Abstract
Reproduction is tightly linked to nutrient availability and metabolic homeostasis, yet how specific metabolic pathways coordinate with cellular signaling to control oogenesis remains unclear. Through a targeted RNAi screen in the Drosophila germline, we identify Acetyl-CoA Carboxylase (Acc), the rate-limiting enzyme in fatty acid synthesis (FAS), as an essential regulator of germline stem cell (GSC) maintenance and oocyte development. Acc loss shifts cellular metabolism toward fatty acid oxidation (FAO), fueling the TCA cycle and electron transport chain, which elevates ATP levels and hyperactivates TOR signaling. This metabolic reprogramming induces excessive protein synthesis, disrupting endosomal trafficking and fusome branching, a germline-specific organelle essential for synchronized cell divisions and oocyte selection. These defects are rescued by inhibiting FAO, suppressing TOR activity, reducing protein synthesis, or restricting dietary protein intake. Our study establishes a direct metabolic–signaling–structural axis in the female germline and highlights Acc as a key metabolic checkpoint that safeguards energy balance, intracellular trafficking, and oocyte fate.
Similar content being viewed by others
Introduction
Lipid homeostasis is a crucial modulator of female reproduction and is highly influenced by nutrient input and the metabolic state of the organism. Diabetes and polycystic ovary syndrome (PCOS) are both linked to dysregulation of lipid metabolism, and women with these conditions often suffer from infertility1,2. PCOS is the most common cause of female infertility. The condition is characterized by dysregulation of oocyte maturation and ovulation3, highlighting the strong connection between lipid metabolism and oocyte development. Despite the known links between lipid metabolism and oogenesis, it remains unclear how lipid metabolism coordinates with cellular signaling pathways at a molecular level to support oogenesis.
The Drosophila ovary is an excellent model for studying the relationship between lipid metabolism, cellular signaling, and oocyte development due to its well-defined cell biology and the wide range of available genetic tools. The functional unit of the ovary, known as an ovariole (Fig. 1A), contains an anterior structure called the germarium, followed posteriorly by a series of progressively more developed egg chambers4. The germarium houses two to three GSCs at its anterior tip (Fig. 1A’), which divide continuously to sustain oogenesis5. Each GSC contains a membrane-enriched organelle (fusome) located adjacent to cap cells, a major component of the GSC niche that maintains GSCs6. Division of a GSC produces a cystoblast (CB), which undergoes four rounds of incomplete divisions to form 2-, 4-, 8-, and 16-cell cysts. During germ cell division, the fusome also grows and branches to connect the germ cells within a cyst. In the 16-cell cyst, one cell located in the posterior region of the cyst becomes an oocyte. The 16-cell cyst is then enveloped by a single layer of follicle cells to form a stage 1 egg chamber, which later matures into an egg4. Mammalian female germ cells also undergo incomplete divisions to form 8-cell cysts, but the cyst cells break apart before entering meiosis7. Given that de novo synthesized fatty acids are vital for generating cell and organelle membranes8, this observation implies a conserved role for lipid metabolism in oogenesis.
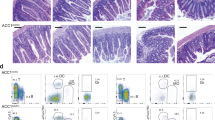
A The ovariole has developing egg chambers. A’ Germline stem cells (GSCs, pink), identified by fusome (red) next to the maintenance niche (cap cells, dark blue). GSC daughter cell cystoblasts (CBs), undergo four incomplete divisions to form 2-, 4-, 8- and 16-cell cysts (light orange). Germ cells within a cyst is connected by a branched fusome. The oocyte (green) is located posterior to each egg chamber. B Schematic of 1st screening: lipid metabolism RNAi lines were individually depleted during development (Dev) and in the adult stage at 29 °C. Female flies were then mated with wild-type (WT) males for egg laying. Sixteen candidates (11.2%) were identified. B’ Relative percentage of pupal numbers produced by females depleted of candidate genes. The number above each bar indicates the relative percentage of pupae. C Schematic of the 2nd screening: Crosses were set up at 18 °C, and newly eclosed nos > RNAi females were mated with WT males at 29 °C. Ovaries immunostained for phenotypic analysis. C’ Heat map showing the frequency of GSC loss and germ cell differentiation defect (diff. def.) in knockdowns of 8 candidates (5.6%). Due to severe GSC loss, diff. def was not determined (black boxes). D Metabolic pathway of Acc. Germaria with LamC (red, cap cell nuclear envelopes), 1B1 (red, fusomes) and DAPI (blue, DNA). Yellow lines indicate GSCs, and yellow dashed circles outline germ cell cysts with unbranched fusomes. E Percentage of germ cell cysts carrying more than 4 germ cells with unbranched or single-branched fusomes in the indicated genotypes. ***, P < 0.001. Statistical analysis was performed using the two-tailed chi-squared test. The numbers represent the cyst counts analyzed. F Ovarioles of the indicated genotypes stained with α-Spectrin (red; fusomes and follicle cell membranes) and Orb (green; oocyte marker). Yellow asterisks indicate posteriorly localized oocytes within egg chambers, denoted as normal localization. Percentages represent the proportion of egg chambers with normal oocyte localization; the number of egg chambers analyzed is shown in parentheses. D7, 7-day-old. Scale bars: 10μm in D; 20μm in F.
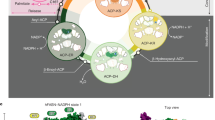
Lipids are a group of water-insoluble organic compounds composed mainly of fatty acids. These molecules play essential roles in energy storage, membrane formation, and signaling9. The two main pathways of lipid metabolism include fatty acid synthesis (FAS) and mitochondrial fatty acid oxidation (FAO, or fatty acid breakdown)8. FAS is mediated by fatty acid synthases (FASN) in the cytoplasm. The process starts with a two-carbon (2 C) acetyl-CoA incorporating a 2 C unit from malonyl-CoA, which is formed via carboxylation of acetyl-CoA by the rate-limiting enzyme Acetyl-CoA Carboxylase (Acc)10. The process continues with more additions of malonyl-CoA units, each of which lengthens the fatty acid carbon chain. Fatty acids can either be stored in the cell as triglycerides (fat) or processed into phospholipids, which form the lipid bilayers of cell membranes. Most of the acetyl-CoA used in FAS is generated from oxidation of pyruvate (glycolysis product) in the mitochondria.
However, acetyl-CoA cannot exit the mitochondria due to its charge. Instead, the acetyl-CoA molecule joins the tricarboxylic acid (TCA) cycle to form citrate, which can cross the mitochondrial membrane. In the cytoplasm, citrate lyase converts citrate back into acetyl-CoA. In contrast to FAS, FAO occurs within the mitochondria11. Fatty acids are converted into fatty acyl-CoA in the cytoplasm and then transported into the mitochondria via Carnitine palmitoyl transferase 1 (CPT1), an FAO rate-limiting enzyme located on the outer mitochondrial membrane12,13. Once inside the mitochondria, fatty acyl-CoA undergoes FAO, breaking down into acetyl-CoA while producing one NADH and one FADH2 per FAO cycle. Acetyl-CoA then enters the TCA cycle to power the electron transport chain (ETC) for ATP production. Meanwhile, NADH and FADH2 directly fuel the ETC, ultimately contributing to ATP generation. Of note, CPT1 activity can be suppressed by malonyl-CoA through a direct interaction14,15. Surprisingly, the significance of the FAS and FAO balance and the cell signaling mechanisms underlying oocyte development remain largely unclear.
One cell signaling factor that is plausibly involved in this process is TOR kinase. TOR kinase is highly conserved across species and plays critical regulatory roles in different processes related to cell growth and proliferation, including FAS, protein synthesis and autophagy. Importantly, TOR-mediated regulatory activities are typically initiated as responses to amino acid availability and energy status16. When nutrients are abundant, TOR is activated and promotes FAS through nuclear translocation of SREBP17, which acts as a transcription factor to increase transcription of SREBP itself, FASN, HMG Coenzyme A synthesis (Hmgs, for steroidogenesis) and Acetyl Coenzyme A synthase (AcCoAs)18,19. TOR also phosphorylates eukaryotic initiation factor 4E binding protein (4E-BP) to promote protein translation20.
In addition, the TOR pathway promotes synthesis of ribosome genes21. Activation of TOR leads to phosphorylation of ribosomal protein S6 Kinase (S6K), which phosphorylates S6 to promote ribosome biogenesis22, thereby facilitating protein translation. Furthermore, the TOR pathway suppresses autophagy23. Studies in Drosophila have shown that TOR signaling can regulate oogenesis by controlling ovarian GSC proliferation and maintenance24, as well as entry into meiosis25. In mammals, TOR signaling controls the growth of primordial follicle26,27, which is formed after germ cell cyst breaks apart, and the individual oocyte is surrounded by a layer of somatic cells28; of note, this mechanism is linked to PCOS29,30. Nevertheless, little is known about the role of TOR signaling in the germ cell cyst and whether it is related to the metabolic balance between FAS and FAO.
In this study, we demonstrate that Acc-mediated lipid metabolism is essential for the maintenance of GSCs and differentiation of the germ cell cyst to support female reproduction. Depletion of Acc boosts FAO, leading to increased production of α-ketoglutarate (α-KG), Glutamine (Gln), and ATP. Elevation of these metabolites activates TOR signaling, which then enhances protein synthesis. The resultant protein excess disrupts endosomal trafficking, impairing fusome branching, germ cell differentiation, and oocyte determination. However, it is possible to mitigate these disruptions by either inhibiting FAO or TOR signaling, reducing protein synthesis, or decreasing dietary protein levels. Overall, our study shows that homeostatic lipid metabolism maintains a balanced energy state to regulate TOR signaling and support oocyte development. Given that TOR hyperactivation and defective oocyte maturation are hallmarks of PCOS31, our findings offer mechanistic insight into how lipid metabolic imbalance in germ cells—distinct from known somatic follicle cell defects—may contribute to PCOS-related infertility.
Results
Two-step RNAi-based screen identifies Acc controls GSC maintenance, fusome branching, and oocyte determination
To understand how lipid metabolism regulates oogenesis, we performed a two-step RNAi-based screen using the TRiP (Transgenic RNAi project) collection32, which targets 83% of the lipid metabolic enzymes (143 out of 172) listed in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. In the initial phase of the screening (Fig. 1B), we set up a rapid screen to identify enzymes potentially involved in oogenesis. For this purpose, we assessed pupae production, as one pupa was resulted from one fertilized egg after knocking down enzymes in the germline during both development and adulthood using the nanos (nos)-GAL4 driver at 29 °C. Female flies with the RNAi-expressing germline were mated with wild-type males, and the resulting pupae were counted (see details in the Materials and Methods section). We identified 16 enzymes (11.2% of those screened) for which depletion reduced pupae production by at least 50% compared to the control (nos>gfpRNAi) (Fig. 1B’ and Supplementary Data. 1). Of these enzymes, eight are involved in synthesizing neutral lipids, such as triacylglycerol for energy storage. The others synthesize polar lipids known for their amphiphilic nature (hydrophilic head and hydrophobic tails), such as phospholipids.
In the second phase of screening (Fig. 1C), we individually knocked down the 16 enzymes in the adult ovarian germline by shifting newly eclosed flies from 18 °C to 29 °C for 7 days to induce GAL4-driven RNAi expression. We then analyzed the ovaries in terms of GSC count and the number of germ cells within a cyst in each indicated genotype. GSCs were identified by their fusome (labeled by 1B1 staining) adjacent to cap cells (labeled by LamC staining) (see Fig. 1D), and germ cell number in a cyst was assessed by counting DAPI-labeled nuclei. GSC count of less than two per germarium indicated a GSC loss phenotype (a healthy germarium houses at least 2 GSCs), while deviations from the typical 2 N pattern in germ cell numbers within a cyst suggested defects in germ cell differentiation. The frequency of these phenotypes was expressed as the percentage of germaria showing GSC loss or differentiation defects, as previously described33. Of the 16 enzymes, four showed GSC loss phenotypes (Fig. 1C’ and Fig. S1). Among the four enzymes associated with GSC loss, Acc carboxylates acetyl-CoA to form malonyl-CoA, a substrate for FAS, while also inhibiting FAO by binding to CPT1, which controls fatty acid import into mitochondria (Fig. 1D)10.
