Abstract
Fibrotic colorectal cancers (CRC) are largely microsatellite-stable and display desmoplastic stroma with poor immune infiltration. Here we identify thrombospondin-2 (THBS2) as a key regulator of the immune-exclusionary phenotype in fibrotic CRC. THBS2 is highly expressed by matrix cancer-associated fibroblasts at the tumor front. In an orthotopic model using desmoplastic tumor organoids, global or fibroblast-specific Thbs2 deletion disrupts the exclusionary barrier and increases intratumoral CD8 T cells. Mechanistically, THBS2 limits recruitment of CXCR3+ CD8 T cells by restraining dendritic- and macrophage-derived CXCL9/10. Depletion of these myeloid cells or blockade of CXCL9/10-CXCR3 signaling abolishes the enhanced CD8 T-cell influx and antitumor efficacy. Spatial profiling demonstrates that THBS2 loss induces proximity between CD8 T cells and myeloid cells and upregulates chemokines. Despite increased infiltration, CD8 T cells manifest exhaustion, rendering tumors highly susceptible to immune checkpoint blockade. THBS2 thus represents a tractable CAF-restricted target to overcome immune exclusion in fibrotic CRCs.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC), a leading cause of cancer-related mortality worldwide, exhibits molecular and clinical heterogeneity, with limited efficacy of immune checkpoint inhibitors (ICIs) in most cases1,2. As in several other malignancies, immune checkpoint inhibitors (ICIs) show remarkable efficacy in CRCs characterized by deficient mismatch repair (dMMR) or high microsatellite instability (MSI-H), including exceptional responses reported in recent neoadjuvant trials (e.g., NICHE)3. However, although dMMR/MSI-H CRCs account for approximately 10–15% of cases and respond exceptionally well to ICIs, the remaining 85–90% are microsatellite stable (MSS) and largely resistant to ICI therapy4,5. This underscores the need to address CRC subtype complexities and the influence of the tumor microenvironment (TME) on therapy.
Molecular classification systems, such as consensus molecular subtypes (CMS) and IMF (based on intrinsic epithelial subtype (I), MSI status (M) and fibrosis (F)), provide insights into the TME and immune landscape of CRC. Fibrotic subtypes (CMS4 and IMF_F) are typically MSS and are associated with cancer-associated fibroblasts (CAFs), poor prognoses, and immune exclusion6,7. CMS2, CMS3, and IMF_NF (non-fibrosis) are also MSS but represent immune-cold subtypes amenable to cytotoxic and targeted treatments like VEGF and EGFR antibodies. In contrast, immune-hot CMS1/MSI tumors, enriched for cytotoxic immune cells, display high CD8+ T cell infiltration and ICI sensitivity5,8. However, fibrotic CMS4/IMF_F CRCs feature immunosuppressive myeloid cells, regulatory T cells (Tregs), and TGF-β-driven desmoplastic stroma, creating an exclusionary environment that prevents CD8+ T cell infiltration9,10,11. Although TGF-β inhibition can sensitize fibrotic CRCs to ICIs12,13,14, such approaches often cause notable side effects9,15, highlighting the need for alternative strategies.
Single-cell transcriptomics have revealed CAF heterogeneity and distinct immune regulatory roles16,17,18, yet targeting CAFs to enhance ICI efficacy remains challenging due to a lack of targetable CAF-specific molecules responsible for the immune-exclusionary environment. Strategies aiming at extracellular matrix (ECM) components (e.g., collagens and hyaluronan) show promise but are limited by systemic toxicity19,20,21. Thus, identifying cancer-specific mechanisms driving immune exclusion is critical.
Thrombospondin-1 (THBS1), a matricellular protein, regulates immune activity and angiogenesis22. Unlike structural ECM components, matricellular proteins are minimally expressed in normal tissues but upregulated in pathological states, including cancer, making them attractive targets23. While blocking THBS1 restores CD8+ T cell function, it does not disrupt the exclusionary barrier, limiting ICI efficacy24. Thrombospondin-2 (THBS2), a structural homolog of THBS1, differs functionally and is implicated in tissue remodeling and collagen organization25,26, with context-dependent roles in tumor biology27,28. Recent work has implicated that CAF-derived THBS2 promotes CRC progression via a CAF-epithelial pathway29. However, specific involvement of CAF-derived THBS2 in the regulation of CRC TMEs, particularly for immune exclusion, has remained unclear.
Here, we identify THBS2-expressing mCAFs at the tumor front as a critical regulator of T-cell exclusion in fibrotic CRCs. Integrating single-cell RNA sequencing (scRNA-seq), spatial profiling, and orthotopic CMS4-like tumor models, we uncover a previously unrecognized CAF–myeloid cell axis in which THBS2 suppresses CXCL9/10–CXCR3-mediated recruitment of effector CD8+ T cells. We further show that loss of THBS2 renders otherwise ICI-refractory fibrotic CRCs sensitive to anti-PD-1/anti-CTLA-4 therapy. These findings extend prior work on THBS2 by revealing a distinct immunological mechanism and highlight THBS2 as a therapeutic target to overcome immune exclusion in CRC.
Results
THBS2 is expressed by tumor fibroblasts within the mCAF subset
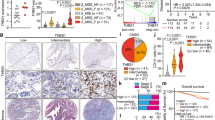
Analysis of the Cancer Genome Atlas (TCGA) CRC dataset demonstrated that THBS2 expression was enriched in CMS4 CRC, with a higher CMS4 proportion in the THBS2-high group (Fig. 1a and Supplementary Fig. 1a). IMF classification confirmed predominant THBS2 expression in fibrotic iCMS2_MSS_F and iCMS3_MSS_F subtypes, particularly the latter (Fig. 1b). THBS2-high CRCs exhibited worse prognosis in the iCMS3 cohort, aligning with its enrichment in the poor-prognosis iCMS3 epithelial subtype (Fig. 1c, d). Positive correlations with stroma-related genes (THBS1, FAP, ACTA2, COL1A1, COL1A2, and VIM) suggests a stroma-associated role of THBS2 (Supplementary Fig. 1b, c). Co-immunofluorescence and RNAscope analyses confirmed the localization of THBS2 in E-cadherin-negative stroma, further supported by the transcriptomic analysis, showing the highest THBS2 expression in tumor stroma (Supplementary Fig. 1d–f).
a, b THBS2 expression in each subtype of CMS (a, n: CMS1 = 85, CMS2 = 132, CMS3 = 78, CMS4 = 184) and IMF (b n: MSI = 82, iCMS2_MSS_NF = 117, iCMS3_MSS_NF = 73, iCMS2_MSS_F = 101, iCMS3_MSS_F = 51) in TCGA-COADREAD dataset. c THBS2 expression in iCMS2 or iCMS3 in TCGA (n: iCMS2 = 298, iCMS3 = 232). d Kaplan-Meier curves for overall survival of iCMS2 or iCMS3 patients according to THBS2 expression in TCGA dataset (n: iCMS2, THBS2 high = 139, THBS2 low = 143; iCMS3, THBS2 high = 118, THBS2 low = 106). e–m scRNA-seq analyses of Colorectal Atlas dataset30 (n = 192). Uniform manifold approximation and projection (UMAP) plot of all cells colored by tissue origin (e) and cell types (f). UMAP feature plot colored by THBS2 expression (g). Violin plots for indicated gene expression in fibroblast subsets (h). UMAP plot of tumor fibroblast subsets colored by cell types (i). Dot plots of indicated gene expression across CAF subtypes (j). Violin plots for indicated gene expression (k) and gene signatures (l) in tumor fibroblast subsets. Scatter plots showing the proportions of THBS2-positive CAFs and mCAF-positive CAFs in individual patients (m). n Co-immunostaining for THBS2 (RNAscope) and CD8 in CMS-annotated human CRC. T: tumor, NT: non-tumor, F: tumor front, I: tumor interior. White lines denote tumor borders. o–t Xenium 5K panel analyses of tumor–non-tumor interfaces in CMS1/MSI and CMS4/MSS CRCs. T tumor, NT non-tumor, F tumor front. Spatial maps (o,q,r) and corresponding pie charts (p) showing annotated cell types. Black dashed lines denote tumor borders. Three-dimensional density maps (s) and quantification of CD8⁺ T-cell density (n = 6, respectively; technical replicates) (t). Dunnett’s test, two-sided (a, b), unpaired Student’s t-test, two-sided (c), Log-rank test (d), Wilcoxon rank sum test, two-sided (k, l), Pearson correlation coefficient, two-sided (m), and Mann-Whitney test, two-sided (t). Mean ± SEM. Scale bars, 200 μm (n, o, q, r). Adjustments for multiple comparisons were made in (a, b), and not in (k, l). Box and whiskers graphs indicate the median and the 25th and 75th percentiles, with minimum and maximum values at the extremes of the whiskers. Source data are provided as a Source Data file.
scRNA-seq analysis of an integrated human CRC cohort30 identified stromal cells, particularly tumor fibroblasts (CAFs), as the primary producers of THBS2 (Fig. 1e–h). CAF subclassification revealed predominant THBS2 expression in matrix CAFs (mCAFs) (Fig. 1i–k)17, co-localizing with the mCAF marker SFRP2 and distinct from the inflammatory CAF (iCAF) marker CXCL14 (Supplementary Fig. 1g–j). The THBS2-high group showed increased enrichment of mCAF signature genes (Fig. 1l). Furthermore, a positive correlation was observed between THBS2 expression and mCAF signature enrichment in patient-based classification (Fig. 1m). These findings establish a strong association between THBS2 and the mCAF subset in CRC, in line with a previously reported finding29.
THBS2-expressing CAFs are predominantly localized at the tumor front of fibrotic CRC
We next examined the relationship between THBS2 expression, mCAF signatures, and immune composition across CMS classes. Immune-active CMS1 showed the highest T-cell infiltration, whereas immunosuppressive fibrotic CMS4/MSS CRCs exhibited the predominant myeloid cell infiltration (Supplementary Fig. 1k). The mCAF signature was upregulated in tumor fibroblasts from both CMS1 and CMS4, with stronger enrichment in CMS4 (Supplementary Fig. 1l). Consistently, Visium spatial transcriptomics illustrated that mCAF signatures were enriched within the tumor core of CMS4/MSS CRC, whereas cytotoxic T-lymphocyte (CTL) signatures were enriched in CMS1/MSI CRC, serving to highlight spatial localization patterns (Supplementary Fig. 1m). RNAscope and immunofluorescence analyses further showed that THBS2+ CAFs were specifically located at the tumor front of CMS4/MSS CRC, where CD8+ T cells were excluded from the tumor periphery (Fig. 1n). Conversely, THBS2 expression was rarely detected in CMS1/MSI CRC, which exhibited abundant CD8+ T cell infiltration (Fig. 1n). Pseudo-bulk analysis of individual cellular components revealed a negative association between the proportion of THBS2+ CAFs and CD8+ T cells at the patient level (Supplementary Fig. 1n,o).
To increase spatial resolution, we profiled tumor–non-tumor interfaces using the Xenium 5 K panel, comparing CMS1/MSI and CMS4/MSS CRCs (Supplementary Fig. 2a,b). This revealed distinct immune TMEs between the two subtypes, with greater enrichment of THBS2+ CAFs at the tumor front of CMS4/MSS CRC (Fig. 1o–q). Spatial domain clustering integrating cellular and neighboring transcriptomes31 identified immune-rich domains (domains 4, 5, and 6) within CMS1/MSI tumor areas, whereas CMS4/MSS tumors were enriched for a fibrotic, immune-low domain (domain 3) (Supplementary Fig. 2c–e). Furthermore, CD8+ T cells in CMS4/MSS CRC were excluded from tumor to adjacent non-tumor regions at the front (Fig. 1r–t).
