Abstract
Circulating metabolites can identify biochemical risk factors related to Alzheimer’s disease (AD). We measured plasma metabolites in 1,068 participants of Caribbean Hispanic ancestry (250 patients with AD and 818 healthy controls) across 2 cohorts and analyzed their relationship with clinical AD, biomarker-supported AD and plasma biomarkers (P-tau181, P-tau217, P-tau231 and Aβ42:Aβ40). Amino acid metabolism pathways were enriched among metabolites associated with P-tau biomarkers, whereas sialic acid and N-glycan pathways were associated with Aβ42:Aβ40. Through several dimensionality reduction approaches, we identified an APOE-ε4 dependent relationship between lysophosphatidylcholines (lysoPCs) carrying polyunsaturated fatty acids and biomarker-supported AD and P-tau biomarkers. In an independent dataset of 110 postmortem brain tissues from non-Hispanic white participants, lysoPCs in the brain were also associated with AD neuropathological features. Our results show that biomarker-based diagnostic criteria identified an APOE-ε4 dependent association with lysoPCs, which play a critical role in the transport of neuroprotective polyunsaturated fatty acids into the brain, and AD.
Similar content being viewed by others
Main
AD is a progressive neurodegenerative disorder characterized by cognitive and memory decline, affecting millions of individuals worldwide. Despite extensive research, there are still many unanswered questions regarding the underlying pathogenic mechanisms of AD, hindering the development of effective therapeutic strategies; however, recent advancements in high-throughput omics technologies have provided a powerful platform to explore the complex molecular landscape of AD1.
Profiling the metabolome using high-resolution mass spectrometry offers a comprehensive analysis of small molecules involved in cellular metabolism. It provides a unique opportunity to study metabolic alterations associated with disease pathogenesis, thus contributing to a better understanding of AD at the molecular level2,3,4,5. Metabolomic studies in AD have revealed a range of altered metabolic signaling. Several studies have demonstrated dysregulation of energy metabolism pathways in AD6,7,8. These include altered glucose metabolism9, reduced glycolysis10,11,12,13,14, impaired mitochondrial function15,16,17 and decreased levels of metabolites such as glucose, lactate and pyruvate18. Alterations in the tricarboxylic acid (TCA) cycle intermediates have also been observed. Studies have reported lower levels of phosphatidylcholine, phosphatidylethanolamine and sphingomyelins in AD19,20,21,22, suggesting disruptions in membrane integrity and signaling pathways as well as altered cholesterol metabolism. Other studies have uncovered alterations in amino acid metabolism in AD23. Reduced levels of certain amino acids, such as tryptophan24, phenylalanine25,26, tyrosine27,28 and branched-chain amino acids (valine, leucine and isoleucine)3,29,30,31,32,33,34,35,36,37,38 may reflect disruptions in neurotransmitter synthesis, neuroinflammation and protein homeostasis. Studies have also shown alterations in the levels of neurotransmitters such as acetylcholine, glutamate and γ-aminobutyric acid (GABA) in patients with AD39,40,41,42,43. These changes may contribute to cognitive dysfunction and synaptic alterations in the disease. Several groups have reported elevated levels of reactive oxygen species and oxidative damage markers, and alterations in antioxidant metabolites and enzymes in whole blood and brains of patients with AD. These findings suggest a role for altered redox status in AD pathogenesis44,45,46,47.
Given that metabolomics is the omics layer closest to the phenotype, it has the potential to uncover critical insights into the disease risk and progression and potentially uncover therapeutic targets; however, previous studies do not resolve the role of metabolites and metabolic pathways in different stages of the disease. By integrating metabolomics data with clinical diagnosis and plasma biomarker levels of AD, we aim to identify metabolic networks underlying the stages of the disease.
In this study, we investigated the associations between metabolites and clinical and biomarker-supported diagnosis of AD, using published plasma P-tau18148 and P-tau217 cutoffs for diagnosis. We also investigated the associations between metabolites and P-tau181, P-tau217, P-tau231 and Aβ42:Aβ40 ratio. Previous studies have shown that cerebrospinal fluid (CSF) and plasma P-tau181 are elevated 15–18 years49 before onset of clinical symptoms. Our analysis of metabolomics in relation to clinical and plasma biomarker-based diagnosis of AD could shed light on early and mid-stage metabolic changes in disease.
Results
Study participants
A total of 717 participants were included in the study, of whom 150 (20.9%) were diagnosed with clinical AD and 567 were cognitively unimpaired controls (Table 1). The study population had a mean ± s.d. age of 69.6 ± 7.6 years, the individuals with clinical AD were slightly older, with a mean ± s.d. age of 73.2 ± 8.3 years), compared to controls who had a mean ± s.d. age of 68.6 ± 7.2 years. Two-thirds of the group were women (65%), and this proportion was similar among patients with AD (67%) and controls (65%). A third of the study group had at least one APOE ε4 allele (38%) and this proportion was only marginally higher in AD (43%) compared to controls (36%). Among AD, 58% were biomarker positive, while 29% of controls were biomarker positive. The mean levels of most plasma-based AD biomarkers were higher in AD than in controls, including P-tau181 (3.02 ± 1.7 pg ml−1 in AD and 2.13 ± 1 pg ml−1 in healthy controls), P-tau217 (0.67 ± 0.56 pg ml−1 in AD and 0.36 ± 0.24 pg ml−1 in healthy controls) and P-tau231 (4.43 ± 2.49 pg ml−1 in AD and 3.4 ± 1.71 pg ml−1 in healthy controls). The mean ratio of Αβ42:Αβ40 was lower in AD cases (0.049 ± 0.01) compared to healthy controls (0.053 ± 0.03). A subset of the study population (n = 113) also had CSF metabolomic data generated (Extended Data Table 1). Among them, 35 were clinically diagnosed with AD and 78 were controls. We also used clinical, plasma metabolomics and biomarker data the Washington Heights, Hamilton Heights and Inwood Community Aging Project (WHICAP) for replication, n = 351. We used data available from participants of Caribbean Hispanic ethnicity, 100 of whom were clinically diagnosed with AD and 251 were healthy controls (Extended Data Table 2). Further, we obtained postmortem brain tissue metabolomic data available in a subset of participants of the Religious Orders Study and Memory and Aging Project (ROSMAP) cohort, n = 110 (Extended Data Table 3). Of them, 71 were diagnosed with AD and had brain pathology information available.
Metabolome-wide association study
We detected 6,445 and 5,827 metabolic features in the HILIC+ and C18− columns, respectively. Restricting to metabolic features detected in at least 70% of all participants, 3,253 and 3,628 features were retained for further analysis. Overall, 442 features were associated with at least one phenotype (clinical diagnosis of AD, biomarker-supported AD, plasma levels of Αβ42:Αβ40 ratio, P-tau181, P-tau217 and P-tau231). Of those, 93 features were annotated with level 1 to level 3 confidence based on Schymanski scale (Fig. 1, Table 2 and Supplementary Table 1). A network visualization of associated metabolic features and feature classes is summarized in Fig. 2a.
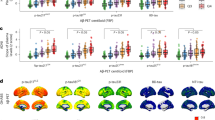
a, Modified Miami plot shows features with positive β values above the zero line and those with negative β values below the zero line. The dark blue points indicate features with FDR q-value < 0.05 for data obtained for each column (C18 and HILIC). NS, not significant. b, Metabolic pathways, with Fisher’s exact test P < 0.05, enriched by features nominally associated with the clinical AD, biomarker-supported AD and plasma biomarker.
a, Network plot showing connections between metabolites (colored by their super class membership) and outcomes (labeled). Solid lines indicate positive associations, and dotted lines indicate negative associations. b, Overlap in features associated with the three P-tau biomarkers and clinical AD at FDR q-value < 0.05 for each column, C18− and HILIC+. c, Levels of metabolites related to the microbial metabolism of tryptophan that were associated with all outcomes in clinical (n = 150 cases and 567 controls) and biomarker-supported AD (n = 191 cases and 492 controls). d, Levels of a lysoPC (20:4) and a phosphatidylcholine (16:1; 22:6) associated with all P-tau biomarkers in clinical and biomarker-supported AD. e, Levels of amino acid metabolites associated with P-tau biomarkers in clinical and biomarker-supported AD. For c–e, the violin plot shows the distribution of the levels, the point indicates the median, the line below the point indicates the 25th percentile and the line above the point indicates the 75th percentile.
