Abstract
Group 2 innate lymphoid cells (ILC2s) directly contribute to local inflammation in type 2 inflammatory airway diseases. Here, we identify ILC2 subsets by single cell RNA sequencing in chronic rhinosinusitis with nasal polyps (CRSwNP) and in a memory inflammatory mouse model. We find that toll-like receptor 4 (TLR4)+ILC2s, with similar markers to their human counterparts, expresse memory cell markers, persist over time, and respond more vigorously to a secondary unrelated antigen challenge in the mouse model. Genetic ablation of TLR4 or blockade by anti-TLR4 antibodies leads to the reduction of IL-13 expression from ILC2s and mucus production in mice. The assay for transposase-accessible chromatin sequencing further confirms the importance of accessible TLR4 gene loci and its down-stream signaling pathway in maintaining trained immunity of TLR4+ILC2s after repeated stimulation by HDM. Taken together, TLR4 has a function in trained immunity maintenance within ILC2s, which may contribute to disease chronicity through a non-specific immunological memory.
Similar content being viewed by others
Introduction
Chronic rhinosinusitis with nasal polyps (CRSwNP) and asthma are chronic airway inflammatory diseases with a type 2 inflammatory signature, which respectively affect approximately 1.1–4.3% and 0.9–29% population globally1,2. In Asian countries, especially in Beijing, China, there has been a notable rise over the past two decades in the type 2 endotype of CRSwNP patients, which is characterized by increased infiltration of eosinophils and elevated release of type 2 cytokines such as IL-4, IL-5, and IL-133. The finding that up to 67% of IL-5+CRSwNP patients have comorbid asthma suggests that disease severity and recurrence rate may be higher in type 2 versus non-type 2 inflammatory diseases4. Thus, further understanding type 2 inflammation is likely to lead to better management of persistent inflammatory airway diseases.
The pathophysiology of type 2 inflammation is predominantly driven by T helper 2 (Th2) cells and group 2 innate lymphoid cells (ILC2s). Upon house dust mite (HDM) re-challenge, tissue resident memory Th2 cells can rapidly proliferate near airways in mice, and induce mucus metaplasia, airway hyperresponsiveness, and airway eosinophil activation5. It has also been shown that CD69+ST2+Th2 memory T cells have long-term memory and can propagate chronic type 2 inflammation in lung tissue of mice, following exposure to ovalbumin (OVA), an experimental allergen6. Similarly, it has been shown that IL-5-producing memory pathogenic Th2 cells in polyp tissue of eosinophilic CRSwNP patients are critical for persistent type 2 inflammation7. Taken together, these findings suggest that memory Th2 cells are involved in disease chronicity of type 2 airway inflammatory diseases in an antigen-specific way. In airways, ILC2s, as a distinct innate lymphocyte subset, initiate a type 2 immune response8. Cumulating evidences suggest that trained immunity of innate immune cells can generate immunological memory towards a secondary unrelated antigen challenge9,10,11. Indeed, allergen-experienced ILC2s in the lungs and lymph nodes of experimental mice have been shown to exhibit a gene expression profile like that of memory T cells and persist for several months, mediating severe allergic inflammation in response to unrelated allergens12. However, it remains unclear whether ILC2s exist trained immunity in human airway diseases, contributing to persistent type 2 inflammation in an antigen-unspecific way. Also, identification and maintenance mechanisms for trained ILC2s vary in different organs of mice and human13. In this regard, characterization of trained ILC2s in human inflammatory airway disease as a potential target would likely be useful in the management of persistent type 2 inflammations.
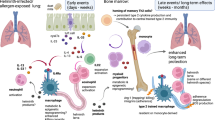
In this work, we investigate ILCs at single-cell resolution in polyp tissue of patients with CRSwNP. Then, we explore the characteristics of trained ILC2s in the polyp tissue and assess whether their presence correlates with disease severity of type 2 inflammatory airway disease. To elucidate the maintenance mechanisms for trained ILC2s, we establish a mouse model with an inflammatory memory response. Mouse ILC2s subsets are identified by single-cell RNA sequencing (scRNA-seq), and the properties of trained ILC2s are further confirmed by assessing production of cytokines from these cells in a mouse model following stimulation with related and unrelated antigens. In addition, we estimate the maintenance of trained ILC2s in the mouse model using genetic ablation and pharmacological inhibition. The underlying mechanisms are implicated using the assay for single-cell transposase-accessible chromatin sequencing (scATAC-seq).
Results
ILC subsets in polyp tissue of CRSwNP patients
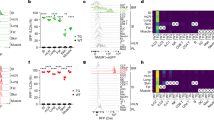
Following scRNA-seq of sorted cells (CD45+Lin⁻) (Lin⁻ defined by the absence of CD1a, CD3, CD11c, CD14, CD16, CD19, CD34, CD94, CD123, CD303, TCRαβ, TCRγδ, and FcεRIα) from five CRSwNP patients using the BD Rhapsody single cell sequencing platform (Fig. 1A), a total of 22,103 cells were clustered for analysis. Overall, natural killer (NK) & ILC1 (NKG7, TBX21, IKZF3, and IFNG), ILC3 (RORC, IL1R1, KIT, and TCF7), ILC2_c01 (ID2, GATA3, and IL7R), and ILC2_c02 (ID2, GATA3, IL17RB, HPGDS, TLR4, IL13, and IL5)14 were found in nasal polyp tissue (Fig. 1B, C). In agreement with these findings, re-analysis of a publicly available scRNA-seq dataset15 consistently demonstrated 2 subsets of ILC2s, ILC2_c01 and ILC2_c02, in polyp tissue (Supplementary Fig. S1A, B). Notably, ILC2_c02 expressed a higher level of memory cell markers and cytokines such as IL17RB, IL1RL1, IL13, TLR4, HPGDS, PTGS2, and CXCL8, when compared to those of ILC2_c01 (Fig. 1D and Supplementary Fig. S1A). The percentage of ILC2_c02 was also lower than ILC2_c01 in both the present and publicly available RNA-seq datasets (Supplementary Fig. S1C, D). Furthermore, the ILC2_c02 cluster was upregulated in patients with eosinophilic CRSwNP, which is characterized by elevated type 2 inflammation and eosinophil infiltration (Supplementary Fig. S1E). Collectively, these suggest that ILC2_c02, with a higher level of cytokines, despite its lower abundance, might experience a specialized form of immune training. Enrichment analysis of differentially expressed genes between ILC2_c01 and ILC2_c02 highlighted that some pathways, including the IL-4 and IL-13 signaling pathway, toll-like receptor cascades, and MYD88-independent toll-like receptor 4 (TLR4) cascade, were significantly more active in ILC2_c02 than in ILC2_c01 (Fig. 1E).
A Schematic diagram of experimental study design for single-cell RNA-sequencing (scRNA-seq) of nasal polyp tissues. Created in BioRender. Wang, Z. (2025) https://BioRender.com/xarrgf5. B Uniform manifold approximation and projection (UMAP) plots of purified and sorted viable CD45+Lin⁻ cells from five chronic rhinosinusitis with nasal polyp (CRSwNP) patients. Each dot represents a single cell, with different innate lymphoid cell (ILC) subsets shown in different colors. C Signature genes for identification of ILC subsets. D The scatter plot manifesting the differentially expressed genes (DEGs) between ILC2_c01 and ILC2_c02 subsets in polyp tissue of CRSwNP patients, colored by adjusted P-values. Adjusted P-values were calculated by a two-tailed Wilcoxon test with Bonferroni correction. E Over-representation analysis showing enriched immune pathways of the ILC2_c02 cluster compared to ILC2_c01 cluster. P-values were calculated by right-tailed Fisher’s Exact Test with Benjamini-Hochberg correction. F RNA velocity analysis of ILC2_c01 and ILC2_c02 subsets with velocity field projected onto the UMAP plot from Fig. 1B. Arrows show the local average velocity evaluated on a regular grid and indicate the extrapolated future states of cells. G Monocle pseudo-time analysis revealing the progression between ILC2_c01 and ILC2_c02 subsets in polyp tissue. H Heatmap of the level of expression for 2621 differentially expressed genes over the pseudo-time trajectory (shown on the left), highlighting specific representative genes in each gene cluster (shown along the right margin).
To trace the origin of the two ILC2 subsets, we performed RNA velocity and pseudo-time analysis. Velocity stream revealed a possible transition from the ILC2_c01 to ILC2_c02 (Fig. 1F); with the majority of the ILC2_c02 subset appearing predominantly distributed towards the trajectory’s terminus (Fig. 1G). Clustering the genes along with pseudo-time showed that differentially expressed genes such as IL13, IL17RB, TLR4 and IL1RL1 in the ILC2_c02 subset tended to increase toward to the terminus of the trajectory (Fig. 1H), suggesting that these differentially expressed genes may facilitate the transition from ILC2_c01 to ILC2_c02.
