cross-linking reaction. After centrifugation and discarding the supernatant, Buffer B, Buffer C and Buffer D/PI mixture (25×) were added sequentially. The DNA was disrupted by sonication on ice and the supernatant (containing chromatin) was transferred to a new EP tube after centrifugation. Add 1× ChIP-Buffer/PI and mix well. 50 µl of the mixture was used as Input and stored in the refrigerator at -20 °C. The rest of the liquid was added with the corresponding IP antibody and mixed at 4 °C overnight. The next day, 1×ChIP buffer (without PI mixture) was added to resuspend the protein beads, and the supernatant was discarded by centrifugation. The overnight antibody/chromatin mixture was centrifuged at 14000 g at 4 °C for 10 min, and the supernatant was added to the EP tube containing protein beads and continued to be incubated overnight in a 4 °C mixer. After incubation, add 1× ChIP buffer to the EP tube and centrifuge to discard the supernatant. DNA purifying was added to the sample tube and incubated at 98 °C for 10 min, followed by 20 min at room temperature. 10,000 g was centrifuged for 10 s, 1 µl of Proteinase K was added and incubated at 55 °C for 30 min, and again at 98 °C for 10 min. After centrifugation at 14000 g for 1 min at room temperature, the supernatant was transferred to a new EP tube, mixed with PCR-grade purified water and used for subsequent PCR assays or frozen at -20 °C for storage. The PCR products were identified by agarose gel electrophoresis. ChIP experiments were performed using the ChIP Assay Kit (ab500, Abcam, Cambridge, USA). Chip assay using manufacturer’s specified antibodies: anti-MAZ (1:100, #39935, Proteintech, Wuhan, China). The procedure for ChIP-qPCR was the same as for RT-qPCR. ChIP-qPCR primers are shown in Table 1.
Dual-luciferase reporter assays
We cloned the sequence of the NDUFS3 promoter region into pGL3-basic dual luciferase. The binding site of MAZ to the NDUFS3 promoter region was mutated. The pGL3-basic dual luciferase reporter vector was purchased from Wuhan Genecreate biological engineering limited company, Wuhan, China. HEK293T cells in 24-well plates were co-transfected with wild-type or mutant plasmids when the cells had grown to 60%-70% fusion. After 48 h, MAZ-OE and its control lentiviral vector were continued to infect HEK293T cells for 24 h. Cells were collected and subjected to dual luciferase reporter assay using the luciferase reporter gene assay kit (T002, Vigorous, Beijing, China). Multimode function measuring instrument (CA95051, Agilent Technologies, USA) was used to detect luciferase activity.
Immunohistochemistry
Tissue chips were purchased from Zhongke Guanghua Intelligent Biotechnology Co. Ltd (MM1601, Xi’an, China). Decolourisation of melanin-containing tissue microarrays using 1% hypochlorous acid (HClO) prior to immunohistochemical staining58. Tissue microarrays were timed for 2 min 30 s by placing them in a sodium citrate antigen repair solution that had been boiled in an autoclave in advance. After treatment with 3% H2O2 for 15 min at room temperature, the tissue microarrays were incubated with Anti-MAZ (1:400, #82852-3-RR, Proteintech, Wuhan, China) antibody at 4 °C overnight. Incubation with secondary antibody was then carried out for 1 h at room temperature and immunity score was performed under a microscope. There are 4 levels of scoring based on the intensity of cellular staining, with no positive staining (negative) scoring 0 points, pale yellow (weakly positive) scoring 1 point, tan (positive) scoring 2 points, and tan (strongly positive) scoring 3 points. The results were graded according to the percentage of positive cells, with ≤25% scored as 1, 26–50% scored as 2, 51–75% scored as 3, and >75% scored as 4. Finally, the two scores were multiplied to obtain the final MAZ score.
MTS assay
Melanoma cells were inoculated in 96-well plates with 2.5 × 103 per well, as well as their controls. Absorbance at 490 nm was measured using an enzyme labeler (51119200, Thermo Fisher Scientific, Waltham, MA, USA) at 0, 24, 48, and 72 h. Absorbance values were assayed by adding 20 µl of MTS assay reagent (CTB169, Promega, WI, USA) per well for 1 h at 37 °C. The relative proliferation rate of the cells was calculated using the formula provided in the kit.
