Abstract
Mitochondria rapidly accumulate mutations throughout a lifetime, potentially acting as a molecular clock for aging and disease. We profiled mitochondrial RNA across 47 human tissues from 838 individuals, revealing rapid development of clonal mosaicism with two distinct tissue-specific aging signatures. Tissues with constant cellular turnover such as the gastrointestinal tract or skin exhibit accelerated accumulation of sporadic mutations and clonal expansions, implicating increased susceptibility to age-related tumorigenesis and dysfunction. By contrast, post-mitotic tissues, such as the heart and brain, accumulate mutations at deterministic hotspots (tissue-specific, recurrently mutated sites), reflecting the cumulative burden of high energy demand and mitochondrial turnover independent of cell division. These findings support a biphasic model of the mitochondrial clock: stochastic clonal expansion of sporadic replication errors in proliferative tissues, versus age-dependent heteroplasmy increases at hotspots in high-metabolic tissues. This mutational landscape provides a map of tissue-specific vulnerabilities during aging and offers potential therapeutic targets.
Similar content being viewed by others
Main
Mitochondria, often described as the powerhouses of eukaryotic cells, probably originated from an α-proteobacterium entering a host cell around 2 billion years ago through an endosymbiotic event1,2. Their circular, double-stranded genome (mitochondrial DNA, mtDNA) bears a resemblance to bacterial DNA and undergoes a mutation rate substantially higher (100–1,000 times) than that of nuclear DNA, attributed to the absence of histone protection, rapid replication cycles and reduced efficiency in DNA damage repair mechanisms3,4. Given their sensitivity to various age-related damages such as replication errors and oxidative damage, mitochondria can act as a sensitive aging clock, recording a tissue-specific chronicle of age-accumulating damage.
The mitochondrial genome, inherited from the maternal germline, populates each cell in multiple copies and exhibits extensive clonal diversity, known as heteroplasmy, both between cells and across different body sites5. This clonal diversity is highly dynamic throughout a lifetime, with mutant clones constantly arising and their abundance undergoing dynamic changes with cell division and mitochondrial turnover (mitophagy, fission and fusion)6,7. These changes are driven by two primary mechanisms: replication errors during cell division in tissues with constant cellular turnover, and mitochondrial turnover, including a process known as relaxed replication8,9,10, which allows mtDNA to replicate independently of the cell cycle, even in post-mitotic cells. The expansion of defective clones can lead to diseases, especially in high-energy-demanding tissues11,12,13,14, such as Alzheimer’s and Parkinson’s diseases15,16,17, macular degeneration18, diabetes19 and heart disease20. Such tissue-specific dynamics position mitochondria as unique markers for uncovering body-wide accumulations of age-related damage.
Clonal mosaicism within mitochondria, characterized by the presence of distinct mitochondrial genotypes within tissues, may be both a driver and indicator of the underlying forces of aging21. Mitochondrial dysfunction, characterized by impaired biogenesis, fission, fusion and mitophagy, leads to increased production of reactive oxygen species (ROS) and genomic instability. These factors coupled with cellular turnover drive the clonal expansion of mutated mitochondrial genomes, contributing to tissue-specific aging patterns. As such, mitochondrial mosaicism reflects the broader genomic instability and cellular decline that underlie aging22.
While previous studies have explored tissue-specific clonal expansions in nuclear DNA during aging23,24,25,26, the mitochondrial genome offers much higher resolution due to its much higher copy number and mutation rate. Existing studies of mitochondrial mutations have focused primarily on animal models (mice27,28,29 and macaques30), tumor samples31 or a limited number of human tissues32,33,34,35. However, a systematic body-wide quantification of mitochondrial clonal mosaicism during aging remains elusive. While recent work has quantified the intrinsic mtDNA mutation rate per replication or turnover event35, the long-term, tissue-specific consequences of these mutational processes across the human lifespan, and how they contribute to the overall aging landscape, remain less well understood. This study aims to utilize the mitochondrial mutational clock to characterize age-accumulated cellular damage over time, specifically focusing on how this clock operates differently in tissues with distinct cellular turnover and metabolic demands. We propose that a biphasic mitochondrial molecular clock exists, with distinct mechanisms driving mutation accumulation in dividing versus post-mitotic tissues. By analyzing RNA reads from 47 tissues in the the Genotype-Tissue Expression (GTEx) project36, we comprehensively measured tissue-specific mutational burden and their dynamics over age. Our findings reveal a striking tissue-specific pattern of mitochondrial genome alteration during aging, driven by both cellular proliferation and mitochondrial turnover, resulting in distinctive aging hallmarks. These dynamics serve as a unique tissue-specific signature that not only delineates the trajectory of cellular clonal expansion and cumulative cellular damage throughout an individual’s lifespan, but also sheds light on the driving forces of aging, such as genomic instability, mitochondrial dysfunction and environmental exposure. Unraveling the intricacies of the mitochondrial clock has the potential to pave the way for early diagnosis and targeted interventions in age-related disorders.
Results
Dynamic mitochondrial clonal mosaicism across human tissues
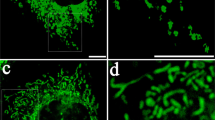
Mitochondrial mutations accumulate and evolve over time, creating intricate clonal structures that vary across different tissues. This dynamic process is driven by the continuous turnover of mitochondria and the renewal or proliferation of the host cells (Fig. 1a). As new mutations arise throughout development and aging, subsequent clonal expansion creates a mosaic pattern of mitochondrial clonal structures (for an example of such structures as revealed by single-cell RNA sequencing (RNA-seq), see Fig. 1b), leading to highly heterogeneous mitochondrial populations across different body sites.
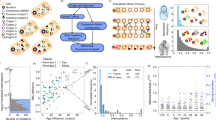
a, A schematic illustrating the accumulation and proliferation of mtDNA variants over time, with each color representing a unique mitochondrial clone. Created with BioRender.com. b, A heatmap showcasing the mitochondrial clonal structure across individual cells of the pancreas (an example derived from reanalysis of data from ref. 64), where each column represents a cell, and row shading indicates variant abundance. The cumulative abundance of each variant across all cells is also shown, reflecting the expected heteroplasmy value in bulk sequencing data. c, A schematic overview of the workflow for calling somatic variants from 47 tissues collected from 838 GTEx donors aged 20–70 years. Created with BioRender.com. d, Typical examples of mitochondrial mutations and clonal expansion among samples from the GTEx cohort. Each row represents a specific mitochondrial variant, and the colored dots indicate the variant’s abundance (heteroplasmy) in different tissues of that individual. The color scheme for tissues follows the GTEx conventions specified in Extended Data Fig. 2. e, The average mitochondrial RNA read coverage across tissues. The plot illustrates the distribution of RNA-seq read coverage across the mitochondrial genome in representative tissues (liver, esophageal mucosa and whole blood), highlighting regions excluded from variant calling owing to low coverage or the presence of known artifacts (tRNA, NUMTs, gene boundaries and so on). f, The number of variant sites by substitution type and functional category.
To comprehensively profile mitochondrial clonal mosaicism, we systematically analyzed mitochondrial RNA (RNA transcribed from mtDNA) from 838 healthy individuals across 47 different tissues from the GTEx cohort (Fig. 1c and Extended Data Fig. 2). After stringent quality control for read depth and sample contamination (Extended Data Fig. 1), a total of 11,802 samples were included in the analysis. We performed variant calling (using Mutect2 tool mitochondrial mode, as per the gnomAD mitochondrial pipeline33) and clonal abundance quantification (heteroplasmy level) at 5,376 sites with detected variants (Fig. 1d,e), encompassing a variety of substitution types (of likely origin of spontaneous deamination and ROS damage) and functional categories (Fig. 1f). RNA coverage over the mitochondrial genome ranged from 2,000× to 8,000× depending on the tissue (Fig. 1e), with all tissues achieving an average of at least 1,000×. Regions with low RNA coverage, including tRNA and gene boundaries, were excluded. Rigorous filtering, in accordance with previously published protocols for variant calling in both mitochondrial genomes33,37 and RNA-seq data24,25,26, was applied to remove potential artifacts, such as nuclear mitochondrial DNA segments (NUMTs)38, strand bias, low base quality and other blacklisted sites, resulting in the exclusion of 2,497 sites (Fig. 1e and Methods). To ensure reliability, only single-nucleotide substitutions were considered, excluding sites with indels or structural variants. In addition, potential sample contamination was thoroughly assessed and eliminated. Notably, mitochondrial RNA may harbor alterations beyond those transcribed from mtDNA, such as RNA modifications39,40. Those sites (2,617 and 13,710; Supplementary Fig. 2) were excluded from subsequent analyses. We also observed a difference between DNA and RNA at position 9,209 (Supplementary Fig. 2), a previously uncharacterized site of currently unknown etiology, which was analyzed separately.