Human TLR4+ILC2s are closely linked to type 2 inflammation
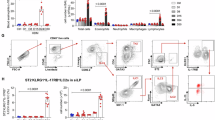
Reportedly, ILC2s express TLR1, 2, 4 and 6, and the activation of ILC2s is partially dependent on TLR ligand stimulation16,17; TLR4 is involved in the regulation of trained immunity of macrophages18. Supplementary Figure S1F illustrated a marked preference for TLR4 expression on ILC2_c02 of nasal polyp tissue, distinguishing from ILC2_c01. Flow cytometric analysis of human nasal single cell suspension showed that ILC2s (CD45+Lin⁻CD127+CD161+CRTH2+) were segregated into two distinct subsets: those with TLR4 expression (TLR4+ILC2s) and those without TLR4 expression (TLR4⁻ILC2s) (Fig. 2A and Supplementary Fig. S1G). Moreover, the expression of IL-17RB, CD44, IL-5, and IL-13 was significantly higher in TLR4+ILC2s than in TLR4⁻ILC2s (Fig. 2B, C). Consistent with the presence of TLR4+ILC2s in polyp tissue, TLR4+ILC2s were also found in induced sputum of CRSwNP patients with comorbid asthma (Supplementary Fig. S1H).
A Representative contour plot analysis for TLR4+ILC2s (CD45+Lin⁻CD127+CRTH2+CD161+TLR4+) in a single cell suspension of nasal polyp tissues from CRSwNP patients. B Representative histogram showing the expression levels of IL-17RB, IL-13, IL-5 and CD44 on TLR4+ILC2s and TLR4⁻ILC2s in polyp tissues of CRSwNP patients. Shading (FMO) indicates isotype control. C Mean fluorescence intensity (MFI) for IL-17RB, IL-13, IL-5, and CD44 detected by flow cytometry on TLR4+ILC2s and TLR4⁻ILC2s in CRSwNP patients (n = 15). Each symbol represents an individual patient, and bars represent mean ± SEM. P-values were calculated by two-tailed paired Wilcoxon tests. D, E Representative contour plot analysis for total ILC2s (CD45+Lin⁻CD127+CRTH2+CD161+ cells) (D), TLR4+ILC2s and TLR4⁻ILC2s (E) in nasal polyp tissues from CRSwNP patients. F Percentage of total ILC2s, TLR4+ILC2s and TLR4⁻ILC2s in nasal polyp tissues of IL-5⁻CRSwNP (n = 9) versus IL-5+CRSwNP patients (n = 29). Each symbol represents an individual patient, and bars represent mean ± SEM. P-values were calculated by two-tailed Mann-Whitney tests. G. Correlation between the frequency of TLR4+ILC2s and percentage of eosinophils, and concentrations of ECP, IL-13 and IL-5 in polyp tissues. Each symbol represents an individual patient. P and r-values were calculated by a two-tailed Spearman correlation test.
CRSwNP patients were divided into two groups, IL-5+CRSwNP and IL-5⁻CRSwNP, according to the presence of type 2 inflammation. As shown in the Table 1, IL-5+CRSwNP patients exhibited increased levels of IL-5, IL-13, eosinophil cationic protein (ECP), and a higher eosinophil count compared to those of IL-5⁻CRSwNP patients. Next, we assessed the relative abundance of TLR4+ILC2s in polyp tissue of CRSwNP patients. The frequency of total ILC2s and TLR4+ILC2s was significantly increased in IL-5+CRSwNP patients compared to IL-5⁻CRSwNP patients, whereas there was no significant difference in the frequency of TLR4⁻ILC2s between IL-5+CRSwNP and IL-5⁻CRSwNP patients (Fig. 2D–F). Furthermore, TLR4+ILC2s, but not TLR4⁻ILC2s, were positively correlated with the number of tissue eosinophils (r = 0.4236; P = 0.0080), expression of tissue IL-13 (r = 0.4471; P = 0.0049), and IL-5 (r = 0.4801; P = 0.0023) (Fig. 2G and Supplementary Fig. S1I). Both TLR4+ILC2s and TLR4⁻ILC2s were positively correlated with the expression of ECP (Fig. 2G and Supplementary Fig. S1I). Taken together, these findings suggest that TLR4+ILC2s might be closely linked to type 2 inflammation in airway diseases.
Mouse TLR4+ILC2s expressing memory cell markers persist for a long period in a type 2 inflammatory model
Given that TLR4+ILC2s with memory markers are associated with type 2 inflammation in airway inflammatory diseases, we developed a mouse type 2 inflammatory model with memory response to evaluate the characteristics and function of TLR4+ILC2s using the Rag1⁻/⁻ mice (lacking T and B cells) (Fig. 3A). Total ILC2s (CD45+Lin⁻ST2+ICOS+) (Lin⁻ defined by the absence of CD3e, CD11b, CD45R/B220, Ly76, Ly6G, Ly6C, and FcεRIα), TLR4+ILC2s, and TLR4⁻ILC2s were present in lung tissue of the mice at day 17 with intra-nasal HDM extracts challenge for 5-consecutive days (Fig. 3B, C). After ceasing the challenge for four weeks, the percentage of total ILC2s, TLR4⁻ILC2s, and IL-13+TLR4⁻ILC2s was significantly decreased in lung tissue at day 45 during the memory phase, along with reduced tissue IL-13 and percentage of MUC5AC+ cells in epithelial layers (Fig. 3B–E and Supplementary Fig. S2A–D). However, re-exposure to HDM led to an increased accumulation of TLR4+ILC2s and IL-13+TLR4+ILC2s in the lung tissues at day 47, accompanied by elevated tissue IL-13 and percentage of MUC5AC+ cells (Fig. 3C, D and Supplementary Fig. S2B–D). Since TLR4+ILC2s were absent in the bronchial epithelial layer, whereas MUC5AC was localized to goblet cells within the surface epithelium, suggesting that TLR4+ILC2s indirectly regulate mucus secretion likely via IL-13 (Supplementary Fig. S2E). HDM re-exposure on day 45 exclusively increased TLR4⁻ILC2s, IL-13+TLR4⁻ILC2s, and IL-13+TLR4+ILC2s in wild-type mice (Supplementary Fig. S3A, B). These findings indicate that, unlike TLR4⁻ILC2s, adaptive immunity plays a limited role in modulating TLR4+ILC2s.
A. Experimental protocol. i.n., intranasal administration; HDM, house dust mite. B–E. Representative contour plot and quantification showing the percentage of total ILC2s (B), TLR4+ILC2s and TLR4⁻ILC2s (C), IL-13+TLR4+ILC2s (D) and IL-13+TLR4⁻ILC2s (E) in lung tissues of Rag1⁻/⁻ mice on days 0, 17, 45, and 47 (n = 8 mice per group). Each symbol indicates one mouse, and bars represent mean ± SEM. Data were pooled from two independent experiments. P-values were calculated by two-tailed one-way ANOVA with Tukey’s Honestly Significant Difference test.
To further characterize ILC2s during the memory phase in mice, we performed scRNA-seq on sorted ILC2s (CD45+Lin⁻ICOS+ST2+) from lung tissue of wild-type mice sensitized in the same way as in Fig. 3A (Fig. 4A). We analyzed 30,277 cells, identifying three distinct ILC2 subsets: ILC2_c01, ILC2_c02, and ILC2_c03 in the lung tissue (Fig. 4B). Among them, ILC2_c01 and ILC2_c02 were present on both day 45 and day 47, while ILC2_c03 was mainly found on day 47 (Fig. 4C). ILC2_c02 subset displayed significantly higher mRNA levels of memory cell markers, type 2 cytokines, and cell cycle-related genes such as Tlr4, Il5, Il13, Il1rl1, Cd44, Bcl2 and Ccr8, compared to ILC2_c01 subset (Fig. 4D–F). This suggests that ILC2_c02 possesses memory characteristics. The ILC2_c03 subset was notable for its elevated levels of Il13, Cd74, Il1r2, and Mki67 (Fig. 4D, E). The velocity stream indicated a transition from the ILC2_c01 subset and ILC2_c03 subset to the ILC2_c02 subset, further supporting the possibility of acquiring memory properties in the ILC2_c02 subset (Fig. 4G). Tlr4 mRNA was primarily found in ILC2_c02, with low expression in both ILC2_c03 and ILC2_c01 subsets (Fig. 4H). Based on TLR4 surface expression analyzed by flow cytometry, we divided ILC2s into TLR4+ILC2s and TLR4⁻ILC2s subsets (Supplementary Fig. S3C). These sorted mouse ILC2 subsets were used for bulk RNA-seq via Smart-seq2. Transcriptomic analysis revealed that sorted TLR4+ILC2s generally reflect the ILC2_c02 cluster, whereas sorted TLR4⁻ILC2s closely match the ILC2_c01 signature (Supplementary Fig. S3D, E). As demonstrated in Fig. 4I, J, mouse TLR4+ILC2s on day 47 exhibited significantly higher levels of IL-5, IL-13, IL-17RB, CD44, and CCR8 than TLR4⁻ILC2s. Mki67 serves as a characteristic differential gene of the ILC2_c03 subset and can be used as an intracellular identification marker. According to flow cytometry analysis, Ki67+TLR4⁻ILC2s expressed more IL-5 and IL-13 than Ki67⁻TLR4⁻ILC2s, but less than TLR4+ILC2s (Supplementary Fig. S3F–H). This possibly indicates that without further distinguishing the ILC2_c03 subset in TLR4⁻ILC2s on day 47, TLR4+ILC2s consistently secreted higher levels of IL-5 and IL-13 than that of TLR4⁻ILC2s. Overall, TLR4 may serve as a surrogate identifier for trained ILC2s (ILC2_c02) in both mice and human airway tissue.
