Abstract
Base excision repair (BER) is initialized by DNA glycosylases, which recognize and flip damaged bases out of the DNA duplex into the enzymes active site, followed by cleavage of the glycosidic bond. Recent studies have revealed that all types of DNA glycosylases repair base lesions less efficiently within nucleosomes, and their repair activity is highly depended on the lesion’s location within the nucleosome. To reveal the underlying molecular mechanism of this phenomenon, we determine the 3.1 Å cryo-EM structure of human 8-oxoguanine-DNA glycosylase 1 (hOGG1) bound to a nucleosome core particle (NCP) containing a common oxidative base lesion, 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo). Our structural analysis shows that hOGG1 can recognize and flip 8-oxodGuo even within NCPs; however, the interaction between 8-oxodGuo and hOGG1 in a NCP context is weaker than in free DNA due to competition for nucleosomal DNA by the histones. Binding of OGG1 and the flipping of 8-oxodGuo by hOGG1 leads to a partial detachment of DNA from the histone core and a ratchet-like inward movement of nucleosomal DNA. Our findings provide insights into how the dynamic structure of nucleosomes modulate the activity of repair enzymes within chromatin.
Similar content being viewed by others
Introduction
Genomic DNA is susceptible to damage from both endogenous and exogenous oxidants, leading to the formation of various oxidative lesions. Accumulation of these lesions may have adverse physiological consequences, such as mutagenesis, genomic instability, and cellular senescence1,2. Notably, among these lesions, 8-Oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo) is highly prevalent and serves as a biomarker of oxidative stress3,4. 8-OxodGuo can form a stable base pair with deoxyadenosine (dA), which results in G → T transversion mutations upon cell division5,6. Additionally, further oxidation of 8-oxodGuo rapidly occurs, generating more mutagenic and harmful lesions such as spiroiminodihydantoin (Sp), guanidinohydantoin (Gh)7,8,9 and DNA-protein crosslinks (DPCs)10,11,12. The deleterious effects of 8-oxodGuo within cells are primarily mitigated through the base excision repair (BER) process. The molecular mechanism of BER for 8-oxodGuo in free double-stranded DNA (dsDNA) has been extensively studied13. The BER is initialized by 8-oxoguanine-DNA glycosylase 1 (OGG1), which specifically recognizes and flips the oxidized nucleobase from the DNA duplex, followed by catalysis of the glycosidic bond cleavage14,15.
In eukaryotic cells, nuclear DNA wraps around histone octamers to form nucleosomes, the fundamental building blocks of chromatin16. Initially, the removal of 8-oxodGuo by OGG1 within nucleosomes was thought to be restricted due to steric hindrance imposed by histones and the bend/rigid structure of nucleosome DNA. However, recent studies have challenged this notion, demonstrating that OGG1 can remove nucleosomal 8-oxodGuo17,18,19,20,21. The efficiency of repair, however, significantly depends on the location and geometric orientation of 8-oxodGuo within the nucleosome18,19,20. These findings suggest a coordinated and adaptable interaction between OGG1 and nucleosomes, allowing OGG1 to recognize and process 8-oxodGuo within nucleosomal DNA. Nevertheless, the molecular mechanism underlying how OGG1 processes 8-oxodGuo in nucleosomes remains unclear.
In this study, we employed single-particle cryo-electron microscopy (cryo-EM) along with biochemical analyses to investigate how human OGG1 (hOGG1) recognizes and initiates the repair of 8-oxodGuo lesions within nucleosome core particles (NCPs). Our findings reveal that hOGG1 can flip and engage an 8-oxodGuo located at superhelical location (SHL) 6.0 of the NCP. The flipping of 8-oxodGuo by hOGG1 results in a partial detachment of DNA from the histone core and a ratchet-like inward movement of nucleosomal DNA. Intriguingly, the strong bending of DNA by OGG1 binding allows formation of histone-DNA contacts on both sides of OGG1 since the DNA is bulged out and not fully displaced from the histones. The concomitant inward movement of DNA, reducing steric hindrance at the dyad region, facilitates another hOGG1 in binding and processing adjacent 8-oxodGuo situated at the dyad position of the NCP. This study provides insights into the general mechanism by which DNA glycosylases recognize and process base lesions to fulfill their repair functions even within nucleosomes.
Results
The affinity of hOGG1 is contingent upon the position of an 8-oxodGuo lesion within NCPs
In our previous work, we prepared three 8-oxodGuo-modified NCPs by reconstituting 8-oxodGuo-modified 145 bp 601 dsDNA with a histone octamer12,20. Specifically, 8-OxodGuo was incorporated to the position 73, 89 and 137 of 601 dsDNA, as illustrated in Fig. 1a. Subsequent treatment of these NCPs with hOGG1 unveiled a pronounced dependency of 8-oxodGuo repair efficiency on its location within the NCP. In NCP-8-oxodGuo-137, where 8-oxodGuo is positioned near the entry/exit region (SHL 6.0), the repair occurred with slightly diminished efficiency compared to its free dsDNA counterpart, dsDNA-8-oxodGuo-137. Notably, repair of NCP-8-oxodGuo-73, with 8-oxodGuo located on the dyad axis (SHL 0), was markedly suppressed, and hOGG1 exhibited minimal or no activity towards NCP-8-oxodGuo-89 (SHL 1.5)20.
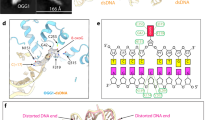
a X-ray crystal structure of a NCP (PDB: 3lz0) showing the locations of the tested 8-oxodGuo. The 8-oxodGuo modifications are represented by means of the red sphere model. b 10% native PAGE depicting the bound of hOGG1K249Q with NCP-8-oxodGuo-137. The red arrow highlights a band corresponding to a 1:1 complex of hOGG1K249Q/NCP-8-oxodGuo-137. c Quantification with fits for the binding of hOGG1K249Q to dsDNA. d Quantification with fits for the binding of hOGG1K249Q to NCPs. The presented data display the individual data points of three independent experiments. NCP nucleosome core particle, SHL superhelical location. Source data are provided as a Source Data file.
