Abstract
Background
Cholangiocarcinoma (CCA) is a rare and highly aggressive malignancy originating in the bile ducts. Owing to limitations involving pathological sampling, the clinical differentiation of CCA from benign biliary diseases remains challenging. This study aimed to evaluate the differences between the bile lipidomes of CCA patients and those of patients with benign disease to develop a bile lipid classifier that can help to differentiate CCA from benign conditions.
Methods
Bile samples were collected by endoscopic retrograde cholangiography (ERCP) from patients with CCA or benign disease. The participants were divided into three cohorts: the first two cohorts underwent untargeted lipidomic analysis, whereas the third cohort was subjected to targeted lipid quantification. Untargeted lipidomic analysis was performed via ultrahigh-performance liquid chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry (UHPLC/IM-QTOF-MS). Targeted lipid quantification was conducted via UHPLC‒MS/MS in multiple reaction monitoring (MRM) mode. Lipid features were screened to construct a bile lipid classifier using the machine learning algorithm, least absolute shrinkage and selection operator (LASSO) regression, followed by cross-validation in two cohorts. The selected lipid features were further validated by targeted quantification in the third cohort. The functions of the significantly differentially abundant lipids in proliferation were validated in CCA cell lines.
Results
In total, 241 bile samples were collected and divided into three cohorts for independent lipidomic analysis: Cohort 1 included 32 CCA samples and 68 benign controls; Cohort 2 included 30 CCA samples and 30 benign controls; and Cohort 3 included 32 CCA samples and 49 benign controls. There were significant differences in the lipid profiles of the bile samples obtained from patients with CCA and individuals with benign disease, with multiple lipid classes, particularly lysophosphatidylcholine (LPC), significantly downregulated in the CCA group. Multimodule correlation networks constructed via weighted lipid coexpression network analysis (WLCNA) revealed significant associations between lipid modules and clinical traits. A machine learning-based bile lipid classifier, termed BileLipid, was developed for CCA diagnosis; this classifier incorporates six lipid features. This classifier achieved areas under the receiver operating characteristic curve (AUCs) of 0.943, 0.956, and 0.828 in Cohorts 1, 2, and 3, respectively. Additionally, the significantly downregulated lipid LPC in CCA bile was found to significantly inhibit the proliferation of CCA cell lines, suggesting its potential role as a protective factor in CCA.
Conclusions
This study not only identified lipidomic alterations in CCA using bile samples but also established and validated a bile lipid classifier with high specificity and sensitivity for distinguishing between CCA and benign bile duct diseases. Our findings highlight the potential of bile lipid biomarkers for improving the differential diagnosis and risk assessment of CCA and preventing potential overintervention in patients with benign biliary disease.
Similar content being viewed by others
Introduction
Cholangiocarcinoma (CCA) is a rare and highly aggressive malignancy that originates in the bile ducts [1, 2]. CCA has a global incidence ranging from 0.3 to 6/100,000 people per year, a mortality rate of 1–6/100,000 people per year, and is the cause of ~2% of all cancer-related deaths worldwide yearly [3]. Despite its rarity, the incidence and mortality of CCA are increasing globally, particularly in Asian countries [3,4,5]. Currently, the diagnosis relies on a combination of serum tumour markers, radiographic imaging, and histological examination [6,7,8]. However, the unique anatomical locations of CCA pose significant challenges for invasive pathological diagnosis [9]. Preoperative confirmation is achieved primarily through brush cytology, forceps biopsy, or fine needle aspiration biopsy [8]. However, the positivity rate of brush cytology is often low [10,11,12], biopsy procedures carry high operational risks and can yield false-negative results [13], and to varying degrees, traditional serum tumour markers and imaging tests suffer from limitations in their sensitivity and specificity [14]. Therefore, developing a safe, efficient, and reliable diagnostic test for CCA is imperative to improve the clinical management of bile duct diseases and improve patient survival outcomes.
Over the last decade, there have been significant strides in the use of various liquid biopsy methods in early diagnosis and precision diagnostics for a wide range of malignant tumours [15]. Most studies in this field have focused on biomarkers derived from blood [16, 17]. However, in certain site-specific diseases, more targeted body fluids (e.g., urine, cerebrospinal fluid, pleural fluid, and peritoneal fluid) can be obtained, providing more biomarkers that carry information about the corresponding diseases [17,18,19]. Theoretically, analysing biomarkers from non-blood sources may provide greater sensitivity than plasma-based assays for certain tumour types or anatomical locations [20, 21]. In this study, we innovatively explored the diagnostic potential of bile samples for CCA. The main advantage of using bile over blood is its ability to exclude interference information from other organs and sites, thereby capturing the unique molecular features of the biliary system in a more targeted manner. This specificity may allow for a more accurate representation of the heterogeneity inherent in biliary tract cancers [22]. Furthermore, bile samples can be collected during endoscopic retrograde cholangiography (ERCP) examinations without the need for additional invasive procedures; because ERCP is a common examination modality for patients with biliary tract diseases, bile has attracted the attention of biliary physicians and scientists as a minimally invasive biospecimen [22,23,24,25].
Lipid metabolism reprogramming, a key characteristic of malignant tumours, has been continuously emphasised in studies, along with advances in lipidomics technology for liquid biopsy of tumours [26]. From the perspective of biological function, lipids are a complex group of biomolecules that serve as structural components of biological membranes and provide and store energy, with specific lipid molecules acting as signalling molecules to regulate cellular activities [27]. The diagnostic value of lipidomics has been validated in various cancers, including lung [28], pancreatic [29], colorectal [30], gastric [31], and endometrial cancers [32], but systematic exploration of lipidomic data in the context of bile duct cancers remains limited. In our previous study, we preliminarily analysed volatile metabolites in bile via gas chromatography‒mass spectrometry and performed comparative analyses between benign and malignant samples [33]. Among the metabolites we detected, there were significant alterations among multiple lipid metabolites, particularly in the free fatty acid families [33]. Building on our previous research foundation and the existing results of lipidomics in liquid biopsy applications in cancer, we determined that bile lipidome analysis could be a pivotal research direction.
In this study, we aimed to evaluate differences in the bile lipidomes of patients with CCA and those with benign bile duct disease to develop a bile lipid classifier capable of distinguishing CCA from benign conditions. The model was trained, validated, and tested in three independent cohorts, utilising both untargeted and targeted lipidomics approaches. Our untargeted lipidomic investigations in Cohort 1 and Cohort 2 identified a bile lipid classifier comprising 6 lipid features that effectively differentiates CCA from benign conditions. To validate the robustness and generalisability of this classifier, we subsequently evaluated its diagnostic performance in an independent cohort (Cohort 3) using a targeted lipid quantification approach. This stepwise validation strategy not only confirmed the reliability of the lipid-based biomarker panel but also underscored its translational potential across diverse clinical settings and analytical platforms. To our knowledge, this study represents the first large-scale lipidomic analysis of bile samples from patients with CCA and benign bile duct disease. We developed and validated a bile lipid classifier consisting of 6 lipids, which demonstrated robust diagnostic performance across three independent cohorts. This highlights the clinical applicability and reliability of bile lipid profiling for CCA diagnosis. Collectively, our findings underscore the potential of bile lipidomics as a promising diagnostic tool for CCA.
Materials and methods
Study design
This was a single-centre, prospective descriptive study of bile samples collected by ERCP from patients with CCA or benign disease. All participants were biliary patients who visited Zhongnan Hospital of Wuhan University from June 2020 to August 2024. The participants were randomly assigned to three cohorts, each meeting the predefined criteria (Cohen’s d = 0.8, α = 0.05, Power = 0.8). Based on power analysis, a minimum of 26 participants per group (with a total sample size of at least 52 participants per cohort) was determined to be sufficient to achieve the required 80% statistical power for detecting the predefined effect size. Each cohort underwent independent lipid extraction and detection, ensuring the reproducibility and robustness of the study results across all groups. Three independent clinical researchers who were blinded to the bile test results collected the clinical data, including demographic information, medical history, imaging results, serum tumour markers, pathology reports, Eastern Cooperative Oncology Group Performance Status (ECOG-PS), disease scoring and tumour staging outcomes. The study was approved by the Medical Ethics Committee of the Clinical Trial Centre at Zhongnan Hospital, Wuhan University (Approval number: Scientific Research Ethics[2020100]). Informed consent for sample collection was obtained from all patients before the operation.
Participants
Bile samples and clinical information were collected from patients with CCA or benign biliary diseases. The inclusion criteria were as follows: (1) benign and malignant biliary tract diseases that required ERCP for examination or treatment and (2) successful completion of the ERCP procedure. The exclusion criteria were as follows: (1) other malignant tumours, such as pancreatic cancer, (2) metastatic cancer, (3) receiving any known anticancer therapy, (4) sepsis due to biliary tract infection, and (5) unclear diagnosis due to lack of clinical information or insufficient follow-up time.
The diagnostic criteria were established according to internationally recognised guidelines and expert consensus, including the National Comprehensive Cancer Network (NCCN), the European Society for Medical Oncology (ESMO), and other authoritative sources [8, 34, 35]. Benign disease was confirmed if any of the following criteria were met: (1) Postoperative pathological examination of resected samples confirming benign characteristics (e.g., biliary lithiasis, chronic inflammation, or adenoma) [8, 34]. (2) Nonsurgical criteria (≥2 criteria required): ① no evidence of tumour progression on CT/MRI scans over a minimum follow-up period of 6 months [34], ② serum CA19-9 levels consistently ≤37 U/mL with no progressive upward trend (after excluding confounding factors such as active biliary obstruction or infection) [8, 34], or ③ clinical follow-up confirming the absence of malignancy-associated symptoms (e.g., progressive jaundice, weight loss) or end-stage events. Benign biliary diseases are further subdivided into inflammatory biliary tract disorders (IBTs, including recurrent pyogenic cholangitis (RPC), primary biliary cholangitis (PBC), primary sclerosing cholangitis (PSC), etc.) and noninflammatory diseases (NIBTs).
