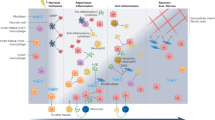
Abstract
Myocardial infarction (MI) mobilizes macrophages, the central protagonists of tissue repair in the infarcted heart. Although necessary for repair, macrophages also contribute to adverse remodeling and progression to heart failure. In this context, specific targeting of inflammatory macrophage activation may attenuate maladaptive responses and enhance cardiac repair. Allograft inflammatory factor 1 (AIF1) is a macrophage-specific protein expressed in a variety of inflammatory settings, but its function after MI is unknown. Here we identify a maladaptive role for macrophage AIF1 after MI in mice. Mechanistic studies show that AIF1 increases actin remodeling in macrophages to promote reactive oxygen species–dependent activation of hypoxia-inducible factor (HIF)-1α. This directs a switch to glycolytic metabolism to fuel macrophage-mediated inflammation, adverse ventricular remodeling and progression to heart failure. Targeted knockdown of Aif1 using antisense oligonucleotides improved cardiac repair, supporting further exploration of macrophage AIF1 as a therapeutic target after MI.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Publicly available single-nuclei RNA sequencing data of human LV myocardium after acute MI were accessed at https://explore.data.humancellatlas.org/projects/e9f36305-d857-44a3-93f0-df4e6007dc97. Publicly available single-cell RNA sequencing data of mouse hearts after acute MI were accessed at https://infection-atlas.org/Rizzo2022/. All other data generated in this study are provided in the Source Data and Supplementary Information sections.
Code availability
The code used to analyze the single-nuclei RNA sequencing dataset is available on GitHub at https://github.com/thorplab.
References
Tsao, C. W. et al. Heart Disease and Stroke Statistics—2022 Update: a report from the American Heart Association. Circulation 145, e153–e639 (2022).
Guillen, I., Blanes, M., Gomez-Lechon, M. J. & Castell, J. V. Cytokine signaling during myocardial infarction: sequential appearance of IL-1 beta and IL-6. Am. J. Physiol. 269, R229–R235 (1995).
Gabriel, A. S., Martinsson, A., Wretlind, B. & Ahnve, S. IL-6 levels in acute and post myocardial infarction: their relation to CRP levels, infarction size, left ventricular systolic function, and heart failure. Eur. J. Intern. Med. 15, 523–528 (2004).
Ridker, P. M. et al. Elevation of tumor necrosis factor-α and increased risk of recurrent coronary events after myocardial infarction. Circulation 101, 2149–2153 (2000).
Huang, S. & Frangogiannis, N. G. Anti-inflammatory therapies in myocardial infarction: failures, hopes and challenges. Br. J. Pharmacol. 175, 1377–1400 (2018).
Dick, S. A. et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20, 29–39 (2019).
Faxon, D. P., Gibbons, R. J., Chronos, N. A. F., Gurbel, P. A. & Sheehan, F. The effect of blockade of the CD11/CD18 integrin receptor on infarct size in patients with acute myocardial infarction treated with direct angioplasty: the results of the HALT-MI study. J. Am. Coll. Cardiol. 40, 1199–1204 (2002).
Thorp, E. B. Cardiac macrophages and emerging roles for their metabolism after myocardial infarction. J. Clin. Invest. 133, e171953 (2023).
DeBerge, M., Chaudhary, R., Schroth, S. & Thorp, E. B. Immunometabolism at the heart of cardiovascular disease. JACC Basic Transl. Sci. 8, 884–904 (2023).
Koelwyn, G. J., Corr, E. M., Erbay, E. & Moore, K. J. Regulation of macrophage immunometabolism in atherosclerosis. Nat. Immunol. 19, 526–537 (2018).
Zhang, S. et al. Efferocytosis fuels requirements of fatty acid oxidation and the electron transport chain to polarize macrophages for tissue repair. Cell Metab. 29, 443–456 (2019).
DeBerge, M. et al. Hypoxia-inducible factors individually facilitate inflammatory myeloid metabolism and inefficient cardiac repair. J. Exp. Med. 218, e20200667 (2021).
