Abstract
Further study of the mechanism of glucocorticoid (GC)-induced osteoblast (OB) apoptosis is highly important for the prevention and treatment of GC-induced osteoporosis and osteonecrosis. Serine/arginine-rich splicing factor 1 (Srsf1) expression was downregulated in a dose-dependent manner during GC-induced OB apoptosis. Knockdown of Srsf1 significantly promotes GC-induced OB apoptosis, while overexpression of Srsf1 significantly inhibits GC-induced OB apoptosis. Mechanistically, GC induces the up-regulation of histone deacetylase 4 (Hdac4) in OB, and inhibits the expression of transcription activator forkhead box C1 (Foxc1) by reducing the levels of histone H3 lysine 9 acetylation (H3K9ac) and H3K27ac in the promoter region of Foxc1, thereby down-regulating Srsf1. Next, SRSF1 regulates GC-induced OB apoptosis by regulating Bcl-2 modifying factor (Bmf) alternative splicing. From the perspective of alternative splicing, this study demonstrates that Srsf1 and its regulatory mechanism may serve as a new target for the prevention and treatment of GC-induced osteoporosis and osteonecrosis.
Similar content being viewed by others
Introduction
Long-term and high-dose glucocorticoid (GC) treatment can destroy the balance of bone metabolism by promoting osteoblast (OB) apoptosis, thus leading to GC-induced osteoporosis and osteonecrosis1,2. The OB, as a functional cell involved in bone formation, is mainly responsible for the synthesis, secretion, and mineralization of the bone matrix to maintain the balance of bone metabolism3,4,5. Therefore, understanding the mechanism involved in preventing GC-induced OB apoptosis is highly important for the prevention and treatment of GC-induced osteoporosis and osteonecrosis.
In in vitro experiments involving GC-induced OB apoptosis, RNA sequence (RNA-Seq) analysis revealed that several members of the serine/arginine (SR)-rich protein family were downregulated. The SR protein family comprises important splicing factors involved in alternative splicing; these proteins can directly bind to precursor mRNAs (pre-mRNAs), function as splicing activators and are closely related to the regulation of apoptosis6,7,8. Thus, we hypothesized that the SR protein family may represent a novel class of targets for preventing GC-induced OB apoptosis. By combining the RNA-seq and real-time quantitative polymerase chain reaction (qPCR) results, we found that among the SR protein family members, serine and arginine-rich splicing factor 1 (SRSF1) were the most downregulated genes related to OB apoptosis induced by GC. SRSF1 is a typical SR protein that contains one or two RNA recognition motifs (RRMs) and an arginine/serine (RS) domain, in which RRM affects apoptosis by binding to pre-mRNAs of apoptosis-related genes to regulate alternative splicing9,10. Olga Anczukow et al.9 reported that SRSF1 was overexpressed in breast cancer cells and that this overexpression promoted cell proliferation and reduced cell apoptosis by enhancing the inclusion of the cancer susceptibility candidate 4 (CASC4) exon 9. Martina Donadoni et al.11 reported that ethanol exposure leads to the downregulation of SRSF1 expression in neural progenitor cells, resulting in the missplicing of myeloid cell leukemia sequence 1 (Mcl-1) pre-mRNA. The ratio of Mcl-1L/Mcl-1S decreased in a dose-and time-dependent manner, which promoted cell apoptosis. Additionally, studies have shown that SRSF1 can regulate cell apoptosis by alternative splicing of apoptosis-related genes, such as B-cell lymphoma extralarge (Bcl-X), cysteinyl aspartate specific proteinase-2 (caspase-2), caspase-9, the inhibitor of caspase-activated DNase (ICAD), the Bcl-2-interacting mediator of cell death (Bim), and the tumor protein P5312. In conclusion, we suggest that SRSF1 may also be involved in GC-induced OB apoptosis by acting as a regulator; however, its specific role and mechanism remain to be elucidated.
This study utilized RNA-seq and bioinformatics analysis to identify SRSF1 as a key regulatory molecule in GC-induced OB apoptosis and to reveal the upstream and downstream regulatory mechanism of Srsf1 in GC-induced OB apoptosis. New targets and mechanisms for the prevention and treatment of GC-induced osteoporosis and osteonecrosis were also explored using these analysis methods.
Results
GC induces OB apoptosis
The morphology of MC3T3-E1 cells cultured in osteogenic induction medium was fibroblast-like. After 3 days of induction, the proliferation of MC3T3-E1 cells reached a plateau (Fig. S1C), and the positive rate of ALP staining showed a trend of rapid increase and then slow decrease, reaching a peak at 15 days (Fig. S1A, B). Alizarin red S staining showed that calcified nodules stained orangered and were scattered (Fig. S1D). These results showed that MC3T3-E1 cells were precursor OBs, which became mature OBs after osteogenic induction culture. Next, we selected mature OBs cultured by osteogenic induction for 15 days as the cell model for this study.
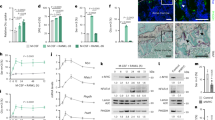
Next, we treated OBs with 0.0, 0.1, 0.2, 0.3, or 0.4 mg/mL GC for 24 h. Compared with those in the control group (0.0 mg/mL), 0.1 and 0.2 mg/mL GC had no significant effect on osteogenic activity and apoptosis of OB (Fig. 1A–D). Moreover, there were no significant changes in the expression of cleaved cysteinyl aspartate-specific proteinase-3 (cleaved-CASP3) or cleaved-CASP9 (Fig. 1G–I). When the concentration of GC was 0.3 mg/mL, the osteogenic activity of OB was inhibited (Fig. 1A, B). The TUNEL-positive (TUNEL*) cell rate detected by TUNEL/DAPI staining was 15.3 ± 1.39% (Fig. 1C, D), and the apoptosis rate detected by flow cytometry was 18.4 ± 1.47% (Fig. 1E, F). Furthermore, the expression of cleaved CASP3 and cleaved CASP9 was upregulated (Fig. 1G–I). When the concentration of GC was 0.4 mg/mL, the osteogenic activity of OB was significantly inhibited (Fig. 1A, B). The percentage of TUNEL* cells detected by TUNEL/DAPI staining was 33.98 ± 3.61% (Fig. 1C, D), and flow cytometry showed that the percentage of apoptotic cells was 34.75 ± 1.48% (Fig. 1E, F). The expression of cleaved CASP3 and cleaved CASP9 was significantly upregulated (Fig. 1G–I), indicating that 0.4 mg/mL GC could induce significant apoptosis in OBs. Next, we detected the expression of mitochondrial apoptotic pathway-related proteins by western blot. The results showed that the expression of B-cell leukemia/lymphoma 2 (BCL2) was downregulated in OBs treated with 0.4 mg/mL GC, while the expression of Bcl2-associated agonist of cell death (BAD), Bcl2-modifying factor (BMF), Bcl2-associated X protein (BAX), cytochrome C (CYT C), and BCL-XL was upregulated (Fig. 1J–P). These results suggest that 0.4 mg/mL GC is an ideal concentration for establishing a model of GC-induced OB apoptosis. Thus, we utilized 0.4 mg/mL GC to treat OBs in subsequent experiments to study the mechanism of GC-induced OB apoptosis.
A, B ALP detection for OBs (n = 4). C, D TUNEL/DAPI staining detection of OBs TUNEL* cell number (n = 3). E, F Apoptosis detected by Annexin V-FITC/PI staining (n = 5). G–I The expression levels of cleaved-CASP3 and cleaved-CASP9 were detected by Western Blot (n = 3). J–P Western Blot was used to detect the expression of BCL2, BAD, BMF, BAX, CYT C, and BCL-XL (n = 3). Data were expressed as mean ± standard deviation in (B, D, F, H, I, K–P), where statistical significance was calculated using Student’s t test in (K–P) and using one-way analysis of variance (ANOVA) with Tukey post hoc test in (B, D, F, H, I). *P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05. 0.0: 0.0 mg/mL, 0.1: 0.1 mg/mL, 0.2: 0.2 mg/mL, 0.3: 0.3 mg/mL, 0.4: 0.4 mg/mL.