Tissue microarray (TMA) analysis showed that the THBS2-high group exhibited enhanced fibrosis, positive associations with SFRP2 expression, and reduced CD8+ T cell infiltration (Supplementary Fig. 2f–j). Exclusive CD8 staining pattern was positively associated with THBS2 expression in CD8-positive CRCs (Supplementary Fig. 2f). Consistent with the aggressive nature of THBS2-high tumors, patients with high THBS2 expression had worse prognosis than those with low THBS2 expression (Supplementary Fig. 2k). Furthermore, serum THBS2 levels were elevated in patients with advanced-stage CRC and positively correlated with the abundance of THBS2⁺ CAFs in matched resected specimens, suggesting that circulating THBS2 levels reflect tumor burden (Supplementary Fig. 2l–o).
Collectively, bioinformatic and clinical sample analyses identified mCAFs as the primary source of THBS2, linking its expression to fibrotic CRC, CD8+ T cell exclusion, and poor prognosis.
THBS2 loss in the TME impairs tumor burdens by enhancing antitumor immunity in mouse fibrotic CRC
To define the in vivo role of THBS2, we performed syngeneic orthotopic implantation of mouse tumor organoids (MTOs) with a CMS4-like mesenchymal phenotype12,32, harboring mutations in Apc, Trp53, Kras, and Tgfbr2, into the rectum of wild-type (WT) mice (Fig. 2a). As in human CMS4 CRC, MTO-derived tumors expressed THBS2 predominantly at the tumor front, where a substantial number of CD8+ T cells were excluded (Fig. 2b). Notably, the number of infiltrating CD8+ T cells decreased over time as tumors became more fibrotic (Fig. 2b). We compared MTO-derived tumors with those from MC38, an immunogenic CMS1/MSI-like mouse colon tumor cell line (Supplementary Fig. 3a). While MC38-derived tumors displayed robust CD8+ T cell infiltration with minimal THBS2 expression, MTO-derived tumors showed high THBS2 expression in both tumor tissue and serum (Supplementary Fig. 3b–d), linking THBS2 to the immune-exclusion phenotype of fibrotic CRCs. MTO-derived tumors also expressed the CAF marker Acta2 and mCAF markers Fap and Sfrp2, but not the iCAF marker Dcn (Supplementary Fig. 3c). Co-immunofluorescence confirmed THBS2 localization to E-cadherin-negative stromal regions, colocalizing with αSMA and FN (Supplementary Fig. 3e).
a Schematic representation of orthotopic implantation of MTO to WT mice. b Immunofluorescence (left) for CD8 and THBS2 in orthotopic MTO tumors, and quantification of CD8+ cells (right, n = 3; biological replicates). Scale bars, 500 μm (top), 100 μm (bottom). White lines denote tumor borders. c Schematic representation of orthotopic implantation of MTO to WT or Thbs2-/- mice. d Macroscopic images (top) and H&E staining (bottom) of orthotopic implantation in WT or Thbs2-/- mice. Scale bars, 1 mm. e, f Volume (e, n: WT = 15, Thbs2-/- = 16) and the change in diameter (f, n: WT = 12, Thbs2-/- = 15) of orthotopic tumors in WT or Thbs2-/- mice. g, h Incidence (g, left), macroscopic numbers (g, right), and H&E staining (h) of liver metastasis (met, arrows) in WT or Thbs2-/- mice (n: WT = 15, Thbs2-/- = 16). Scale bars, 1 mm. i Kaplan-Meier curves after MTO implantation (n: WT = 17, Thbs2-/- = 19). j, Immunofluorescence for CD8, THBS2, and αSMA in orthotopic tumors in WT or Thbs2-/- mice 5-weeks post-injection of MTOs. Scale bars, 500 μm. k, Volcano plot of upregulated genes in Thbs2-/- mice versus WT mice after MTO implantation (n = 3 biological replicates). l–n Anti-CD8 antibody (αCD8 ab) treatment on MTO-bearing WT or Thbs2-/- mice (n: WT control = 6, WT αCD8 ab = 6, Thbs2-/- control = 7, Thbs2-/- αCD8 ab = 8). Schematic representation (l), macroscopic images (m), and volumes and weights (n) of tumors. o,p Immunohistochemistry for indicated proteins (o) and quantification (p) in orthotopic tumors of WT or Thbs2-/- mice (n = 3). Red dashed lines denote tumors (d, m). Scale bars, 50 μm. Tukey’s test, two-sided (b), Unpaired Student’s t-test, two-sided (e, g), two-way ANOVA (f), Log-rank test (i), Fisher’s exact test, two-sided (g), Fisher’s LSD test, two-sided (n, p). Mean ± SEM. Adjustments for multiple comparisons were made in (b). Source data are provided as a Source Data file.
To evaluate the effect of THBS2 loss in non-epithelial cells, we analyzed orthotopic MTO tumors in Thbs2-/- mice (Fig. 2c). Negligible THBS2 expression in these tumors suggests that THBS2 is primarily derived from non-epithelial cells (Supplementary Fig. 3f). Strikingly, Thbs2-/- mice developed smaller tumors, fewer liver metastases, and had improved prognosis compared to WT mice (Fig. 2d–i). Tumors in Thbs2-/- mice lacked THBS2 expression at the tumor front and instead displayed pronounced CD8+ T cell infiltration into the tumor interior, without changes in αSMA levels (Fig. 2j), suggesting that THBS2 loss disrupts the exclusionary barrier for CD8+ T cells. Despite equivalent proliferation, apoptosis was increased in Thbs2-/- tumors (Supplementary Fig. 3g). Transcriptomic analyses revealed upregulation of genes and signatures associated with antitumor immunity, inflammation, and apoptosis (Fig. 2k and Supplementary Fig. 3h–j). Importantly, anti-CD8 antibody treatment in Thbs2-/- mice, that effectively depleted CD8+ T cells, restored tumor growth and concomitantly reduced apoptosis (Fig. 2l–p), confirming the cytotoxic activity of the infiltrating CD8+ T cells in Thbs2-/- tumors.
Given that colon cancers differ from rectal cancers in biological behavior, metastatic potential, and therapeutic response33 and that the CMS4 subtype is more prevalent in colon cancer34, we tested a colon cancer model by injecting MTOs into the cecum of WT and Thbs2-/- mice (Supplementary Fig. 3k). As in the rectal model, Thbs2-/- mice developed smaller cecum tumors with increased CD8+ T cell infiltration (Supplementary Fig. 3l, m).
Together, these results indicate that TME-derived THBS2 promotes the progression of fibrotic CRCs by establishing an exclusionary barrier that limits antitumor CD8⁺ T-cell responses.
CAF-derived THBS2 promotes CD8+ T cell exclusion and the progression of fibrotic CRC
To specifically investigate the role of fibroblast-derived THBS2, we generated floxed mice for Thbs2 (Thbs2f/f) by targeting exon 3 (Fig. 3a and Supplementary Fig. 4a,b). Thbs2f/f mice were crossed with Col1a2-cre mice to obtain fibroblast-specific Thbs2 knockout mice (Thbs2ΔFibro) for orthotopic rectal implantation of MTOs (Fig. 3b). Tumors from Thbs2ΔFibro mice exhibited markedly reduced THBS2 expression (Fig. 3c), confirming fibroblasts as the primary source of THBS2. Notably, Thbs2ΔFibro mice developed smaller tumors, lacked the exclusionary barrier, had increased CD8+ T cell infiltration, and showed enhanced apoptosis, mirroring Thbs2-/- mice (Fig. 3d–h).
a Gene targeting strategy for Thbs2. b Schematic representation of orthotopic implantation of MTO to Thbs2f/f or fibroblast-specific Thbs2 knockout mice (Thbs2ΔFibro). c Thbs2 expression in MTO-derived orthotopic tumors from Thbs2f/f or Thbs2ΔFibro mice analyzed by qRT-PCR (n = 3). d Macroscopic images of orthotopic tumors in Thbs2f/f or Thbs2ΔFibro mice. e Tumor diameter in Thbs2f/f or Thbs2ΔFibro mice (n = 11). f Immunofluorescence for CD8 and THBS2 in orthotopic MTO tumors. F: tumor front, I: tumor interior. Magnified views (middle and right) correspond to the yellow and magenta boxed regions in the left panels. g Immunohistochemistry for cleaved-caspase 3 (C-Cas3) in orthotopic MTO tumors. h Quantification of CD8+ cells in (f) and C-Cas3+ cells in (g; n = 3; biological replicates). i, Schematic representation of orthotopic co-implantation of MTO and WT CAF or Thbs2-/- CAF into WT or Thbs2-/- mice. Comparisons were made between tumors in WT mice implanted with WT CAF, tumors in Thbs2-/- mice implanted with Thbs2-/- CAF, and tumors in Thbs2-/- mice implanted with WT CAF. j, k Macroscopic images (j) and volumes (k) of orthotopic tumors in (i)(n: MTO & WT CAF → WT = 12, MTO & Thbs2-/- CAF →Thbs2-/- = 10, MTO & WT CAF→Thbs2-/- = 10). l–o Immunofluorescence (l) and immunohistochemistry (m) for indicated proteins in orthotopic tumors from (i), with quantifications (n, o) (n = 3; biological replicates). F: tumor front, I: tumor interior. Magnified views (middle and bottom) correspond to the yellow and magenta boxed regions in the top panels (l). Scale bars: 1 mm (d, j), 200 μm (f, l), 50 μm (g, m). Unpaired Student’s t-test, two-sided (c, e, h), Šídák’s multiple comparison test, two-sided (k), Tukey’s test, two-sided (n, o). Mean ± SEM. Adjustments for multiple comparisons were made in (n, o, k). Source data are provided as a Source Data file.
We next isolated CAFs from the orthotopic rectal tumors of WT and Thbs2-/- mice. CAFs isolated from WT and Thbs2-/- tumors showed comparable growth and migration, but Thbs2-/- CAFs exhibited suppressed epithelial-mesenchymal transition (EMT) and TGF-β signaling with enrichment of an iCAF signature (Supplementary Fig. 4c–e), indicating a shift from mesenchymal to inflammatory phenotypes. Subsequently, we performed co-injection experiments of these CAFs with MTOs (Fig. 3i). As expected, co-injecting CAFs into host mice of the same genotype did not alter tumor phenotypes (Fig. 3j,k). Co-injection of MTOs with WT CAFs into WT mice yielded tumors with high THBS2 expression and CD8+ T cell exclusion at the tumor front (Fig. 3l). Conversely, co-injection with Thbs2-/- CAFs into Thbs2-/- mice produced smaller tumors with pronounced CD8+ T cell infiltration (Fig. 3j–l). In contrast, co-injection of MTOs with WT CAFs into Thbs2-/- mice restored tumor size, THBS2 expression, and the exclusionary barrier for CD8+ T cells, with reducing apoptosis (Fig. 3j–o). These findings emphasize the pivotal function of CAF-derived THBS2 in CD8+ T cell exclusion.
We further tested whether adding THBS2+ CAFs to the immune-hot MC38 model could induce T cell exclusion. For this, we co-injected THBS2+ CAFs with MC38 cells into WT mice. Although THBS2 expression was detectable in these tumors, its distribution was irregular and did not resemble the organized fibrotic front observed in MTO-derived CMS4-like tumors (Supplementary Fig. 4f, g). Consequently, a robust immune-exclusionary barrier failed to form; although CD8+ T cell infiltration was significantly reduced, substantial numbers of CD8+ T cells remained. This suggests that CAF activation and the structured fibrotic architecture driven by mesenchymal CRC cells such as MTOs are essential for establishing a robust immunosuppressive barrier, and CMS1-like MC38 cells are insufficient to fully recapitulate this effect, even in the presence of THBS2⁺ CAFs.
Collectively, these results provide insight into the unique features of fibrotic CMS4 CRC and highlight the requirement for tumor–CAF crosstalk in generating an exclusionary TME.
