Abstract
Cholestasis, a bile flow disorder common to many liver diseases, currently lacks effective treatments. Emerging evidence links gut microbiota disturbances to cholestatic liver injury. Here, an antibiotic cocktail (ABX)-treated mouse model confirmed the indispensable role of the intestinal microbiota, with marked shifts including increased Alistipes putredinis (A. putredinis) and decreased Clostridium spp. (C. spp.). In vitro, ferulic acid and wogonin effectively modulated the gut flora, and in vivo they alleviated liver injury. Administration of A. putredinis exacerbated hepatic inflammation by disrupting intestinal barrier integrity and facilitating bacterial translocation, an effect reversed by ferulic acid. Conversely, treatment with C. spp. and wogonin enhanced bile salt hydrolase activity and bile acid excretion. Notably, combined treatment with ferulic acid and wogonin or C. spp. significantly ameliorated cholestatic liver injury. These findings underscore the critical role of gut microbiota in cholestasis and suggest therapeutic potential for microbiota-targeted and natural compound-based interventions.
Similar content being viewed by others
Introduction
Cholestasis occurs when bile flow from the liver to the small intestine is disrupted1. often resulting from viral infections, medications, or metabolic and genetic conditions2. The accumulation of bile acids (BAs), particularly hydrophobic BAs, in the liver parenchyma is pivotal in the pathogenesis of cholestatic liver damage. This causative factor is typically associated with bile duct destruction and fibrosis3,4. Despite ursodeoxycholic acid being the primary treatment for various cholestatic disorders aimed at enhancing bile cholestasis, the majority of patients only exhibit a partial response, with many progressing to liver failure and cirrhosis5,6. Thus, there is an urgent need to discover an effective medication for treating intrahepatic cholestasis.
In the last two decades, increasing attention has been given to the bidirectional interaction between BAs and the microbiota7,8,9. BAs affect the intestinal microbiota composition by modulating the host’s intestinal immune landscape and possessing intrinsic antimicrobial activity10. In turn, intestinal microbiota play a role in deconjugating, dehydroxylating, and enzymatically modifying the pool of BAs, and they also modulate bile acid transporters such as apical sodium-dependent BAs transporter11,12,13. Recent studies have indicated that dysbiosis in microbial communities and their metabolites, such as BAs, can influence the liver immune microenvironment and contribute to liver diseases14,15. Therefore, treatments that adjust the crosstalk within the gut-liver bile acid system and microbial modulation of BA metabolism offer more effective interventions and therapeutic strategies for cholestasis-related diseases16,17.
As microbial research technology has advanced, the crucial role of traditional Chinese medicine (TCM) in influencing host metabolism by regulating the composition of the microbiota has been confirmed18,19. Certain probiotic bacteria, such as Lactobacillus rhamnosus GG and Lactobacillus acidophilus, have also demonstrated therapeutic properties for cholestatic liver disease20,21. Research on the gut’s commensal bacteria helps to accurately comprehend the complex dynamics at the host–microbiota interface. However, the intricate relationships among TCM, microbial communities, and the host require further study.
Yinzhihuang (YZH) is a hospital prescription comprising Artemisia capillaris Thunb., Gardenia jasminoides Ellis, Scutellaria baicalensis Georgi and Lonicera japonica Thunb. Clinically, YZH has been used to treat various types of liver injury, reduce alanine aminotransferase, and enhance bilirubin clearance in newborns22,23. Phytochemical analyses have identified ferulic acid (FA) and wogonin (Wog) as abundant in YZH24. FA, a phenolic acid, exhibits antioxidant, anti-inflammatory, anti-fibrotic, and endothelial-protective activities25. Wog, a flavonoid from Scutellaria baicalensis Georgi, suppresses proinflammatory cytokine production, reduces oxidative stress, and demonstrates anticancer and neuroprotective effect26,27. However, the potential beneficial effects of FA and Wog on cholestasis remain unclear.