Srsf1 is involved in GC-induced OB apoptosis
The gene expression profile of GC-treated OBs was examined by RNA sequencing (RNA-seq) after the OBs were treated with a 0.4 mg/mL concentration of GC (Fig. 2A). GO and KEGG enrichment pathway analyses of DEGs (Fig. 2B, C) revealed that 67 genes were related to apoptosis (Fig. 2D), 35 genes were related to RNA splicing, and the expression of the splicing-related SR protein family members Srsf1, Srsf2, Srsf3, and Srsf7 was downregulated. Among these genes, Srsf1 exhibited the most significant change in expression among the RNA splicing-related genes (Fig. 2E). Combined with the findings of previous studies reporting the role of the SR protein family in the regulation of apoptosis12, we believe that GC-induced OB apoptosis may be related to RNA splicing and may be closely related to Srsf1.
A Heat map showing differentially expressed genes (n = 3). B Gene ontology (GO) analysis (n = 3). C Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway analysis (n = 3). D Heat map showing differentially expressed apoptosis-related genes (n = 3). E Heat map showing differentially expressed RNA splice-related genes (n = 3). F–K The relative expression of Srsf1, Srsf2, Srsf3, Srsf7, Bmf, and Bcl2l1 was detected by qPCR (n = 4). L–M The expression level of SRSF1 was detected by western blot (n = 4). N The correlation between SRSF1 expression and OB apoptosis rate was analyzed by Pearson correlation (n = 4). In (F–K, M), the data are expressed as mean ± SD, where statistical significance was calculated using Student’s t test in (F–K) and using one-way ANOVA with Tukey post hoc test in (M). Control control group, GC glucocorticoid treatment group. *P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05.
We utilized qPCR to validate the expression of Srsf1, Srsf2, Srsf3, Srsf7, Bmf, and Bcl2l1 in the RNA-seq gene expression profiles. The results showed that Srsf1, Srsf2, Srsf3, and Srsf7 were downregulated in GC-treated OBs. In contrast, Bmf and Bcl2l1 were upregulated, which is consistent with the RNA-seq data (Fig. 2F–K). Western blot analysis further showed that SRSF1 expression was downregulated in a dose-dependent manner with increasing GC concentration (Fig. 2L, M). Next, GC induction was applied to construct a mouse OB apoptosis model (Figs. S2A, B and S7), and the ratio of SRSF1-positive OB was decreased compared with the control group (Fig. S2C, D). Pearson correlation analysis of the relative expression of SRSF1 and the OB apoptosis rate showed that SRSF1 expression was negatively correlated with OB apoptosis (r = −0.801, P < 0.001) (Fig. 2N). Therefore, we suggest that Srsf1 is involved in regulating GC-induced OB apoptosis. However, the specific role of Srsf1 in GC-induced OB apoptosis remains unclear.
Role of Srsf1 in GC-induced OB apoptosis
We utilized an Srsf1-overexpressing lentivirus to transfect OBs to investigate the effect of Srsf1 on GC-induced OB apoptosis. OBs were treated with GC after successful overexpression of Srsf1, as determined by qPCR and western blot (Fig. 3A–C). TUNEL/DAPI and Annexin V-FITC/PI staining revealed that the percentage of TUNEL* cells in the Srsf1-overexpressing group was lower (Fig. 3D, E), and the percentage of apoptotic cells was lower (Fig. 3I, J) than those in the control and empty vector groups. Western blot analysis revealed a significant decrease in cleaved CASP3 and cleaved CASP9 expression (Fig. 3F–H), and the expression of anti-apoptotic protein BCL2 increased, whereas the expression of pro-apoptotic proteins BAX and BAD decreased (Fig. S3A), indicating that overexpression of Srsf1 could inhibit GC-induced OB apoptosis. In contrast, after successful knockdown of Srsf1, OBs were treated with GC to induce apoptosis via RNA interference (RNAi) lentivirus (Fig. 4A–C). Compared with those in the control and empty vector groups, the percentage of TUNEL* cells in the Srsf1 knockdown group was greater (Fig. 4D, E), the apoptosis rate was greater (Fig. 4I, J), and the expression of cleaved CASP3 and cleaved CASP9 was greater (Fig. 4F–H), and the expression of anti-apoptotic proteins BCL2 and BCL-XL decreased, whereas the expression of pro-apoptotic proteins BAX and BAD increased (Fig. S3A), indicating that knockdown of Srsf1 promoted GC-induced OB apoptosis.
A qPCR to verify Srsf1 overexpression efficiency (n = 4), GV492: lentiviral vector, OE-Srsf1: overexpression of Srsf1. B, C The overexpression efficiency of SRSF1 was detected by western blot (n = 3). D, E TUNEL/DAPI staining detection of OBs TUNEL* cell number (n = 3). F–H Expression levels of cleaved-CASP3 and cleaved-CASP9 were detected by western blot (n = 3). I, J Apoptosis detected by Annexin V-FITC/PI staining (n = 4). In (A, C, D, G, H, and J), the data were expressed as mean ± SD, and statistical significance was calculated using one-way ANOVA with Tukey post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05.
A qPCR to verify the knockdown efficiency of Srsf1 (n = 4), CV279: the control lentivirus vector, KD-Srsf1: knockdown of Srsf1. B, C Western Blot detection of SRSF1 knockdown efficiency (n = 4). D, E TUNEL/DAPI staining detection of OBs TUNEL* cell number (n = 3). F–H Expression levels of cleaved-CASP3 and cleaved-CASP9 were detected by western blot (n = 3). I, J Apoptosis detected by Annexin V-FITC/PI staining (n = 4). In (A, C, D, G, H, and J), the data were expressed as mean ± SD, and statistical significance was calculated using one-way ANOVA with Tukey post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05.
SRSF1 regulates alternative splicing of Bmf pre-mRNA and participates in GC-induced OB apoptosis
Studies have shown that SRSF1 can affect cell apoptosis by regulating the alternative splicing of precursor mRNAs (pre-mRNAs) of apoptosis-related genes12. Therefore, we utilized the catRAPID database to predict the apoptosis-related genes that might be regulated by SRSF1. The catRAPID omics v2.1 (protein(s) VS transcriptome) module predicted that SRSF1 might bind to Bmf RNA. The results of the catRAPID signature analysis revealed that the interaction score of SRSF1 with RNA was 0.82 (a score greater than 0.50 suggests a propensity for binding), and the three RNA-binding regions of SRSF1 were PF13893, PF00076, and PF1425 (Fig. S4A). The presence of the catRAPID fragment further revealed the possible fragments involved in the binding of SRSF1 to Bmf pre-mRNA. SRSF1 had the highest interaction propensity with Bmf pre-mRNA at nucleotide sites 7807–8006 (Fig. S4B). Next, to determine the relationship between SRSF1 and Bmf, we performed RIP experiments and found that SRSF1 could bind to Bmf RNA (Fig. S4C, D). Then, we designed specific primers at nucleotide positions 7807–8006 of Bmf to amplify the corresponding DNA fragments, and the in vitro transcribed RNA was labeled with biotin. RNA pull-down assays revealed that biotinylated Bmf RNA oligomers could interact with SRSF1 (Fig. S4E, F). These results suggest that SRSF1 can bind to Bmf pre-mRNA at nucleotide sites 7807–8006 and may be involved in the regulation of alternative splicing of Bmf.