Notably, only Acc depletion resulted in fusome branching abnormalities (Fig. 1D, and Fig. S1). In cysts with more than four germ cells (n = 137), 55% showed unbranched or single-branched fusomes, termed “flower cysts,” in which germ cells surrounded a defective fusome. In contrast, the remaining cysts displayed multiple-branched fusomes, similar to those in control cysts (n = 130) (Fig. 1E). The efficiency of the AccRNAi line was demonstrated by a significant reduction in Acc expression in Acc-depleted ovaries at both the mRNA and protein levels (Fig. S2A and B). Additionally, Acc1 null mutant GSCs (absence of GFP), generated using the FLP-FRT-mediated recombination system34, were rapidly lost, with only a few mutant cysts displaying fusome branching defects, likely due to early cell death before reaching the 16-cell stage (Fig. S2C and C’).
To validate the role of Acc in the germline, we knocked down Holocarboxylase synthetase (Hcs), a biotin ligase essential for the activation of carboxylases, including Acc35, as well as the other four carboxylases and two biotinidases, which recycle biotin to maintain cellular levels36. Strikingly, only Hcs-depletion fully replicated the defects observed in Acc-depleted germ cells (Fig. 1D and E; Fig. S3). These results indicate a specific role for Acc-mediated lipid metabolism in fusome branching, a process that is critical for oocyte development37. Indeed, oocytes, as labeled by Orb38, were not observed in posterior of Acc-depleted or Hcs-depleted egg chambers, in sharp contrast to the control (Fig. 1F). These results indicate that germline Acc is essential for oogenesis.
Acc loss in GSCs reduced niche-GSC attachment and unbranched fusomes in germ cell cysts
To investigate how Acc impacts the female germline, we analyzed Acc-depleted germ cells using various markers. Drosophila ovarian GSCs are maintained in the niche through BMP stemness signaling and E-cadherin-mediated GSC-niche attachment5. Furthermore, GSC differentiation is driven by Bag of marbles (Bam), a master differentiation factor39. When germ cells enter meiosis, Bam expression is suspended, and the cells exhibit meiosis-induced double-strand breaks (DSBs)40, which are later repaired when germ cells enter the follicle cell zone40. We found that Acc depletion reduced E-cadherin expression at the GSC-niche junction (Fig. S4A), while BMP signaling (assessed by pMad expression) remained unaffected (Fig. S4B). In contrast, Bam expression (revealed by Bam-GFP expression) was gone from flower cysts (Fig. S4 C), indicating disrupted germ cell differentiation.
Consistent with our previous findings41, ~50% of Acc-depleted germ cells (n = 53) exhibited multiple spindle poles (Fig. S4D). Together with the observation that these cells failed to follow the 2 N rule, this indicates defective mitotic division upon Acc depletion. In addition, Acc-depleted germ cells displayed an abnormal DSB pattern (labeled by γH2AV), which covered the entire nucleus, in contrast to the punctate signals observed in the control (Fig. S4E). Furthermore, apoptotic germ cell cysts were present in 60% of Acc-depleted germaria (n = 60), compared to 16% of control germaria (n = 56) with apoptotic germ cell cysts. (Fig. S4F), indicating cell death occurred within these cysts.
To determine whether the fusome branching defect resulted from Acc depletion in GSCs or differentiated cysts, we selectively knocked down Acc in each population (Fig. S4G). Knocking down Acc in germ cell cysts with bam-GAL410 did not lead to unbranched fusomes. However, Acc knockdown in GSCs, using nos-GAL4 and suppression of GAL4 activity in germ cell cysts with bam-GAL8042, led to unbranched fusomes. This suggests the defect may arise from Acc-depleted GSCs, although we cannot exclude AccRNAi expression in 16-cell cysts due to limited bam-GAL80 activity or insufficient bam-GAL4 strength in germ cell cysts.
Acc depletion causes unbranched fusomes through elevated FAO
Acc depletion is expected to reduce FAS while enhancing FAO, based on its known metabolic function (see Fig. 1D). To investigate the contribution of these metabolic changes to the germline phenotypes observed upon Acc depletion, we mimicked reduced FAS by suppressing FASN and alleviated the phenotypes by inhibiting FAO. In Drosophila, there are three FASNs, including FASN1, FASN2 and FASN343; of these three, FASN2 is minimally expressed in the ovary44. To inhibit FAS, we knocked down FASN1 and FASN3 in the germline and introduced a loss-of-function allele for FASN2 (FASN2EP695/ + )45, hereafter referred to as FASN[Dep]. FAO suppression was achieved by co-knocking down Acc and Withered, Drosophila CPT1 (dCPT1)46, which facilitates the import of fatty acids into mitochondria.
We validated the suppression of FAS and FAO by assessing neutral lipid levels using BODIPY staining (Fig. 2A and B). Lipid droplets were dramatically reduced in germ cells depleted of Acc, Hcs, or FASN, indicating impaired FAS. Co-knockdown of and dCPT1 partially restored lipid droplet levels, confirming effective FAO suppression. Quantification (Fig. 2B) showed that control and dCPT1-knockdown GSCs had an average of about three lipid droplets, whereas Acc- or Hcs-depleted GSCs had nearly none. Co-depletion of Acc and dCPT1, or Hcs and dCPT1, as well as FASN depletion, resulted in approximately half the number of lipid droplets in GSCs compared to controls. FAS disruption modestly reduced GSC numbers, while FAO suppression only slightly improved GSC numbers in Acc- or Hcs-depleted germlines. (Fig. 2C).
A Germaria with LamC (red, cap cell nuclear envelopes), 1B1 (red, fusomes), and BODIPY (green, neutral lipid droplets). Yellow lines indicate GSCs, and the yellow dashed circle outlines cysts with unbranched fusomes. B Number (no) of lipid droplets (LDs) per GSC. Each dot represents a GSC. C GSC number in the germaria in the indicated genotypes. The left y axis shows the proportion of germaria carrying the indicated GSC number. The right y axis shows the average GSC number per germarium (D) Percentage of germ cell cysts carrying at least 4 germ cells with unbranched or single-branched fusomes. E Ovarioles with α-spectrin (red, fusomes, and follicle cell membranes) and Orb (green, an oocyte marker). Yellow asterisks indicate posteriorly located oocyte in the egg chamber, denoted as normal localization. Percentages represent the proportion of egg chambers with normal oocyte localization; parentheses show egg chamber numbers. F Ovaries carrying mature eggs (white and big eggs, asterisks), denoted as normal. Percentages represent the proportion of normal ovaries; the number of ovaries imaged and analyzed is shown in parentheses. G Number of eggs produced per female of the indicated genotype on the fourth day after mating. N = 4 biological replicates were performed. Orange-filled and unfilled circles on the x axis in (B, D, and G) represent the expression dCPT1RNAi and gfpRNAi, respectively. Error bars indicate mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Asterisks indicate significant differences compared with control (gfpRNAi & mCherryRNAi), or comparisons between two groups indicated by a line. Statistical analysis in B, C, and G were performed using the unpaired two-tailed Student’s t-test, and in (D) using the two-tailed chi-squared test. The numbers above each bar in C or beside each bar in D represent the number of germaria or germ cell cysts analyzed, respectively. UAS-RNAi expression was driven by nos-GAL4; D7, 7-day-old. Scale bars: 10 μm in A, 20 μm in E, and 1 mm in F.
Notably, FAS disruption did not affect fusome branching, but FAO suppression in Acc- or Hcs-depleted germlines significantly restored defects of fusome branching (Fig. 2D) and oocyte determination (Fig. 2E). Similar rescue effects were observed with the FAO inhibitor etomoxir (Fig. S5). Additionally, co-knockdown of Acc with Hepatocyte nuclear factor 4 (Hnf4), a master transcription factor of FAO genes47, also suppressed the unbranched fusome phenotype (Table S1). While Acc- or Hcs-depleted ovaries were smaller and sterile, FAS suppression reduced ovary size and egg production to half that of the controls (Fig. 2F and G). In contrast, FAO suppression increased ovary size and fertility in Acc- or Hcs-depleted ovaries (Fig. 2F and G). These findings demonstrate that both FAS and FAO are essential for GSC maintenance and fecundity, with elevated FAO specifically affecting fusome morphology and oocyte determination.
Elevated FAO in Acc-depleted germline hyperactivates TOR signaling
To explore how Acc regulates oogenesis, we performed RNA-sequencing (RNA-seq) on RNA isolated from 7-day-old control (nos>mCherryRNAi) and Acc-depleted ovaries (nos>AccRNAi) (Fig. S6). We identified 2355 genes with significant expression changes ( > 1.5-fold, q < 0.05); 1459 genes were upregulated, and 896 genes were downregulated in Acc-depleted ovaries. Gene set enrichment analysis (GSEA) revealed that genes involved in fatty acid biosynthesis, oocyte determination, and cyst formation were downregulated, in line with the observed phenotypes. Interestingly, genes encoding ribosomal units were upregulated, suggesting an increase in ribosome biogenesis. Correspondingly, nucleoli (labeled by Fibrillarian), where ribosome biogenesis occurs48, were larger in the Acc-depleted germline (Fig. 3A), suggesting increased ribosomal biogenesis49.
A Control (mCherryRNAi) and ACC-knockdown (KD) germaria with α-spectrin (red, fusomes and follicle cell membranes) and Fibrillarin (yellow, nucleoli) labeling. B Germaria expressing the autophagy flux sensor GFP-mCherry-Atg8a (green and red) in combination with a controlRNAi or ACCRNAi. Arrows indicate autolysosomes (red), and arrowheads indicate autophagosomes (bule). The percentages represent the proportion of germaria exhibiting large puncta of Fibrillarin (A) and yellow (B) signals in germaria (outlined by dashed lines), respectively. The number of germaria analyzed is indicated in parentheses. [Note that endoreplicating nurse cells in the egg chamber also exhibit large Fibrillarin puncta.] C Representative immunoblot shows expression of p4E-BP (Thr37/46) and pS6K (Thr398) in control and Acc-KD ovaries.N = 3. Actin was used as a loading control. Molecular weight markers are indicated to the right of the blots. D Germaria of the indicated genotypes with LamC (red, cap cell nuclear envelopes), 1B1 (red, fusomes), and p4E-BP (green, TOR signaling reporter) labels. E Germaria with the indicated genotypes with LamC (red), 1B1 (red), and OPP (green, translation rate) labels. F p4E-BP intensity per germarium in the indicated genotypes. G, G’ OPP intensity per GSC (G) and germarium (G’) in the indicated genotypes. Orange-filled and unfilled circles on the x axis in (F, G and G’) the expression of dCPT1RNAi and gfpRNAi lines, respectively, along with the indicated genotypes. Each dot in F and G’ represents a germarium and in G represents a GSC. Error bars indicate mean ± SD. ***, P < 0.001. Asterisks indicate significant differences compared with the control, or the comparisons between two groups indicated by a line. Statistical analyses were performed using the unpaired two-tailed Student’s t-test. UAS-RNAi expression was driven by nos-GAL4. D7, 7-day-old. Scale bars are 10 μm.
As Acc suppression has been shown to repress autophagy in yeast50, we examined autophagy in Acc-depleted germ cells using the mCherry-GFP-Atg8 reporter (Fig. 3B). In control germ cells, Atg8-labeled autophagosomes appeared yellow (positive for both GFP and mCherry), while autolysosomes appeared red due to the fusion of autophagosome with lysosomes, where the acidic environment quenches GFP51. In contrast, Acc-depleted germ cells showed an accumulation of large yellow puncta, indicating impaired autophagic flux. Increased ribosomal biogenesis, suppressed autophagy, and upregulation of ribosome genes and TOR signaling downstream targets (FASN1, SREBP, Hmgs, and AcCoAs)18,19 in Acc-depleted ovaries (Fig. S7A) collectively suggest elevated TOR signaling.