THBS2 loss in the TME provokes an antitumor immune landscape
Although THBS2 has been implicated in collagen fibrillogenesis and assembly25,35, collagen-related gene sets in both Thbs2-/- tumors and CAFs remained unchanged (Supplementary Fig. 5a, b), and the amount and alignment of collagen fibers were comparable between WT and Thbs2-/- tumors (Supplementary Fig. 5c). Thus, immune activation in THBS2-deficient tumors is unlikely due to altered stromal integrity, suggesting involvement of other mechanisms.
scRNA-seq analysis of orthotopic tumors revealed a marked increase in immune cell and decrease in epithelial/stromal cell proportions in Thbs2-/- tumors (Fig. 4a–c), indicating a shift toward an immune-rich TME. Epithelial cells from Thbs2-/- tumors showed enrichment of inflammation, IFN signaling, and apoptosis gene sets (Supplementary Fig. 5d–g), consistent with earlier findings that tumor epithelial cells are more prone to antitumor immune-mediated cell death in the absence of THBS2 (Fig. 2). Reclassification of stromal cells revealed that Thbs2 expression closely overlapped with the mCAF signatures and Sfrp2 expression, with many double-positive cells detected (Supplementary Fig. 5h–k), consistent with human CRC data (Fig. 1).
a,b UMAP of single-cell RNA sequencing (scRNA-seq) data of orthotopic MTO tumors in WT or Thbs2-/- mice. all tumor cells colored by host mouse genotypes (a) and major cellular components (b). c Pie charts showing the proportions of indicated cell types in each genotype. d, e UMAP of immune cells colored by cell types (d) and proportion of each cell type (e). f UMAP of T and natural killer cells (T/NK cells) colored by cell types. g Proportion of T/NK cell types. h, i UMAP of CD8+ T cells colored by cell subtypes (h) and cell counts of each subtype (i). j, k Violin plots of CD8+ T cells (j) and monocytes/macrophages (Mono/Macs) (k) for indicated gene signatures. l Immunofluorescence staining of indicated proteins in orthotopic tumors. Scale bars, 100 μm. m Violin plots of Mono/Mac for indicated gene signatures. n UMAP of Mono/Mac colored by cell subtypes. o–q Proportion (o) and violin plots for indicated gene signatures and gene expression (p, q) of Mono/Mac subtypes. r–t UMAP of DCs colored by DC subtypes (r). UMAP feature plots of DCs in WT and Thbs2-/- mice for indicated gene expression (s). t Violin plots of cDCs for indicated genes signatures. u, v UMAP of all tumor cells (u) and immune cells (v) colored by cell types (left) and feature plots for Cxcl10 (right). w UMAP of T/NK cells colored by cell types (left) and feature plots for Cxcr3 (right). x Schematic representation of models for CD8+ T cell recruitment regulated by THBS2+ CAF. Wilcoxon rank sum test, two-sided (j,k,m,p,q,t). Source data are provided as a Source Data file.
Within the Immune compartment, Thbs2-/- tumors exhibited increased T/natural killer (T/NK) cells, monocytes/macrophages (Mono/Macs), and dendritic cells (DCs) (Fig. 4d, e and Supplementary Fig. 5l). CD8+ T cells were the predominant T/NK subtype, with an elevated CD8/Treg ratio (Fig. 4f, g). All CD8+ T cell subpopulations, including naïve, stem-like (Stem-like Tex), effector-like (Effector-like Tex), and terminal exhausted (Terminal Tex) cells, were expanded, with Terminal Tex cells most enriched (Fig. 4h, i and Supplementary Fig. 5m). These results demonstrate that THBS2 loss not only abolishes the CD8+ T cell exclusion barrier but also promotes effector-like and exhausted phenotypes via persistent antigen stimulation, as evidenced by elevated effector and dysfunction/exhaustion signatures (Fig. 4j).
Macrophages and DCs in the THBS2-deficient TME display features consistent with CD8+ T cell recruitment
Given their established roles in shaping T cell infiltration36,37, we next focused on myeloid populations. Mono/Macs from Thbs2-/- tumors displayed increased M1 signatures and IFN-γ responses, with reduced M2 signatures and lower expression of the M2 marker CD206 (Fig. 4k, l). Pathways related to “T cell chemotaxis” and “proliferation” were also enriched in these cells (Fig. 4m). Reclassification revealed a decrease in the TAM3 subset (high M2 signatures) and an increase in TAM2, characterized by elevated M1 signatures and CD8+ T cell recruitment chemokines such as Cxcl10, along with T cell proliferation/migration programs (Fig. 4n–q and Supplementary Fig. 5n).
DCs were also more abundant in Thbs2-/- tumors, with higher proportion of all subtypes, including plasmacytoid DCs (pDCs), conventional DC1 (cDC1), and cDC2 (Fig. 4r). All subsets exhibited elevated expression of CXCL9 and CXCL1036,37,38,39 in Thbs2-/- tumors (Fig. 4s). cDC populations showed upregulation of IFN-γ response and T cell chemotaxis signatures (Fig. 4t). These features in macrophages and DCs point toward a myeloid compartment favorable for CD8+ T cell recruitment.
Given these findings in myeloid cells, we next examined whether CAFs themselves might contribute to this program. Comparison of T cell recruitment–related transcriptional signatures in CAFs from WT and Thbs2-/- tumors revealed no significant differences (Supplementary Fig. 6a), suggesting that CAF-derived recruitment signals are unlikely to account for the observed CD8⁺ T cell influx. Instead, these results point to other cell types, notably macrophages and DCs, as potential contributors. Consistently, ligand–receptor analysis suggested more frequent interactions between Mono/Macs or DCs and T cells in Thbs2-/- tumors, as well as increased contacts between these myeloid cells and stromal cells (Supplementary Fig. 6b, c). Bulk tumor transcriptomic analyses further showed enrichment in “T cell chemotaxis,” “proliferation,” “migration,” and “CD8+ T cell recruitment” pathways (Supplementary Fig. 6d), identifying Cxcl10, Il4ra, and Irf1 as leading-edge genes commonly found in top-ranked immune-related signatures, including inflammatory responses, IFN signaling, and IL6/JAK/STAT3 pathways (Supplementary Fig. 6e, f). qRT-PCR analysis confirmed increased expression of Cxcl9, Cxcl10, and Cxcr3 in Thbs2-/- tumors (Supplementary Fig. 6g). scRNA-seq analysis further demonstrated that immune cells, particularly Mono/Macs and DCs, expressed higher levels of Cxcl10, Il4ra, and Irf1, while Cxcr3 was enriched in T/NK cells (Fig. 4u–w and Supplementary Fig. 6h).
Together, these results indicate that in the THBS2-deficient TME, macrophages and DCs acquire transcriptional programs that promote CD8⁺ T cell recruitment, potentially via CXCL9/10–CXCR3 interactions (Fig. 4x), a possibility further evaluated in subsequent spatial and in vivo depletion studies.
Spatial dynamics of CD8+ T cell recruitment in Thbs2 -/- tumors
To investigate the spatial dynamics of CD8+ T cell recruitment at the tumor front, we performed multiplex imaging mass cytometry (mIMC) on MTO-derived tumors from WT and Thbs2-/- mice. No significant changes were observed in αSMA expression, collagen deposition, or vascularity (CD31+) in Thbs2-/- tumors, suggesting that THBS2 loss in CAFs does not affect CAF maintenance, collagen assembly, or angiogenesis (Supplementary Fig. 7a). In contrast, infiltration of CD8+ T cells, CD4+ T cells, macrophages (F4/80+), and DCs (CD11c+) was markedly enhanced in Thbs2-/- tumors, whereas the spatial distribution of B cells (B220+) and Treg cells (Foxp3+) remained unchanged (Fig. 5a and Supplementary Fig. 7a). These findings align with the notion that THBS2 loss in CAFs skews fibrotic tumors into an immunogenic state.
a Imaging mass cytometry (IMC) images of orthotopic tumors in WT or Thbs2-/- mice at two weeks post-injection of MTO. White lines denote tumor borders. Scale bars, 100 μm. T: tumor, NT: non-tumor. b–d Neighborhood composition analyses using CytoMAP. Region prevalence (b) and dimensionality reduction UMAP plots (c, d) of neighborhoods involving CD8+ T cells, F4/80+ TAMs, and CD11c+ DCs in tumors of WT or Thbs2-/- mice. Regions are color-coded based on types defined in Supplementary Fig. 7b. e–g CD8+ T cell infiltration analysis. Representative images showing infiltrating CD8⁺ T cells; inset images highlight infiltration bands at the tumor–non-tumor interface (e). Green lines indicate tumor borders. Histograms (f) and scatter plots (g) show distances of CD8+ T cells from borders (n: WT = 261, Thbs2-/- = 2459). h–m Proximity analyses between CD8+ T cells, F4/80+ macrophages, and CD11c+ DCs. Green lines indicate tumor borders. Representative images (h) histograms (i) and scatter plots (j) show the nearest distances between CD8+ T cells and CD11c+ DCs (n: WT = 706, Thbs2-/- = 880). Representative images (k) histograms (l) and scatter plots (m) show the nearest distances between CD8+ T cells and F4/80+ macrophages (n: WT = 960, Thbs2-/- = 1022). Green lines denote tumor borders. Mann-Whitney U-test, two-sided (g, j, m). Mean ± SEM. Source data are provided as a Source Data file.
CytoMAP analysis40 demonstrated that regions enriched for CD8+ T cells, macrophages, and DCs were significantly expanded in Thbs2-/- tumors compared to WT controls (Fig. 5b–d and Supplementary Fig. 7b–d), suggesting increased interactions among these cell types, consistent with scRNA-seq results (Supplementary Fig. 6). Notably, CD8⁺ T cells dramatically crossed the tumor boundary in Thbs2⁻/⁻ tumors (Fig. 5e–g). Neighbor analysis revealed that CD8+ T cells were in close proximity to DCs and macrophages in Thbs2-/- tumors, implying an enhanced potential for these myeloid cells to facilitate CD8+ T cell recruitment into the tumor interior (Fig. 5h–m). Sustained increases in CD8+ T cell infiltration and persistent proximity between CD8+ T cells, DCs, and macrophages were observed at five weeks post-injection (Supplementary Fig. 7e–h), indicating a lasting disruption of the CD8+ T cell exclusionary barrier.
We next asked whether similar spatial patterns occur in human CRC using Xenium spatial transcriptomics. In line with mIMC findings from mouse CMS4-like MTO tumors, proximity between infiltrating CD8+ T cells and macrophages or cDCs was reduced in THBS2-high CMS4/MSS CRC compared with THBS2-low CMS1/MSI CRC, particularly within the tumor interior (Fig. 6a, b). CXCL9 and CXCL10 were mainly expressed by cDCs and macrophages, while CXCR3 expression was upregulated in CD8+ T cells (Fig. 6c). Close spatial associations between CXCL9/10-expressing cDCs or macrophages and CXCR3-expressing CD8+ T cells were more frequent at the tumor front in CMS1/MSI CRC (Fig. 6d–f). These observations further support a model in which CXCL9/10 produced by DCs and macrophages mediates CD8+ T cell recruitment in THBS2-low conditions.
a Spatial maps of the indicated cell types in Xenium 5 K data at tumor (T)–non-tumor (NT) interfaces in CMS1/MSI and CMS4/MSS CRCs. F: tumor front. White dashed lines denote tumor borders. b Violin plots showing the nearest distances of macrophages or cDCs to CD8+ T cells. c Dot plots of indicated gene expression across cell types. d Violin plots showing the nearest distances of CXCL9/10+ macrophages (Mac) or cDCs to CXCR3+ CD8+ T cells. e Proportions of indicated cells in CMS1/MSI and CMS4/MSS CRCs. f Spatial expression patterns of CXCL9 (orange), CXCL10 (red), and CXCR3 (green). Arrows indicate the corresponding cell types. g, h CellChat analysis of cell-cell interactions in Xenium data. The heatmap (g) shows the differential number of pairwise interactions between CMS1/MSI and CMS4/MSS. Top colored bar indicates the total incoming signaling and the right colored bar indicates the total outgoing signaling for each cell cluster. Red and blue bars denote an increase or decrease, respectively, in CMS4/MSS compared to CMS1/MSI. The chord diagram illustrates ligand-receptor pairs upregulated in CMS1/MSI (h, left) or CMS4/MSS (h, right), with edge weights reflecting their contribution to signaling among indicated cell types. Scale bars, 100 μm (a), 50 μm (f). Šídák’s multiple comparison test, two-sided (b); Mann-Whitney U-test, two-sided (d). Mean ± SEM. Adjustments for multiple comparisons were not made in (b). Source data are provided as a Source Data file.