We identified 77 metabolic features associated (false discovery rate (FDR) q-value < 0.05) with both clinical AD and biomarker-supported AD. Metabolites associated with biomarker-supported AD were enriched in pathways related to amino acid metabolism and the urea cycle. Tryptophan metabolism and drug metabolism (other enzymes) pathways were enriched by metabolites associated with clinical AD. The top metabolites associated with clinical AD included phenylacetylglutamine (q-value = 0.04, level 1 confidence in annotation), dodecanoyl carnitine (q-value = 0.01, level 3) and tryptophan (q-value = 0.04, level 3). The top metabolites associated with biomarker-supported AD included creatinine (q-value = 0.01, level 1), valyl-serine (q-value = 0.03, level 1) and phenylacetylglutamine (q-value = 0.03, level 1). 143 metabolites with a level 1–3 confidence score for annotation were associated (q-value < 0.05) with at least one measured plasma biomarker (Fig. 1 and Supplementary Table 1). The top metabolites associated with Αβ42:Αβ40 ratio included 3-oxododecanoic acid (q = 0.01, level 3) and pyridoxamine 5-phosphate (q = 6.3 × 10−6, level 5). Metabolic features associated with Αβ42:Αβ40 ratio were enriched in sialic acid metabolism, N-glycan degradation, TCA cycle, glycosphingolipid, glycerophospholipid and galactose metabolism (Fig. 1b).
Valyl-serine, creatinine and citrulline were among the 24 metabolic features, annotated with level 1 confidence, that were associated with plasma levels of P-tau181. Several lysoPCs, including lysoPC (22:6), lysoPC (18:0) and lysoPC (20:4) were negatively associated with plasma P-tau181 levels after multiple testing correction (Supplementary Table 1). Metabolic features associated with P-tau181 levels were enriched in several essential amino acid metabolism pathways including tyrosine, arginine, proline, methionine, cysteine, glycine, serine, threonine, alanine, aspartate, asparagine and lysine metabolism. Other pathways included urea cycle/amino group metabolism, selenoamino metabolism and glutathione metabolism (Fig. 1b).
Comparing metabolites associated with AD and its biomarkers
Metabolites were exclusively associated with Αβ42:Αβ40 ratio with no overlap with metabolites associated with AD or P-tau biomarkers. Tryptophan was associated with clinical AD, P-tau181 and P-tau217, and phenylacetylglutamine was associated with both clinical AD and P-tau181. Several metabolites that derive from microbial metabolism of tryptophan in the gut were associated with P-tau181 and P-tau217, including 1H-indole-3-carboxaldehyde (level 3), indoleacetaldehyde (level 5) and indoleacrylic acid (level 5). Indoxyl sulfate (level 1), a bacterial co-metabolite that is a known uremic toxin50, was associated with P-tau181, P-tau217 and P-tau231 (Fig. 2c). Other metabolites associated with all three P-tau biomarkers included creatinine (level 1), leucyl-proline (level 1) and 2-aminoadipic acid (level 1). LysoPC (20:4) (level 3), with an arachidonic acid side chain, and a phosphatidylcholine (16:1, 22:6) (level 3), with palmitoleic acid and docosahexaenoic acid side chains, were associated with both P-tau181 and P-tau217 (Fig. 2c).
Co-abundance analysis of metabolites
We clustered co-abundant metabolic features using weighted gene coexpression network analysis (WGCNA) independently on metabolic features detected in the HILIC and C18 columns. WGCNA identified 18 and 15 co-abundant metabolic modules in the HILIC and C18 columns, respectively (Supplementary Fig. 4). We then tested the association of each module with clinical and biomarker-supported AD and levels of the plasma biomarkers (Fig. 3a and Extended Data Fig. 1a). The purple module was negatively associated with biomarker-supported AD and P-tau181. The green-yellow module was negatively associated with P-tau181, P-tau217 and P-tau231 levels, and the black module was negatively associated with P-tau181 and P-tau217. The salmon module was positively associated with biomarker-supported AD, P-tau181, P-tau217 and P-tau231 levels. Enrichment analysis of the metabolites co-abundant in the purple module found that fatty amides (q-value = 5 × 10−3), glycerophosphocholines (q-value = 5 × 10−3) and sphingoid bases (q-value = 5 × 10−3) were over-represented in the module (Fig. 3b). Glycerophosphocholines (q-value = 3 × 10−22) were also significantly enriched in the green-yellow module, while amino acids and peptides were the top group over-represented in the black (q-value = 1.31 × 10−16) and salmon modules (9.22 × 10−7).
a, Volcano plot shows metabolic modules significantly associated with clinical and biomarker-supported AD, and AD biomarkers using Bonferroni adjusted P value. b, Chemical classes enriched by module member metabolic features present at a proportion of at least 6.5%. Other, not within any of the above classes of metabolites. c–g, The levels of the top three hub metabolites, based on eigengene-based connectivity, of the yellow (c), purple (d), salmon (e), green-yellow (f) and black (g) module in clinical (n = 150 cases and 567 controls) and biomarker-supported AD (n = 191 cases and 492 controls) are also shown. In plots c–g, the violin plot shows the distribution of the levels, the point indicates the median, the line below the point indicates the 25th percentile and the line above the point indicates the 75th percentile.
We then identified the hub metabolites that were most connected to other metabolites in the purple, salmon, green-yellow and black modules (Fig. 3c and Supplementary Table 2). Notably, three lysoPCs were hub metabolites in the purple module and all of them were more abundant in biomarker-negative participants compared to individuals who were defined as having biomarker-supported AD (Fig. 3d). Phosphatidylcholines and lysoPCs were also the hub metabolites in the green-yellow module (Fig. 3f and Supplementary Table 2) and were also more abundant in biomarker-negative participants compared to those with biomarker-supported AD. Creatinine was the most connected metabolite in the salmon module and was higher in those with biomarker-supported AD compared to biomarker-negative participants (Fig. 3e).
LysoPCs are associated with AD biomarkers
Both the metabolome-wide association study (MWAS) and WGCNA detected lysoPCs to be significantly associated with biomarker-supported AD, P-tau181, P-tau217 and P-tau231 levels. Thus, we tested the joint association of all lysoPCs with clinical and biomarker-supported AD by constructing lysoPC principal components (PCs). We constructed PCs for the 47 metabolic features annotated as lysoPCs (21 unique lysoPCs; Supplementary Table 3) detected by HILIC and C18 columns and found that the first five PCs explained ~65% of the variance (Supplementary Fig. 5). We tested the association of the first five PCs together in a regression model adjusted for age and sex (Fig. 4a). PC1 was negatively associated with biomarker-supported AD whereas PC5 was positively associated with clinical diagnosis of AD. We found a significant interaction between APOE ε4 allele carrier status and PC4, with clinical AD (P for interaction = 0.03) and biomarker-supported AD (P for interaction = 0.001), and performed an analysis stratified by APOE ε4 allele status. We found the same pattern of APOE-ε4-specific lysoPC association when we used the P-tau181, P-tau217 and P-tau231 levels as the outcome (Fig. 4b). There was no significant interaction between lysoPCs and APOE ε4 allele carrier status when considering the association between each lysoPC, individually, and clinical or biomarker-supported AD (Extended Data Fig. 2).
a, The odds ratio (point) and 95% CI (lines around the point) of PC1–5 in relation to biomarker-supported AD (n = 191 cases and 492 controls) and clinical AD (n = 150 cases and 567 controls) estimated using logistic regression adjusted for age and sex. b, The results from analysis stratified by APOE ε4 allele status. c, The loadings of lysoPCs (all adducts detected, adduct number indicated by ‘a’ followed by adduct number for the lysoPC) on the four PCs (PC1, PC2, PC4 and PC5) significantly associated with biomarker-positive status or clinical AD.
The stratified analysis (Fig. 4a) revealed that PC1 was protective of biomarker-supported AD and PC5 was risk-inducing for clinical AD only in APOE ε4 noncarriers, whereas the risk conferred by PC2 and PC4 were restricted to APOE ε4 carriers. We observed similar results when biomarker-supported AD was defined using P-tau217 (Extended Data Fig. 3). We investigated the loadings of the lysoPCs on PCs 1, 2, 4 and 5 and particularly focused on lysoPCs that have polyunsaturated fatty acid (PUFA) side chains (Fig. 4c, Supplementary Fig. 6 and Extended Data Fig. 4c). PC1 had positive loadings from all lysoPCs, which indicated that all lysoPCs were negatively associated with biomarker-supported AD, particularly among APOE ε4 noncarriers. PC4 had negative loadings from lysoPCs that carry docosahexaenoic acid (DHA; lysoPC (22:6)) and arachidonic acid (AHA; lysoPC (20:4)).