Malignancy diagnosis required the following: (1) malignancy confirmed by ultrasound-guided core needle biopsy or ERCP-guided forceps biopsy (≥3 sites) or surgical specimens, independently verified by two senior pathologists [8, 34]; (2) Nonsurgical cases (≥3 criteria required): ① dynamic contrast-enhanced imaging (CT/MRI) showing progressive invasive features (e.g., capsular penetration, vascular invasion) [8, 34], ② PET-CT demonstrating FDG uptake (SUVmax ≥3.0) [34], ③ serum CA19-9 elevation (increase across three consecutive tests at 4-week intervals, excluding inflammatory interference), ④ multidisciplinary-team-confirmed tumour progression during 6-12-month follow-up [8, 34], or ⑤ malignancy-related end-stage outcomes (biliary obstructive liver failure/tumour-associated mortality) [8]. Clinical staging of CCA was performed according to the TNM staging criteria from the 8th edition of the American Joint Committee on Cancer (AJCC) Hepatobiliary and Pancreatic Cancer Staging Manual [36].
Sample collection and preparation
Fasting bile samples were collected from patients via the ERCP method during diagnostic or therapeutic biliary drainage. Bile was aspirated following bile duct intubation and prior to the injection of contrast medium. Fresh bile samples were centrifuged at 12,000×g for 10 minutes, and the supernatant was filtered through a 0.45-μm sterile filter before being stored at -80°C.
Before analysis, the bile samples were thawed, and lipids were extracted via an adjusted extraction method (MTBE/methanol) [28, 29]. All reagents were obtained from Sigma‒Aldrich® (Merck). Briefly, 200 μL of bile was mixed with 250 μL of ice-cold methanol (34860) and 750 μL of methyl tert-butyl ether (MTBE, 34875). For targeted lipid quantification, three classes of lipid internal standards (5 Cers, 4 LPCs, 3 TGs) were spiked into the extract. The mixture was vortexed for 1 min and then centrifuged at 16000 × g for 5 min at 4 °C. Subsequently, 800 μL of the upper organic phase was transferred to a new centrifuge tube, filtered through a 0.22-μm oil filter, and evaporated to dryness under a vacuum centrifugal concentrator (12,000 × g, 4 °C). The dry extract was reconstituted in 50 μL of methanol, vortexed, and transferred to a glass amber vial with a microinsertion tube for lipidomic analysis. Pooled quality control (QC) samples were created by taking 5 μL aliquots of the supernatant from each sample for QC and generating an annotated lipid database.
Lipidomic analysis
For untargeted lipidomics, an Agilent 1290 Infinity UHPLC coupled to a quadruple time‒of‒flight mass spectrometer equipped with an ion mobility drift tube (Agilent DTIM-QTOF-MS 6560, Agilent Technologies, USA) was used for LC–IM–MS/MS data acquisition. Chromatographic separations were performed on an Agilent Infinity Poroshell 120 EC-C18 column (3.0 × 100 mm, 2.7 µm) with the column temperature maintained at 50 °C. The mobile phases were composed of (A) H2O/MeOH (9:1, v/v) with 10 mM NH4AC and 0.2 mM NH4F and (B) ACN/MeOH/IPA (2:3:5, v/v) with 10 mM NH4AC and 0.2 mM NH4F. The elution gradient was carried out as follows: 0–1 min, 70% B; 1–3.5 min, 70% B to 86% B; 10–11 min, 86% B to 100% B; 11–17 min, 100% B; 17–17.1 min, 100% B to 70% B; and 17.1–19 min, 70% B. The flow rate was 0.6 mL/min. For positive and negative mode analyses, the injection volumes were 3 μL and 6 μL, respectively. The sample was maintained at 4°C throughout the entire analysis.
The mass spectrometry parameters were configured as follows. Electrospray ionisation (ESI) was performed in positive ion mode with a mass scan range of 50–1000 Da, and in negative ion mode with a range of 50–1700 Da. The sheath gas temperature was maintained at 370 °C with a flow rate of 12 L/min, while the drying gas temperature was set to 300 °C with a flow rate of 10 L/min. The nebuliser pressure was 50 psi. The capillary voltage was set to 3500 V in positive mode and 3000 V in negative mode. The collision cross section (CCS) values were measured with the single-field method with nitrogen as the drift gas. The maximum drift time was set to 60 ms, and the scan rate was set to 1 frame per second. A drift gas upgrade kit was used to maintain both the drift tube and trap funnel pressures at 3.9 and 3.8 Torr, respectively. The entrance and exit voltages of the drift tube were set to 1200 V and 250 V, respectively. The trap filling and trap release times were set to 20,000 μs and 150 μs, respectively. All the ion mobility‒mass spectrometry (IM‒MS) fragmentation spectra were acquired in “alternating frames” mode with frame 2 fixed at 20 V. The MS1 spectra were acquired in “single frame–not fragmented” mode. All data acquisition was carried out with MassHunter Workstation Data Acquisition Software (version B.09.00; Agilent Technologies, USA).
For targeted lipid quantification, the sample extracts were analysed using an LC‒ESI‒MS/MS system (UPLC, ExionLC AD, https://sciex.com.cn/; MS, QTRAP® 6500+ System, https://sciex.com/). The analytical conditions were as follows: UPLC: column, Thermo Accucore™C30 (2.6 μm, 2.1 mm×100 mm i.d.); solvent system, A: acetonitrile/water (60/40, V/V, 0.1% formic acid, 10 mmol/L ammonium formate), B: acetonitrile/isopropanol (10/90 VV/V, 0.1% formic acid, 10 mmol/L ammonium formate); gradient programme, A/B (80:20, V/V) at 0 min, 70:30 V/V at 2.0 min, 40:60 V/V at 4 min, 15:85 V/V at 9 min, 10:90 V/V at 14 min, 5:95 V/V at 15.5 min, 5:95 V/V at 17.3 min, 80:20 V/V at 17.3 min, 80:20 V/V at 20 min; flow rate, 0.35 ml/min; temperature, 45°C; and injection volume, 2 μL. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (QTRAP)-MS. LIT and triple quadrupole (QQQ) scans were acquired with a QQQ–linear ion trap mass spectrometer (QTRAP), QTRAP® 6500 + LC‒MS/MS System, equipped with an ESI Turbo Ion-Spray interface, operating in positive ion mode and controlled by Analyst 1.6.3 software (Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature, 500°C; ion spray (IS) voltage, 5500 V; and ion source gas 1 (GS1), gas 2 (GS2), and curtain gas (CUR), which were set at 45, 55, and 35 psi, respectively. Instrument tuning and mass calibration were performed with 10 and 100 μmol/L polypropylene glycol solutions in QQQ and LIT modes, respectively. QQQ scans were acquired as MRM experiments with the collision gas (nitrogen) set to 5 psi. DP and CE for individual MRM transitions were performed with further DP and CE optimisation. A specific set of MRM transitions was monitored for each period according to the metabolites eluted within this period.
Lipid annotation
For untargeted lipidomics, after the IM‒MS data were acquired, mass and single-field CCS calibrations were first performed via the IM–MS Reprocessor and IM–MS Browser (version B.08.00; Agilent Technologies, USA), respectively. The lipids in all the ion IM‒MS data files of the QC samples were subsequently annotated with Lipid Annotator (Version 1.0; Agilent Technologies, USA), which generated a database that contains accurate mass, retention time (RT) and CCS values. The database can be exported to a PCDL file for later use with the ID browser. The method parameters used for lipid annotation were as follows: feature finding parameters with a Q score ≥ 30.0; library parameters with positive ions ([M + NH4]+, [M + H]+, [M+Na]+) and negative ions ([M + CH3COO]- and [M-H]-); identification parameters: sum composition level, mass deviation≤10.0 ppm; multiple lipid IDs for the same feature, reporting the top candidate only; score threshold, fragment score≥30 and total score≥60; and constituent level, reporting the dominant constituent if the relative abundance differential was ≥10%. Third, the compound features in the MS1 data of each sample were extracted with a mass profiler (version B.08.01, Agilent Technologies, USA). Feature finding was performed for signals having a minimum ion count of 100 using the maximum ion volume as a measure of abundance. The isotope model was selected for common organic molecules, and the charge states were limited to a range of 1−1. The features were subsequently aligned with an RT tolerance of 0.2 min, a CCS tolerance of 5%, and a mass of 10 ppm. The final filter was based on a Q score > 80.0. The frequency of sample occurrence in at least one group was equal to or greater than 50%. Finally, the annotated database with accurate mass, RT and CCS values was used to identify the MS1 features by launching an ID browser followed by returning the feature list to the mass profiler. Lipid annotation was completed on the basis of the RT, CCS, and m/z values of the lipid features. Lipid features from the Cohort 1 and Cohort 2 libraries were simultaneously compared to determine the correspondence between isoforms and different ion forms, and lipids with the same molecular formula were numbered according to RT and abundance.
For targeted lipid quantification, lipid identification was performed utilising RT, precursor‒product ion pairs, and tandem mass spectrometry (MS/MS) spectral matching. Quantitative analysis was conducted using multiple reaction monitoring (MRM) mode on a QQQ mass spectrometer. The raw MS data were processed with Analyst software (version 1.6.3).
Lipidomic data processing
The raw lipidomic data obtained after annotation and peak area extraction were normalised by taking the natural logarithm. To reduce potential interbatch variations for untargeted lipidomic data, each feature peak of the study samples was divided by the average abundance of that lipid from the same cohort to obtain the relative abundance. MetaboAnalyst 6.0 (https://www.metaboanalyst.Ca/) and the R package ropls v.1.36.0 were used for univariate analysis of lipid abundance, and multivariate analysis included principal component analysis (PCA), partial least squares–discriminant analysis (PLS–DA) and orthogonal partial least squares–discriminant analysis (OPLS–DA). The false discovery rate (FDR) is controlled via the Benjamini‒Hochberg (BH) procedure, which adjusts raw p values to account for multiple hypothesis testing. For differential expression analysis, the log2-fold change (FC) was used to quantify the relative change in expression levels between the two conditions. Differential features between two groups were identified by the p value adjusted by the FDR and FC significance thresholds (FDR-adjusted p < 0.05, Wilcoxon rank-sum test; |log2(FC)| ≥ 1).
Weighted lipid correlation network analysis (WLCNA)
In the weighted coexpression analysis of lipids detected in untargeted lipidomic cohorts, the correlation matrix between lipids in the dataset represents a fully connected weighted network, where the weight on each edge corresponds to the correlation between the quantitative profiles of two lipids. A soft threshold of 9 was selected, and the adjacency matrix was transformed into a topological overlap matrix (TOM) with the TOMsimilarity function. Module identification was performed using dynamic tree cutting with deepSplit = 1, and hierarchical clustering of the coexpression network identified 10 modules (R package: WGCNA v.1.73). The Spearman correlations between modules and clinical traits were calculated, significantly correlated modules and clinical traits were identified, key lipids within the identified modules were analysed, and lipid coexpression networks within each module were constructed (R packages: WGCNA v.1.73, psych v.2.4.6.26; GraphPad Prism 9.0.0; Cytoscape 3.10.2).