In this study, using a broad-spectrum antibiotic cocktail (ABX)-treated mouse model, we found that the therapeutic effects of YZH on cholestatic liver damage were linked to alterations in gut microbiota. YZH treatment decreased the abundance of Alistipes and increased the abundance of Clostridium. Additionally, we identified the effective compounds of YZH, including FA and Wog, which influence the growth of characteristic gut microbiota in vitro, thereby improving outcomes in cholestatic liver disease patients. Furthermore, we performed in vivo experiments to confirm the mechanism by which the characterized bacteria and the active compounds interact with cholestasis. Finally, the combination of FA and Wog or Clostridium spp. also improved cholestasis. Thus, our findings provide insights into the gut microbiota associated with cholestatic liver injury and highlight the therapeutic potential of probiotics and active compounds.
Results
YZH treatment protects the liver against intrahepatic cholestasis
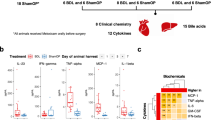
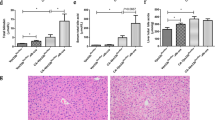
We first evaluated the therapeutic effects of YZH in alpha-naphthyl isothiocyanate (ANIT)-induced mice. Compared with ANIT-induced cholestatic mice, mice treated with 2.7 g/kg and 5.4 g/kg YZH presented decreased levels of alanine aminotransferase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), direct bilirubin (D-BIL) and total bilirubin (T-BIL) (Figs. 1A–C and S1A–B). Liver tissue staining further demonstrated that YZH markedly ameliorated liver tissue injury, collagen deposition, and hepatic fibrosis (Figs. 1D–G and S1C–F).
A Serum ALT in each group. B Serum AST in each group. C Serum ALP in each group. D Representative images of liver sections after staining with H&E (100 μm). E Quantification of histological score. F Representative images of liver sections after staining with Masson (200 μm). G Quantification of collagen volume fraction (%). Data are represented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. YZH Yinzhihuang, ANIT alpha-naphthyl isothiocyanate, ALT alanine aminotransferase, AST aspartate transaminase, ALP alkaline phosphatase, H&E hematoxylin and eosin.
Gut microbiota is essential for the protective effects of YZH on cholestatic liver damage in ANIT mice
To further investigate the role of gut microbiota in mediating the beneficial effects of YZH on ANIT-induced cholestatic liver injury, the original gut microbiota was depleted using a broad-spectrum ABX cocktail for 5 days, followed by 2 weeks of YZH treatment. ABX was administered daily throughout the experiment to ensure effective microbiota clearance. Elimination of the gut microbiota abolished the protective effects of YZH, as evidenced by serum levels of liver injury markers ALT, AST, ALP, D-BIL and T-BIL (Figs. 2A–C and S2A–B). Furthermore, microbiota depletion prevented YZH-mediated improvements in ANIT-induced hepatic pathology and collagen deposition (Figs. 2D–G and S2C–F).
A Serum ALT in each group (n = 5). B Serum AST in each group (n = 5). C Serum ALP in each group (n = 5). D Representative images of liver sections after staining with H&E (100 μm). E Representative images of liver sections after staining with Masson (200 μm). F Quantification of histological score (n = 5). G Quantification of collagen volume fraction (%) (n = 5). H Alpha diversity of gut microbial communities assessed by Simpson and the Observed_OTUs (n = 5). I NMDS plot of weighted UniFrac analysis (n = 5). J LEfSe analysis identifying differentially abundant taxa between ANIT and 5.4 g/kg YZH-treated mice. The criteria for feature selection is Log LDA Score > 2. K Relative abundance of Alistipes in human feces (n = 10–19). L Relative abundance of Clostridium in human feces (n = 10–19). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. YZH Yinzhihuang, ANIT alpha-naphthyl isothiocyanate, ALT alanine aminotransferase, AST aspartate transaminase, ALP alkaline phosphatase, H&E hematoxylin and eosin, NMDS non-metric multidimensional scaling, LDA linear discriminant analysis, LEfSe LDA Effect Size, qPCR quantitative polymerase chain reaction.