LysoPCs and polysaturated fatty acids are correlated in plasma and CSF
We tested the correlation between lysoPC PCs 1, 2, 4 and 5 with circulating PUFAs in CSF and in plasma (annotated at level 1 confidence), stratified by APOE ε4 allele status (Extended Data Figs. 4a and 5). PC1 was positively correlated with circulating levels of eicosapentaenoic acid (EPA) and AHA, in both APOE ε4 carriers and noncarriers. PC2 was negatively correlated with circulating levels of EPA in carriers and noncarriers and negatively correlated with AHA only among carriers of at least one APOE ε4 allele. It was positively correlated with arachidic acid, linolenic acid (precursor to EPA and DHA) and octadecadienoic acid only among APOE ε4 noncarriers. PC4 was negatively correlated with most measured plasma PUFAs, among carriers and noncarriers, and negatively correlated with CSF levels of DHA. PC5 was negatively correlated with plasma EPA and positively correlated with plasma AHA in carriers and noncarriers and it was also negatively correlated with linolenic acid (precursor to EPA and DHA) only among those with no APOE-ε4 allele (Extended Data Fig. 5), in which group PC5 was associated with an increased risk of AD. We also found that lysoPCs that carry EPA and DHA were positively correlated with the CSF levels of their respective PUFAs, whereas this correlation for AHA was negligible (Extended Data Fig. 5b). This indicates that lysoPCs that are known to transport long-chain PUFAs into the central nervous system51 might play a role in AD biology in an APOE ε4-dependent way.
Replication of LysoPCs association with AD biomarkers in the WHICAP cohort
To validate and generalize our findings, we examined the association between lysoPCs and clinical and biomarker-supported AD in participants of Caribbean Hispanic ethnicity in the WHICAP cohort that reside in northern Manhattan, New York and have different exposure profiles from the Genetic Studies of Alzheimer’s disease in Caribbean Hispanics (EFIGA) residents in the Dominican Republic. We used identical methods and parameters for high-resolution mass spectrometry-based metabolomics data in 351 plasma samples to identify and test association of lysoPCs with clinical AD and biomarker-supported AD. We detected 45 metabolic features annotated as lysoPCs (25 unique, and 20 features were overlapping with the EFIGA cohort) that were used to generate PCs. We then tested the association of the first five PCs with clinical and biomarker-supported AD, stratified by APOE ε4 allele carrier status (Extended Data Fig. 6). PC3 had a significant interaction with APOE ε4 allele carrier status (P for interaction = 0.04). We found that PC1, with positive loadings from all lysoPCs investigated, was negatively associated with biomarker-supported AD and this effect was only seen among those with no APOE-ε4 allele, similar to our finding in the EFIGA cohort. PC3, with large negative loadings from lysoPCs with AHA, was negatively associated with clinical AD among those with no APOE-ε4 allele. PC5, with positive loadings from lysoPC with DHA, was negatively associated with biomarker-supported AD among APOE-ε4 noncarriers. We examined the similarity of the loadings on PCs 1–5 for the 37 lysoPCs detected in both the cohorts, EFIGA and WHICAP, and found that loadings on PC1 were highly correlated (spearman rho = 0.92, P < 2.2 × 10−16; Supplementary Fig. 7).
LysoPC replication in the ROSMAP cohort
To further validate our results and determine the effect of lysoPCs on disease stage, we examined the association between lysoPCs and AD pathology in the ROSMAP cohort. We used the metabolomics data derived from 110 brain samples in the ROSMAP cohort, collected postmortem, to test the association between lysoPCs and phosphatidylcholines with pathological definition of AD, amyloid burden, tangle density and global pathology. There were 14 lysoPCs and 13 phosphatidylcholines measured in the ROSMAP cohort. We constructed PCs from the lysoPCs and tested the association of the first five PCs with AD pathology (Extended Data Fig. 7). We identified that PC3 was negatively associated with increased tangle density, global pathology and a pathological diagnosis of AD. Of the lysoPCs carrying PUFAs, we only detected AHA in the ROSMAP cohort. PC3 had positive loadings from AHA, although not an important contributor to PC3. Three PCs were negatively associated with amyloid burden and tau tangles implying that lysoPCs and PCs were reduced in the postmortem AD brains (Supplementary Table 4). These findings are consistent with plasma lysoPCs observation in the EFIGA cohort.
Discussion
We investigated the association of metabolites with plasma biomarkers and clinical diagnoses in a cohort of Caribbean Hispanic individuals to identify metabolic pathways associated with hallmarks of AD pathology. Specifically, we compared metabolic pathways enriched in clinical AD and biomarker-based diagnosis of AD. As plasma biomarkers change several years before clinical symptoms49, comparing clinical and biomarker-based diagnosis could clarify pathways that precede clinical manifestation of disease, potentially identifying early interventions. Two of the most notable findings in our study were that metabolite profiles differed when a clinical diagnosis was used versus a validated plasma biomarker-based diagnosis and that lysoPCs, which have been previously associated with AD51,52, have an APOE ε4 dependent risk of AD in our unbiased approach in a Hispanic population. This finding was replicated in a separate cohort of Caribbean Hispanic and non-Hispanic white individuals and in plasma and postmortem brain tissue.
LysoPCs were associated with quantitative levels of plasma P-tau181, P-tau217 and with biomarker-supported AD (defined by P-tau181). Co-abundance analysis revealed associations between P-tau181 and P-tau217 and metabolic modules that harbor several lysoPCs as hub metabolites, potentially suggesting an important role in disease pathogenesis. Several studies have reported lower levels of lysoPCs in the brains, CSF and plasma of patients wth AD53,54,55,56,57,58,59,60,61,62,63,64. These changes often involve a decrease in levels of lysoPC species, particularly those that bind anti-inflammatory PUFAs, in patients with AD. Some lysoPC species have been implicated in promoting neurotoxicity and inflammation65,66,67. They can induce oxidative stress, impair mitochondrial function and activate immune cells, leading to neuronal damage and death. LysoPCs are also involved in dysregulation of lipid metabolism. The breakdown of phosphatidylcholine, a major lipid component of cell membranes, can generate lysoPCs. Disruptions in enzymes involved in this process, such as phospholipase A2 (PLA2), have been observed in AD and may contribute to altered lysoPC levels68,69,70. Further validation in the frontal cortex from postmortem brain tissue in ROSMAP strengthens the biological relevance of the lysoPCs in AD etiology. Further research using animal models should explore this possible mechanism behind the observed association.
We observed a differential effect of lysoPCs within APOE ε4 carriers and noncarriers. The risk conferred by lysoPCs was restricted to APOE ε4 carriers, whereas the protective effects were significant within APOE ε4 noncarriers. We previously showed significant differences in metabolic profiles in a small multiethnic AD cohort and these differences remained when the analysis was restricted to APOE ε4 carriers71. APOE ε4 carriers tend to exhibit higher levels of specific lysoPC species, including DHA and AHA in CSF, plasma and brain tissue compared to noncarriers72,73,74,75. Elevated levels of certain lysoPCs in APOE ε4 carriers have been linked to increased Aβ deposition, tau phosphorylation and neuroinflammation. In our study, we observed distinct patterns of lysoPC alterations in APOE ε4 carriers compared to noncarriers.
We also found that metabolism of tryptophan (an essential amino acid) was associated with clinical and plasma P-tau231. Urea cycle/amino group metabolism was associated with biomarker-supported AD and all P-tau biomarkers. Tyrosine metabolism was associated with biomarker-supported AD, P-tau181 and P-tau217. Limited research has focused on measuring tyrosine levels specifically in the brains of patients with AD but administering tyrosine orally can enhance memory and cognitive function76. Tryptophan is an essential amino acid and a precursor for the synthesis of serotonin, a neurotransmitter involved in mood regulation and cognition. Alterations in tryptophan metabolism may impact serotonin availability in the brain and contribute to AD pathophysiology, particularly Aβ pathology24. Aβ accumulation can disrupt tryptophan metabolism, leading to altered levels of tryptophan and its metabolites. Conversely, tryptophan metabolites, such as kynurenic acid, can affect Aβ aggregation and clearance, potentially influencing disease progression77,78. Of note, tryptophan levels in plasma were associated with clinical diagnosis of AD and were also mildly correlated with CSF levels (correlation of 0.24; Table 2). Several microbial metabolites of tryptophan were also associated with P-tau biomarkers and indole was a hub metabolite in a module associated with P-tau181 and P-tau217.