Kido, M. et al. Hypoxia-inducible factor 1-alpha reduces infarction and attenuates progression of cardiac dysfunction after myocardial infarction in the mouse. J. Am. Coll. Cardiol. 46, 2116–2124 (2005).
Rowley, A. H. et al. Allograft inflammatory factor-1 links T-cell activation, interferon response, and macrophage activation in chronic Kawasaki disease arteritis. J. Pediatric Infect. Dis. Soc. 6, e94–e102 (2017).
Chen, Z.-W. et al. Identification, isolation, and characterization of daintain (allograft inflammatory factor 1), a macrophage polypeptide with effects on insulin secretion and abundantly present in the pancreas of prediabetic BB rats. Proc. Natl Acad. Sci. USA 94, 13879–13884 (1997).
Schluesener, H. J., Seid, K., Kretzschmar, J. & Meyermann, R. Allograft-inflammatory factor-1 in rat experimental autoimmune encephalomyelitis, neuritis, and uveitis: expression by activated macrophages and microglial cells. Glia 24, 244–251 (1998).
Utans, U., Arceci, R. J., Yamashita, Y. & Russell, M. E. Cloning and characterization of allograft inflammatory factor-1: a novel macrophage factor identified in rat cardiac allografts with chronic rejection. J. Clin. Invest. 95, 2954–2962 (1995).
Li, Y. et al. Upregulation of allograft inflammatory factor‑1 expression and secretion by macrophages stimulated with aldosterone promotes renal fibroblasts to a profibrotic phenotype. Int. J. Mol. Med. 42, 861–872 (2018).
Harney, S. M. J. et al. Fine mapping of the MHC class III region demonstrates association of AIF1 and rheumatoid arthritis. Rheumatology 47, 1761–1767 (2008).
Mishima, T. et al. Allograft inflammatory factor-1 augments macrophage phagocytotic activity and accelerates the progression of atherosclerosis in ApoE−/− mice. Int. J. Mol. Med. 21, 181–187 (2008).
Postler, E., Rimner, A., Beschorner, R., Schluesener, H. J. & Meyermann, R. Allograft-inflammatory-factor-1 is upregulated in microglial cells in human cerebral infarctions. J. Neuroimmunol. 104, 85–91 (2000).
Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022).
Lavine, K. J. et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc. Natl Acad. Sci. USA 111, 16029–16034 (2014).
Rizzo, G. et al. Dynamics of monocyte-derived macrophage diversity in experimental myocardial infarction. Cardiovasc. Res. 127, e232–e249 (2022).
Autieri, M. V. et al. Allograft inflammatory factor-1 expression correlates with cardiac rejection and development of cardiac allograft vasculopathy. Circulation 106, 2218–2223 (2002).
Kelemen, S. E. & Autieri, M. V. Expression of allograft inflammatory factor-1 in T lymphocytes: a role in T-lymphocyte activation and proliferative arteriopathies. Am. J. Pathol. 167, 619–626 (2005).
Lindsey, M. L. et al. Reperfused vs. nonreperfused myocardial infarction: when to use which model. Am. J. Physiol. Heart Circ. Physiol. 321, H208–H213 (2021).
Elizondo, D. M., Brandy, N. Z. D., da Silva, R. L., de Moura, T. R. & Lipscomb, M. W. Allograft inflammatory factor-1 in myeloid cells drives autoimmunity in type 1 diabetes. JCI Insight 5, e136092 (2020).
Heng, T. S. P. et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Egaña-Gorroño, L. et al. Allograft inflammatory factor-1 supports macrophage survival and efferocytosis and limits necrosis in atherosclerotic plaques. Atherosclerosis 289, 184–194 (2019).
Heidt, T. et al. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ. Res. 115, 284–295 (2014).
Jiang, W. et al. Necrotic cardiac myocytes skew macrophage polarization towards a classically activated phenotype. PLoS ONE 18, e0282921 (2023).
Hayashi, T. et al. The programmed death-1 signaling axis modulates inflammation and LV structure/function in a stress-induced cardiomyopathy model. JACC Basic Transl. Sci. 7, 1120–1139 (2022).