To pinpoint the microbial drivers of this effect, we performed 16S ribosomal RNA (16S rRNA) gene sequencing on cecal samples collected from control, ANIT-treated, and YZH-treated mice. Analysis of α-diversity and β-diversity revealed that YZH treatment did not significantly alter the overall microbial diversity (Fig. 2H, I). Linear discriminant analysis effect size28,29,30 (LEfSe) revealed distinct differences in the faecal microbiota composition between the ANIT and YZH groups. At the genus level, Alistipes and Serratia were predominant in ANIT-treated mice, whereas Lachnospira, Clostridium and Ruminiclostridium were enriched in YZH-treated mice (Fig. 2J). To further validate these findings, we assessed the abundance of these genera in faecal samples from healthy individuals and patients with cholestasis. Compared with controls, the cholestatic group exhibited significantly increased Alistipes abundance and decreased Clostridium as determined by qPCR (Figs. 2K, L and S2G). Consistently, in the ANIT-induced mouse model, Alistipes abundance was markedly elevated relative to controls, while YZH treatment notably restored Clostridium abundance (Fig. S2H–I). Together, these findings indicate that the therapeutic effects of YZH on cholestatic liver damage rely on the presence and modulation of the gut microbiota.
Alistipes putredinis and Clostridium spp. are key strains affecting cholestatic liver injury
To identify specific strains within the genus Alistipes, we cultured fecal samples from ANIT-treated mice under anaerobic conditions. Since Alistipes is a gram-negative anaerobe, we supplemented the culture medium with 7.5 μg/mL vancomycin and 100 μg/mL kanamycin to inhibit the growth of gram-positive bacteria and facultative anaerobes31. A total of 432 monoclonal colonies were obtained through isolation and cultivation. Given that Alistipes possess tryptophanase32, an enzyme that catalyzes the conversion of tryptophan to indole, we employed p-dimethylaminobenzaldehyde (PDL) as an indole indicator. PDL reacts with indole to form a red-colored rose indole33. This colorimetric assay enables the screening of bacterial colonies capable of metabolizing tryptophan. Of the 432 bacterial colonies isolated, 68 colonies exhibited a positive indole reaction. One bacterial colony was confirmed as Alistipes putredinis (A. putredinis) through 16S rRNA species identification (Fig. 3A).
A Schematic overview of the procedures used for screening A. putredinis microbes. B The experimental design scheme. C Relative abundance of A. putredinis in stool samples measured by qPCR (n = 6). D Relative abundance of C. spp. in stool samples measured by qPCR (n = 6). E Serum ALT in each group (n = 5). F Serum AST in each group (n = 5). G Serum ALP in each group (n = 5). H Representative images of liver sections after staining with H&E (100 μm). I Quantification of histological score (n = 5). J Representative images of liver sections after staining with Masson (100 μm (top) ann 200 μm (bottom). K Quantification of collagen volume fraction (%) (n = 5). Data are represented as mean ± SEM. #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001 vs. ANIT’. *p < 0.05, **p < 0.01, ***p < 0.001 vs. ANIT. ANIT alpha-naphthyl isothiocyanate, ANIT’ single ANIT dose, 16S rRNA 16S ribosomal RNA, A. putredinis Alistipes putredinis, C. spp.Clostridium spp., CFU colony forming unit, ALT alanine aminotransferase, AST aspartate transaminase, ALP alkaline phosphatase, H&E hematoxylin and eosin, qPCR quantitative polymerase chain reaction.
To assess the roles of A. putredinis and C. spp. in cholestasis, we first depleted the gut microbiota with antibiotics, then gavaged mice with 2.0 × 108 colony forming units (CFUs) of each strain every three days. In the two-hit ANIT model (ANIT group), mice received ANIT (62.5 mg/kg) on days 1 and 13 to induce chronic cholestasis and evaluate the hepatoprotection conferred by C. spp. In the single-hit model (ANIT′ group), a single ANIT dose on day 1 elicited acute cholestasis without transporter adaptations to probe A. putredinis pathogenicity34,35 (Fig. 3B). The colonization of A. putredinis and C. spp. was confirmed by qPCR (Fig. 3C, D). Although there was no notable difference in the levels of D-BIL and T-BIL between the control and A. putredinis groups, we observed a marked increase in the activity of ALT, AST and ALP in A. putredinis-colonized mice. Furthermore, we identified that colonization of C. spp. significantly reduced ALT, AST, ALP, D-BIL and T-BIL levels (Figs. 3E–G and S3A and B). In addition, histological alterations, collagen deposition and hepatic fibrosis in liver tissues were significantly mitigated by C. spp. treatment and exacerbated by A. putredinis (Figs. 3H–K and S3C–F). Furthermore, in a bile duct ligation (BDL) model, mice were observed for 9 days post-ligation, with A. putredinis supplementation at the start and every three days thereafter. BDL induced acute liver injury, elevated ALT, AST, ALP, D-BIL and T-BIL, and portal microfibrosis compared to sham controls. A. putredinis further elevated serum AST and D-BIL levels in BDL mice, and exacerbated hepatic pathological injury and fibrosis (Fig. S3G–O). Overall, these results suggest that A. putredinis could aggravate intrahepatic cholestasis, while C. spp. has a therapeutic effect on cholestatic diseases.