To explore whether SRSF1 affects GC-induced OB apoptosis by regulating Bmf alternative splicing, we found, using qPCR, that there was no significant difference in Bmf variant 1 (V1) expression (P > 0.05). However, Bmf V2 expression was upregulated, and Bmf V3 expression was downregulated in the GC treatment group compared with the control group. Among them, the expression of Bmf V2 was the highest in the control group, and the expression of Bmf V2 changed most significantly after GC treatment (Fig. 5A). The in vivo results of GC-induced OB apoptosis also showed that the number of BMF-positive OB was increased (Fig. S2E, F). Compared with those in the GC treatment group, the expression of Bmf V1 was not significantly different in the Srsf1-overexpressing OBs treated with GC, while the expression of Bmf V2 and Bmf V3 was downregulated (Fig. 5A). Compared with those in the GC treatment group, the expression of Bmf V1 was not significantly different in the Srsf1 knockdown OB treated with GC, but the expression of Bmf V2 and Bmf V3 was upregulated (Fig. 5A). Therefore, SRSF1 did not affect the expression of Bmf V1 but was negatively correlated with the expression of Bmf V2 and Bmf V3 in GC-treated OBs. BMF is a proapoptotic member of the BCL-2 family of proteins that contains only the BH3 domain and is essential for heterodimerizing prosurvival members of the BCL-2 family and proapoptotic functions13,14. Using the National Center for Biotechnology Information (NCBI) and UniProt databases, we found that human BMF V1 contains a BH3 domain (IARKLQCIADQFHRL) and has been shown to promote apoptosis, while BMF V2 and BMF V3 do not contain a BH3 domain and have no effect on apoptosis15 (Fig. 5B). However, all three of the mouse BMF variants contain the same BH3 domain (IARKLQCIADQFHRL) as does humans (Fig. 5C), which led us to speculate that they may have the ability to promote apoptosis. We knocked down Bmf via an RNAi lentivirus (Fig. 5D–F) and then induced OB apoptosis by GC. The results showed that knockdown of Bmf reduced the apoptosis rate (Fig. 5G–J) and decreased the expression of cleaved CASP3 and cleaved CASP9 (Fig. 5K–M), in addition, the expression of anti-apoptotic proteins BCL2 and BCL-XL increased, while the expression of pro-apoptotic protein BAX decreased (Fig. S3B). To further demonstrate that SRSF1 affects GC-induced OB apoptosis by regulating Bmf expression, we knocked down Srsf1 again in Bmf-knockdown OBs and then induced OB apoptosis with GC. Knockdown of Bmf significantly inhibited GC-induced OB apoptosis, but further knockdown of Srsf1 had no effect on OB apoptosis (Fig. S5A–G). These results suggest that in GC, low expression of SRSF1 promotes OB apoptosis by splicing Bmf V2 and Bmf V3, among which BMF V2 is the major regulator of apoptosis.
A qPCR was utilized to detect the expression changes in Bmf variants (n = 4). B, C Structure diagram of human and mouse BMF protein subtypes. D The knockdown efficiency of Bmf was detected by qPCR (n = 4), sh-Bmf: knockdown of Bmf. E, F Western Blot detection of BMF knockdown efficiency (n = 3). G, H Apoptosis detected by Annexin V-FITC/PI staining (n = 4). I, J TUNEL/DAPI staining detection of OBs TUNEL* cell number (n = 3). K–M Expression levels of cleaved-CASP3 and cleaved-CASP9 were detected by western blot (n = 3). In (A, D, F, H, I, L, M), the data were expressed as mean ± SD, and statistical significance was calculated using one-way ANOVA with Tukey post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05.
FOXC1 regulates Srsf1 expression by binding to the Srsf1 promoter
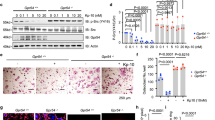
In response to GC, Srsf1 is downregulated at the transcriptome level (Fig. 2F); therefore, we hypothesized that the regulatory effect of GC on Srsf1 downregulation may be related to transcription factor regulation. Using the AnimalTFDB 4.0 database, we found that the transcription factor forkhead box C1 (FOXC1), a transcription activator that promotes gene transcription by binding to gene promoters, can bind to the Srsf1 promoter region16,17. After treatment with GC, the expression of Foxc1 was downregulated in the OB (Fig. 6A–C), and the in vivo results of GC-induced OB apoptosis showed that the number of FOXC1-positive OB decreased (Fig. S2G, H). CHIP-qPCR further showed that FOXC1 could bind to nucleotide sites −1800 ~ −1601, −1600 ~ −1401, and −800 ~ −601 in the Srsf1 promoter region (Fig. 6D). Therefore, we hypothesized that GC may regulate Foxc1 expression to suppress Srsf1 expression. To test this hypothesis, we overexpressed Foxc1 (Fig. 6E–G) or knocked down Foxc1 (Fig. 6H–J) using different lentiviruses and subsequently treated OBs with GC. The results showed that cells overexpressing Foxc1 were resistant to GC-induced downregulation of Srsf1 expression (Fig. 6K–M). The expression of anti-apoptotic proteins BCL2 and BCL-XL increased, and the expression of pro-apoptotic proteins BAX and BAD decreased (Fig. S3C). Conversely, knockdown of Foxc1 further promoted GC-induced downregulation of Srsf1 expression (Fig. 6N–P), and the expression of anti-apoptotic proteins BCL2 and BCL-XL decreased. Expression of the pro-apoptotic proteins BAX and BAD increased (Fig. S3C). These results suggest that GC regulates GC-induced OB apoptosis by inhibiting the expression of transcription activator FOXC1, thereby down-regulating the expression of Srsf1.
A Relative expression of Foxc1 detected by qPCR (n = 4). B, C Expression of FOXC1 was detected by western blot (n = 3). D CHIP-qPCR was utilized to detect the binding sites of FOXC1 and Srsf1 promoter region. E Relative expression of Foxc1 by qPCR (n = 4). F, G Western Blot was utilized to detect the expression of FOXC1 (n = 3). H Relative expression of Foxc1 by qPCR (n = 5). I, J The expression level of FOXC1 was detected by western blot (n = 3). K Relative expression of Srsf1 by qPCR (n = 4). L, M Western blot was utilized to detect the expression of SRSF1 (n = 3). N Relative expression of Srsf1 detected by qPCR (n = 4). O, P Western blot was utilized to detect the expression level of SRSF1 (n = 3). In (A, C, D, E, G, H, J, K, M, N, P), the data are expressed as mean ± SD, where statistical significance was calculated using Student’s t test in (A, C, D) and using one-way ANOVA with Tukey post hoc test in (F, G, I, J, L, M, O). *P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05.
HDAC4 inhibits the expression of Foxc1 by reducing the levels of H3K9ac and H3K27ac in the Foxc1 promoter region
Studies have shown that the histone deacetylase4/5 (Hdac4/5) inhibitor LMK-235 can regulate cell proliferation, differentiation and apoptosis by affecting the expression of many genes18,19,20. Our study found that treatment of OB with LMK-235 upregulated Foxc1 expression levels in cells (Fig. 7A–C). LMK-235-mediated inhibition of Hdac family members is nonspecific and inhibits the activity of Hdac1, Hdac2, Hdac4, Hdac5, Hdac6, Hdac8, and Hdac11. Therefore, it is unclear which specific Hdac family member regulates Foxc1 expression. After treatment of OBs with GC, the qPCR results showed that GC promoted the upregulation of Hdac4, and the in vivo results of GC-induced OB apoptosis showed that the number of HDAC4-positive OB was increased (Fig. S2I, J), while the expression of other Hdac family members was not significantly different (Fig. 7D). After Hdac4 knockdown lentivirus was used to specifically knock down Hdac4 in the OB (Fig. 7E–G), ChIP–qPCR revealed that the levels of histone H3 lysine 9 acetylation (H3K9ac) and H3K27ac in the Foxc1 promoter region increased (Fig. 7H). After the cells were treated with GC, the results showed that Hdac4 knockdown reversed the GC-induced downregulation of Foxc1 expression (Fig. 7I–K), and GC-induced OB apoptosis rate decreased (Fig. S6A, B), the expression of anti-apoptotic proteins BCL2 and BCL-XL increased, and the expression of pro-apoptotic proteins BAX and BAD decreased (Fig. S3D). In contrast, the Hdac4-overexpressing lentivirus specifically increased Hdac4 expression in OBs (Fig. 7L–N) and reduced H3K9ac and H3K27ac levels in the Foxc1 promoter region (Fig. 7O). After the cells were treated with GC, the results showed that the overexpression of Hdac4 further promoted the GC-induced downregulation of Foxc1 expression (Fig. 7P–R), increased GC-induced OB apoptosis rate (Fig. S6A, B), decreased expression of anti-apoptotic proteins BCL2 and BCL-XL, and increased expression of pro-apoptotic proteins BAX and BAD (Fig. S3D). Increased FOXC1 expression has been reported to be associated with increased H3 acetylation in the Foxc1 promoter region. H3K9ac and H3K27ac are common forms of H3 acetylation that function in the regulation of gene expression to activate transcription; conversely, decreased levels of H3K9ac and H3K27ac inhibit gene transcription21,22,23. Therefore, the above results suggest that GC regulates GC-induced OB apoptosis by upregulating HDAC4 expression and reducing H3K9ac and H3K27ac levels in the Foxc1 promoter region, thereby inhibiting Foxc1 expression.