Heparan sulfate and chondroitin sulfate degradation processes were associated with P-tau217 and P-tau231. Heparan sulfate and chondroitin sulfate are types of glycosaminoglycans (GAGs), or sulfated carbohydrates, that are found in the extracellular matrix of cells. GAGs have been reported in accumulation and clearance of Aβ79,80,81 and aggregation of tau protein82,83. GAGs can interact with various inflammatory molecules, including cytokines and chemokines, and modulate neuroinflammatory processes in AD84,85. Chondroitin sulfate and heparan sulfate chains present on proteoglycans can act as binding sites for inflammatory molecules, contributing to the activation of immune cells and the generation of a proinflammatory environment in the brain86. The Aβ42:Aβ40 ratio was associated with sialic acid metabolism and N-glycan degradation, pathways that have previously been associated with Aβ production, clearance and aggregation87,88,89,90.
Taken together these results suggest that it is essential to combine biochemical analysis with biomarkers of disease to achieve a better understanding of the metabolic heterogeneity in AD pathogenesis and enable identification of biological mechanisms of the disease. Specifically, identification of metabolic pathways associated with plasma biomarkers might indicate biological mechanisms underlying AD pathology at different stages of the disease.
There are some limitations of the analysis presented here. First, the untargeted metabolomics analysis identified several features that were eventually excluded from analyses and interpretation because of sample missingness or lack of reliable annotation. Excluding metabolites missing in 30% of the sample could potentially remove features that are not expressed or have very low levels in AD cases, affecting power of discovery. Second, pathway and functional analyses using methods such as MetaboAnalyst rely on reference databases for metabolite annotation, the accuracy and comprehensiveness of which may be affected by the coverage and quality of these databases. Third, in a limited resource environment, the diagnosis of AD based on P-tau181 might not be precise. Further investigation, specifically with longitudinal measures of biomarkers and metabolic assessments, are needed to disentangle the metabolic cascades in the different stages of disease progression.
In summary, we show that several lysoPCs are robustly associated with early amyloidosis in AD as measured by P-tau181 and P-tau217. Given that lysoPCs play an important role in the transport of long-chain PUFAs into the brain, our findings show that changes in fatty acid metabolism in the brain occur early in AD and suggest that levels of lysoPCs/PUFAs in circulation may influence disease onset or progression. This study demonstrates the utility of high-resolution mass spectrometry-based untargeted metabolomics to reveal biochemical differences in participants with aberrant plasma biomarker profiles and to identify metabolic perturbations in different stages of the disease. Furthermore, it may be possible to use metabolomic profiling to monitor these critical biochemical processes during therapeutic interventions
Methods
The Estudio Familiar de Influencia Genetica en Alzheimer cohort
This cohort has been recruiting individuals of Caribbean Hispanic ancestry with suspected sporadic or familial AD and healthy controls, similar in age, through advertisements in local newspapers and radio stations, and through clinical referrals in the Dominican Republic and in the Washington Heights neighborhood of New York City48. Participants in this study provided informed consent under protocols approved by the Columbia University Irving Medical Center Institutional Review Board, and the National Health Bioethics Committee of the Dominican Republic. They underwent medical and neurological history, detailed examinations, neuropsychological testing and collection of blood for plasma and DNA processing. CSF was collected in a subgroup of participants. The clinical diagnosis of AD was based on the National Institute on Aging and Alzheimer’s Association (NIA-AA) criteria91. All clinical diagnoses were determined in a consensus conference attended by a neurologist, a neuropsychologist and an internist with expertise in dementia and geriatrics. In brief, individuals with clinical AD must have had a history of progressive cognitive decline in the absence of other brain disorders (including conditions such as rare epilepsies and malignant neoplasms of the nervous system) and objective evidence of a decline in memory and in at least two other cognitive domains such as verbal fluency or executive function. If patients presented with a history of comorbidities such as cerebrovascular disease, stroke-related dementia and neurological disorders, a diagnosis of AD was not assigned. For this study only participants with a clinical diagnosis of AD were included. Healthy controls showed no evidence of cognitive decline or signs of other neurodegenerative diseases. For the analyses in this manuscript, only biological samples and data from individuals recruited between 1 January 2018 and 30 April 2022, were considered.
The Washington Heights, Inwood Columbia Aging Project
This project has been recruiting participants from the Washington Heights neighborhood for over two decades in a community aging study. All participants provided informed consent under protocols approved by the Columbia University Irving Medical Center Institutional Review Board. Within the cohort there are individuals who are non-Hispanic white (24%), African American (28%), Caribbean Hispanic (48%) from the Dominican Republic and Puerto Rico, and 68% are women. During each assessment, participants receive the neuropsychological test battery, medical interview and are re-consented for sharing of genetic information and autopsy. Blood is drawn, barcoded and brought to our laboratory within 2 h of collection. All the medical, neurological, psychiatric and neuropsychological data collected are reviewed at a weekly consensus conference. Diagnosis of AD is based on accepted criteria91.
Sample collection
Blood was collected in dipotassium ethylenediaminetetraacetic acid (K2EDTA) tubes by standard venipuncture and transported to a laboratory for centrifugation, preparation of plasma and storage at −80 ˚°C within 2 h of collection. CSF was obtained by standard aseptic technique, distributed into aliquots of 400 µl each in polypropylene tubes, frozen and stored at −80 °C (ref. 48).
Plasma and CSF metabolomics data generation
Plasma and CSF metabolites were extracted using internal standards-fortified acetonitrile and the extracts were injected on two chromatographic columns: a hydrophilic interaction column (HILIC) under positive ionization (HILIC+) and a C18 column under negative ionization (C18−), coupled to a Thermo Orbitrap HFX Q-Exactive mass spectrometer, scanning for molecules within 85–1,250 kDa. The methodological details have been previously published92,93. Samples were randomized to ensure equal representation of cases–controls and male–female sex across the batches. This was performed using a randomization algorithm. For each sample, triplicate injections were performed sequentially for each sample, alternating between the HILIC and C18 columns. This produced three technical replicates per sample per column. The untargeted mass spectral data were processed through a computational pipeline that leverages open source feature detection and peak alignment software, apLCMS94 and xMSanalyzer95. The feature tables were generated containing information on the mass-to-charge (m/z) ratio, retention time and median summarized abundance/intensity of each ion for each sample. The extraction pipeline adequately dealt with drift over samples run and we did observe any evidence of drift in intensity (Supplementary Fig. 1). Correction for batch effects was performed using ComBat, which uses an empirical Bayesian framework to adjust for known batches in which the samples were run96. Each of these ions are referred to as metabolic features. For the analysis, metabolic features detected in at least 70% of all the samples were retained, leaving 3,253 features from the HILIC+ column and 3,628 features from the C18− column for plasma samples. Metabolic features missing in 70% of the participants did not show any differential missingness by case status (Supplementary Fig. 2). Zero-intensity values were considered below the detection limit of the instrument and were imputed with half the minimum intensity observed for each metabolic feature. The intensity of each metabolic feature was log10-transformed, quantile normalized and autoscaled for normalization and standardization. Principal-component analysis identified 29 individuals (Supplementary Fig. 3) as outliers, who were excluded from association analysis.
Metabolite annotation
Annotations were assigned in two stages: first, using an internal reference library curated using authentic chemical standards run using the same method and instrument, and second by matching features unassigned in the first step to the Human Metabolome Database (HMDB) using the R package xMSannotator (v.1.3.2)95. This uses a multistage clustering algorithm that uses metabolic pathway associations, intensity profiles, retention time, mass defect and isotope/adduct patterns to assign putative annotations to metabolic features. When a feature had multiple matches, we used the following rules to assign an annotation: first, we screened features based on the confidence score assigned by xMSannotator, and the annotation with the highest score was used. Second, if all annotations had the same score, we chose the annotation with the lowest difference in expected and observed mass (delta parts per million (ppm)). Finally, if all features had the same score and delta ppm, we indicated the identity as ‘multiple matches’ as we could not decipher a unique putative annotation. If a feature did not match any database entries, it was denoted as ‘unknown’ (33% from HILIC+ column and 40% from C18− column). The confidence in annotation was based on criteria defined by Schymanski et al.97, where level 1 corresponds to a confirmed structure identified through MS/MS and/or comparison to an authentic standard; level 2 to a probable structure identified through spectral matches to a database; level 3 to a putative identification with a speculative structure; level 4 to an unequivocal molecular formula but with insufficient evidence to propose a structure; and level 5 to an exact mass but not enough information to assign a formula. Of the 3,253 features retained for analysis from the HILIC column, 2,165 (66%) were annotated, 100 at level 1 confidence (4.6 % of annotated), 970 at level 3 confidence (45% of annotated) and 1,095 at level 5 (50% of annotated). Of the 3,628 retained from the C18 column, 2,187 (60%) were annotated, 151 at level 1 (6.9% of annotated), 1,189 at level 3 (54%) and 847 at level 5 confidence (39%).