Consistent with qRT-PCR data (Supplementary Fig. 6g), RNAscope analyses demonstrated elevated CXCL9 and CXCL10 expression in orthotopic Thbs2-/- tumors, which was confirmed at the protein level by ELISA (Supplementary Fig. 8a, b). Double immunofluorescence identified CD11c+ DCs and F4/80+ macrophages as the primary sources of these chemokines (Supplementary Fig. 8c, d), supporting their role in CD8+ T cell recruitment in the THBS2-deficient TME. Analyses of a large-scale scRNA-seq dataset further confirmed Mono/Macs and DCs as major sources of CXCL9 and CXCL10 and revealed a correlation between their gene expression in Mono/Macs or DCs and CXCR3 expression in CD8+ T cells (Supplementary Fig. 8e–g). CellChat ligand-receptor analyses of Xenium data predicted strong interactions between THBS2⁺ CAFs and both macrophages and DCs (Fig. 6g and Supplementary Fig. 8h). Importantly, THBS2 itself was highlighted as a potential ligand specifically in CMS4/MSS CRCs mediating interactions between CAFs and macrophages/DCs (Fig. 6h).
Inhibition of CXCL9/10-CXCR3 signaling axis suppresses enhanced recruitment of CD8+ T cells induced by THBS2 loss in CAFs
Given our spatial and transcriptomic analyses implicating DCs and macrophages as major sources of CXCL9 and CXCL10 in the THBS2-deficient TME, we next tested their functional roles in recruiting CD8+ T cells into the tumor interior. We selectively depleted either macrophages or DCs in MTO-derived Thbs2-/- tumors (Supplementary Fig. 9a). Both depletions significantly reduced intratumoral CXCL9 and CXCL10 expression, confirming that these myeloid populations are primary sources of the chemokines (Supplementary Fig. 9b, c). Consistent with this, CD8⁺ T cell infiltration was significantly diminished in both settings (Supplementary Fig. 9d, e). These results support macrophages and DCs as key mediators reprogramming the TME from immune-suppressive to immune-active in the absence of THBS2. Notably, DC depletion, but not macrophage depletion, abolished the antitumor effect of THBS2 loss, suggesting a potential central role for DCs in maintaining the immune-active phenotype of the Thbs2-/- MTO model (Supplementary Fig. 9f, g). This difference may reflect the bipotential nature of tumor-associated macrophages (TAMs), which can exert both pro- and anti-tumor functions, as suggested by our scRNA-seq data (Fig. 4p).
To determine whether THBS2 directly modulates chemokine production by these myeloid cells, we co-cultured a mouse dendritic cell line (DC2.4) or bone marrow–derived macrophages (BMDMs) with MTOs. In both systems, co-culture increased CXCL10 expression, which was abolished in the presence of recombinant THBS2 (Supplementary Fig. 9h). Together with spatial interaction data from CRC samples (Fig. 6h), these findings support a model in which THBS2 directly suppresses chemokine production by DCs and macrophages, thereby limiting CD8⁺ T cell infiltration.
To validate the functional relevance of the CXCL9/10–CXCR3 axis, we treated MTO-bearing WT and Thbs2-/- mice with a combination of anti-CXCL9 and anti-CXCL10 antibodies (Fig. 7a). This treatment reduced CD8+ T cell infiltration and restored tumor burden in Thbs2-/- mice, while causing minimal effects in WT mice (Fig. 7b–e). Administration of an anti-CXCR3 antibody produced similar effects, reversing the enhanced CD8⁺ T cell infiltration, increased apoptosis, and reduced tumor burden in orthotopic Thbs2-/- tumors, without altering the numbers of CD11c+ DCs or F4/80+ macrophages (Fig. 7f–h and Supplementary Fig. 10a–c). Spatial mIMC analyses confirmed that CXCR3 blockade eliminated CD8⁺ T cell penetration across the tumor boundary and abolished the reduced distances between DCs/macrophages and CD8⁺ T cells (Fig. 7i–o and Supplementary Fig. 10d–f).
a–e Combined blockage of CXCL9 (αCXCL9 ab) and CXCL10 (αCXCL10 ab) in MTO-bearing WT or Thbs2-/- mice (n: WT control = 8, WT αCXCL9 ab/CXCL10 ab = 9, Thbs2-/- control = 6, Thbs2-/- αCXCL9 ab/CXCL10 ab = 9). Schematic representation (a), macroscopic images (b), and volumes and weights (c) of tumors. Immunofluorescence for CD8 (d) and its quantification (e). Bottom images show magnified views of the yellow dashed boxes in the top panels (d). f–o Anti-CXCR3 antibody (αCXCR3 ab) treatment in MTO-bearing WT or Thbs2-/- mice (n: WT control = 10, WT αCXCR3 ab = 10, Thbs2-/- control = 7, Thbs2-/- αCXCR3 ab = 8). Schematic representation (f) macroscopic images (g) and volumes and weights (h) of tumors. i Representative IMC images of orthotopic tumors. T: tumor, NT: non-tumor. j, k CD8+ T cell infiltration analysis from (f). Histograms (j) and scatter plots (k) show distances of CD8+ T cells from tumor borders. l–o, Proximity analyses between CD8+ T cells and F4/80+ macrophages or CD11c+ DCs in (f). Histograms (l) and scatter plots (m) show the nearest distances between CD8+ T cells and CD11c+ DCs. Histograms (n) and scatter plots (o) show the nearest distances between CD8+ T cells and F4/80+ macrophages. Red dashed lines indicate tumor regions in (b,g). White lines denote tumor borders in (i). Scale bars, 1 mm (b, g), 500 μm (d, top), 50 μm (d, bottom), 100 μm (i). Fisher’s LSD test (c,e,h), Mann-Whitney U-test, two-sided (k,m,o). Mean ± SEM. Source data are provided as a Source Data file.
Collectively, these findings establish that THBS2 maintains an exclusionary barrier for CD8+ T cells in fibrotic CRCs by suppressing the CXCL9/10-CXCR3 signaling axis through its inhibitory effects on DCs and macrophages.
Inhibition of THBS2 strongly renders fibrotic CRCs sensitive to ICIs
Despite an immune-active TME, THBS2-deficient tumors continued to grow (Fig. 2f, i). Time-course studies revealed that Thbs2-/- tumors exhibited increased numbers of CD8+ T cells in both the tumor interior and front at all time points, but their numbers gradually declined by nine weeks (Fig. 8a–f). CD8+ T cells in the interior displayed higher PD-1 and CTLA-4 positivity than those at the tumor front (Fig. 8a–f and Supplementary Fig. 11a–d). These data suggest that continuous stimulation by tumor antigens and signals from surrounding cells induces exhaustion of CD8+ T cells in the tumor interior, aligning with scRNA-seq data showing enhanced dysfunction and exhaustion signatures in Thbs2-/- tumors (Fig. 4j). Additionally, gene signatures related to T cell receptor (TCR) signaling, a critical regulator of PD-1 and CTLA-4 expression, were enriched in T cells from Thbs2-/- tumors (Fig. 8g). Likewise, Cd274 (PD-L1) and its inducer Irf1 were significantly upregulated (Supplementary Fig. 5g and 6f). TCGA analysis demonstrated that CMS1/MSI CRCs also exhibited enrichment in CTL, dysfunction, and exhaustion signatures (Fig. 8h and Supplementary Fig. 11e). These findings underscore the shared characteristics between Thbs2-/- tumors and CMS1/MSI CRCs, indicating that, akin to MSI CRCs, Thbs2-/- tumors rely on immune checkpoints to evade antitumor immunity despite high CD8+ T cell infiltration. Supporting this, THBS2 loss-associated gene signature was enriched in CMS1/MSI CRCs compared to other subtypes, including fibrotic CMS4/MSS CRCs (Fig. 8i, j and Supplementary Fig. 11f).
a–f Immunofluorescence for CD8 and PD-1 in MTO-derived orthotopic tumors from WT or Thbs2-/- mice at 2-weeks (a), 5-weeks (c), and 9-weeks (e) post-injection of MTO. White lines denote the tumor borders. Scale bars, 100 μm. Quantification of PD-1+ and PD-1- CD8+ T cells at 2-weeks (b), 5-weeks (d), and 9-weeks (f) post-injection of MTO. F tumor front, I tumor interior. Magnified views (right panels for WT and Thbs2-/-) correspond to the yellow and magenta boxed regions in the left panels. g Violin plots of T cells for indicated gene signatures in tumors from WT and THBS2-/- mice. h, i Box plots of GSVA values for indicated gene signatures in each CMS (n: CMS1 = 85, CMS2 = 132, CMS3 = 78, CMS4 = 184) subtype of CRC in TCGA-COADREAD dataset. j GSEA for Thbs2 KO signatures comparing CMS1 CRCs vs. CMS4 CRCs in TCGA. k–n Anti-PD-1 antibody (αPD-1 ab) or anti-CTLA-4 antibody (αCTLA-4 ab) treatment in MTO-bearing WT or Thbs2-/- mice (n: WT control = 12, WT αCTLA-4 ab = 11, WT αPD-1 ab = 12, Thbs2-/- control = 11, Thbs2-/- αCTLA-4 ab = 11, Thbs2-/- αPD-1 ab = 13). Schematic representation (k) changes in tumor diameter (l) tumor weights and volumes (m) and H&E images of tumors (n). Scale bars, 1 mm (n). Wilcoxon rank sum test, two-sided (g) Dunnett’s test, two-sided (h, i) Two-way ANOVA (l) Unpaired t-test, two-sided (m). Mean ± SEM. Adjustments for multiple comparisons were made in (h, i) and not in (m). Box and whiskers graphs indicate the median and the 25 and 75th percentiles, with minimum and maximum values at the extremes of the whiskers. Source data are provided as a Source Data file.
To build on the efficacy of targeting THBS2, we investigated whether the increased CD8+ T cell infiltration could sensitize fibrotic CRCs to ICI therapies. For this purpose, we administered anti-PD-1 or anti-CTLA-4 antibodies to MTO-bearing Thbs2-/- mice (Fig. 8k). Strikingly, anti-PD-1 antibody treatment exhibited profound and sustained reduction in tumor size, leading to complete remission in all treated Thbs2-/- mice both macroscopically and histologically (Fig. 8l–n and Supplementary Fig. 11g). Treatment with anti-CTLA-4 antibody, although less effective than anti-PD-1, also achieved a substantial tumor reduction in Thbs2-/- mice (Fig. 8l–n and Supplementary Fig. 11g).
These findings demonstrate that targeting THBS2 not only enhances CD8+ T cell infiltration but also creates a permissive immune environment that dramatically improves ICI efficacy, supporting the rationale for combined approaches in treating fibrotic CRCs.
Discussion
Fibroblast-derived THBS2 is a key regulator of the immune-exclusionary TME in fibrotic CRCs. Highly expressed in mCAFs at the tumor front, THBS2 establishes a barrier that prevents CD8⁺ T-cell entry. Its inhibition disrupts this barrier, increases CD8⁺ infiltration, reprograms the TME toward an immune-active state, and sensitizes tumors to ICIs, supporting THBS2 as a therapeutic target in fibrotic CRC.