A Relative expression of Foxc1 detected by qPCR (n = 5). B, C Expression of FOXC1 was detected by western blot (n = 3). D Relative expression of Hdac family members by qPCR (n = 5). E Relative expression of Hdac4 detected by qPCR (n = 5). F, G Western blot was utilized to detect the expression level of HDAC4 (n = 3). H CHIP-qPCR was utilized to detect H3K9ac and H3K27ac in the Foxc1 promoter region (n = 4). I Relative expression of Foxc1 detected by qPCR (n = 5). J, K The expression of FOXC1 was detected by western blot (n = 4). L Relative expression of Hdac4 detected by qPCR (n = 5). M, N Western blot was utilized to detect the expression of HDAC4 (n = 3). O CHIP-qPCR was utilized to detect H3K9ac and H3K27ac in the Foxc1 promoter region (n = 4). P Relative expression of Foxc1 detected by qPCR (n = 5). Q, R Western blot was utilized to detect the expression level of FOXC1 (n = 4). In (A, C, D, E, G, H, I, K, L, N, O, P, R), the data are expressed as mean ± SD, where statistical significance was calculated using Student’s t test in (A, C, D, O) and using one-way ANOVA with Tukey post hoc test in (E, G, H, I, K, L, N, P, R). *P < 0.05, **P < 0.01, ***P < 0.001, nsP > 0.05.
Conclusions
Our work uncovers that GC negatively regulates Srsf1 expression and that knockdown of Srsf1 significantly promotes GC-induced OB apoptosis, while overexpression of Srsf1 significantly inhibits GC-induced OB apoptosis. Mechanistically, GC induces the upregulation of Hdac4 and decreases the levels of H3K9ac and H3K27ac in the Foxc1 promoter region to inhibit the expression of Foxc1, thereby reducing the expression of Srsf1. SRSF1 can bind to the 7807–8006 nucleotide sites of Bmf pre-mRNA to regulate the alternative splicing of Bmf. Overexpression of Srsf1 reduced the expression of Bmf V2 and Bmf V3 variants, thereby inhibiting GC-induced OB apoptosis. Conversely, knockdown of Srsf1 increased the expression of Bmf V2 and V3 variants, thereby promoting GC-induced OB apoptosis (Fig. 8). Therefore, Srsf1 and its regulatory mechanism may lead to new targets for the prevention and treatment of GC-induced osteoporosis and osteonecrosis.
GC induces the upregulation of Hdac4 and decreases the level of H3K9ac and H3K27ac in the Foxc1 promoter region to inhibit the expression of FOXC1, thereby reducing the expression of Srsf1. SRSF1 can bind to the 7807–8006 nucleotide sites of Bmf pre-mRNA to regulate the alternative splicing of Bmf. Overexpression of Srsf1 reduced the expression of Bmf V2 and Bmf V3 variants, thereby inhibiting GC-induced OB apoptosis; conversely, knockdown of Srsf1 increased the expression of Bmf V2 and V3 variants, thereby promoting GC-induced OB apoptosis.
Discussion
It has been shown that long-term and high-dose GC use can lead to steroid-induced osteoporosis and osteonecrosis, which is due to the harmful effects of excessive GC on OB, osteocytes and osteoclasts, disrupting the balance of bone formation and bone resorption. GC can inhibit the proliferation and differentiation of OB, enhance the apoptosis of OB and osteocyte, and inhibit bone formation. On the contrary, the effect of GC on osteoclasts is mainly manifested as promoting osteoclast proliferation, reducing osteoclast apoptosis, and increasing bone resorption24,25,26. In this study, we aimed to explore the role and mechanism of GC-induced OB apoptosis in order to explore new methods for the prevention and treatment of steroid-induced osteoporosis and osteonecrosis. At present, the regulatory mechanism of alternative splicing in GC-induced OB apoptosis remains unclear. In this study, we identified the splicing factor SRSF1 as a key regulator of GC-induced OB apoptosis. GC downregulates Srsf1 expression through the Hdac4/Foxc1 axis, which further regulates the alternative splicing of the mitochondrial apoptosis-related gene Bmf, thereby affecting GC-induced OB apoptosis.
Alternative splicing is involved in the posttranscriptional modification of genes, and the SR family is an important class of splicing factors that regulate alternative splicing27,28. Our RNA-seq and qPCR results showed that the expression of Srsf1, Srsf2, Srsf3, and Srsf7 was downregulated in response to GC treatment. Additionally, Srsf6, Srsf9, and Srsf10 were downregulated, with Srsf1 being the most significantly downregulated. Srsf2, Srsf3, Srsf6, and Srsf9 have been shown to be highly expressed in tumor cells and to have antiapoptotic effects on tumor cells29,30,31,32. Like in most SR family members, Srsf1 is upregulated in many cancers, such as breast cancer, acute lymphoblastic leukemia, and lung cancer. This not only increases the proliferation ability, anti-autophagy ability, and antiapoptotic ability of cancer cells but also increases the resistance of cancer cells to chemotherapy and radiotherapy33,34,35,36. Srsf1 has a deleterious effect on carcinogenesis, but these findings indirectly indicate that Srsf1 may have a strong antiapoptotic effect. In addition to its role in tumor cells, Srsf1 has been implicated in the regulation of apoptosis in neural progenitor cells and T cells from patients with systemic lupus erythematosus11,37. Like the effects of Srsf1 found in the above studies, we found that during GC-induced OB apoptosis, Srsf1 was downregulated in a dose-dependent manner with increasing GC concentration, resulting in OB apoptosis, while overexpression of Srsf1 significantly inhibited OB apoptosis.
The regulation of apoptosis by Srsf1 involves many regulatory pathways. For example, Martina Donadoni et al.11 reported that SRSF1 affects neural progenitor cell apoptosis by regulating the Mcl-1L/Mcl-1S ratio. Takayuki Katsuyama et al.37 reported that low expression of SRSF1 in T cells from systemic lupus erythematosus patients promoted apoptosis by reducing the expression of the antiapoptotic gene Bcl-XL. Jacqueline et al.38 reported that in non-small cell lung cancer, SRSF1 promotes the synthesis of the proapoptotic gene caspase-9a by binding to intron six of caspase-9. We found that SRSF1 could bind to Bmf RNA to regulate alternative splicing during GC-induced OB apoptosis. Overexpression of Srsf1 reduced the expression of Bmf V2 and Bmf V3 variants, thereby inhibiting GC-induced OB apoptosis, while knockdown of Srsf1 increased the expression of Bmf V2 and V3 variants; however, there was no significant difference in the expression of Bmf V1 when Srsf1 was overexpressed or knocked down. We knocked down Srsf1 again in OBs with Bmf knockdown and then induced OB apoptosis in GC. Knockdown of Bmf significantly inhibited GC-induced OB apoptosis, but further knockdown of Srsf1 had no effect on OB apoptosis, indicating that the regulatory effect of Srsf1 on GC-induced OB apoptosis requires the mediation of Bmf. This study showed for the first time that Srsf1 affects GC-induced OB apoptosis not only by regulating the expression of apoptosis-related genes such as Mcl-1, Bcl-X, and caspase-9 but also by regulating alternative splicing of Bmf. However, this study has shortcomings in that the detailed molecular mechanism by which SRSF1 regulates alternative splicing of Bmf has not been elucidated.
