Abstract
The progressive accumulation of amyloid beta (Aβ) plaques is a hallmark of Alzheimer’s disease (AD). However, the biochemical mechanisms of their formation and the consequences associated with plaque formation remain elusive. In female 5xFAD and APPNL-G-F mice, we map region-specific, plaque-associated lipids with large molecular coverage including isomers. We describe a multimodal framework that integrates matrix assisted laser desorption/ionization with laser-induced postionization (MALDI-2) mass spectrometry imaging, trapped ion mobility spectrometry, and fluorescence microscopy. Our approach improves detectability and spatial-chemical resolution. We couple these measurements with a computational pipeline for multimodal image coregistration and discovery of plaque-altered lipids. Here, we show the lipids in and around Aβ plaques are highly heterogeneous. Integration of our data with existing spatial transcriptomics data suggests that region-specific accumulation of simple gangliosides is likely driven by lysosomal degradation of complex species. Together, this work provides a generalizable framework to understand lipid alterations within the Aβ plaque microenvironment.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the most prevalent form of dementia1. The hallmark pathologies of AD include amyloid beta (Aβ) plaque formation and neurofibrillary tangles, which are associated with cell death and brain atrophy1,2. Recently, spatial transcriptomic studies have revealed the rich and complex molecular alterations in the Aβ plaque microenvironment3. In concert, there is a rising interest to investigate the connections between brain lipids, which cannot be directly measured from gene expression data, and their relation to the progression of AD pathologies4. For example, the most significant genetic risk factor for late-onset AD is the presence of the E4 allele of apolipoprotein (APOE E4), a lipid transport protein5. It has been suggested that APOE4 promotes neuroinflammatory responses6, including the formation of reactive astrocytes and activated microglia. Further evidence suggests that cholesterol-rich lipid rafts may be the primary domain in which processing of amyloid precursor protein takes place7. Interestingly, several classes of lipids appear to be spatially altered in the vicinity of Aβ plaques. These include gangliosides, an abundant class of glycolipids, and sulfatides (ST), specialized components of myelin sheaths, which are decreased early in AD pathogenesis8. Taken together, these findings highlight a strong link between AD pathology and the lipidome.
Matrix assisted laser desorption/ionization mass spectrometry imaging is a tool of choice for the label-free mapping of hundreds of lipids and other endogenous molecules simultaneously and in situ, offering unmatched spatial-chemical specificity9,10,11. The ability of MALDI to map lipids, as well as Aβ peptides, in rodent models of AD has been demonstrated12,13,14,15,16,17,18,19,20,21, revealing colocalization of several lipids to Aβ plaques, including simple gangliosides, concomitant with the depletion of ST in the same areas. Further, lipid and peptide imaging has been applied to postmortem human AD tissue from both familial22 and sporadic AD patients23,24,25, demonstrating that many of the same lipid species observed to be spatially altered in animal models are also dysregulated in humans.
Despite progress, the molecular portrait of the lipidome in AD is incomplete due to limited chemical detail, spatial resolution, and brain coverage offered by existing approaches. Particularly, reliance on targeted lipid analysis and manual peak selection inherently limits the obtainable lipid coverage, leading to potential biases. Second, previously reported spatial resolutions are insufficient to fully characterize the plaque microenvironment. Third, large brain coverage is necessary to understand global lipid alterations between different key anatomical regions, especially those more susceptible to AD pathology versus less resilient areas. Finally, multiple isomeric forms of lipids are functionally important but nontrivial to characterize, requiring enhanced instrumentation and methods.
Here we present a combined instrumental and computational workflow for unbiased mapping and nontargeted discovery of AD-associated lipids with high sensitivity and molecular specificity, using the 5xFAD mouse model of AD as an evaluation platform26,27. Specifically, MALDI with post-ionization (MALDI-2)28,29 is employed to enhance detectability and facilitate imaging at high-spatial resolutions while maintaining chemical detail. We then introduce a computational pipeline for coregistration of MSI data with Aβ-stained fluorescence images, automatic Aβ-plaque-localized pixel selection in the hyperspectral MS images, and unsupervised clustering of plaque-associated lipid profiles across brain regions. This workflow follows the concepts of our image-guided mass spectrometry approach, microMS30 used for single cell10,31 and subcellular measurements32. Here, instead of dissociating the tissue, we use optical microscopy to guide the data analysis of MS images, targeting extracellular plaques rather than cells33. By comparison of pixels associated with plaques and control pixels, lipids enriched or depleted within and near Aβ plaques are highlighted. Multimodal acquisition and coregistration25,34 also revealed heterogeneity in lipid contents among plaques in different brain regions. Finally, we employ trapped ion mobility spectrometry (TIMS) to resolve lipid isomers exhibiting differential distribution in AD tissue. Although TIMS MSI has been applied to study AD models, identification of lipid isomers was not achieved35. To expand on this work, we confirm the structure of the several lipid isomers by sequential TIMS–collision-induced dissociation. This approach enables spatial lipidomic profiling of AD brains with combined high spatial resolution and isomer separation, providing a powerful tool to study biochemistry in the heterogeneous Aβ plaque microenvironment.
Results
Non-targeted discovery of plaque-associated lipids
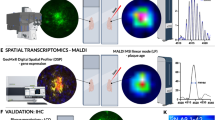
To probe the lipid landscape within the Aβ plaque microenvironment, we developed a high-spatial-resolution MALDI-2 MSI approach with multimodal data analysis (Fig. 1). Here, the use of 5 µm spatial resolution with excellent detectability14, was achieved by leveraging the signal enhancement afforded by MALDI-2. A multimodal, computational approach was used to discover plaque-associated and plaque-depleted lipids via accurate coregistration of Thioflavin S-stained fluorescence images and MSI data (details in Fig. S1), non-targeted lipid discovery, and atlas-informed brain region segmentation and lipid analysis (Fig. 1).
Obtained from extracted mouse brain, cryosections are imaged using MALDI-2 TIMS MS, followed by Aβ staining on the same-section and fluorescence imaging. The images are coregistered and plaque-associated pixels and brain regions are automatically extracted from MSI data. In our data analysis workflow, plaque-enriched and depleted lipids are determined from multiple hypothesis testing and visualized by volcano plots and subtraction spectra. Plaque-association is confirmed by inspection of ion images for discovered features. Next, we applied machine learning techniques to determine the extent of heterogeneity within plaque pixels across and within different brain regions. We then integrated MSI data with publicly available spatial transcriptomic data to provide insight into the mechanisms of plaque-accumulation in Aβ plaques.