Mutant clones display distinct characteristics based on their likely origin, underscoring the complex underlying forces of mutational gain and clonal expansion. Figure 1d shows typical examples of germline variants and somatic mutations. Germline variants are shared across all tissues but can exhibit widely different clonal abundances (heteroplasmy levels) among tissues due to random drift, as seen with the variants chrM_228_G_A in GTEx-1C64N and chrM_12389_C_T in GTEx-WY7C. Somatic mutations mostly occur in a single tissue spot, with a few instances of early developmental origin shared across multiple tissues. Although somatic mutations acquired later in life usually exhibit low abundance, some can reach high abundance over time. For example, the somatic missense variant chrM_3392_G_A in the individual GTEx-1GZ4H attains nearly 100% heteroplasmy level in the thyroid but is undetectable in other tissues. Notably, pathogenic mutations can reach high levels of heteroplasmy in specific tissues, such as chrM_11778_G_A in GTEx-15CHQ, associated with Leber hereditary optic neuropathy (LHON, Fig. 1d). This evidence highlights the critical potential for clonal expansion and the importance of examining tissue-specific clonal dynamics. Moreover, mutations of likely early developmental origin are shared across multiple tissues and are more likely to originate from the same germ layer, as in the case of chrM_76_C_T in GTEx-1F6I4, all from the mesoderm. Our subsequent analyses focus on somatic mutations, defined as those mutations present in one or more, but not all, tissues of an individual (that is, not germline). These mutations provide valuable insights into the tissue-specific dynamics of mutation accumulation and clonal expansion during aging.
Age drives tissue-specific mitochondrial mutation rates
The accumulation rate of mitochondrial mutations varies widely across tissues, driving distinct clonal dynamics. Most tissues exhibit a continuous gain of somatic mutations, with a maximum rate of 0.3 somatic mutations per year observed in the liver (Fig. 2a,b). Notably, tissues with constantly renewing epithelial cells6,41 (for example, the gastrointestinal tract, typical of endodermal origin) and nondividing cells with high energy demands10 (for example, muscle and the heart, typical of mesodermal origin) show particularly high rates of mutation accumulation. This observation suggests that both replicative stress and metabolic demands play important roles in mitochondrial mutagenesis, with potential implications for tissue-specific aging and disease susceptibility. Interestingly, a small fraction of mutations are shared across different tissues (Fig. 2a), suggesting they may originate either during early development or from recurrent mutations at specific hotspot sites (explored further in subsequent sections). Notably, these shared mutations tend to occur within tissues derived from the same germ layer (Extended Data Fig. 5), further implying that shared cellular lineage influences mutational patterns. After excluding hotspot mutations (which are more likely to arise independently in different tissues rather than through shared lineage), approximately 20% of all somatic mutations appear to stem from early developmental lineages, with less than 5% shared across all three germ layers—probably arising before gastrulation (Extended Data Fig. 6). This relatively small percentage aligns with previous findings35, suggesting that, while shared lineage plays a role, the majority of somatic mutations in these tissues arise later in development or during adulthood.
a, The number of tissue-private mutations (somatic mutations found only in a single tissue sample) per individual across 15 representative tissues, stratified by age group. The average number for the remaining tissues within each germ layer origin (endoderm, mesoderm and ectoderm) is also presented. The stacked bars at the bottom illustrate the number of mutations shared across multiple tissues within an individual, categorized by the number of germ layers in which they occur (1, 2 or all 3). b, Mitochondrial mutations and clonal expansion with age in the liver. The number of mutations represents the average across individuals within each age group. Total heteroplasmy reflects the summation of the heteroplasmy of all variants in an individual. Linear regression shows a significant increase in mutation count with age (β = 0.31 mutations per year, P = 4 × 10−16, n = 190 individuals). c, The rate of mutation accumulation (β, mutations per year) assessed by linear regression across six representative tissues. The shaded areas represent the 95% confidence interval (CI) for the linear regression line β. d, Differential rates (β) of age-related mutation accumulation across all tissues, derived from linear regression (sample sizes per tissue indicated in d). The error bars represent the 95% CI, which is based on the t-statistic of the corresponding mutation rate β and total heteroplasmy μ estimates. The asterisks indicate significant age-dependent mutation rate β (P < 0.001, FDR <0.05 after Bonferroni correction). Sample sizes (n, number of individuals) are marked on top of β estimates of each tissue. Detailed statistics for each tissue are provided in Supplementary Table 2. Tissues in this panel are color-coded to represent different tissue types, following the standard GTEx conventions (Extended Data Fig. 2). e, The relationship between age-related mutation accumulation (mutations per 10 years) with the total number of somatic mutations and their total heteroplasmy at an advanced age (60 years old), with the circle size representing the total heteroplasmy of somatic mutations. In b and d, box plots display the median (center line), interquartile range (IQR, box limits) and whiskers (extending to 1.5× IQR); notches indicate 95% CI for the median.
We further examined the pattern of mutational accumulation in each individual tissue. Among the 47 tissues analyzed from the GTEx cohort, 21 exhibited a significant positive correlation between the number of mutations and age (linear regression β > 0, P < 0.001, false discovery rate (FDR) <0.05 after Bonferroni correction), suggesting a progressively accumulating mitochondrial mutations throughout life (Fig. 2d and Extended Data Figs. 3 and 4). However, the dynamics of accumulation vary significantly between different tissues. The liver exhibits the most rapid accumulation of mutations with age (β = 0.31 per year, P = 4 × 10−16; Fig. 2b), followed by the left ventricle (β = 0.16 per year, P = 3 × 10−23) and esophageal mucosa (β = 0.11 per year, P = 8 × 10−24; Fig. 2c). This differential accumulation pattern probably reflects the interplay between tissue-specific factors, suggesting that both cellular turnover rate and environmental exposure have a substantial impact on mutational burden and clonal expansion. Further comparing esophageal mucosa (primarily epithelial) with esophageal muscularis (smooth muscle) highlights the profound impact of cellular composition on mutational burden (βmucosa: 0.11 per year > βmuscularis: 0.03 per year, P = 5.6 × 10−11; Fig. 2c). The higher mutation rate observed in the epithelial layer suggests that the constant turnover of these cells, with their continuous replication and division, drives increased clonal dynamics and heightened accumulation of mutations over time, with implications for understanding tissue-specific vulnerabilities to age-related dysfunction and, potentially, the development of malignancies in epithelial tissues. In addition, sun-exposed skin exhibits a much higher rate of mitochondrial mutations compared with nonexposed skin (βexposed: 0.09 per year > βnonexposed: 0.03 per year, P = 2 × 10−6; Fig. 2c), which underscores the influence of environmental factors, such as ultraviolet (UV) radiation, on mutational dynamics.
Both the number of distinct mitochondrial mutations and the total abundance of those mutations (total heteroplasmy) increase with age, suggesting a continuous acquisition of mutations and subsequent clonal expansions of these mutations (Fig. 2d,e). This expansion, resulting from the proliferation of cells carrying specific mitochondrial mutations, drives the overall increase in clonal abundance. The liver, exhibiting the highest rate of mutation accumulation, reaches an average mutant heteroplasmy level of 15% by the age of 60 (Fig. 2b,d). Overall, while age is the primary driver of mitochondrial clonal diversity, the rate of mutation accumulation and clonal expansion varies significantly across tissues (Fig. 2d,e). This variation probably reflects the influence of tissue-specific factors, such as cellular turnover rates, metabolic demands and exposure to environmental stressors.