HDAC4, a member of the class IIa HDAC family with specific amino acid sequences for lysine acetylation, regulates chromatin remodeling and subsequent gene transcription by controlling histone acetylation status39,40,41. HDAC4 turns off gene transcription by deacetylating histone lysines, resulting in condensation of positively charged chromatin42,43,44. It has been reported that GC can recruit HDAC4 by activating glucocorticoid receptor (GR), promote GR-HDAC4 protein interaction, reduce H3K9ac and H3K27ac levels of target genes, and thus inhibit the expression of target genes45,46. Consistent with our results, we found that Hdac4 expression was upregulated in OB under GC treatment, and the upregulated HDAC4 suppressed Foxc1 expression by reducing H3K9ac and H3K27ac levels in the Foxc1 promoter region. Studies have shown that HDAC4 often negatively regulates the transcription factor myocyte enhancer factor (MEF2) to regulate gene expression, thereby affecting the development and function of a variety of cell types47,48,49. However, our results show that under the action of GC, HDAC4 regulates gene expression by negatively regulating the expression of transcription factor FOXC1. At present, we have not explored whether HDAC4 regulates MEF2 expression during GC-induced OB apoptosis, which is a deficiency of this study. As a transcription factor in the FOX family, FOXC1 is characterized by a conserved “forkhead” DNA-binding domain and functions to regulate cell growth, survival, and metabolism50,51. It has been shown that FOXC1 regulates gene transcription through its N-terminal and C-terminal activation domains and phosphorylated transcriptional repression domains17,52,53,54. In this study, we demonstrated that FOXC1 regulates Srsf1 expression by binding to nucleotide sites at –1800 ~ –1601, –1600 ~ –1401, and –800 ~ –601 in the Srsf1 promoter region. In the presence of GC, upregulated Hdac4 inhibits Srsf1 expression by inhibiting Foxc1 expression. In the future, we will focus on investigating the role of Srsf1 in GC-induced osteoporosis and osteonecrosis in vivo.
Methods
Animals
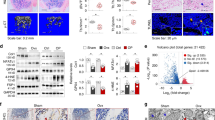
Twenty 6-month-old SPF C57BL/6J male mice were used to construct the animal model of glucocorticoids induced apoptosis in osteoblasts (GOA). The mice were provided by Experimental Animal Center of Guizhou Medical University (ethics approval No.2402798), and we have complied with all relevant ethical regulations for animal use. Methylprednisolone (60 mg/kg; Pfizer, USA) was injected alternately into both gluteal muscles of mice once a day for 4 weeks. Four weeks after methylprednone administration, the mice were killed after anesthesia, and bilateral femoral heads were removed under aseptic operation. The osteonecrosis was evaluated by micro-CT and hematoxylin and eosin (HE) staining, and OB apoptosis and key molecular expression were detected by immunofluorescence staining.
Cells
The cells were digested and collected by centrifugation (1000 revolutions per minute for 5 min) when the growth density of the third-generation MC3T3-E1 cells (PYTHONBIO, Guangzhou, China) reached 90%. The cell density was adjusted to 1 × 104 cells/mL with complete culture medium (89% MEM alpha medium, 10% fetal bovine serum, 1% penicillin/streptomycin solution), and the cells were seeded in 6-well plates at 2 × 104 cells/well and cultured in a 5% CO2 incubator at 37 °C for 2 days. The complete culture medium was discarded when the cell density reached 80%, and osteogenic induction medium (100 mL of complete culture medium, 78.6 µL of 5 mg/mL dexamethasone sodium phosphate injection, 306 mg of β-glycerol phosphate, 20 µL of 0.25 g/mL vitamin C injection) was added. The osteogenic induction medium was changed every 2 days, and the cells were cultured in an incubator for 15 days to induce the maturation of MC3T3-E1 cells into OBs. Twenty-four hours after reseeding the dishes with mature OBs, the OBs were treated with 20 nM LMK-235. In addition, OBs were treated with different concentrations of hydrocortisone (the active form of GC that binds directly to GRs) for 12 h to induce apoptosis. Annexin V-FITC/PI staining and TUNEL/DAPI staining were used to detect apoptosis.
Alkaline phosphatase staining
When the cell density in the six-well plate reached 90%, the cells were washed three times with phosphate-buffered saline (PBS), 1 mL of 4% paraformaldehyde was added to each well, the cells were fixed at room temperature for 20 min, and the cells were subsequently washed three times with distilled water. According to the instructions of the BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime, Shanghai, China), 1 mL of alkaline phosphatase (ALP) staining working solution (alkaline phosphatase color development buffer 3 mL + 300× BCIP solution 10 μL + 150× NBT solution 20 μL) was added to every well, and the plate was incubated at room temperature in the dark for 30 min. The cells were then washed twice with distilled water and observed under a microscope.
Alizarin red S staining
When the cell density in the plate reached 90%, the cells were washed three times with PBS, 1 mL of 4% paraformaldehyde was added to each well, the cells were fixed at room temperature for 30 min and washed twice with PBS, 1 mL of alizarin red S staining solution was added to each well (Solarbio, Beijing, China), and the cells were dyed at room temperature for 10 min, washed twice with PBS, and observed under a microscope.
CCK-8 assay
According to Cell Counting Kit-8 (Beyotime, Shanghai, China), 2000 cells with 100 µL and 10 µL CCK-8 solution are added to 96-well plates. After mixing, the 96-well plates are placed in cell incubators for incubation. The absorbance values of different culture times were measured at 450 nm.
Annexin V-FITC/PI staining
OBs were treated with different concentrations of hydrocortisone, the active form of GC that can directly bind to GRs to exert its effects, for 24 h to induce apoptosis. The adherent cells were digested with trypsin without EDTA (Biological Industries, Shenzhen, China) after the cells were washed twice with precooled PBS. The cells were collected and centrifuged, the culture medium was removed, and the cells were resuspended in 100 μL of 1 × binding buffer and subsequently transferred to 5 mL flow assay tubes. In each flow assay tube, 5 μL of Annexin V-fluorescein isothiocyanate (FITC) and 5 μL of propidium iodide (PI) (Becton, Dickinson and Company, Shanghai, China) were added, and the cells were gently vortexed and incubated at room temperature in the dark for 20 min. Seven hundred microliter of 1 × binding buffer was added to every flow assay tube and detected by flow cytometry (Beckman, USA, Navios) within 1 h.
TUNEL/DAPI staining
Adherent cells were washed twice with PBS, dried, and fixed by adding 1 mL of 4% paraformaldehyde for 30 min. The cells were washed once with PBS, 1 mL of PBS containing 0.3% Triton X-100 was added, and the cells were subsequently incubated for 5 min at room temperature. The cells were washed twice with PBS, 100 μL of terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick-end labeling (TUNEL) detection solution (10 μL of TdT enzyme + 90 μL of fluorescent labeling solution) was added according to the instructions of the One Step TUNEL Apoptosis Assay Kit (Beyotime, Shanghai, China), and the cells were incubated at 37 °C in the dark for 60 min. The cells were washed three times with 1 mL of PBS and incubated with 10 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime, Shanghai, China) for 8 min at room temperature in the dark. The plates were washed three times with PBS, sealed with anti-fluorescence quenching solution, and observed by a laser confocal microscope.