Although CAFs are known to shape antitumor immunity41,42, their contribution to spatial T cell exclusion has been incompletely defined. Desmoplastic/CAF-rich CMS4 CRCs associate with immunosuppression, yet a subset-resolved causal link between the mCAF and CD8⁺ T-cell exclusion has been largely correlative16,17,18,43,44. Here we delineate a mechanistically distinct, non-redundant function of CAF-derived THBS2 in immune exclusion. Whereas Liu et al. primarily linked mCAF-derived THBS2 to an epithelial CD47/MAPK axis29, our work interrogates the immune-exclusionary front in vivo. Using a CMS4-like tumor organoid (MTO) model, that recapitulates desmoplasia and CD8+ T cell exclusion and combining genetically modified mice (global and fibroblast-specific Thbs2 deletion) with a syngeneic orthotopic implantation system, we show that THBS2 restrains CXCL9/10 production from DC/macrophage, thereby limiting CXCR3-dependent CD8⁺ T cell entry at the tumor front. Causality is supported by convergent perturbations—DC/macrophage depletion, CXCL9/10 neutralization, and CXCR3 blockade—together with patient-level spatial mapping. The dependence on a fibrotic, CMS4-like context is underscored by co-injecting THBS2⁺ CAFs with MC38 cells, which reduced intratumoral CD8⁺ T cells yet failed to build a robust exclusionary front, highlighting the requirement for mesenchymal tumor–CAF crosstalk and organized fibrosis.
CAFs are one of the key components in the TME exerting both tumor-promotive and tumor-suppressive effects in a context dependent manner. Reflecting their high plasticity and marked heterogeneity, various CAF subtypes have been proposed across different cancers by using single cell transcriptomic analyses45. While some CAF subtypes are unique to certain disease settings, mCAFs, iCAFs, and antigen-presenting CAFs (apCAFs) are considered as core CAF subtypes46. mCAFs are primarily characterized by their role in ECM remodeling and upregulated TGFβ signaling, whereas iCAFs are defined by their capacity to produce cytokines and chemokines mediating inflammatory response in the TME. apCAFs express MHCII and CD74 enabling crosstalk with CD4+ T cells. In addition to these subtypes, our reanalysis of a large-scale human CRC dataset30 identified reticular CAFs (rCAFs) resembling reticular fibroblasts in lymphoid tissues, vascular CAFs (vCAFs) associated with angiogenesis and endothelial cells, and tumor-associated CAFs (tCAFs) expressing NDRG1 and ENO1. Among these CAF subtypes THBS2 was predominantly expressed in mCAFs, in good agreement with the recent work by Liu et al29.
Complementary spatial and single-cell analyses localize THBS2 to mCAFs at tumor fronts and link its loss to increased CD8⁺–myeloid proximity and immune-activating program without changes in collagen content/alignment or αSMA expression. Exploratory in vitro assays suggesting that recombinant THBS2 can dampen CXCL10 induction in DCs/BMDMs exposed to tumor cues, together with cell-cell interaction analysis of human Xenium data indicating enriched THBS2⁺ CAF to DC/macrophage communication and nominating THBS2 among candidate ligands, are consistent with a direct CAF-to-myeloid axis. Overall, these data indicate that THBS2 enforces exclusion primarily by tuning myeloid chemokine output, rather than by altering stromal architecture, suggesting a safer therapeutic profile than broad ECM-targeting approaches36,37,47.
THBS2 differs functionally from THBS1. While THBS1 produced by monocyte-like cells suppresses CD8+ T cell activity24. CAF-derived THBS2 creates a spatial barrier, by limiting CXCL9/10 production from DCs/macrophages, central to immune resistance in fibrotic CRCs. Furthermore, CD8+ T cells in THBS2-deficient tumors exhibit increased exhaustion markers such as PD-1 and CTLA-4, driven by chronic antigen stimulation. Elevated PD-L1 levels associated with increased IRF1 expression further indicate a reliance on immune checkpoints for tumor progression. These MSI-like features rationalize the strong synergy between THBS2 inhibition and ICIs, with anti-PD-1 inducing complete pathological remission.
Fibrotic CRCs, comprising 25-30% of cases, are aggressive, desmoplastic, and resistant to current cytotoxic and immunotherapy regimens6,7. Because most stromal-targeting strategies lack CAF specificity and are often toxic9,15, a CAF-restricted target is needed. THBS2 fits this profile: it is prominently expressed by CAFs with minimal expression in normal tissues48, and its inhibition reprograms the TME without disrupting stromal architecture. Additionally, serum THBS2 levels, circulating THBS2 levels correlate with tumor burden and advanced stage in our cohort, suggesting utility for patient stratification and response monitoring. Thus, THBS2 provides a tractable and potentially safer entry point to extend ICI benefit to fibrotic, MSS CRC.
In summary, inhibiting THBS2 dismantles the CD8⁺ T cell exclusionary front, enhances recruitment via the CXCL9/10–CXCR3 axis, and amplifies ICI efficacy, providing a coherent strategy to overcome immune resistance in fibrotic CRCs. This study establishes THBS2 as a promising therapeutic target and supports its development for monotherapy and combination treatments to improve CRC outcome.
Methods
This study was approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine and the Ethics Committee of Osaka Metropolitan University.
Mice
C57BL/6 J (Stock No. 000664) mice were purchased from Japan SLC, Inc. Thbs2-/- mice (B6;129S4-Thbs2tm1Bst/J; Stock No. 006238) were obtained from the Jackson Laboratory. Col1a2-cre mice49 were generously provided by the National Institute of Biomedical Innovation, Health and Nutrition. Thbs2 floxed (Thbs2f/f) mice were generated through outsourced services at Macrogen, Inc. (Seoul, Korea) using the CRISPR/Cas9 system. Briefly, PMSG and HCG were administered to C57BL/6 N female mice, and after 48 h, these mice were mated with C57BL/6 N stud males. Vaginal plug-positive females were sacrificed the following day, and fertilized embryos were harvested. Synthesized sgRNA, Cas9 nuclease, and dsDonor were mixed and microinjected into one-cell embryos. The microinjected embryos were incubated at 37 °C for 1–2 h before being transplanted into the oviducts of pseudopregnant ICR recipient mice. After birth, genotyping of the founders was conducted using the primer set F1 (GAGACCAATGCACTGCATCTAG) and R1 (CAATGAAGGTTTGGGGATGGG), yielding a 2422 bp PCR product. Positive samples were identified via digestion with KpnI and NheI restriction enzymes, flowed by TA cloning and sequencing for confirmation. Thbs2ΔFibro mice were generated by crossing Thbs2f/f mice with Col1a2-cre mice. Genotyping was performed by PCR, with primer sequences listed in Supplementary Table 1. All animals were bred in specific pathogen-free environments. Experimental/control animals were bred separately. CO2 inhalation was applied for euthanasia. All the experimental procedures conformed to institutional guidelines and Guidelines for Proper Conduct of Animal Experiments of Science Council of Japan and were approved by the animal research committee of Kyoto University (Kyoto, Japan).
Human samples
Paraffin-embedded tissue sections of CRC were obtained from surgically resected CRC samples. Serum samples were collected from 102 patients (male, n = 47, average age = 67, range 40–91; female, n = 55, average age = 70, range 34–91) with CRCs and 10 patients (male, n = 7, average age = 71, range 63–68; female, n = 3, average age = 63, range 57–75) with benign tumors (adenoma: n = 6, sessile serrated lesion: n = 2, both adenoma and sessile serrated lesion: n = 2). None of the patients had undergone preoperative radiation or chemotherapy. Written informed consent was obtained from these patients under a protocol approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine. We analyzed a tissue microarray (TMA) containing 147 surgically resected human CRC samples (male, n = 77, average age at diagnosis = 64, range 29–85; female, n = 70, average age at diagnosis = 66, range 26–90). Among these TMA samples, 53 CRCs were previously annotated as MSI-H, and 77 were annotated as MSS based on immunostaining for MLH1, MSH2, MSH6, and PMS2. To classify CD8 staining patterns, we selected only tissue cores with a clearly defined tumor–non-tumor interface, since the definition of the “exclusive” pattern requires exclusion of CD8⁺ T cells from the tumor core relative to its margin. From the 147 CRC samples represented on the TMA, 60 cores met this criterion (diffuse = 28; exclusive = 32). The use of patient samples for this experiment was approved by the Ethics Committee of Kyoto University and Osaka Metropolitan University without requiring written informed consent. Instead, informed consent was obtained via an opt-out process on the institutions’ websites.
Cell lines
MTO (kindly provided by Dr. Battle’s laboratory) were cultured as previously described12. The human embryonic kidney (HEK) 293 T cell line was purchased from the American Type Culture Collection (ATCC, Rockville, USA: catalog number CRL-3216). The murine cell line MC38 was purchased from Kerafast (MA, USA: catalog number ENH204-FP). MC38 and HEK293T were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS, 1X L-glutamine and 1% penicillin-streptomycin. DC2.4 (SCC142) were purchased from Sigma Aldrich and cultured in RPMI-1640 supplemented with 10% FBS, 1X L-glutamine, 1X HEPES, 1X non-essential amino acids and 0.0054×2-mercaptoethanol. All cell lines were maintained at 37°C in a humidified atmosphere with 5% CO2.
Co-culture experiment
Bone marrow-derived macrophages (BMDMs) were established as previously described50. Briefly, bone marrow cells were collected from femur and humerus of C57BL/6 mice. After red blood cells lysis, cells were cultured in RPMI-1640 supplemented with 10% FBS and macrophage colony-stimulating factor (M-CSF; Miltenyi Biotec, 130-101-704) for 5 days to differentiate into BMDM. BMDMs or DC2.4 cells were seeded at a density of 105/well in a 24-well plate, and cultured in RPMI supplemented with 10% FBS and L-glutamine for 12 h. After co-cultured with Matrigel-removed MTOs for 48 h with or without 50 ng/mL recombinant THBS2 (R&D; 1635-T2), total medium was aspirated. Cells were rinsed with PBS twice and lysed for RNA extraction. Since MTOs were barely attached to tissue culture plate, remaining cells could be considered as BMDM or DC cell line.
ELISA
Human serum samples were collected from whole blood of patients with adenomas and CRCs by using EDTA tubes, followed by centrifugation. The concentration of THBS2 in the serum was measured using the Human Thrombospondin-2 Quantikine ELISA Kit (R&D systems; DTSP20). Mouse serum samples were collected by standing still whole blood for 30 min followed by centrifugation. The concentration of THBS2 in the serum was measured using THBS2 ELISA Kit (Mouse) (Aviva Systems Biology; OKCD00921). Mouse tumor lysate was collected by homogenizing orthotopic implanted MTO tumors in PBS and 1 mM PMSF (CST; 8553) by using TissueLyser LT (QIAGEN). CXCL9 and CXCL10 concentration was measured by using DuoSet ELISA kit (R&D; DY492-05 and DY466-05).
Orthotopic injection of organoids to murine rectum and cecum
Mouse tumor organoids were injected orthotopically as previously described21,24,32,51. Eight-to-ten-week-old male mice, weighing 19–28 g, were anaesthetized using isoflurane inhalation and placed in a supine position. For rectal injections, the rectal mucosa was exposed by inserting two blunt-ended hemostats into the anus. A 20 µL cell suspension containing 105 MTO or MC38 cells, mixed with Matrigel (Corning; 356231), was directly injected into the rectal mucosa. For orthotopic injections into cecum, mice were exteriorized their cecum by a laparotomy. A 10 µL cell suspension containing 5 × 105 MTO was injected per mouse. For co-injection experiments, a mixture of 105 MTO cells or MC38 cells and 105 CAFs per mouse was combined with 20 µL of Matrigel and injected. Injections were performed using a 30 G needle under stereo microscope guidance. Mice implanted with MTOs were euthanized at the time points indicated in the main article. The maximal tumor burden permitted by the ethics committee was 25 mm in diameter. Tumor diameter was measured with a caliper once a week after implantation and at the time of euthanasia. None of the tumors had exceeded this limit. Orthotopic primary lesions and liver metastases were evaluated both macroscopically and histologically.