Sporadic versus hotspot mutations define aging hallmarks
The distinct clonal dynamics observed across tissues potentially arise from two primary mutational processes: deamination-derived replication errors and oxidative (ROS) damage. However, their contributions differ significantly, leading to unique aging hallmarks across tissues. Replication errors, primarily C > T (β = 0.03 per year, P = 8.6 × 10−202) and T > C (β = 0.02 per year, P = 9.7 × 10−94) transitions characteristic of spontaneous deamination, are the dominant contributor to age-related mitochondrial mutation accumulation and clonal expansion (Fig. 3a). This is evident in the significant increase of these mutations with age, compared with minimal expansion of transversions, including ROS-related C > A and C > G.
a, Rate of mutational accumulation (β, mutations per year) by different substitution types, averaged over all 47 tissues. The lines represent linear regression fits of mutation count against age. The shaded areas represent the 95% CI for the slope (β). b, Tissue-specific aging hallmarks. The heatmap highlights the unique mutational signatures characteristic of aging in different tissues. Age-accelerated mutational hotspots are defined as sites with significant age-dependent increases in heteroplasmy (identified via linear regression; details in Supplementary Table 3). c, Non-ROS mutation accumulation across tissues. From top to bottom: the accumulation rate of hotspot and sporadic mutations (estimates from linear regression), total heteroplasmy at age 60, total number of mutations at age 60, and cell type enrichment of each tissue. The asterisks indicate a significant age-dependent mutation rate β assessed by linear regression (P < 0.001, FDR <0.05 after Bonferroni correction). The error bars represent 95% CI, which is based on the t-statistic of the corresponding slope β and mean μ estimates. Sample sizes (n, number of individuals) are marked on top of β estimates of each tissue. Detailed statistics for each estimate (slope β, 95% CI, exact P value) are provided in Supplementary Tables 2 and 3. For total heteroplasmy, box plots display the median (center line), interquartile range (IQR, box limits) and whiskers (extending to 1.5× IQR); the notches provide an approximate 95% CI around the median.
Further examination reveals that the accumulation of replication errors (non-ROS mutations) is not uniform across tissues but is instead shaped by two distinct mechanisms: sporadic mutations and mutations at hotspots. Sporadic mutations, occurring randomly throughout the mitochondrial genome, are primary contributors to mutagenesis and subsequent clonal expansion in tissues with continuous cellular turnover, such as the gastrointestinal tract (Fig. 3c). This is supported by the enrichment of constantly renewing cell types (epithelial cells, hepatocytes and keratinocytes) in tissues with high clonal expansion (Fig. 3c, bottom). Even within the brain, regions with a higher proportion of dividing glia display increased clonal diversity compared with neuron-rich regions (Fig. 3c, bottom), further emphasizing the role of cellular turnover in propagating these sporadic mutations.
By contrast, post-mitotic tissues such as the heart, brain and muscle exhibited a distinct pattern of mutation accumulation, concentrated primarily at specific hotspot sites (Fig. 3b,c). While hotspot mutations were observed across tissues of high metabolic activity, their relative contribution to the overall mutational burden was significantly higher in post-mitotic tissues. We defined hotspots as mitochondrial positions exhibiting both high prevalence (Supplementary Fig. 3, mutations present in over 10% of individuals by the age of 60, which is significantly higher than the overall mutation rate across all sites, P < 1 × 10−9, Fisher’s exact test, FDR <0.001 after Bonferroni correction) and a significant increase in frequency with age (Supplementary Fig. 4). We identified 22 such sites with significant age-accumulating effects (Fig. 3b and Supplementary Fig. 4). These hotspots, representing a distinct aging hallmark in these tissues (Fig. 3b), are probably a consequence of the cumulative burden of high energy metabolism and rapid mitochondrial turnover, coupled with the inherent error tendencies of DNA polymerase γ during relaxed replication associated with this turnover42. Relaxed replication allows individual mtDNA molecules to replicate independently of the cell cycle8,9,10. This is particularly relevant at hotspot sites, which are more susceptible to the same mutations across different cells, leading to their accumulation (increase of heteroplasmy) within a tissue. Given that this accumulation is expected to occur in a quasi-deterministic manner regardless of the individual, we further validated the age-dependent increase in heteroplasmy at these hotspot sites through targeted deep sequencing (average coverage >10,000×) of mitochondrial genomes from independent human testicular tissue samples (Extended Data Fig. 10). Furthermore, we replicated this age-dependent increase of heteroplasmy at these hotspots using independent data from the Encyclopedia of DNA Elements (ENCODE) and The Cancer Genome Atlas (TCGA, nontumor samples) (Supplementary Figs. 5 and 6). While linear regression was used to model the rate of mutation accumulation for ease of interpretation, this may not fully capture the complexity of the process, particularly in tissues like the heart, where mutation accumulation may exhibit a nonlinear pattern, potentially accelerating at advanced ages (Supplementary Fig. 7).
ROS-related damage (C > A and C > G) is also primarily localized to a few hotspot lesions (Fig. 4a). While ROS damage is prevalent in energy-intensive tissues (Fig. 4a), it does not significantly accumulate with age (Fig. 4b), consistent with previous reports in mice27,28,29, suggesting efficient repair or removal mechanisms.
a, ROS damage in all tissues. The relative mitochondrial copy number (reflecting higher energy demand), estimated from RNA data, is displayed to illustrate the correlation between a tissue’s energy demand and its susceptibility to ROS damage. Sample sizes (n, number of individuals) are marked on top of box plots of each tissue. To account for potential bias due to varying mitochondrial copy numbers across tissues, which could affect mutation detection sensitivity, we reanalyzed the data after subsampling all tissues to a uniform sequencing depth (Extended Data Fig. 7). Tissues are color-coded to represent different tissue types, following the standard GTEx conventions. b, Mutation accumulation in representative tissues. Top: the age-dependent increase in non-ROS mutation number for four selected tissues, exemplifying the varying rates and patterns of mutation accumulation and clonal expansion. Bottom: the various extent of ROS-induced damage in those tissues, which remains relatively constant with age. The rate of mutation accumulation (β, mutations per year) was assessed by linear regression across six representative tissues. For total heteroplasmy in a and b, box plots display the median (center line), interquartile range (IQR, box limits) and whiskers (extending to 1.5× IQR); the notches provide an approximate 95% CI around the median.
In conclusion, tissue-specific mitochondrial aging hallmarks are shaped by the complex interplay between sporadic and hotspot mutations, associated with distinct processes: cellular turnover, which leads to clonal expansion of sporadic mutations in proliferative tissues, and mitochondrial turnover (including relaxed replication), which underlies increased heteroplasmy of hotspot mutations, particularly in high-metabolic, post-mitotic tissues. This duality defines the biphasic nature of the mitochondrial molecular clock.
Widespread accumulation of deleterious somatic mutations
In contrast to germline mitochondrial variants, which presumably undergo strong purifying selection, sporadic somatic mutations are much enriched for potentially deleterious effects (Fig. 5a, P < 1 × 10−15, Fisher’s exact test). Deleterious variants are defined as mutations in conserved sites, including loss-of-function (LoF), conserved missense and conserved rRNA sites as predicted by conservation metrics derived from population studies33. These deleterious somatic mutations are rarely observed at the population level (Extended Data Fig. 8) and are found predominantly at low heteroplasmy within individual tissue sites (Fig. 5c).
a, The functional profile of somatic versus germline variants. The bin width is scaled to reflect the typical number of germline and somatic variants (as sampled across 15 body sites as in Fig. 2a) in an individual. b, The age accumulation of somatic mutations by variant type in representative tissues. Variant types are colored as in a. c, The tissue-specific accumulation of deleterious mutations (defined as mutations in conserved sites, including LoF, conserved missense and conserved rRNA sites). This panel demonstrates the preferential accumulation of deleterious mutations in tissues with high cellular turnover, particularly in constantly renewing epithelial compartments. By contrast, high-energy-demanding tissues primarily accumulate mutations at hotspots within the noncoding D-loop region. Age-dependent effects (β) are assessed using linear regression. The error bars represent the 95% CI, which is based on the t-statistic of the corresponding mutation rate β and mean heteroplasmy μ estimates. Tissues in c are color-coded to represent different tissue types, following the standard GTEx conventions. Sample sizes (n, number of individuals) are marked on top of β estimates of each tissue. Detailed statistics for each estimate (slope β, 95% CI, exact P value) are provided in Supplementary Table 2. For total heteroplasmy, box plots display the median (center line), interquartile range (IQR, box limits) and whiskers (extending to 1.5× IQR); the notches provide an approximate 95% CI around the median.