Because the utility of MALDI-2 to profile neurodegenerative disease models has not yet been shown, we first evaluated if MALDI-2 could provide signal enhancement for lipids previously shown to be dysregulated in AD. In positive ion mode, significant signal enhancement was obtained for known AD-associated lipids, including Vitamin A1, cholesterol, and lyso-phosphatidylcholines (Figs. S2 and S3). In negative mode, enhancement of AD plaque-associated phosphatidylethanolamines (PE) and ST was observed (Fig. S4). In contrast, we observed a decrease in signal of monogangliosides (GM) GM1(36:1) and GM(38:1), which were associated with the generation of asialo-ganglioside (GA1) fragment ions, in agreement with the observations of Soltwisch et al.28 (Fig. S4). Overall, we found that MALDI-2 enhanced the signal of many low-abundance species in both positive and negative ion modes. Encouraged by the signal enhancement observed in these characterization experiments, we next generated a non-targeted workflow to find plaque-associated lipids using the detection enhancement from MALDI-2. Because a majority of lipid classes known to be implicated in AD are better detected in negative ion mode, we focused on negative mode data for the remainder of this work.
To assess the viability of our multimodal computational approach, we compared fluorescence images with MALDI ion images of lipids previously reported to be plaque localized. Individual Aβ plaques were readily detected by fluorescence imaging following Thioflavin S staining of coronal tissue sections from a 5xFAD mouse brain (Fig. 2a). Comparison of fluorescence images to MSI images of monoganglioside GM3(36:1) (Fig. 2b), a lipid known to accumulate in Aβ plaques22,25,36, obtained at 5-µm spatial resolution from the same section post-MSI show a high degree of colocalization (Fig. 2c), validating this approach. Additional comparison to a lipid species known to be depleted in Aβ plaques, the sulfatide ST(42:1)22 provides further confirmation (Fig. 2d–f). In principle, direct imaging of Aβ peptides via MALDI post-lipid MSI can be done. However, since peptide imaging on the same section requires tissue rinsing, matrix reapplication, and an additional MSI run, we chose a multimodal MSI and fluorescence approach to facilitate lipid discovery due to higher throughput as well as higher spatial resolution of Aβ plaques. However, to confirm the specificity of Thioflavin S to Aβ plaques, we compared fluorescence images to direct detection of Aβ1-40 and Aβ1-42 via MALDI (Supporting Information and Fig. S5) and observed excellent correlation.
a Fluorescence images of tissue stained for Aβ plaques by Thioflavin S post-MSI. b MSI image of the same region shows GM3(36:1), a monoganglioside. c Merged fluorescence and MSI data show colocalization of GM3(36:1) to Aβ plaques. d Colocalization analysis by scattering normalized Thioflavin S intensity against GM3(36:1) intensity. e Thioflavin S and Hoechst highlights cell nuclei size and location relative to plaques, arrows indicate regions of Thioflavin S+ plaques f MSI image of ST(42:1) shows depletion of sulfatides around Aβ plaques, g merged fluorescence and MSI image of ST(42:1) and Thioflavin S. h Colocalization analysis of Thioflavin S and ST(42:1). M1 and M2 denote Mander’s overlap coefficients. All n = 6 5xFAD animals were stained, representative images are shown. Localization trends were consistent across all animals.
Building on these results, we next developed a computational approach to select pixels from Aβ plaques and compare them to control pixels for nontargeted discovery of altered lipids. Briefly, MSI data and fluorescence data (obtained post-MSI) were coregistered via affine transformation (Fig. S1). Pixels in the MSI data that aligned with Aβ plaques were extracted (Fig. 3a, b), while an equal number of control pixels were extracted from regions that did not stain for Thioflavin S (Fig. 3c). Specifically, control pixels were selected by first dilating a region of pixels around each plaque within a 200 µm diameter. Any pixels in the plaque-surrounding region that were positive for Thioflavin S, indicative of a nearby plaque, were excluded and replaced with the next-closest Thioflavin S negative pixels to maintain an equal number of pixels in each group. We adopted this dilation strategy as a conservative approach to ensure that each set of “plaque pixels” fully encompassed the plaque considering the resolution differences between MSI and fluorescence microscopy (Fig. 2).
a Representative ion image at m/z 1179.73, representative of all measurements from 5xFAD animals. b Extraction of plaque-localized pixels from the ion image. c Extraction of control pixels from the same ion image. d Volcano plot highlighting discovery of plaque enriched and depleted lipids. Data were compared with a two-sided t-test (no FDR correction). e Average difference mass spectrum highlights discovered enriched and depleted lipids. f A zoomed-in region of the difference plot from m/z 1100 to 1600 is included to highlight ganglioside differences. Measurements in all panels were obtained from n = 3 animals, two sections per animal. Source data are provided as a Source data file.
To avoid redundant features from isotopic peaks as well as matrix and fragment ions, we first refined m/z features to a list of putative lipids. To accomplish this, we searched all m/z features on LIPID MAPS37 for negative ions with a mass tolerance of 0.01 m/z. This feature list was further refined by ensuring that each putative lipid also exhibited at least one expected isotopic peak in addition to the monoisotopic m/z. Next, we tested each of the refined lipid features for significance to identify putative plaque-associated lipids. A parametric t-test was used as a majority (79%) of lipid features were normally distributed (p > 0.05 via Shapiro–Wilk test).
The results are readily visualized in volcano plots (Fig. 3d) and averaged subtraction plots (Fig. 3e). We observed plaque-colocalization of monogangliosides, including GM3 and GM2, PE, phosphatidylinositols (PI), and lyso-phosphatidylinositols (LPI). In contrast, ST, including oxidized (ST(OH)) was depleted within plaques. To provide additional confidence in lipid identifications beyond mass matching, on-tissue MS/MS was obtained for selected altered lipids (Figs. S6–S8, and Table S1). We next sought to investigate how lipids were altered across and within major brain regions.
Brain-regional analysis
To contextualize the discovered plaque-associated lipids, we investigated lipid alterations across larger anatomical regions between 5xFAD and wild type (WT) animals by comparing plaque pixels to an equal number of randomly selected pixels from matched brain regions in WT animals (n = 6 for each group). Specifically, we investigated the hippocampus, cerebral cortex, entorhinal cortex, and midbrain (Fig. 4a). We observed that many plaque-associated lipids were differentially altered across brain regions with trends that approximately followed their natural abundance in the brain. For instance, LPI(18:0), GM3(36:1), and GM2(36:1) were significantly enriched in 5xFAD mice relative to WT animals (p ≤ 0.05 via unpaired t-test) in all brain regions investigated with region-dependent differences further identified (Fig. 4b–f, exact statistics are provided in the Source data). Higher levels of these lipids in the hippocampus relative to the cerebral cortex were observed, which is consistent with previous work showing that different plaque-enriched lipids, detected via DESI, were enriched in the hippocampus relative to the cerebral cortex38. In contrast, ST were significantly decreased across all major brain regions (Fig. 4e, f). The regions more susceptible to pathologies (e.g., hippocampus and cerebral cortex) exhibited greater lipid alterations than other regions, with the exception of a more significant decrease of ST in the midbrain—the region where this lipid is most abundant.