Real-time quantitative polymerase chain reaction
Total RNA was extracted from cells with TRIzol (Invitrogen, California, USA), and the RNA concentration and purity were determined via a Nanodrop 2000. The RNA samples were diluted in 100 ng/μL solution. cDNA was synthesized according to the instructions of the M-MuLV First Strand cDNA Synthesis Kit (Sangon Biotech, Shanghai, China), and real-time qPCR was performed based on the instructions of 2× SG Fast qPCR Master Mix (High Rox) (Sangon Biotech, Shanghai, China). The primer sequences are shown in Table 1.
Western blot
Cells were lysed in RIPA buffer (Solarbio, Beijing, China) to extract cellular proteins, which were quantified with a BCA protein assay kit (Solarbio, Beijing, China). Then, SDS‒PAGE gels were prepared, and the samples were added. Electrophoresis was performed at a constant voltage of 80 V, the maintenance current was 200 mA to transfer the proteins to the PVDF membrane, and 5% skim milk powder was used for blocking. Anti-cleaved caspase-3 (ab32042, Abcam, UK), cleaved caspase-9 (Asp353) (9509, Cell Signaling Technology, USA), anti-Bmf antibody (ab9655, Abcam, UK), Bax (E4U1V) rabbit mAb (41162, Cell Signaling Technology, USA), Bcl-2 (124) mouse mAb (15071, Cell Signaling Technology, USA), Cystatin C (D6U3E) rabbit mAb (24840, Cell Signaling Technology, USA), recombinant anti-Bcl-XL antibody (ab32370, Abcam, UK), recombinant anti-Bad antibody (ab32445, Abcam, UK), SF2/ASF (D6S7V) rabbit mAb (14908, Cell Signaling Technology, USA), and β-actin (13E5) rabbit mAb (4970, Cell Signaling Technology, USA) were incubated as primary antibodies overnight at 4 °C. The sections were incubated with anti-mouse IgG, HRP-linked antibody (7076, Cell Signaling Technology, USA), and anti-rabbit IgG, HRP-linked antibody (7074, Cell Signaling Technology, USA) secondary antibodies for 1 h at room temperature. A gel documentation system (Clinx, Shanghai, China) was utilized for exposure.
Lentivirus infection
The gene overexpression or knockdown lentiviruses utilized in this study were purchased from GeneChem (Shanghai, China). The optimal number of cells in a six-well plate was 1 × 105 per well. After 24 h of culture at 37 °C, the confluence of the cells was 20–30%. Different volumes of negative control virus and HitransG P/A infection boost were added to determine the optimal multiplicity of infection (MOI) and the appropriate increase in virus infection. The optimal MOI was defined as an efficiency of virus infection greater than 80%, and no influence on cell status was observed via fluorescence microscopy. After the cells were inoculated in six-well plates and cultured for 24 h, an appropriate volume of lentivirus and 40 μL of HitransG P infection solution were added according to the optimal MOI, resulting in a final volume of 1 mL. After 16 h of infection, the medium was replaced with fresh complete medium, and the culture was continued. The solution was changed every 2 days thereafter. After 72 h of infection, 10 μg/mL puromycin was added for 72 h. The cells were seeded in 25 cm2 culture flasks by digestion and centrifugation, 5 μg/mL puromycin was added to maintain screening for 2 weeks, and the cells were collected for qPCR and western blot analysis to determine the gene expression levels.
RNA-sequence
Total RNA was extracted from the GC-treated group and the control group by the TRIzol method. RNA was quantified, RNA purity was detected by a NanoDrop 2000 spectrophotometer (A260/280 between 1.8 and 2.2), and RNA integrity was detected by an Agilent 2100 bioanalyzer. Messenger RNA (mRNA) with a polyA tail was enriched by oligo(dT) magnetic beads, and ribosomal RNA was removed from total RNA to obtain mRNA. The mRNAs were randomly disrupted with divalent cations in NEB Fragmentation Buffer, and libraries were constructed utilizing the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs, Beijing, China). The library was sequenced after quality inspection. Transcriptome sequencing and analysis were performed by GeneChem (Shanghai, China). HISAT2 software was used to align the clean reads after quality control with the mouse reference genome, and the HTSeq-count tool was used to calculate the gene expression level according to the alignment results.
Differential expression results between groups were obtained using DESeq2. Differentially expressed genes (DEGs) were obtained with the screening criteria of P < 0.05 and an absolute fold change ≥ 2. Using the Metascape tool, with P < 0.01 as the standard, the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathways were analyzed for the DEGs.
RNA-binding protein immunoprecipitation
Cells were lysed with RIPA cell lysis buffer (Solarbio, Beijing, China) according to the instructions of the Imprint® RNA Immunoprecipitation (RIP) Kit (Merck Millipore, Darmstadt, Germany). A/G magnetic beads (MCE, Sovizzo Vicenza, Italia) were prepared, mixed with 900 µL of RIP immunoprecipitation buffer and 100 µL of cell lysis supernatant and then incubated overnight at 4 °C. After the A/G magnetic beads were washed six times with 500 µL of RIP Wash Buffer, the RNA was purified and diluted to a 10 ng/μL concentration. After cDNA synthesis and amplification, the purified RNA was subjected to agarose gel electrophoresis and photographed by an 1800 series gel imaging system. The Bmf primer sequences are shown in Table 1.
RNA-pull down assay
RNA pull-down analysis was performed with synthetic biotinylated Bmf RNA (Tsingke, Beijing, China) as a probe, and cell lysates were prepared with RIPA lysis buffer according to the instructions of the PierceTM Magnetic RNA‒protein Pull-down Kit (Thermo Fisher Scientific, Massachusetts, USA). The cells were incubated with cell lysates for 60 min at 4 °C with agitation after the binding of biotinylated Bmf RNA to streptavidin magnetic beads. The RNA-binding protein complexes were eluted, collected, and subjected to western blot analysis. The primer sequences for the Bmf probe are shown in Table 1.
Chromatin immunoprecipitation
The MAGnify™ Chromatin Immunoprecipitation System (492024, Thermo Fisher Scientific, Massachusetts, USA), H3K27ac (8173, Cell Signaling Technology, USA), H3K9ac (9649, Cell Signaling Technology, USA), and FOXC1 (PA1-807, Thermo Fisher Scientific, USA) specific antibodies were utilized for chromatin immunoprecipitation. Normal rabbit IgG (2729; Cell Signaling Technology, USA) was utilized as a negative control. Purified DNA was utilized in the qPCRs, and the primer sequences are shown in Table 1.
micro-CT
The mice were killed after anesthesia. After disinfection, both femoral heads were removed, the residue was washed and removed, and the mice were fixed in 4% paraformaldehyde (Solarbio, Beijing, China) for 2 days. Bruker micro-CT was used to scan the femoral head (scanning time 10 min, exposure time 300 ms, resolution 20 μm). NRecon software (V1.7.4.2, Bruker) was used to reconstruct the image of the femoral head. CT Analyzer software (1.18.8.0, Bruker) was used to analyze the ratio of bone volume to total volume (BV/TV), bone mineral density, trabecular number (Tb.N) and trabecular thickness (Tb.Th).
HE staining
Bilateral femoral heads of mice were removed, fixed with 4% paraformaldehyde (Solarbio, Beijing, China) for 2 days, dehydrated with gradient ethanol (SINOPHARM, Beijing, China), and the tissues were soaked in xylene for transparency for 1 h. Staining was performed according to the instructions of HE staining kit (Solarbio, Beijing, China), and photographs were taken with a biological microscope (CX-31, Olympus Corporation, Tokyo, Japan).