To investigate whether varied binding affinity of hOGG1 contributes to the differing repair efficiencies of 8-oxodGuo at distinct positions, we determined the dissociation constants (Kd) of the catalytically inactive hOGG1K249Q mutant22 with 8-oxodGuo-modified 145 bp 601 dsDNA and NCPs using electrophoretic mobility shift assays (EMSAs). Incubation of dsDNA-8-oxodGuo-137, dsDNA-8-oxodGuo-89, or dsDNA-8-oxodGuo-73 with limited hOGG1K249Q resulted in a discernible band (marked by a red arrow, Supplementary Fig. 1b-d) migrating slightly slower than the untreated dsDNA counterpart. This band disappeared as the concentration of hOGG1K249Q increased, accompanied by the appearance of smeared and slower-migrating bands in the gel. In contrast, EMSA of hOGG1K249Q with unmodified dsDNA showed only a few dispersive bands (Supplementary Fig. 1a).
The Kd values of hOGG1K249Q bound to 8-oxodGuo-modified dsDNA varied slightly, ranging from 0.24 μM to 0.42 μM (Table 1 and Fig. 1c), roughly one order of magnitude smaller than that of hOGG1K249Q bound to unmodified dsDNA (1.31 μM). These results confirmed that hOGG1 is fundamentally able to bind the native dsDNA and exhibits significantly higher affinity for the 8-oxodGuo lesion. Thus, the observed clear band in the EMSA likely corresponds to the 1:1 complex of hOGG1K249Q with 8-oxodGuo-modified dsDNA, in which hOGG1K249Q flips and tightly engages the 8-oxodGuo14. In the presence of an excess of hOGG1K249Q, multiple hOGG1K249Q molecules nonspecifically bind along the dsDNA, as confirmed by the cryo-EM study. A detailed discussion of this observation will be presented later in the manuscript.
The aforementioned findings suggest that hOGG1 has high specificity for 8-oxodGuo in free dsDNA, with the position of 8-oxodGuo in dsDNA playing only a minor role in hOGG1’s binding affinity. The binding affinity of hOGG1K249Q to NCP-8-oxodGuo-137, NCP-8-oxodGuo-73, and NCP-8-oxodGuo-89 decreases sequentially (Table 1 and Fig. 1d). Notably, a clear band was observed in the EMSA of hOGG1K249Q with NCP-8-oxodGuo-137 (Fig. 1b, marked by an arrow), but it was much weaker in the EMSAs of hOGG1K249Q with NCP-8-oxodGuo-89 and especially with NCP-8-oxodGuo-73 (Supplementary Fig. 1f, g). These results suggest that 8-oxodGuo lesions in nucleosomal DNA can also be specifically recognized by hOGG1. However, the location of 8-oxodGuo in the NCP significantly influences its interaction with hOGG1, confirming that efficient binding of 8-oxodGuo is a crucial factor in modulating the repair efficiency of hOGG1 in NCP. Consequently, NCP-8-oxodGuo-137, which exhibits the highest binding affinity to hOGG1, was selected as the substrate for the structural study of hOGG1 repair within NCP.
hOGG1K249Q can flip and engage 8-oxodGuo at SHL 6.0 of the NCP
We employed single-particle cryo-electron microscopy (cryo-EM) to scrutinize the complex formed by hOGG1K249Q and NCP-8-oxodGuo-137. Two datasets were recorded, enabling the generation of a cryo-EM 3D map with a global resolution of 3.1 Å (Fig. 2a and b, Table 2, Supplementary Fig. 2). As expected, hOGG1K249Q is bound at SHL 6.0 in a 1:1 complex with the NCP, at the site where the NCP contains the 8-oxodGuo. With a notable exception (see below), the observed NCP-hOGG1K249Q particles were very homogeneous in 2D and 3D classification, consistent with the well-defined high-affinity species in our EMSA experiments, showing specific binding to the lesion. Intriguingly, the cryo-EM map reveals that hOGG1K249Q induces a bulge in the nucleosomal DNA at SHL 6.0, allowing access to and flipping of the 8-oxodGuo out of the DNA duplex. We do not observe any contacts between hOGG1K249Q and the histones, thus binding is solely governed by the lesion and flanking DNA. The 8-oxodGuo is flipped out from its base-pairing and resides within the catalytic pocket of hOGG1K249Q, while the overall interaction mode including the bending of DNA closely resembles that observed in the crystal structure of hOGG1K249Q bound to a 8-oxodGuo-modified 15 bp dsDNA (PDB: 1ebm, Supplementary Fig. 3a and b)15,23.
a DeepEM-enhanced post-processed cryo-EM reconstruction of hOGG1K249Q bound to NCP-8-oxodGuo-137 at 3.1 Å resolution. Light gray: histone octamer, dark gray: DNA, orange: hOGG1. b Side and top views of the derived structural model (ribbon representation) illustrating hOGG1 binding to the 8-oxodGuo lesion without direct interactions with the histone core. c Side view of the structural model with close-up views on the hOGG1 active site overlaid by the density map. Interface I demonstrates interactions between hOGG1K249Q and the 8-oxodGuo lesion in the enzyme’s catalytic pocket. Interface II reveals interactions in the cytosine recognition pocket. Cyan represents 8-oxodGuo-137, dark gray corresponds to DNA, and orange represents hOGG1. Nucleotides and interacting residues are depicted as sticks with color-coded oxygens (red), sulfurs (yellow) and nitrogens (blue).
A detailed analysis of the two structures reveals subtle differences. In the crystal structure of the hOGG1K249Q/dsDNA complex (PDB: 1 ebm), the 8-oxodGuo directly interacts with five residues, Phe 319, Cys 253, Asp 268, Gln 315 and Gly 42. Phe 319 and Cys 253 sandwich the 8-oxodGuo base within the catalytic pocket. The side-chain carbonyl of Gln 315 forms hydrogen bonds with both N1-H and N2-H of 8-oxodGuo. Gly 42, a pivotal residue responsible for distinguishing between 8-oxodGuo and native guanine, establishes a crucial hydrogen bond with the N7-H of 8-oxodGuo15. In the structure of the hOGG1K249Q/NCP-8-oxodGuo-137 complex, the entire catalytic pocket of hOGG1K249Q shift by >1 Å towards the O6/N7 face of 8-oxodGuo. This shift leads to a reduced interaction between Gln 315 and the N1-H of 8-oxodGuo, as well as a weakening of the π-π interaction between the aromatic nucleobase of 8-oxodGuo and the phenyl group of Phe 319 (Fig. 2c and Supplementary Fig. 3c, d). Notably, due to the relatively low local resolution of the 8-oxodGuo binding pocket in the hOGG1K249Q/NCP-8-oxodGuo-137 complex, biases during model building may partially account for the subtle differences observed between the two structures.