Establishment, validation and evaluation of diagnostic models
The optimal lipid biomarker model for predicting CCA was built via binary logistic regression with the least absolute shrinkage and selection operator (LASSO, R package: glmnet v.4.1.8). The probability of CCA was calculated as follows: probability = 1/(1+e^(-(α0 + (∑θ0Xk)), where α0 is the intercept, θ0 is the final logistic regression coefficient for the kth lipid, and Xk is the natural logarithm of the normalised concentration level. The performance of the discriminant model in both the training and validation cohorts was evaluated by estimating the area under the curve (AUC) according to receiver operating characteristic (ROC) analysis (R package: pROC v.1.18.5, GraphPad Prism 9.0.0). Clinical decision curve analysis (DCA), clinical impact curve (CIC) analysis, and calibration analyses were used for multiple evaluations of the model diagnostic performance and clinical application (R packages: car v.3.1.2, rmda v.1.6, rms v.6.8.1, ResourceSelection v.0.3.6).
Cell culture and stimulation with LPC
Human CCA cells and normal bile duct epithelial cells (TFK1, QBC939 and HIBEpiC; Cell Bank of Type Culture Collection, Shanghai, China) were cultured at Roswell Park Memorial Institute (RPMI) 1640 medium (11875093 Gibco, USA) containing 10% foetal bovine serum (A5669701, Gibco, USA), 100 units/mL penicillin and 100 μg/mL streptomycin (15140122, Gibco, USA) in an incubator at 37°C with 5% CO2. Exogenous LPC (Sigma–Aldrich, 62962) was dissolved in ethanol and added to preheated medium at 37°C (ethanol to medium volume ratio of 0.1%) to prepare a stimulating environment.
Cell proliferation assay
The CCK-8 assay was used to measure cell proliferation according to the instructions provided with the kit (BS350A, Biosharp, China). Briefly, 5000 cells/well were plated in 96-well plates. After the cells adhered, exogenous lipids were introduced at concentrations of 50, 100, and 200 μM. The assays were conducted at 24, 48, 72, and 96 hours. For the EdU cell proliferation analysis (ab222421, Abcam, UK), the cells were incubated with 10 μM EdU after stimulation with 100 μM lipids for 48 hours. The cells were then fixed, permeabilized, and stained with Hoechst, and the number of EdU-positive cells in five random fields was counted under a fluorescence microscope. For the clonogenic assay, cells stably stimulated with lipids and control cells in the logarithmic growth phase were seeded into 6-well plates. After 2 weeks of incubation, the colonies were fixed with 4% paraformaldehyde and stained with crystal violet, after which the number of colonies was determined. Cell cycle analysis was conducted with a cell cycle staining kit in accordance with the manufacturer’s instructions (CCS012, MultiSciences, China). The proportions of stained cells in the G0/G1, S, and G2/M phases were visualised and calculated with a CytoFLEX flow cytometer.
RNA sequencing and transcriptional analysis
The CCA cell line QBC939 was cultured with or without 100 μM LPC for 48 hours. Total RNA was isolated with an RNeasy mini kit (Qiagen, Germany). One microgram of total RNA was used for library preparation. mRNA was isolated with oligo(dT) beads, fragmented, and primed using random primers. cDNA synthesis was followed by adaptor ligation and size selection using DNA Clean Beads. Libraries were amplified by PCR, validated, and sequenced on an Illumina instrument with a 2×150 paired-end configuration.
Cutadapt (v1.9.1) was used to process the data, removing adapters and low-quality bases. HISAT2 (v2.2.1) was used to index and align the clean data to the reference genome. HTSeq (v0.6.1) was used to estimate gene expression levels. Differential expression analysis was conducted using the DESeq2 package, with a significance threshold of a p value ≤ 0.05 and a log2FC > 1. Gene Ontology (GO) terms were identified using GOSeq (v1.34.1), and KEGG pathway enrichment was performed using in-house scripts.
Statistical analysis
Count data are expressed as frequencies (percentages), normally distributed data are expressed as the mean ± standard deviation (SD), and skewed data are expressed as the median (25th and 75th percentiles). The chi-square test (count data), independent samples t test (normally distributed), or Mann‒Whitney U test (skewed distribution) was used for univariate analysis. The choice between parametric and nonparametric methods was determined by the data distribution and sample size. If the data lacked a normal distribution, logarithmic transformation was performed.
Results
Study populations and patient characteristics
This observational analysis included 94 patients with CCA and 147 benign controls. The tumour group included 19 intrahepatic CCA (iCCA) patients, 30 perihilar CCA (pCCA) patients, and 45 distal CCA (dCCA) patients. The benign control group included 68 patients with simple biliary stones, 60 patients with RPC, 11 patients with PSC, 4 patients with PBC, and 4 patients with benign biliary tumours (2 patients with adenomas, 1 patient with a papillary mucinous tumour, and 1 patient with polypoid hyperplasia). Benign controls were further subdivided into those with IBTs and those with NIBTs.
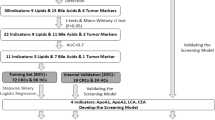
These patients were randomised into 3 cohorts: Cohort 1 consisted of 68 benign controls (34 IBT patients and 34 NIBT patients) and 32 CCA patients (9 iCCA patients, 10 pCCA patients, and 13 dCCA patients), Cohort 2 included 30 benign controls (14 IBT patients and 16 NIBT patients) and 30 CCA patients (6 iCCA patients, 14 pCCA patients, and 10 dCCA patients), and Cohort 3 included 30 benign controls (27 IBT patients and 22 NIBT patients) and 30 CCA patients (6 iCCA patients, 6 pCCA patients, and 22 dCCA patients). There were no significant differences in sex, age, or body mass index (BMI) between the CCA and control groups in the 3 cohorts. The flowchart of patient recruitment, cohort allocation, data analysis, and diagnostic modelling for this study is shown in Fig. 1. The demographic characteristics of the 241 subjects are shown in Supplementary Table S1.
a Study design workflow: a Study participants were stratified into three independent cohorts. Cohorts 1 and 2 underwent independent untargeted lipidomic analyses for diagnostic model development and cross-validation. Cohort 3 was subsequently analysed using targeted lipid quantification to verify candidate lipids identified via untargeted lipidomics analysis and validate the diagnostic model. b Demographic Characteristics: A multilayer circular chart systematically presents the clinical and demographic information across six hierarchically structured levels. The innermost layer (1) denotes cohort allocation (Cohorts 1–3), followed by group classification (control vs. tumour) in the second layer. The third layer defines the clinical stratification criteria: the control group is subdivided into inflammatory and noninflammatory biliary tract disorders, whereas the tumour group is categorised according to TNM stage (I-IV). The fourth layer specifies disease subtypes. The control group includes simple biliary stones (SBS), recurrent pyogenic cholangitis (RPC), primary sclerosing cholangitis (PSC), primary biliary cholangitis (PBC), and benign biliary tumours. The tumour group was further classified into the following CCA subtypes: intrahepatic (iCCA), perihilar (pCCA), and distal (dCCA). The fifth and sixth layers summarise sex distribution and age stratification, respectively.
Bile lipid composition
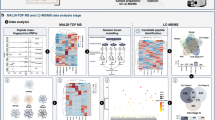
In Cohort 1 and Cohort 2, untargeted lipidomic profiling employing IM‒MS was conducted to characterise the bile lipidome comprehensively in the first two cohorts. Lipid extraction, library construction (incorporating isomer-resolved databases), and LC‒IM‒MS analysis were performed independently for each cohort to assess analytical reproducibility. The total ion chromatogram (TIC) plots and PCA plots of the QC samples in the two cohorts showed good concordance, indicating that the assays in both batches of this study had strong performance stability (Supplementary Fig. S1, 2). In +ESI mode, a total of 202 lipid features were identified in Cohort 1. The lipid with the highest percentage was lysophosphatidylcholines (LPCs), with 74 profiles (36.43%), followed by ceramides (Cers, 17.82%), acylcarnitines (ACars, 10.89%), and triglycerides (TGs, 10.40%). A total of 180 lipid features were identified in Cohort 2, with the predominant species being LPCs (43.89%) and TGs (16.67%) (Fig. S3a, c). Negative electrospray ionisation mode (-ESI) identified 243 lipids in Cohort 1 and 196 lipids in Cohort 2, with Cers and phosphatidylcholines (PCs) being the most prevalent in both cohorts (Fig. S3b, d). The RT, CCS, and mass-to-charge ratio (m/z) for the corresponding lipid features are in the Supplementary Materials (Supplementary Tables S2-5).
Bile lipidomic profiling of CCA patients and benign controls
To compare the differences in lipidomic profiles between CCA patients and benign controls, we performed differential lipid analysis, lipid profile clustering and chemometric analysis on the results from both cohorts analysed in positive and negative ion modes.
In positive ion mode, we identified 82 differential features between tumours and controls in Cohort 1, with 22 upregulated and 60 downregulated features (FDR < 0.05). In Cohort 2, we identified 109 differential features, 35 of which were upregulated and 74 of which were downregulated (Fig. S4). The types of lipids that were upregulated and downregulated in both cohorts were essentially the same, with the most significantly upregulated lipid type being TGs and the most significantly downregulated lipid type being LPCs. We then performed hierarchical clustering by means of Ward’s method on the basis of the top 25 most significantly differential lipids (Wilcoxon rank-sum test). The CCA and control samples in both cohorts exhibited hierarchical clustering, with a more pronounced clustering effect observed in Cohort 2 (Fig. S5). PCA, PLS–DA and OPLS–DA score plots revealed partial discrimination between tumours and controls, with more pronounced separation observed in Cohort 2, especially in positive ion mode (Figs. S6, 7).
Similarly, in negative ion mode, the two cohorts had similar lipidomic differences. The differential lipids detected were predominantly downregulated, with the major species being LPCs, fatty acids (FAs), and Cers (Figs. S4–7).
To evaluate the diagnostic accuracy of our method on the basis of lipidomic profiles, we used a random forest classification algorithm because of its robustness in high-dimensional data analysis. In both positive and negative ion modes, the algorithm identified similar features that differentiated CCA patients from benign controls. The out-of-bag error rates for Cohort 1 and Cohort 2 were 0.18 and 0.117, respectively, on the basis of the positive ion mode data and 0.232 and 0.1, respectively, on the basis of the negative ion mode data (Fig. S8).