a Anatomical annotations for brain regions studied and optical image showing location of region of interest (ROI) for high-spatial-resolution imaging. The left-hemisphere image is derived from the Allen Mouse Brain Atlas (CCFv3), atlas.brain-map.org. b–f (left) Ion image at 5 µm spot size and pitch of plaque-associated lipids and (right) their relative amounts in various brain regions determined from whole-brain-slice imaging at 50 µm spot size and pitch. Each point is averaged from all sections imaged for a given animal (n = 6 per group). Box plots are shown with center line as median, boxes across the 25th–75th percentile (the interquartile range, or IQR), and whiskers extending to the lowest and highest non-outlier points, where outliers are points more than 1.5 times the interquartile range. Error bars in box plots represent the minimum and maximum non-outlier point. Statistics, including exact p values are provided in the Source data. g Performance evaluation of models for predicting region-of-origin for plaque-associated lipids. Each dot is from one five-fold cross validation using data from all animals (n = 6 per group). Box plots are shown with center line as median, extending to the IQR, with whiskers to the highest and lowest non-outlier points. h Confusion matrix from the random forest model, showing prediction power for different brain regions. i Effect of retained lipid features on the random forest model performance. The shaded area denotes ±1 standard deviation from 5-fold cross validation. j The first two dominant features from t-SNE analysis of lipid profiles from all plaque-localized pixels showing regional heterogeneity, color-coded by brain region. k 14 “plaque-pixel” clusters were identified via DBSCAN. l Heatmaps comparing normalized feature intensity (with each feature scaled between −1 and 1) of plaque-associated lipids organized by cluster ID (left) and brain region (right). Data in box plots, t-SNE, and heat maps are obtained from n = 6 animals per group, with 2–3 sections imaged per animal. *p < 0.01, **p < 0.001 as determined from two-sided, unpaired t-tests. Source data are provided as a Source data file.
We further hypothesized that plaques, which appear identical in Thioflavin S-stained microscopy images may undergo region-specific lipid accumulation. To investigate this, we developed a series of machine learning models and dimensionality reduction to uncover spatial patterns in plaque-lipid biochemistry. First, we trained a random forest (RF) model to identify lipids that best classify pixels in WT animals by brain region. In parallel, we trained a second RF model to identify lipids that best distinguish plaque-containing pixels in 5xFAD animals from non-plaque pixels that were randomly selected from WT animals. We achieved 99% classification accuracy in both models (Fig. S9). The models revealed largely distinct sets of discriminating lipid features, with only four overlapping between the top 20 from each model.
We then asked whether the lipid profiles of plaque-containing pixels alone could be used to predict the brain region of origin. Using the top 30 features from the plaque vs. non-plaque model (ensuring we only looked at plaque-associated features) and using all plaque MSI pixels from 5xFAD animals (n = 6) we trained a new RF tasked with classifying plaque pixels by region. To evaluate performance, we benchmarked the RF model against additional classifiers via five-fold cross validation (Fig. 4h). Based on its performance, we selected RF for all classification tasks (Fig. 4h, i). A confusion matrix using the final RF classifier is shown in Fig. 4h, highlighting that the plaque pixels from the hippocampus had the highest classification success. These results provide strong, quantitative evidence that lipid accumulation in plaques is region-specific. These region-specific lipid patterns may reflect regional differences in cellular type or immune activation across brain regions affected by amyloid pathology.
To uncover latent structure in plaque-lipid profiles without predefined class labels, we applied t-SNE on plaque-associated pixels after retaining 16 lipid features (Fig. 4i). While RF models leverage absolute intensity for classification, we applied z-score normalization prior to t-SNE to control for baseline regional variation and highlight changes due to region-specific lipids in plaques (Fig. S10). This analysis revealed distinct regional structure in the biochemical environments for the plaques (Fig. 4j). We obtained 14 “plaque-pixel” clusters, defined by lipid content, which correlated well to different anatomical regions (Fig. 4k, l).
Pixels from all brain regions were associated with multiple clusters (Fig. 4k), suggesting further heterogeneity within each broadly defined brain structure. For instance, cluster 13—composed primarily of hippocampal plaque pixels—was associated with high levels of PI and LPI, while hippocampal clusters 1 and 11 contained lower levels of PI, LPI, and ST, but higher levels of GM3(36:1) and GM1(36:1) (Fig. 4i). We also compared the Thioflavin S intensity in each brain region to normalized lipid intensities (Fig. S11), which further underscored regional differences, such as the decreased relative abundance of GM3(36:1) in the midbrain, despite the number of plaques. Whole-coronal-section fluorescence and ion images at 10 μm resolution are provided as visual confirmation of the heterogenous distribution of plaque-associated lipids across the brain (Fig. S12). These observations were uniquely enabled by the large brain coverage achieved by our imaging data and computational analysis.
Observing multiple clusters per brain region prompted us to investigate plaque heterogeneity across hippocampal subregions (Fig. 5). In the hippocampus, plaque pixels grouped according to hippocampal region (Fig. 5b). An RF classifier trained to predict the subregion a plaque originated from achieved 84% accuracy, providing additional evidence for heterogeneity in plaque lipid contents within a brain region of major interest in AD. We further observed that the intensities of lipids in plaques depended on the hippocampal region (Fig. 5c). For instance, LPIs and PIs were most abundant within the subiculum. This heterogeneity is further highlighted by intensity box plots, ion images, and feature attribution maps from the RF model (Fig. 5d–f). Taken together, these findings demonstrate striking regional heterogeneities in the composition of lipids associated with AD pathologies.
a Anatomical annotations for hippocampal subregions. The schematic is derived from the Allen Mouse Brain Atlas (CCFv3), atlas.brain-map.org. b t-SNE analysis of lipid profiles for plaque pixels in the hippocampus displays clustering by subregion. c Intensities of features highlight differential accumulation of lipids in different plaque populations. d–f Boxplots of feature intensities across hippocampal subregions (ion identity shown on top), and corresponding ion images (10 μm spatial resolution) and feature attribution images from a random forest classifier. Box plots are shown with center line as median, boxes across the 25th–75th percentile (IQR), and whiskers extending to the lowest and highest non-outlier points, where outliers are points more than 1.5 times the IQR. Data are derived from one specimen, with n (pixels): DG = 485 CA3 = 291, CA1 = 291, subiculum = 1456. Source data are provided as a Source data file.