The binding of hOGG1 to 8-oxodGuo induces a localized structural distortion in nucleosomal DNA
From the comprehensive dataset acquired, we also observed distinct particles of NCP-8-oxodGuo-137, unaccompanied by the hOGG1K249Q. Subsequent to meticulous reconstruction and refinement procedures, we successfully generated a cryo-EM 3D map of NCP-8-oxodGuo-137, achieving an average resolution of 2.8 Å (Supplementary Fig. 4). Notably, the canonical NCP structural model (PDB: 3lz0)24 aligns exceptionally well with the density map of NCP-8-oxodGuo-137 (Supplementary Fig. 4b). In addition, DNase I footprinting of dsDNA-8-oxodGuo-137 and NCP-8-oxodGuo-137 revealed very similar digestion patterns to those of WT dsDNA and WT NCP, respectively (Supplementary Fig. 5). These observations align perfectly with the findings reported by Shigdel’s research23 and indicate that the presence of the 8-oxodGuo-137 modification induces minimal structural alterations within the NCP. Therefore, we adopted the canonical NCP model as our reference to reveal how the binding of of hOGG1 to 8-oxodGuo distorts the overall structure of nucleosomal DNA.
A comparative analysis between the structure of the hOGG1K249Q/NCP-8-oxodGuo-137 complex and the crystal structure of a canonical NCP (PDB: 3lz0) reveals a noticeable disruption in the nucleosomal DNA upon hOGG1K249Q binding to 8-oxodGuo within NCPs (Fig. 3). Following the flipping of 8-oxodGuo by hOGG1K249Q, a small DNA segment containing 8-oxodGuo-137 slightly detaches from the histone core, widening the minor groove of the downstream 3 base pairs by an average of 10.6 Å (Fig. 3c).
a Alignment of the hOGG1K249Q/NCP-8-oxodGuo-137 complex structure with that of a canonical NCP (PDB: 3lz0). The close-up view highlights the pronounced widening of the minor groove upon hOGG1 binding and the resulting ejection of 8-oxodGuo-137 from the DNA helix. b DNA registry shifts at SHL 5.5 to SHL 7.0 due to local distortions of nucleosomal DNA upon hOGG1 binding. In comparison to a canonical NCP model (PDB: 3lz0), the DNA is pushed inwards from the nearby DNA entry/exit site toward the dyad axis and partially lifted off the NCP core. c Distance plot depicting the minor groove width of the nucleosomal DNA segment spanning from SHL 5.5 to SHL 7.0. The minor groove width was measured between DNA backbone phosphates. Light gray: histone octamer of hOGG1K249Q/NCP-8-oxodGuo-137 complex, dark gray: DNA of hOGG1K249Q/NCP-8-oxodGuo-137 complex, light blue: histone octamer of canonical NCP, dark blue: DNA of canonical NCP, orange: hOGG1. NCP nucleosome core particle. Source data are provided as a Source Data file.
An electrostatic surface potential analysis of the canonical NCP crystal structure (3lz0)24, conducted using ChimeraX25 and Bluues26, revealed that the Arg-rich histone H3 fragment spanning from His 39 to Tyr 54 is electrostatically attracted to the negatively charged DNA segment at SHL 6.0, displaying overall surface potential of -0.66 kJ/(mol•q) (Supplementary Fig. 6). Conversely, in the hOGG1K249Q/NCP-8-oxodGuo-137 complex, a more extensive cavity is observed between H3 and the DNA segment at SHL 6.0, resulting in the restoration of the positive surface potential of 0.13 kJ/(mol•q) for this histone fragment (Supplementary Fig. 6). These findings collectively confirm that hOGG1 binding induces the partial detachment of DNA from the histone core, consequently weakening the local electrostatic interactions between the negatively charged DNA phosphate backbone and the positively charged histone.
The partial detachment of DNA results in a 2 bp registry shift towards the dyad axis for the DNA segment flanking 8-oxodGuo-137 at the NCP entry/exit side (SHL 6.5 – SHL 7.0), a 1 bp shift at SHL 6.0, and a 0.5 bp shift at SHL 4.5 (Fig. 3b and Supplementary Fig. 3f-g). It is noteworthy that the end of the nucleosomal DNA (SHL 7.0) regains contacts to the histone core in the hOGG1K249Q/NCP-8-oxodGuo-137 complex, thus hOGG1 binding does not completely detach one side of the nucleosomal DNA from the histones, but induces a bulge that allows flanking interactions with histones. Additionally, the ratchet-like motion of DNA leads to no obvious disturbance in the interactions between the histone core and nucleosomal DNA, except in the region from SHL 4.5 to SHL 7.0 of the NCP (Supplementary Fig. 7). A MatchMaker BLOSUM-62 comparison analysis27,28 of the histone core of the hOGG1K249Q/NCP-8-oxodGuo-137 complex with that of a canonical NCP revealed an RMSD of only 0.6 Å across all 750 pairs, using an iteration cutoff as 2 Å (Supplementary Fig. 7). Comparative analysis of the H3 N-Tail, which binds to the entry and exit ends of nucleosomal DNA, and the H2B N-tail and C-tail, which themselves bind to DNA located on SHL 4.5, in these two structures shows an overall good match. This indicates that structural alternations in the histone octamer core are very minor, regardless of hOGG1 binding (Supplementary Fig. 7b, c). Taken together, hOGG1 binding to 8-oxodGuo-137 in NCP produces a DNA bulge structure, ensuring the recognition and repair of the lesion by the repair enzyme. However, this structural distortion is locally limited and does not alter the overall structure of NCP.