To test this idea, we assessed the phosphorylation of TOR substrates S6K and 4E-BP (Thor in fly)20,22, and S6K substrate (S6)22, in control and Acc-depleted ovaries. Western blot showed increased levels of pS6K and p4E-BP in Acc-depleted ovaries (Fig. 3C). In germaria, p4E-BP was expressed throughout the germline, consistent with a previously report52, while pS6 was detected in a subset of germ cells but was not restricted to a specific germ cell stage (Fig. 3D, Fig. S7B and Supplementary Data 1). Both p4E-BP and pS6 expressions were significantly decreased by treatment with rapamycin (Fig. S7B and C), a TOR inhibitor53, confirming TOR dependence.
Consistently, both p4E-BP and pS6 expressions were increased in Acc- or Hcs-depleted germlines (Fig. 3D and F, and Fig. S7D), indicating elevated TOR signaling. This increase was disrupted by FAO suppression via dCPT1 knockdown (Fig. 3D and F, and Fig. S7D). In contrast, FAS suppression did not affect pS6 expression in the germline (Fig. S7D). Increased pS6 and p4E-BP levels suggest enhanced TOR-dependent protein translation. Thus, we analyzed protein translation rates in Acc-depleted germ cells using an O-propargyl-puromycin (OPP) incorporation assay54. Germ cells with depleted Acc exhibited higher OPP incorporation compared to controls, and this increase was suppressed by FAO inhibition (Fig. 3E, G and G’). Taken together, our results demonstrate that germline depletion of Acc activates TOR signaling and protein synthesis via increased FAO.
Acc depletion activates TOR-S6K-dependent protein synthesis, causing excess protein burden that disrupts fusome branching
To investigate whether activated TOR signaling in the Acc-depleted germline leads to GSC loss, unbranched fusomes and oocyte determination failure, we treated control and Acc-depleted flies with rapamycin to suppress TOR signaling. Rapamycin treatment reversed GSC loss and fusome unbranching phenotypes in Acc-depleted ovaries, but it had no effect on control flies (Fig. 4A, B and D). Similarly, reducing TOR or Insulin receptor (InR) expression, which mediates insulin signaling upstream of TOR20, in Acc-depleted ovaries rescued these defects to levels comparable to rapamycin treatment (Table S1). These findings indicate that Acc depletion activates TOR signaling, disrupting GSC maintenance and fusome branching.
A Germaria of indicated genotypes with or without rapamycin treatment (rapa) were labeled with LamC (red, cap cell nuclear envelopes) and 1B1 (red, fusomes and follicle cell membranes) or α-spectrin (red, fusomes and follicle cell membranes). Yellow lines indicate GSCs and yellow dashed circles outline germ cell cysts with unbranched fusomes. B, C Number of GSCs in the germaria in the indicated genotypes. The left y axis indicates the proportion of germaria with the indicated GSC number. The right y axis shows the average GSC number per germarium. Error bars indicate mean ± SEM. D Percentage of germ cell cysts (carrying at least 4 germ cells) in the indicated genotypes carrying single-branched or unbranched fusomes. Statistical analysis was performed using two-tailed chi-squared test. Orange-filled and-unfilled circles on the x axis in (B–D) indicate the presence or absence of the indicated RNAi. The number of germaria analyzed in B and C and germ cell cysts analyzed in D are shown above each bar. Yellow background indicates the flies were treated with rapamycin. *, P < 0.05; **, P < 0.01; ***, P < 0.001. NS, not significant. Black asterisks in B and C indicate significant differences compared with control, in D compared with Acc-KD group, or comparisons between two groups as indicated by a line. Red asterisks in C indicate significant differences compared to ACC-KD group. Statistical analysis in (B, C) were performed using the unpaired two-tailed Student’s t-test, and in D by the chi-squared test. E Ovarioles of the indicated genotypes with α-spectrin (red, fusomes and follicle cell membranes) and Orb (green, oocyte marker). Yellow asterisks indicate posteriorly located oocyte in the egg chamber, denoted as normal localization. The percentage shows the proportion of egg chambers with normal localization, and the number of egg chambers analyzed is shown in parentheses. Scale bars, 10 μm in A, 20 μm in E. RNAi expression was driven by nos-GAL4. D7, 7-day-old.
Interestingly, while TSC1 depletion, which increases TOR activity caused GSC loss24, it did not lead to fusome branching defects (Table S1), likely due to differing TOR signaling strength in the Acc-depleted germline. While we cannot rule out the possibility that Acc depletion affects other metabolic or signaling pathways, our data support a model in which elevated TOR activity, in combination with additional Acc-regulated processes, disrupts fusome morphology and oocyte development.
To understand how TOR signaling disrupts germline homeostasis upon Acc depletion, we examined TOR downstream effects. Since TOR signaling suppresses autophagy, we tested whether this suppression contributes to the phenotypes observed. To mimic autophagy suppression, we knocked down Atg8a, a critical regulator of autophagy55, in the germline. Atg8a-depletion slightly decreased GSC number without affecting fusome branching, whereas co-knockdown Atg8a with Acc did not worsen Acc depletion phenotypes (Fig. 4A, B and D). However, co-knockdown reduced the effectiveness of rapamycin in rescuing GSC numbers (Fig. 4B), aligning with the role of autophagy in maintaining E-cadherin levels essential for GSC maintenance56. Similarly, autophagy suppression by chloroquine treatment57 also produced comparable results (Fig. S8). These results indicate that functional autophagy is required for GSC maintenance but not for fusome branching.
TOR signaling also induces the nuclear localization of SREBP, which activates the FASN and Hmgs transcription to promote FAS and steroidogenesis17, respectively. However, co-knockdown of Acc with SREBP or Hmgs failed to rescue GSC loss or fusome branching defects (Fig. 4A, C and D). This suggests that the TOR-dependent activation of FAS and steroidogenesis pathways is not required for these phenotypes—particularly since malonyl-CoA, the key substrate for FAS, is still lacking in Acc-depleted germ cells. Strikingly, co-knockdown of Acc and S6K fully rescued fusome branching defects but not GSC loss (Fig. 4A, C and D). While knocking down 4E-BP, an inhibitor of protein translation58, did not induce flower cyst formation (Table S1), suppressing protein translation with cycloheximide treatment restored fusome branching defects in Acc-depleted germline (Fig. S9). This suggests that excess protein synthesis driven by TOR-S6K signaling disrupts fusome branching in the Acc-depleted germline, and increased translation via 4E-BP) depletion alone is insufficient to cause fusome defects.
Furthermore, while Acc depletion completely disrupted oocyte determination, rapamycin treatment rescued oocyte formation in 62% of egg chambers (n = 480. In contrast, co-knockdown of Acc and S6K rescued oocyte formation only in 22% of egg chambers (n = 468) (Fig. 4E), suggesting additional factors influence oocyte formation under Acc depletion. In summary, our results demonstrate that under Acc depletion, increased FAO activates TOR signaling, and the resulting TOR–S6K-mediated protein synthesis is a predominant factor disrupting fusome branching and oocyte formation.
ACC depletion-induced TOR-S6K–dependent protein synthesis disrupts fusome branching, at least in part by impairing endosomal trafficking
Protein overload can stress the endosomal trafficking system59, which transports proteins into various cellular compartments, and may contribute to the fusome branching defects induced by Acc depletion.
This system comprises different types of endosomes60. Early endosomes, enriched and regulated by Rab5, receive internalized materials from the plasma membrane, serving as a primary sorting station. These early endosomes can mature into late endosomes (enriched and regulated by Rab7), which fuse with lysosomes expressing Lamp-1 for cargo degradation. Alternatively, materials can be sent to recycling endosomes (enriched and regulated by Rab11), which return essential molecules back to the plasma membrane. Our RNA-seq results showed that genes involved in protein transport were downregulated in Acc-depleted ovaries (see Fig. S6C), suggesting that endosomal trafficking was disrupted. We thus examined the expression patterns of Rab5, Rab7, Rab11, and Lamp1 using a GFP knock-in line (Lamp-1-GFP).
Acc depletion did not significantly affect Rab5 expression (Fig. S10A). However, the expression levels of Rab7 and Lamp1-GFP were elevated, accompanied by increased punctate signals compared to controls (Fig. 5A and B). Strikingly, reducing FAO (via dCPT1 knockdown) or inhibiting S6K-dependent protein synthesis (via S6K knockdown) in the Acc-depleted germline restored Rab7 expression to the control pattern (Fig. 5A), along with the rescue of fusome branching defects. In contrast, Rab11 expression was reduced in the Acc depletion germline (Fig. 5C). Acc depletion causes a global disruption of endosomal trafficking in the germline.
A Germaria of the indicated genotypes with α-spectrin (red, fusomes and follicle cell membranes) and Rab7 (green, late endosomes). B Control and ACC-knockdown (KD) germaria expressing Lamp1-GFP (green, lysosomes) were labeled with LamC (red), 1B1 (red). Yellow dashed circles in A outline germ cell cysts with unbranched fusomes. The percentages shown indicate the proportion of germaria displaying large punctate Rab7 (A) and Lamp1-GFP (B) signals, respectively; the number of germaria analyzed is provided in parentheses. C Control and ACC-KD germaria were labeled with α-spectrin (blue, fusomes), Rab11 (green, recycling endosomes), and E-cadherin (E-cad, red). Enlarged views of the square in germaria are shown to the right with or without the Rab11 channel. Yellow lines indicate GSCs; arrows indicate the presence of E-cad in Rab11-labeled endosomes. D Pearson’s correlation coefficient values for E-cad and Rab11 colocalization in the germarium in the indicated genotypes. Each dot represents a germarium. Orange-filled and-unfilled circles on the x axis indicate the presence of S6KRNAi and gfpRNAi, respectively. Error bars indicate the mean ± SD. **, P < 0.01; ***, P < 0.001. NS, not significant. Asterisks indicate significant differences compared with the control, or comparisons between two groups indicated by a line. Statistical analyses were performed using the unpaired two-tailed Student’s t-test. Scale bars, 10 μm. UAS-RNAi expression was driven by nos-GAL4. D7, 7-day-old.
Rab11 is known to mediate E-cadherin recycling in the germline61; we found that Acc depletion decreased the colocalization of E-cadherin in Rab11-labeled recycling endosomes (Fig. 5C). Suppressing S6K expression alone did not affect Rab11/E-cadherin colocalization, whereas Acc co-knockdown with S6K rescued the reduction of Rab11 and Rab11/E-cadherin colocalization in the germline, as compared to Acc depletion alone (Fig. 5C). Furthermore, a previous study showed that Rab11 mutant female germ cells display unbranched fusomes, deformed oocytes, and E-cadherin recycling failure61, mimicking Acc depletion phenotypes. However, overexpression of wild-type or constitutively active Rab11 (Rab11CA)62, did not rescue fusome branching defects (Fig. S10B and Table S1). These results indicate that Acc depletion disrupts fusome branching, at least in part via the disruption of endosomal trafficking, which are caused by elevated FAO and TOR-dependent protein synthesis.
Lysosomes can also fuse with autophagosomes (positive for Atg8a) to form autolysosomes (positive for both Atg8a and Lamp1) that mediate degradation of macromolecules51. We found that both autophagosomes and autolysosomes were increased in the Acc-depleted germline (Fig. S10C and D), consistent with our previous finding that autophagic flux is disrupted when Acc is depleted (see Fig. 3B, and Fig. S10E). Rapamycin treatment or co-knockdown of dCPT1 reduced the accumulation of Atg8a-labeled autophagosomes and autophagic flux (Fig. S10C’-E’). Taken together, these results indicate that FAO-TOR activation in the germline disrupts global endosomal trafficking and protein degradation.
Increased FAO-TCA-ETC flux activates TOR signaling in Acc-depleted germ cells
We next asked how increased FAO activates TOR signaling. Acetyl-CoA is generated in mitochondria, either from pyruvate (glycolysis product) or FAO, and is then converted into citrate, which fuels the TCA cycle to supply reducing equivalents to the ETC chain for ATP production (Fig. 6A). α-KG is an intermediate in the TCA cycle. Glutamine (Gln) is converted from Glutamate (Glu), which is derived from α-KG. α-KG and Gln are known activators of the TOR pathway63. To understand how Acc depletion alters metabolism in the germline, we performed metabolomic analysis on control and Acc-depleted ovaries (Fig. 6A). Our results showed that acetyl-CoA, citrate, α-KG and Gln levels were all elevated in Acc-depleted ovaries. However, levels of Glu and malate, another TCA intermediate that is converted into oxaloacetate (OAA), remained unchanged. Interestingly, fumarate (converted into malate) levels were significantly reduced in Acc-depleted ovaries.