Plate cloning experiment
Melanoma cell line with 1 × 103 cells/well was inoculated into 6-well plates and cultured continuously for 7–14 d. Discard the medium of the six-well plate and wash with PBS. The plates were then fixed with 4% paraformaldehyde for 30 min at room temperature and stained with crystal violet staining solution for 15 min at room temperature. Finally, cell colonies were counted under the microscope.
EdU assay
Culture overnight in 24-well plates with 5 × 103 melanoma cell lines per well. After incubation with 1 μl EdU working solution per well for 2 h, cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Then, 0.3% Triton X-100 was added to each well to permeabilize the cells for 15 min. Next, each well was incubated for 30 min with 100 µl of Click reaction solution, protected from light. Finally, fluorescence was detected and counted by fluorescence microscopy (DM4B, Leica, Germany). The DNA synthesis rate was assessed using the EdU assay kit (C0078S, Beyotime, Beijing, China) according to the manufacturer’s instructions.
Flow cytometry
The count of cells for each cycle was detected using the Cell Cycle Assay Kit (550825, Biotech, New Jersey, USA). In the experiment, cells were harvested at a growth density of 50–60%. Cells were fixed in 75% alcohol at 4 °C overnight, followed by PI staining in the dark. The number of cells per cycle was finally detected by flow cytometry (PARTEC CyFlow Space).
Wound healing assay
Cells were seeded in six-well plates and allowed to adhere overnight. Cells were delineated with a 20 ml spiking lance tip. Fluorescence microscopy was used to take 0 h and 24 h images, and the healing area was subsequently counted using ImageJ software.
Transwell migration assay and Transwell invasion assay
The Transwell (3524, Corning, USA) was supplemented with a suspension of cells in serum-free medium in the upper layer of the chambers. The upper half of the Transwell invasion chamber was pre-coated with a 1:8 dilution of Matrigel gel (356234, Biotech, NJ, USA). Serum-containing DMEM medium was included in the lower chamber. Cells were stabilized with 4% paraformaldehyde after 24 h. Crystalline violet staining and then photographed with a fluorescence microscope.
Melanoma cell line implantation tumor model
BALB/c nude mice (1–2 months old, male, weighting about 20 g) were obtained to construct an implantable tumor model from the Department of Laboratory Animals of Kunming Medical University. PBS 100 µl containing 5 × 106 cells were injected subcutaneously into the axillae of both sides of the mice in the NC-KD and KD groups (NC-OE and OE groups were inoculated with 2 × 106 per inoculum, 6 mice per group). Tumor volumes of mice were observed and recorded daily. Mice were euthanized 30 d after tumor injection site edema disappeared. Tumors of mice were collected and weighed, and divided into two parts for Western Blot and qPCR as needed. All animal experiments were approved by the Institutional Animal Care and Utilization Committee of Kunming Medical University. (The maximum tumor measurement diameter allowed by the Animal Care and Use Committee of Kunming Medical University was 1.5 cm, and all experiments did not exceed these limits.).
Seahorse experiments
Cellular oxygen consumption rate (OCR) was measured using the Seahorse XF Cellular Mitochondrial Stress Assay Kit (103016, Agilent Technologies, USA). Inoculate 3 × 104 melanoma cells per well in Seahorse XF 24 cell culture microplates and incubate overnight. The OCR of the cells were measured using the Seahorse XF 24 cell culture microplate (S7801B, Agilent Technologies, USA) when the cell fusion reached more than 90%. During the OCR assay, 1.5 μM oligomycin, 2 μM p-trifuoromethoxy carbonyl cyanide phenylhydrazone (FCCP, reversible oxidative phosphorylation inhibitor) and 0.5 μM rotenone plus antimycin A (Rote/AA, mitochondrial complex I inhibitor and mitochondrial complex III inhibitor) were injected sequentially. The results were analyzed using Seahorse XF-96 Wave software and normalized to total protein.
ATP content testing
The ATP content assay was performed using the ATP content assay kit (BC0305, Solarbio Life Sciences, Beijing, China). Firstly, 5 × 106 cells were collected and 1 mL of extraction solution was added. And the cells were broken by ultrasonic cell pulverizer (SCIENTZ-IID, Ningbo, China). The supernatant is then extracted by centrifugation at 10,000 g for 10 min and added to 500 μl of chloroform. The supernatant was retained after centrifugation again at 10,000 g. Finally, the working solution was added and the absorbance values were measured using an enzymelabeled instrument (Multiskan GO, Thermo Fisher Scientific, MA, USA) for 10 s and 3 min 10 s, respectively. ATP content was calculated according to the formula provided by the manufacturer.