Non-specific binding of hOGG1 molecules to free dsDNA and nucleosomal DNA devoid of 8-oxodGuo lesions
During cryo-EM data collection, 2D classes were obtained for a subset of particles where two or three hOGG1K249Q molecules bound to a single stretch of free dsDNA-8-oxodGuo-137 (Fig. 4a). The resulting 2D averages clearly depict a distinctive kinking pattern at each hOGG1K249Q binding site on the free dsDNA (Fig. 4b). By fitting the model of hOGG1K249Q/dsDNA (PDB: 1ebm) to the limited particles of the [hOGG1K249Q]2/[dsDNA-8-oxodGuo-137]1 complex, we generated a low-resolution 3D reconstruction volume confirming that the DNA strand is significantly kinked at each hOGG1 binding site, with ~12 base pairs between two adjacent hOGG1 binding sites (Supplementary Fig. 8). Next, we constructed a model of a 145 bp 601 dsDNA bound with multiple hOGG1 using the composite tool integrated into the x3dna webserver (http://web.x3dna.org)29. The theoretical molecular model suggests that DNA is kinked several times to afford a spiral-like structure under the random combination of multiple hOGG1 binding events (Fig. 4c).
a 2D classes depicting particles with multiple hOGG1K249Q molecules binding to a dsDNA molecule in 2:1 and 3:1 ratios. b Selected 2D class averages of particles displaying a distinctive kinking pattern at each hOGG1K249Q binding site on the dsDNA stretch. c Top view of an in-silico model illustrating six hOGG1 molecules bound to a single free 601 DNA stretch in a random distribution. The model was generated utilizing the crystal structure of hOGG1 bound to free dsDNA (PDB: 1ebm) and the 601 DNA sequence through the x3dna webserver. (http://web.x3dna.org/). Gray: DNA, orange: hOGG1.
Verdine et al. have systematically studied how 8-oxoguanine-DNA glycosylases interrogate and recognize 8-oxodGuo in free dsDNA using X-ray crystallography and molecular dynamics simulations23,30,31,32,33,34. They found that upon glycosylase binding, the DNA backbone structures around the target nucleotide are slightly bent. This local structural change un-stacks and buckles the target base pair, allowing the enzyme to further interact with and interrogate the target base. If 8-oxodGuo is recognized, it is extruded from the helix, accompanied by DNA kinking. Flipping native guanine base is kinetically unfavorable23. However, when the glycosylase is covalently linked to the dsDNA, native guanine can also be flipped out from the DNA helix, resulting in a kinked DNA strand31. Thus, interrogative binding of glycosylases to dsDNA only slightly bends the strand, while significant DNA kinking occurs alongside the extrusion of the target base. It is noteworthy that the 145 bp DNA substrate (dsDNA-8-oxodGuo-137) features only a single 8-oxodGuo modification. The multiple kinked structure induced by the binding of multiple hOGG1 molecules along dsDNA-8-oxodGuo-137 (Fig. 4a) suggests that, in addition to 8-oxodGuo-137, hOGG1 may also non-specifically recognize and flip native nucleobases from the dsDNA strand. We propose a synergistic effect to explain this unusual observation. The flipping of 8-oxodGuo by hOGG1 result in DNA kinking and structural distortion for ~10 bp DNA flanking both sides of the lesion, such as minor grooving broadening. As another hOGG1 slides and approaches the hOGG1/8-oxodGuo binding site, its movement may be impeded or even halted by the structure distortion. This creates a sufficient time interval for the second hOGG1 molecule to flip a native guanine base and kink the DNA strand. This synergistic effect propagates along the DNA strand bidirectionally, eventually forming a spiral-like structure (Fig. 4c).
A comprehensive analysis of the cryo-EM data also showed some particles in which two hOGG1K249Q molecules bind to a single NCP-8-oxodGuo-137 (Fig. 5a, Supplementary Fig. 9). Structural examination of the 2D classes of the [hOGG1K249Q]2/[NCP-8-oxodGuo-137]1 complex suggests that one hOGG1K249Q is bound to the 8-oxodGuo-137 at SHL 6.0, as depicted in the near-atomic resolution structure of the hOGG1K249Q/NCP-8-oxodGuo-137 complex, while the second one is positioned near SHL -2.0 (Fig. 5b). Of note, the DNA stretch at SHL -2.0 is in the central region of the NCP and devoid of 8-oxodGuo. Whether hOGG1 only interrogatively binds at SHL -2.0 or has already flipped a native nucleobase from the DNA strand still remains elusive, however, this structure provides further evidence that hOGG1 can non-specifically bind to and interrogate nucleosomal DNA.
a Illustration of representative 2D classification. b Crude map depicting the binding of hOGG1K249Q in conjunction with NCP-8-oxodGuo-137 at a ratio of 2:1, obtained from the same data collection as presented in Fig. 2. Orange represents hOGG1K249Q bound on 8-oxodGuo-137 (SHL 6.0), Pink corresponds to hOGG1K249Q bound at SHL −2.0. NCP, nucleosome core particle. SHL, superhelical location.
Discussion
Base excision repair (BER) is the first DNA repair pathway which was discovered five decades ago. The molecular mechanism of repairing common damaged nucleobases, such as 8-oxodGuo, U, and alkylated purines, within free dsDNA have been extensively studied. BER typically commences with DNA glycosylases, each specifically targeting a damaged nucleobase along the DNA strand. Upon encountering the damaged site, the DNA glycosylase flips the targeted base from the DNA strand. Subsequently, the flipped base is excised from the DNA strand, generating an apurinic/apyrimidinic (AP) site. This site is then processed by subsequent BER enzymes to restore the correct DNA sequence35. Recent scientific interest has focused on the repair of common nucleobase lesions within chromatin DNA via BER. Utilizing reconstituted nucleosomes—the fundamental building blocks and repeating units of chromatin—as an in vitro model, we and others have demonstrated significant activity decrease of various DNA glycosylases in nucleosomes compared to free dsDNA. The repair efficiency is largely dependent on the location and geometric orientation of damaged bases within nucleosomes36,37,38,39,40,41. However, the molecular mechanism of damage processing by DNA glycosylases within nucleosomes has been rarely reported42,43. Recently, Gao et al. incubated AAG (alkyladenine DNA glycosylase) with NCPs containing a single deoxyinosine modification at various locations and obtained the Cryo-EM structures of AAG-NCP complexes. However, all the structures are in post-catalytic states, with an AP site remaining in the catalytic pocket of the glycosylase43. In the present study, we employed cryo-EM to attain a global 3.1 Å-resolution structure of a complex consisting of a catalytically inert DNA glycosylase mutant, hOGG1K249Q, and a NCP containing 8-oxoGuo located at SHL 6.0 (NCP-8-oxodGuo-137). To the best of our knowledge, this represents the first high-resolution structure illustrating how a DNA glycosylase processes nucleobase damage within a nucleosome.