A Schematic diagram of experimental study design for single-cell (sc) RNA-seq of lung tissues. B Uniform manifold approximation and projection (UMAP) plots of ILC2 subsets from HDM-challenged mice at day 45 and day 47. Each dot represents a single cell, with different ILC2 subsets shown in different colors. C UMAP plots displaying sorted ILC2 subsets at different time points. D Signature genes used for ILC2s subsets identification. E Heatmap displaying the differential gene expression among three ILC2 clusters. The right panel displays enriched pathways associated with the marker genes of each cluster. Enriched pathways of different ILC2 subsets are shown in different colors. P-values were calculated by right-tailed Fisher’s Exact Test with Benjamini-Hochberg correction. F. The scatter plot manifesting the differentially expressed genes (DEGs) between ILC2_c01 and ILC2_c02 subsets, colored by adjusted P-values. Adjusted P-values were calculated by a two-tailed Wilcoxon test with Bonferroni correction. G RNA velocity analysis of ILC2_c01, ILC2_c02 and ILC2_c03 subsets with velocity field projected onto the UMAP plot from Fig. 4B. Arrows show the local average velocity evaluated on a regular grid and indicate the extrapolated future states of cells. H UMAP showing the gene expression intensity of Tlr4 on ILC2 clusters. I Representative histogram showing the expression levels of IL-5, IL-13, CCR8, IL-17RB, and CD44 on TLR4+ILC2s and TLR4⁻ILC2s detected by flow cytometry in lung tissues of HDM-sensitized mice at day 47. J Mean fluorescence intensity (MFI) of IL-5, IL-13, CCR8, IL-17RB, and CD44 on TLR4+ILC2s and TLR4⁻ILC2s detected in the lung tissues of HDM-sensitized mice at day 47 (n = 6 mice per group). Data were pooled from two independent experiments. Each symbol indicates one mouse, and bars represent mean ± SEM. P-values were calculated by two-tailed Wilcoxon tests.
To determine which components of HDM extracts induce TLR4 expression on ILC2s, mouse ILC2s were stimulated with HDM extracts, Der p1, and lipopolysaccharide (LPS) in vitro. Both Der p1 and HDM extracts, unlike endotoxin LPS, induced TLR4 expression on ILC2s (Supplementary Fig. S4A). Stimulation with Der p1 and HDM also upregulated phosphorylation of transcription factor c-Fos in ILC2s, as confirmed by the inhibitor T-5224 (Supplementary Fig. S4B, C). Changes in p-Jun and PU.1 expression were observed only with HDM extracts, not Der p1 (Supplementary Fig. S4B, D). Thus, allergens in HDM extracts, rather than endotoxin LPS, predominantly induced TLR4 expression in ILC2s via activation of c-Fos.
Mouse TLR4+ILC2s respond more vigorously than TLR4⁻ILC2s to a secondary antigen challenge
To continue investigating the properties of TLR4+ILC2s, the Rag1⁻/⁻ mice undergone intranasal HDM challenge for 5-consecutive days, were challenged again with HDM or saline after 4 weeks at day 45 (Fig. 5A). Overall, total cells in bronchoalveolar lavage fluid (BALF), lung tissue TLR4+ILC2s, IL-13 expression, and MUC5AC+ cells were significantly increased in HDM-sensitized Rag1⁻/⁻ mice with the recall HDM challenge compared with saline challenged mice (Fig. 5B–E). The number of TLR4⁻ILC2s, however, was not significantly altered in the mice with the recall HDM challenge (Fig. 5E).
A Experimental protocol for HDM + HDM sensitization in Rag1⁻/⁻mice. i.n., intranasal administration; HDM, house dust mite. B Representative image of MUC5AC immunohistochemistry staining picture. C, D Representative contour plots analysis of total ILC2s (C), TLR4+ILC2s and TLR4⁻ILC2s (D). E Number of total cells in the bronchoalveolar lavage fluid (BALF), percentage of total ILC2s, TLR4+ILC2s, TLR4⁻ILC2s, concentration of tissue IL-13 and percentage of MUC5AC+cells among the epithelial cells (n = 5 mice per group). Each symbol indicates one mouse, and bars represent mean ± SEM. F Experimental protocol for HDM + papain sensitization in Rag1⁻/⁻mice. G Representative image of MUC5AC immunohistochemistry staining picture. H, I Representative contour plots analysis of total ILC2s (H), TLR4+ILC2s and TLR4⁻ILC2s (I). J Number of total cells in the BALF, percentage of total ILC2s, TLR4+ILC2s, TLR4⁻ILC2s, concentration of tissue IL-13, and percentage of MUC5AC+cells among the epithelial cells (n = 5 mice per group). Each symbol indicates one mouse, and bars represent mean ± SEM. Data for each experimental protocol were pooled from two independent experiments. P-values were calculated by two-tailed one-way ANOVA with Tukey’s Honestly Significant Difference test.
To assess the trained immunity of TLR4+ILC2s, we examined HDM-sensitized Rag1⁻/⁻ mice that received a single intranasal papain challenge on day 45 (Fig. 5F). Compared to saline challenged controls, the papain challenge in HDM-sensitized Rag1⁻/⁻ mice significantly increased total cells in BALF, TLR4+ILC2s, MUC5AC+ cells, and the concentration of IL-13, but not the number of TLR4⁻ILC2s in the lung tissues (Fig. 5G–J). Collectively, these results indicate that HDM-experienced TLR4+ILC2s can vigorously respond to a secondary unrelated antigen challenge and induce more severe lung inflammation than that of TLR4⁻ILC2s.
To confirm the role of TLR4+ILC2s in eliciting severe inflammation, sorted TLR4+ILC2s or TLR4⁻ILC2s from HDM/HDM-sensitized wild type mice were intravenously transferred into Rag2⁻/⁻Il2rg⁻/⁻ mice (lacking mature T cells, B cells, and ILCs), which were then sensitized by intranasal HDM challenged as indicated (Fig. 6A). Assessment of lung inflammation in these mice revealed that the percentage of eosinophils and MUC5AC+ cells, IL-13 expression, and airway hyperreactivity were significantly higher in mice receiving TLR4+ILC2s than those of mice receiving TLR4⁻ILC2s (Fig. 6B–F), indicating that TLR4+ILC2s, acting as trained ILC2s, are able to enhance type 2 inflammation upon a recall challenge.
A Experimental protocol. i.n., intranasal administration; i.v., tail intravenous injection; HDM, house dust mite. B Representative contour plots analysis eosinophils in lung tissues of different groups. C–E Percentage of eosinophils in lung tissues (C), concentration of tissue IL-13 (D) and airway hyperreactivity (E) in Rag2⁻/⁻Il2rg⁻/⁻C57BL/6 J mice following transfer of ILC2 subsets from HDM-sensitized wild type C57BL/6 J mice (TLR4+ILC2 or TLR4-ILC2 group, n = 4 mice per group; other group, n = 5 mice per group). F Representative image of MUC5AC immunohistochemistry staining picture (left) and statistics summarizing (right) in lung tissue of Rag2⁻/⁻Il2rg⁻/⁻C57BL/6 J mice (TLR4+ILC2 or TLR4-ILC2 group, n = 4 mice per group; other group, n = 5 mice per group). Each dot indicates one mouse, and bars represent mean ± SEM. Results for (C–F) were confirmed in two independent experimental replicates, with representative data shown. P-values were calculated by two-tailed one-way ANOVA with Tukey’s Honestly Significant Difference test.