The accumulation of these deleterious mutations is not uniform across tissues. They occur sporadically, particularly in tissues with continuous cellular turnover (Fig. 5b,c), such as the liver (β = 0.098 per year, P = 1 × 10−8), esophageal mucosa (β = 0.029 per year, P = 4.2 × 10−8), and adrenal glands (β = 0.035 per year, P = 9.8 × 10−9). This leads to a higher burden of deleterious mutations in constantly renewing epithelial tissues, potentially contributing to age-related dysfunction.
By contrast, in high-energy-demand tissues such as the heart and kidney, mutations tend to cluster at specific hotspots as previously described, predominantly within the noncoding D-loop region (Fig. 5b,c). This concentration in a noncoding region suggests a potentially less detrimental impact compared with sporadic mutations in renewing tissues. However, the functional consequences of these hotspot mutations on cellular and tissue physiology require further investigation. These findings further highlight the complex interplay of sporadic and hotspot mutations in shaping the landscape of deleterious mitochondrial mutations.
Intriguingly, some predicted LoF variants exhibited unexpectedly high heteroplasmy (>0.5) and population frequency (>0.1%). These included start-codon loss in MT-ND1 (chrM_3308_T_C/G), stop-codon loss in MT-CO1 (chrM_7444_G_A) and another start-codon loss in MT-ATP6 (chrM_8527_A_G). However, further analysis revealed alternative downstream sequences that could potentially compensate for these mutations (Supplementary Fig. 8). In MT-ND1, a second AUG codon downstream could serve as an alternative start site. Similarly, in MT-CO1, the downstream AGG can act as a nonstandard stop codon in mitochondria. In MT-ATP6, GUG has been reported as an alternative start codon43. Thus, these variants might not result in protein function loss, which highlights the unique ability of mitochondria to tolerate certain mutations due to the presence of alternative codons.
Environment and sex affect mitochondrial mutation patterns
Beyond age and tissue, environmental factors and biological sex significantly influence mitochondrial mutational burden and clonal expansion. Our analysis reveals a striking interaction between mitochondrial haplogroup and sun exposure in shaping the mutational landscape of skin. Individuals with non-L haplogroups, typically associated with non-African ancestry and lighter skin pigmentation, exhibit a substantially higher mutation burden in sun-exposed skin compared to those with the L haplogroup (βnon-L: 0.09 per year > βL: 0.02 per year, P = 0.01; Fig. 6a). This disparity probably arises from increased susceptibility to UV-induced damage in individuals with lighter skin, rather than a direct genetic effect of the haplogroup itself. Notably, the age-dependent accumulation of mutations is consistently high in sun-exposed skin across all non-L haplogroups, but not in non-sun-exposed skin (Fig. 6c). Interestingly, while nuclear genomes show a strong UV mutational signature (SBS7a/b) in sun-exposed skin, particularly in non-L haplogroups (Extended Data Fig. 9), mitochondria exhibit very unique mutational spectra across tissues. This pattern is dominated by C > T and T > C transitions, with no typical tobacco (SBS4) or UV (SBS7a/b) mutational signatures even in lung and skin cancer (Extended Data Fig. 9), as previously reported by the TCGA study31. Considering the potential impact of increased cell turnover and clonal expansion35,44 in sun-exposed skin, which could increase the abundance of mutations arising from normal mitochondrial mutagenesis, further research is needed to definitively establish the mechanisms involved in the link between UV exposure and mitochondrial mutation burden in individuals with lighter skin pigmentation.
a, The differential accumulation of mutations in sun-exposed skin by mitochondrial haplogroup (L versus non-L). The fitted regression line shows the rate of mutation accumulation (slope β) in sun-exposed skin between individuals with the L haplogroup (typically associated with African ancestry and higher melanin content) and those of the non-L haplogroup. Shaded areas around the regression lines represent 95% CI of the mutation rate β. b, The mutational burden in mammary and reproductive tissues by sex. The lines show linear regression fits; the shaded areas represent the 95% CI for slope. A comparison was performed via interaction terms (sex and tissue) in the linear regression model. Detailed regression statistics are provided in Supplementary Table 2. c, Mutation rates (β, mutations gained per 10 years, derived from linear regression) of sun-exposed and non-sun-exposed skin across haplogroups, annotated with self-reported ethnicity makeup. The error bars represent the 95% CI of the mutation rate β. Sample sizes (n, number of individuals) are marked on top of β estimates of each group. d, The dosage sensitivity of mitochondrial verus nuclear genetic information processing machinery. LOEUF measures tolerance to LoF mutations, while allelic fold change (aFC) measures tolerance to expression altering cis-regulatory variants, with larger values indicating higher tolerance to dosage change.
Sex also plays a role in shaping the mitochondrial mutational landscape (Supplementary Fig. 9). We observe a substantially higher mutation burden in the breast tissue of females compared with males (female > male, P = 1 × 10−7; Fig. 6b), a difference probably established early in life, possibly during adolescence, as it plateaus after the age of 20. In addition, distinct mutational patterns are evident in male and female reproductive organs, with a notable age-dependent accumulation in the testis but not in the ovary (βtestis: 0.14 per year > βovary: 0.0 per year, P = 1 × 10−8; Fig. 6b). Sex differences in nondimorphic tissues are generally less pronounced than those in sexually dimorphic tissues; minor differences in mutational burdens (P < 0.05, not significant after multiple testing correction; Supplementary Fig. 9) were also observed in heart (male > female), adipose and skin (female > male) tissue, a trend similar to previous findings in nuclear genomes25.
Notably, mitochondrial genetic information processing machinery (replication, transcription and translation) demonstrates remarkable resilience to dosage alterations, contrasting sharply with the sensitivity of its nuclear counterparts. This resilience is indicated by the tolerance of LoF mutations, the predominantly recessive nature of mitochondrial diseases and the presence of large-effect expression quantitative trait loci (eQTLs) at nuclear genes coding those mitochondrial machinery components (Fig. 6d). These findings suggest that mitochondrial quality control mechanisms are robust and less susceptible to dosage-related disruptions. Nevertheless, while dosage may be well buffered, the functional consequences of the specific, age-accumulated mutations we observe across different tissues, and their potential role in age-related decline, warrant further study.
Overall, our findings emphasize the resilience of mitochondrial genetic information processing machinery to genetic variations and gene dosage changes, underscoring the unique robustness of mitochondria compared with nuclear systems. However, environmental factors such as UV exposure and biological sex significantly influence the accumulation and clonal expansion of mitochondrial mutations, highlighting their importance in shaping the tissue-specific landscape over age.
Discussion
Our findings support a biphasic model of mitochondrial molecular clock that governs the accumulation of mutations across human tissues, with distinct mechanisms operating in proliferative and post-mitotic cells. Mitochondria, as ancient endosymbionts within eukaryotic cells, possess their own genomes and exhibit a high degree of clonal diversity due to their large copy numbers and constant turnover. This diversity, shaped by replication errors and ROS damage, has been implicated in aging and disease. Our systematic examination of mitochondrial RNA from 838 individuals reveals that mitochondria indeed function as a mutational clock, recording and potentially driving the aging process across various tissues.