The structure of the hOGG1K249Q/NCP-8-oxoGuo-137 complex unmistakably reveals the extrusion and flipping of 8-oxodGuo from the nucleosomal DNA, with subsequent engagement in the catalytic pocket of hOGG1. Although the local resolution of the 8-oxodGuo binding pocket is much lower than the global 3.1 Å resolution, a comparative analysis between the structure of the hOGG1K249Q/NCP-8-oxoGuo-137 complex and the previously reported crystal structure of hOGG1K249Q/dsDNA-8-oxoGuo shows that in the former complex, the entire catalytic pocket of hOGG1K249Q has a slight shift towards the O6/N7 face of 8-oxodGuo. Consequently, interactions between 8-oxodGuo with hOGG1 are slightly weaker in the hOGG1K249Q/NCP-8-oxoGuo-137 compared to the hOGG1K249Q/dsDNA-8-oxoGuo complex. The reduction in interactions resulting from this catalytic pocket shift elucidates the lower binding affinity and repair activity of hOGG1 towards NCP-8-oxodGuo-137 compared to dsDNA-8-oxodGuo-137. Notably, in the hOGG1K249Q/NCP-8-oxoGuo-137 structure, the N-terminal tail of H3 was not fully resolved. This tail typically interacts with DNA situated at both SHL 7.0 and SHL -1.0 (Supplementary Fig. 10). The steric hindrance exerted by the H3 tail and DNA at SHL -1.0 may elucidate the observed shift in the catalytic pocket of hOGG1K249Q towards the O6/N7 face of 8-oxodGuo. In addition, core histones compete with hOGG1 for DNA binding (Supplementary Fig. 6), which would also weaken the interaction between hOGG1 and the base lesion. However, considering that the 601 DNA, selected from a vast pool of chemically synthesized random DNA molecules, exhibits remarkably high affinity for histone octamers44, the competitive effect of core histones might be less pronounced within NCPs composed of physiological DNA.
The Kd of hOGG1K249Q bound to an unmodified NCP is comparable to that of hOGG1K249Q bound to unmodified dsDNA (Table 1), suggesting that hOGG1’s capacity for non-specific binding to and interrogation of nucleosomal DNA is not significantly impaired by the NCP structure. This conclusion is further supported by the observation of the [hOGG1K249Q]2/[NCP-8-oxodGuo-137]1 complex, in which one hOGG1K249Q binds to the 8-oxodGuo-137 at SHL 6.0, while the second one is positioned at SHL −2.0, devoid of 8-oxodGuo (Fig. 5). Numerous studies have consistently shown that lesions in the outer strand of nucleosomal DNA are repaired at a faster rate by DNA glycosylases compared to counterparts in the inner strand45,46. The outer strand is more solvent-accessible, making it easier for the repair enzyme to encounter.
In this context, hOGG1 can efficiently interrogate nucleosomal DNA. Once an 8-oxodGuo located on the outer face of an NCP is encountered, an NCP-8-oxodGuo/hOGG1 encounter complex is formed. Next, the base lesion is flipped out, and the local nucleosomal DNA is stripped from the histone core and kinked, resulting in the formation of a stable lesion recognition complex. This step must overcome a rather high energy barrier, thus determining the repair kinetics23. In the hOGG1K249Q/NCP-8-oxoGuo-137 complex, the flipping of 8-oxoGuo-137 results in gradual decreases in the DNA shift register, from 2 bp for DNA at SHL 7.0 to 0.5 bp for DNA at SHL 4.5. This suggests that the DNA segment downstream of 8-oxodGuo-137 (from position 138 – 145), situated at the entry/exit region of NCP, can be more easily detached from the histone core than the segment upstream of 8-oxodGuo-137. Notably, both 8-oxodGuo-73 and 8-oxodGuo-89 are located in the central region of nucleosome DNA, requiring a larger energy barrier to be overcome for flipping the lesion and kinking DNA at these two locations. This elucidates why hOGG1 exhibits lower affinity and repair activity for NCP-8-oxodGuo-73 and NCP-8-oxodGuo-89. This observation also provides a general explanation for the location-dependent DNA repair efficiency in NCPs.
In summary, we have acquired a high-resolution cryo-EM structure of hOGG1/NCP-8-oxodGuo-137 complex, providing a detailed insight into how a base lesion is bound and processed by a DNA glycosylase with the nucleosomal context. The overall interaction between DNA glycosylase and the base lesion within a NCP closely resembles that observed in free DNA. Despite inducing local distortion in the DNA, the overall structure of the NCP remains intact. This challenges the prevailing notion that dynamic dissociation of nucleosomal DNA from the histone core is a prerequisite for initiating BER by DNA glycosylases. Our findings support an alternative proposition that a base lesion inside nucleosomes can be processed by DNA glycosylase without disassembling the intact nucleosome. Furthermore, this study also provides insights into why DNA glycosylases’ activity is suppressed in NCP and is significantly dependent on the location of base lesions within the nucleosome.
Methods
NCP Preparation
NCPs were reconstituted following the procedures outlined in our prior publication20. Synthetic oligonucleotides were sourced from Sangon Biotech Co. Detailed information regarding the oligonucleotides sequence and ligation strategy can be found in Supplementary Fig. 11 and Supplementary Table 1.
hOGG1 expression and purification
The hOGG1K249Q mutant and the wild-type hOGG1 were cloned into a pET-DUET1 vector and expressed in E. coli Rosetta (DE3) competent cells. Protein expression was induced at OD 600 = 0.75 by adding 1 mM IPTG, followed by incubation at 18 °C for 20 h. Subsequently, cells were harvested and lysed via sonication on ice in the binding buffer containing 300 mM NaCl, 50 mM K2HPO4/KH2PO4 (pH 8.0), 10 mM BME. The collected lysate underwent purification using a His-Trap FF (Cytiva, #11000458) affinity chromatography column. Bound protein was eluted with a linear gradient of the elution buffer containing 300 mM NaCl, 50 mM K2HPO4/KH2PO4 (pH 8.0), 10-600 mM imidazole, 10 mM BME. Fractions containing the target protein were pooled and further purified by a HILOAD 16/600 Superdex 75 PG (Cytiva, #28989333) size exclusion chromatography (SEC) with SEC buffer containing 20 mM Tris HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 1 mM BME. The resulting pure protein fractions were combined, concentrated to ~100 µM and flash-frozen in liquid nitrogen before storage at −80 °C. The purity and integrity of expressed proteins were confirmed by SDS-PAGE and mass spectroscopy analysis (Supplementary Fig. 12 and Supplementary Table 2).