A Diagram of acetyl-CoA-related metabolic pathways and the relative abundances of intracellular metabolites in ACC-knockdown (KD) ovaries. Metabolic enzymes are shown in blue; increased and decreased metabolites in Acc-depleted ovaries are shown in green and red, respectively. I, II, III, IV and V represent the corresponding five complexes of the ETC chain. The conversion of α-KG to succinyl-CoA produces NADH, which donates electrons to complex I, where the conversion of succinate to fumarate generates FADH2, which donates electrons to complex II. Graphs show the relative levels of the metabolites in ACC-KD ovaries (red circles) compared to controls (black circles); each red symbol represents a biological replicate. B Control and ACC-KD germaria expressing mRuby3-iATPSnFR (ratio-metric ATP sensor) with LamC (blue, cap cell nuclear envelopes) and 1B1 (blue, fusomes) labels. mRuby3 (red) is a reference fluorescent signal; iATPSnFR (green) indicates ATP levels. The images on the right show the ATP levels (GFP/mRuby3 ratio) from low (blue) to high (yellow). White dashed lines outline germaria. C GFP/mRuby3 ratio per germarium with the indicated genotype. D Germaria of the indicated genotypes with LamC (red), 1B1 (red) and p4E-BP (green, TOR signal reporter) labels. Yellow dashed circles outline germ cell cysts with unbranched fusomes. E Expression of p4E-BP per germarium in the indicated genotypes. Orange-filled and unfilled circles on the x axis indicate the presence of ACCRNAi and gfpRNAi, respectively, along with the indicated genotype. Each dot in C and E represents a germarium. Error bars, mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. NS, not significant. Black asterisks indicate significant differences compared with the control (gfpRNAi &mcherryRNAi), and red asterisks in E indicate significant differences compared with the ACC-KD alone group. Statistical analyses were performed using the unpaired two-tailed Student’s t-test. Scale bars, 10 μm. UAS-RNAi was driven by nos-GAL4; D7, 7-day-old.
To determine whether FAO elevation in Acc-depleted germ cells increases ATP production, we used an intensity-based ATP sensing Fluorescent Reporter, iATPSnFR64. This reporter contains a superfolder GFP inserted into the ATP-binding region of the epsilon subunit of the F0-F1 ATPase from Bacilus subtilis; upon ATP binding, the reporter undergoes a conformational change, and shows increased fluorescence. Depletion of ATP synthase subunits, ATPsynE, in the germline caused a dramatic reduction in GFP fluorescence intensity (Fig. 6B and C), confirming the reliability of the ATP reporter. As expected, the Acc-depleted germline displayed about 1.2-fold GFP fluorescence compared to controls. These results indicate that Acc depletion increases FAO-TCA-ETC flux for ATP generation.
To validate the role of the FAO-TCA-ETC flux in TOR activation, we conducted a series of genetic manipulations to eliminate the elevated metabolites observed in Acc-depleted ovaries, using either RNAi lines or heterozygous mutants. We co-depleted Acc along with several different enzymes including: AcCoAS, which converts acetate to acetyl-CoA65; Citrate synthase 1 (CS1) or CS2, enzymes responsible for converting acetyl-CoA to citrate66; Isocitrate dehydrogenase 3 non-catalytic subunit beta (Idh3b), which helps convert isocitrate to α-KG67; Glutamine synthase 1 (GS1), which converts Glu to Gln68; and NADH dehydrogenase (ubiquinone) 19 kDa subunit (ND-19) and NP15.6, subunits of Complex I69. We first examined if these genetic manipulations could rescue the fusome branching defect in the Acc-depleted germline (Fig. S11), given that elevated TOR signaling disrupted fusome branching.
Strikingly, these genetic manipulations greatly reduced the fusome branching defect (from 51% to 4% of analyzed germ cell cysts), except that Acc and CS1, and Acc and CS2 co-knockdown only rescued the unbranched fusome phenotype in 11% (P = 0.06, n = 76 germ cell cysts) and 14% of analyzed cysts (P < 0.05, n = 66 germ cell cysts), respectively. This weaker rescue is likely due to the redundancy of CS1 and CS2 for citrate synthesis66. Indeed, the elevated expression of p4E-BP in the Acc-depleted germline was suppressed by these genetic manipulations (Fig. 6D and E). These results indicate that the elevated FAO-TCA-ETC flux activates TOR signaling in the germline when Acc is depleted.
Protein starvation restores Acc depletion phenotypes in the germline
Dietary composition profoundly influences metabolism. Since Acc depletion results in excessive energy in the germline, we fed Acc-depleted flies a protein-poor diet, in which protein only comprised 5% of the calories provided by sucrose70,71. Acc-depleted flies fed with the protein-poor diet for 7 days showed significant rescue of GSC loss and unbranched fusome phenotypes compared to flies fed with a protein-rich (normal diet) diet (Fig. 7A–C). In contrast, protein starvation did not noticeably affect GSC number and fusome morphology in control flies (Fig. 7A–C). Additionally, protein starvation rescued oocyte formation in 37% of egg chambers (n = 327) in Acc-depleted ovaries, compared to no oocytes in Acc-depleted ovaries under a protein-rich diet formation (336 egg chambers examined) (Fig. 7D). These results indicate that protein starvation can suppress the phenotypes caused by Acc-depletion.
A Germaria with LamC (gray, cap cell nuclear envelopes), and 1B1 (gray, fusomes) under protein-rich or protein-poor diets. B Number of GSCs in germaria. The left y axis indicates the proportion of germaria with the indicated GSC number. The right y axis shows the average GSC number. C Percentage of cysts carrying single branched and unbranched fusomes. D ACC-depleted ovarioles on protein-rich or -poor diet with α-spectrin (red, fusomes and follicle cell membranes) and Orb (green, oocyte marker). Yellow asterisks indicate the posteriorly located oocyte (NL) in the egg chamber. The percentage shows the proportion of egg chambers with normal localization of oocytes, parentheses shows egg chambers number. E ACC-depleted germaria from flies on protein-rich or -poor diet with LamC (red) and 1B1 (red). F Percentage of remaining GSCs represents the average number of GSCs at each time point normalized to D0 of same genotype. Flies on a protein-rich diet for the first 4 days, and by a protein-poor diet for 4 days. G Percentage of germ cell cysts with single-branched and unbranched fusomes. The number of germaria analyzed in B and cysts analyzed in C and G is shown above each bar. Error bars in B are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant. Asterisks indicate significant differences compared with the control, or comparisons between two groups indicated by a line. Yellow outlines on symbols in F indicate that the percentage of remaining GSCs was significantly different than D0 of same genotype. Statistical analysis in (B, F) were performed using the unpaired two-tailed Student’s t-test, in C and G using the two-tailed chi-squared test. Orange-filled and unfilled circles on the x axis in (B, C, and G) indicate with and without a protein-poor diet, respectively. Yellow lines indicate GSCs, and yellow dashed circles outline cysts with unbranched fusomes. Scale bars: 10 μm in (A, E) and 20 μm in (D). UAS-RNAi was driven by nos-GAL4. D0, D4, D7 and D8 are newly eclosed, 4-, 7- and 8-day-old.
We next asked whether protein starvation could reverse Acc-depleted phenotypes. Depletion of Acc in the adult germline for 4 days caused both GSC loss and fusome branching defects (Fig. 7E and F), similar to those observed with 7 days of knockdown. Strikingly, Acc-depleted flies cultured on a protein-rich diet for 4 days and then switched on a protein-poor diet for an additional 4 days showed a 29% rescue of GSC number, as compared to Acc-depleted flies maintained on a protein-rich diet for 4 days (Fig. 7E and F). In addition, switching from a protein-rich to a protein-poor diet rescued the unbranched fusome phenotype (Fig. 7E and G). Thus, we concluded that a protein-poor diet could both prevent and reverse germline phenotypes caused by Acc depletion.
Interestingly, while Acc depletion in the male germline caused GSC loss, it did not affect fusome branching or male fertility (Fig. S12). We speculated that male and female germ cells may employ different lipid metabolism strategies. Furthermore, overexpression of the human orthologue of fly Acc, ACACA, in the Acc-depleted germline, largely rescued the unbranched fusome phenotype (Fig. S13), suggesting functional conservation between fly Acc and human ACACA, and a potential dietary therapeutic approach for metabolic disorders associated with ACACA deficiency.
Discussion
Acc sustains energy homeostasis for oogenesis
Lipid metabolism is essential for germ cell development and reproduction, yet its specific role in oogenesis remains incompletely understood. While one in vitro study in mice has suggested that FAO is critical for oocyte meiotic resumption72, in vivo evidence has been limited. In this study, we uncover the central role of Acc in balancing FAS and FAO to maintain energy homeostasis during oocyte determination. This balance is crucial for maintaining physiological TOR signaling, which regulates protein synthesis and quality control. In Acc-depleted female germ cells, loss of malonyl-CoA reduces FAS while relieving CPT1 inhibition, thereby promoting FAO and enhancing TCA cycle flux. This metabolic shift leads to increased levels of α-KG, its downstream metabolite Gln, and ATP, ultimately driving protein overproduction. These findings are consistent with a study in Aspergillus niger, where FAO enzyme overexpression increased both ATP and protein synthesis73.
The increased levels of α-KG, Gln, and ATP collectively hyperactivate TOR signaling in Acc-depleted germ cells. In addition, recent evidence suggests that malonyl-CoA directly inhibits TOR by binding its catalytic domain74; thus, malonyl-CoA reduction in Acc-depleted cells may further contribute to TOR hyperactivation. Hyperactivated TOR signaling drives protein overproduction, which disrupts endosomal trafficking, interferes with the transport of oocyte determinants, and ultimately impairs oocyte determination. Additionally, hyperactivated TOR signaling suppresses autophagy, a key process for GSC maintenance that potentially acts via preservation of E-cadherin levels56. Moreover, reduced FAS impairs GSC maintenance and egg production, highlighting the role of de novo FAS during oogenesis.
Under nutrient-limiting conditions, AMPK is activated and phosphorylates Acc to suppress its activity, shifting metabolism toward FAO to meet energy demands75. However, in the context of Acc depletion under nutrient-replete conditions, excessive ATP and protein accumulation create a metabolic burden, leading to disrupted trafficking and compromised oogenesis. In summary, our findings demonstrate that Acc-mediated metabolic balance is essential not only for adapting to nutrient stress but also for maintaining energy homeostasis during oocyte development and GSC maintenance under physiological conditions. Given that TOR signaling elevation is a hallmark of PCOS, our findings may also provide mechanistic insight into the metabolic dysfunctions underlying PCOS-related infertility.
Acc regulates fusome branching in the female germline
The branched fusome bridges germ cells within a cyst, acting as a channel for cell-cell communication that is essential for germ cell differentiation and oocyte specification76. Fusome branching is established through four rounds of incomplete divisions of the originating from the CB, the intermediate daughter cell of the GSC. During the first incomplete division, the fusome in the CB interacts with one centrosome to anchor the mitotic spindle pole (see Fig. 2 in Huynh, 2006). In subsequent divisions, newly formed fusomes in daughter cells extend through arrested ring canals and fuse with the original fusome, generating a branched network. Proper fusome morphology depends on microtubule organization, as evidenced by unbranched fusome phenotypes in mutants for Orbit/CLASP, Abnormal Spindle77,78, and Lissencephaly-1, a dynein motor regulator79.