Altered bile lipidome of CCA patients
To further delineate the metabolic alterations of distinct lipid species in CCA patients, we integrated lipid profiles and their classifications obtained from both positive and negative ion modes within the same cohort. Upon data integration, PCA score plots demonstrated enhanced discrimination between the CCA and control groups (Fig. 2a, b). On the basis of the top 50 most significantly differential lipid features (Wilcoxon rank-sum test), hierarchical clustering was performed to generate lipid abundance heatmaps. Among the top 50 differential lipids, a predominance of features derived from the positive ion mode was observed (Fig. 2c, d).
a, b Principal component analysis (PCA) score plots showing separation between CCA patients and benign controls in cohorts 1 and 2. c, d Hierarchical clustering heatmaps displaying the top 50 most significantly differentially abundant annotated lipids between the control and tumour groups. Sample information, including group, sex, age, and tumour stage, is annotated on the columns, whereas lipid types and ESI models are annotated on the rows for each lipid feature.
By combining these data with the metabolic relationships between different lipid species, we created a visualisation network to highlight the most dysregulated lipid species in CCA (Fig. 3). In the lipidomic profile of CCA patients, multiple lipids appeared to be downregulated, with LPCs being the most significantly downregulated. Conversely, TGs were the most significantly upregulated species. This difference between LPCs and TGs was even more pronounced in Cohort 2 (Fig. 3b). In addition, a variety of Cers were also downregulated in the CCA group, with this downregulation being more pronounced in Cohort 1 (Fig. 3a). The altered bile lipidome of CCA patients suggests the possible accumulation of TGs and depletion of lipids such as LPCs, Cers, and FAs in the CCA tumour microenvironment.
Lipid metabolism involves a complex network of interrelated pathways in which different lipid classes undergo conversions through specific enzymatic reactions. The present diagram illustrates the dysregulated lipid metabolism observed in cholangiocarcinoma (CCA) via the mapping of these metabolic pathways. Graphs showing lipidomic pathways clustered into individual lipid classes in Cohort 1 (a) and Cohort 2 (b). Differential features between two groups were identified by the p value adjusted by the false discovery rate (FDR) and fold change (FC) significance thresholds (FDR-adjusted p < 0.05, Wilcoxon rank-sum test, Benjamini‒Hochberg (BH) procedure; |log2(FC)| ≥ 1). Circles represent the detected lipid species, where the circle size expresses the significance according to -log10(FDR), whereas the colour darkness defines the degree of up/downregulation (red/blue) according to the log2(FC). The most discriminating lipids are annotated.
Effects of disease status on the lipidomic characterisation of CCA
On the basis of the integrated data from the positive and negative ion modes, we analysed the impact of different tumour locations on the lipidome of CCA. Regardless of whether the tumour was iCCA, pCCA, or dCCA, consistent lipidomic alterations were observed, indicating that tumour location is not a decisive factor for lipid changes in bile (Fig. S9). We further stratified tumour patients into early (stage I/II) and intermediate/advanced (stage III/IV) stages. The differences between CCA patients and benign controls appeared to be associated with tumour stage, as the significantly differential lipids exhibited more pronounced changes in the intermediate/advanced group. However, the distinctions between benign and malignant samples remained significant overall. Both the PCA and PLS–DA plots clearly distinguished between the benign and malignant samples, while a high degree of overlap was observed between the early and intermediate/advanced tumour stages. In the PLS–DA score plot, only a few early-stage samples overlapped with the elliptical boundary of the benign control group (Figs. 4a–d, S10).
a–d compare early-stage (I/II) and intermediate/advanced-stage (III/IV) CCA with benign controls in Cohorts 1 (a, b) and 2 (c, d). e–h contrast the inflammatory biliary tract disease (IBT) and noninflammatory biliary tract disease (NIBT) groups with the CCA patients in Cohorts 1 (e, f) and 2 (g, h). The results are visualised through two complementary analyses: (1) partial least squares–discriminant analysis (PLS–DA) score plots, illustrating group separation on the basis of the top two latent components, and (2) lipid clustering heatmaps displaying hierarchical relationships and abundance patterns of the 25 most statistically significant differential lipids. Keywords: C Benign controls, I/II early-stage CCA (TNM stages I-II), III/IV intermediate/advanced-stage CCA (TNM stages III-IV), IBT inflammatory biliary tract disease, NIBT noninflammatory biliary tract disease.
Considering that IBTs are significant risk factors and crucial differential diagnoses for CCA, we similarly analysed the lipidomic differences among IBTs, NIBTs, and CCA. Consistent with the differences observed between tumour location and tumour stage subgroups, the primary distinction in bile lipidomics lies in the malignant versus benign characteristics. However, a trend of transition from IBTs to CCA was observed. In the PLS–DA plots for both cohorts, IBTs are positioned at the junction of NIBTs and CCA. The concentration differences of differential lipids between IBTs and CCA are not as pronounced as those between NIBTs and CCA. These findings suggest that bile lipidomic profiles may be influenced by disease progression and malignancy status (Fig. 4e–h, Fig. S10).
Effects of demographic and clinical indicators on the lipidomic characteristics of CCA
Previous studies in lipidomics have highlighted the impact of sex differences on study subgroups, suggesting that sex should be an important consideration in lipidomic analyses [29, 37, 38]. To investigate the effect of sex on the benign/malignant classification, we performed OPLS–DA with sex stratification. The results indicated that the sex stratification model improved the accuracy of correctly assigning samples to the benign and malignant categories (Fig. S11). Specifically, in Cohort 1, the discrimination efficiency between benign cases and CCA cases was better for both males (Q2 = 0.498) and females (Q2 = 0.512) after sex stratification than before sex stratification (Q2 = 0.394). In Cohort 2, the discriminatory effect in the female group was greater (Q2 = 0.82 vs. 0.66) than that in the male group, whose Q2 was lower after sex stratification than before (Q2 = 0.61), although it remained above 0.6. The visualisations of CCA metabolic alterations after sex stratification in the two cohorts are shown in the Supplementary Materials (Fig. S12-13).
Next, to further analyse their variations across the different age groups, BMI categories, liver function (Child‒Pugh grade), and performance status (ECOG-PS) subgroups, we selected the lipids with the highest levels of significance among the three lipid classes. Significant differences in the relative concentrations of these lipids were consistently noted between the control and tumour groups, and these differences were independent of the other covariates (Fig. S14).
Weighted lipid correlation network analysis (WLCNA)
Because lipid extraction and analysis were independently conducted in the two cohorts, we selected 91 lipid features identified in both cohorts in positive ion mode and 124 lipid features identified in both cohorts in negative ion mode as candidate lipids for WLCNA and diagnostic modelling. A total of 215 lipids from the two cohorts were used to identify 10 modules (soft threshold >9, scale-free topological fit index >0.8; Fig. 5a–c, Fig. S15). Analysis of module‒trait relationships revealed that 9 modules, most notably the turquoise module, were inhibited in CCA, and a significant increase in lipids was associated with the gold module (Fig. S15). We analysed the relationships between lipid coexpression modules and clinical traits, including the age-adjusted Charlson Comorbidity Index (aCCI), BMI, Child‒Pugh grade, ECOG score, and various blood test indicators, and the results revealed significant correlations (Fig. 5d).
a Network heatmap based on the topological overlap matrix (TOM) showing hierarchical clustering of lipids, where bile lipids are assigned to 10 distinct modules, each with a unique colour. Lipids not assigned to any coexpression pattern are marked in gray. b, c Clustering relationships and correlation heatmap of the 10 modules. d Heatmap displaying module‒clinical trait correlations. The colour scale ranges from blue to red, indicating Spearman’s correlation, while the circle size represents statistical significance. e, f Lipid‒module correlations (x-axis) and lipid‒clinical trait correlations (y-axis) were assessed to identify potential key lipids. Lipids with the highest correlation to both the module and clinical traits are labelled, along with F values, p values, and 95% confidence intervals from simple slope tests. g, h Coexpression networks of lipids within the gold and turquoise modules, where circle size represents betweenness centrality, colour intensity indicates degree, and label font size reflects closeness centrality.
Notably, the gold and turquoise modules had strong associations with several clinical phenotypes, with correlation coefficients of 0.454 (p < 0.001) and -0.497 (p < 0.001) for aCCI and 0.433 (p < 0.001) and -0.477 (p < 0.001) for total bilirubin (TBIL), respectively (Fig. 5d). Lipids in the gold and turquoise modules were further analysed to assess lipid-module correlations and lipid-clinical trait correlations. This analysis aimed to identify potential key lipids related to six clinical indicators, including the aCCI, Child‒Pugh grade, TBIL, alkaline phosphatase (ALP), phospholipid (PLIP), and CA19-9. The gold module showed positive correlations with these metrics, with all lipids in the module being triglycerides (TGs) and TG 50:1 [M + NH4]+ identified as the key lipid. In contrast, the turquoise module exhibited negative correlations, consisting predominantly of LPCs and several lysophosphatidylethanolamines (LPEs), with LPC 18:3 [M + H]+ identified as the key lipid in the turquoise module (Fig. 5e, f). Finally, the coexpression network of lipids inside the gold and turquoise modules was mapped (Fig. 5g, h).
Together, these results highlight a striking association between clinical traits and bile lipid species/classes in the disease characteristics of CCA, suggesting the potential value of bile lipid profiling for clinical diagnosis.