Electrophoretic mobility shift assay (EMSA)
Free 601 dsDNA or NCP (0.1 µM, fluorescently labeled at the 5’ end of the forward DNA strand) was combined with hOGG1K249Q at varying concentrations (0, 0.1, 0.2, 0.5, 1, 1.5, 2.0, 3.0, 4.0 µM) in EMSA buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 1 mM DTT). The mixtures were incubated on ice for 1 h and subsequently analyzed using 10% native PAGE. Gel imaging is achieved through fluorescence (FAM) scanning, with an excitation wavelength of 488 nm and an emission wavelength of 526 nm. Quantification analysis of the gel was performed using Amersham ImageQuant. The percentages of NCP/dsDNA binding were plotted against the concentration of hOGG1K249Q, and the resulting sigmoidal curves were fitted using Saturation Binding/Specific binding with Hill slope equation [Y = Bmax*X^h/(Kd^h + X^h)] to yield Kd values.
Cryo-EM sample preparation
NCP-8-oxodGuo-137 (1.4 µM) and hOGG1K249Q (5.1 µM) were mixed in EM Buffer (10 mM HEPES [pH 7.5], 25 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT) in a total volume of 9.5 µL and incubated on ice for 4 h. To crosslink the sample, glutaraldehyde was added to a final concentration of 0.1% (w/v) and incubated on ice for 10 min prior to cryo-EM grid preparation. Quantifoil Cu 200, R2/1 grids were glow-discharged for 7 s at 20 mA (GloQube, Quorum) before application of 4.5 µl of the aforementioned sample. The sample was blotted 2.3 s and vitrified in liquid ethane using a Leica EM GP plunging freezer.
Cryo-EM data collection and processing
Cryo-EM data were collected using a FEI Titan Krios G3 transmission electron microscope (Thermo Fisher Scientific) operating at 300 kV. The microscope was equipped with a Gatan K2 Summit direct electron detector and a GIF quantum energy filter, operated with a slit width of 20 eV. Two datasets were collected, consisting of 2,624 movies for dataset 1 and 11,070 movies for dataset 2. The defocus values ranged from −1.1 – −2.9 μm, and the respective total doses were 43.76 and 39.35 e-/Å2 over 40 frames, with a magnified pixel size of 1.045 Å (Table 2).
Micrograph movies were first subjected to motion correction and dose-weighting using MotionCor247, and then CTF parameter were estimated using CTFFIND448. All further cryo-EM data processing steps were performed using cryoSPARC v4.0.149 and following releases, as well as Relion v3.150 (Supplementary Fig. 2). Initially, the two collected datasets were processed individually in cryoSPARC, following the steps of blob picking with an applied diameter of 100–300 Å and particle extraction with a box size of 256 pixel. The extracted particle subsets were subjected to multiple rounds of 2D classification, resulting in an initial cleaned subset of 2D classes comprising 144,688 particles for dataset 1 and 673,536 particles for dataset 2. The initial 2D classes served as templates for a reference-based template picking approach on all 13,694 micrographs from the combined two datasets, using a particle diameter set at 220 Å. The initially picked 8,279,033 particles were manually curated and again subjected to multiple rounds of 2D classification. This process resulted in a refined particle subset consisting of 1,134,348 particles. This refined particle subset underwent an ab-initio reconstruction with 4 classes. The best-resolved 3D volume was used to generate a mask specifically for the hOGG1 portion, which was employed in a masked 3D classification of all 1,134,348 particles into 5 classes. The most well-resolved class, featuring a nominal resolution of 3.3 Å and comprising 107,630 particles, was utilized in the subsequent processing steps and transferred to Relion using the pyEM script51. Within Relion, the particle subset underwent additional polishing through 2D classification, reducing the size to 100,662 particles. This was followed by two iterative rounds of CTF refinement and Bayesian particle polishing. Subsequently, the polished particles were reintroduced into cryoSPARC and subjected to another round of ab-initio reconstruction, followed by heterogeneous refinement with 2 classes. This step further removed another subset of 34,292 poorly resolved particles, resulting in a final particle subset of 66,370 high-quality particles. Conducting 2 iterative rounds of 3D flexible refinement52 with a cropped box size of 220 pixels and an applied training box size of 128 pixels resulted in the final reconstruction of hOGG1K249Q bound to an NCP-8-oxodGuo-137 at 3.1 Å. Subsequently, the refined cryo-EM map was further post-processed using the deep-learning-based algorithm DeepEMhancer (Supplementary Fig. 2)53. A directional FSC plot for the final, unsharpened 3D reconstitution was performed using the 3DFSC software54.
For the NCP-8-oxodGuo-137 structure, particles were initially picked on dataset 2 (11,070 micrographs) using blob picker in cryoSPARC (Supplementary Fig. 4). The particles were filtered by 2D classification, ab-initio reconstruction and heterogeneous refinement using 336-pixel box, Fourier-cropped to 84 pixels. The yielded classes of 1,584,400 particles with clearly defined features were further sorted by 3D classification and heterogeneous refinement, leaving a new particle set of 815,588 particles, which were re-extracted using 336-pixel box without Fourier crop. The particles were further refined using cryoSPARC local CTF refinement, yielded a final map of NCP-8-oxodGuo-137 after non-uniform refinement at 2.8 Å resolution.
For [hOGG1K249Q]2/[NCP-8-oxodGuo-137]1 complex structure, initial analysis was performed on dataset 1 (2624 micropgrahs). In cryoSPARC, particles were sorted by blob picking (Supplementary Fig. 9), followed by multiple rounds of 2D classification with an extraction box size of 336 pixel, Fourier-cropped to 84 pixels. Selection of [hOGG1K249Q]2/[NCP-8-oxodGuo-137]1 classes were used as input for Topaz training. The particles were subject to further 2D classification, ab-initio, and heterogeneous refinement. The final selection of 22,295 particles were re-extracted with 336-pixel box without Fourier-crop. Non-uniform refinement (C1) and local refinement were performed, resulted in a final map of [hOGG1K249Q]2/[NCP-8-oxodGuo-137]1 at 4.4 Å resolution.