Zhang, W. et al. Necrotic myocardial cells release damage‐associated molecular patterns that provoke fibroblast activation in vitro and trigger myocardial inflammation and fibrosis in vivo. J. Am. Heart Assoc. 4, e001993 (2015).
Cramer, T. et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112, 645–657 (2003).
Autieri, M. V. & Chen, X. The ability of AIF-1 to activate human vascular smooth muscle cells is lost by mutations in the EF-hand calcium-binding region. Exp. Cell Res. 307, 204–211 (2005).
Brown, D. M. et al. Calcium and ROS-mediated activation of transcription factors and TNF-α cytokine gene expression in macrophages exposed to ultrafine particles. Am. J. Physiol. Lung Cell. Mol. Physiol. 286, L344–L353 (2004).
Schappe, M. S. et al. Chanzyme TRPM7 mediates the Ca2+ influx essential for lipopolysaccharide-induced Toll-like receptor 4 endocytosis and macrophage activation. Immunity 48, 59–74 (2018).
Autieri, M. V., Kelemen, S. E. & Wendt, K. W. AIF-1 is an actin-polymerizing and Rac1-activating protein that promotes vascular smooth muscle cell migration. Circ. Res. 92, 1107–1114 (2003).
Price, L. S. et al. Calcium signaling regulates translocation and activation of Rac. J. Biol. Chem. 278, 39413–39421 (2003).
Wheeler, A. P. et al. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J. Cell Sci. 119, 2749–2757 (2006).
Geng, J. et al. TLR4 signalling via Piezo1 engages and enhances the macrophage mediated host response during bacterial infection. Nat. Commun. 12, 3519 (2021).
Bosco, E. E., Mulloy, J. C. & Zheng, Y. Rac1 GTPase: a ‘Rac’ of all trades. Cell. Mol. Life Sci. 66, 370 (2008).
Hirota, K. & Semenza, G. L. Rac1 activity is required for the activation of hypoxia-inducible factor 1. J. Biol. Chem. 276, 21166–21172 (2001).
Sumbayev, V. V. LPS-induced Toll-like receptor 4 signalling triggers cross-talk of apoptosis signal-regulating kinase 1 (ASK1) and HIF-1α protein. FEBS Lett. 582, 319–326 (2008).
Zimmerman, M. C. et al. Requirement for Rac1-dependent NADPH oxidase in the cardiovascular and dipsogenic actions of angiotensin II in the brain. Circ. Res. 95, 532–539 (2004).
Bouin, A.-P., Grandvaux, N., Vignais, P. V. & Fuchs, A. p40phox is phosphorylated on threonine 154 and serine 315 during activation of the phagocyte NADPH oxidase: implication of a protein kinase C-type kinase in the phosphorylation process. J. Biol. Chem. 273, 30097–30103 (1998).
Piotrowska, K. et al. Over-expression of allograft inflammatory factor-1 (AIF-1) in patients with rheumatoid arthritis. Biomolecules 10, 1064 (2020).
Dick Sarah, A. & Epelman, S. Chronic heart failure and inflammation. Circ. Res. 119, 159–176 (2016).
Yang, Z. F. et al. Allograft inflammatory factor-1 (AIF-1) is crucial for the survival and pro-inflammatory activity of macrophages. Int. Immunol. 17, 1391–1397 (2005).
Li, Q. et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat. Commun. 11, 1456 (2020).
Baillet, A. et al. Unexpected function of the phagocyte NADPH oxidase in supporting hyperglycolysis in stimulated neutrophils: key role of 6-phosphofructo-2-kinase. FASEB J. 31, 663–673 (2017).
Yuan, X. et al. Aldosterone promotes renal interstitial fibrosis via the AIF‑1/AKT/mTOR signaling pathway. Mol. Med. Rep. 20, 4033–4044 (2019).
Knoops, B. et al. Specific interactions measured by AFM on living cells between peroxiredoxin-5 and TLR4: relevance for mechanisms of innate immunity. Cell Chem. Biol. 25, 550–559 (2018).
Bolognese, L. & Cerisano, G. Early predictors of left ventricular remodeling after acute myocardial infarction. Am. Heart J. 138, S79–S83 (1999).