In our study, Acc depletion in the germline led to mitotic defects associated with multiple centrosomes, primarily in early germ cells (GSCs/CBs) (Fig. S4D). Interestingly, unbranched fusome phenotypes were most prominent in more developed cysts containing over eight germ cells (Fig. S3E). Furthermore, the mitotic defect has also been reported in Acc mutant fission yeast80. Based on these observations and previous findings that microtubules are essential for fusome morphology, we propose that fusome branching defects arise as a cumulative result of mitotic failures, processes that are closely linked to microtubule function. Notably, the unbranched fusome phenotype caused by Acc depletion resembles the phenotype caused by germline mutation of Rab11, a key regulator of recycling endosomes61. Studies in C. elegans and mammals have shown that Rab11-dependent recycling endosomes carry centrosome components to the spindle pole for spindle pole organization81. In the Acc-depleted germline, expression of Rab11 is reduced, and Rab11-mediated E-cadherin recycling is disrupted (Fig. 5C). However, supplementing Rab11 in Acc-depleted germline did not rescue the unbranched fusome phenotype (Fig. S5B). Additionally, other steps of endosomal trafficking, such as early late endosomes (Rab7) and lysosomes (Lamp1), are also defective in Acc-depleted germline (Fig. 5A and B). These results suggest that Acc depletion causes a global endosomal trafficking defect, in agreement with a previous study showing that Acc controls protein trafficking in the Drosophila wing imaginal disc82.
Although reduced FAS has been reported to alter the composition of the plasma membrane83, and impaired lipid droplet formation has been shown to disrupt the membrane integrity of late endosomes and autophagosomes84, both contributing to endosomal trafficking defects in cultured cells, our results suggest that protein overproduction may play a more prominent role in the germline context. In our study, suppressing FAO (Fig. 2), suppressing FAO (Fig. 2), TOR-dependent S6K signaling (Fig. 4) or protein synthesis using cycloheximide (Fig. S9) substantially suppressed fusome unbranched phenotypes. Furthermore, germline-specific disruption of FASN did not result in unbranching fusome phenotypes (Fig. 2), implying that while altered membrane composition cannot be ruled out, elevated protein synthesis is likely a key contributor to the trafficking defects observed upon Acc depletion. Taking all these observations together, our study suggests that Acc depletion leads to energy imbalance, causing a metabolic shift toward FAO and subsequent protein overload, which disrupts protein endosomal trafficking and results in mislocalization of proteins essential for microtubule establishment, ultimately causing the unbranched fusome phenotype.
These trafficking and fusome defects likely have broader implications for oocyte development and organelle localization during later stages of cyst maturation. Previous studies have shown that fusomes in 8- and 16-cell cysts are more enriched in ER components and are more closed connected to ER than those in GSCs and CBs85,86. This structural distinction supports the idea that cell-cell transport of newly synthesized, secretory-destined proteins may be more active in later-stage cysts, particularly in the 16-cell cysts. In this context, nutrient restriction—by limiting available energy and reducing global protein synthesis—could relieve the stress placed on the endomembrane system and partially rescue fusome branching defects in Acc-depleted germ cells. In addition, mitochondria are known to be transported along the fusome into the Balbiani body87, which plays a key role in oocyte specification. Given that Acc depletion leads to defective fusome branching and endosomal trafficking, it is plausible that this disrupts mitochondrial transport, preventing their proper enrichment in the Balbiani body. Disruption of fusome architecture and endosomal trafficking in Acc-depleted germ cells may therefore impair mitochondrial transport and localization, compromising oocyte maturation and metabolic preparedness.
Acc regulates germ cell development in a sex-dependent manner
In males, the fusome is present but dispensable for normal germ cell development88; however, it appears to mediate intercellular connectivity and synchronize germ cell behaviors under stress89. Surprisingly, depletion of Acc or Hcs in male germ cells does not cause unbranched fusome phenotype nor does it cause reduced fertility (Fig. S12). A similar result was reported in a recent study66 that showed knocking down Acc does not reduce fertility, whereas suppressing Acetyl-CoA converted from ATP-citrate lyase (ACLY) leads to sterility. Furthermore, François et al. has proposed that cytosolic acetyl-CoA in male germ cells is not utilized as a fuel source but instead supports protein acetylation for spermatid individualization and subsequent sperm maturation66. These studies therefore suggest that male and female germ cells may adopt distinct metabolic strategies for utilizing FAS and FAO to meet energy demands, consistent with the fact that a female germ cell cyst produces only one oocyte, whereas a male germ cell cyst generates 64 sperm cells. Indeed, male germ cells with dCPT1 knockdown to inhibit FAO exhibit lipid droplet accumulation in GSCs and GSC loss90, whereas female germ cells with dCPT1 knockdown do not show these effects (Fig. 2B, C). These differences suggest that females may rely less on FAO during early germ cell development.
Elevated FAO-associated to TOR signaling activation may be conserved in flies and mammals
Although the lipid metabolic pathway is conserved between flies and mammals, mammals have two ACC isoforms, ACC1 (ACACA in humans) and ACC2 (ACACB in humans). ACC1 is located in the cytoplasm and produces malonyl-CoA for FAS, while ACC2 is found on the mitochondrial outer membrane and generates malonyl-CoA to inhibit CPT1 and suppress FAO91,92. Mice lacking both ACC1 and ACC2 exhibit reduced lipogenesis, decreased liver triglycerides, and elevated plasma triglycerides (Kim et al., 2017). The rise in plasma triglycerides is attributed to the activation of Sterol Regulatory Element-Binding Protein-1c (SREBP-1c), which increases expression of Glycerol-3-Phosphate Acyltransferase 1 (GPAT1), enhancing Very Low-Density Lipoprotein (VLDL) secretion. The activation of SREBP-1c requires TOR to trigger its proteolytic cleavage in the ER93. This cleavage releases the active N-terminal fragment, which translocates to the nucleus and functions as a transcription factor. Therefore, it is likely that TOR signaling is activated in ACC1/ACC2 double-knockout liver, similar to its activation in the Acc-depleted female germline of flies. ACC1 and ACC2 share high amino acid identity, except that ACC2 contains a mitochondrial targeting sequence at its N-terminus. As a result, pharmacological drugs designed to inhibit both ACC1 and ACC2 (e.g., MK-4704) are currently undergoing Phase 1 clinical trials (Kim et al., 2017). Thus, our findings may provide valuable insights into the effects of ACC inhibitors currently being developed for therapeutic applications.
Limitations of the study
The TRiP RNAi lines used in our screen are robustly expressed in the female germline; however, most target genes are represented by only a single TRiP line, limiting our ability to validate the results using two independent publicly available RNAi lines.
Our research suggests that the unbranched fusome phenotype is a cumulative consequence of mitotic failure. However, it takes approximately six days for a GSC to develop into a 16-cell cyst, and technical challenges have prevented us from successfully culturing germaria in vitro over this extended period for use in live imaging experiments. Our research also shows that Acc depletion leads to increased acetyl-CoA, which may enhance protein acetylation and contribute to the phenotypes observed in Acc-depleted germ cells. However, this aspect was not explored here, as it lies beyond the current scope of the study.
Methods
Contact for reagent and resource sharing
Further information and reagent requests may be directed to and will be fulfilled by the Lead Contact, Dr. Hwei-Jan Hsu, at the Institute of Cellular and Organismic Biology, Academia Sinica (cohsu@gate.sinica.edu.tw).
Experimental model and subject detail
Drosophila strains and culture
Drosophila stocks were maintained at 22–25 °C on standard medium unless otherwise indicated. w1118 was used as a wild-type control. The 143 RNAi lines used for screening were obtained from Bloomington Drosophila Stock Center and are described in Supplementary Data 1. The fly mutant lines, reporter lines, and RNAi lines used to study the role of Acc in the germline are listed in Table S3-Key Source Information. nos-Vp16-GAL4 was used to express transgenes in the germline, unless otherwise noted. Other genetic elements are described in FlyBase (http://flybase.bio.indiana.edu).
For the screen to identify lipid metabolism-associated enzymes that control oogenesis, flies expressing RNAi driven by nos-GAL4 were cultured at 29 °C throughout all developmental stages. Seven RNAi-expressing virgin female flies were crossed with five wild-type male flies and incubated for two days at 29 °C. Then, the parent flies were removed. Vials without parent flies were allowed to grow for another 4-5 days at 29 °C. The number of pupae in the vial was counted and compared to the number produced by the control flies (nos>gfpRNAi); lipid metabolic enzymes with less than 50% expression after knockdown compared to controls were selected as candidates. At least two replicates were performed for each candidate.
For the second-step screen, the cross (nosGAL4 and UAS-candidateRNAi) was cultured at 18 °C to limit GAL4 expression during development. Then, female virgin flies expressing RNAi were collected and cultured with wild-type (yw) males at 29 °C to activate GAL4 expression for 7 days. nos>gfpRNAi was used as a control. Ovaries from 10 females were then dissected for immunostaining; 5 pairs were mounted to assess phenotypes by direct observation, and 20 germaria were randomly selected for imaging and detailed analysis, following methodologies commonly used in female germline research (see more detail in the section on Sampling, imaging, image analysis and quantification). GSC number loss was identified as GSC number less than two in the germarium. Differentiation defect was identified as germ cell number in cyst cells that does not follow the 2 N rule. The percentage of germaria exhibiting GSC loss or not following the 2 N rule was calculated and presented in the heat map (see Fig. 1C′). Each experiment was independently replicated twice. Seven-day-old female flies (cultured in 18 °C during development and 29 °C during adult stage) were used for ovary experiments unless otherwise indicated.
For adult knockdown experiments, flies were fed with a normal diet with dry yeast powder unless indicated; food was changed daily until dissection.
The phenotype of each genetic manipulation is summarized in Table S1; genotypes of the flies used in each figure are listed in Supplementary Data 2.
Fly transgenic line generation
The ACCRNAi line (Bloomington stock #77408) was generated through the insertion of the pVALIUM20-attB-ACCRNAi construct (Stock ID HMS01230) and used for RNAi screening; the insert is located on the third chromosome. For genetic studies, the same RNAi plasmid (kindly provided by Dr. Norbert Perrimon, Harvard Medical School, USA) was injected to w1118 embryos bearing an attp40 landing site on the second chromosome to generate the ACCRNAi line on the second chromosomes (performed by WellGenetics Inc, Taiwan).
To generate the UASp-mRuby3-iATPSnFR1.0 line, the cyto-iATPSnFR1.0 plasmid was obtained from Addgene (plasmid #102550). The iATPSnFR1.0 sensor was amplified with primers bearing Not1 and BamH1 restriction enzyme sites and subcloned into the UASp-attB vector (Drosophila Genomics Resource Center, DGRC, stock #1358) to create the UASp-iATPSnFR1.0 construct. The Ruby3 fragment was amplified from pKanCMV-mClover3-mRuby3 (Addgene plasmid #74254, kindly provided by Dr. Shu-Ling Chiu, Institute of Cellular and Organismic Biology, Academia Sinica) using primers containing a NotI site. The fragment was then inserted upstream of iATPSnFR1.0 in the UASp-iATPSnFR1.0 construct to generate the UASp-mRuby3-iATPSnFR1.0 construct. The final plasmid was injected into embryos bearing an attP2 landing site on the third chromosome (performed by WellGenetics Inc, Taiwan).
To generate the UASz-human (h) ACACA-HA line, the human ACACA gene was amplified from the human ACACA MGC cDNA clone (#168907) using a forward primer containing a BamHI site and the Drosophila Kozak sequence (CCAAA) to enhance protein translation, along with a reverse primer containing a 3× HA tag sequence and an XbaI site. The Kozak-hACACA-3HA fragment was subcloned into the UASz1.0 vector (DGRC Stock #1431). The construct was made by the RNAi core (Academia Sinica, Taiwan). The final plasmid was injected into embryos bearing attP2 landing site on the third chromosome (performed by WellGenetics Inc, Taiwan). The primers are listed in Table S2.
Protein-poor diet preparation
A protein-poor diet was prepared by adding 2 g of agar (#190923001, Geneteks) to 200 ml of 5% molasses (Golden A Molasses unsulphured, Cane molasses, GROEB FARMS, MI, United States) and boiling. The mixture was cooled to 60 °C, and ~5 ml of molasses-agar medium was added into each empty plastic vial. After allowing the food to solidify, it was stored at 4 °C for up to 7 days until use.
Etomoxir-, rapamycin-, chloroquine-, cycloheximide-containing food preparation
The surface of the food was coated with 150 µl ddH2O (control) or ddH2O containing 200 mM Etomoxir (E1905-5MG, Sigma Aldrich), 50 µM rapamycin (#37094, Sigma-Aldrich) with and without 50 mM chloroquine (C6628-25G, Sigma Aldrich; a generous gift from Dr. Guang-Chao Chen, Institute of Biochemistry, Academia Sinica, Taiwan), or 1 mM cycloheximide (C1988, Sigma Aldrich). Then, the food was air-dried for 1 h; the prepared food was stored at 4 °C for up to 7 days until use.