TLR4 is essential for trained immunity of ILC2s
To further demonstrate the role of TLR4 in eliciting trained immunity of ILC2s, the HDM-sensitized Tlr4⁻/⁻ mice were challenged again with HDM or saline after 4 weeks at day 45 (Fig. 7A). The data showed that TLR4 deficiency led to a reduction in the percentage of total ILC2s, IL-13+ILC2s, MUC5AC+ cells, and tissue IL-13 expression (Fig. 7B–E). And then, we intraperitoneally administrated neutralizing TLR4 antibody on day 45 in HDM-sensitized Rag1⁻/⁻ mice 2 h before the recall HDM challenge (Fig. 7F). This treatment also inhibited the recall challenge-induced elevation of total inflammatory cells in BALF, the percentage of total ILC2s and MUC5AC+ cells, and the expression of IL-13 in lung tissue (Fig. 7G–J). Next, we sorted TLR4+ILC2s from HDM/HDM-sensitized wild-type mice, and then adoptively transferred them into Rag2⁻/⁻Il2rg⁻/⁻ mice (Fig. 7K). Two hours before intranasal HDM challenge, the recipient mice received an intraperitoneal injection of the TLR4-neutralizing antibody (Fig. 7K). The anti-TLR4 antibody blockade reduced total inflammatory cells in BALF, tissue eosinophils, MUC5AC+ cells, and airway hyperreactivity in Rag2⁻/⁻Il2rg⁻/⁻ mice that had received TLR4+ILC2s (Fig. 7L–O).
A Experimental protocol employing HDM-sensitized Tlr4⁻/⁻ mice. i.n., intranasal administration; HDM, house dust mite. B, C Representative contour plots analysis of total ILC2s (B) and IL-13+ILC2s (C). D Representative image of MUC5AC immunohistochemistry staining picture to evaluate mucus. E Percentage of total cells in the bronchoalveolar lavage fluid (BALF), percentage of total ILC2s and IL-13+ILC2s in lung tissues, concentration of IL-13 in lung tissues, and the percentage of MUC5AC+cells among the epithelial cells of mice (wild-type group, n = 6 mice; Tlr4⁻/⁻group, n = 5 mice). F Experimental protocol employing HDM-sensitized Rag1⁻/⁻ mice treated with TLR4 antibody. i.n., intranasal administration; i.p., intraperitoneally injection; HDM, house dust mite; Ab, antibody. G, H Representative contour plots analysis of total ILC2s (G) and IL-13+ILC2s (H). I Representative image of MUC5AC immunohistochemistry staining picture to evaluate mucus. J Percentage of total cells in the bronchoalveolar lavage fluid (BALF), percentage of total ILC2s and IL-13+ILC2s in lung tissues, concentration of IL-13 in lung tissues, and the percentage of MUC5AC+cells among the epithelial cells of mice (control group, n = 6 mice; anti-TLR4 group, n = 6 mice). K Experimental protocol employing Rag2⁻/⁻Il2rg⁻/⁻C57BL/6 J mice. i.n., intranasal administration; i.v., tail intravenous injection; i.p., intraperitoneally injection; HDM, house dust mite; Ab, antibody. L–O Representative contour plots analysis of eosinophils in lung tissues (L), representative image of MUC5AC immunohistochemistry staining (M), percentage of total cells in the bronchoalveolar lavage fluid (BALF), eosinophils in lung tissues, the percentage of MUC5AC+cells among the epithelial cells of mice (N), and airway hyperreactivity (O) in Rag2⁻/⁻Il2rg⁻/⁻C57BL/6 J mice injected with purified TLR4+ILC2s from HDM-sensitized wild type C57BL/6 J mice and then treated with TLR4 antibody before the following HDM challenge (n = 5 mice in each group). Each dot indicates one mouse, and bars represent mean ± SEM. Data for E and J were pooled from two independent experiments. Results for N to O were confirmed in two independent experimental replicates, with representative data shown. P-values calculated by a two-tailed unpaired t test.
To further confirm the role of TLR4 in trained immunity of ILC2s, we established an in vitro trained ILC2 model by stimulating mouse TLR4⁻ILC2s with HDM as indicated in the Fig. 8A. Following HDM training, ILC2s exhibited higher expression of TLR4 and IL-17RB and greater intracellular IL-5 and IL-13 accumulation than untreated controls (Fig. 8B). Consistent with this finding, stimulation with the HDM component LPS similarly enhanced IL-5 and IL-13 expression in trained ILC2s (Fig. 8C). When TLR4⁻ILC2s were pre-treated with the TLR4 antagonist TAK242 for 1 h before training, we observed a significant reduction in IL-13 expression in trained ILC2s, irrespective of HDM re-exposure (Fig. 8D, E). Collectively, TLR4 signaling may play a role in mediating ILC2 memory formation, not be always regarded as a surface marker.
A Schematic diagram for in vitro inducing trained ILC2s from sorted mouse TLR4⁻ILC2s. HDM, house dust mite; LPS, lipopolysaccharide. B, C Mean fluorescence intensity (MFI) of TLR4, IL-5, IL-13, Ki67 and IL-17RB in in vitro-generated trained ILC2s derived from TLR4⁻ILC2s. Data were pooled from three independent experiments. P-values were calculated by two-sided one-way ANOVA with Tukey’s Honestly Significant Difference test (B) or two-sided unpaired t test (C). D, E IL-5, IL-13 and Ki67 MFI in in vitro-generated trained ILC2s, which were pretreated with TAK242 (TLR4 antagonist, 1 h). Data were pooled from three independent experiments. P-values were calculated by a two-sided unpaired t test.
Epigenetic reprogramming of trained ILC2s
To investigate the underlying epigenetic reprograming of TLR4+ILC2s, we analyzed the profiles of DNA accessibility using scATAC-seq in sorted cells (CD45+Lin⁻ICOS+ST2+) from lung tissue of wild-type mice, which sensitized in the same way as in Fig. 3A (Fig. 9A). As shown in Fig. 9B, a clear separation between ILC2s at day 45 and at day 47 was visible. Regarding ILC2_c02, there was an increase in chromatin accessibility for genes including Il1rl1, Runx2, Ptgs2, Tlr4, Gata3, and Il5 at day 47 compared to day 45 (Fig. 9C). Moreover, motif analysis indicated that transcription factors such as JDP2, JunB, Fos, BACH2, BATF, and GATA3 were involved in driving the chromatin accessibility of genes at day 47 (Fig. 9D). By genome visualization, we found that the accessibility of Tlr4 and Il1rl1 promoter in ILC2_c02 on day 47 was greater than that on day 45 (Fig. 9E). Notably, the open chromatin region in the Tlr4 promoter contained JunB and JunD motifs, followed by Fos motif and PU.1 motif (Fig. 9F). Our predictions suggested an increased binding activity of the AP-1 family (consists of Fos and JunB) contribute to enhanced accessibility of Tlr4 in ILC2_c02 on day 47 (Fig. 9G). In addition, the binding activity of MafK, which is downstream of Tlr4, was also elevated (Fig. 9G). Collectively, the epigenetic reprogramming regulates the chromatin accessibility of Tlr4 and Il1rl1, contributing to the trained immunity of ILC2s.
A Schematic diagram of the experimental study design for scATAC-seq of lung tissues from wide-type HDM sensitized mice. B Uniform manifold approximation and projection (UMAP) plots of sorted viable CD45+Lin⁻ICOS+ST2+cells from HDM-sensitized mice at day 45 and day 47. Each dot represents a single cell, with different ILC2 subsets shown in different colors. C Alteration of chromatin accessibility in the ILC2_c02 subset at day 47 versus day 45. The Y axis presents the log2 fold change of gene activity at day 47 versus day 45 and in library 2, while the X axis shows the log2 fold change in library 1. D Transcription factor binding motifs significantly enriched in the ILC2_c02 subset at day 47 versus day 45. P-values were calculated by a right-tailed hypergeometric test. E DNA accessibility tracing of the Tlr4 and Il1rl1 of different ILC2 subsets. Shaded boxes delineate increased chromatin accessibility. F Predicted motifs within Tlr4 promoter. G Motif footprinting trace showing binding activities of transcription factor JunB, Fos, PU.1 and MafK.
Discussion
In the present study, we demonstrated the presence of two subsets of ILC2s (ILC2_c01 and ILC2_c02) in human type 2 inflammatory airway diseases, in which TLR4+ILC2s with memory cell markers were elevated in polyp tissue of CRSwNP, and positively correlated with persistent markers of type 2 inflammation. Similarly, TLR4+ILC2s with memory cell characteristics were found to be the major ILC2s subset in a mouse model of type 2 inflammation, producing type 2 cytokines on recall allergen exposure during the memory stage. Indeed, the mouse TLR4+ILC2s were also able to respond to exposure with an unrelated antigen during the recall memory stage, suggesting “trained immunity” properties of these cells. Supporting this observation, blocking or silencing TLR4 signaling also dramatically reduced type 2 inflammation and the number of IL-13+ILC2s in the lungs of these experimental animals. Further, Tlr4 gene loci accessibility and its downstream signaling were linked to trained immunity of ILC2s from the mouse model of type 2 inflammation. Collectively, these findings indicate that TLR4 is involved in the trained ILC2s contributing to the chronicity of type 2 airway inflammatory diseases.