Mitochondrial respiratory chain complex I activity assay
A total of 5 × 106 melanoma cells were collected and then mixed with reagents. The supernatant was extracted by centrifugation at 600 g for 5 min. Next, the extracted supernatant was centrifuged at 11,000 g for 10 min, and then broken with an ultrasonic cell pulverizer. Finally, the absorbance value A1 at 340 nm and the absorbance value A2 after 2 min were measured with an enzymelabeled instrument. The enzyme activity was calculated according to the formula provided in the mitochondrial respiratory chain complex I assay kit (SK006-2, Baiao Leibo, Beijing, China).
Statistics and Reproducibility
GraphPad Prism v9.0 and R software v4.3.0 were used for experimental data analysis. Results are presented as mean ± standard error (SEM) or standard deviation (SD). Differences with a p-value < 0.05 were considered as significant statistically. Statistical analyses included one-way analysis of variance (ANOVA), Student’s t-test for unpaired or paired samples, and mixed ANOVA.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data involved in the bioinformatics analysis in this study are available in TCGA (https://portal.gdc.cancer.gov/) and GEO public databases. Source data covered in this study are presented in the supplementary data 1. Uncropped and unedited blot and gel images can be reviewed from the supplementary data 2. Experimentally relevant data can be obtained by contacting the corresponding author.
Change history
23 August 2025
A Correction to this paper has been published: https://doi.org/10.1038/s42003-025-08660-1
References
Long, G. V., Swetter, S. M., Menzies, A. M., Gershenwald, J. E. & Scolyer, R. A. Cutaneous melanoma. Lancet 402, 485–502 (2023).
Garbe, C. et al. European consensus-based interdisciplinary guideline for melanoma. Part 1: Diagnostics: Update 2022. Eur. J. Cancer 170, 236–255 (2022).
Gosman, L. M., Țăpoi, D. A. & Costache, M. Cutaneous melanoma: a review of multifactorial pathogenesis, immunohistochemistry, and emerging biomarkers for early detection and management. Int J. Mol. Sci. 24, 15881 (2023).
Rodríguez-Enríquez, S. et al. Transcriptional regulation of energy metabolism in cancer cells. Cells 8, 1225 (2019).
Jen, J. & Wang, Y. C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 23, 53 (2016).
Kim, S. & Shendure, J. Mechanisms of interplay between transcription factors and the 3D genome. Mol. Cell 76, 306–319 (2019).
Chen, A. & Koehler, A. N. Transcription factor inhibition: lessons learned and emerging targets. Trends Mol. Med. 26, 508–518 (2020).
Zhao, J., Wen, D., Zhang, S., Jiang, H. & Di, X. The role of zinc finger proteins in malignant tumors. FASEB J. 37, e23157 (2023).
Wang, M. et al. Comprehensive analysis of the tumor-promoting effect and immune infiltration correlation MAZ from pan-cancer to hepatocellular carcinoma. Int. Immunopharmacol. 115, 109660 (2023).
Song, J. et al. Human genes for KNSL4 and MAZ are located close to one another on chromosome 16p11.2. Genomics 52, 374–377 (1998).
Song, J. et al. Genomic organization and expression of a human gene for Myc-associated zinc finger protein (MAZ). J. Biol. Chem. 273, 20603–20614 (1998).
Parks, C. L. & Shenk, T. Activation of the adenovirus major late promoter by transcription factors MAZ and Sp1. J. Virol. 71, 9600–9607 (1997).
Song, J., Ugai, H., Kanazawa, I., Sun, K. & Yokoyama, K. K. Independent repression of a GC-rich housekeeping gene by Sp1 and MAZ involves the same cis-elements. J. Biol. Chem. 276, 19897–19904 (2001).
Bossone, S. A., Asselin, C., Patel, A. J. & Marcu, K. B. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. USA 89, 7452–7456 (1992).
Wang, W. et al. G-quadruplexes promote the motility in MAZ phase-separated condensates to activate CCND1 expression and contribute to hepatocarcinogenesis. Nat. Commun. 15, 1045 (2024).
Roberts, G. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26, 5568–5572 (1998).
Gromak, N., West, S. & Proudfoot, N. J. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell Biol. 26, 3986–3996 (2006).