Frangogiannis, N. G. Matricellular proteins in cardiac adaptation and disease. Physiol. Rev. 92, 635–688 (2012).
Park, J. S. et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 578, 621–626 (2020).
Previtera, M. L. & Sengupta, A. Substrate stiffness regulates proinflammatory mediator production through TLR4 activity in macrophages. PLoS ONE 10, e0145813 (2016).
Dehn, S. & Thorp, E. B. Myeloid receptor CD36 is required for early phagocytosis of myocardial infarcts and induction of Nr4a1-dependent mechanisms of cardiac repair. FASEB J. 32, 254–264 (2018).
De Zoysa, M. et al. Allograft inflammatory factor-1 in disk abalone (Haliotis discus discus): molecular cloning, transcriptional regulation against immune challenge and tissue injury. Fish Shellfish Immunol. 29, 319–326 (2010).
Chiaramonte, M. et al. Allograft inflammatory factor AIF-1: early immune response in the Mediterranean sea urchin Paracentrotus lividus. Zoology 142, 125815 (2020).
da Silva, R. L. et al. Leishmania donovani infection suppresses allograft inflammatory factor-1 in monocytes and macrophages to inhibit inflammatory responses. Sci. Rep. 11, 946 (2021).
Lai, Y., Wang, Y., Fan, X. & Zhao, Y. Allograft inflammatory factor-1 stimulates inflammatory properties of peripheral blood leukocytes and increases cell viability via enhancing mitochondrial function in Ctenopharyngodon idellus. Fish Shellfish Immunol. 127, 412–418 (2022).
Rohde, D. et al. S100A1 is released from ischemic cardiomyocytes and signals myocardial damage via Toll-like receptor 4. EMBO Mol. Med. 6, 778–794 (2014).
Timmers, L. et al. Toll-like receptor 4 mediates maladaptive left ventricular remodeling and impairs cardiac function after myocardial infarction. Circ. Res. 102, 257–264 (2008).
Olsen, A.-M. S. et al. Duration of treatment with nonsteroidal anti-inflammatory drugs and impact on risk of death and recurrent myocardial infarction in patients with prior myocardial infarction. Circulation 123, 2226–2235 (2011).
Abbate, A. et al. Effects of interleukin-1 blockade with anakinra on adverse cardiac remodeling and heart failure after acute myocardial infarction [from the Virginia Commonwealth University-Anakinra Remodeling Trial (2) (VCU-ART2) pilot study]. Am. J. Cardiol. 111, 1394–1400 (2013).
Broch, K. et al. Randomized trial of interleukin-6 receptor inhibition in patients with acute ST-segment elevation myocardial infarction. J. Am. Coll. Cardiol. 77, 1845–1855 (2021).
Yi, S. et al. Surface engineered polymersomes for enhanced modulation of dendritic cells during cardiovascular immunotherapy. Adv. Funct. Mater. 29, 1904399 (2019).
Zhang, X. et al. CC chemokine receptor 2-targeting copper nanoparticles for positron emission tomography-guided delivery of gemcitabine for pancreatic ductal adenocarcinoma. ACS Nano 15, 1186–1198 (2021).
van der Laan, A. M. et al. Monocyte subset accumulation in the human heart following acute myocardial infarction and the role of the spleen as monocyte reservoir. Eur. Heart J. 35, 376–385 (2013).
Casimiro, I., Chinnasamy, P. & Sibinga, N. E. S. Genetic inactivation of the allograft inflammatory factor-1 locus. Genesis 51, 734–740 (2013).
Wan, E. et al. Enhanced efferocytosis of apoptotic cardiomyocytes through myeloid-epithelial-reproductive tyrosine kinase links acute inflammation resolution to cardiac repair after infarction. Circ. Res. 113, 1004–1012 (2013).
Crooke, S. T., Witztum, J. L., Bennett, C. F. & Baker, B. F. RNA-targeted therapeutics. Cell Metab. 27, 714–739 (2018).
Shen, W. et al. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 37, 640–650 (2019).