Integrated MALDI and spatial transcriptomics
Having established that lipid accumulation is region-specific, we next asked what biological mechanisms were responsible for this heterogeneity. Of the altered lipid classes, the accumulation of simple gangliosides such as GM2 and GM3 was particularly striking. This raised the question of whether their enrichment was due to increased synthesis, lysosomal degradation of more complex gangliosides, or both (Fig. 6a). While pathway analysis is generally challenging for many classes of lipids, the synthetic and catabolic pathways for gangliosides are well characterized39, making them ideal for integration with transcriptomic data. Using a publicly available spatial transcriptomics dataset of 5xFAD mice40, we first investigated 20 genes (Table S4) involved in ganglioside synthesis and catabolism (Fig. 6b). Among the most upregulated genes were Hexa, Hexb, and Ctsa, all involved in lysosomal degradation. To verify that this result was not specific to the particular study or the 5xFAD model, we considered the same set of genes from a publicly available spatial transcriptomic dataset of APPNL-G-F mice3. We observed that the same genes–Hexa, Hexb, and Ctsa were among the most upregulated, along with other lysosomal genes, including Glb1 and Asah1. The agreement between these two datasets and models provides strong evidence that lysosomal degradation is not the result of gene overexpression in the transgenic 5xFAD model.
a Pathways for simple ganglioside synthesis and degradation. b Volcano plot of genes involved in ganglioside synthesis and degradation in 5xFAD mice from a publicly available dataset40. c Volcano plot of ganglioside genes in APPNL-G-F mice from a publicly available dataset3. d Coenrichment (log2FC gene × log2FC lipid) by region. All possible lipid gene pairs are considered and are plotted in descending order. e Corresponding coenrichment in APPNL-G-F mice. Data in c, d were compared with a two-sided t-test (no FDR correction). Source data are provided as a Source data file.
To relate gene expression with lipid dysregulation, we calculated gene-lipid coenrichment scores, defined as log2FC(gene) × log2FC(lipid), for gene lipid pairs. Here, we consider all ganglioside-related genes (Table S3) and all detected gangliosides. In addition to our lipid MSI data set from 5xFAD mice, we analyzed a cohort of APPNL-G-F mice (n = 7) to align with the spatial transcriptomics data for this model. Overall, trends in lipid dysregulation were consistent across models, with a lower log2FC for some lipids in APPNL-G-F mice, likely due to its slower pathological development.
Gene-lipid coenrichment is shown in Fig. 6c, d for 5xFAD and APPNL-G-F mice, respectively. Notably, several gene-lipid pairs were highly coenriched between both independent datasets. For instance, Hexa/GM3(36:1), Hexb/GM3(36:1), and Ctsa/GM2(36:1) were highly coenriched in both datasets. The consistency in both upregulated genes and lipids between these models provides strong evidence that lysosomal degradation of complex gangliosides contributes to the accumulation of simple gangliosides in Aβ plaques. Although their fold changes were much smaller, we also observed upregulation in genes responsible for ganglioside synthesis. This suggests that there may be a compensatory increase in ganglioside synthesis in response to lysosomal degradation, indicating that simple ganglioside accumulation in AD may arise from both processes.
Trapped ion mobility separation of AD-associated glycolipid isomers
Motivated by transcriptomic evidence suggesting lysosomal mechanism in ganglioside accumulation, we next investigated if resolving isomer-level differences in gangliosides could further distinguish spatial patterns in plaque-lipid networks. To this end, we enabled TIMS in order to determine which plaque-associated lipids consisted of multiple isomers and then to separate them and map their spatial distribution. We observed that GM1(36:1) and GM1(38:1) both produced two peaks with unique mobilities at the same m/z. Gangliosides are glycolipids composed of a ceramide chain and a glycan, and we putatively assigned the isomers as GM1a and GM1b, which differ in the position of the sialic acid, a charged sugar, on the glycan (Fig. 7a, c). Performing MSI with TIMS engaged revealed unique spatial distributions for each peak (Fig. 7). Notably, both a- and b-isomers were associated with Aβ plaques, despite being generated from different biosynthetic pathways. The differences in their localization was observed in other regions, for instance, GM1b(36:1) was less enriched in the white matter of the corpus callosum relative to GM1a(36:1) (Fig. 7b, d), as illustrated by the red color in this region.
a Extracted ion mobilogram (EIM) at m/z 1544.87 shows the TIMS-enabled separation of GM1b(36:1) and GM1a(36:1) in wild type and 5xFAD mice. b Ion images are shown for mobility-selected GM1b(36:1) in blue, GM1a(36:1) in red, and a merged image of both. In c the EIM at m/z 1572.90 shows separation of the a and b isomers of a longer-chain version of GM1, GM1(38:1) with corresponding ion images in (d).
To confirm the identity of each isomer, we performed on-tissue TIMS with MS/MS (Fig. S14). Because the TIMS separation occurs prior to the collision cell, fragment ions share the mobility of their precursor, such that diagnostic fragment ions can be assigned to each mobility resolved peak. Based on diagnostic fragment ions, we identified the earlier eluting peak as GM1b and later eluting peak as GM1a. These results build off previous MALDI TIMS separation of ganglioside isomers41,42, shown here for the first time in a neurodegenerative disease model.
Discussion
We have shown that advanced MSI tools, combined with fluorescence imaging, machine learning, and transcriptomic integration, can greatly expand our understanding of lipid alterations in AD. By jointly studying plaque-associated lipids and their regional alterations, we discovered previously unseen rich heterogeneities in lipid profiles associated with individual plaques across the brain. To investigate the biological mechanisms underlying this heterogeneity, we integrated lipid MSI data from both 5xFAD and APPNL-GF mice with two independent spatial transcriptomic data sets obtained from each model. In both cases, we observed upregulation of lysosomal genes, including Hexa, Hexb, and Ctsa, along with elevated simple gangliosides. This consistency between both transgenic and knock-in models, at a disease relevant stage of 5–8 months old, demonstrates that the lysosomal signature is not model-dependent or a late-stage pathology due to aging.
Our results are consistent with the findings of Wang et al., who used qPCR to investigate a panel of genes in the cortex and hippocampus of 12-month-old APP/PS1 mice43. Based on a targeted panel of four genes, they observed a significant increase in Hexa in both brain regions. In addition, they report an “unexpected” increase in the synthetic gene B4galnt1. We propose upregulation reflects a compensatory mechanism to lysosomal catabolism and supports a dual mechanism of ganglioside accumulation.
In addition, we demonstrated ion mobility-enabled resolution of a- and b-series ganglioside isomers. This capability is critical to understanding Alzheimer’s lipid pathology given their unique synthetic pathways. For instance, previous work, such as that Taki et al.43 showed alterations in b-series gangliosides (e.g., GD1b and GT1b), but not in a-series gangliosides in postmortem AD tissue, suggesting they play distinct roles. However, the separation of GM1a and GM1b isomers has not previously been shown in AD tissue—an advance facilitated by our methodology. Resolving more subtle forms of isomerization, such as C=C geometry and sn-position may require the addition of derivatization, ion/ion reactions and/or ion multiplexing techniques44,45. Further, many of the plaque-associated lipids identified in our study, including gangliosides GM1 and GM3, also show altered concentrations in plasma46,47 suggesting that MSI-guided findings can be used to refine lipid biomarker panels for early AD diagnosis.