Immunofluorescence staining
The femoral head was decalcified by EDTA, dehydrated by ethanol, transparent by xylene and embedded by paraffin. The bone wax was sectioned using a microtome (RM2016, Leica, Germany), and the sections were baked at 60 °C for 3 h. The baked sections were deparaffinized and dehydrated. Sections were stained according to the instructions of the one-step TUNEL apoptosis detection kit (green fluorescence) (Beytime, China). In addition, the deparaffinized sections were subjected to high temperature and pressure for antigen repair and blocked with 10% goat serum (Solarbio, China) at 37 °C for 30 min. Recombinant anti-SF2 antibody [EPR8240] (ab133689, Abcam, UK), anti-Bmf antibody (ab9655, Abcam, UK), osteocalcin antibody(DF12303, Affinity Biosciences, China), recombinant anti-FOXC1 antibody [EPR20685] (ab227977, Abcam, UK) and anti-HDAC4 antibody [HDAC-144] (ab12171, Abcam, UK) for primary antibody reactions (incubated overnight at 4 °C). Goat anti-rabbit IgG H&L (HRP) (ab205718, Abcam, UK) was combined with FITC-Tyramide (green fluorescence) and Cy5-Tyramide (red fluorescence) for secondary antibody reactions (incubation for 2 h at 37 °C). After DAPI staining solution was incubated at room temperature in the dark for 5 min, the slides were blocked with anti-fluorescence quench agent, and images were acquired by laser confocal microscope.
Statistics and reproducibility
All the data were analyzed using SPSS 22.0 statistical software, and GraphPad Prism 9 software was used to construct the statistical plots. The Kolmogorov–Smirnov test was used to analyze the normality of the data to assess statistical significance, and the Levene test was used to analyze the homogeneity of variance of the data between groups. Normally distributed and homogeneous variance data are presented as the mean ± standard deviation (SD). A two-tailed unpaired Student’s t test was used for comparisons involving only two groups, one-way analysis of variance (ANOVA) was used for comparisons involving three or more groups, and Tukey’s post hoc test was utilized for pairwise comparisons. P < 0.05 was considered to indicate statistical significance. The experiments in this study were reproducible, with biological replicates greater than or equal to three.
Ethics oversight
All experiments in this study were supervised, examined and approved by the Experimental Animal Ethics Committee of Guizhou Medical University (Approval number: 2402798).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All raw RNA-seq data generated in this study were deposited in NCBI’s GEO under the accession number GSE291858. The numerical source data for graphs are available in Supplementary Data 1. Uncropped and unedited blots are available in Supplementary Information (Supplementary Fig. 8). Other data are available from the corresponding authors on reasonable request.
Code availability
No custom codes/software were generated in this study. All the data analysis in this study were performed using open-source software tools.
References
Zhao, R. et al. Melatonin rescues glucocorticoid-induced inhibition of osteoblast differentiation in MC3T3-E1 cells via the PI3K/AKT and BMP/Smad signalling pathways. Life Sci. 257, 118044 (2020).
Chotiyarnwong, P. & McCloskey, E. V. Pathogenesis of glucocorticoid-induced osteoporosis and options for treatment. Nat. Rev. Endocrinol. 16, 437–447 (2020).
Amarasekara, D. S., Kim, S. & Rho, J. Regulation of osteoblast differentiation by cytokine networks. Int. J. Mol. Sci. 22, 2851 (2021).
Abuna, R. P. F. et al. Osteoporosis and osteoblasts cocultured with adipocytes inhibit osteoblast differentiation by downregulating histone acetylation. J. Cell Physiol. 236, 3906–3917 (2021).
Hu, Y. et al. Acvr1 deletion in osteoblasts impaired mandibular bone mass through compromised osteoblast differentiation and enhanced sRANKL-induced osteoclastogenesis. J. Cell Physiol. 236, 4580–4591 (2021).
Hao, C. et al. Identification of heterogenous nuclear ribonucleoproteins (hnRNPs) and serine- and arginine-rich (SR) proteins that induce human papillomavirus type 16 late gene expression and alter L1 mRNA splicing. Arch. Virol. 167, 563–570 (2022).
Zheng, X. et al. Serine/arginine-rich splicing factors: the bridge linking alternative splicing and cancer. Int. J. Biol. Sci. 16, 2442–2453 (2020).
Liu, Y. & Shi, S. L. The roles of hnRNP A2/B1 in RNA biology and disease. Wiley Interdiscip. Rev. RNA 12, (2021). e1612.
Anczuków, O. et al. SRSF1-regulated alternative splicing in breast cancer. Mol. Cell 60, 105–117 (2015).
Lv, Y. et al. SRSF1 inhibits autophagy through regulating Bcl-x splicing and interacting with PIK3C3 in lung cancer. Signal Transduct. Target Ther. 6, 108 (2021).
Donadoni, M., Cicalese, S., Sarkar, D. K., Chang, S. L. & Sariyer, I. K. Alcohol exposure alters pre-mRNA splicing of antiapoptotic Mcl-1L isoform and induces apoptosis in neural progenitors and immature neurons. Cell Death Dis. 10, 447 (2019).
Kędzierska, H. & Piekiełko-Witkowska, A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 396, 53–65 (2017).
Jenkins, L. J. et al. Genotype-tailored ERK/MAPK pathway and HDAC inhibition rewires the apoptotic rheostat to trigger colorectal cancer cell death. Mol. Cancer Ther. 22, 52–62 (2023).
Li, M., Liang, W., Zhu, C. & Qin, S. Smad4 mediates Bmf involvement in sheep granulosa cell apoptosis. Gene 817, 146231 (2022).
Morales, A. A. et al. Expression and transcriptional regulation of functionally distinct Bmf isoforms in B-chronic lymphocytic leukemia cells. Leukemia 18, 41–47 (2004).
Li, M. et al. Loss of FOXC1 contributes to the corneal epithelial fate switch and pathogenesis. Signal Transduct. Target Ther. 6, 5 (2021).
Chen, H. Y. et al. ELAVL1 is transcriptionally activated by FOXC1 and promotes ferroptosis in myocardial ischemia/reperfusion injury by regulating autophagy. Mol. Med. 27, 14 (2021).
Mazzocchi, M. et al. LMK235, a small molecule inhibitor of HDAC4/5, protects dopaminergic neurons against neurotoxin- and α-synuclein-induced degeneration in cellular models of Parkinson’s disease. Mol. Cell Neurosci. 115, 103642 (2021).
Trazzi, S. et al. HDAC4: a key factor underlying brain developmental alterations in CDKL5 disorder. Hum. Mol. Genet. 25, 3887–3907 (2016).
Gao, Y. et al. HDAC5-mediated Smad7 silencing through MEF2A is critical for fibroblast activation and hypertrophic scar formation. Int. J. Biol. Sci. 18, 5724–5739 (2022).
Forma, E. et al. Impact of OGT deregulation on EZH2 target genes FOXA1 and FOXC1 expression in breast cancer cells. PLoS ONE 13, e0198351 (2018).
Wang, J. et al. Blockage of CacyBP inhibits macrophage recruitment and improves anti-PD-1 therapy in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 42, 303 (2023).
Finke, D. et al. Histone deacetylase 4 deletion broadly affects cardiac epigenetic repression and regulates transcriptional susceptibility via H3K9 methylation. J. Mol. Cell Cardiol. 162, 119–129 (2022).
Chen, M., Fu, W., Xu, H. & Liu, C. J. Pathogenic mechanisms of glucocorticoid-induced osteoporosis. Cytokine Growth Factor Rev. 70, 54–66 (2023).
Li, W. et al. PINK1/Parkin-mediated mitophagy inhibits osteoblast apoptosis induced by advanced oxidation protein products. Cell Death Dis. 14, 88 (2023).