Mosaic clonal analysis
Mosaic clones were generated using the FLP/FRT-mediated mitotic recombination method34. Two- to three-day-old flies of the genotypes hs-flp/+; FRT42B/ubi-gfpFRT42B and hs-flp/+; ACC1FRT42B/ubi-gfpFRT42B were generated from standard crosses and subjected to heat shock at 37 °C for 1 h, twice a day with a 6 h interval, for 3 days. Flies were maintained at 25 °C after heat shock, with daily food changes until dissection. Homozygous mutant cells were identified by the absence of GFP. To analyze GSC maintenance, the percentage of germaria containing at least one GSC clone at 14 days after clone induction (aci) was normalized to that at 7 days aci for each genotype. Three biological replicates were performed, with ~100 germaria analyzed per genotype at each time point in each replicate.
Egg production assay
Flies were grown at 18 °C; upon eclosion, seven newly eclosed females for each genotype were collected and cultured with five wild-type males in vials at 25 °C overnight for mating. Following mating, the flies were shifted to 29 °C for the egg laying assay. Flies were transferred into plastic bottles with small holes, capped with a plastic plate containing 5% molasses agar with a layer of wet yeast paste. The bottles were inverted to allow egg-laying on the plate, which was changed daily, and the number of eggs was recorded. The experiment was performed with 4 replicates.
Male fertility assay
Three 14-day-old males with the indicated genotypes were mated with a single 7-day-old virgin w1118 female at 25 °C. The females were pre-fed with wet yeast for 2 days prior to mating. After one day of mating, the males were removed. The mated females were then allowed to lay eggs on fresh food, which was changed daily. The total number of eggs was quantified within 72 h. A minimum of 10 crosses were tested for each genotype.
Immunostaining and fluorescence microscopy
Ovaries were dissected, fixed, and immunostained as described previously94. In brief, ovaries were dissected in pre-warmed Grace’s insect medium (GIM), Lot:2323315, Gibco) and fixed with 5.3% paraformaldehyde (Cat No 043368.9 M, Thermo Scientific) in GIM for 13 min with gentle rotation at room temperature. After fixation, samples were washed three times with PBST (0.1% Triton in 1X PBS) for 20 min each. Ovaries were then teased apart using a syringe needle and incubated in blocking solution for 3 h at room temperature or overnight at 4 °C. Blocked ovaries were incubated with primary antibody for 3 h at room temperature or overnight at 4 °C, followed by three PBST washes (20 min each). Samples were then incubated with secondary antibody under the same conditions. Details of the primary and secondary antibodies used are provided in Table S3-Key Source Information.
BODIPY staining was performed as previously described, with minor modifications90. First, BODIPY 493/503 (D3922, Thermo Fisher) was dissolved in DMSO at a stock concentration of 5 mM and stored at −20 °C. Ovaries were dissected and fixed as described above. After primary and secondary antibody staining, samples were incubated in PBST containing BODIPY (1:500) for 15 min, followed by a PBST wash.
OPP staining was performed using the Click-iT Plus OPP Alexa-Fluor-488™ Protein Synthesis Assay Kit (C10456, Molecular Probes) as previously described95, with minor modifications. Briefly, ovaries were dissected in pre-warmed GIM. Dissected ovaries were then incubated with 20 µM Click-iT OPP solution in pre-warmed GIM for 30 min on a rotator at room temperature, followed by fixation in 5.3% PFA for 13 min. Samples were washed with PBST, and then the immunostaining protocol was performed as described above. After immunostaining, ovaries were incubated in freshly prepared Click-IT reaction buffer (from the kit) for 30 min in the dark with rotation. The samples were then washed with the Click-iT reaction buffer twice, and then washed three times with PBST for 20 min each.
Apoptotic cells were detected using the Apoptag® Fluorescein In Situ Apoptosis Detection Kit (S7110, Merck)96. Ovaries of 7-day-old flies on a protein-poor diet were used as a positive control, as this diet induces apoptosis of escort cells in the germarium94. In brief, ovaries were dissected in Grace’s medium, fixed, and washed as described previously. The samples were incubated with 300 µl of equilibration buffer for 5 min on the rotator twice at room temperature. Ovaries were then incubated with a reaction mix consisting of 76 µl reaction buffer and 32 µl TdT enzyme for 1 h at 37 °C in a dark chamber. The reaction was stopped by adding 500 µl STOP/ WASH solution, and samples were subsequently rinsed with PBST and immunostained as previously described.
Ovaries expressing GFP-Mcherry-Atg8a were dissected and fixed without immunostaining.
Ovaries expressing Ruby3-iATPSnFR1.0 were dissected, fixed, and labeled with 1B1 and LamC.
Immunofluorescence staining of Drosophila testes was performed according to a previously established protocol97. Testes were dissected in 1x PBS and fixed in 4% PFA diluted in 1x PBS for 30 min. Following fixation, samples were washed twice for 30 min at 25 °C in 1x PBS supplemented with 0.5% Triton X-100. Testes were then blocked with PBTB (1x PBS, 0.2% Triton X-100, and 0.2% bovine serum albumin) for 1 h at 25 °C. Primary antibody incubation was carried out overnight at 4 °C. After primary antibody incubation, testes were washed twice for 30 min in PBTB and then incubated for 2 h at 25 °C with the appropriate secondary antibodies diluted in PBTB.
After washing thoroughly, samples were stained with 0.5 μg/ml DAPI (32248, Sigma) for 15 min followed by mounting in mounting solution [80% glycerol containing 20 µg/mL N-propyl gallate] (P3130, Sigma). Images of fixed ovaries were obtained using Zeiss LSM 700 and 900 Laser Scanning confocal microscopes equipped with a 63x objective lens.
Quantitative real-time PCR (qRT-PCR)
Total RNA from freshly dissected ovaries was extracted using a standard Trizol extraction protocol. Briefly, 30 pairs of ovaries were dissected from each genotype and frozen at −80 °C for at least 2 h. The frozen ovaries were homogenized in 500 µl Trizol (NTRR- 0200, NovelGene), followed by the addition of another 500 μl Trizol. Next, 200 μl of 1-Bromo-3-chloropropane (BCP, B9673-200ml, Sigma) was added to the sample, and the tube was centrifuged at 9391× g (rcf) for 15 min at 4 °C. The aqueous phase was transferred to a new tube, and 500 μl isopropanol (I9516-500ml, Sigma) was added for RNA precipitation. After centrifugation, the pellet was washed with 75% ethanol, and dissolved in 30 μl RNase-free ddH₂O. The RNA samples were stored at −80 °C until use.
For RT-PCR, 1 μg of total RNA was reverse-transcribed to cDNA using the Transcriptor First Strand cDNA Synthesis Kit (04379012001, Roche). The resulting cDNA was then mixed with gene specific primers and TaqMan probes (Roche) and 2X LightCycler 480 Probes Master (04707494001, Roche) for PCR reactions. Gene expression was analyzed using the Roche LightCycler 480 machine (05015243001). RpL32 expression was used as an internal control. Each gene expression measurement was performed on biological triplicates. Primer sequences and probe details are provided in Table S2.
RNA-sequencing analysis
RNA extraction from ovary samples was performed as described above. Then, RNA-seq analysis was performed as reported previously98. RNA quantification was conducted to prepare libraries using the SureSelect XT HS2 mRNA Library Preparation kit (Agilent), followed by size selection with AMPure XP beads (Beckman Colter, USA). Sequencing was accomplished using Illumina’s sequencing-by-synthesis technology. Sequencing data (FASTQ reads) were processed using Illumina’s base calling program bcl2fastq v2.20. Adapter clipping and sequence quality trimming were performed using TRimmomatic v0.36 with a sliding-window approach. Transcriptome alignments were made using the HISAT2 tool. Subsequently, differential expression analysis was performed with StringTie (version 2.1.4) and DEseq (version 1.39.0) or DEseq2 (version 1.28.1) with genome bias correction through Welgene Biotech’s in-house pipeline. Additionally, Gene Set Enrichment Analysis (GSEA)99 was performed as needed. Functional enrichment analysis on differentially expressed genes was carried out using ClusterProfiler (version 3.6). Gene expression levels were quantified as fragments per kilobase of transcript per million mapped reads (FPKM), with genes exhibiting low expression (Transcripts per million, TPM < 0.3) being excluded. Data graphing was performed using SRplot100.
Western blot
Protein extraction solution was prepared by adding 2x Proteinase inhibitor (cOmplete™, EDTA-free protease inhibitor cocktail, Sigma Aldrich), and 200 µM Phenylmethylsulfonyl Fluoride (PMSF, 329-98-6, Sigma Aldrich) to freshly prepared RIPA buffer (50 mM TRIS-HCl pH 6.8, 150 mM NaCl, 1 mM EDTA, 0.5% Deoxycholate, 1% NP-40) on ice. For each genotype, 10 pairs of ovaries were dissected in GIM. For control ovaries, vitellogenic egg chambers were removed using forceps, leaving only the transparent part of the ovaries for subsequent experiments. Total protein was extracted from homogenizing ovaries in 70 µl of freshly prepared extraction solution. The ovaries were homogenized for 2 min on ice using a grinder, then vortexed and spun down. The tissues were homogenized again for an additional 1 min on ice. Samples were then centrifuged at 13523 × g (rcf) for 5 min, and the supernatant was transferred into a new tube and stored at −20 °C for up to 2 days. Protein concentration was measured using Bio-Rad protein assay (500-0006, BioRad) with BSA standard. The samples were heat-denatured at 95°C for 5 min.
A total of 30 µg protein was loaded onto a 10% SDS-PAGE, followed by transfer of the proteins to a PVDF membrane. The membrane was blocked in 5% BSA for 1 h, then incubated with the primary antibody diluted in 5% BSA at 4°C overnight, and washed three times in TBST for 10 min each. The following primary antibodies were used: rabbit anti- Drosophila pT398 S6K (1:1000, #9209, Cell Signaling; a gift from Dr. Guang-Chao Chen, Institute of Biochemistry, Academia Sinica, Taiwan), rabbit anti-pT37/46 4E-BP (1:1000, 236B4, Cell Signaling Technology), and mouse anti-beta-Actin (1:1000, ab8224, Abcam).
The membrane was then incubated with HRP-conjugated secondary antibodies: anti-mouse IgG-HRP (1:10000, lot#2194079, Millipore) and anti-rabbit IgG-HRP (1:10000, lot#107487, Jackson Immuno Research) diluted in 5% BSA at room temperature for 1 h, followed by three washes for 10 min each in PBST. HRP-signals were detected using 1X ECL reagent (JJ96-K004M, T-Pro Biotechnology) on Image Quant LAS4000 biomolecular imager (GE, ImageQuant LAS4000). For unprocessed and uncropped scans, please refer to Supplementary Fig. 14.
Metabolite profiling
Fifty pairs of ovaries from each genotype were dissected in cold 1xPBS. Since nos > ACCRNAi ovaries lacked vitellogenic egg chambers, the vitellogenic egg chambers from control ovaries were removed using forceps, leaving the transparent regions for metabolite extraction. Metabolites from each sample (containing about 12.5 µg DNA) were extracted using 200 µl metabolite extraction solvent (2:2:1 acetonitrile: methanol: ddH2O) and stored at −20 °C overnight. Samples were homogenized by vortexing for 5 s followed by cold bath sonication for 5 min, repeated twice. Tissue debris was pelleted by centrifugation at 15871 × g (rcf) for 10 min at 4 °C. A total of 200 µl metabolite-containing supernatant was transferred into a new tube. The supernatant was freeze-dried for at least 3 h using a freeze drier (VirTis BenchTop K). Dried samples were kept at −80 °C until analyzed for amino acids and TCA cycle byproducts by GC-MS (Agilent 7250 GC/Q-TOF) or for acetyl-CoA by UPLC-MS (Waters ACQUITY UPLC/ Xevo TQ-XS).