Traditionally, immunological memory is regarded as an exclusive hallmark for adaptive immunity. Long-lived antigen-specific memory T and B lymphocytes from adaptive immune memory persist for a long time and can promptly respond to the specific antigen19. Nevertheless, the innate immune system also displays trained immunity, whereby innate immune cells such as macrophages, monocytes, and natural killer cells acquire immunological memory and enhance responsiveness of cells when they reencounter unrelated pathogens20. In this regard, ILC2s are the main innate resource for type 2 cytokines in persistent type 2 inflammation. It is well-known that the induction of trained ILC2s during helminth infection facilitates the clearance of helminth21, however, the role of trained ILC2s in inflammatory airway diseases is not fully understood. Although trained ILC2s have been shown to be induced in vivo from the peripheral blood of asthmatic patients22, there is no evidence for the presence of trained ILC2s at the local airway site affected in asthma. The present study has demonstrated the presence of local trained ILC2s in both polyp tissue of CRSwNP patients and induced sputum of CRSwNP patients with comorbid asthma. Furthermore, our data have demonstrated that TLR4+ILC2s may play a role as trained ILC2s in both human and mouse airways. Unlike the antigen-specific nature of adaptive immune memory, the induction of trained immunity in ILC2s allows for a rapid response to non-specific antigens and amplifies persistent type 2 inflammation in the airways. This may help us explain why asthma patients often react to unrelated antigens in the clinic. Data derived from the mouse model of type 2 inflammation also showed that total ILC2s and TLR4+ILC2s in the lungs of these animals were increased from baseline levels following both specific and non-specific allergen challenge, and that IL-13+TLR4+ILC2s remained at a high level even four weeks following cessation of allergen challenge. Collectively, the findings suggest that trained immunity of ILC2s is likely to persist over the long term in airway inflammatory diseases.
Verma et al.22 have shown that mouse-trained ILC2s express several genes, including Nr4a1, CD44, Fhl2 and Stat6, but not Il17rb. Contrary to these findings, Martinez-Gonzalez et al.12 have reported an increased expression of IL-17RB in trained ILC2s in the lung tissue of mouse. CD25lowILC2s exhibit long-term survival, particularly in the resolution phase of HDM-driven allergic airway inflammation23. Taken together, these studies suggest that markers identifying trained ILC2 may vary due to the activation mode, the tissue environment, the nature of the stimulus, or species-specific differences. It is reported that activation of ILC2s is partially dependent on TLR1, 2, 4, and 616,17. Moreover, ILC2s can directly recognize antigens and pathogens by expressing pattern recognition receptors, which provide the foundation for their role in trained immunity of innate cells18,24. As expected, our results confirmed that TLR4 was preferentially expressed on the ILC2_02 subset with upregulated memory cell markers. Indeed, in the inflammatory mouse model, TLR4+ILC2s showed a more robust response to a secondary, unrelated antigen challenge than that of TLR4⁻ILC2s. As genetic ablation and pharmacological inhibition of TLR4 blocked the infiltration of inflammatory cells and reduce type 2 inflammation in mouse lung tissue. In addition, TLR4 is also crucial for maintaining trained ILC2s and acts as a marker for their trained state. Our scATAC-seq analysis confirms that epigenetic reprogramming, as defined by chromatin accessibility, regulates the trained immunity of ILC2s.
Finally, scRNA-seq in human polyp tissue and mouse lung tissue in the present study showed that TLR4+ILC2s express the increase of IL1RL1 (ST2), the receptor for IL-33. Notably, upregulation of IL-17RB, the receptor for IL-25, was evident in both human-trained TLR4+ILC2s as well as mouse-trained TLR4+ILC2s. Steer et al. have reported that activation of mouse lung ILC2s by endogenous IL-33 in the neonatal period may train ILC2s25. It has been found that macrophages trained by HDM exhibit IL-17RB expression26, however, it is uncertain whether these trained macrophages exhibit increased responsiveness to IL-25 in allergic conditions. The question of whether IL-25 or IL-33 plays a role involving in the formation of trained ILC2s still awaits further investigations.
Our data support the concept that TLR4+ILC2 could be regarded as trained ILC2s in inflammatory airway diseases, contributing to persistent type 2 inflammation. Obviously, future studies are required to elucidate how the TLR4+ILC2 subset and TLR4⁻ILC2 subset develop in vivo. On the other hand, some studies have shown that trained ILC2s could remain at the local site or migrate into the draining lymph node12,22. In this point of view, the differing migration patterns between TLR4+ILC2 subset and TLR4⁻ILC2 subset should be warrant further investigations. Although the fact that genetic ablation of TLR4 during the memory stage alleviates inflammation and restores lung function, highlighting the role of TLR4 in airways, it is worth to note that we cannot rule out the role of toll-like receptor expression on epithelial cell, as well as that the toll-like receptors play a protective role against pathogen infection27,28. To obtain more definitive evidence, ILC2-specific TLR4 conditional knockout mice would be essential. Nevertheless, blocking TLR4 using monoclonal antibodies or a specific inhibitor may be taken as an auxiliary strategy to manage persistent airway inflammation under the condition of controlling infection.
Methods
Ethical statements
This study complies with all relevant ethical regulations. Human study protocol was performed according to the Declaration of Helsinki and approved by the Medical Ethics Committees of Beijing TongRen Hospital (TRECKY2019-027). All patients provided written informed consent. All experiments involving mice were performed according to the governmental and institutional guidelines for animal welfare and approved by the Institutional Animal Care and Use Committee of the Capital Medical University, Beijing, China (AEEI-2022-207).
Participants
A total of 43 CRSwNP patients (7 females and 36 males), aged 18–75 years and scheduled for endonasal sinus surgery, were randomly recruited from 2022 to 2025 in the Department of Rhinology and Allergy, Beijing TongRen Hospital. All patients were consecutively enrolled without preselection for sex or age. The sex of patients was determined by self-report. Patients with immunodeficiency, fungal sinusitis, coagulation disorder, neoplasia, and pregnancy were excluded. None of the patients had been treated with oral corticosteroids, antibiotics, or biologics within the 4-week period before enrollment. Five patients were included in the scRNA-seq study, while 38 patients were enrolled for flow cytometry analysis. The demographic characteristics of participants in each study are presented in Table 1 and Supplementary Table S1.
The diagnosis of CRSwNP was made according to the European Position Paper on Rhinosinusitis and Nasal Polyps 2020 guidelines2, and the diagnosis of asthma was based on history and the physician’s diagnosis according to Global Initiative for Asthma29. CRSwNP patients were further grouped according to type 2 inflammation in polyp tissue. A cut-off value of 12.98 pg/mL for IL-530 was used. Sections of nasal polyp tissues were also stained with hematoxylin and eosin (H&E) stain for calculating infiltrated eosinophils. The randomly selected stained samples were assessed by two independent pathologists. The number of eosinophils were calculated in three randomly selected high-power fields (HPF; magnification × 400) and expressed as the percentage of inflammatory cells per HPF.
Mice
Female Rag1⁻/⁻C57BL/6 J (strain no. T004753), Rag2⁻/⁻Il2rg⁻/⁻C57BL/6 J (strain no. T059630), and Tlr4⁻/⁻C57BL/6 J (strain no. T051714) mice were purchased from GemPharmatech (Nanjing, China). Wild-type (WT) female C57BL/6 J mice were purchased from the Vital River Laboratories (Beijing, China). All mice were housed in a specific pathogen-free facility under controlled conditions, including a constant temperature of 22–26 °C, humidity levels of 40–70%, and a 12 h light/dark cycle. All mice had free access to water and food (standard chow). All animals were used for experimentation at 8–10 weeks of age. All animals were euthanized by intraperitoneal injection of an overdose of sodium pentobarbital (150 mg/kg). Because female mice have been reported to be developed a more pronounced type of allergic airway inflammation than male mice31,32, only female mice are used in this study.
Preparation of single cell suspensions from tissues of human nasal polyps and mouse lungs
Fresh human nasal polyp tissues were transferred to tubes and cut into small pieces using scissors. These fragments were further dissociated in the Gentle MACS tissue dissociator (Miltenyi Biotec, Germany). After dissociation, the tissues were digested in RPMI 1640 medium containing 2 mg/mL collagenase (Worthington, USA) and 0.04 mg/mL DNase I (Roche, Switzerland). Following a 30 min incubation at 37 °C, the digested tissues were processed again in the GentleMACS. Next, each cell suspension was passed through a 70 μm cell strainer, and the red blood cells were lysed using Versalyse Lysing Solution (Beckman Colter, USA).
Lungs of euthanised mice were harvested following cardiac perfusion with phosphate-buffered saline (PBS). The lungs were cut into small pieces, and then digested for 30 min at 37 °C in PBS containing 50 µg/mL Liberase (Roche) and 100 µg/mL DNase I (Sigma-Aldrich, USA). The cell suspensions were filtered through a 70 μm cell strainer, and red blood cells (RBC) were lysed using RBC lysing buffer (Solarbio, China). Then, the cell suspension was centrifuged at 300 × g for 8 min and then resuspended in RPMI 1640 medium with 2% fetal bovine serum (FBS, Gibco, USA).