Last, we note that different cell types in the brain, such as astrocytes and neurons have distinct lipids48 and that there are alterations in cell type and composition around Aβ plaques, such as accumulation of disease-associated astrocytes49 and microglia50 are likely contributors to the lipid heterogeneity observed. We hypothesize regional differences in cell-type composition may be a major driver in the heterogeneity of plaque-associated lipids. Recent evidence suggests the accumulation of lipid droplets in microglia, and their transport to neurons, could play a major role in AD pathology51. These cellular changes, along with their structural alterations, are expected to have a direct impact on the lipid profiles within Aβ plaque regions. The emerging fields of single cell proteomics52,53 and single cell lipidomics48,54 hold great potential for uncovering the cell-type specific contributions to lipid biosynthesis and distribution. High spatial resolution MSI, supported by proteomics and immunofluorescence, offers a powerful strategy to unravel the intricate relationship between cell type and lipid alterations.
We introduced a pipeline for unbiased discovery of Aβ plaque-associated lipids and mapped them with high spatial resolution and isomer separation. This was enabled by the integration of fluorescence microscopy, MALDI-2 ion mobility/mass spectrometry, and computational tools. Our approach can be extended to achieve spatial lipidomics and metabolomics in various disease models and tissue types. Work is ongoing to extend this approach to characterize the lipidome in other AD models and human samples in concert with other spectroscopic methods and functional measures38.
Beyond AD, this pipeline is well-suited for examining regional lipid differences at defined brain locations, including extracellular plaques, rare cells, and targeted locations containing diseased tissue. The requirement is that these locations are highlighted via morphology or fluorescence-imaging. The incorporation of photocleavable mass-tags55,56 is an intriguing approach to make the pipeline fully MS-compatible. Photocleavable mass tags provide multiplexed tagging, which could allow more cell types and key protein markers to be detected than traditional fluorescence microscopy. This approach can also correlate changes in cell composition to plaque morphology, which should allow details on the brain response to the plaques to be inferred.
Methods
Animals
All animal procedures were approved by the Illinois Institutional Animal Care and Use Committee (IACUC, protocol 23228) and were performed under strict adherence to national and ARRIVE standards for the ethical treatment and care of animals. We analyzed n = 6 five-month-old female 5xFAD mice26 (C57BL/6J × SJ/LJ), along with n = 6 age- and sex- matched WT controls (C57BL/6J) obtained from The Jackson Laboratory. In addition, we also studied the knock-in model APPNL-G-F (n = 7, 5–8 months old females on a C57BL/6N-derived, C57BL/6J-maintained background), raised from in-house colonies at UIC originally obtained from RIKEN BRC. Because there are significant differences in amyloid pathology between male and female animals, and because we did not seek to study the influence of sex, only female animals were studied. Prior to sacrifice, animals were managed by the Division of Animal Resources at UIUC with a 12 h light-dark cycle, at 22 °C and 55% ambient humidity, and were fed ad libitum. Euthanasia of animals was conducted via slow asphyxiation using CO2 in accordance with the IACUC protocol. Animals were processed in three batches of n = 6–7 animals at a time.
Chemicals and reagents
Thioflavin S, 2,5-dihydroxyacetophenone (DHAP), methanol, acetonitrile, ethanol, and paraformaldehyde (PFA) were purchased from Sigma Aldrich. Hoechst 33342, tris-buffered saline (TBS), and phosphate-buffered saline (PBS) were obtained from Thermo Fisher Scientific.
Sample preparation
Immediately after euthanasia, animals were transcardially perfused with 60 mL of ice-cold modified Gey’s balanced salt solution57. Brains were quickly surgically dissected and frozen on dry ice. To obtain thin tissue sections for MSI, brain regions from all six animals were adhered onto the chuck of a cryostat-microtome (Leica Biosystems) with deionized water. Coronal sections of 12 µm thickness were obtained at bregma −2.80 ± 0.2 mm and thaw mounted onto ITO-coated glass slides (Delta Technologies, Loveland, CO). To help mitigate batch effects all animals studied were sectioned simultaneously and thaw-mounted together onto a given slide.
Tissue sections were coated with DHAP at 10 mg/mL in 8:1:1 MeOH:ACN: H2O using an HTX-M5 Sprayer (HTX Technologies, Chapel Hill, NC). Matrix was applied with 10 passes using a flow rate of 125 µL/min and a nozzle temperature of 60 °C. The sprayer was set to a distance of 60 mm from the sample and applied at 10 psi with a velocity of 1200 mm/min. Ten passes of matrix were applied with 15 s drying time between passes.
For high-spatial-resolution tissue imaging of lipids, DHAP was applied with a laboratory fabricated sublimation setup. The setup consisted of a glass sublimation apparatus (Wilmad Glass) placed onto a heated sand bath. Briefly, ~500 mg of DHAP was evenly spread onto the bottom of the sublimation chamber using a stainless-steel tea strainer. An ITO-coated glass slide was attached to the bottom of the cold finger with double-sided copper tape and sealed within the apparatus with an O-ring. Vacuum was established using a rough pump. The cold finger was then filled with an ice-water slush. Matrix was sublimed by placing the apparatus onto a sand bath heated to 170 °C for 2 min applying a matrix density of 50 µg/cm2.
For TIMS imaging, DHAP matrix was applied with a newly acquired HTX SubliMATE (HTX Technologies) by aliquoting 40 mg of matrix into the ceramic wafer. Next, vacuum pressure was stabilized to ca. 40 mtorr while the top of the apparatus was cooled with an ice-water slush for 5 min. Matrix was sublimed by heating the ceramic wafer to 200 °C for 5 min, yielding a matrix density of 120 µg/cm2.
Mass spectrometry imaging
Lipid MSI was performed on a timsTOF fleX MALDI-2 trapped ion mobility quadrupole time-of-flight mass spectrometer (Bruker Corp., Billerica, MA). The instrument was equipped with a Bruker SmartBeam 3D Laser consisting of two Nd: YAG lasers operating at 355 nm (frequency tripled) and 266 nm (frequency quadrupled) for MALDI and MALDI-2, respectively. The instrument was configured and controlled using timsControl (Client version 5.1.2) and flexImaging (Version 7.4, Bruker Daltonics GmbH & Co. KG). Detailed instrument settings are provided in Table S2. For high spatial-resolution imaging, tissue sections with sublimed matrix application were imaged using a “single M2” beam with either a 5 × 5 µm2 or 10 × 10 µm2 (TIMS off) or 20 × 20 µm2 (TIMS on) spot size. To profile global changes, 2, 3 tissue sections from each animal (n = 6, 7 animals per group, n = 2, 3 sections per animal) were imaged with the “M5 M2” laser beam generating a 50 × 50 µm2 spot size. Spectra were collected at 1 kHz with 100 shots and a post-ionization delay of 10 µs. Before every acquisition, mass calibration was performed using red phosphorus in LDI mode. Lock mass calibration was performed using three abundant lipid ions.