Isolation of CAFs
CAFs were isolated from orthotopic tumors derived from MTOs. Briefly, MTOs were injected into the rectum of eight-to-ten-week-old male mice as described above. After euthanasia, tumors weighing 0.2–1.0 g were excised from the rectum. Tumors were minced using a scalpel and dissociated with the Tumor Dissociation Kit (Miltenyi Biotec; 130-096-730) and GentleMACS Dissociator (Miltenyi Biotec). Red blood cells were removed using Red Blood Cell Lysis Solution (Miltenyi Biotec; 130-094-183). CAFs were then isolated using the Tumor-associated Fibroblast Isolation Kit (Miltenyi Biotec; 130-116-474). Primary cells were seeded onto fibronectin-coated 4-well plates (Thermo Fisher, 176740; fibronectin from Corning; 354008). CAFs were cultured in DMEM supplemented with 10% FBS, 1% L-glutamine, and 1% penicillin-streptomycin (Thermo Fisher; 15140-122).
Immortalization by retroviral transduction
The pBABE-zeo large T genomic plasmid (Addgene #1778) was introduced into JM109 competent cells (RBC Bioscience; RH717). After selection on ampicillin-containing media, colonies were processed using a midiprep kit. Purified plasmids were co-transfected with pGP and pE-eco into actively growing HEK293T cells using the Retrovirus Packaging Kit Eco (Takara Bio; 6160). Virus-containing supernatants were collected 48 h post-transfection and filtered to remove cellular debris. Target cells were infected with the virus in the presence of 8 µg/mL polybrene. Following infection, cells were selected using 100 µg/mL Zeocin (InvivoGen; ant-zn-05).
MTS assay
Cells were seeded on a 96-well plate at a density of 500 cells per well and incubated. CellTiter 96 reagent (Promega; G4000) was added to the medium at 20% of the total volume. The plate was then incubated at 37 °C for 1 h. The absorbance was measured using a microplate reader (Tecan Japan) every 24 h.
Migration assay
CAFs were tested for their migratory ability using polyethylene terephthalate membrane inserts with 8 µm pores in a 24-well format (Corning; 353097). A total of 2.5 × 104 cells per well were seeded into the inner chamber containing serum-free DMEM, while the outer chamber contained DMEM supplemented with 10% FBS to serve as a chemoattractant. After 8 h of incubation at 37 °C with 5% CO₂, cells that migrated through the pores to the underside of the insert were fixed in cold methanol for 20 minutes and stained with crystal violet for 20 min.
In vivo treatment
MTO was orthotopically implanted into the rectum of 8–10-week-old male C57BL/6 and Thbs2-/- mice. For CD8+ T cell depletion, mice were intraperitoneally injected with anti-CD8 antibody (200 µg; Bio X Cell, BE0061) or control IgG (200 µg; Bio X Cell, BE0090) twice per week. For macrophage depletion, mice were intraperitoneally injected with clodronate liposome (250 µg; Hygieia Bioscience, 16004473) once per week. For DC depletion, mice were intraperitoneally injected with anti-CD11c antibody (200 µg; Bio X Cell, BE0038) twice per week. For CXCL9/10 inhibition, mice were intraperitoneally injected with anti-CXCL9 antibody (300 µg; Bio X Cell, BE 0309) and anti-CXCL10 antibody (300 µg; Bio X Cell, BE 0440) twice per week. For CXCR3 inhibition, mice were intraperitoneally injected with anti-CXCR3 antibody (200 µg; Bio X Cell, BE0249) twice per week. For neutralizing antibody treatment, mice were treated with intraperitoneal injections of anti-PD-1 antibody (200 µg; Bio X Cell, BE0146), anti-CTLA-4 antibody (200 µg; Bio X Cell, BE0032), or control IgG (200 µg; Bio X Cell, BE0089) twice per week. Each treatment continued for 3 weeks, starting 1 week after MTO implantation. Before initiating treatment, successful implantation was confirmed by palpation and luciferase assay imaging.
Histology, immunohistochemistry, and immunofluorescence
Tissues were fixed in masked formalin 2 A (Japan Tanner;NTS 00109) for 2–3 days and embedded in paraffin. Paraffin-embedded tissues were sectioned into 4 µm slices for human samples and 5 µm slices for mouse samples. Hematoxylin and eosin (H&E) staining was performed using standard protocols. Masson’s trichrome staining was carried out according to the manufacturer’s instructions (Sigma, HT15-1KT). For IHC analysis, tissue sections were deparaffinized, rehydrated, and subjected to antigen retrieval by boiling in a microwave for 15 min or autoclaving in citrate buffer (pH 6.0) or EDTA buffer (pH 9.0). Sections were incubated overnight at 4 °C with primary antibodies, followed by incubation with HRP-conjugated secondary antibodies (Vector Laboratories; VECTASTAIN Elite ABC HRP kits) for 1 h at room temperature. Visualization was performed using DAB (Dako; K3467). For immunofluorescence analysis, tissue sections were processed similarly for deparaffinization, rehydration, and antigen retrieval (microwave boiling in citrate buffer, pH 6.0, or EDTA buffer, pH 9.0). Sections were incubated with primary antibodies for 1 h at room temperature, followed by HRP-conjugated secondary antibodies (Vector Laboratories; VECTASTAIN ImmPRESS). Fluorescent signals were generated using the Opal Multiplex IHC system (Akoya Biosciences) with OPAL520 or OPAL570 diluted in 1× Amplification Diluent. The primary-secondary-HRP complex was stripped through microwave reboiling, and subsequent target proteins were tagged using different OPAL signals. Finally, sections were counterstained with DAPI using Fluoro-KEEPER (Nakalai Tesque; 12745-74) and mounted. Primary antibodies used were listed on Supplementary Table 2. Images were acquired using a BZ-X710 microscope (KEYENCE). The total number of cells and the number of positively stained cells were quantified using QuPath ver.0.3.2. or BZ-X Analyzer.
Imaging mass cytometry (IMC)
Images were acquired using a Hyperion Imaging System (Standard BioTools). All operations were conducted following the manufacturer’s procedure. Briefly, based on hematoxylin and eosin (HE)-stained serial tissue sections, we randomly selected regions of interest (ROIs) at the tumor border region. Images were laser ablated at 200 Hz, and raw data were acquired using a commercial acquisition software (Hyperion Imaging System, Standard BioTools). The state of Hyperion Imaging System was monitored by the interspersed acquisition of data from the tuning slide (Standard BioTools). Images were analyzed using HALO software (Version 3.6, Indica Labs, Albuquerque, NM, USA). Nuclear segmentation was performed using HALO Highplex FL (Version 4.2) based on DAPI staining. Positivity thresholds for each marker were set based on staining intensity and were reviewed across all samples to ensure consistency. Data for each cell, including marker expression and x/y coordinates, were exported for spatial analysis. Cells were phenotyped based on marker expression as follows: CD8⁺ T cells, tumor-associated macrophages (F4/80⁺), and dendritic cells (DCs; CD11c⁺).
Spatial analyses of IMC images
To evaluate spatial relationships between immune cells, HALO Nearest Neighbor Analysis module was applied. The distance between each immune cell subtype (macrophages and DCs) and their nearest neighboring CD8⁺ T cell was measured, providing insights into the spatial proximity of immune cells within the tissue microenvironment. CD8⁺ T cell infiltration into tumor regions was assessed using the HALO Infiltration Analysis module. The distance of each CD8⁺ T cell from the tumor border was calculated to quantify infiltration depth. Data obtained from HALO analyses were further processed using CytoMAP (RRID:SCR_021227). Neighborhoods were defined within a 30 µm radius around each cell, and the staining intensity of CD8, F4/80, and CD11c markers was calculated for cells within each neighborhood. The neighborhood compositions were analyzed using Self-Organizing Maps (SOM) clustering, generating a heatmap that depicted the percentage of each marker intensity per neighborhood. This clustering approach enabled the visualization of spatially distinct immune microenvironments.
RNA in situ hybridization
To assess the spatial expression of THBS2 gene on FFPE sections of human CRC and TMA, RNAscope (Advanced Cell Diagnostic, USA) was outsourced to Morphotechnology, Japan. The probe used for human THBS2 was #555471, and the signal was visualized using Fast Red dye. Nuclei were counterstained with hematoxylin. For RNAscope-IHC double staining, the manufacturer’s protocol for FFPE samples was followed (https://acdbio.com/system/files_force/UM%20322452%20FFPE%20SamplePrep%20Rev%2003152017.pdf, https://acdbio.com/sites/default/files/322360-USM%20RNAscope%202.5%20HD%20RED%20Pt2_11052015.pdf). The probes used for mouse Cxcl9 and Cxcl10 were #489341 and # 408921, respectively. After the objective RNA signal was developed with Fast Red, sections proceed to IF steps as previously described.
RNA extraction and qRT-PCR analysis
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN). For human CRC and mouse orthotopic tumor samples, tissues were first homogenized with TRIzol reagent (Thermo Fisher; 15596018) and a beads homogenizer (Bertin Technologies Precellys 24 tissue homogenizer) before RNA extraction. Complementary DNA (cDNA) was synthesized using the ReverTra Ace qPCR RT Kit (TOYOBO; FSQ-201). Real-time PCR was performed in duplicate using the SYBR Green PCR Master Mix (Roche Diagnostics) on a LightCycler 96 system (Roche Diagnostics). Gene expression levels for each sample were normalized to Gapdh RNA. The qRT-PCR primers used in this study are listed in Supplementary Table 1.
RNA Sequencing (RNA-seq) and Analysis
RNA-seq was outsourced to Macrogen, Inc., and performed using the NovaSeq 6000 platform with 2 × 100 base pair paired-end sequencing, prepared with the TruSeq RNA Sample Prep Kit v2. Adaptors and low-quality bases were trimmed from the reads using Trimmomatic version 0.39 (RRID:SCR_011848) with default parameters. The reads were aligned to the Mus musculus reference genome (build mm10) using STAR version 2.7.3a (RRID:SCR_004463) and quantified using RSEM version v1.3.1 (RRID:SCR_000262). Read count data were normalized using the iDEGES/edgeR method. Differentially expressed genes (DEG) were determined with the TCC package (RRID:SCR_001779), applying a false discovery rate (FDR) cutoff value < 0.3. Gene set enrichment analysis (GSEA) was performed using the GSEA software (http://www.gsea-msigdb.org/gsea/index.jsp). The gene sets used included Hallmarks (h.all.v7.4.symbols.gmt), Reactome (c2.cp.reactome.v7.5.1.symbols.gmt), GOBP (c5.all.v7.5.1.symbols.gmt), and custom signatures (Supplementary Data 1). “mCAF”, “iCAF” gene sets were derived from a previous report17. “CD8 T cell recruitment chemokines” was compiled based on several references52,53,54,55. CMS classification of human CRC samples was determined using the R package CMScaller.