Sputum induction and processing
Sputum induction was performed in asthmatic patients following established protocols33,34. Briefly, patients inhaled nebulized 4.5% saline solution for 10 min and expectorated sputum into a sterile specimen cup. The sputum samples were treated with 5% solution of dithiothrietol (DTT; Sigma-Aldrich) in PBS to digest mucus, and then centrifuged at 300 × g for 10 min. Afterwards, the cell pellet was re-suspended in 5.0 mL PBS, and then filtered through a 70 μm cell strainer before centrifugation as above. Subsequently, the cell pellet was resuspended in RPMI 1640 with 2% FBS and processed for analysis by flow cytometry.
Inflammatory mouse models
The WT female C57BL/6 J mice, Rag1⁻/⁻C57BL/6 J mice or Tlr4⁻/⁻C57BL/6 J mice were anesthetized with 3% isoflurane and sensitized intranasally with 50 µg / 50 µL of HDM (Cosmo Bio Co. LTD, Japan), followed by an intranasal challenge with 25 µg of HDM. To clarify the memory function of ILC2s, mice were rested for 4 weeks, and then given a recall challenge of HDM (25 µg) or papain (25 µg) on day 45. Some mice pre-challenged with HDM also received 20 µg of TLR4 monoclonal antibody 2 h before recall challenge on day 45 to investigate the effect of blocking TLR4 on recall memory.
Flow cytometry analysis
To stain human ILC2s and mouse ILC2s, single cell suspensions prepared from nasal polyp tissues, induced sputum and mouse lung tissues were treated with a human BD Fc Block (BD Bioscience, USA) or mouse BD Fc Block anti-mouse CD16/CD32 mAb 2.4G2 (BD bioscience), and then stained with specific cell surface markers in staining buffer (PBS supplemented with 1% FBS and 5 mM EDTA) for 30 min on ice. For further intracellular staining, cells were incubated for another 4 h at 37 °C in the presence of 2 nmol/mL monensin (BD Biosciences) and Brefeldin A (1000 ×, eBioscience, USA) in the cell stimulation cocktail (500 ×, eBioscience). The stained cells were fixed and permeabilized using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit (554714, BD Biosciences). For transcription factor staining in ILC2s, cells were fixed and permeabilized using the FOXP3/Transcription factor staining buffer (eBioscience). Afterwards, cells were then incubated for 30 min at 4 °C in the presence of commercially available monoclonal antibodies. Staining reagents included: FITC-anti-human CD11c (B-ly6), FITC-anti-human CD1a (HI149), FITC-anti-human CD3 (UCHT1), FITC-anti-human CD19 (HIB19), FITC-anti-human CD14 (M5E2), FITC-anti-human CD16 (3G8), FITC-anti-human CD94 (HP-3D9), FITC-anti-human CD123(7G3), Bv421-anti-human CD45 (HI30), Bv786-anti-human CD161 (DX12), FITC-anti-human TCRαβ (T10B9.1A-31), PE-anti-human IL-13 (JES10-5A2), APC-Cy7-anti-mouse CD45 (30-F11), and APC-Lineage antibody cocktail from BD Biosciences; FITC-anti-human CD34 (581), FITC-anti-human CD303 (201 A), PE-Cy5.5-anti-human CD44 (IM7), FITC-anti-human FcεRIα (AER-37), APC-Cy7-anti-human CD127 (A019D5), PerCP-anti-human CD45 (HI30), Bv605-anti-human CRTH2 (BM16), APC-anti-mouse FcεRIα (MAR-1), PE-anti-mouse ICOS (7E.17G9), PE-Cy7-anti-mouse TLR4 (SA15-21), Bv711-anti-human/mouse/rat ICOS (C398.4 A), AF700-anti-mouse CCR8 (SA214G2), PE-anti mouse PU.1 (7C2C34), AF647-anti-phospho-Jun (ser73) antibody (A19012A/R/C), and APC-anti-mouse CD170 (S17007L) from Biolegend (USA); PE-Cy7-anti-human TLR4 (HTA125), eFluor450-anti-human/mouse IL-5 (TRFK5), PerCP-eFluor710-anti-mouse ST2 (RMST2-33), AF700-anti-human/mouse CD44 (IM7), PE-anti-mouse IL-25R (MUNC33), FITC-anti-phospho-c-Fos (Ser32) antibody (cFosS32-BA9), FITC-anti Ki67 (SolA15), and PE-anti-mouse IL-13 (eBio13A) from eBioscience; APC-anti-human IL-17RB (Arg18-Gly289) from R&D systems; PerCP-Cy5.5-anti-human/mouse CD11b (M1/70), PE-Cy7-anti-mouse CD11c (N418), and PE-anti-mouse Ly6G (RB6-8C5) from Tonbo bioscience (USA). Details of antibodies are listed in Supplementary Table S2. Cell aggregates were excluded from analysis on the basis of FSC-H/FSC-A and SSC-H/SSC-A characteristics. Data were acquired on a Cytek Aurora (Cytek, USA) or BD LSR Fortessa (BD Biosciences), and analyzed using FlowJo software (v.10, Tree Star Inc., USA).
Inflammatory cytokine measurements
Tissue homogenates were prepared from nasal polyp samples, which had been frozen on collection from the individuals during nasal surgery. Nasal polyp tissues and mouse lung tissues were weighted and snap-frozen. For every 0.1 g of tissue, 1 mL of 0.9 % NaCl containing a Complete Protease Inhibitor Cocktail (Roche) was added. Samples were then mechanically homogenized for 5 minutes using a TissueLyser LT (Qiagen, Germany). After homogenization, the suspensions were centrifuged, and the supernatants were collected and stored at – 80 °C until analysis. Cytokines IL-5, IFN-γ, IL-17, and IL-13 were analyzed in human tissue homogenate using Luminex xMAP suspension array technology (LXSAHM-04, R&D, USA) in a Bio-Plex 200 system (Bio-Rad, USA), following the manufacturer’s instructions. ECP was determined by using the UniCAP system (10-9321-41 and 14-4515-41, Phadia, Sweden). Mouse IL-13 ELISA (88-7137-88, Thermo Fisher Scientific, USA) was utilized to assess the level of IL-13 in lung tissues of the experimental mice.
Stimulation of ILC2s
To obtain a sufficient number of ILC2s, mice were intranasally administered with recombinant mouse IL-33 (mrIL-33, Novoprotein, China) at a dosage of 400 ng in 50 μL of saline for 5 days. On day 6, the mice were sacrificed to harvest ILC2s. Lin⁻ cells were enriched from the lungs using the Mouse Hematopoietic Progenitor Cell Enrichment Set-DM (558451, BD Biosciences) according to the manufacturer’s guidelines. To the end, the sorted ILC2s (CD45+Lineage⁻ICOS+ST2+) were then cultured in RPMI 1640 medium, supplemented with 10 % FBS, 1 % penicillin/streptomycin, 0.1% β-mercaptoethanol, 10 ng/mL rmIL-7 (R&D Systems), and 10 ng/mL rmIL-2 (R&D Systems) for 6 days. On day 7, the ILC2s were stimulated separately with HDM extracts (Cosmo Bio Co., LTD.), Der p1 (Greer Labs, USA), and LPS (Sigma) for 24 h at different concentrations. Afterwards, flow cytometry was conducted to analyze the expression of TLR4, phosphorylated(p)-c-Fos, p-Jun, and PU.1 in the ILC2s. To confirm the role of transcriptional factor in induction of TLR4 on ILC2s, the ILC2s were pre-treated with the transcription factor c-Fos / AP-1 inhibitor T-5224 (20 μM, MCE) and the PU.1 inhibitor DB1976 (100 nM, MCE) for 1 h prior to stimulation.
To generate in vitro-trained ILC2s, TLR4⁻ILC2s were isolated from IL-33 challenged WT C57BL/6 J mice. On day 1, TLR4⁻ILC2s were stimulated with 100 µg/mL HDM or medium for 2 d, washed to remove the allergen, and cultured in the media alone. After a 6-day resting period, cells were re-stimulated with either a low dose of HDM (25 µg/mL) or LPS (1 ng/mL) for 16 h. Subsequently, we measured the expression of TLR4, IL-17RB, IL-5, IL-13 and Ki67 in ILC2s by flow cytometry. To evaluate the role of TLR4 in trained immunity of ILC2s, TLR4 antagonist TAK242 (10 µM; MCE) were used to pretreat ILC2s for 1 h before HDM stimulation on day 1.