Acknowledgements
This work was supported by the American Heart Association (grant CDA34110032 to M.D.) and the National Institutes of Health (grants R01HL122309 to E.B.T. and R01HL163635 to N.S.). These studies used sequencing services provided by the NUSeq Core and imaging services provided by the Center for Advanced Microscopy & Nikon Imaging Center at Northwestern University. Figure schematics were created with Servier Medical Art and BioRender. Copies of the licenses to use these programs are provided in Supplementary Table 8. Publication costs for this research were also supported by the Sidney & Bess Eisenberg Memorial Fund.
Author information
Authors and Affiliations
Contributions
M.D., K.G. and E.B.T. designed the research studies. M.D., K.G., D.P.S., S.P., B.R.L., M.I.T., X.L., N.E.S.S. and E.B.T. performed and analyzed experiments. C.L. analyzed single-nuclei RNA sequencing data. Z.-D.G. and S.H. performed echocardiography. A.M.v.d.L. and H.W.M.N. collected and provided human specimens. A.M. and S.Y. prepared and characterized ASOs. N.E.S.S. provided Aif1-deficient mice. M.D. and E.B.T. wrote the manuscript, and all authors contributed to manuscript revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Benjamin L. Prosser, Rong Tian and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
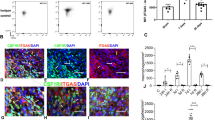
Extended Data Fig. 1 Allograft inflammatory factor 1 (Aif1) is enriched within recruited inflammatory macrophage clusters, but not lymphocytes, after myocardial infarction (MI) in mice.
A Using the Infection Atlas (Rizzo 2022) to visualize publicly available single cell RNA sequencing data (GSE135310, GSE197441, GSE197853), Aif1 expression was analyzed among monocytes and macrophages isolated from the hearts of mice at various time points after MI. Inflammatory myeloid subsets include Ly6Chi monocytes (Ly6Chi mo) and recruited macrophage clusters (Res MHCII, MHCII + IL1B + , and Isg15). B Aif1 gene expression in cardiac T and B cells 5 days after MI as visualized using the Infection Atlas. C AIF1 expression in peripheral blood lymphocytes 5 days after MI. Dashed lines represent fluorescence minus one (FMO) staining controls (n = 4 naïve and n = 4 day 5 MI, two-tailed unpaired t test, pooled from two independent experiments). Data are presented as mean ± SEM.
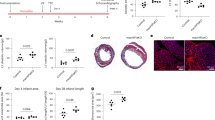
Extended Data Fig. 2 Allograft inflammatory factor 1 (Aif1) worsens cardiac repair after myocardial infarction (MI).
A Mice with whole body deletion of Aif1 (Aif1-/-) or controls (Aif1 + /+) were subjected to permanent occlusion MI. Percent infarct (INF)/left ventricle (LV), percent area-at-risk (AAR)/LV, and percent INF/AAR were measured one week after MI (n = 5 Aif1 + /+ and n = 6 Aif1-/-, two-tailed unpaired t test, pooled from two independent experiments). B Gene expression in whole cardiac extracts from Aif1 + /+ or Aif1-/- bone marrow chimera mice (n = 3 naïve and n = 5 day 3 MI, two-way ANOVA with Tukey’s test, pooled from two independent experiments). Data are presented as mean ± SEM.
Extended Data Fig. 3 Allograft inflammatory factor 1 (AIF1) in macrophages promotes glycolytic metabolism and inflammatory activation in response to necrotic myocardial cells but is dispensable for macrophage efferocytosis and efferocytic production of IL-10.
A Gene expression in bone marrow-derived macrophages (BMDM) after treatment with necrotic myocardial cell (NMC) extracts or lipopolysaccharide (LPS) (n = 4 untreated, n = 3 NMC or LPS, two-way ANOVA with Tukey’s test, two independent experiments). B Extracellular acidification rate (ECAR) in BMDMs after treatment with NMC extracts (n = 6 for all groups, two-way ANOVA with Tukey’s test, two independent experiments). C Percent efferocytosis of apoptotic Jurkat cells co-cultured with BMDMs. Scale bar, 10μm. D IL-10 production by efferocytic BMDMs (for C and D, n = 4 for all groups, two-tailed unpaired t test, two independent experiments). Data are presented as mean ± SEM.