Model building
For the hOGG1K249Q bound to an NCP-8-oxodGuo-137 model (PDB: 9eoz), we performed rigid body fitting in UCSF Chimera55, utilizing the available crystal structures of DNA-bound hOGG1 (PDB: 1 ebm) and the NCP composed of the Widom 601 DNA sequence (PDB: 3lz0). The model building process was conducted manually in COOT56, incorporating secondary structure restraints for protein and libg restraints for DNA. This was followed by alternating rounds of real-space refinement in PHENIX 1.1957,58 and molecular dynamics simulations in ISOLDE v1.2.259. The 3D Flex refined cryo-EM density map was utilized for refinements, and for manual building in COOT, structural information from the DeepEMhancer post-processed map was also considered.
Construction of a protein-bound DNA model by Web 3DNA 2.0 webserver
The models depicting 6 hOGG1 molecules bound to one 145 bp 601 DNA were constructed using composite component of Web 3DNA 2.0 (http://web.x3dna.org). We selected the binding site number as 6 and input sequence of 145 bp 601 DNA with a B-form conformation. In the subsequent step, hOGG1K249Q (PDB: 1ebm) was employed as a template, loaded onto the DNA at various locations with random or fixed base-pair steps.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The structural model of the hOGG1/NCP-8-oxodGuo-137 complex generated in this study has been deposited in the Protein Data Bank (PDB) under accession code 9EOZ. The corresponding cryo-EM reconstructions of hOGG1/NCP-8-oxodGuo-137 complex have been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-19870. The crystal structures of canonical NCP and hOGG1K249Q/dsDNA complex used in this study are available in Protein Data Bank (PDB) under accession codes 3LZ0 and 1EBM, respectively. The sequences of oligonucleotides and protein utilized in this study are provided in the Supplementary Information. Source data are provided with this paper.
References
Cadet, J. & Davies, K. J. A. Oxidative DNA damage & repair: an introduction. Free Radic. Biol. Med. 107, 2–12 (2017).
Poetsch, A. R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 18, 207–219 (2020).
Burrows, C. J. & Muller, J. G. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 98, 1109–1151 (1998).
Cadet, J., Douki, T. & Ravanat, J. L. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc. Chem. Res. 41, 1075–1083 (2008).
Kouchakdjian, M. et al. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry 30, 1403–1412 (1991).
Shibutani, S., Takeshita, M. & Grollman, A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434 (1991).
Duarte, V. et al. Oxaluric acid as the major product of singlet oxygen-mediated oxidation of 8-oxo-7,8-dihydroguanine in DNA. J. Am. Chem. Soc. 122, 12622–12628 (2000).
Luo, W. C., Muller, J. G. & Burrows, C. J. The ph-dependent role of superoxide in riboflavin-catalyzed photooxidation of 8-oxo-7,8-dihydroguanosine. Org. Lett. 3, 2801–2804 (2001).
Fleming, A. M., Muller, J. G., Dlouhy, A. C. & Burrows, C. J. Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2’-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing. J. Am. Chem. Soc. 134, 15091–15102 (2012).
Hickerson, R. P., Chepanoske, C. L., Williams, S. D., David, S. S. & Burrows, C. J. Mechanism-based DNA-protein cross-linking of muty via oxidation of 8-oxoguanosine. J. Am. Chem. Soc. 121, 9901–9902 (1999).
McKibbin, P. L. et al. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J. Am. Chem. Soc. 135, 13851–13861 (2013).
Bai, J., Zhang, Y., Xi, Z., Greenberg, M. M. & Zhou, C. Oxidation of 8-oxo-7,8-dihydro-2′-deoxyguanosine leads to substantial DNA-histone cross-links within nucleosome core particles. Chem. Res. Toxicol. 31, 1364–1372 (2018).
Rogacheva, M. V. & Kuznetsova, S. A. Repair of 8-oxoguanine in DNA. The mechanisms of enzymatic catalysis. Russ. Chem. Rev. 77, 817–843 (2008).
Nash, H. M., Lu, R., Lane, W. S. & Verdine, G. L. The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hogg1: direct identification, ablation and chemical reconstitution. Chem. Biol. 4, 693–702 (1997).
Bruner, S. D., Norman, D. P. G. & Verdine, G. L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403, 859–866 (2000).
McGinty, R. K. & Tan, S. Nucleosome structure and function. Chem. Rev. 115, 2255–2273 (2015).
Menoni, H., Shukla, M. S., Gerson, V., Dimitrov, S. & Angelov, D. Base excision repair of 8-oxog in dinucleosomes. Nucleic Acids Res. 40, 692–700 (2012).
Bilotti, K., Kennedy, E. E., Li, C. & Delaney, S. Human OGG1 activity in nucleosomes is facilitated by transient unwrapping of DNA and is influenced by the local histone environment. DNA Repair 59, 1–8 (2017).
Bilotti, K., Tarantino, M. E. & Delaney, S. Human oxoguanine glycosylase 1 removes solution accessible 8-oxo-7,8-dihydroguanine lesions from globally substituted nucleosomes except in the dyad region. Biochemistry 57, 1436–1439 (2018).
Ren, M., Shang, M., Wang, H., Xi, Z. & Zhou, C. Histones participate in base excision repair of 8-oxodguo by transiently cross-linking with active repair intermediates in nucleosome core particles. Nucleic Acids Res. 49, 257–268 (2021).
Ren, M., Greenberg, M. M. & Zhou, C. Participation of histones in DNA damage and repair within nucleosome core particles: mechanism and applications. Acc. Chem. Res. 55, 1059–1073 (2022).
Dalhus, B. et al. Separation-of-function mutants unravel the dual-reaction mode of human 8-oxoguanine DNA glycosylase. Structure 19, 117–127 (2011).
Shigdel, U. K. et al. The trajectory of intrahelical lesion recognition and extrusion by the human 8-oxoguanine DNA glycosylase. Nat. Commun. 11, 4437 (2020).
Vasudevan, D., Chua, E. Y. D. & Davey, C. A. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010).
Pettersen, E. F. et al. UCSF chimerax: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Fogolari, F. et al. Bluues: A program for the analysis of the electrostatic properties of proteins based on generalized born radii. BMC Bioinforma. 13, S18 (2012).