Establishing, tuning and validating the classification model with differential bile lipids
As illustrated in the workflow in Fig. 6a, we conducted diagnostic modelling using data from both Cohort 1 and Cohort 2. Variable selection and parameter regularisation were performed by penalising likelihood maximisation via parameter λ, and the optimal combinations of bile lipid markers for diagnosing CCA were identified through the LASSO binary logistic regression model. The developed diagnostic models were then validated in the alternate cohort, with the best diagnostic model chosen on the basis of cross-validation results across the two cohorts. Given that the previous section indicated improved accuracy in distinguishing between benign and malignant cases after sex stratification, we simultaneously conducted modelling and validation processes in male and female patients.
a The establishment workflow of the CCA classifier. Diagnostic modelling was conducted using data from Cohort 1 and Cohort 2. Optimal combinations of bile lipid markers were identified through the LASSO binary logistic regression model. These diagnostic models were then validated in another cohort. The best diagnostic model was selected on the basis of cross-validation results across the two cohorts. Additionally, the modelling and validation process was performed with sex stratification to account for potential sex-specific differences in lipid profiles. b–d Diagnostic performance of the serum markers for CCA. The figure highlights the optimal cutoff points for each marker in this study cohort, along with the sensitivity (SEN), specificity (SPE), and area under the curve (AUC) values, including the 95% confidence interval (CI) for the AUC. e-l Diagnostic performance was assessed by modelling with the positive ion mode datasets of Cohort 1 (e, f) and Cohort 2 (g, h) and the negative ion mode datasets of Cohort 1 (i, j) and Cohort 2 (k, l). The other cohort served as a validation set. This assessment was conducted both with and without sex stratification. Waterfall plots for predicting the probability of CCA using sex-neutral (m), male (n), and female (o) optimal diagnostic models; the optimal cutoff points, SEN, SPE and AUC are labelled. Each stream represents the predicted probability of disease according to the classifier, and the colours indicate the gold standard of diagnosis: red represents CCA, and blue represents benign cases. p, q SEN, SPE and AUC for the BileScreen, BileMet and the bile lipid classifier (BileLipid).
We also assessed the diagnostic efficacy of conventional serum hepatobiliary tumour biomarkers as clinical benchmarks in all 241 patients from the three cohorts, with their optimal cutoff values determined based on Youden’s index. Among these biomarkers, carbohydrate antigen 19-9 (CA19-9) demonstrated the highest discriminative power for CCA (AUC = 0.778), followed by carcinoembryonic antigen (CEA; AUC = 0.716) and carbohydrate antigen 125 (CA125; AUC = 0.707) (Fig. 6b–d).
When modelling was performed with the Cohort 1 positive ion mode dataset without differentiating between sexes, LASSO regression identified 6 variables at the optimal λ (Fig. S16). Using the regression coefficients of these 6 lipid features, a predictive model was constructed for ROC analysis, yielding an AUC of 0.943 in the training set (Cohort 1) and 0.956 in the validation set (Cohort 2) (Fig. 6e). After stratification by sex, 3 and 7 lipid features were identified, and sex-related models were constructed for the male and female sets, respectively (Fig. S17). The male diagnostic model achieved AUCs of 0.950 and 0.942 in the training and validation sets, respectively, whereas the female diagnostic model achieved AUCs of 0.955 and 0.975 in the training and validation sets, respectively (Fig. 6f).
Subsequently, diagnostic modelling and validation for both sex-agnostic and sex-stratified groups were performed on the positive ion mode dataset of Cohort 2 (Fig. 6g, h), the negative ion mode dataset of Cohort 1 (Fig. 6i, j) and the negative ion mode dataset of Cohort 2 (Fig. 6k, l). The results demonstrated that the best diagnostic efficacy was achieved by modelling with the positive ion mode dataset of Cohort 1, both with and without sex stratification. Using this model, we calculated the probability of CCA incidence for all samples. The optimal diagnostic model of bile lipids achieved an AUC of 0.948 without sex stratification, 0.948 for males and 0.966 for females (Fig. 6m–o).
Diagnostic performance of the optimal bile lipid classifier for CCA
Despite modest performance gains in sex-stratified models, the sex-agnostic bile lipid classifier was selected for clinical validation given its cross-sex generalizability. We compared this bile lipid classifier with other high-quality bile marker studies to contextualise its efficacy. One such study, known as BileScreen, analysed 23 gene mutations and 44 genes with methylations identified from bile samples. BileScreen developed a multilevel biomarker-based malignancy detection model and validated its diagnostic ability for biliopancreatic malignancies in a cohort of 338 patients [20]. Another study, called BileMet, focused on metabolomic markers from bile samples. BileMet developed a metabolic marker-based biliary tract cancer detection model and validated its diagnostic ability in 336 patients [25].
Compared with the classifiers from BileScreen and BileMet, the bile lipid classifier in this study demonstrated comparable diagnostic performance, with certain metrics outperforming the aforementioned models. In the training set, the bile lipid classifier achieved an AUC of 0.943, compared with 0.953 for BileScreen and 0.891 for BileMet. In the validation set, the bile lipid classifier achieved an AUC of 0.956, whereas BileScreen and BileMet achieved AUCs of 0.950 and 0.912, respectively. Furthermore, the sensitivity of the bile lipid classifier was better than that of BileMet and BileScreen, while its specificity was similar to that of BileMet but lower than that of BileScreen (Fig. 6p, q).
In addition, we conducted multiple evaluations of the bile lipid classifier using clinical DCA, CIC analysis, and calibration analyses (Fig. S18). The DCA results demonstrated that the net benefit of clinical decision-making for all three models was superior to that of CA19-9. The CIC analysis indicated that good overlap between the model predictions and real cases was achieved after a risk threshold of 0.4. In some models and datasets, overlap was observed at a risk threshold of 0.2 or even lower, highlighting the model’s value for both high-risk patients and early diagnosis. The calibration curves also showed good agreement, with the Hosmer–Lemeshow test p values for all the models being greater than 0.05, indicating that there was no statistically significant difference between the predicted probability and the true value.
We further evaluated the ability of the bile lipid classifier to identify CCA in patients with normal CA19-9 levels. In this study, 60 patients, including 9 patients with CCA, had CA19-9 levels below the upper limit of the normal range (37.0 U/mL). The bile lipid classifier demonstrated strong discriminative performance for CCA in this population, achieving an AUC of 0.954, a sensitivity of 1, and a specificity of 0.882. The optimal CA19-9 cutoff, determined from the pooled dataset of 241 patients across the three cohorts, was 68.17 U/mL—higher than the conventional clinical threshold of 37 U/mL (Fig. 6d). This observation is consistent with previous studies reporting elevated thresholds associated with improved diagnostic performance [39]. On the basis of this threshold, we performed stratified diagnostic evaluations of the patients. For those with CA19-9 levels less than 68.17 U/mL, the bile lipid classifier achieved an AUC of 0.931, with a sensitivity of 1 and a specificity of 0.797 (Fig. S19).
Independent validation and recalibration of the BileLipid classifier through targeted lipid quantification
The diagnostic evaluation confirmed the clinical translational value of the bile lipid classifier, termed BileLipid. To increase data robustness and analytical precision, we conducted LC‒MS/MS–MRM-based targeted quantification of 6 candidate lipids in the biomarker panel of BileLipid in Cohort 3, which included Cer d40:2, LPC 18:0, LPC 18:2, LPC 20:3, TG 50:1, and TG 54:4 (Table S6). Lipid abundance profiles mirrored trends in previous cohorts: Cer and LPC species were downregulated, whereas TG levels were elevated (Fig. 7a, b). The diagnostic performance of individual lipids for CCA was evaluated across all cohorts. LPC 18:2 exhibited the highest single-metabolite discriminatory capacity, with AUC values of 0.817, 0.960, and 0.858 in Cohorts 1, 2, and 3, respectively (Fig. S20).
a Heatmap of the normalised abundance of the 6 lipid features across the 3 cohorts. b Intergroup comparison of the relative concentrations of the 6 lipids in Cohort 3 using targeted lipid quantification (Wilcoxon rank-sum test). c Diagnostic performance (ROC curves) of the original BileLipid classifier and the recalibrated adj.BileLipid model (refitted using Cohort 3 quantification data). Waterfall plots displaying predicted CCA probabilities for Cohort 3 patients using (d) BileLipid and (e) adj.BileLipid classifiers.
We subsequently evaluated the diagnostic performance of BileLipid in Cohort 3, achieving an AUC of 0.828, a sensitivity of 62.5%, and a specificity of 91.8% (Fig. 7c, d). Given that BileLipid was derived from untargeted lipidomics data, we recalibrated the lipid weighting coefficients using targeted quantification results from Cohort 3. This optimised model, adj.BileLipid (Fig. S21), showed enhanced diagnostic performance in Cohort 3, with an AUC of 0.897, sensitivity of 75.0%, and specificity of 91.8% (Fig. 7c, e).
Application of the BileLipid classifier in real-world clinical patients
Four samples from patients with biliary obstruction and similar clinical presentations (aged 63-73) were selected to further illustrate the performance of the BileLipid classifier in predicting CCA. All 4 patients were admitted to the hospital with obstructive jaundice. Imaging revealed dilatation of the upper portion of the common bile duct, with stenosis or nodules in the middle and lower portions of the duct, raising clinical suspicion of malignant bile duct tumours.
Patient 1 had nodules at the distal end of the common bile duct and dilation of the proximal duct. ERCP endoscopy revealed an obstruction in the lower part of the common bile duct, suggesting a possible tumour lesion. Cytological examination of the lower common bile duct revealed a relatively high number of bile duct epithelial cells. Most of these cells were well differentiated, although a small number showed slight disorganisation and mild heterogeneous hyperplasia (Fig. 8a). The patient did not undergo surgery or antitumor therapy; instead, biliary stenting was performed to alleviate jaundice symptoms. Three years after discharge, the patient remained alive and well with no signs of malignancy, indicating a benign biliary obstruction.
a Imaging of Patient 1 revealed nodules at the distal end of the common bile duct and dilation of the proximal duct. Cytological examination of the lower common bile duct revealed a relatively high number of bile duct epithelial cells. Most of these cells are well differentiated, although a small number exhibit slight disorganisation and mild heterogeneous hyperplasia. b Patient 2 presented with dilated intrahepatic bile ducts and localised stenosis at the lower end of the common bile duct. Postoperative pathology revealed localised intestinal epithelial hyperplasia with inflammatory cell infiltration in the biliary epithelium and localised pancreatic follicular atrophy. c, d Patients 3 and 4 exhibited similar imaging findings of marked dilatation of the intra- and extrahepatic bile ducts and stenosis of the middle and lower portions of the common bile duct. Postoperative pathology confirmed adenocarcinoma of the bile ducts. e Sex, age, CA19-9, and the bile lipid classifier-predicted probability of CCA in 4 patients.
Patient 2 presented with dilated intrahepatic bile ducts and localised stenosis at the lower end of the common bile duct. ERCP endoscopy revealed an abnormal biliopancreatic duct confluence combined with lower bile duct stenosis. Owing to the severe progression of jaundice, the patient underwent pancreaticoduodenal surgery. Postoperative pathology revealed localised intestinal epithelial hyperplasia with inflammatory cell infiltration in the biliary epithelium and localised pancreatic follicular atrophy (Fig. 8b), confirming a diagnosis of benign biliary obstruction.