Extended Data Fig. 4 Allograft inflammatory factor 1 (AIF1) promotes Hypoxia Inducible Factor (HIF)-1α-dependent glycolysis after myocardial infarction (MI).
A HIF-1α activation in cardiac macrophages 3 days after MI (n = 3 day 0 MI and n = 6 day 3 MI, two-way ANOVA with Tukey’s test, pooled from two independent experiments). B Isolation of cardiac macrophages 3 days after MI with representative enrichment assessed by flow cytometry. C Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) by cardiac macrophages isolated from the infarct 3 days after MI (n = 4 for all groups, two-way ANOVA with Tukey’s test, two independent experiments). D Publicly available microarray data (GSE112630) of cytokine and glycolytic gene expression among CCR2+ and CCR2− macrophages isolated from the hearts of patients during ischemic (ICM) or dilated cardiomyopathy (DCM). Data are presented as mean ± SEM.
Extended Data Fig. 5 Calcium influx is required for inflammatory glycolytic reprogramming of macrophages after TLR4 stimulation.
A Calcium influx in bone marrow-derived macrophages (BMDM) after TLR4 or ionomycin (IM) stimulation. Images are representative of two independent experiments. Scale bar, 20 μm. B Cytokine production by BMDMs treated with a cell-permeable, calcium chelator (BAPTA) (n = 4 for IL-6 and TNF-α and n = 3 for IL-10, two-way ANOVA with Tukey’s test, three independent experiments). Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) in BMDMs after treatment with calcium chelators, C BAPTA or D EGTA. Vehicle treated groups are the same in C and D (n = 6 vehicle and n = 8 for all other groups, two-way ANOVA with Tukey’s test, two independent experiments). E ECAR and OCR in BMDMs treated with ionomycin (n = 5 for all groups, two-way ANOVA with Tukey’s test, two independent experiments). Data are presented as mean ± SEM.
Extended Data Fig. 6 Allograft inflammatory factor 1 (AIF1) is dispensable for calcium influx in macrophages after TLR4 stimulation.
A Mean fluorescent intensity (MFI) of the calcium indicator dye, Fluo-4, in bone marrow-derived macrophages (BMDM) 30 minutes after TLR4 stimulation as measured by fluorescent microscopy. Scale bar, 10μm (n = 10 for all groups, two-way ANOVA with Tukey’s test, two independent experiments). B Kinetics of Fluo-4 MFI in BMDMs after TLR4 stimulation as measured by a fluorescent plate reader (n = 3 for all groups, two-way ANOVA with Tukey’s test, two independent experiments). Data are presented as mean ± SEM.
Extended Data Fig. 7 Actin polymerization is required for inflammatory glycolytic reprogramming of macrophages after TLR4 stimulation.
A Rac1 knockdown efficiency in bone marrow-derived macrophages (BMDM) (n = 4 for all groups, two-tailed unpaired t test, two independent experiments. B Cytokine production by BMDMs treated with a cell-permeable, actin polymerization inhibitor, Cytochalasin D (CytoD) (n = 4 for all groups, two-way ANOVA with Tukey’s test, three independent experiments). C Extracellular acidification rate (ECAR) in BMDMs after treatment with CytoD (n = 3 for all groups, two-way ANOVA with Tukey’s test, two independent experiments). D Actin polymerization in cardiac macrophages 3 days after MI (n = 3 day 0 MI and n = 6 day 3 MI, two-way ANOVA with Tukey’s test, pooled from two independent experiments). Data presented as mean ± SEM.
Extended Data Fig. 8 Role of Hypoxia inducible factor 1 (HIF)-1α in Rac1 activation, membrane ruffling, glycolysis, and transcription after TLR4 stimulation.