Blood-based biomarker analyses
The methods have been previously described in detail48. In brief, blood samples were obtained through standard venipuncture using K2EDTA tubes. Plasma was isolated via centrifugation at 2000g for 15 min at 4 °C within 2 h of collection and aliquoted into polypropylene tubes, frozen and stored at −80 °C. Plasma biomarker assessments were carried out utilizing the Quanterix Simoa single molecule array technology HD-X platform. Samples were diluted and assayed in duplicate following package insert protocols using the following Quanterix kits: Simoa Neurology 3-Plex A (N3PA, cat. no. 101995) for Aβ42 and Aβ40, Simoa P-tau181 Advantage V2 (cat. no. 103714) for phosphorylated-tau181, ALZpath Simoa P-tau217 v.2 (cat. no. 104371) for phosphorylated-tau217 and Simoa P-tau231 (cat. no. 102292) for phosphorylated-tau231. Mean coefficients of variation were ≤5%. The ratio of Aβ42:Aβ40 was also computed. Biomarker levels were log10-transformed and autoscaled before statistical analysis.
Biomarker-supported AD
A previous analysis in the EFIGA cohort48 found that plasma P-tau181 and the P-tau181:Aβ42 ratio were the most discriminatory biomarkers between patients with AD and healthy controls. This was determined using established laboratory CSF cutoff points in a subset of individuals of the EFIGA cohort with CSF biomarker data available. Then, using plasma biomarker data available in the same individuals with CSF biomarkers, they determined the classification performance and optimal cutoff points for each plasma biomarker. Elevated P-tau181 is highly correlated with AD98,99,100,101 and decreased plasma Aβ42 is correlated with AD; however, due to extracerebral sources of Aβ42, it is not as reliable a measure in plasma as P-tau181, so we chose P-tau181 over P-tau181:Aβ42 ratio to identify AD pathology in this cohort. In this analysis, participants with plasma P-tau181 < 2.63 pg ml−1 were considered biomarker status negative (BM− or healthy controls) and those with plasma P-tau181 ≥ 2.63 pg ml−1 were considered to have biomarker-supported AD (BM+). We use ‘biomarker-supported AD’ throughout the manuscript and individuals having biomarker-supported AD were considered as biomarker positive. To test whether our findings were robust to P-tau isoforms, we also used a cutoff with plasma P-tau217, where levels <0.39 pg ml−1 were considered healthy controls and those with plasma P-tau217 ≥ 0.39 pg ml−1 were considered as BM+. This cutoff was chosen using the same approach as that described for P-tau181 above.
Statistics and reproducibility
We used two approaches to find circulating metabolic features associated with outcomes of interest (1) a MWAS framework with correction for multiple comparisons by controlling the FDR at 5%; and (2) a co-abundance analysis to find modules of metabolic features associated with outcomes, providing a means of unsupervised dimensionality reduction based on correlation between the metabolic features. Both analyses were conducted separately for data from each column. All analyses were conducted in R (v.4.4.1).
Metabolome-wide association study
An MWAS was conducted using multiple linear models, adjusted for age and sex. The analyses were conducted separately for data from each column. We corrected for multiple comparisons using an FDR of 5% and q-values were estimated using the Benjamini–Hochberg method102.
Network plot
To visualize the significant associations, we constructed a network plot for all outcomes and their significantly associated features. Restricting the results to features with a q-value < 0.05 and to those with annotations assigned, we used RefMet103 to obtain metabolite classes. We then plotted the network with nodes representing the outcome and metabolic features, colored by metabolite class, and edges representing the direction of association using the igraph package (v.1.2.11) in R.
Metabolite co-abundance analysis
Co-abundance modular analysis was conducted using weighted gene correlation network analysis104 using the WGCNA R package (v.1.69). Using normalized intensity values for each metabolic feature from each sample, we first constructed a metabolic feature co-abundance network using pairwise Pearson correlations between each metabolic feature. We used a soft threshold of 4 for the HILIC+ data and 3 for the C18− data, which were chosen based on saturation of the R2 at 0.9. This correlation network, where the nodes were metabolic features and edges were the scaled correlation coefficients, was used to create the topological overlap matrix, which provides a measure of similarity between a given pair of metabolic features in the network. This similarity matrix was used to create a dendrogram to assign metabolic features into modules based on their co-abundance pattern. We used the following parameters: minimum module size of 30, merge cutHeight of 0.25, an unsigned network, and a reassign threshold of 0. After network and dendrogram construction, modules were defined using the moduleEigengenes function in WGCNA. The module eigengene is a quantitative representation of a module derived from a principal-component analysis (PCA) as the first PC, constructed using only those metabolic features that were part of the module. Association analyses were conducted to find modules associated with outcomes in linear regression models, adjusted for age and sex. We used the Bonferroni method to correct for multiple comparisons.
Pathway analysis
To determine the biological relevance of the metabolic features associated with AD and biomarkers, we conducted pathway analysis using the R package MetaboAnalystR105 (v.4.0.0). We used the MWAS results from both columns and applied a nominal P value cutoff of 0.01 to determine metabolic pathway enrichment using the mummichog algorithm and the human MFN reference database106. We present results for pathways with a Fisher’s exact test P value < 0.05.
Chemical class enrichment
This approach was used to determine the different chemical classes represented by metabolic feature members of WGCNA modules significantly associated with outcomes. All metabolites in WGCNA modules that were associated with AD and biomarkers were used as inputs for the enrichment analysis. The main chemical classes enriched were determined using the Enrichment Analysis module in MetaboAnalyst (v.5.0) using HMDB IDs as input type and metabolites as feature type for features with an annotation confidence score ≤ 3.
Construction of plasma lysoPC principal components and stratified analysis by APOE ε4 status
Based on the findings described, we performed a PCA using features that were annotated as lysoPCs from both columns (41 from HILIC+ and 6 from C18−). As PCs 1–5 explained ~65% of the variance in the data, we used the first five PCs in logistic regression models to find the association with clinical diagnosis of AD and biomarker-supported AD, adjusted for age and sex. We tested for the presence of an interaction between the combinations of the lysoPCs and the presence of at least one APOE ε4 allele and performed a stratified analysis as there was a significant interaction term between APOE ε4 allele and PC4 for both clinical (P value for interaction = 0.03) and biomarker-supported AD (P value for interaction = 0.001).
Correlation between plasma and CSF metabolites
Among people with both plasma and CSF metabolomics data available (n = 113), plasma metabolites with level 1 confidence that were significantly associated with any outcome were tested for their correlation with the same metabolite identified in CSF using spearman correlation.
Association between lysoPCs and blood-based biomarkers in Caribbean Hispanic participants of a different cohort
To provide validation of our findings, we used plasma metabolomics data and plasma biomarkers levels available in participants of Caribbean Hispanic ethnicity in the WHICAP cohort. The metabolomic and biomarker data were generated using the same methods as in the study samples of the EFIGA cohort, as described above. We identified 45 metabolites annotated as lysoPCs (39 from HILIC+ and 6 from C18− columns) and computed PCs as described above. We tested association of the first five PCs (explained ~60% of the variance) with clinical and biomarker-supported AD, adjusted for sex and age. This analysis was also conducted stratified by APOE ε4 allele carrier status as we found a significant interaction between PC3 and biomarker-supported AD (P value for interaction = 0.04).