The tissue debris was used for extraction of genomic DNA. In brief, tissue debris was ground in 200 μl of Buffer A (100 mM Tris-HCl pH 7.5, 100 mM EDTA, 100 mM NaCl, 1% SDS) and incubated at 65 °C for 30 min. Next, 800 μl of Buffer B (1-part 5 M KAc and 2.5-parts 6 M LiCl) was added, and the sample was incubated on ice for 10 min to precipitate DNA. After centrifugation, the pellet was washed with 70% ethanol, air dried, and dissolved in 50 μl of ddH2O. The DNA amount was measured using a Nanodrop 8000 spectrophotometer (Thermo Scientific). Each experiment was done in at least 3 biological replicates.
Sampling, imaging, image analysis, reproducibility and quantification
To ensure reproducibility, we performed sampling and imaging following procedures wildly used for female germline analysis101,102,103.
Desired genetic crosses were established for each genotype using 15 females and 10 males. Between 50 and 100 progenies were collected over a 2-day period at the designated temperature. From these, 10 female flies were randomly selected for dissection, yielding a total of 20 ovaries (2 per fly). To minimize sample variability, all incorporation assays and immunostaining experiments were conducted in parallel on the same bench using identical reagent mixtures.
For imaging, 5–6 ovaries from different individuals were randomly chosen and mounted, resulting in ~100–120 germaria per condition. During mounting, older egg chambers were removed to isolate the germaria, which were then pooled and evenly distributed on slides for imaging and analysis. All germaria were examined to assess phenotypes, and 15–30 germaria displaying representative phenotypes were imaged for detailed analysis (Fig. 1D and Fig. 4A).
Confocal imaging was performed using consistent acquisition settings across all experimental groups, with control samples used to define baseline parameters. All experiments were independently repeated in at least two biological replicates and validated using either two independent RNAi lines or a combination of genetic and pharmacological approaches.
GSCs in germaria were identified by their anteriorly anchored fusome (membranous cytoskeletal structure, labeled by 1B1) and by their juxtaposition to cap cells (labeled by LamC). GSCs in testes were identified as Vasa and 1B1 double-positive cells located near hub cells.
For fusome morphology, we scored the fusome branching phenotypes in the cysts containing more than four germ cells. Each germarium typically contains 6-8 such cysts.
For fluorescence intensity measurements, ImageJ (NIH) was used to analyze signal intensity (arbitrary units) and colocalization. For pMad expression, the average fluorescence intensity in confocal z-sections at the largest GSC nuclear area was measured.
For p4E-BP and pS6 quantification, five z-sections from the middle of each germarium were merged to generate a projection. The mean fluorescence intensity was measured the germline, and background signal was subtracted from the total intensity for each image. For intensity quantification, 15 images were used for each genotype.
For OPP intensity in GSCs, a single z-section containing the GSC with the largest nucleus was selected. For OPP intensity in germaria, five central z-sections of each germarium were merged. The mean fluorescence intensity was measured the germline, and background signal was subtracted from the total intensity for each image. For intensity quantification, 15 images were used for each genotype.
For ATP level quantification, a z-section showing the largest cross-sectional area of the germarium expressing nos>mRuby3-iATPSnFR1.0 was selected. Mean fluorescence intensities of the GFP channel (ATP signal) and the mRuby3 channel (internal control) were measured within the germline region, and the ATP/mRuby3 ratio was calculated using ImageJ.
For E-cadherin measurements, the z-section with the highest E-cadherin expression in the junction between cap cells and GSC was selected, and mean fluorescence intensities were measured.
Colocalization between Rab11 and E-cadherin, as well as between Lamp1-GFP and Atg8a, was quantified in each z-section of the germarium using Pearson’s correlation coefficient calculated with the JACoP plugin in ImageJ.
The number of lipid droplets was determined by manually counting BODIPY-positive puncta within the cytoplasm of GSCs. For quantification, 10-15 germaria were analyzed (Fig. 2A).
For ovary morphology analysis, 10 images were captured for each genotype and visually assessed for morphological features. One representative image per genotype is shown in the figure.
Statistical analysis and graphing
Quantitative data were collected and visualized using Excel (Microsoft) or GraphPad Prism 8 software (version 8.3.0). The number of GSCs or germ cell cysts is indicated above each bar in the graphs. The number of germaria is presented in dot plot graphs or annotated directly in the figure panels. P-values were computed using a two-tailed unpaired Student’s t-test or chi-squared test for comparisons between two groups. The type of statistical test used, and the meaning of asterisks/P-values are consistently stated in each figure legend.
Representative images were selected from at least two technical replicate experiments showing consistent results or from independent biological samples with similar phenotypes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA sequencing data has been deposited in SRA under accession number PRJNA1322194. Metabolomic data has been submitted to Metabolomics Workbench under accession numbers ST004284 and ST004252. Source data are provided with this paper. The remaining data that support the findings of this study are available upon request. Source data are provided with this paper.
References
Liu, Q., Xie, Y. -j, Qu, L. -h, Zhang, M. -x & Mo, Z. -c Dyslipidemia involvement in the development of polycystic ovary syndrome. Taiwan. J. Obstet. Gynecol. 58, 447–453 (2019).
Wang, Y. & Wang, Y. Accelerated ovarian aging among type 2 diabetes patients and its association with adverse lipid profile. Front. Endocrinol. 13, 780979 (2022).
Collée, J., Mawet, M., Tebache, L., Nisolle, M. & Brichant, G. Polycystic ovarian syndrome and infertility: overview and insights of the putative treatments. Gynecol. Endocrinol.: J. Int. Soc. Gynecol. Endocrinol. 37, 869–874 (2021).
Spradling, A. C. Germline cysts: communes that work. Cell 72, 649–651 (1993).
Kirilly, D. & Xie, T. The Drosophila ovary: an active stem cell community. Cell Res. 17, 15–25 (2007).
Xie, T. & Spradling, A. C. A niche maintaining germ line stem cells in the Drosophila ovary. Sci. (N. Y.) 290, 328–330 (2000).
Lei, L. & Spradling, A. C. Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Development (Cambridge, England) 140, 2075–2081 (2013).
Chandel, N. S. et al. Lipid metabolism. Cold Spring Harb. Perspect. Biol. 13, a040576 (2021).
Flores, J., White, B. M., Brea, R. J., Baskin, J. M. & Devaraj, N. K. Lipids: chemical tools for their synthesis, modification, and analysis. Chem. Soc. Rev. 49, 4602–4614 (2020).
Chen, L. et al. Acetyl-CoA carboxylase (ACC) as a therapeutic target for metabolic syndrome and recent developments in ACC1/2 inhibitors. Expert Opin. Invest. drugs 28, 917–930 (2019).
Schulz, H. in Encyclopedia of Biological Chemistry (Second Edition) (eds William J. Lennarz & M. Daniel Lane) 281–284 (Academic Press, 2013).
Wang, M. et al. Carnitine palmitoyltransferase system: a new target for anti-inflammatory and anticancer therapy? Front. Pharmacol. 12, 760581 (2021).
McGarry, J. D. & Brown, N. F. The mitochondrial carnitine palmitoyltransferase system. from concept to molecular analysis. Eur. J. Biochem. 244, 1–14 (1997).
McGarry, J. D., Leatherman, G. F. & Foster, D. W. Carnitine palmitoyltransferase I. The site of inhibition of hepatic fatty acid oxidation by malonyl-CoA. J. Biol. Chem. 253, 4128–4136 (1978).
Liang, K. Mitochondrial CPT1A: Insights into structure, function, and basis for drug development. Front. Pharmacol. 14, 1160440 (2023).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Porstmann, T. et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8, 224–236 (2008).
Madison, B. B. Srebp2: A master regulator of sterol and fatty acid synthesis. J. lipid Res. 57, 333–335 (2016).
Mariano, V. et al. SREBP modulates the NADP+/NADPH cycle to control night sleep in Drosophila. Nat. Commun. 14, 763 (2023).
Hay, N. & Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 (2004).
Iadevaia, V., Liu, R. & Proud, C. G. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Devel. Biol. 36, 113–120 (2014).
Nakashima, A. & Tamanoi, F. in The Enzymes 28, (eds Fuyuhiko Tamanoi & Michael N. Hall) 167–187 (Academic Press, 2010).
Mugume, Y., Kazibwe, Z. & Bassham, D. C. Target of rapamycin in control of autophagy: puppet master and signal integrator. Intl. J.Mol. Sci. https://doi.org/10.3390/ijms21218259 (2020).
LaFever, L., Feoktistov, A., Hsu, H. J. & Drummond-Barbosa, D. Specific roles of target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Dev. (Camb., Engl.) 137, 2117–2126 (2010).
Wei, Y. et al. TORC1 regulators Iml1/GATOR1 and GATOR2 control meiotic entry and oocyte development in Drosophila. Proc. Natl. Acad. Sci. USA 111, E5670–E5677 (2014).
Adhikari, D. et al. Disruption of Tsc2 in oocytes leads to overactivation of the entire pool of primordial follicles. Mol. Hum. Reprod. 15, 765–770 (2009).
Adhikari, D. et al. Tsc/mTORC1 signaling in oocytes governs the quiescence and activation of primordial follicles. Hum. Mol. Genet. 19, 397–410 (2010).
O’Connell, J. M. & Pepling, M. E. Primordial follicle formation - some assembly required. Curr. Opin. Endocr. Metab. Res. 18, 118–127 (2021).
Yaba, A. & Demir, N. The mechanism of mTOR (mammalian target of rapamycin) in a mouse model of polycystic ovary syndrome (PCOS). J. Ovarian Res. 5, 38 (2012).
Liu, J., Wu, D. C., Qu, L. H., Liao, H. Q. & Li, M. X. The role of mTOR in ovarian Neoplasms, polycystic ovary syndrome, and ovarian aging. Clin. Anat. (N. Y., N. Y.) 31, 891–898 (2018).
Liu, A. L. et al. New insights into mTOR signal pathways in ovarian-related diseases: polycystic ovary syndrome and ovarian cancer. Asian Pac. J. Cancer Prev. 17, 5087–5094 (2016).
Ni, J. Q. et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. methods 8, 405–407 (2011).
Sanchez, C. G. et al. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 18, 276–290 (2016).
Xu, T. & Rubin, G. M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Dev. (Camb., Engl.) 117, 1223–1237 (1993).
Solórzano-Vargas, R. S., Pacheco-Alvarez, D. & León-Del-Río, A. Holocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells. Proc. Natl. Acad. Sci. USA 99, 5325–5330 (2002).
Hymes, J. & Wolf, B. Biotinidase and its roles in biotin metabolism. Clin. Chim. Acta 255, 1–11 (1996).
Hinnant, T. D., Merkle, J. A. & Ables, E. T. Coordinating proliferation, polarity, and cell fate in the drosophila female germline. Front Cell Dev. Biol. 8, 19 (2020).
Barr, J., Gilmutdinov, R., Wang, L., Shidlovskii, Y. & Schedl, P. The Drosophila CPEB protein orb specifies oocyte fate by a 3’UTR-dependent autoregulatory loop. Genetics 213, 1431–1446 (2019).
Chen, D. & McKearin, D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 13, 1786–1791 (2003).
Jang, J. K., Sherizen, D. E., Bhagat, R., Manheim, E. A. & McKim, K. S. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116, 3069–3077 (2003).
Fang, C. T. et al. Inhibition of acetyl-CoA carboxylase impaired tubulin palmitoylation and induced spindle abnormalities. Cell Death Discov. 9, 4 (2023).
Matias, N. R., Mathieu, J. & Huynh, J. R. Abscission is regulated by the ESCRT-III protein shrub in Drosophila germline stem cells. PLoS Genet 11, e1004653 (2015).
Parvy, J. P. et al. Drosophila melanogaster Acetyl-CoA-carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLoS Genet 8, e1002925 (2012).
Li, H. et al. Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Sci. (N. Y., N. Y.) 375, eabk2432 (2022).
Vos, M. et al. Cardiolipin promotes electron transport between ubiquinone and complex I to rescue PINK1 deficiency. J. cell Biol. 216, 695–708 (2017).