A Rac1-GTP levels in bone marrow derived macrophages (BMDMs) after 1 hour of TLR4 stimulation with lipopolysaccharide (LPS). Dashed line represents background absorbance level (n = 4 for all groups, two-way ANOVA with Tukey’s test, two independent experiments). B F-actin immunostaining of BMDMs after 1 hour of TLR4 stimulation with quantification of membrane ruffling area (μm2) (n = 30 for all groups, whiskers 10-90 percentile, two-way ANOVA with Tukey’s test, two independent experiments). C Extracellular acidification rate (ECAR) in BMDMs after TLR4 stimulation with LPS (n = 12 vehicle and n = 16 LPS, two-way ANOVA with Tukey’s test, two independent experiments). D ECAR of Hif1 + /+ and Hif1-/- BMDMs after TLR4 stimulation (n = 14 unstimulated and n = 16 LPS, two-way ANOVA with Tukey’s test, two independent experiments). E Gene expression in BMDMs after TLR4 stimulation (n = 4 for all groups, one-way ANOVA with Dunnett’s test, two independent experiments). F Gene expression in Hif1 + /+ and Hif1-/- BMDMs after 3 hours of TLR4 stimulation (n = 3 unstimulated and n = 4 LPS, two-way ANOVA with Tukey’s test, two independent experiments). Data presented as mean ± SEM.
Extended Data Fig. 9 Allograft inflammatory factor 1 (Aif1) antisense oligonucleotide (ASO) dose response.
ASOs were formulated in PBS, administered at doses of 1.6, 8, 40, and 75 mg/kg, and injected subcutaneously on days 0, 6, and 10. Aif1 tissue expression and general ASO tolerability was evaluated on day 14. Gene expression levels of Aif1 in A heart and B liver. Gene expression levels of Ccl2 in C heart and D liver. E Liver or F spleen weight as a percent of total body weight. Plasma levels of G alanine transaminase or H creatinine (n = 6 PBS, n = 3 for 1.6, 8, and 40 mg/kg, and n = 2 for 75 mg/kg, one-way ANOVA with Dunnett’s test, pooled from two independent experiments). Data presented as mean ± SEM.
Extended Data Fig. 10 Mechanical regulation of macrophage metabolism by Allograft Inflammatory Factor 1 (AIF1) leads to adverse remodeling after cardiac injury.
Myocardial infarction leads to necrotic death of cardiomyocytes and release of damage associated molecular patterns (DAMPs). Recognition of DAMPs on macrophages by Toll-like Receptors (TLR), including TLR4, leads to calcium influx and activation of Allograft Inflammatory Factor 1 (AIF1). AIF1 interacts with RAC1 to promote actin polymerization and mechanical stiffness. This activates NADPH oxidase (NOX) and increases reactive oxygen species (ROS) production. Elevated ROS levels stabilize Hypoxia Inducible Factor (HIF)-1α, leading to its nuclear translocation and transcription of genes involved in glycolysis and inflammation. This switch to glycolytic metabolism fuels increased production of proinflammatory cytokines, including interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α, which contribute to adverse left ventricular remodeling and progression to heart failure (HF).
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Table
Supplementary Tables 1–8. Lists of antibodies, chemicals, peptides and recombinant proteins, commercial assays, cell lines, mouse strains, qPCR primers, recombinant DNA and software.
Source data
Source Data Fig. 1
Source data Fig. 1d–f.
Source Data Fig. 2
Source data Fig. 2b–f.
Source Data Fig. 3
Source data Fig. 3b–d.
Source Data Fig. 4
Source data Fig. 4a–h.
Source Data Fig. 5
Source data Fig. 5a–i.
Source Data Fig. 6
Source data Fig. 6a–h.
Source Data Fig. 7
Source data Fig. 7a–f.
Source Data Fig. 8
Source data Fig. 8b–h.
Source Data Extended Data Fig. 1
Source data Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Source data Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Source data Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Source data Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Source data Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Source data Extended Data Fig. 6.
Source Data Extended Data Fig. 7
Source data Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Source data Extended Data Fig. 8.
Source Data Extended Data Fig. 9
Source data Extended Data Fig. 9
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
DeBerge, M., Glinton, K., Lantz, C. et al. Mechanical regulation of macrophage metabolism by allograft inflammatory factor 1 leads to adverse remodeling after cardiac injury. Nat Cardiovasc Res 4, 83–101 (2025). https://doi.org/10.1038/s44161-024-00585-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44161-024-00585-y