Single-cell RNA sequencing (scRNA-seq)
Tumors generated by orthotopic implantation of MTO in the rectum of WT and Thbs2-/- mice (n = 2 per group) were processed into single-cell suspensions using the Tumor Dissociation Kit (Miltenyi Biotec; 130-096-730). Red blood cells were removed from the dissociated tumors using the Red Blood Cell Lysis Solution (Miltenyi Biotec; 130-094-183), and dead cells were eliminated with the EasySep Dead Cell Removal (Annexin V) Kit (STEMCELL Technologies). scRNA-seq libraries were prepared using the Chromium Next GEM Single Cell 3’ v3.1Kit (10X Genomics; 1000269). Cells were loaded onto the 10X Chromium Controller (10X Genomics) with the Chromium Next GEM Chip G Single Cell kit (10X Genomics; 1000127) at a concentration of 1000 cells per µL, as described in the manufacturer’s protocol (10X Chromium Single Cell 3’ Reagent Kits User Guide (v3.1 Chemistry Dual Index)). A total of 16,500 cells from each mouse were loaded. Generation of gel beads in emulsion (GEMs), barcoding, GEM-RT clean-up, complementary DNA amplification and library construction were performed according to the manufacturer’s protocol. Individual sample quality was checked using a TapeStation (Agilent Technology). The final library pool was sequenced using the DNBSEQ-G400RS (MGI). The Cell Ranger software suite (version 7.1.0; 10X Genomics) was used to process, align, and summarize unique molecular identifier (UMI) counts. The total cell recovery for the orthotopic tumor in Thbs2-/- mice was 16,327 cells captured at a mean depth of 25,436 reads per cell and 1286 mean genes per cell. The total cell recovery for the orthotopic tumor in WT was 15,487 cells captured at a mean depth of 27,438 reads per cell and 1495 mean genes per cell.
Analysis of scRNA-seq
For scRNA-seq data from orthotopically implanted mouse tumors, additional data processing and analysis were performed using Seurat (version 4.3.0) in R (version 4.3.1). Cells expressing fewer than 200 genes, cells with over 20% mitochondrial gene expression, and the genes expressed in fewer than five cells were excluded. After filtering, the expression data for 12,474 cells from Thbs2-/- and 9,821 cells from WT mice were converted into Seurat objects. Each sample was normalized using the SCTransform, with sample ID and mitochondrial gene percentages regressed out. The normalized objects were merged, and principal component analysis was conducted using Seurat. For data integration, the Harmony R package was employed. Cells were visualized according to UMAP embeddings, and all cells were classified into three compartments: epithelium (Epcam), immune (Ptprc), or stroma (Pecam1, Dcn, or Rgs5). After extraction and reclustering, fast pre-ranked gene set enrichment analysis named fgsea (v1.32.0, R package) with default parameters was used for the epithelial compartment to perform functional enrichment analysis for hallmark gene sets of cluster-specific differentially expressed genes. The immune compartment was extracted and re-normalized. It was further divided into B cells, T/NK cells, DCs, Mono/Macs, and TANs. Key marker genes for each subset are described in Supplementary Fig. 4j. The CD8a-expressing T cell cluster was extracted, re-normalized and further clustered. Key marker genes for the subclusters of CD8+ T cells are described in Supplementary Fig.4k. After extraction and reclustering, canonical marker genes were used to annotate monocyte-like cells (Ly6c2, Plac8, and Thbs1) and CD3+ tumor-associated macrophage (TAM) (Cd3g) in the Mono/Mac compartment. For DC subclustering, Xcr1 was used to identify cDC1, Cd74 for cDC2, and Siglech and Ly6d for plasmacytoid DCs (pDCs). The stromal compartment was extracted and re-normalized. It was then divided into CAF (Dcn), Endothelial cells (Pecam1), Pericytes (Rgs5). For unauthorized clustering, k-nearest neighbor and Louvain algorithms were applied using Seurat. Signature scores were calculated by Seurat’s AddModuleScore function. Signature genes used are listed in Supplementary Data 1. ‘Dysfunction’56, ‘Exhaustion’57, ‘M1 signature’, and ‘M2 signature’58 gene sets were derived from previous reports. Additionally, ‘Effector’, ‘T cell proliferation’, ‘MHC class I’, ‘TCR signaling’ and ‘TCR downstream’ signatures were obtained from MSigDB (“GOLDRATH_NAIVE_VS_EFF_CD8_TCELL_DN”, “GOBP_POSITIVE_REGULATION_OF_T_CELL_PROLIFERATION”, “GOBP_ANTIGEN_PROCESSING_AND_PRESENTATION_OF_PEPTIDE_ANTIGEN_VIA_MHC_CLASS_I”, “REACTOME_TCR_SIGNALING” and “REACTOME_DOWNSTREAM_TCR_SIGNALING”). Ligand-receptor interaction analysis is performed by Python 3.10 and CellPhoneDB version 4. After identifying different cell types in our scRNA-seq as described above, we followed recommended procedures for the preparation of input files using CellPhoneDB. “means” analysis was performed at threshold of percentage of cells expressing a gene of 0.5. All interaction score was summed in each pair of cell types and plotted.
Raw gene expression (count) data and metadata of single cell transcriptomic dataset of human CRCs (SMC) were directly accessed through the GEO website (GSE132465) or figshare (https://doi.org/10.6084/m9.figshare.25323397.v1) and converted into Seurat object. Original author’s annotation30,43 was used to T cells and stromal cells. Tumor Fibroblast was extracted and used in further analysis according to marker gene expression (MMP11, COMP, FAP, and SFRP2 for mCAF; PDGFRA, CXCL14, and CFD for iCAF; NDRG1 and ENO1 for tCAF; RGS5 for Pericyte; RGS5 and MYH11 for vCAFl; CD74 and CD52 for apCAF; CCL19 and CCL21 for rCAF). Double-positive, single-positive, or double-negative cell expression was identified for gene expression and/or gene signatures with the WhichCells function as previously described59. Differential gene expression analysis between clusters was run with the FindMarkers function in Seurat. Cells were visualized according to UMAP embeddings. Signature scores were calculated using Seurat’s AddModuleScore function.
Spatial transcriptome analysis
Spatial Transcriptomics (ST) slides were made from FFPE blocks of surgically resected CRC specimens which were determined as CMS1/MSI, CMS2, or CMS4 by using RNA-seq data (for Visium) or staining (for Xenium) based on previous reports60. Visium ST analysis was performed by Takara Bio, Japan. Thin slice of FFPE blocks of identical area to ST slides were made and stained with hematoxylin and eosin. Sequence library of the ST slides was created using Visium Spatial for FFPE Gene Expression Kit and Dual Index Kit TS Set A (10X genomics). Raw sequencing reads of spatial transcriptomics were quality checked and mapped by Space Ranger v1.3.1. The gene-spot matrices and corresponding image of H&E stain were analyzed with the Seurat package (versions 5.1.0) in R version 4.4.1. Spots were filtered for minimum detected gene count of 200 genes. All objects were normalized using the SCTransform v2 function and integrated. Dimensionality reduction and clustering were performed with principal component analysis (PCA) at a resolution of 1.2 using the first 20 PCs. Signature scoring, including the analysis of a predefined mCAF signature, and a CTL signature43, was performed using the scGSVA function with default parameters. Spatial feature expression plots were generated with the SpatialFeaturePlot function in Seurat.
ST platform of Xenium 5 K was generated using Xenium Prime 5 K Human Pan-Tissue & Pathways pre-designed panel and performed by KOTAI Bio, Japan. Spots with nCount <50 or nFeature <5 were discarded. Raw UMI counts were log-normalized. As a single-cell reference we re-processed the public colorectal-cancer dataset of Lee et al61. The original single-cell annotations were consolidated into 13 broad classes after removing the low-abundance “Unknown” and “Mast-cell” categories. Deconvolution was performed with RCTD (spacexr v2.2.1) using default parameters except UMI_min = 50, DOUBLET_THRESHOLD = 50, and doublet_mode = “doublet“62. Spots labeled as “reject” (spot_class = “reject”) by RCTD were excluded from downstream analyses. Fibroblast spots whose log-normalized THBS2 expression exceeded 2 were labelled THBS2+ CAF.
Spatial neighborhoods were inferred with BANKSY v1.2.031. We identified the top 2,000 variable genes from each Xenium field of view (FOV), and used the union of these gene sets as a shared feature set for downstream integration. BANKSY graphs were constructed with k_geom = 15 and lamda = 0.8. Dimensionality reduction and clustering were performed with principal component analysis (PCA) on the concatenated BANKSY matrices, followed by Harmony correction (default parameters). Clustering was performed using the first 20 PCs at a resolution of 0.15 with the Leiden algorithm (FindClusters, algorithm = 4).
For generation of 3-D density maps of CD8⁺ T cells and THBS2+ CAF, spots annotated as CD8⁺ T cells or THBS2-CAFs were extracted from each Xenium FOV. Cell coordinates were converted to 500 × 500 kernel density maps using kde2d from the MASS R package (v7.3-61). The greyscale tissue image was placed at z = 0, and the two density surfaces were scaled to a shared global maximum and visualized in 3D using plotly (v4.11.0).
Cell–cell signalling was inferred with CellChat v 2.1.263. For each Xenium FOV, the log-normalized RNA matrix and spatial coordinates were imported with createCellChat (datatype = “spatial”). Only spots labelled “singlet” (spot_class = “singlet”) were retained. Genes detected in ≥ 10 cells were kept, and the human ligand-receptor database (CellChatDB.human) was assigned. Communication probabilities were computed with a truncated-mean model (computeCommunProb, trim = 0.1), adjusting for cluster size (population.size = TRUE). Spatial constraints were set to ≤ 10 µm for contact-dependent and ≤ 200 µm for secreted interactions. Significance was assessed with 100 permutations, and interactions involving <10 cells were discarded (filterCommunication). FOV-level CellChat objects were then merged, and differential interaction networks between the two conditions were visualized with netVisual_diffInteraction (weight.scale = TRUE). Differential signaling was identified with identifyOverExpressedGenes (thresh.pc = 0.1, thresh.fc = 0.05, thresh.p = 0.05); ligand–receptor pairs with |log₂FC | ≥ 0.05 were retained and shown as pathway-level chord diagrams using netVisual_chord_gene.
Bioinformatics analysis of clinical data
Data for TCGA-COADREAD was accessed through cBioportal (https://www.cbioportal.org). CRC samples in TCGA were stratified according to CMS subtypes (n: CMS1 = 85, CMS2 = 132, CMS3 = 78, CMS4 = 184) based on previous report using CMScaller6. MSI-H and MSS groups in TCGA were stratified by MSIsensor (n: MSI-H = 88, MSS = 494). Bulk iCMS classification was performed according to previous report7 using CMScaller. Briefly, gene expression quantities standardized with FPKM was log transformed and iCMS was determined using nearest template prediction. Adjusted p-value < 0.05 was considered as significant. IMF classification was conducted with applying iCMS, MSI, and fibrotic transcriptome (CMS4), and CRC samples in TCGA were stratified into five IMF classes: i2_MSS_NF (n = 117), i2_MSS_F (n = 101), i3_MSI (n = 81), i3_MSS_NF (n = 73) and 966 i3_MSS_F (n = 51). Raw gene expression data CRC patient dataset (GSE35602) was directly accessed through the GEO website (NCBI). GSVA was performed with R package GSVA. Gene signatures used are listed in Supplementary Data 1. The ‘Thbs2 KO signature’ was generated based on the top 20 DEGs that were upregulated in MTO-derived orthotopic tumors of Thbs2-/- mice, as identified from RNA-seq data.
Statistical analysis
All samples represent biological replicates. Data are presented as the mean ± SEM. Statistical analyses were performed using GraphPad Prism 10 or the R software environment. Pairwise differences were evaluated using two-tailed independent Student’s t-tests. When the data did not meet this test, a Mann-Whitney U-test was applied. For comparisons between three or more groups, statistical significance was determined using one-way or two-way ANOVA, followed by multiple comparison tests described in Figure Legends. Differences in Kaplan-Meier survival curves were analyzed using the Log-rank test. The Fisher’s exact test was used to evaluate the significance of differences between categorial covariates. P-values less than 0.05 (P < 0.05) were considered statistically significant.
Materials availability
This study did not generate new unique reagents.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets generated during this study have been deposited in the GEO repository on the NCBI website and are accessible under the following accession codes: RNA-seq_mouse_CAF (GSE285500 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE285500]), RNA-seq_mouse_tumor (GSE285501 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE285501]), RNA-seq_human_CRC (GSE285503 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE285503]), scRNA-seq (GSE285504 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE285504]) and Visium (GSE285505 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE285505]). Xenium spatial expression data from human CRC have been deposited in the GEA under accession number E-GEAD-1117. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided in Source Data File with this paper. Source data are provided with this paper.