A key finding is the steady accumulation of clonal diversity with age across the body, albeit at varying rates and under distinct driving mechanisms. This observation aligns with previous studies in mice and macaques, which have consistently demonstrated that mitochondrial mutations accumulate with age in various tissues27,29,30. This age-related accumulation is driven primarily by deamination-associated replication errors rather than ROS damage, suggesting that the fidelity of DNA replication may be a critical factor in the aging process28. The pace of clonal expansion mirrors the rate of cellular turnover, particularly evident in rapidly regenerating epithelial tissues such as the gastrointestinal tract and skin, where mutant clones can constitute up to 15% of the mitochondrial population by age 60. This rapid accumulation of genetic errors and subsequent clonal expansions underscore the vulnerability of these tissues to age-related dysfunction and potentially tumorigenesis. This observation is consistent with previous studies in cancer genomes, which have highlighted an age-dependent accumulation of mitochondrial mutations in tumor samples31.
By contrast, nondividing tissues such as the heart and brain exhibit age-accumulated mutations primarily at specific hotspots. These hotspots, probably a consequence of high mitochondrial turnover in energy-intensive tissues8,29, represent a unique aging hallmark. Interestingly, while these tissues also show elevated ROS damage at specific lesions, it does not accumulate with age, which aligns with previous findings in mice28,45. This suggests a robust mitochondrial machinery capable of clearing ROS-induced damage.
Environmental factors and biological sex significantly influence mitochondrial mutational dynamics. Sun exposure increases the mutation burden in lightly pigmented skin (non-L haplogroup), highlighting an interaction between environment and genetics. This elevated mutational burden aligns with findings on nuclear genomes, where UV exposure induces characteristic mutational signatures25,26. However, as mitochondria exhibit distinct mutational spectra dominated by C > T and T > C transitions, with no typical UV-associated signature, it is unclear whether UV radiation directly causes these mitochondrial mutations or indirectly increases the burden through elevated cellular turnover and clonal expansion, which warrants further investigation. Distinct sex-specific patterns are also evident in breast and reproductive tissues, emphasizing the importance of considering sex in understanding tissue-specific aging.
Our findings establish the mitochondrial genome as an informative mutational clock, intricately recording the diverse stressors encountered by different tissues throughout an individual’s lifespan. Previous studies have estimated the average mitochondrial mutation rate to be around 5 × 10−8 per base pair per replication35. Our work further demonstrates that the accumulation of mutations with age varies significantly across tissues. This variation is driven by factors such as cellular mitotic activity, the prevalence of mutational hotspots and the inherent replication dynamics of mitochondria independent of cell division6, creating distinct mutational landscapes in different tissues over time. The finding of this clock’s biphasic nature—with distinct mechanisms operating in dividing versus nondividing tissues—offers unique insight for understanding the multifaceted process of aging, highlighting varying degrees of vulnerability to age-related pathologies. The distinct mutational signatures observed in proliferative and energy-intensive tissues underscore the importance of considering both replicative stress and metabolic demand in understanding the etiology of age-related diseases. Moreover, the identification of age-accelerated mutational hotspots opens avenues for early detection and potential intervention strategies. Further elucidating the precise etiology and functional consequences of these accumulated mutations, and their contribution to cellular and tissue dysfunction, is also of critical importance and demands further investigation, ideally using single-cell multi-omics approaches8,46 to link specific mitochondrial genotypes to cellular phenotypes and aging. By harnessing the power of the mitochondrial clock, we can potentially develop personalized approaches to diagnose, monitor and mitigate age-related decline and dysfunction, ultimately promoting healthier aging and longevity.
Methods
Ethical compliance
The analysis of publicly available data from the GTEx project (accessed via dbGaP accession phs000424.v8.p2), ENCODE project and TCGA project was conducted in accordance with their respective data access policies and the ethical guidelines under which the data were originally collected, including appropriate institutional review board approvals and participant consent obtained by the respective consortia. The analysis of human testicular tissue samples was approved by the Ethics Committee of Shanghai Changhai Hospital (approval no. CHEC2021-086).
Study samples and datasets
Our analysis primarily utilized mitochondrial RNA-seq data from the GTEx project v8 dataset (dbGaP accession phs000424.v8.p2). Tissues with fewer than 70 individuals, as well as cultured fibroblasts and Epstein–Barr virus-transfected lymphocytes, were excluded. Our final dataset encompassed 11,802 samples (after quality control) from 838 individuals, spanning 47 distinct tissues. To validate and compare our findings, we incorporated data from the ENCODE project (https://www.encodeproject.org/) and TCGA project (https://portal.gdc.cancer.gov/) to assess the presence and accumulation of mitochondrial mutation hotspots in an independent cohort.
Statistics and reproducibility
This cross-sectional study analyzed public RNA-seq data from 47 tissues (838 individuals, aged 20–70) from the GTEx v8 cohort (dbGaP: phs000424.v8.p2). No statistical method was used to predetermine sample size; sample sizes were based on data availability for public cohorts (GTEx) and are comparable to those reported in previous studies25,36. Findings were replicated using ENCODE and TCGA data and were validated with targeted mtDNA sequencing from eight human testicular samples (Ethics Committee of Shanghai Changhai Hospital, CHEC2021-086). Samples and variants were excluded on the basis of quality control filters, including checks for contamination, sequencing coverage, error-prone sites and variant call quality. Randomization and blinding were not applied to this observational and computational study design. Statistical analyses, performed using R, primarily involved linear regression to model age effects (assessed via t-statistic for slope β, 95% confidence intervals reported) and Fisher’s exact test for proportions. For linear regression analyses, data distribution was assumed to be normal, but this was not formally tested; data distribution is shown graphically in relevant figures. Significance was generally defined as P < 0.05, with multiple testing correction (Bonferroni or FDR) applied where appropriate. All statistical tests were two-sided, unless noted otherwise.
Mitochondrial variant calling and annotation
Mitochondrial variant calling and annotation were conducted using RNA-seq data from the GTEx project, following a workflow similar to the mtDNA variant calling pipeline utilized in the Genome Aggregation Database (gnomAD)33. Raw sequencing reads were initially aligned to the human reference genome GRCh38 with STAR v2.5.3a, in accordance with the GTEx Consortium pipeline. Reads mapping to the mitochondrial genome (chrM) were extracted and realigned to chrM using the Burrow-Wheeler Aligner47, retaining only uniquely mapped reads to mitigate the impact of nuclear sequences of mitochondrial origin (NUMTs). After marking duplicates, the Mutect248 mitochondrial mode was applied for variant calling, requiring a minimum base quality of 18. Considering the circular nature of mtDNA and the limitation of BWA-MEM supporting mapping to the linear genome, which results in low read coverage in the D-loop region and less accurate variant calling, we implemented a two-step mapping and calling process. First, reads were directly mapped to the rCRS mtDNA sequence49 to identify variants in chrM:576–16024. Second, the mtDNA sequence was split at the 8,000th base to create a new linear genome, and reads were remapped to this modified reference to identify variants in chrM:1–575 and chrM:16025–16569. These two sets of variants were then combined and subjected to flagging and filtering using GATK FilterMutectCalls.
Population frequencies of variants were obtained from the gnomAD database33. Variants were annotated utilizing Ensembl’s Variant Effect Predictor (VEP) v10150. Variants classified as stop_gained, start_lost and stop_lost were designated as LoF. Local sequence constraint metrics (MLC) were derived from gnomAD, considering scores higher than 0.7 as conserved. Moreover, pathogenic variants were annotated according to the MITOMAP51 database. Haplogroups were assigned using Haplocheck52.
Removal of contaminated samples
To minimize the risk of erroneous variant calls due to potential cross-contamination between samples, we implemented a multistep filtering strategy. Initially, samples flagged as potentially contaminated by Haplocheck were removed. In addition, we used a negative binomial regression model to identify and remove outliers in mutation counts, accounting for covariates such as tissue, age, sex, haplogroup, ischemic time and cohort as follows:
Samples with residuals exceeding the population norm (P < 0.001) were considered potentially contaminated and excluded. Finally, to ensure sufficient sequencing depth for accurate variant detection, samples with inadequate coverage were removed. While most samples had adequate RNA coverage across protein-coding, rRNA and D-loop regions, those with less than 200× coverage at over 10% of the 16,569 mitochondrial genome positions were excluded from further analysis.