Association between lysoPCs and other metabolites, and brain pathology in the ROSMAP cohort
To provide external validation of our findings with lysoPCs, we obtained data from the ROSMAP cohort and examined associations between lysoPCs in brain samples and brain pathology5. We also investigated plasma metabolomics data in ROSMAP published recently107 but could not identify any lysoPCs in the dataset. We computed PCs with all brain lysoPCs identified, as described above. We tested association of the first five PCs with amyloid, tangles, total global pathology, clinical and pathological diagnosis of AD, adjusted for sex and age at death. We also analyzed the relationship between all metabolites measured in brain tissue and brain pathology using linear regression, adjusted for sex and age at death.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All intermediate and final results are included in the text and the supplementary materials of the paper. The metabolomics and phenotype data from the EFIGA and WHICAP cohorts are deposited in the Synapse AD knowledge portal and can be accessed at https://www.synapse.org/Synapse:syn70083418/wiki/635413. The ROSMAP dataset (https://www.synapse.org/Synapse:syn10235595) can be accessed through Synapse and the Rush Alzheimer’s Disease Center by submitting a data request at https://www.radc.rush.edu/requests/data.htm.
Code availability
We used standard packages implemented in R for association, coexpression and PCAs. R code used for the analyses in this study is available from the corresponding authors upon request. Interested researchers are encouraged to contact the corresponding authors with a brief description of the intended use. Access will be provided for noncommercial purposes and solely for reproducing the results presented in this article.
References
Badhwar, A. et al. A multiomics approach to heterogeneity in Alzheimer’s disease: focused review and roadmap. Brain 143, 1315–1331 (2020).
Johnson, C. H., Ivanisevic, J. & Siuzdak, G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17, 451–459 (2016).
Horgusluoglu, E. et al. Integrative metabolomics-genomics approach reveals key metabolic pathways and regulators of Alzheimer’s disease. Alzheimers Dement. 18, 1260–1278 (2022).
Wilkins, J. M. & Trushina, E. Application of metabolomics in Alzheimer’s disease. Front. Neurol. 8, 719 (2017).
Huo, Z. et al. Brain and blood metabolome for Alzheimer’s dementia: findings from a targeted metabolomics analysis. Neurobiol. Aging 86, 123–133 (2020).
Yu, L., Jin, J., Xu, Y. & Zhu, X. Aberrant energy metabolism in Alzheimer’s disease. J. Transl. Int. Med. 10, 197–206 (2022).
Minhas, P. S. et al. Restoring metabolism of myeloid cells reverses cognitive decline in ageing. Nature 590, 122–128 (2021).
Bartke, A., Brannan, S., Hascup, E., Hascup, K. & Darcy, J. Energy metabolism and aging. World J. Mens. Health 39, 222–232 (2021).
Cunnane, S. C. et al. Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 19, 609–633 (2020).
Hoyer, S., Nitsch, R. & Oesterreich, K. Predominant abnormality in cerebral glucose utilization in late-onset dementia of the Alzheimer type: a cross-sectional comparison against advanced late-onset and incipient early-onset cases. J. Neural Transm. Park Dis. Dement. Sect. 3, 1–14 (1991).
An, Y. et al. Evidence for brain glucose dysregulation in Alzheimer’s disease. Alzheimers Dement. 14, 318–329 (2018).
Madeira, C. et al. D-serine levels in Alzheimer’s disease: implications for novel biomarker development. Transl. Psychiatry 5, e561 (2015).
Wu, L., Zhang, X. & Zhao, L. Human ApoE isoforms differentially modulate brain glucose and ketone body metabolism: implications for Alzheimer’s disease risk reduction and early intervention. J. Neurosci. 38, 6665–6681 (2018).
Hashimoto, K. et al. Possible role of D-serine in the pathophysiology of Alzheimer’s disease. Prog. Neuropsychopharmacol. Biol. Psychiatry 28, 385–388 (2004).
Wang, W., Zhao, F., Ma, X., Perry, G. & Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol. Neurodegener. 15, 30 (2020).
Quntanilla, R. A. & Tapia-Monsalves, C. The role of mitochondrial impairment in Alzheimer s disease neurodegeneration: the tau connection. Curr. Neuropharmacol. 18, 1076–1091 (2020).
Jadiya, P. et al. Impaired mitochondrial calcium efflux contributes to disease progression in models of Alzheimer’s disease. Nat. Commun. 10, 3885 (2019).
Zhang, X., Alshakhshir, N. & Zhao, L. Glycolytic metabolism, brain resilience, and Alzheimer’s disease. Front. Neurosci. 15, 662242 (2021).
Kosicek, M. & Hecimovic, S. Phospholipids and Alzheimer’s disease: alterations, mechanisms and potential biomarkers. Int J. Mol. Sci. 14, 1310–1322 (2013).
Whiley, L. et al. Evidence of altered phosphatidylcholine metabolism in Alzheimer’s disease. Neurobiol. Aging 35, 271–278 (2014).
Yin, F. Lipid metabolism and Alzheimer’s disease: clinical evidence, mechanistic link and therapeutic promise. FEBS J. 290, 1420–1453 (2023).
Patrick, R. P. Role of phosphatidylcholine-DHA in preventing APOE4-associated Alzheimer’s disease. FASEB J. 33, 1554–1564 (2019).
Griffin, J. W. & Bradshaw, P. C. Amino acid catabolism in Alzheimer’s disease brain: friend or foe? Oxid. Med. Cell Longev. 2017, 5472792 (2017).
Savonije, K. & Weaver, D. F. The role of tryptophan metabolism in Alzheimer’s disease. Brain Sci. https://doi.org/10.3390/brainsci13020292 (2023).
Liu, P. et al. Phenylalanine metabolism is dysregulated in human hippocampus with Alzheimer’s disease related pathological changes. J. Alzheimers Dis. 83, 609–622 (2021).
Whiley, L. et al. Metabolic phenotyping reveals a reduction in the bioavailability of serotonin and kynurenine pathway metabolites in both the urine and serum of individuals living with Alzheimer’s disease. Alzheimers Res. Ther. 13, 20 (2021).
McCann, A. et al. Serum tyrosine is associated with better cognition in Lewy body dementia. Brain Res. 1765, 147481 (2021).
Hajjar, I., Liu, C., Jones, D. P. & Uppal, K. Untargeted metabolomics reveal dysregulations in sugar, methionine, and tyrosine pathways in the prodromal state of AD. Alzheimers Dement. 12, e12064 (2020).
Basun, H. et al. Amino acid concentrations in cerebrospinal fluid and plasma in Alzheimer’s disease and healthy control subjects. J. Neural Transm. Park Dis. Dement. Sect. 2, 295–304 (1990).
Fonteh, A. N., Harrington, R. J., Tsai, A., Liao, P. & Harrington, M. G. Free amino acid and dipeptide changes in the body fluids from Alzheimer’s disease subjects. Amino Acids 32, 213–224 (2007).
Gonzalez-Dominguez, R., Garcia-Barrera, T. & Gomez-Ariza, J. L. Metabolite profiling for the identification of altered metabolic pathways in Alzheimer’s disease. J. Pharm. Biomed. Anal. 107, 75–81 (2015).
Ibanez, C. et al. Toward a predictive model of Alzheimer’s disease progression using capillary electrophoresis-mass spectrometry metabolomics. Anal. Chem. 84, 8532–8540 (2012).
Larsson, S. C. & Markus, H. S. Branched-chain amino acids and Alzheimer’s disease: a Mendelian randomization analysis. Sci. Rep. 7, 13604 (2017).
Li, H. et al. Defect of branched-chain amino acid metabolism promotes the development of Alzheimer’s disease by targeting the mTOR signaling. Biosci. Rep. https://doi.org/10.1042/bsr20180127 (2018).
Ruiz, H. H. et al. Increased susceptibility to metabolic dysregulation in a mouse model of Alzheimer’s disease is associated with impaired hypothalamic insulin signaling and elevated BCAA levels. Alzheimers Dement. 12, 851–861 (2016).
Toledo, J. B. et al. Metabolic network failures in Alzheimer’s disease: a biochemical road map. Alzheimers Dement. 13, 965–984 (2017).
Trushina, E., Dutta, T., Persson, X. M., Mielke, M. M. & Petersen, R. C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE 8, e63644 (2013).
Tynkkynen, J. et al. Association of branched-chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer’s disease: a prospective study in eight cohorts. Alzheimers Dement. 14, 723–733 (2018).
Kandimalla, R. & Reddy, P. H. Therapeutics of neurotransmitters in Alzheimer’s disease. J. Alzheimers Dis. 57, 1049–1069 (2017).