Yue, C. et al. Aucubin prevents steroid-induced osteoblast apoptosis by enhancing autophagy via AMPK activation. J. Cell Mol. Med. 25, 10175–10184 (2021).
González-Rodríguez, P., Klionsky, D. J. & Joseph, B. Autophagy regulation by RNA alternative splicing and implications in human diseases. Nat. Commun. 13, 2735 (2022).
Sciarrillo, R. et al. The role of alternative splicing in cancer: from oncogenesis to drug resistance. Drug Resist. Updat. 53, 100728 (2020).
Kędzierska, H. et al. Decreased expression of SRSF2 splicing factor inhibits apoptotic pathways in renal cancer. Int. J. Mol. Sci. 17, 1598 (2016).
Zhang, L., Li, J. & Zhang, L. SRSF3 restriction eases cervical cancer cell viability and metastasis via adjusting PI3K/AKT/mTOR signaling pathway. Contrast Media Mol. Imaging 2022, 8497078 (2022).
Yang, X. et al. SRSF6 regulates alternative splicing of genes involved in DNA damage response and DNA repair in HeLa cells. Oncol. Rep. 44, 1851–1862 (2020).
Zhang, Q., Lv, R., Guo, W. & Li, X. microRNA-802 inhibits cell proliferation and induces apoptosis in human cervical cancer by targeting serine/arginine-rich splicing factor 9. J. Cell Biochem. 120, 10370–10379 (2019).
Jiang, L. et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLoS Genet. 12, e1005895 (2016).
Anczuków, O. et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19, 220–228 (2012).
Zou, L. et al. Correlation of SRSF1 and PRMT1 expression with clinical status of pediatric acute lymphoblastic leukemia. J. Hematol. Oncol. 5, 42 (2012).
Sheng, J. et al. SRSF1 modulates PTPMT1 alternative splicing to regulate lung cancer cell radioresistance. EBioMedicine 38, 113–126 (2018).
Katsuyama, T., Martin-Delgado, I. J., Krishfield, S. M., Kyttaris, V. C. & Moulton, V. R. Splicing factor SRSF1 controls T cell homeostasis and its decreased levels are linked to lymphopenia in systemic lupus erythematosus. Rheumatology (Oxford) 59, 2146–2155 (2020).
Shultz, J. C. et al. SRSF1 regulates the alternative splicing of caspase 9 via a novel intronic splicing enhancer affecting the chemotherapeutic sensitivity of non-small cell lung cancer cells. Mol. Cancer Res. 9, 889–900 (2011).
Federspiel, J. D., Greco, T. M., Lum, K. K. & Cristea, I. M. Hdac4 interactions in huntington’s disease viewed through the prism of multiomics. Mol. Cell Proteom. 18, S92–S113 (2019).
Cheng, C. et al. HDAC4 promotes nasopharyngeal carcinoma progression and serves as a therapeutic target. Cell Death Dis. 12, 137 (2021).
Zhang, X., Qi, Z., Yin, H. & Yang, G. Interaction between p53 and Ras signaling controls cisplatin resistance via HDAC4- and HIF-1α-mediated regulation of apoptosis and autophagy. Theranostics 9, 1096–1114 (2019).
Kong, Q. et al. HDAC4 in ischemic stroke: mechanisms and therapeutic potential. Clin. Epigenetics 10, 117 (2018).
Helmstadter, K. G. et al. CaMKII and PKA-dependent phosphorylation co-regulate nuclear localization of HDAC4 in adult cardiomyocytes. Basic Res. Cardiol. 116, 11 (2021).
Zang, W. J. et al. HDAC4 promotes the growth and metastasis of gastric cancer via autophagic degradation of MEKK3. Br. J. Cancer 127, 237–248 (2022).
Shi, H. et al. Intrauterine programming of cartilaginous 11β-HSD2 induced by corticosterone and caffeine mediated susceptibility to adult osteoarthritis. Ecotoxicol. Environ. Saf. 239, 113624 (2022).
Qi, Y. et al. H3K9ac of TGFβRI in human umbilical cord: a potential biomarker for evaluating cartilage differentiation and susceptibility to osteoarthritis via a two-step strategy. Stem Cell Res. Ther. 12, 163 (2021).
Richter, E. A. & Hargreaves, M. Exercise, GLUT4, and skeletal muscle glucose uptake. Physiol. Rev. 93, 993–1017 (2013).
He, T. et al. Cyclic AMP represses pathological MEF2 activation by myocyte-specific hypo-phosphorylation of HDAC5. J. Mol. Cell Cardiol. 145, 88–98 (2020).
Clocchiatti, A., Di Giorgio, E., Demarchi, F. & Brancolini, C. Beside the MEF2 axis: unconventional functions of HDAC4. Cell Signal 25, 269–276 (2013).
Han, B. et al. FOXC1: an emerging marker and therapeutic target for cancer. Oncogene 36, 3957–3963 (2017).
Elian, F. A., Yan, E. & Walter, M. A. FOXC1, the new player in the cancer sandbox. Oncotarget 9, 8165–8178 (2017).
Koo, H. Y. & Kume, T. FoxC1-dependent regulation of vascular endothelial growth factor signaling in corneal avascularity. Trends Cardiovasc. Med. 23, 1–4 (2013).
Lin, Z. et al. FOXC1 promotes HCC proliferation and metastasis by Upregulating DNMT3B to induce DNA Hypermethylation of CTH promoter. J. Exp. Clin. Cancer Res. 40, 50 (2021).
Sun, C. C. et al. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway. Genome Med. 12, 77 (2020).
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 82260429 and 82260434); Science and Technology Foundation of Guizhou Provincial Health Committee (Grant No. gzwkj2025-362); Guizhou Provincial Basic Research Plan (Natural Science) Talent Team Support Project in 2025 (Grant No. Qiankehe Basis QNB [2025] 006); Guizhou Provincial Natural Science Foundation (Grant No. Qiankehe basis-ZK [2022] general 399, Qiankehe basis-ZK [2023] general 345, and Qiankehe basis-ZK [2022] general 447); Discipline Outstanding Reserve Talent Program of the Affiliated Hospital of Guizhou Medical University (Grant No. gyfyxkrc-2023-07); Cultivation Project of NSFC General Program of the Affiliated Hospital of Guizhou Medical University (Grant No. gyfynsfc[2023]-02); Cultivation Project of National Natural Science Foundation of Guizhou Medical University (Grant No. 21NSFCP08); Science and Technology Foundation of Guizhou Provincial Health Committee (Grant No. gzwkj2021-234); Start-up Fund for Doctoral Research of the Affiliated Hospital of Guizhou Medical University (Grant No. gyfybsky-2022-14); The Eighth Batch of High-Level Innovative Talent Project in Guizhou Province (Grant No. Qian Wei Ren Ling Ban Fa [2024] No. 3); Medical Research Joint Fund for High-Quality Development of Health in Guizhou Province (Grant No. 2024GZYXKYJJXM0129). We thank the Clinical Research Center of Guizhou Medical University for providing the experimental platform and guidance and LetPub (http://www.letpub.com.cn) for providing the English-assisted revision.
Author information
Authors and Affiliations
Contributions
Hong Luo designed the study, performed the experiments, analyzed the data, and wrote the manuscript; Zhihong Xie performed the experiments and analyzed the data; Wang Tao and Fanchao Li performed the experiments and revised the manuscript; Chengyou Yang, Wentao Dong, Jianhua Wu, Qiang Wang, Fengyang Xu, and Jiong Liu performed the experiments and collected the data; and Wuxun Peng and Fei Zhang designed the study and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Mubashir Ahmad, Weiwei Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Martina Rauner and Joao Valente. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, H., Wang, T., Xie, Z. et al. Glucocorticoids regulate the expression of Srsf1 through Hdac4/Foxc1 axis to induce apoptosis of osteoblasts. Commun Biol 8, 566 (2025). https://doi.org/10.1038/s42003-025-07989-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07989-x