Removal of error-prone sites
We applied multiple filters to eliminate sites vulnerable to technical artifacts: (1) exclusion of blacklisted regions, encompassing positions 66–71, 300–316, 513–526, 566–573, 3,106–3,107, 12,418–12,425 and 16,181–16,194 (refs. 32,33,53) (these regions harbor homopolymer tracts that may induce polymerase slippage during polymerase chain reaction, leading to sequencing errors); (2) exclusion of known RNA modification sites39,40(Supplementary Fig. 2), including positions 295, 2,617 and 13,710; (3) exclusion of sites that may be affected by NUMTs38 (these sites were identified by mapping NUMT sequences to the mitochondrial genome and subsequently removed37); and (4) exclusion of all tRNA regions and gene boundary regions that are inadequately covered by polyA RNA libraries. To account for the low coverage observed at gene boundaries in RNA-seq, we trimmed bases from the ends of gene regions where coverage fell below 200×.
Quality control of mitochondrial variant calling
To enhance the reliability of our variant calls, we implemented several filtering steps. First, we excluded all indels, multiallelic sites and variants located within 3 bp of read ends. Moreover, variant calls exhibiting strand bias were filtered out. Strand bias was assessed using Fisher’s exact test, comparing allele-specific forward and reverse read counts. If the lower bound of confidence interval of odds ratio exceeded 2, the variant was flagged for strand bias.
In addition, to further differentiate variants from background sequencing errors, we adopted the same statistical approach used in previous studies24. We estimated an empirical error rate by counting the frequency of alternative alleles at all sites using BCFtools mpileup. Low-frequency alternative alleles (<1%), primarily attributed to sequencing errors, were modeled using a beta-binomial distribution. This analysis revealed a background error rate of approximately 5 × 10−4 in the GTEx data (Supplementary Fig. 1), consistent with estimates by previous GTEx studies25. Variants output by Mutect2 were then further filtered on the basis of their deviation from this distribution. To minimize false-positive calls, we applied an allele fraction cutoff of 0.005. Only variants with an allele fraction >0.005, representing a significant deviation from the sequencing error rate of 5 × 10−4 (P < 1 × 10−7, FDR <0.05 after Benjamini–Hochberg multiple testing correction), were considered reliable calls.
Assessing factors influencing mitochondrial clonal dynamics
To determine the factors contributing to clonal dynamics, we used a linear regression model with mutation count or heteroplasmy level as the response variable. The model included covariates such as tissue type, age, sex, macro-haplogroup, tissue ischemic time and cohort group (postmortem versus organ donor). The significance of the interaction between each covariate (sex, tissue type, environmental exposure and so on) and age on the rate of mutation accumulation was evaluated using the interaction term in the linear regression model. Reported confidence intervals are based on the t-statistic of the corresponding mutation rate β and mean heteroplasmy μ estimates.
Recognizing the substantial variation in read coverage across tissues, we repeated the analysis by subsampling the coverage to a uniform depth of 1,000×. This step ensured that observed differences in mutation counts between tissues were not simply artifacts of varying sequencing depths.
We assessed mutational signatures and their proposed etiologies using the SigProfileAssignment54 tool to identify matches to known COSMIC signatures55.
Cell type enrichment assessment among bulk tissue samples
To investigate the association between cell type composition and mitochondrial mutation burden, we used xCell56, a method used by the GTEx consortium for cell-type-specific eQTL analysis, to assess cell type enrichment across tissues (excluding the brain). xCell enables enrichment analysis for 64 immune and stromal cell types based on gene expression data. We focused on the ten most abundant cell types, including endothelial cells, epithelial cells, fibroblasts, hepatocytes, keratinocytes, mesenchymal stem cells, myocytes, sebocytes, adipocytes and neutrophils.
For brain tissues, we utilized CIBERSORT57, a deconvolution algorithm using support vector regression, to achieve a more granular breakdown of specific cell types. CIBERSORT, using customizable reference gene expression signatures, allowed us to infer the composition of astrocytes, endothelia, microglia, neurons and oligodendrocytes within the original expression profile of the brain tissues.
Dosage constraint in genetic information processing genes
We evaluated gene dosage constraint using the LoF observed/expected upper-bound fraction (LOEUF) score58. For a tissue-specific assessment, we utilized our recently developed metric, MoDs (tissue-specific dosage constraint)59, which quantifies the sensitivity of gene expression to gene dosage changes based on the presence of large-effect cis-regulatory variants. Genes involved in genetic information processing were curated from the Kyoto Encyclopedia of Genes and Genomes (KEGG)60 and Mitocarta61 databases. Specifically, mitochondrial genetic information processing genes were selected on the basis of their classification in Mitocarta (‘mtDNA maintenance’, ‘mtRNA metabolism’ and ‘Translation’) and KEGG (‘Mitochondrial transcription and translation factors’ and ‘Mitochondrial DNA replication factors’). Nuclear genetic information processing genes were identified on the basis of their categorization in KEGG (‘DNA replication proteins’, ‘DNA repair and recombination proteins’, ‘Translation factors’, ‘Messenger RNA biogenesis’ and ‘Transcription machinery’). Inheritance modes associated with genes linked to Mendelian diseases were determined using the Online Mendelian Inheritance in Man (OMIM) database.
Validation of mitochondrial mutation hotspots in testicular tissue samples
To independently validate our findings from the analysis of bulk RNA-seq data, we performed targeted mtDNA sequencing on testicular tissue samples obtained from eight individuals. These tissues were collected with ethical approval (approval number CHEC2021-086) from patients undergoing orchiectomy for various medical conditions at Shanghai Changhai Hospital between 2021 and 2022. Each patient provided informed consent for the use of their discarded testicular tissue. Genomic DNA was extracted from the tissues using the QIAamp DNA Micro Kit (Qiagen). The complete mitochondrial genome was enriched using the xGen Human mtDNA Hyb Panel (Integrated DNA Technologies), which utilizes biotinylated probes to capture the entire mitochondrial genome. Captured DNA was then prepared for sequencing using the KAPA Hyper Prep Kit (Roche), following the manufacturer’s protocol. Sequencing was performed on the GeneMind SURFSeq 5000 platform, generating high-coverage data (>10,000×) for accurate variant calling. Samples were assigned deidentified study IDs (format: TST-R-NN; Supplementary Table 3). The original sequencing data are deposited at the National Omics Data Encyclopedia (NODE) at https://www.biosino.org/node, under the accession number OEP00006064.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The GTEx project v8 RNA-seq and WGS data are available from dbGaP (dbGaP: phs000424.v8.p2). GTEx (v8) eQTL summary statistics were obtained from the GTEx Portal (https://gtexportal.org/home/datasets). RNA-seq data from the ENCODE project used for validation are publicly available via the ENCODE portal (https://www.encodeproject.org/). RNA-seq data from nontumor samples from TCGA used for validation are publicly available via the GDC portal (https://portal.gdc.cancer.gov/). The human testicular tissue targeted mtDNA sequencing data generated in this study have been deposited in the National Omics Data Encyclopedia (NODE, https://www.biosino.org/node) under accession number OEP00006064.
Code availability
Custom code used for the analysis presented in this study is publicly available via GitHub and has been archived with persistent identifiers. The code pipeline used for mitochondrial variant calling, subsequent analyses and figure generation (mitoclock) is available via GitHub at https://github.com/xlilab/mitoclock and via Zenodo at https://doi.org/10.5281/zenodo.15256270 (ref. 62). The code for calculating the tissue-specific dosage sensitivity metric (MoDs) is available via GitHub at https://github.com/xlilab/MoDs and via Zenodo at https://doi.org/10.5281/zenodo.15256295 (ref. 63).
References
Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 14, 255–274 (1967).
Anderson, S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981).
Khrapko, K. et al. Mutational spectrometry without phenotypic selection: human mitochondrial DNA. Nucleic Acids Res. 25, 685–693 (1997).