Trapped ion mobility imaging
TIMS with high resolving power was performed to resolve ganglioside isomers using a 25 ms accumulation time with a 1400 ms ramp time. The TIMS in N2 pressure was increased to ~2.55 mbar in order to increase the resolution for high m/z ions58. Concurrently, the Δt6 (ramp start -> accumulation exit) voltage was increased to −175 V to access higher mobilities. A detailed set of TIMS parameters are provided in Table S3. Mobility calibration was performed using the ESI source with ESI-L low concentration tuning mix (Agilent Technologies, USA).
Post-MALDI histological staining
To correlate lipid distributions with amyloid plaque accumulation, the same brain sections analyzed by MSI were subsequently stained with Thioflavin S based on previous protocols59 and modified for MSI. First, DHAP matrix was removed by washing sections in 50% EtOH for 5 min. Next, tissues were fixed in 4% PFA in 1× PBS for 20 min. After fixation, sections were washed for 2 min in 1× in TBS with 0.1% Tween 20 (TBST), followed by a quick wash in TBS and then incubated with 1% w/w Thioflavin S, filtered by 0.22 µm, for 8 min in the dark. After amyloid staining, sections were washed with 80% EtOH (2× for 3 min), 95% EtOH (3 min) and then stained for nuclei in Hoechst 33342 (0.5 µg/mL in 1× TBS) for 10 min. Slides were briefly dried under N2 and mounted with ProLongGold™ Antifade. Tiled fluorescence images were obtained on an Axio M2 imager (Zeiss, Jena, Germany) equipped with an AxioCam ICc5 camera and a 0.63× camera adapter and an X-cite Series 120 Q mercury lamp (Lumen Dynamics, Mississauga, Canada) using Zen 3.1 acquisition software. Images were obtained with a 10× air objective with 20% tile overlap using GFP (ex. 450–490 nm; em. 500–550 nm) and DAPI (ex. 335–383 nm; em. 420–470 nm) filter cubes for Thioflavin S and Hoechst, respectively. Images were exported as .jpeg files for downstream processing.
Data analysis
Raw binary Bruker .d data files were parsed into Python using pyTDF-SDK60 and subsequently written as .mat files for processing in MATLAB 2022b. To mitigate inter-sample technical variability, data were quantile normalized. To address batch effects from analyzing animals collected and measured at three distinct times, the data for experiments that used all 12 animals were first quantile normalized at each m/z and subsequently batch corrected via ComBat61 for all studies. Coregistration of fluorescence and MSI data was performed using affine transformation with at least 8 support points between images. Anatomical regions were manually extracted with reference to the Allen Brain Atlas62.
Plaque-containing MSI pixels were extracted from Thioflavin S+ regions and segmented by region. To provide a quantitative metric for heterogeneity, we assessed how well machine learning models could classify plaque pixels by brain region. We evaluated linear discriminant analysis, support vector machines, RF, and linear regression. Each model was assessed with five-fold cross validation using MATLAB’s “cvpartition”. For high-spatial-resolution regional classification within the hippocampus, we trained an additional RF model on hippocampal plaque pixels, with hippocampal regions as labels. Feature importance was computed using predictorImportance, normalized, and then applied as weights to generate spatially resolved feature attribution maps.
For dimensionality reduction, t-SNE was performed in MATLAB 2022b with the function “t-sne” using Euclidean distance as the distance metric and with all other parameters as default. To account for regional variability, data input to t-SNE were z-score normalized using the region-wise mean and standard deviation calculated from WT animals.
Unsupervised clustering was performed using DBSCAN63 with an epsilon of 4.4 and a minimum cluster size of 100 points.
Integration of spatial transcriptomics data
Publicly available spatial transcriptomics data sets were obtained for 5xFAD mice40 and APPNL-G-F mice3. Detailed information on data acquisition is available in the associated references, relevant steps are briefly summarized as follows. The 5xFAD data set consisted of n = 10 5xFAD and n = 10 WT animals at 7 months of age. One section per animal was analyzed using the 10× Genomix Visium platform, with approximately ~5000 capture spots per slides. The data (as output from Space Ranger) was downloaded from the publicly available dataset at https://zenodo.org/records/10404412. The APPNL-G-F animal data set consisted of n = 5 APPNL-G-F and n = 5 WT animals spanning 3, 6, 12, and 18 months of age. Coronal sections were processed using Spatial Transcriptomics (a predecessor to 10× Genomics Visium platform) with approximately 500–1000 spots per section. All spatial transcriptomics data were imported into MATLAB 2022b using custom scripts for integration with lipid MSI data.
Statistics and reproducibility
Data were checked for normality using a Shapiro–Wilk test. Suitably normal data (p>0.05 fromShapiro–Wilk) were compared with a twotailed t-test. A sample size of n ≥ 6 animals per group was chosen for robust statistics. No statistical method was employed to predetermine sample size. No data were excluded. Sections were selected at random for imaging. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry data in this study have been deposited in the publica MassIVE repository under accession code MSV000099585. Microscopy data and processed mass spectrometry data (.mat) have been deposited in the Illinois Data Bank under accession code IDB-4907703. The complete raw data for all 48 tissue sections imaged is not included due to file size limitations (>2 TB), but is available on request. The spatial transcriptomics data were published previously and are publicly available from Lee et al.40 and Chen et al.3. Source data are provided with this paper.
Code availability
The scripts used to perform image registration, regional analysis, and parsing transcriptomic data are publicly available on GitHub (https://github.com/timtk1/MALDI2Alz)64
References
Masters, C. L. et al. Alzheimer’s disease. Nat. Rev. Dis. Prim. 1, 15056 (2015).
Long, J. M. & Holtzman, D. M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179, 312–339 (2019).
Chen, W.-T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991.e19 (2020).
Wong, M. W. et al. Dysregulation of lipids in Alzheimer’s disease and their role as potential biomarkers. Alzheimers Dement. 13, 810–827 (2017).
Martens, Y. A. et al. ApoE cascade hypothesis in the pathogenesis of Alzheimer’s disease and related dementias. Neuron 110, 1304–1317 (2022).
Candore, G. et al. Inflammation, cytokines, immune response, apolipoprotein E, cholesterol, and oxidative stress in Alzheimer disease: therapeutic implications. Rejuvenation Res. 13, 301–313 (2010).