Henikoff, S. & Henikoff, J. G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA 89, 10915–10919 (1992).
Eddy, S. R. Where did the blosum62 alignment score matrix come from? Nat. Biotechnol. 22, 1035–1036 (2004).
Li, S., Olson, W. K. & Lu, X.-J. Web 3dna 2.0 for the analysis, visualization, and modeling of 3d nucleic acid structures. Nucleic Acids Res. 47, W26–W34 (2019).
Fromme, J. C. & Verdine, G. L. DNA lesion recognition by the bacterial repair enzyme mutm*. J. Biol. Chem. 278, 51543–51548 (2003).
Banerjee, A., Yang, W., Karplus, M. & Verdine, G. L. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 434, 612–618 (2005).
Banerjee, A., Santos, W. L. & Verdine, G. L. Structure of a DNA glycosylase searching for lesions. Science 311, 1153–1157 (2006).
Blainey, P. C., van Oijen, A. M., Banerjee, A., Verdine, G. L. & Xie, X. S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl Acad. Sci. USA 103, 5752–5757 (2006).
Qi, Y. et al. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 462, 762–766 (2009).
Baute, J. & Depicker, A. Base excision repair and its role in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 43, 239–276 (2008).
Cook, J. C. & Delaney, S. The domino effect: nucleosome dynamics and the regulation of base excision repair enzymes. DNA 2, 248–263 (2022).
Rodriguez, Y., Hinz, J. M. & Smerdon, M. J. Accessing DNA damage in chromatin: preparing the chromatin landscape for base excision repair. DNA Repair 32, 113–119 (2015).
Rodriguez, Y. & Smerdon, M. J. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. J. Biol. Chem. 288, 13863–13875 (2013).
Maher, R. L., Wallace, S. S. & Pederson, D. S. The lyase activity of bifunctional DNA glycosylases and the 3′-diesterase activity of ape1 contribute to the repair of oxidized bases in nucleosomes. Nucleic Acids Res. 47, 2922–2931 (2019).
Prasad, A., Wallace, S. S. & Pederson, D. S. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase nth1. Mol. Cell. Biol. 27, 8442–8453 (2007).
Kennedy, E. E., Li, C. & Delaney, S. Global repair profile of human alkyladenine DNA glycosylase on nucleosomes reveals DNA packaging effects. ACS Chem. Biol. 14, 1687–1692 (2019).
Weaver, T. M. et al. Structural basis for ape1 processing DNA damage in the nucleosome. Nat. Commun. 13, 5390 (2022).
Zheng, L., Tsai, B. & Gao, N. Structural and mechanistic insights into the DNA glycosylase aag-mediated base excision in nucleosome. Cell Discov. 9, 62 (2023).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Olmon, E. D. & Delaney, S. Differential ability of five DNA glycosylases to recognize and repair damage on nucleosomal DNA. ACS Chem. Biol. 12, 692–701 (2017).
Hinz, J. M., Rodriguez, Y. & Smerdon, M. J. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proc. Natl Acad. Sci. USA 107, 4646–4651 (2010).
Zheng, S. Q. et al. Motioncor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. Ctffind4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. Cryosparc: Algorithms for rapid unsupervised cryo-em structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. Relion: Implementation of a bayesian approach to cryo-em structure determination. J. Struct. Biol. 180, 519–530 (2012).
Asarnow, D., Palovcak, E. & Cheng, Y. UCSF pyem v0.5. Zenodo. https://doi.org/10.5281/zenodo.3576630 (2019).
Punjani, A. & Fleet, D. J. 3dflex: Determining structure and motion of flexible proteins from cryo-em. Nat. Methods 20, 860–870 (2023).
Sanchez-Garcia, R. et al. Deepemhancer: A deep learning solution for cryo-em volume post-processing. Commun. Biol. 4, 874 (2021).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-em through tilting. Nat. Methods 14, 793–796 (2017).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Real-space refinement in phenix for cryo-em and crystallography. Acta crystallogr. D. Struct. biol. 74, 531–544 (2018).
Liebschner, D. et al. Macromolecular structure determination using x-rays, neutrons and electrons: recent developments in phenix. Acta crystallogr. D. Struct. biol. 75, 861–877 (2019).
Croll, T. I. Isolde: A physically realistic environment for model building into low-resolution electron-density maps. Acta crystallogr. D. Struct. biol. 74, 519–530 (2018).
Acknowledgements
This work was supported by National Key Research and Development Program of China (no. 2023YFA0913800 to C.Z.), the National Natural Science Foundation of China (no. 22377059 to C.Z.), by the Deutsche Forschungsgemeinschaft (SFB1361-project ID 393547839, TRR237-project ID 369799452, HO2489/11-1 and the Gottfried Wilhelm-Leibniz Prize to K.-P.H.), by Haihe Laboratory of Sustainable Chemical Transformations and Frontiers Science Center for New Organic Matter, Nankai University (no. 63181206 to C.Z.). Mengtian Ren was supported by the China Scholarship Council to study for 1 year as a visiting Ph.D. student at LMU. The authors thank Dr. Katja Lammens and Dr. Michael Kugler (Gene Center and Department of Biochemistry, Ludwig-Maximilians-Universität München) for their valuable suggestions on structural refinement.
Author information
Authors and Affiliations
Contributions
M.R. performed most of the biochemical experiments, as well as sample preparation for cryo-EM and data collection. F.G. and Y.F. carried out the cryo-EM data processing and structure refinement. J.M. and X.S. participated in the biochemical experiments. A.Y., M.L., Y.F. contributed advice and assistance to M.R. for biochemical experiments and Cryo-EM. Y.F. oversaw the structure deposition and submission process for the PDB database. C.Z. and K.-P.H. designed and supervised the project. M.R., F.G., Y.F., K.-P.H. and C.Z. wrote the manuscript. All authors have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ren, M., Gut, F., Fan, Y. et al. Structural basis for human OGG1 processing 8-oxodGuo within nucleosome core particles. Nat Commun 15, 9407 (2024). https://doi.org/10.1038/s41467-024-53811-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53811-3
This article is cited by
-
Structural basis of gap-filling DNA synthesis in the nucleosome by DNA Polymerase β
Nature Communications (2025)
-
Contributing factors to the oxidation-induced mutational landscape in human cells
Nature Communications (2024)