Ren, L. X. et al. Myc-associated zinc-finger protein promotes clear cell renal cell carcinoma progression through transcriptional activation of the MAP2K2-dependent ERK pathway. Cancer Cell Int. 21, 323 (2021).
He, J. et al. SIPL1, regulated by MAZ, promotes tumor progression and predicts poor survival in human triple-negative breast cancer. Front Oncol. 11, 766790 (2021).
Jiang, Q., Guan, Y., Zheng, J. & Lu, H. TBK1 promotes thyroid cancer progress by activating the PI3K/Akt/mTOR signaling pathway. Immun. Inflamm. Dis. 11, e796 (2023).
Triner, D. et al. Myc-associated zinc finger protein regulates the proinflammatory response in colitis and colon cancer via STAT3 signaling. Mol. Cell Biol. 38, e00386–18 (2018).
Zhang F. et al. Transcription factor MAZ potentiates the upregulated NEIL3-mediated aerobic glycolysis, thereby promoting angiogenesis in hepatocellular carcinoma. Curr. Cancer Drug Targets. https://doi.org/10.2174/0115680096265896231226062212 (2024).
Tasdogan, A. et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120 (2020).
Harel, M. et al. Proteomics of melanoma response to immunotherapy reveals mitochondrial dependence. Cell 179, 236–250.e18 (2019).
Rodrigues, T. & Ferraz, L. S. Therapeutic potential of targeting mitochondrial dynamics in cancer. Biochem Pharm. 182, 114282 (2020).
Loeffen, J. et al. cDNA sequence and chromosomal localization of the remaining three human nuclear encoded iron sulphur protein (IP) subunits of complex I: the human IP fraction is completed. Biochem. Biophys. Res. Commun. 247, 751–758 (1998).
Procaccio, V. et al. Human NDUFS3 gene coding for the 30-kDa subunit of mitochondrial complex I: genomic organization and expression. Mamm. Genome 11, 808–810 (2000).
Samanta, D., Veerapandiyan, A., Burrow, T. A. & Gokden, M. Mitochondrial Ultrastructural Defects in NDUFS3-Related Disorder. J. Pediatr. Neurosci. 16, 299–302 (2021).
Luo, N. et al. Reduced electron transport chain complex I protein abundance and function in Mfn2-deficient myogenic progenitors lead to oxidative stress and mitochondria swelling. FASEB J. 35, e21426 (2021).
D’Angelo, L. et al. NDUFS3 depletion permits complex I maturation and reveals TMEM126A/OPA7 as an assembly factor binding the ND4-module intermediate. Cell Rep. 35, 109002 (2021).
Chanthammachat, P. & Dharmasaroja, P. Metformin restores the mitochondrial membrane potentials in association with a reduction in TIMM23 and NDUFS3 in MPP+-induced neurotoxicity in SH-SY5Y cells. EXCLI J. 18, 812–823 (2019).
Kurelac, I. et al. NDUFS3 knockout cancer cells and molecular docking reveal specificity and mode of action of anti-cancer respiratory complex I inhibitors. Open Biol. 12, 220198 (2022).
Putignani, L. et al. Preliminary evidences on mitochondrial injury and impaired oxidative metabolism in breast cancer. Mitochondrion 12, 363–369 (2012).
Suhane, S., Berel, D. & Ramanujan, V. K. Biomarker signatures of mitochondrial NDUFS3 in invasive breast carcinoma. Biochem. Biophys. Res. Commun. 412, 590–595 (2011).
He, X. & Cao, X. Identification of alternatively spliced GRIM-19 mRNA in kidney cancer tissues. J. Hum. Genet. 55, 507–511 (2010).
Wang, P. et al. Reduced expression of NDUFS3 and its clinical significance in serous ovarian cancer. Int J. Gynecol. Cancer 23, 622–629 (2013).
Song, B., Du, J., Feng, Y., Gao, Y. J. & Zhao, J. S. Co-expressed differentially expressed genes and long non-coding RNAs involved in the celecoxib treatment of gastric cancer: An RNA sequencing analysis. Exp. Ther. Med. 12, 2455–2468 (2016).
Peng, W. F. et al. The key genes underlying pathophysiology association between the Type 2-diabetic and Colorectal cancer. J. Cell Physiol. 233, 8551–8557 (2018).