Khrapko, K. et al. Mitochondrial mutational spectra in human cells and tissues. Proc. Natl Acad. Sci. USA 94, 13798–13803 (1997).
Wai, T. et al. The role of mitochondrial DNA copy number in mammalian fertility. Biol. Reprod. 83, 52–62 (2010).
Lawless, C., Greaves, L., Reeve, A. K., Turnbull, D. M. & Vincent, A. E. The rise and rise of mitochondrial DNA mutations. Open Biol. 10, 200061 (2020).
Pickles, S., Vigie, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 (2018).
Glynos, A. et al. High-throughput single-cell analysis reveals progressive mitochondrial DNA mosaicism throughout life. Sci. Adv. 9, eadi4038 (2023).
Elson, J. L., Samuels, D. C., Turnbull, D. M. & Chinnery, P. F. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am. J. Hum. Genet. 68, 802–806 (2001).
Chinnery, P. F. & Samuels, D. C. Relaxed replication of mtDNA: a model with implications for the expression of disease. Am. J. Hum. Genet. 64, 1158–1165 (1999).
Stewart, J. B. & Chinnery, P. F. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16, 530–542 (2015).
Durham, S. E., Samuels, D. C., Cree, L. M. & Chinnery, P. F. Normal levels of wild-type mitochondrial DNA maintain cytochrome c oxidase activity for two pathogenic mitochondrial DNA mutations but not for m.3243A–>G. Am. J. Hum. Genet. 81, 189–195 (2007).
Greaves, L. C. et al. Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 10, e1004620 (2014).
Stewart, J. B. & Chinnery, P. F. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat. Rev. Genet. 22, 106–118 (2021).
Dolle, C. et al. Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat. Commun. 7, 13548 (2016).
Grimm, A. & Eckert, A. Brain aging and neurodegeneration: from a mitochondrial point of view. J. Neurochem. 143, 418–431 (2017).
Lin, M. T., Simon, D. K., Ahn, C. H., Kim, L. M. & Beal, M. F. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum. Mol. Genet. 11, 133–145 (2002).
Kenney, M. C. et al. Characterization of retinal and blood mitochondrial DNA from age-related macular degeneration patients. Invest. Ophthalmol. Vis. Sci. 51, 4289–4297 (2010).
Nomiyama, T. et al. Accumulation of somatic mutation in mitochondrial DNA extracted from peripheral blood cells in diabetic patients. Diabetologia 45, 1577–1583 (2002).
Matam, K. et al. Evidence for the presence of somatic mitochondrial DNA mutations in right atrial appendage tissues of coronary artery disease patients. Mol. Genet. Genomics 289, 533–540 (2014).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Lima, T., Li, T. Y., Mottis, A. & Auwerx, J. Pleiotropic effects of mitochondria in aging. Nat Aging 2, 199–213 (2022).
Kakiuchi, N. & Ogawa, S. Clonal expansion in non-cancer tissues. Nat. Rev. Cancer 21, 239–256 (2021).
Garcia-Nieto, P. E., Morrison, A. J. & Fraser, H. B. The somatic mutation landscape of the human body. Genome Biol. 20, 298 (2019).
Rockweiler, N. B. et al. The origins and functional effects of postzygotic mutations throughout the human life span. Science 380, eabn7113 (2023).
Yizhak, K. et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science https://doi.org/10.1126/science.aaw0726 (2019).
Arbeithuber, B. et al. Age-related accumulation of de novo mitochondrial mutations in mammalian oocytes and somatic tissues. PLoS Biol. 18, e3000745 (2020).
Sanchez-Contreras, M. et al. The multi-tissue landscape of somatic mtDNA mutations indicates tissue-specific accumulation and removal in aging. eLife https://doi.org/10.7554/eLife.83395 (2023).
Serrano, I. M. et al. Mitochondrial haplotype and mito-nuclear matching drive somatic mutation and selection throughout ageing. Nat. Ecol. Evol. 8, 1021–1034 (2024).
Arbeithuber, B. et al. Advanced age increases frequencies of de novo mitochondrial mutations in macaque oocytes and somatic tissues. Proc. Natl Acad. Sci. USA 119, e2118740119 (2022).
Yuan, Y. et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 52, 342–352 (2020).
Li, M., Schroder, R., Ni, S., Madea, B. & Stoneking, M. Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc. Natl Acad. Sci. USA 112, 2491–2496 (2015).
Laricchia, K. M. et al. Mitochondrial DNA variation across 56,434 individuals in gnomAD. Genome Res. 32, 569–582 (2022).
Kauppila, T. E. S., Kauppila, J. H. K. & Larsson, N. G. Mammalian mitochondria and aging: an update. Cell Metab. 25, 57–71 (2017).
An, J. et al. Mitochondrial DNA mosaicism in normal human somatic cells. Nat. Genet. 56, 1665–1677 (2024).
Consortium, G. T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Battle, S. L. et al. A bioinformatics pipeline for estimating mitochondrial DNA copy number and heteroplasmy levels from whole genome sequencing data. NAR Genom. Bioinform. 4, lqac034 (2022).
Wei, W. et al. Nuclear-embedded mitochondrial DNA sequences in 66,083 human genomes. Nature 611, 105–114 (2022).
Bar-Yaacov, D. et al. RNA–DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA. Genome Res. 23, 1789–1796 (2013).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Sender, R. & Milo, R. The distribution of cellular turnover in the human body. Nat. Med. 27, 45–48 (2021).
Zheng, W., Khrapko, K., Coller, H. A., Thilly, W. G. & Copeland, W. C. Origins of human mitochondrial point mutations as DNA polymerase gamma-mediated errors. Mutat. Res. 599, 11–20 (2006).
Dubot, A. et al. GUG is an efficient initiation codon to translate the human mitochondrial ATP6 gene. Biochem. Biophys. Res. Commun. 313, 687–693 (2004).
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Kennedy, S. R., Salk, J. J., Schmitt, M. W. & Loeb, L. A. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 9, e1003794 (2013).
Lareau, C. A. et al. Mitochondrial single-cell ATAC-seq for high-throughput multi-omic detection of mitochondrial genotypes and chromatin accessibility. Nat. Protoc. 18, 1416–1440 (2023).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Andrews, R. M. et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147 (1999).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Brandon, M. C. et al. MITOMAP: a human mitochondrial genome database–2004 update. Nucleic Acids Res. 33, D611–D613 (2005).
Weissensteiner, H. et al. Contamination detection in sequencing studies using the mitochondrial phylogeny. Genome Res. 31, 309–316 (2021).
Wei, W. et al. Germline selection shapes human mitochondrial DNA diversity. Science https://doi.org/10.1126/science.aau6520 (2019).
Diaz-Gay, M. et al. Assigning mutational signatures to individual samples and individual somatic mutations with SigProfilerAssignment. Bioinformatics https://doi.org/10.1093/bioinformatics/btad756 (2023).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017).
Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M. & Alizadeh, A. A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 1711, 243–259 (2018).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Dong, D. et al. An RNA-informed dosage sensitivity map reflects the intrinsic functional nature of genes. Am. J. Hum. Genet. 110, 1509–1521 (2023).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Wang, Z. xlilab/mitoclock: v1.0.0 (v1.0.0). Zenodo https://doi.org/10.5281/zenodo.15256270 (2025).
Dong, D. xlilab/MoDs: v1.0.0 (v1.0.0). Zenodo https://doi.org/10.5281/zenodo.15256295 (2025).
Enge, M. et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330 (2017).
Acknowledgements
X.L. is supported by National Key R&D Program of China (grant nos. 2021YFA0805200 and 2024YFC2707000) and National Natural Science Foundation of China (grant no. 32470637). C.Y. is supported by grants from the National Natural Science Foundation of China (grant no. 82373011). We thank L. Ma and X. Zheng from the Institutional Center for Shared Technologies and Facilities of SINH, CAS for their assistance with computational resources and support.