Cheng, H. et al. Mechanisms of disease: new therapeutic strategies for Alzheimer’s disease—targeting APP processing in lipid rafts. Nat. Clin. Pract. Neurol. 3, 374–382 (2007).
Han, X. Potential mechanisms contributing to sulfatide depletion at the earliest clinically recognizable stage of Alzheimer’s disease: a tale of shotgun lipidomics. J. Neurochem. 103, 171–179 (2007).
Gode, D. & Volmer, D. A. Lipid imaging by mass spectrometry–a review. Analyst 138, 1289 (2013).
Xie, Y. R. et al. Multiscale biochemical mapping of the brain through deep-learning-enhanced high-throughput mass spectrometry. Nat. Methods https://doi.org/10.1038/s41592-024-02171-3 (2024).
Croslow, S. W., Trinklein, T. J. & Sweedler, J. V. Advances in multimodal mass spectrometry for single-cell analysis and imaging enhancement. FEBS Lett. 598, 591–601 (2024).
Carlred, L. et al. Probing amyloid-β pathology in transgenic Alzheimer’s disease (tgArcSwe) mice using MALDI imaging mass spectrometry. J. Neurochem. 138, 469–478 (2016).
Kaya, I. et al. Delineating amyloid plaque associated neuronal sphingolipids in transgenic Alzheimer’s disease mice (tgArcSwe) using MALDI imaging mass spectrometry. ACS Chem. Neurosci. 8, 347–355 (2017).
Kaya, I. et al. Novel trimodal MALDI imaging mass spectrometry (IMS3) at 10 μm reveals spatial lipid and peptide correlates implicated in Aβ plaque pathology in Alzheimer’s disease. ACS Chem. Neurosci. 8, 2778–2790 (2017).
Kaya, I., Zetterberg, H., Blennow, K. & Hanrieder, J. Shedding light on the molecular pathology of amyloid plaques in transgenic Alzheimer’s disease mice using multimodal MALDI imaging mass spectrometry. ACS Chem. Neurosci. 9, 1802–1817 (2018).
Ge, J. et al. Tetramodal chemical imaging delineates the lipid–amyloid peptide interplay at single plaques in transgenic Alzheimer’s disease models. Anal. Chem. 95, 4692–4702 (2023).
Stoeckli, M., Staab, D., Staufenbiel, M., Wiederhold, K.-H. & Signor, L. Molecular imaging of amyloid b peptides in mouse brain sections using mass spectrometry. Anal. Biochem. 311, 33–39 (2002).
Hong, J. H. et al. Global changes of phospholipids identified by MALDI imaging mass spectrometry in a mouse model of Alzheimer’s disease. J. Lipid Res. 57, 36–45 (2016).
Ollen-Bittle, N. et al. Co-registration of MALDI-MSI and histology demonstrates gangliosides co-localize with amyloid beta plaques in Alzheimer’s disease. Acta Neuropathol. 147, 105 (2024).
Caughlin, S. et al. Membrane-lipid homeostasis in a prodromal rat model of Alzheimer’s disease: characteristic profiles in ganglioside distributions during aging detected using MALDI imaging mass spectrometry. Biochim. Biophys. Acta BBA Gen. Subj. 1862, 1327–1338 (2018).
Kaya, I. et al. Brain region-specific amyloid plaque-associated myelin lipid loss, APOE deposition and disruption of the myelin sheath in familial Alzheimer’s disease mice. J. Neurochem. 154, 84–98 (2020).
Michno, W. et al. Spatial neurolipidomics at the single amyloid-β plaque level in postmortem human Alzheimer’s disease brain. ACS Chem. Neurosci. 15, 877–888 (2024).
Mendis, L. H. S., Grey, A. C., Faull, R. L. M. & Curtis, M. A. Hippocampal lipid differences in Alzheimer’s disease: a human brain study using matrix-assisted laser desorption/ionization-imaging mass spectrometry. Brain Behav. 6, e00517 (2016).
Kakuda, N. et al. Distinct deposition of amyloid-β species in brains with Alzheimer’s disease pathology visualized with MALDI imaging mass spectrometry. Acta Neuropathol. Commun. 5, 73 (2017).
Enzlein, T., Lashley, T., Sammour, D. A., Hopf, C. & Chávez-Gutiérrez, L. Integrative single-plaque analysis reveals signature Aβ and lipid profiles in the Alzheimer’s brain. Anal. Chem. https://doi.org/10.1021/acs.analchem.3c05557 (2024).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Mishra, R. et al. Augmenting neurogenesis rescues memory impairments in Alzheimer’s disease by restoring the memory-storing neurons. J. Exp. Med. 219, e20220391 (2022).
Soltwisch, J. et al. Mass spectrometry imaging with laser-induced postionization. Science 348, 211–215 (2015).
Niehaus, M., Soltwisch, J., Belov, M. E. & Dreisewerd, K. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat. Methods 16, 925–931 (2019).
Comi, T. J., Neumann, E. K., Do, T. D. & Sweedler, J. V. MicroMS: a Python platform for image-guided mass spectrometry profiling. J. Am. Soc. Mass Spectrom. 28, 1919–1928 (2017).
Neumann, E. K., Ellis, J. F., Triplett, A. E., Rubakhin, S. S. & Sweedler, J. V. Lipid analysis of 30 000 individual rodent cerebellar cells using high-resolution mass spectrometry. Anal. Chem. 91, 7871–7878 (2019).
Castro, D. C., Xie, Y. R., Rubakhin, S. S., Romanova, E. V. & Sweedler, J. V. Image-guided MALDI mass spectrometry for high-throughput single-organelle characterization. Nat. Methods 18, 1233–1238 (2021).
Enzlein, T. et al. Computational analysis of alzheimer amyloid plaque composition in 2D- and elastically reconstructed 3D-MALDI MS images. Anal. Chem. 92, 14484–14493 (2020).
Wehrli, P. M. et al. Correlative chemical imaging and spatial chemometrics delineate Alzheimer plaque heterogeneity at high spatial resolution. JACS Au 3, 762–774 (2023).
Michno, W. et al. Structural amyloid plaque polymorphism is associated with distinct lipid accumulations revealed by trapped ion mobility mass spectrometry imaging. J. Neurochem. 160, 482–498 (2022).
Kaya, I. et al. Spatial lipidomics reveals region and long chain base specific accumulations of monosialogangliosides in amyloid plaques in familial Alzheimer’s disease mice (5xFAD) brain. ACS Chem. Neurosci. 11, 14–24 (2020).
Conroy, M. J. et al. LIPID MAPS: update to databases and tools for the lipidomics community. Nucleic Acids Res. 52, D1677–D1682 (2024).
Huang, H. X. et al. Mass spectrometry imaging highlights dynamic patterns of lipid co-expression with Aβ plaques in mouse and human brains. J. Neurochem. 168, 1193–1214 (2024).