Strub, B. et al. Mutations of the withered (whd) gene in Drosophila melanogaster confer hypersensitivity to oxidative stress and are lesions of the carnitine palmitoyltransferase I (CPT I) gene. Genome / Natl. Res. Counc. Can. = Génome / Cons. Natl. de. Rech. Can. 51, 409–420 (2008).
Palanker, L., Tennessen, J. M., Lam, G. & Thummel, C. S. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 9, 228–239 (2009).
Chen, D. & Huang, S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. cell Biol. 153, 169–176 (2001).
Grewal, S. S., Li, L., Orian, A., Eisenman, R. N. & Edgar, B. A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7, 295–302 (2005).
Gross, A. S. et al. Acetyl-CoA carboxylase 1-dependent lipogenesis promotes autophagy downstream of AMPK. J. Biol. Chem. 294, 12020–12039 (2019).
Mauvezin, C., Ayala, C., Braden, C. R., Kim, J. & Neufeld, T. P. Assays to monitor autophagy in Drosophila. Methods (San. Diego, Calif.) 68, 134–139 (2014).
Wei, Y. et al. The GATOR complex regulates an essential response to meiotic double-stranded breaks in Drosophila. Elife https://doi.org/10.7554/eLife.42149 (2019).
Lamming, D. W. et al. Inhibition of the Mechanistic Target of Rapamycin (mTOR)-rapamycin and beyond. Cold Spring Harbor Perspect. Med. https://doi.org/10.1101/cshperspect.a025924 (2016).
Gui, J., Samuels, T. J., Grobicki, K. Z. A. & Teixeira, F. K. Simultaneous activation of Tor and suppression of ribosome biogenesis by TRIM-NHL proteins promotes terminal differentiation. Cell Rep. 42, 112181 (2023).
Nieto-Torres, J. L., Leidal, A. M., Debnath, J. & Hansen, M. Beyond Autophagy: The Expanding Roles of ATG8 Proteins. Trends in Biochemical Sciences 46, 673-686 (2021).
Murmu, N. & Shravage, B. V. Autophagy slows the aging of Germline stem cells in Drosophila through modulation of E-cadherin. Preprint at bioRxiv, https://doi.org/10.1101/2022.03.31.486570 (2022).
Mauthe, M. et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455 (2018).
Gingras, A. C. et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864 (2001).
Son, S.-H. et al. Metabolite trafficking enables membrane-impermeable-terpene secretion by yeast. Nat. Commun. 13, 2605 (2022).
Elkin, S. R., Lakoduk, A. M. & Schmid, S. L. Endocytic pathways and endosomal trafficking: a primer. Wien. medizinische Wochenschr. (1946) 166, 196–204 (2016).
Bogard, N., Lan, L., Xu, J. & Cohen, R. S. Rab11 maintains connections between germline stem cells and niche cells in the Drosophila ovary. Dev. (Camb., Engl.) 134, 3413–3418 (2007).
Zhang, J. et al. Thirty-one flavors of drosophila rab proteins. Genetics 176, 1307–1322 (2007).
Takahara, T., Amemiya, Y., Sugiyama, R., Maki, M. & Shibata, H. Amino acid-dependent control of mTORC1 signaling: a variety of regulatory modes. J. Biomed. Sci. 27, 87 (2020).
Lobas, M. A. et al. A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat. Commun. 10, 711 (2019).
Karan, D., Lesbats, M., David, J. R. & Capy, P. Evolution of the AMP-forming Acetyl-CoA synthetase gene in the drosophilidae family. J. Mol. Evolution 57, S297–S303 (2003).
François, C. M., Pihl, T., Dunoyer de Segonzac, M., Hérault, C. & Hudry, B. Metabolic regulation of proteome stability via N-terminal acetylation controls male germline stem cell differentiation and reproduction. Nat. Commun. 14, 6737 (2023).
Duncan, D. M., Kiefel, P. & Duncan, I. Mutants for drosophila isocitrate dehydrogenase 3b are defective in mitochondrial function and larval cell death. G3 Genes|Genomes|Genet. 7, 789–799 (2017).
Caizzi, R., Bozzetti, M. P., Caggese, C. & Ritossa, F. Homologous nuclear genes encode cytoplasmic and mitochondrial glutamine synthetase in Drosophila melanogaster. J. Mol. Biol. 212, 17–26 (1990).
Teixeira, F. K. et al. ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat. Cell Biol. 17, 689–696 (2015).
Drummond-Barbosa, D. & Spradling, A. C. Stem Cells and Their Progeny Respond to Nutritional Changes during Drosophila Oogenesis. Developmental Biol. 231, 265–278 (2001).
Su, Y. H. et al. Diet regulates membrane extension and survival of niche escort cells for germline homeostasis via insulin signaling. Development (Cambridge, England) https://doi.org/10.1242/dev.159186 (2018).
Downs, S. M., Mosey, J. L. & Klinger, J. Fatty acid oxidation and meiotic resumption in mouse oocytes. Mol. Reprod. Dev. 76, 844–853 (2009).
Chen, X., Pan, B., Yu, L., Wang, B. & Pan, L. Enhancement of protein production in Aspergillus niger by engineering the antioxidant defense metabolism. Biotechnol. Biofuels Bioprod. 17, 91 (2024).
Nicastro, R. et al. Malonyl-CoA is a conserved endogenous ATP-competitive mTORC1 inhibitor. Nat. Cell Biol. 25, 1303–1318 (2023).
Ren, Y. & Shen, H. M. Critical role of AMPK in redox regulation under glucose starvation. Redox Biol. 25, 101154 (2019).
Huynh, J.-R. in Cell-Cell Channels (eds Frantisek Baluska, Dieter Volkmann, & Peter W. Barlow) 217–235 (Springer New York, (2006).
Miyauchi, C., Kitazawa, D., Ando, I., Hayashi, D. & Inoue, Y. H. Orbit/CLASP is required for germline cyst formation through its developmental control of fusomes and ring canals in Drosophila males. PLoS One 8, e58220 (2013).
Riparbelli, M. G., Massarelli, C., Robbins, L. G. & Callaini, G. The abnormal spindle protein is required for germ cell mitosis and oocyte differentiation during Drosophila oogenesis. Exp. Cell Res 298, 96–106 (2004).
Liu, Z., Xie, T. & Steward, R. Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Dev. (Camb., Engl.) 126, 4477–4488 (1999).
Saitoh, S. et al. Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J. cell Biol. 134, 949–961 (1996).
Das, S., Hehnly, H. & Doxsey, S. A new role for Rab GTPases during early mitotic stages. Small GTPases 6, 11–15 (2015).
Sasamura, T., Matsuno, K. & Fortini, M. E. Disruption of Drosophila melanogaster lipid metabolism genes causes tissue overgrowth associated with altered developmental signaling. PLoS Genet 9, e1003917 (2013).
Wei, X. et al. Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature 539, 294–298 (2016).
Peng, W. et al. Endosomal trafficking participates in lipid droplet catabolism to maintain lipid homeostasis. Nat. Commun. 16, 1917 (2025).
Röper, K. Rtnl1 is enriched in a specialized germline ER that associates with ribonucleoprotein granule components. J. Cell Sci. 120, 1081–1092 (2007).
Snapp, E. L., Iida, T., Frescas, D., Lippincott-Schwartz, J. & Lilly, M. A. The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol. Biol. Cell 15, 4512–4521 (2004).
Cox, R. T. & Spradling, A. C. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Dev. (Camb., Engl.) 130, 1579–1590 (2003).
Kaufman, R. S. et al. Drosophila sperm development and intercellular cytoplasm sharing through ring canals do not require an intact fusome. Development (Cambridge, England) https://doi.org/10.1242/dev.190140 (2020).
Lu, K. L. & Yamashita, Y. M. Germ cell connectivity enhances cell death in response to DNA damage in the Drosophila testis. eLife 6, e27960 (2017).
Senos Demarco, R., Uyemura, B. S., D’Alterio, C. & Jones, D. L. Mitochondrial fusion regulates lipid homeostasis and stem cell maintenance in the Drosophila testis. Nat. Cell Biol. 21, 710–720 (2019).
Wang, Y. et al. Acetyl-CoA carboxylases and diseases. Front Oncol. 12, 836058 (2022).
Abu-Elheiga, L. et al. Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal. Proc. Natl. Acad. Sci. USA 102, 12011–12016 (2005).
Laplante, M. & Sabatini, D. M. mTORC1 activates SREBP-1c and uncouples lipogenesis from gluconeogenesis. Proc. Natl. Acad. Sci. USA 107, 3281–3282 (2010).
Yang, S. A. et al. FOXO/Fringe is necessary for maintenance of the germline stem cell niche in response to insulin insufficiency. Dev. Biol. 382, 124–135 (2013).
Morita, S., Ota, R. & Kobayashi, S. Downregulation of NHP2 promotes proper cyst formation in Drosophila ovary. Dev. Growth Differ. 60, 248–259 (2018).
Tseng, C. Y. et al. Notch signaling mediates the age-associated decrease in adhesion of germline stem cells to the niche. PLoS Genet 10, e1004888 (2014).
Tseng, C. Y. et al. chinmo-mutant spermatogonial stem cells cause mitotic drive by evicting non-mutant neighbors from the niche. Dev. Cell 57, 80–94 e87 (2022).
Rastegari, E. et al. WD40 protein Wuho controls germline homeostasis via TRIM-NHL tumor suppressor Mei-p26 in Drosophila. Development (Cambridge, England) https://doi.org/10.1242/dev.182063 (2020).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Tang, D. et al. SRplot: a free online platform for data visualization and graphing. PLoS One 18, e0294236 (2023).
Ayyub, C., Banerjee, K. K. & Joti, P. Reduction of Cullin-2 in somatic cells disrupts differentiation of germline stem cells in the Drosophila ovary. Developmental Biol. 405, 269–279 (2015).
Banisch, T. U., Maimon, I., Dadosh, T. & Gilboa, L. Escort cells generate a dynamic compartment for germline stem cell differentiation via combined Stat and Erk signalling. Development 144, 1937–1947 (2017).
Duan, T., Thyagarajan, S., Amoiroglou, A., Rogers, G. C. & Geyer, P. K. Analysis of a rare progeria variant of Barrier-to-autointegration factor in Drosophila connects centromere function to tissue homeostasis. Cell Mol. Life Sci. 80, 73 (2023).
Acknowledgements
We thank C.K.Y., J.M., G.-C.C., C.Y., A.T., E.B., D.M.M., H.W.P., S.-L.C., the Bloomington Drosophila Stock Center, the Vienna Drosophila Resource Center, the Developmental Studies Hybridoma Bank for Drosophila stocks, plasmids and antibodies. We also thank the Taiwan fly core for ordering fly lines and reagents, the Bioinformatics core and the Image core in the Institute of Cellular and Organismic Biology, National RNAi Core Facility at Academia Sinica in Taiwan for providing shRNA reagents and related services, as well as the Small Molecule Metabolomics Core Lab at the Institute of Plant and Microbial Biology, Academia Sinica for their assistance with RNA seq analysis, image analysis, and metabolomics analysis, M.C. for English editing, B.S. for helping graphic abstract, M.-C.L. and C.-H.C. for valuable comments on this article. We would like to thank Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools (2016). This work was supported by the thematic grant (AS-CDA-108-L03) and the Investigator grant of Academia Sinica (AS-IA-113-L05 HJH).
Author information
Authors and Affiliations
Contributions
O.A., L.-H.Y., W.-D.W. and H.-J.H. designed and interpreted the experiments, and wrote the paper. Y.-T.K. performed the 1st screening for lipid metabolism. O.A. and H.-J. Lee performed metabolomics analysis. C.-H.L. generated the iATPSnFR fly line and performed ATP measurement. C.-Y.T. and T.-L.H. performed testis experiments. T.M.L. performed RNA seq analysis. K.K. performed cycloheximide experiment. O.A. performed the remaining experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare that there is no conflict of interest regarding the publication of this article.
Peer review
Peer review information
Nature Communications thanks Todd Strochlic and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Amartuvshin, O., Lin, CH., Ke, YT. et al. Acetyl-CoA carboxylase maintains energetic balance for functional oogenesis. Nat Commun 16, 10677 (2025). https://doi.org/10.1038/s41467-025-65708-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65708-w