Author information
Authors and Affiliations
Contributions
X.L. conceived and supervised the study. Z.W. performed the primary computational analyses, curated the data, developed analysis software, created visualizations and co-wrote the initial paper draft. Z.L. and H.L. contributed to methodology development, software validation and data analysis support. C.Y. provided resources (clinical samples for validation) and contributed to the related investigation. All authors contributed to the review and editing of the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Workflow of mitochondrial variant calling from RNA-seq.
This flowchart outlines the step-by-step process for identifying and quantifying mitochondrial variants from RNA sequencing data, including alignment, variant calling (a), quality control, and filtering procedures (b).
Extended Data Fig. 2 Age and tissue distribution of analyzed samples.
Overview of sample characteristics for the 838 GTEx v8 donors included in the study across 47 tissues. The figure displays the distribution of donor age (years), sex (female/male ratio), tissue ischemic time (hours), and donor cohort type (surgical/organ donor/postmortem) for each tissue analyzed. The number of individuals available for each tissue is indicated. Violin plots illustrate the density distribution of the data, while overlaid box plots indicate the median (center line), interquartile range (IQR, box limits), and whiskers (extending to 1.5x IQR). Tissues are colored according to the GTEx project conventions.
Extended Data Fig. 3 Number of somatic mitochondrial mutations across tissues and age groups.
The figure presents scatter plots showing the distribution of the total count of somatic mitochondrial mutations identified in each tissue of an individual, stratified by age. Each dot represents the mutation count for an individual. Linear regression lines are shown for each plot, with the corresponding regression coefficient (β) and P-value (P) indicated. Shaded areas around the regression lines represent the confidence intervals (CI) for β.
Extended Data Fig. 4 Total heteroplasmy of somatic mitochondrial mutations across tissues and age groups.
Each scatter plot shows the combined abundance (total heteroplasmy) of all detected somatic mitochondrial mutations for individuals within a specific tissue stratified by age. Each dot represents one individual. Linear regression lines are shown for each plot, with the corresponding regression coefficient (β) and P-value (P) indicated. Shaded areas around the regression lines represent the confidence intervals (CI) for β.
Extended Data Fig. 5 Proportions of variants shared by tissues from the same germ layer.
This figure displays the proportion of shared mitochondrial variants among tissues originating from the same germ layer (endoderm, mesoderm, ectoderm). These proportions are compared against a null distribution (gray violins) generated by 1000 random permutation of tissue labels, demonstrating a significant enrichment of shared variants within tissues of the same developmental origin. Violin plots illustrate the density distribution of random permutations (null distribution), while overlaid box plots indicate the median (center line), interquartile range (IQR, box limits), and whiskers (extending to 1.5x IQR).
Extended Data Fig. 6 Distribution of shared somatic mitochondrial mutations across tissues.
This figure displays the number of somatic mutations found in two or more tissues. To investigate the potential developmental origins of these shared mutations, they were categorized based on whether the tissues arise from the same germ layer (endoderm, mesoderm, or ectoderm) or from different germ layers. This analysis helps to understand the potential developmental stage at which these mutations first appeared. Hotspot mutations were analyzed separately, as they may arise independently in different tissues rather than reflecting a shared lineage.
Extended Data Fig. 7 ROS damage in all tissues after read depth subsampling.
This figure illustrates the prevalence of ROS-induced damage across various tissues, with sequencing depth subsampled to 1000x to account for potential discovery bias due to read depth variation. While the estimation is noisier due to reduced read depth, the overall trend aligns with the observation in Fig. 4a: tissues with higher energy demands exhibit a greater degree of ROS damage. Sample sizes (n, number of individuals) are marked on top of each tissue. Boxplots display the median (center line), interquartile range (IQR, box limits), and whiskers (extending to 1.5x IQR); The notches provide an approximate 95% confidence interval around the median.
Extended Data Fig. 8 Population allele frequencies versus maximum heteroplasmy across tissues.
This figure compares the population allele frequencies of mitochondrial variants in gnomAD with their maximum heteroplasmy observed across all GTEx tissues. Stacked bar charts show the distribution of maximum body-wide heteroplasmy levels (observed across 47 GTEx tissues for 838 individuals) for mitochondrial variants within different population allele frequency bins (derived from gnomAD data). Variants are categorized by functional impact (D-loop, rRNA, Synonymous, Missense, LoF).
Extended Data Fig. 9 Comparison of mutational signatures in nuclear and mitochondrial genomes across normal and tumor tissues.
COSMIC mutational signatures were determined using the SigProfileAssignment tool on TCGA tumor samples and GTEx normal tissues. For GTEx sun-exposed skin, samples were stratified into L and non-L groups based on their mitochondrial haplogroup. While nuclear genomes exhibit established COSMIC signatures, reflecting various mutagenic processes, mitochondrial genomes display distinct mutational spectra dominated by C > T and T > C transitions, with no typical tobacco and UV mutational signatures even in lung and skin cancer.
Extended Data Fig. 10 Validation of age-associated mitochondrial mutation hotspots via targeted DNA sequencing.
a, Scatter plots depict the relationship between chronological age and heteroplasmy levels for five mitochondrial DNA hotspot mutations (chrM:13369, chrM:13676, chrM:152, chrM:2623, chrM:368) in human testicular tissue samples (n = 8 individuals), analyzed via targeted DNA sequencing ( > 10,000x coverage; see Methods). These hotspots were previously identified as showing significant age-related increases in heteroplasmy in the GTEx testis samples (see Fig. 3 and Supplementary Table 3). Linear regression analysis was performed to evaluate the trend of heteroplasmy change with age at each site. Hotspot chrM:9209 (potentially due to post-transcriptional RNA modification or other non-DNA mechanisms as suggested by previous findings, see Supplementary Fig. 2), was not detected among the testicular DNA samples. b, Heteroplasmy levels and corresponding read counts for the five validated hotspot mutations and associated metadata for the testicular tissue samples. Data are presented as: heteroplasmy (reference allele read count, alternative allele read count). Samples are identified by de-identified study IDs (TST-R-NN; see Methods for details). Variants identified as germline (chrM:152_T_C in samples TST-K-01, TST-B-02 and TST-F-06) are indicated with an asterisk and were excluded from the age-heteroplasmy analysis in Panel a.
Supplementary information
Supplementary Information
Supplementary Figs. 1 (background error rate), 2 (RNA–DNA differences), 3 (hotspot frequency heatmap), 4 (age versus heteroplasmy at hotspots), 5 (hotspot replication ENCODE), 6 (hotspot replication TCGA), 7 (nonlinear accumulation), 8 (rescued LoF) and 9 (sex differences) with associated legends
Supplementary Table 1
Complete list of mitochondrial variants called across all samples. This supplementary table provides a comprehensive catalog of all identified mitochondrial variants, including their genomic coordinates, heteroplasmy levels and functional annotations.
Supplementary Table 2
Summary statistics for the age-accumulating effect of mitochondrial somatic mutations. This table provides a comprehensive summary of the statistical analyses examining the relationship between age and mitochondrial somatic mutations across different tissues and mutation types. β: indicates the estimated rate of change in mutation count or heteroplasmy per year of age. P value: represents the statistical significance of the age effect. 95% CI: provides the 95% confidence interval for β.
Supplementary Table 3
Heteroplasmy levels at tissue-specific mitochondrial mutation hotspots in GTEx, ENCODE, TCGA and testicular tissue datasets. This table provides the heteroplasmy levels observed at the 22 tissue-specific mitochondrial mutation hotspots identified in our study (see Fig. 3b for hotspot definitions) across four independent datasets: GTEx (v8, RNA-seq data), ENCODE (RNA-seq data), TCGA (RNA-seq data from nontumor samples) and the testicular tissue samples (targeted DNA sequencing). Testicular tissue samples are identified by deidentified study IDs (format: TST-R-NN; see the Methods for details). For GTEx, ENCODE and TCGA, sample IDs and tissue types are provided according to each database’s original metadata.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Z., Li, Z., Liu, H. et al. Mitochondrial clonal mosaicism encodes a biphasic molecular clock of aging. Nat Aging 5, 1637–1651 (2025). https://doi.org/10.1038/s43587-025-00890-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-00890-6