Code availability
All data analysis and processing were executed through the use of published software or packages, the details of which were previously described and are referred to within the Methods. No novel code or mathematical algorithms were generated by this study.
References
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet 394, 1467–1480 (2019).
Arnold, M. et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 66, 683–691 (2017).
Chalabi, M. et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 26, 566–576 (2020).
Le, D. T. et al. PD-1 Blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Ganesh, K. et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 16, 361–375 (2019).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Joanito, I. et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 54, 963–975 (2022).
Gelsomino, F., Barbolini, M., Spallanzani, A., Pugliese, G. & Cascinu, S. The evolving role of microsatellite instability in colorectal cancer: a review. Cancer Treat. Rev. 51, 19–26 (2016).
Tauriello, D. V. F., Sancho, E. & Batlle, E. Overcoming TGFbeta-mediated immune evasion in cancer. Nat. Rev. Cancer 22, 25–44 (2022).
Dienstmann, R. et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 17, 268 (2017).
Ros, J. et al. The evolving role of consensus molecular subtypes: a step beyond inpatient selection for treatment of colorectal cancer. Curr. Treat. Options Oncol. 22, 113 (2021).
Tauriello, D. V. F. et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543 (2018).
Nakanishi, Y. et al. Simultaneous loss of both atypical protein kinase C genes in the intestinal epithelium drives serrated intestinal cancer by impairing immunosurveillance. Immunity 49, 1132–1147 (2018).
Nakanishi, Y., Diaz-Meco, M. T. & Moscat, J. Serrated colorectal cancer: the road less travelled? Trends Cancer 5, 742–754 (2019).
Colak, S. & Ten Dijke, P. Targeting TGF-beta signaling in cancer. Trends Cancer 3, 56–71 (2017).
Kasashima, H. et al. Stromal SOX2 upregulation promotes tumorigenesis through the generation of a SFRP1/2-expressing cancer-associated fibroblast population. Dev. Cell 56, 95–110 (2021).
Cords, L. et al. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat. Commun. 14, 4294 (2023).
Grout, J. A. et al. Spatial positioning and matrix programs of cancer-associated fibroblasts promote T-cell exclusion in human lung tumors. Cancer Discov. 12, 2606–2625 (2022).
Karamanos, N. K. et al. Extracellular matrix-based cancer targeting. Trends Mol. Med 27, 1000–1013 (2021).
Huang, J. C. et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct. Tar. 6, 153 (2021).
Martinez-Ordoñez, A. et al. Hyaluronan driven by epithelial aPKC deficiency remodels the microenvironment and creates a vulnerability in mesenchymal colorectal cancer. Cancer Cell 41, 252–271 (2023).
Sweetwyne, M. T. & Murphy-Ullrich, J. E. Thrombospondin1 in tissue repair and fibrosis: TGF-beta-dependent and independent mechanisms. Matrix Biol. 31, 178–186 (2012).
Gerarduzzi, C., Hartmann, U., Leask, A. & Drobetsky, E. The matrix revolution: matricellular proteins and restructuring of the cancer microenvironment. Cancer Res. 80, 2705–2717 (2020).
Omatsu, M. et al. THBS1-producing tumor-infiltrating monocyte-like cells contribute to immunosuppression and metastasis in colorectal cancer. Nat. Commun. 14, 5534 (2023).
Kyriakides, T. R. et al. Mice that lack thrombospondin 2 display connective tissue abnormalities that are associated with disordered collagen fibrillogenesis, an increased vascular density, and a bleeding diathesis. J. Cell Biol. 140, 419–430 (1998).
Kyriakides, T. R., Zhu, Y. H., Yang, Z., Huynh, G. & Bornstein, P. Altered extracellular matrix remodeling and angiogenesis in sponge granulomas of thrombospondin 2-null mice. Am. J. Pathol. 159, 1255–1262 (2001).
Streit, M. et al. Thrombospondin-2: a potent endogenous inhibitor of tumor growth and angiogenesis. Proc. Natl. Acad. Sci. USA 96, 14888–14893 (1999).
Nan, P. et al. Tumor-stroma TGF-beta1-THBS2 feedback circuit drives pancreatic ductal adenocarcinoma progression via integrin alpha(v)beta(3)/CD36-mediated activation of the MAPK pathway. Cancer Lett. 528, 59–75 (2022).
Liu, Z. et al. THBS2-producing matrix CAFs promote colorectal cancer progression and link to poor prognosis via the CD47-MAPK axis. Cell Rep. 44, 115555 (2025).
Chu, X. et al. Integrative single-cell analysis of human colorectal cancer reveals patient stratification with distinct immune evasion mechanisms. Nat. Cancer 5, 1409–1426 (2024).
Singhal, V. et al. BANKSY unifies cell typing and tissue domain segmentation for scalable spatial omics data analysis. Nat. Genet 56, 431–441 (2024).
Muta, Y. & Nakanishi, Y. Mouse colorectal cancer organoids: Lessons from syngeneic and orthotopic transplantation systems. Eur. J. Cell Biol. 104, 151478 (2025).
Nicolas, A. M. et al. Inflammatory fibroblasts mediate resistance to neoadjuvant therapy in rectal cancer. Cancer Cell 40, 168–184.e113 (2022).
Stintzing, S. et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann. Oncol. 30, 1796–1803 (2019).
Bornstein, P., Kyriakides, T. R., Yang, Z., Armstrong, L. C. & Birk, D. E. Thrombospondin 2 modulates collagen fibrillogenesis and angiogenesis. J. Investig. Dermatol Symp. Proc. 5, 61–66 (2000).
Spranger, S., Dai, D., Horton, B. & Gajewski, T. F. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–723 (2017).
House, I. G. et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. 26, 487–504 (2020).
Pascual-Garcia, M. et al. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8(+) T cell tumor-infiltration impairing anti-PD1 therapy. Nat. Commun. 10, 2416 (2019).
Lim, R. J. et al. CXCL9/10-engineered dendritic cells promote T cell activation and enhance immune checkpoint blockade for lung cancer. Cell Rep. Med 5, 101479 (2024).
Stoltzfus, C. R. et al. CytoMAP: a spatial analysis toolbox reveals features of myeloid cell organization in lymphoid tissues. Cell Rep. 31, 107523 (2020).
Baker, A. T., Abuwarwar, M. H., Poly, L., Wilkins, S. & Fletcher, A. L. Cancer-associated fibroblasts and T cells: from mechanisms to outcomes. J. Immunol. 206, 310–320 (2021).
Jenkins, L. et al. Cancer-associated fibroblasts suppress CD8+ T-cell infiltration and confer resistance to immune-checkpoint blockade. Cancer Res. 82, 2904–2917 (2022).
Lee, H. O. et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 52, 594 (2020).
Chen, S. et al. Identification of the “Collagen-Macrophage” sub-category of patients with colorectal cancer as an extension of the CMS4 subtype with THBS2 as a therapeutic target. BMC Gastroenterol. 25, 342 (2025).
Cheng, P. S. W., Zaccaria, M. & Biffi, G. Functional heterogeneity of fibroblasts in primary tumors and metastases. Trends Cancer 11, 135–153 (2025).
Kennel, K. B., Bozlar, M., De Valk, A. F. & Greten, F. R. Cancer-associated fibroblasts in inflammation and antitumor immunity. Clin. Cancer Res 29, 1009–1016 (2023).
Chow, M. T. et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 50, 1498–1512.e1495 (2019).
Gopinath, P., Natarajan, A., Sathyanarayanan, A., Veluswami, S. & Gopisetty, G. The multifaceted role of matricellular Proteins in health and cancer, as biomarkers and therapeutic targets. Gene 815, 146137 (2022).
Florin, L. et al. Cre recombinase-mediated gene targeting of mesenchymal cells. Genesis 38, 139–144 (2004).
Hamada, K. et al. Retinoblastoma-binding Protein 9 suppresses intestinal inflammation and inflammation-induced tumorigenesis in mice. Cell Mol. Gastroenterol. Hepatol. 19, 101435 (2025).
Kasashima, H. et al. Mouse model of colorectal cancer: orthotopic co-implantation of tumor and stroma cells in cecum and rectum. STAR Protoc. 2, 100297 (2021).
Romero, J. M. et al. Chemokine expression predicts T cell-inflammation and improved survival with checkpoint inhibition across solid cancers. npj Precis. Oncol. 7, 73 (2023).
Jorapur, A. et al. EBV+ tumors exploit tumor cell-intrinsic and -extrinsic mechanisms to produce regulatory T cell-recruiting chemokines CCL17 and CCL22. PLoS Pathog. 18, e1010200 (2022).
Allen, F. et al. CCL3 augments tumor rejection and enhances CD8(+) T cell infiltration through NK and CD103(+) dendritic cell recruitment via IFNgamma. Oncoimmunology 7, e1393598 (2018).
Gao, Q. et al. Cancer-cell-secreted CXCL11 promoted CD8(+) T cells infiltration through docetaxel-induced-release of HMGB1 in NSCLC. J. Immunother. Cancer 7, 42 (2019).
Li, H. et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 176, 775–789.e718 (2019).
Yost, K. E. et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019).
Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459 (2020).
Muta, Y. et al. Enhanced SREBP2-driven cholesterol biosynthesis by PKCλ/ι deficiency in intestinal epithelial cells promotes aggressive serrated tumorigenesis. Nat. Commun. 14, 8075 (2023).
Li, X. et al. A modified protein marker panel to identify four consensus molecular subtypes in colorectal cancer using immunohistochemistry. Pathol. Res Pr. 220, 153379 (2021).
Lee, H. O. et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet 52, 594–603 (2020).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 40, 517–526 (2022).
Jin, S., Plikus, M. V. & Nie, Q. CellChat for systematic analysis of cell-cell communication from single-cell transcriptomics. Nat. Protoc. 20, 180–219 (2025).
Acknowledgements
Research was supported in part by Grants-in-Aid from the JSPS KAKENHI (23K21432, 23H02891, 23K27582, 24K02438, 25 K02638); the Japan Agency for Medical Research and Development (AMED), P-PROMOTE (23ama221326h0001, 24ama221326h0002, 24ama221515h0003, 24ama221139h0001, 25ama221444h0001, 25ama221139h0002, 25ama221326h0001); the Moonshot Research and Development Program (JPMJMS2022, JP22zf0127009); COI-NEXT (JPMJPF2018); the Fusion Oriented Research for disruptive Science and Technology (FOREST, 23719768); the foundations of Takeda Science, Princess Takamatsu Cancer Research, Astellas, Daiichi Sankyo of Life Science, Yasuda, Mochida Memorial, Uehara, Naito, and Kyoto University.
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.N., Y.M. and K.I.; methodology, Y.N., Y. and K.; investigation, K.I., Y. M., J.C., K.Y., M.O., N. A., M.I., Y. M., L.C., G.Y., K.H., K.M., M.K., K.I., S.N., N.A., T.U., M.N., T.M., Y.H., M.F., N.K., M.M.N., T.F., Y.K., H.K., A.F. and S.O.; resources, J.M., M.T.D-M, T.K., K.K., M.Y., K.M., Y.I. and K.O.; writing original draft, Y.N., Y.M. and K.I.; writing – review & editing, all authors; supervision, Y.N., Y.M. and H.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Phyllis Cheung, Jens Siveke, Zlatko Trajanoski and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Iwane, K., Nakanishi, Y., Muta, Y. et al. Targeting fibroblast derived thrombospondin 2 disrupts an immune-exclusionary environment at the tumor front in colorectal cancer. Nat Commun 16, 11590 (2025). https://doi.org/10.1038/s41467-025-66485-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66485-2