Xiao, Y. et al. Emerging therapies in cancer metabolism. Cell Metab. 35, 1283–1303 (2023).
Deng, C. et al. Refactoring transcription factors for metabolic engineering. Biotechnol. Adv. 57, 107935 (2022).
Kant, R. et al. Deregulated transcription factors in cancer cell metabolisms and reprogramming. Semin Cancer Biol. 86, 1158–1174 (2022).
Lambert, M., Jambon, S., Depauw, S. & David-Cordonnier, M. H. Targeting transcription factors for cancer treatment. Molecules 23, 1479 (2018).
Zhang, H. et al. NOP2-mediated m5C modification of c-Myc in an EIF3A-dependent manner to reprogram glucose metabolism and promote hepatocellular carcinoma progression. Res. (Wash. D. C.) 6, 0184 (2023).
Zheng, C. et al. Roles of Myc-associated zinc finger protein in malignant tumors. Asia Pac. J. Clin. Oncol. 18, 506–514 (2022).
Martínez-Reyes, I. & Chandel, N. S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11, 102 (2020).
Vasan, K., Werner, M. & Chandel, N. S. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 32, 341–352 (2020).
Huang, C., Radi, R. H. & Arbiser, J. L. Mitochondrial metabolism in melanoma. Cells 10, 3197 (2021).
Okoye, C. N., Koren, S. A. & Wojtovich, A. P. Mitochondrial complex I ROS production and redox signaling in hypoxia. Redox Biol. 67, 102926 (2023).
Zhou, Y. et al. Recent advances of mitochondrial complex I inhibitors for cancer therapy: current status and future perspectives. Eur. J. Med. Chem. 251, 115219 (2023).
Yan, L. R., Wang, A., Lv, Z., Yuan, Y. & Xu, Q. Mitochondria-related core genes and TF-miRNA-hub mrDEGs network in breast cancer. Biosci. Rep. 41, BSR20203481 (2021).
Pereira, C. V. et al. Myopathy reversion in mice after restauration of mitochondrial complex I. EMBO Mol. Med. 12, e10674 (2020).
Liao, W. et al. Predicting the future risk of lung cancer: development, and internal and external validation of the CanPredict (lung) model in 19·67 million people and evaluation of model performance against seven other risk prediction models. Lancet Respir. Med. 11, 685–697 (2023).
Farach, L. S. et al. Epilepsy risk prediction model for patients with tuberous sclerosis complex. Pediatr. Neurol. 113, 46–50 (2020).
Kim, H. C. et al. Risk prediction model in rheumatoid arthritis-associated interstitial lung disease. Respirology 25, 1257–1264 (2020).
Raskin, L. et al. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. J. Invest Dermatol. 133, 2585–2592 (2013).
Yan, B. Y. et al. Novel immune signatures associated with dysplastic naevi and primary cutaneous melanoma in human skin. Exp. Dermatol 28, 35–44 (2019).
Zeng, F., Liu, X., Wang, K., Zhao, Z. & Li, G. Transcriptomic profiling identifies a DNA repair-related signature as a novel prognostic marker in lower grade gliomas. Cancer Epidemiol. Biomark. Prev. 28, 2079–2086 (2019).
Wang, L. & Wang, G. Depigmentation of melanin-containing tissues using hypochlorous acid to enhance hematoxylin-eosin and immunohistochemical staining. Appl. Immunohistochem. Mol. Morphol. 32, 53–59 (2024).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 31960200; 82160540) and Yunnan Revitalization Talent Support Program.
Author information
Authors and Affiliations
Contributions
Y.F. participated in the completion of most of the experiments and was supervised in the manuscript writing. Q.N., N.W., Y.G. and E.L. performed animal experiments. T.X. was involved in the analysis of the data, and F.Y., X.Z., L.G., X.Y. and J.X. provided experimental technical support. Q.Z., Z.Y., B.S. and Y.K. were assigned to the evaluation of the results, Y.Z. has guided the experimental designs and reviewed and revised the manuscript. All authors have reviewed and accepted the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
All animal procedures were conducted according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee of Kunming Medical University (permit number: K2020-0006).
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, Y., Ni, Q., Wu, N. et al. Molecular mechanisms of MAZ targeting up-regulation of NDUFS3 expression to promote malignant progression in melanoma. Commun Biol 7, 1491 (2024). https://doi.org/10.1038/s42003-024-07209-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07209-y