Patients 3 and 4, who had similar imaging findings of marked dilatation of the intra- and extrahepatic bile ducts and stenosis of the middle and lower portions of the common bile duct, underwent pancreaticoduodenal surgery. Postoperative pathological examinations confirmed adenocarcinoma of the bile ducts (Fig. 8c, d). These 4 patients had highly similar clinical presentations and low sensitivity to CA19-9 (17.55–21.62 U/ml, all below the upper limit of the normal range of 37.00 U/ml). CCA was suspected prior to treatment, and the benign or malignant nature of the conditions was confirmed only through subsequent clinical findings.
According to the predictions of the BileLipid classifier in this study, the predicted probabilities of CCA in Patients 1 and 2 were less than 0.05, whereas the predicted probabilities in Patients 3 and 4 were both greater than 0.8 (Fig. 8e). This consistency with the final clinical results demonstrates the model’s strong discriminatory performance and highlights the potential value of bile lipid biomarkers in the differential diagnosis of CCA, thereby aiding in the identification and management of patients with biliary obstruction in a clinical setting.
LPC is protective against CCA in vitro
The results of the bile lipidomic differential analysis revealed that LPC levels were significantly lower in the bile samples of CCA patients than in those of patients with benign disease and that LPC was the lipid with the most significant difference among all lipid types (Figs. 2, 3). The turquoise module, identified by WLCNA as having significant clinical relevance, was also predominantly enriched with the LPC class of lipids (Fig. 5). To understand the functional importance of these significantly depleted lipids in CCA, we examined the effects of LPC on the proliferation of the CCA cell lines (TFK1, QBC939) and normal biliary epithelial cell (HiBEC). The cells were stimulated with an LPC mixture consisting of 69% C16:0 chains, 27% C18:0 chains, and 3% C18:1 chains. We found that LPC significantly inhibited cell proliferation in a dose-dependent manner (Fig. 9a). This growth-inhibitory effect was confirmed by colony formation (Fig. 9b) and EdU (Fig. 9c) assays in the QBC939 and TFK1 cell lines. Moreover, LPC delayed the G1/S transition in these two cell lines (Fig. 9d). These findings indicate that LPC suppresses cell proliferation and delays the G1/S transition in CCA cells, suggesting its role as a protective factor against CCA.
a LPC decreased the viability of CCA cell lines (TFK1, QBC939) and normal biliary epithelial cell (HiBEC), as determined by the CCK-8 assay. b LPC significantly suppressed the colony formation of QBC939 and TFK1 cells. c EdU assays indicated that the proliferation of QBC939 and TFK1 cells was suppressed after 48 h of culture with LPC. d Flow cytometry analyses indicated that treatment with LPC delayed the G1/S transition. e The GO enrichment results revealed that multiple lipid metabolism pathways were altered after LPC treatment. *p < 0.05; **p < 0.01; ***p < 0.001. ns: not significant.
We then performed RNA sequencing analysis on LPC-treated CCA cells to explore potential mechanisms of action. The GO enrichment results revealed that multiple lipid metabolism pathways were altered after LPC treatment, with lipid catabolic processes, small molecule catabolic processes, and fatty acid metabolic processes being the most significantly changed (Fig. 9e). KEGG enrichment analysis revealed that the neuroactive ligand‒receptor interaction, MAPK signalling, and multiple lipid metabolism pathways were the most significantly altered cellular processes in CCA cells following LPC treatment (Fig. S22). Among the genes involved in these pathways, PDK4 (FC = 2.3, p = 7.20E−16), LYPD6 (FC = 2.7, p = 3.43E−08), and FADS2 (FC = 2.3, p = 1.55E−05) presented the most significant differences. Both PDK4 and FADS2 are involved in fatty acid metabolism, which has been widely reported to influence CCA development. These findings suggest that LPC may affect CCA cell proliferation by regulating fatty acid metabolism.
Discussion
In this study, we developed an efficient bile lipidomic assay using IM–MS to differentiate the lipidomic profiles of bile samples from patients with CCA and those with benign bile duct diseases. We conducted difference analysis, machine learning modelling, and diagnostic efficacy assessment to evaluate the potential of this bile lipidomic assay in clinical applications. This study established and validated a new bile lipid classifier, BileLipid, which consists of six lipids. The classifier achieved AUCs of 0.975 and 0.978 for the CCA in the training and validation sets, respectively. This classifier was then validated in a third cohort via LC‒MS/MS, and the six lipids were further targeted for quantitative analysis with MRM mode. Furthermore, we investigated the function of the downregulated lipid LPC, and in vitro experiments confirmed the inhibitory effect of LPC on the proliferation of CCA cell lines. This work not only broadens the application field of bile liquid biopsy technology but also provides new insights for clinical translation.
The importance of bile as a characteristic body fluid in diagnosing CCA and differentiating benign from malignant biliary strictures is gradually being emphasised [22]. Several studies have validated the unique advantages of bile samples in diagnosing biliopancreatic malignancies [20, 22, 23, 25, 40, 41]. In addition to genomics, transcriptomics, and proteomics, metabolomics—which assesses the end products of gene expression and environmental influences—has emerged as a highly promising field in clinical diagnostics [25, 42,43,44]. Notably, existing studies have focused primarily on ctDNA in bile because challenges such as the reproducibility and comprehensiveness of small molecule analysis in complex body fluids limit the acceptance of metabolite analysis in liquid biopsy. The BileMet study compared the performance of metabolic fingerprints in bile and plasma for the diagnosis of biliary tract cancers and revealed that bile metabolic diagnostic markers had better diagnostic performance than did plasma metabolic markers (AUC = 0.891 vs. 0.717) [25]. BileMet further validated the diagnostic advantages of bile in the field of metabolic markers, and our results suggest that lipid metabolites in bile have a diagnostic value comparable to that of ctDNA and metabolomics.
We observed significant downregulation of several lipids in the CCA bile samples. This overall suppression of lipid metabolism in cancer body fluids has also been reported in other tumours [24, 28, 29, 31, 32]. In a study by Wang G et al., changes in lipid profiles in the plasma of lung cancer patients were analysed. They reported that the downregulated lipids in lung cancer patients were mainly LPCs, which aligns with the findings of the present study. However, Wang G et al. reported that the upregulated lipids in the plasma of lung cancer patients were primarily PCs. Their diagnostic panel included 3 LPCs, 5 PCs, and 1 TG. In their panel, the LPCs were all downregulated, whereas the PCs and the TG were upregulated in lung cancer patients. All these lipids are involved in glycerophospholipid and glycerolipid metabolism [28]. Similarly, Wolrab D et al. reported the downregulation of LPCs and the upregulation of TGs in pancreatic cancer, which is consistent with our findings in CCA. This similarity may be attributed to the close anatomical proximity of the origin sites of CCA and pancreatic cancer. Additionally, they reported that the most significantly downregulated lipid in pancreatic cancer was sphingomyelin (SM) [29]. In contrast, our study revealed that there was little difference in SM between CCA patients and benign controls. Our findings also revealed a decrease in free fatty acid levels in bile, which is consistent with our previous results [33].
Sharma et al. analysed the lipidome profile of bile from gallbladder cancer patients and gallbladder stones. Although they similarly reported lipid inhibition in cancer bile, the predominant lipid profiles in bile detected in that study were not exactly identical to those reported in the present study. In Sharma’s study, PC and phosphatidylethanolamine (PE) were the predominant lipids in bile, whereas in gallbladder cancer, PC, PE, phosphatidylserine (PS), and phosphatidylinositol (PI) were the main lipid species whose expression was downregulated [24]. The predominant lipids in bile identified in this study were PC, LPC, and Cer, with LPC and Cer being the main lipid species whose expression was downregulated. These differences highlight the heterogeneity of lipid metabolism in CCA compared with that in gallbladder cancer, and this heterogeneity may also be influenced by other factors, such as sampling methods and assay platforms. In the study by Sharma et al., bile was obtained primarily via aspiration from the gallbladder or bile ducts via a 24-G needle during surgical interventions. In contrast, in our study, bile samples were collected during ERCP examinations. This method is less invasive and traumatic than surgery, allowing bile to be captured under conditions that are closer to the physiological environment prior to clinical intervention. Consequently, this ensures that the sample retains information that is more representative of the original microenvironmental ecology.
The differences in bile lipidomic profiles between patients with CCA and those in the benign control group suggest the potential for early and accurate diagnosis of CCA. However, translating this method into clinical practice requires consideration of real-world clinical conditions. Therefore, we comprehensively evaluated the clinical information of the enroled patients, including sex, age, BMI, liver function, comorbidities, tumour stage, and other relevant indicators, before diagnostic modelling. This approach aims to minimise potential biases in laboratory testing and sample grouping. After categorising CCA patients into early- and intermediate/advanced-stage groups, we observed distinct differences between benign and malignant cases that were independent of tumour stage. Although some lipids showed more pronounced concentration changes in intermediate/advanced-stage tumours, the lipid profiles of early- and intermediate/advanced-stage tumours were highly similar and markedly different from those of benign diseases. These findings suggest that bile lipidome analysis could be instrumental in the early diagnosis of CCA. Early detection through this method allows timely therapy and intervention, potentially enhancing the survival benefits for patients.
Notably, the bile lipidomic profile of patients with IBTs more closely resembles that observed in CCA. In contrast, the differences between NIBTs and CCA were more pronounced than those between IBTs and CCA. This finding underscores the clinical importance of distinguishing between IBTs and CCA, as it is often more challenging to differentiate between these two conditions than between CCA and NIBTs such as simple biliary stones. However, while the results of this study suggest that the bile lipidomic profiles of IBTs and CCA share more similarities than the bile lipidomic profiles of NIBTs and CCA, the differences between IBTs and CCA are still significant enough to allow for differentiation. This finding highlights the potential advantage of using bile lipidomic profiling in the diagnostic process for CCA.
Furthermore, differences in specific lipids across age, BMI, Child‒Pugh grade, and ECOG-PS persisted between the benign and malignant groups. This persistence suggests that variations in biliary lipid profiles are specific characteristics of CCA rather than effects of demographic or clinical factors.
The integration of biliary lipid groups and clinical phenotypes using WLCNA revealed significant differences in lipid modules between CCA patients and benign controls. Module‒trait relationship analysis revealed key regulatory lipid modules and critical lipids involved in CCA pathogenesis. These findings highlight the associations between lipid species regulation and clinical traits. Notably, significant correlations were observed between specific lipid modules and clinical indicators such as the aCCI, Child‒Pugh grade, hepatic and renal function parameters (bilirubin, albumin, ALP, creatinine, and uric acid), blood lipids (total cholesterol, triglycerides, high-density lipoprotein, and PLIPs), and tumour markers (CA19‒9). This preliminary analysis supports the hypothesis that unique lipid coexpression patterns are linked to specific clinical features.