Sandhoff, R. & Sandhoff, K. Emerging concepts of ganglioside metabolism. FEBS Lett. 592, 3835–3864 (2018).
Lee, E. J. et al. Spatial transcriptomic brain imaging reveals the effects of immunomodulation therapy on specific regional brain cells in a mouse dementia model. BMC Genom. 25, 516 (2024).
Djambazova, K. V. et al. MALDI TIMS IMS of disialoganglioside isomers─GD1a and GD1b in murine brain tissue. Anal. Chem. 1176–1183 https://doi.org/10.1021/acs.analchem.2c03939 (2022).
Djambazova, K. V. et al. MALDI TIMS IMS reveals ganglioside molecular diversity within murine S. aureus kidney tissue abscesses. J. Am. Soc. Mass Spectrom. 35, 1692–1701 (2024).
Taki, T. An approach to glycobiology from glycolipidomics: ganglioside molecular scanning in the brains of patients with Alzheimer’s disease by TLC-blot/matrix assisted laser desorption/ionization-time of flight MS. Biol. Pharm. Bull. 35, 1642–1647 (2012).
Bonney, J. R. et al. Relative quantification of lipid isomers in imaging mass spectrometry using gas-phase charge inversion ion/ion reactions and infrared multiphoton dissociation. Anal. Chem. 95, 17766–17775 (2023).
Xu, S. et al. Spatially and temporally probing distinctive glycerophospholipid alterations in Alzheimer’s disease mouse brain via high-resolution ion mobility-enabled sn-position resolved lipidomics. Nat. Commun. 15, 6252 (2024).
Liu, L., Zhang, K., Tan, L., Chen, Y.-H. & Cao, Y.-P. Alterations in cholesterol and ganglioside GM1 content of lipid rafts in platelets from patients with Alzheimer disease. Alzheimer Dis. Assoc. Disord. 29, 63–69 (2015).
Chua, X. Y. et al. Lipidomics profiling reveals distinct patterns of plasma sphingolipid alterations in Alzheimer’s disease and vascular dementia. Alzheimers Res. Ther. 15, 214 (2023).
Neumann, E. K., Comi, T. J., Rubakhin, S. S. & Sweedler, J. V. Lipid heterogeneity between astrocytes and neurons revealed by single-cell MALDI-MS combined with immunocytochemical classification. Angew. Chem. 131, 5971–5975 (2019).
Pike, C. J., Cummings, B. J. & Cotman, C. W. Early association of reactive astrocytes with senile plaques in Alzheimer’s disease. Exp. Neurol. 132, 172–179 (1995).
Mattiace, L. A., Davies, P., Yen, S.-H. & Dickson, D. W. Microglia in cerebellar plaques in Alzheimer’s disease. Acta Neuropathol. 80, 493–498 (1990).
Haney, M. S. et al. APOE4/4 is linked to damaging lipid droplets in Alzheimer’s disease microglia. Nature 628, 154–161 (2024).
Ghatak, S. et al. Single-cell patch-clamp/proteomics of human Alzheimer’s disease iPSC-derived excitatory neurons versus isogenic wild-type controls suggests novel causation and therapeutic targets. Adv. Sci. 11, 2400545 (2024).
Savas, J. N. et al. Amyloid accumulation drives proteome-wide alterations in mouse models of Alzheimer’s disease-like pathology. Cell Rep. 21, 2614–2627 (2017).
Saunders, K. D. G. et al. Single-cell lipidomics using analytical flow LC-MS characterizes the response to chemotherapy in cultured pancreatic cancer cells. Anal. Chem. 95, 14727–14735 (2023).
Yagnik, G., Liu, Z., Rothschild, K. J. & Lim, M. J. Highly multiplexed immunohistochemical MALDI-MS imaging of biomarkers in tissues. J. Am. Soc. Mass Spectrom. 32, 977–988 (2021).
Claes, B. S. R. et al. MALDI-IHC-guided in-depth spatial proteomics: targeted and untargeted MSI combined. Anal. Chem. 95, 2329–2338 (2023).
Nemes, P., Rubakhin, S. S., Aerts, J. T. & Sweedler, J. V. Qualitative and quantitative metabolomic investigation of single neurons by capillary electrophoresis electrospray ionization mass spectrometry. Nat. Protoc. 8, 783–799 (2013).
Silveira, J. A., Ridgeway, M. E. & Park, M. A. High resolution trapped ion mobility spectrometery of peptides. Anal. Chem. 86, 5624–5627 (2014).
Lazarov, O. et al. Environmental enrichment reduces Aβ levels and amyloid deposition in transgenic mice. Cell 120, 701–713 (2005).
Luu, G. T. et al. TIMSCONVERT: a workflow to convert trapped ion mobility data to open data formats. Bioinformatics 38, 4046–4047 (2022).
Zhang, Y., Parmigiani, G. & Johnson, W. E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2, lqaa078 (2020).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Schubert, E., Sander, J., Ester, M., Kriegel, H. P. & Xu, X. DBSCAN revisited, revisited: why and how you should (still) use DBSCAN. ACM Trans. Database Syst. 42, 1–21 (2017).
Trinklein, T. J. et al. Multimodal mass spectrometry imaging for plaque- and region-specific neurolipidomics in Alzheimer’s disease mouse models. Github Maldi2Alz https://doi.org/10.5281/zenodo.17363890 (2025).
Acknowledgements
Research reported in this publication was supported by the National Institute on Aging of the National Institutes of Health under Award Number R01AG078797 (J.V.S., F.L., O.L.), by the Office of the Director, National Institutes of Health of the National Institutes of Health under Award Number S10OD032242 (J.V.S.), and by the National Institute on Drug Abuse of the National Institutes of Health under Award Number P30DA018310 (J.V.S.). Through acceptance of this federal funding, NIH has been given a right to make this manuscript publicly available in PubMed Central upon the Official Date of Publication, as defined by NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
T.J.T., J.V.S., F.L. and O.L. conceptualized the project. O.L. and K.R.S. provided animals. S.S.R. prepared the animal samples. T.J.T. and. S.W.C. wrote the code for data analysis. T.J.T. and S.O. performed mass spectrometry imaging. T.J.T. and M.A. performed fluorescence microscopy. T.J.T., S.W.C., F.L. and J.V.S. contributed to data analysis and interpretation. T.J.T. wrote an initial draft of the manuscript. T.J.T., J.V.S., F.L. and O.L. contributed to editing the manuscript. All authors contributed to the discussion of the project and have reviewed and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
.Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Trinklein, T.J., Rubakhin, S.S., Okyem, S. et al. Multimodal mass spectrometry imaging for plaque- and region-specific neurolipidomics in Alzheimer’s disease mouse models. Nat Commun 16, 10969 (2025). https://doi.org/10.1038/s41467-025-65956-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65956-w