Adoptive transfer of TLR4+ILC2s and TLR4⁻ILC2s
TLR4+ILC2s and TLR4⁻ILC2s were isolated from HDM-recall challenged WT C57BL/6 J mice (47 d, Fig. 6A). Briefly, Lin⁻ cells were enriched, and then TLR4+ILC2s and TLR4⁻ILC2s were sorted using a BD FACS Aria. Overall, about 5000 TLR4+ILC2s and TLR4⁻ILC2s were then transferred by intravenous injection into Rag2⁻/⁻Il2rg⁻/⁻C57BL/6 J mice, and these recipient mice were administered intranasal HDM on three consecutive days. On day 5, the animals assessed for airway hyperreactivity as described below, before being sacrificed for assessment of lung inflammation, and the presence of type 2 cytokines. Some Rag2⁻/⁻Il2rg⁻/⁻C57BL/6 J mice injected with purified TLR4+ILC2s from HDM-sensitized WT C57BL/6 J mice also received 20 µg of TLR4 monoclonal antibody 2 h before intranasal HDM challenge on day 1 to investigate the effect of blocking TLR4 on airway inflammation.
Airway hyperreactivity measurement
After anaesthetizing the mice with 80 mg/kg pentobarbital sodium, followed by tracheotomy and endotracheal intubation, airway responsiveness was determined by measuring lung resistance in response to increasing dose of aerosolized methacholine (0–48 mg/mL) using the FlexiVent system (Scireq Inc. Canada). Group averages were expressed as the fold increase over baseline resistance35.
Immunohistochemistry and Immunofluorescence staining
Paraffin-embedded tissue sections (4 μm thick) were dewaxed and rehydrated using standard procedures. Sections were then treated with 3% H2O2 at room temperature for 30 min and blocked with blocking buffer (Solarbio) for 1 h to prevent non-specific staining. Primary antibodies were incubated overnight at 4 °C. After washing, tissue sections were incubated corresponding secondary antibodies for 1 hour. Finally, sections were washed and stained with diaminobenzidine or 4’,6-diamidino-2-phenylindole (DAPI). The primary antibodies were included: anti-MUC5AC (1:500, 45M1, Abcam), PE-anti-TLR4 antibody (1:50, SA15-21, Biolegend), and anti-GATA3 antibody (1:25, D13C9, CST). The secondary antibodies were included: HRP-conjugated anti-mouse IgG secondary antibody (1:1000, Abcam), Alexa 488-conjugated or Alexa 647-conjugated secondary antibodies (1:1000, Invitrogen). Details of antibodies are listed in Supplementary Table S2. For quantification, MUC5AC+ cells were calculated in five randomly selected areas of the airway epithelial layers and expressed as a percentage of total epithelial cells.
Single-cell RNA sequencing and analysis in nasal polyp tissues
A BD Rhapsody system was used to capture the transcriptomic information of sorted CD45+Lin⁻ immune cell populations in nasal polyp tissues from five patients with CRSwNP, and transcriptome libraries were prepared using the BD Rhapsody single-cell whole-transcriptome amplification kit (633801, BD Biosciences) according to the manufacturer’s instructions. Sequencing was performed using the Illumina NovaSeq 6000 platform with the PE150 protocol at Capital Bio Technology Co., Ltd (Beijing, China). Raw base call files were demultiplexed into FASTQ files, and the BD WTA Rhapsody Analysis Pipeline (v.2.0) was used to perform alignment based on the reference genome (GRCh38).
Gene-by-cell matrices were imported into R (v.4.0.3) using the Seurat package (v.4.3.0)36. Cells, in which 500-3000 unique genes were detected and the mitochondrial genome transcript ratio was < 20%, were further analyzed. Raw counts were normalized and transformed into logarithmic scales. Highly variable genes were selected, and a principal component analysis (PCA) matrix was then calculated. With the batch effects adjusted using the Harmony algorithm, cell clusters were identified by the Louvain algorithm. The uniform manifold approximation and projection (UMAP) was used to visualize the cells. Different expressed genes (DEGs) were calculated using the Seurat FindClusters function.
Based on the DEGs for each cluster, contaminating cells were removed, and ILCs based on high expression of ID2, IL7R were subjected to further analysis. Re-clustering of the cells provided four separate clusters. An unbiased enrichment analysis of the MSigDB Hallmark pathways37 of differentially expressed genes was performed using the clusterProfiler package (v.4.10.0)38. RNA velocity was calculated using velocyto (v.0.17)39, and subsequent analyses were performed by scVelo (v.0.3.3). With the function scv.pl.velocity embedding stream, the velocity graph was then used to embed RNA velocities in the UMAP plot generated by Seurat in the form of streamlines. Monocle 3 (v.1.3.5) was used to infer the pseudo-time trajectories of ILC2s, followed by the ClusterGVis package (v.0.1.1) to visualize pseudo-time-dependent gene expression changes. The scRNA-seq dataset obtained for the present study was further validated using the scRNA-seq dataset of sinus tissue obtained by Wang et al.15. Gene-by-cell matrices were generated by Cell Ranger (v.7.1.0) with default parameters, and processed using the Seurat v4 workflow with the same strategy.
Single-cell RNA sequencing and analysis in mouse lung tissues
Lung ILC2s sorted from lung tissues of WT mice and were loaded into Chromium microfluidic chips with 3’ v3 chemistry and barcoded with 10 x Chromium Controller (10 x Genomics). RNA from the barcoded cells was subsequently reverse-transcribed, and sequencing libraries were constructed with reagents from a Chromium Single Cell 3’ v3 reagent kit (PN-1000075, 10 x Genomics) according to the manufacturer’s instructions. Sequencing was performed using the Illumina NovaSeq 6000 platform with the PE150 protocol at Novogene Co., Ltd. Raw base call files were demultiplexed into FASTQ files, and the Cell Ranger Suite (10 x Genomics, v.7.1.0) was used to perform alignment based on the reference genome GRCm38. Cells with 200–4000 unique genes detected and mitochondrial genome transcript ratio was < 20% were kept for downstream analysis. Data matrices were proceeded with Seurat v4 standard workflow. T cells, B cells, fibroblasts and endothelial cells were removed according to the expression of Cd3e, Iglc1, Pdgfra and Clde5. Gene expression density was plotted using plot_density function in the Nebulosa package (v.1.18.0). RNA velocity, enrichment analysis and DEGs analysis was performed with the same procedure.
scATAC-seq and analysis
Nuclei were isolated from sorted ILC2s from lung tissues, counted, and processed Chromium Nuclei Isolation Kit (PN-1000493, 10 x Genomics). In brief, the cell suspension was pelleted and resuspended in freshly prepared lysis buffer, and incubated on ice for 10 min. After centrifugation, the nuclei were pelleted, washed and resuspended in Resuspension Buffer (10 x Genomics). ScATAC-seq libraries were prepared with the Chromium single-cell ATAC reagent kits v2 (PN-1000390, 10 x Genomics) according to the manufacturer’s instructions. To gather more information about TLR4+ILC2s, we also prepared a separate library that mixed sorted TLR4+ILC2s with sorted TLR4⁻ILC2s in a 1:6 ratio. Sequencing was also performed using the Illumina NovaSeq 6000 platform, and Cell Ranger ATAC (v.2.1.0) was used to demultiplex the library and perform alignment based on the reference genome (GRCm38). We then processed the scATAC-seq dataset using Signac (v.1.12.0)40, following the protocol outlined in the Signac vignette “Analyzing adult mouse brain scATAC-seq” (https://stuartlab.org/signac/articles/mouse_brain_vignette). Cells containing the following quality measures were retained: peaks between 3000 and 50,000, fragments in peaks > 50%, and TSS enrichment > 3. Data normalization and dimensionality reduction were performed using latent semantic indexing (LSI). The first LSI component captured sequencing depth across the samples and was not utilized in downstream analyses. Gene activity scores were generated by quantifying unique molecular identifiers (UMIs) mapped to the gene coordinates and the 2 kb upstream region. Then, we utilized the TransferData function to assign cells from the scATAC-seq experiment to ILC2 clusters c01, c02, or c03. Transcription factor motif information was added using the AddMotifs function using Jaspar motifs. Motif in promoter of Tlr4 was predicted using the JASPAR database.
Bulk RNA sequencing and analysis
Lung TLR4+ and TLR4⁻ILC2s were sorted from lung tissues of WT mice on day 45, with 200 cells collected per group in triplicate. Sorted cells were directly lysed in 0.2% Triton X-100 containing ribonuclease inhibitor (Takara). Total cDNA was reverse-transcribed, and sequencing libraries were prepared following the standard Smart-seq2 protocol41. Libraries were sequenced using Illumina platforms with a PE150 strategy in Novogene Bioinformatics Technology Co., Ltd (Beijing, China). Low-quality reads and adapter sequences were removed using Trimmomatic42. The resulting clean reads were then aligned to the mouse genome (GRCm38) using HISAT2 (v.2.2.1)43 with default parameters. Transcript abundances were estimated with featureCounts (v.2.0.3)44. Differential expression analyses were performed by DESeq2 (v.1.48.1).