Sexual asymmetries in metabolic homoeostasis, driven by evolutionary adaptations, result in differential mechanisms for energy storage and expenditure between men and women. These differences, which include variations in antioxidant damage capacity, resistance to starvation, and susceptibility to obesity, significantly influence the structure of lipid metabolism [45]. Wolrab D et al. reported that TGs were upregulated in pancreatic cancer patients. Notably, this upregulation was primarily concentrated in male patients and not evident in female patients, in whom some TGs were even downregulated [29]. The results of the present study similarly demonstrated sex differences in lipid profiles. These differences between benign and malignant samples were more evident when the lipid profiles of male and female patients were compared separately than when they the comparison did not control for sex. Our results revealed that the upregulation of TGs was more pronounced in male CCA patients, whereas in female CCA patients, the downregulation of LPCs was more significant. However, the sex differences in the trend of Cer changes were not consistent across the two cohorts, indicating that the findings related to sex stratification should be interpreted with caution and require further validation (Fig. S13-14). Larger sample sizes and in-depth studies may be needed to further explore the sexual dimorphism of lipid metabolism in CCA.
Although sex-stratified diagnostic models demonstrated superior performance—with the female-specific model achieving a higher AUC than the sex-agnostic 6-feature model and the male-specific model attaining a comparable AUC with only 3 features—suggesting a potential enhancement in diagnostic value through sex stratification, this approach inherently reduces the training data volume and introduces additional confounders. Moreover, lipid features that exhibit consistent importance across sexes may hold broader biological importance. Therefore, we ultimately retained the sex-agnostic bile lipid classifier for subsequent validation and translation. In independent Cohort 3, targeted quantitative analysis was performed on the 6 lipids included in the classifier, further verifying its diagnostic performance and clinical promotability.
We subsequently reviewed the clinical presentation of the enroled patients using the probability of CCA incidence predicted by the bile lipid classifier. We found that this classifier not only plays a valuable role in the diagnosis and early management of CCA but also aids in identifying benign cases, helping to avoid the risk of overintervention. The identification of benign and malignant bile duct stenosis has always been a significant challenge for biliary surgeons. Traditionally, clinicians rely mainly on imaging and endoscopy for diagnosis. The current gold standard for preoperative diagnosis is cytobrush or clamp biopsy under ERCP. Despite its high specificity (90–100%), the sensitivity (62–88%) and accuracy (63–93.7%) of this method are unsatisfactory [46,47,48,49]. This leads to a high rate of resection of benign stenoses [46], highlighting the need for more sensitive diagnostic methods. Patient 2 in this study underwent pancreaticoduodenal surgery due to the rapid progression of obstructive jaundice and suspected biliary tract cancer. If preoperative diagnostic conditions have been clarified, the issue faced by the patient could have been resolved with less invasive manoeuvres, such as biliary drainage or stenting, without resorting to major surgery. This is particularly important for the development of patient management protocols in real-world clinical situations. The bile lipid classifier proposed in this study may be promising for assisting in addressing such issues in clinical practice.
As one of the most significantly differentiated classes of lipids in CCA bile, LPCs were explored in vitro to evaluate their roles and functions. The results of our cell proliferation and cell cycle assays demonstrated that LPC inhibits the proliferation of CCA and normal biliary epithelial cells. Given its significant reduction in CCA bile, we hypothesise that this class of lipids may play a potential protective role against the progression of CCA. The perturbation of acyl chains at the sn-2 site of LPC can affect biofilm fluidity and membrane protein function, potentially serving as the structural basis for its mediated biological functions [50]. On the other hand, LPC can act as a signalling molecule by binding to G protein-coupled receptors and toll-like receptors, exerting biological effects such as inducing oxidative stress, promoting inflammatory responses, and triggering apoptosis [51]. LPC-treated transcriptome analysis in this study revealed that the G protein-coupled receptor signalling pathway was similarly enriched. Several clinical lipidomic studies have reported the downregulation of LPC in cancer plasma, including in lung cancer [28], hepatocellular carcinoma [52], and pancreatic cancer [29]. Zhang L and colleagues reported that LPC can inhibit lung cancer cell proliferation by inducing mitochondrial dysfunction, altering lipid metabolism, increasing fatty acid oxidation, and reprogramming the ACSL5/phosphatidylinositol 3-kinase/extracellular signal-regulated kinase-regulated triacylglycerol–LPC balance [53]. In our study, LPC-treated CCA cells presented significant alterations in lipid metabolism, with notable changes observed in fatty acid metabolic processes. Therefore, targeting LPC and lipid metabolism might be potential therapies for cancer treatment. However, in vitro experimental validation is only a preliminary exploration of the function of LPC in our study, and further studies are needed to verify its specific mechanism of action, biological function, and relationship with clinical phenotypes. Taken together, our findings suggest the protective role of LPC against CCA, which, combined with the findings of previous studies, demonstrates the value of biliary lipid biomarkers in the clinical management of CCA.
The limitations of this study include potential biases and constraints inherent to its design. First, the focus of this study was solely on bile samples without the extension of observations to blood or tissue samples. As a result, fully elucidating the lipidomic characteristics of CCA across different biological matrices is not yet possible. Second, although we employed a three-cohort independent analysis to ensure the reproducibility of our results, the clinical translation of the bile lipid classifier still requires further validation. To do this, larger sample sizes and multicenter studies are needed to continuously improve and adjust the diagnostic model and address the limitations posed by the single-centre sample. Furthermore, comprehensive clinical data, such as treatment responses and survival outcomes in patients with CCA, are lacking, which may warrant further in-depth investigation. Finally, additional studies are necessary to validate the current findings and explore the potential clinical implications of bile lipid profiling in greater detail.
In summary, this study not only identified lipidomic alterations in CCA on the basis of bile samples but also established and validated a bile lipid classifier with high specificity and sensitivity for distinguishing between CCA and benign bile duct diseases. Additionally, we provide an in-depth evaluation and discussion of this diagnostic model in real-world clinical conditions. On the basis of this study, we recommend the use of bile lipid profiling to assist in the clinical assessment of biliary diseases and propose a bile lipid classifier-based disease management strategy (Fig. 10). This classifier has the potential to assist in the clinical management of biliary diseases on the basis of ERCP. These findings highlight the value of bile lipid biomarkers for improving the differential diagnosis and risk assessment of CCA while preventing potential overintervention in patients with benign biliary diseases. The focus of subsequent research may be to develop more sophisticated detection technologies targeting differentially abundant lipids and verify their performance in a larger sample cohort. Additionally, the reprogramming rules of abnormal lipid metabolism in CCA and the specific mechanisms by which differentially abundant lipids, such as LPC, affect the occurrence and development of CCA require in-depth attention.
The diagnosis of benign and malignant biliary diseases is typically conducted through CT, MRCP, blood tumour markers, and ERCP. Inconclusive results can lead to underdiagnosis or delays in the treatment of malignant diseases and unnecessary interventions or overtreatment of benign conditions. Integrating the bile lipid classifier with clinical diagnostic tools may enhance risk assessment and improve the subsequent management of biliary diseases.
Data availability
All source data, experimental details, results and materials are available in the text and supplementary file. Other relevant data can be obtained from the corresponding author.
Change history
10 October 2025
The original online version of this article was revised: In the Conclusions section, the article originally read: “This study not only identified lipidomic alterations in CCA using bile samples but also established and validated a sex-related bile lipid classifier with high specificity and sensitivity for distinguishing between CCA and benign bile duct diseases.” The correct sentence is as follows (“sex-related” removed): “This study not only identified lipidomic alterations in CCA using bile samples but also established and validated a bile lipid classifier with high specificity and sensitivity for distinguishing between CCA and benign bile duct diseases.”
21 October 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41416-025-03237-5
References
Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261–80.
Brindley PJ, Bachini M, Ilyas SI, Khan SA, Loukas A, Sirica AE, et al. Cholangiocarcinoma. Nat Rev Dis Prim. 2021;7:65.
Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557–88.
Vithayathil M, Khan SA. Current epidemiology of cholangiocarcinoma in Western countries. J Hepatol. 2022;77:1690–8.
Valle JW, Kelley RK, Nervi B, Oh D-Y, Zhu AX. Biliary tract cancer. Lancet. 2021;397:428–44.
Cadamuro M, Al-Taee A, Gonda TA. Advanced endoscopy meets molecular diagnosis of cholangiocarcinoma. J Hepatol. 2023;78:1063–72.
Macias RIR, Kornek M, Rodrigues PM, Paiva NA, Castro RE, Urban S, et al. Diagnostic and prognostic biomarkers in cholangiocarcinoma. Liver Int. 2019;39:108–22.
Vogel A, Bridgewater J, Edeline J, Kelley RK, Klümpen HJ, Malka D, et al. Biliary tract cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:127–40.
Navaneethan U, Hasan MK, Lourdusamy V, Njei B, Varadarajulu S, Hawes RH. Single-operator cholangioscopy and targeted biopsies in the diagnosis of indeterminate biliary strictures: a systematic review. Gastrointest Endosc 2015;82:608–14
Khamaysi I, Firman R, Martin P, Vasilyev G, Boyko E, Zussman E. Mechanical perspective on increasing brush cytology yield. ACS Biomater Sci Eng. 2024;10:1743–52.
Karsenti D, Privat J, Charissoux A, Perrot B, Leblanc S, Chaput U, et al. Multicenter randomized trial comparing diagnostic sensitivity and cellular abundance with aggressive versus standard biliary brushing for bile duct stenosis without mass syndrome. Endoscopy. 2023;55:796–803.
Navaneethan U, Njei B, Lourdusamy V, Konjeti R, Vargo JJ, Parsi MA. Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: a systematic review and meta-analysis. Gastrointest Endosc. 2015;81:168–76.
Zhang C, Li Y, Song M, Sun Z, Han X, Ren J, et al. False-negative factors of percutaneous transluminal clamp biopsy for suspected malignant biliary stricture: 194 cases analyzed from a single center. Insights Imaging. 2024;15:108.
Izquierdo-Sanchez L, Lamarca A, La Casta A, Buettner S, Utpatel K, Klümpen H-J, et al. Cholangiocarcinoma landscape in Europe: Diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J Hepatol. 2022;76:1109–21.
Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131.
Markou A, Tzanikou E, Lianidou E. The potential of liquid biopsy in the management of cancer patients. Semin Cancer Biol. 2022;84:69–79.
Tivey A, Church M, Rothwell D, Dive C, Cook N. Circulating tumour DNA - looking beyond the blood. Nat Rev Clin Oncol. 2022;19:600–12.
Chen CK, Liao J, Li MS, Khoo BL. Urine biopsy technologies: Cancer and beyond. Theranostics. 2020;10:7872–88.
Bao Y, Zhang D, Guo H, Ma W. Beyond blood: Advancing the frontiers of liquid biopsy in oncology and personalized medicine. Cancer Sci. 2024;115:1060–72.
He S, Zeng F, Yin H, Wang P, Bai Y, Song Q, et al. Molecular diagnosis of pancreatobiliary tract cancer by detecting mutations and methylation changes in bile samples. EClinicalMedicine. 2023;55:101736.
Wu J, Lin Y, Yang K, Liu X, Wang H, Yu T, et al. Clinical effectiveness of a multitarget urine DNA test for urothelial carcinoma detection: a double-blinded, multicenter, prospective trial. Mol Cancer. 2024;23:57.
Arechederra M, Rullán M, Amat I, Oyon D, Zabalza L, Elizalde M, et al. Next-generation sequencing of bile cell-free DNA for the early detection of patients with malignant biliary strictures. Gut. 2022;71:1141–51.
Vedeld HM, Grimsrud MM, Andresen K, Pharo HD, von Seth E, Karlsen TH, et al. Early and accurate detection of cholangiocarcinoma in patients with primary sclerosing cholangitis by methylation markers in bile. Hepatology. 2021;75:59–73.
Sharma N, Yadav M, Tripathi G, Mathew B, Bindal V, Falari S, et al. Bile multi‐omics analysis classifies lipid species and microbial peptides predictive of carcinoma of the gallbladder. Hepatology. 2022;76:920–35.
Yang S, Fu J, Qin W, Wang R, Gu M, Huang Y, et al. Bile metabolic fingerprints distinguish biliary tract cancer from benign biliary diseases. Hepatology. 2025;81:476–91
Jin H-R, Wang J, Wang Z-J, Xi M-J, Xia B-H, Deng K, et al. Lipid metabolic reprogramming in tumor microenvironment: from mechanisms to therapeutics. J Hematol Oncol 2023;16:103
Jeon YG, Kim YY, Lee G, Kim JB. Physiological and pathological roles of lipogenesis. Nat Metab. 2023;5:735–59.
Wang G, Qiu M, Xing X, Zhou J, Yao H, Li M, et al. Lung cancer scRNA-seq and lipidomics reveal aberrant lipid metabolism for early-stage diagnosis. Sci Transl Med. 2022;14:eabk2756.
Wolrab D, Jirásko R, Cífková E, Höring M, Mei D, Chocholoušková M, et al. Lipidomic profiling of human serum enables detection of pancreatic cancer. Nat Commun. 2022;13:124.
Ecker J, Benedetti E, Kindt ASD, Höring M, Perl M, Machmüller AC, et al. The colorectal cancer lipidome: identification of a robust tumor-specific lipid species signature. Gastroenterology 2021;161:910–23
Liu Z-C, Wu W-H, Huang S, Li Z-W, Li X, Shui G-H, et al. Plasma lipids signify the progression of precancerous gastric lesions to gastric cancer: a prospective targeted lipidomics study. Theranostics. 2022;12:4671–83.
Yan X, Zhao W, Wei J, Yao Y, Sun G, Wang L, et al. A serum lipidomics study for the identification of specific biomarkers for endometrial polyps to distinguish them from endometrial cancer or hyperplasia. Int J Cancer. 2022;150:1549–59.
Liu FS, Liu YY, Hao XY, Liu B, Yan XY, Li AL, et al. Altered bile metabolome and its diagnostic potential for biliopancreatic malignancies. Clin Chim Acta. 2024;554:117777.
Benson AB, D’Angelica MI, Abrams T, Abbott DE, Ahmed A, Anaya DA, et al. NCCN Guidelines® Insights: Biliary Tract Cancers, Version 2.2023. J Natl Compr Canc Netw. 2023;21:694–704.
Téllez L, Payancé A, Tjwa E, Del Cerro MJ, Idorn L, Ovroutski S, et al. EASL-ERN position paper on liver involvement in patients with Fontan-type circulation. J Hepatol. 2023;79:1270–301.
Chun YS, Pawlik TM, Vauthey J-N. 8th Edition of the AJCC Cancer Staging Manual: Pancreas and hepatobiliary cancers. Ann Surg Oncol. 2018;25:845–7.
Audano M, Maldini M, De Fabiani E, Mitro N, Caruso D. Gender-related metabolomics and lipidomics: From experimental animal models to clinical evidence. J Proteom. 2018;178:82–91.
Tabassum R, Widén E, Ripatti S. Effect of biological sex on human circulating lipidome: An overview of the literature. Atherosclerosis. 2023;384:117274
Patel AH, Harnois DM, Klee GG, LaRusso NF, Gores GJ. The utility of CA 19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am J Gastroenterol. 2000;95:204–7.
Gou Q, Zhang CZ, Sun ZH, Wu LG, Chen Y, Mo ZQ, et al. Cell-free DNA from bile outperformed plasma as a potential alternative to tissue biopsy in biliary tract cancer. ESMO Open. 2021;6:100275
Ohyama H, Hirotsu Y, Amemiya K, Miura Y, Hirose S, Oyama T, et al. Comparison of genomic profiling of circulating tumor DNA in pancreaticobiliary malignancies in plasma and bile. Cancer. 2023;129:1714–22.
Mayerle J, Kalthoff H, Reszka R, Kamlage B, Peter E, Schniewind B, et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut. 2018;67:128–37.
Yao Y, Wang X, Guan J, Xie C, Zhang H, Yang J, et al. Metabolomic differentiation of benign vs malignant pulmonary nodules with high specificity via high-resolution mass spectrometry analysis of patient sera. Nat Commun. 2023;14:2339.
Liu ZX, Wang L, Gao SG, Xue Q, Tan FW, Li ZL, et al. Plasma metabolomics study in screening and differential diagnosis of multiple primary lung cancer. Int J Surg. 2023;109:297–312.
Mauvais-Jarvis F. Sex differences in energy metabolism: natural selection, mechanisms and consequences. Nat Rev Nephrol. 2023;20:56–69.
Otsuka S, Ebata T, Yokoyama Y, Igami T, Mizuno T, Yamaguchi J, et al. Benign hilar bile duct strictures resected as perihilar cholangiocarcinoma. Br J Surg. 2019;106:1504–11.
Jeon TY, Choi MH, Yoon SB, Soh JS, Moon S-H. Systematic review and meta-analysis of percutaneous transluminal forceps biopsy for diagnosing malignant biliary strictures. Eur Radio. 2022;32:1747–56.
Giovannini D, Bailly A, Seigneurin A, Fior-Gozlan M, Eyraud P-Y, Roth G, et al. Biliary cytology: A diagnostic tree for adenocarcinoma based on a cohort of 135 patients with endoscopic retrograde cholangiopancreatography for stenosis of the extrahepatic bile duct. Cancer Cytopathol. 2022;130:433–42.
Ko SW, Lee SS, So H, Hwang JS, Song TJ, Lee SK, et al. A novel method of biopsy for indeterminate pancreaticobiliary strictures: tube-assisted biopsy. Endoscopy. 2020;52:589–94.
Zhang Q, Yao D, Rao B, Jian L, Chen Y, Hu K, et al. The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3. Nat Commun. 2021;12:6869.
Liu P, Zhu W, Chen C, Yan B, Zhu L, Chen X, et al. The mechanisms of lysophosphatidylcholine in the development of diseases. Life Sci. 2020;247:117443.
Caponigro V, Tornesello AL, Merciai F, La Gioia D, Salviati E, Basilicata MG, et al. Integrated plasma metabolomics and lipidomics profiling highlights distinctive signature of hepatocellular carcinoma in HCV patients. J Transl Med. 2023;21:918.
Zhang L, Liu X, Liu Y, Yan F, Zeng Y, Song Y, et al. Lysophosphatidylcholine inhibits lung cancer cell proliferation by regulating fatty acid metabolism enzyme long-chain acyl-coenzyme A synthase 5. Clin Transl Med. 2023;13:e1180.
Funding
This work was supported by the research funds from Cancer Research and Translational Platform Project of Zhongnan Hospital of Wuhan University (ZLYNXM202004); and the research funds from the medical Sci-Tech innovation platform of Zhongnan Hospital of Wuhan University (PTXM2023008).
Author information
Authors and Affiliations
Contributions
FL and YL participated in the lipidomic experimental and data analysis. SZ participated in the cell experimental and data analysis. JZ, XP, YC, WF, TY, and XY collected the samples. BL, XC and TW collected the clinical data and completed various scores and staging interpretations. MZ involved in code processing for R analysis. JL, XL, BL, XC, TW, WM, and PJ participated in the diagnosis and clinical treatment of all included patients. FL, SG, and SL participated in the writing and revising of the article. YY, SG, SL and WH designed, revised and guided the whole process of the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Medical Ethics Committee of the Clinical Trial Centre at Zhongnan Hospital, Wuhan University (Approval number: Scientific Research Ethics[2020100]). Informed consent for sample collection was obtained from all patients, and the study procedures followed the Declaration of Helsinki Guidelines.
Consent for publication
All the authors agree to the publication of the manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In the Conclusions section, the article originally read: “This study not only identified lipidomic alterations in CCA using bile samples but also established and validated a sex-related bile lipid classifier with high specificity and sensitivity for distinguishing between CCA and benign bile duct diseases.” The correct sentence is as follows (“sex-related” removed): “This study not only identified lipidomic alterations in CCA using bile samples but also established and validated a bile lipid classifier with high specificity and sensitivity for distinguishing between CCA and benign bile duct diseases.”
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, FS., Liu, YY., Zhang, SK. et al. Lipidomic profiling of human bile distinguishes cholangiocarcinoma from benign bile duct diseases with high specificity and sensitivity: a prospective descriptive study. Br J Cancer 133, 1565–1582 (2025). https://doi.org/10.1038/s41416-025-03144-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41416-025-03144-9
This article is cited by
-
An update on schistosomicide and tumoricide in the future: arachidonic acid
Lipids in Health and Disease (2025)