Francis, P. T. The interplay of neurotransmitters in Alzheimer’s disease. CNS Spectr. 10, 6–9 (2005).
Yang, Z., Zou, Y. & Wang, L. Neurotransmitters in prevention and treatment of Alzheimer’s disease. Int. J. Mol. Sci. https://doi.org/10.3390/ijms24043841 (2023).
Li, Y. et al. Implications of GABAergic neurotransmission in Alzheimer’s disease. Front. Aging Neurosci. 8, 31 (2016).
Xu, Y. et al. Neurotransmitter receptors and cognitive dysfunction in Alzheimer’s disease and Parkinson’s disease. Prog. Neurobiol. 97, 1–13 (2012).
Martinez de Toda, I., Miguelez, L., Vida, C., Carro, E. & De la Fuente, M. Altered redox state in whole blood cells from patients with mild cognitive impairment and Alzheimer’s disease. J. Alzheimers Dis. 71, 153–163 (2019).
Holubiec, M. I., Gellert, M. & Hanschmann, E. M. Redox signaling and metabolism in Alzheimer’s disease. Front. Aging Neurosci. 14, 1003721 (2022).
Huang, W. J., Zhang, X. & Chen, W. W. Role of oxidative stress in Alzheimer’s disease. Biomed. Rep. 4, 519–522 (2016).
Butterfield, D. A. & Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 20, 148–160 (2019).
Honig, L. S. et al. Evaluation of plasma biomarkers for A/T/N classification of Alzheimer disease among adults of Caribbean Hispanic ethnicity. JAMA Netw. Open 6, e238214 (2023).
Jia, J. et al. Biomarker changes during 20 years preceding Alzheimer’s disease. N. Engl. J. Med 390, 712–722 (2024).
Niwa, T. Uremic toxicity of indoxyl sulfate. Nagoya J. Med. Sci. 72, 1–11 (2010).
Semba, R. D. Perspective: the potential role of circulating lysophosphatidylcholine in neuroprotection against Alzheimer disease. Adv. Nutr. 11, 760–772 (2020).
Law, S. H. et al. An updated review of lysophosphatidylcholine metabolism in human diseases. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20051149 (2019).
Schmerler, D. et al. Targeted metabolomics for discrimination of systemic inflammatory disorders in critically ill patients. J. Lipid Res. 53, 1369–1375 (2012).
Mulder, C. et al. Decreased lysophosphatidylcholine/phosphatidylcholine ratio in cerebrospinal fluid in Alzheimer’s disease. J. Neural Transm. 110, 949–955 (2003).
Fan, L. et al. Identification of metabolic biomarkers to diagnose epithelial ovarian cancer using a UPLC/QTOF/MS platform. Acta Oncol. 51, 473–479 (2012).
Chan, R. B. et al. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem. 287, 2678–2688 (2012).
Grimm, M. O. et al. From brain to food: analysis of phosphatidylcholins, lyso-phosphatidylcholins and phosphatidylcholin-plasmalogens derivates in Alzheimer’s disease human post mortem brains and mice model via mass spectrometry. J. Chromatogr. A 1218, 7713–7722 (2011).
Villamil-Ortiz, J. G. et al. Differential pattern of phospholipid profile in the temporal cortex from E280A-familiar and sporadic Alzheimer's disease brains. J. Alzheimers Dis. 61, 209–219 (2018).
Ross, B. M., Moszczynska, A., Erlich, J. & Kish, S. J. Phospholipid-metabolizing enzymes in Alzheimer’s disease: increased lysophospholipid acyltransferase activity and decreased phospholipase A2 activity. J. Neurochem. 70, 786–793 (1998).
Li, N. J. et al. Plasma metabolic profiling of Alzheimer’s disease by liquid chromatography/mass spectrometry. Clin. Biochem. 43, 992–997 (2010).
Liu, Y., Li, N., Zhou, L., Li, Q. & Li, W. Plasma metabolic profiling of mild cognitive impairment and Alzheimer’s disease using liquid chromatography/mass spectrometry. Cent. Nerv. Syst. Agents Med. Chem. 14, 113–120 (2014).
Cui, Y. et al. Lysophosphatidylcholine and amide as metabolites for detecting Alzheimer disease using ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry-based metabonomics. J. Neuropathol. Exp. Neurol. 73, 954–963 (2014).
Gonzalez-Dominguez, R., Garcia-Barrera, T. & Gomez-Ariza, J. L. Combination of metabolomic and phospholipid-profiling approaches for the study of Alzheimer’s disease. J. Proteom. 104, 37–47 (2014).
Casanova, R. et al. Blood metabolite markers of preclinical Alzheimer’s disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement. 12, 815–822 (2016).
Inose, Y., Kato, Y., Kitagawa, K., Uchiyama, S. & Shibata, N. Activated microglia in ischemic stroke penumbra upregulate MCP-1 and CCR2 expression in response to lysophosphatidylcholine derived from adjacent neurons and astrocytes. Neuropathology 35, 209–223 (2015).
Sheikh, A. M., Michikawa, M., Kim, S. U. & Nagai, A. Lysophosphatidylcholine increases the neurotoxicity of Alzheimer’s amyloid β1-42 peptide: role of oligomer formation. Neuroscience 292, 159–169 (2015).
Knuplez, E. & Marsche, G. An updated review of pro- and anti-inflammatory properties of plasma lysophosphatidylcholines in the vascular system. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21124501 (2020).
Gattaz, W. F., Maras, A., Cairns, N. J., Levy, R. & Forstl, H. Decreased phospholipase A2 activity in Alzheimer brains. Biol. Psychiatry 37, 13–17 (1995).
Sanchez-Mejia, R. O. et al. Phospholipase A2 reduction ameliorates cognitive deficits in a mouse model of Alzheimer’s disease. Nat. Neurosci. 11, 1311–1318 (2008).
Sanchez-Mejia, R. O. & Mucke, L. Phospholipase A2 and arachidonic acid in Alzheimer’s disease. Biochim. Biophys. Acta 1801, 784–790 (2010).
Vardarajan, B. et al. Differences in plasma metabolites related to Alzheimer’s disease, APOE epsilon4 status, and ethnicity. Alzheimers Dement. 6, e12025 (2020).
Coughlan, G. et al. APOE epsilon4 alters associations between docosahexaenoic acid and preclinical markers of Alzheimer’s disease. Brain Commun. 3, fcab085 (2021).
Lim, W. L. et al. Effects of a high-fat, high-cholesterol diet on brain lipid profiles in apolipoprotein E epsilon3 and epsilon4 knock-in mice. Neurobiol. Aging 34, 2217–2224 (2013).
Kariv-Inbal, Z. et al. The isoform-specific pathological effects of apoE4 in vivo are prevented by a fish oil (DHA) diet and are modified by cholesterol. J. Alzheimers Dis. 28, 667–683 (2012).
Miranda, A. M. et al. Effects of APOE4 allelic dosage on lipidomic signatures in the entorhinal cortex of aged mice. Transl. Psychiatry 12, 129 (2022).
van de Rest, O., van der Zwaluw, N. L. & de Groot, L. C. Literature review on the role of dietary protein and amino acids in cognitive functioning and cognitive decline. Amino Acids 45, 1035–1045 (2013).
Maitre, M., Klein, C., Patte-Mensah, C. & Mensah-Nyagan, A. G. Tryptophan metabolites modify brain Aβ peptide degradation: a role in Alzheimer's disease? Prog. Neurobiol. 190, 101800 (2020).
Pathak, S. et al. The influence of kynurenine metabolites on neurodegenerative pathologies. Int. J. Mol. Sci. https://doi.org/10.3390/ijms25020853 (2024).
Suzuki, K., Katzman, R. & Korey, S. R. Chemical studies on Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 24, 211–224 (1965).
Snow, A. D. & Wight, T. N. Proteoglycans in the pathogenesis of Alzheimer’s disease and other amyloidoses. Neurobiol. Aging 10, 481–497 (1989).
Snow, A. D., Cummings, J. A. & Lake, T. The unifying hypothesis of Alzheimer’s disease: heparan sulfate proteoglycans/glycosaminoglycans are key as first hypothesized over 30 years ago. Front. Aging Neurosci. 13, 710683 (2021).
Hasegawa, M., Crowther, R. A., Jakes, R. & Goedert, M. Alzheimer-like changes in microtubule-associated protein tau induced by sulfated glycosaminoglycans. Inhibition of microtubule binding, stimulation of phosphorylation, and filament assembly depend on the degree of sulfation. J. Biol. Chem. 272, 33118–33124 (1997).