To better compare single-cell and bulk RNA sequencing library, pseudo-bulk expression profiles were constructed from single-cell RNA-seq data using the “AggregateExpression” function. Each pseudo-bulk sample contained 800 cells to achieve a sequencing depth comparable to that of the bulk RNA-seq libraries. Expression matrices were merged using ComBat-seq45 and normalized with a variance-stabilizing transformation. UMAP embeddings were computed using the tidydr package (v.0.0.5) to illustrate the similarity among samples.
Statistical analysis
Statistical analyses were performed using Prism software (v.8; GraphPad) and R (v.4.0.3). Differences between 2 groups involving the data from CRSwNP patients were analyzed using a two-tailed Mann-Whitney U test, and differences between the paired samples were analyzed using two-tailed paired Wilcoxon tests. Associations between variables were evaluated using two-tailed Spearman correlation analysis. Data from mice were presented as means ± SEM and analyzed using a two-tailed unpaired Student’s t test for two-group comparison. Significant differences among multiple groups were determined by two-tailed one-way ANOVA with Tukey’s Honestly Significant Difference test. Differentially expressed genes calculated by the two-tailed Wilcoxon test with Bonferroni correction. Pathway analysis was performed using right-tailed Fisher’s Exact Test with Benjamini-Hochberg correction.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data generated in this study have been deposited in public repositories: human RNA-seq data in the Genome Sequence Archive (GSA) under accession HRA005687, and mouse data in GSA under accession CRA014947. For human sequencing data, access is restricted to protect participant privacy in compliance with ethical regulations. Requests for these data will be reviewed by the corresponding author to verify the applicant’s identity and research purpose. Requests following the guidelines of GSA for non-commercial use will be processed within one month, and approved access will be valid for one year. The published scRNA-seq data reused in this study are available at https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000772. All other data are available in the article and its supplementary files of from the corresponding author upon request. Source data are provided in this paper.
Code availability
Analysis scripts have been deposited at Zenodo: (https://doi.org/10.5281/zenodo.15803048).
References
Mortimer, K. et al. The burden of asthma, hay fever and eczema in adults in 17 countries: GAN Phase I study. Eur. Respir. J. 60, (2022).
Fokkens, W. J. et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology 58, 1–464 (2020).
Wang, W. et al. Changes in the clinical and histological characteristics of Chinese chronic rhinosinusitis with nasal polyps over 11 years. Int. Forum Allergy Rhinol. 9, 149–157 (2019).
Bachert, C., Bhattacharyya, N., Desrosiers, M. & Khan, A. H. Burden of disease in chronic rhinosinusitis with nasal polyps. J. Asthma Allergy 14, 127–134 (2021).
Rahimi R. A., Nepal K., Cetinbas M., Sadreyev R. I., Luster A. D. Distinct functions of tissue-resident and circulating memory Th2 cells in allergic airway disease. J. Exp. Med. 217, https://doi.org/10.1084/jem.20190865 (2020).
Kobayashi, T., Iijima, K., Matsumoto, K., Lama, J. K. & Kita, H. Lung-resident CD69(+)ST2(+) T(H)2 cells mediate long-term type 2 memory to inhaled antigen in mice. J. Allergy Clin. Immunol. 152, 167–181.e166 (2023).
Endo, Y. et al. The interleukin-33-p38 kinase axis confers memory T helper 2 cell pathogenicity in the airway. Immunity 42, 294–308 (2015).
Mjösberg, J. M. et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12, 1055–1062 (2011).
Netea, M. G. et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020).
Fanucchi, S., Dominguez-Andres, J., Joosten, L. A. B., Netea, M. G. & Mhlanga, M. M. The intersection of epigenetics and metabolism in trained immunity. Immunity 54, 32–43 (2021).
Divangahi, M. et al. Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat. Immunol. 22, 2–6 (2021).
Martinez-Gonzalez, I. et al. Allergen-Experienced Group 2 Innate Lymphoid Cells Acquire Memory-like Properties and Enhance Allergic Lung Inflammation. Immunity 45, 198–208 (2016).
Spits, H. & Mjösberg, J. Heterogeneity of type 2 innate lymphoid cells. Nat. Rev. Immunol. 22, 701–712 (2022).
Mazzurana, L. et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res. 31, 554–568 (2021).
Wang, W. et al. Single-cell profiling identifies mechanisms of inflammatory heterogeneity in chronic rhinosinusitis. Nat. Immunol. 23, 1484–1494 (2022).
Hardman, C. S. et al. CD1a presentation of endogenous antigens by group 2 innate lymphoid cells. Sci. Immunol. 2, eaan5918 (2017).
Hardman, C. S. et al. IL-6 effector function of group 2 innate lymphoid cells (ILC2) is NOD2 dependent. Sci. Immunol. 6, eabe5084 (2021).
Bhattarai, S. et al. TLR4 is a regulator of trained immunity in a murine model of Duchenne muscular dystrophy. Nat. Commun. 13, 879 (2022).
Soerens, A. G. et al. Functional T cells are capable of supernumerary cell division and longevity. Nature 614, 762–766 (2023).
Netea, M. G. et al. Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016).
Hartung, F. & Esser-von Bieren, J. Trained immunity in type 2 immune responses. Mucosal Immunol. 15, 1158–1169 (2022).
Verma, M. et al. The molecular and epigenetic mechanisms of innate lymphoid cell (ILC) memory and its relevance for asthma. J. Exp. Med. 218, e20201354 (2021).
Li, B. W. S. et al. Group 2 innate lymphoid cells exhibit a dynamic phenotype in allergic airway inflammation. Front. Immunol. 8, 1684 (2017).
Owen, A. M. et al. MyD88-dependent signaling drives toll-like receptor-induced trained immunity in macrophages. Front. Immunol. 13, 1044662 (2022).
Steer, C. A., Mathä, L., Shim, H. & Takei, F. Lung group 2 innate lymphoid cells are trained by endogenous IL-33 in the neonatal period. JCI Insight 5, e135961 (2020).
Lechner, A. et al. Macrophages acquire a TNF-dependent inflammatory memory in allergic asthma. J. Allergy Clin. Immunol. 149, 2078–2090 (2022).
Kawai, T. & Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011).
Horn, K. J., Schopper, M. A., Drigot, Z. G. & Clark, S. E. Airway Prevotella promote TLR2-dependent neutrophil activation and rapid clearance of Streptococcus pneumoniae from the lung. Nat. Commun. 13, 3321 (2022).
Bateman, E. D. et al. Global strategy for asthma management and prevention: GINA executive summary. Eur. Respir. J. 31, 143–178 (2008).
Tomassen, P. et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J. Allergy Clin. Immunol. 137, 1449–1456 (2016).
Takeda, M. et al. Gender difference in allergic airway remodelling and immunoglobulin production in mouse model of asthma. Respirology 18, 797–806 (2013).
Weiss, K. et al. Barrier housing and gender effects on allergic airway disease in a murine house dust mite model. Immunohorizons 5, 33–47 (2021).
NE, A. lexis et al. The glutathione-S-transferase Mu 1 null genotype modulates ozone-induced airway inflammation in human subjects. J. Allergy Clin. Immunol. 124, 1222–1228.e1225 (2009).
Burbank, A. J. et al. Gamma tocopherol-enriched supplement reduces sputum eosinophilia and endotoxin-induced sputum neutrophilia in volunteers with asthma. J. Allergy Clin. Immunol. 141, 1231–1238.e1231 (2018).
Verma, M. et al. Experimental asthma persists in IL-33 receptor knockout mice because of the emergence of thymic stromal lymphopoietin-driven IL-9(+) and IL-13(+) type 2 innate lymphoid cell subpopulations. J. Allergy Clin. Immunol. 142, 793–803 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Wu, T. et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Zhang, Y., Parmigiani, G. & Johnson, W. E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2, lqaa078 (2020).
Acknowledgements
We express our gratitude to Dr. Di Yu from the University of Queensland for providing valuable suggestions for this study. This work was supported by grants from the National Natural Science Foundation of China (82271140 to F.L., 82025010 to C.W. and 82371115 to L.Z.); Beijing Nova Program (202204844198 and 20250484974 to F.L.); Beijing Municipal Public Welfare Development and Reform Pilot Project for Medical Research Institutes (JYY2021-2 to L.Z. and JYY2023-1 to C.W.).
Author information
Authors and Affiliations
Contributions
Y.L. and F.L. conducted all experiments and drafted the manuscript with input from all authors. Z.W. conducted the bioinformatic analysis. Y.Z., S.D., and X.W. helped with the sample collection of human samples. W.W. and S.Y. helped with the design of the mouse experiments. N.Z. and C.B. helped with paper correction. L.Z., C.W., and F.L. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Bernard Mari, who co-reviewed with Marine Gautier-Isola, Charles Pilette and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Y., Wang, Z., Duan, S. et al. TLR4+group 2 innate lymphoid cells contribute to persistent type 2 immunity in airway diseases. Nat Commun 16, 7108 (2025). https://doi.org/10.1038/s41467-025-62532-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62532-0