Rauch, J. N. et al. Tau internalization is regulated by 6-O sulfation on heparan sulfate proteoglycans (HSPGs). Sci. Rep. 8, 6382 (2018).
Smith, P. D., Coulson-Thomas, V. J., Foscarin, S., Kwok, J. C. & Fawcett, J. W. ‘GAG-ing with the neuron’: the role of glycosaminoglycan patterning in the central nervous system. Exp. Neurol. 274, 100–114 (2015).
Evans, A. D., Pournoori, N., Saksala, E. & Oommen, O. P. Glycosaminoglycans' for brain health: harnessing glycosaminoglycan based biomaterials for treating central nervous system diseases and in-vitro modeling. Biomaterials 309, 122629 (2024).
Stephenson, E. L. & Yong, V. W. Pro-inflammatory roles of chondroitin sulfate proteoglycans in disorders of the central nervous system. Matrix Biol. 71-72, 432–442 (2018).
Zhao, J. & Lang, M. New insight into protein glycosylation in the development of Alzheimer’s disease. Cell Death Discov. 9, 314 (2023).
Haukedal, H. & Freude, K. K. Implications of glycosylation in Alzheimer’s disease. Front. Neurosci. 14, 625348 (2020).
Rawal, P. & Zhao, L. Sialometabolism in brain health and Alzheimer’s disease. Front. Neurosci. 15, 648617 (2021).
Patel, D. A., Henry, J. E. & Good, T. A. Attenuation of β-amyloid-induced toxicity by sialic-acid-conjugated dendrimers: role of sialic acid attachment. Brain Res. 1161, 95–105 (2007).
McKhann, G. M. et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 263–269 (2011).
Liu, K. H. et al. High-resolution metabolomics assessment of military personnel: evaluating analytical strategies for chemical detection. J. Occup. Environ. Med. 58, S53–S61 (2016).
Walker, D. I. et al. Metabolome-wide association study of anti-epileptic drug treatment during pregnancy. Toxicol. Appl. Pharm. 363, 122–130 (2019).
Yu, T., Park, Y., Johnson, J. M. & Jones, D. P. apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics 25, 1930–1936 (2009).
Uppal, K., Walker, D. I. & Jones, D. P. xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal. Chem. 89, 1063–1067 (2017).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Schymanski, E. L. et al. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Technol. 48, 2097–2098 (2014).
Giacomucci, G. et al. Plasma p-tau181 as a promising non-invasive biomarker of Alzheimer’s disease pathology in subjective cognitive decline and mild cognitive impairment. J. Neurol. Sci. 453, 120805 (2023).
Karikari, T. K. et al. Blood phosphorylated tau 181 as a biomarker for Alzheimer’s disease: a diagnostic performance and prediction modelling study using data from four prospective cohorts. Lancet Neurol. 19, 422–433 (2020).
McGrath, E. R. et al. Blood phosphorylated tau 181 as a biomarker for amyloid burden on brain PET in cognitively healthy adults. J. Alzheimers Dis. 87, 1517–1526 (2022).
Mielke, M. M. et al. Plasma phospho-tau181 increases with Alzheimer’s disease clinical severity and is associated with tau- and amyloid-positron emission tomography. Alzheimers Dement. 14, 989–997 (2018).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995).
Fahy, E. & Subramaniam, S. RefMet: a reference nomenclature for metabolomics. Nat. Methods 17, 1173–1174 (2020).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Chong, J. & Xia, J. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34, 4313–4314 (2018).
Li, S. et al. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 9, e1003123 (2013).
Batra, R. et al. The landscape of metabolic brain alterations in Alzheimer’s disease. Alzheimers Dement. 19, 980–998 (2023).
Acknowledgements
Data collection for this project was supported by EFIGA funded by the NIA and by the National Institutes of Health (NIH) (R56AG063908, R01AG067501 and RF1AG015473). We acknowledge the services of CEDIMAT for collaborating with sample collection and processing in the EFIGA cohort. WHICAP is funded by the NIA and the NIH (WHICAP, R01AG072474 and R01AG066107). The metabolomics core that generated the metabolomics data for the project is supported by the National Center for Advancing Translational Sciences grant- 5UL1TR001873. ROSMAP is supported by P30AG10161, P30AG72975, R01AG15819, R01AG17917, U01AG46152 and U01AG61356.
Author information
Authors and Affiliations
Contributions
Conceptualization: R.M., G.W.M. and B.N.V. Methodology: G.W.M., B.N.V. and R.N. Participant enrollment and sample collection: D.R.-D., M.M., D.R., R.L., L.S.H. and R.M. Data generation and analysis and interpretation: V.K., R.N., S.D., H.U., A.J.L., L.S.H., R.M., G.W.M. and B.N.V. Funding acquisition: R.M., G.W.M. and B.N.V. Project administration: D.R.-D., R.L. and R.M. Supervision: B.N.V., G.W.M. and R.M. Writing—original draft: V.K., B.N.V. and G.W.M. Writing—review and editing: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Rui Pinto and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Results from coexpression analysis using data from the C18 column.
In a, the volcano plot shows metabolic modules significantly associated with clinical AD, biomarker-supported AD and AD biomarkers using Bonferroni adjusted p value. In b, the chemical classes enriched by module member metabolic features present at a proportion of at least 6.5%. The levels of the top three hub metabolites, based on eigengene-based connectivity, of the yellow (c), brown (d), salmon (e), green-yellow (f), and cyan (g) module in clinical and biomarker-supported AD, are also shown.
Extended Data Fig. 2 Associations between individual lysoPCs and AD and biomarkers.
The * indicates FDR q-value < 0.05 and the # indicates an FDR q-value for interaction with APOE ε4 carrier < 0.05. There was no significant interaction with APOE ε4 in the relationship between clinical AD or biomarker-supported AD and any of the individual lysoPCs (all adducts detected, adduct number indicated by a*).
Extended Data Fig. 3 Lysophosphatidylcholines (lysoPCs) are associated with clinical AD and biomarker-supported AD (defined by either P-tau181 or P-tau217).
The odds ratio (point) and confidence interval (whiskers) of PC1–5 in relation to clinical AD and biomarker-supported AD, defined as P-tau181 supported AD or P-tau217 supported AD, stratified by APOE ε4 allele status. An * indicates p value < 0.05. Similar trends in association are seen between lysoPC constructs and P-tau181 or P-tau217 supported AD within each APOE ε4 strata.
Extended Data Fig. 4 Correlation of lysoPCs with polyunsaturated fatty acids in plasma and CSF.
In a, the correlation between plasma lysoPC profiles and fatty acids in CSF (top panel) and plasma. In b, the correlation between lysoPCs that carry fatty acids of interest (DHA, EPA, AHA) and the levels of these fatty acids measured in CSF. In c, the characteristics of fatty acids associated with circulating lysoPCs. * indicates p value < 0.05.
Extended Data Fig. 5
Correlation of lysoPCs with polyunsaturated fatty acids in plasma stratified by APOE ε4 allele status.
Extended Data Fig. 6 Lysophosphatidylcholines (lysoPCs) constructs associated with clinical AD and biomarker-supported AD in the WHICAP cohort.
In a, the odds ratio (point) and confidence interval (whiskers) of PC1–5 in relation to biomarker-supported AD and clinical AD. In b, the results from analysis stratified by APOE \({\rm{\varepsilon }}\)4 allele status. In c, the loadings of lysoPCs (all adducts detected, adduct number indicated by a*) on the four PCs (PC1, PC2, PC3, and PC5) significantly associated with biomarkers positive status or clinical AD.
Extended Data Fig. 7 Brain lysoPC association in ROSMAP cohort.
In a, the first five principal components explained over 95% of the variance. In b, PC3 was nominally associated with pathological definition of AD, tau tangles, general pathology and neurofibrillary tangles. In c, lysoPCs detected in the ROSMAP cohort. EPA and DHA carrying lysoPCs were not detected but lysoPC.a.20.4, which carries AHA, was measured.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7
Supplementary Tables 1–4
Four supplementary tables with a descriptive cover page.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kalia, V., Reyes-Dumeyer, D., Dubey, S. et al. Lysophosphatidylcholines are associated with amyloidosis in early stages of Alzheimer’s disease. Nat Aging 6, 221–234 (2026). https://doi.org/10.1038/s43587-025-01025-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-01025-7