Ferulic acid and wogonin ameliorate cholestasis by regulating Alistipes putredinis and Clostridium spp
We previously showed that YZH formula alleviates cholestatic liver injury in an intestinal microbiota-dependent manner36. To explore the material basis underlying this effect, we queried the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database to retrieve YZH’s principal compounds and evaluate their pharmacokinetic profiles. Given oral bioavailability (OB) dictates whether a compound acts systemically or remains within the gut lumen37,38, we applied an OB < 40% cutoff to prioritize compounds likely to engage directly with intestinal microbes or be converted by them into more bioactive derivatives39. Accordingly, we selected 19 YZH compounds meeting the OB < 40% criterion from TCMSP and conducted in vitro co culture assays with two characteristic flora, A. putredinis and C. spp. to directly probe their microbiota interactions (Fig. 4A). The results indicated that FA exhibited the strongest inhibitory effect on the growth of A. putredinis, whereas Wog most significantly enhanced both the growth and bile salt hydrolase (BSH) activity of C. spp. (Figs. 4B–D and S4A). Furthermore, we administered 50 mg/kg and 100 mg/kg FA and 20 mg/kg and 40 mg/kg Wog by gavage to ANIT mice or BDL mice in vivo (Fig. 4E, F). FA and Wog treatment decreased levels of liver damage indices, including ALT, AST, ALP, D-BIL and T-BIL (Figs. 4G–L and S4B–E). Compared with ANIT or BDL mice, a lower histological score, collagen volume fraction (%), sirius red positive area (%) and α-smooth muscle actin (α-SMA) positive area (%) were observed after FA and Wog treatment (Figs. 4M–T, S4F–M and S5A–I). As expected, FA and Wog administration decreased the mRNA expression of Il-6 and Tnf-a in the liver (Fig. S4N–Q).
A Schematic flow diagram for screening active compounds. B Effect of ferulic acid on the growth of A. putredinis in vitro (n = 5). C Effect of wogonin on the growth of C. spp. in vitro (n = 5). D The indicated compounds on the BSH activity of C. spp. in vitro (n = 4). D–F The experimental design scheme. G–I Serum ALT, AST and ALP in control, ANIT and FA treatment groups (n = 5). J–L Serum ALT, AST and ALP in control, ANIT and Wog treatment groups (n = 5). M, N Representative images of liver sections after staining with H&E (100 μm). O, P Quantification of histological score (n = 5). Q, R Representative images of liver sections after staining with Masson (Q: 100 μm, R: 200 μm). S, T Quantification of collagen volume fraction (%) (n = 5). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. OB oral bioavailability, DMSO dimethyl sulfoxide, ANIT alpha-naphthyl isothiocyanate, A. putredinis Alistipes putredinis, C. spp. Clostridium spp., BSH bile salt hydrolase, FA ferulic acid, Wog wogonin, i.g. intragastric administration, ALT alanine aminotransferase, AST aspartate transaminase, ALP alkaline phosphatase, H&E hematoxylin and eosin.
Ferulic acid reverses A. putredinis-induced intestinal mucosal barrier damage and bacterial translocation
To further investigate the potential role of A. putredinis in intrahepatic cholestasis, stool samples were subjected to shotgun metagenomic sequencing. Alpha diversity analysis, including Chao1 and Shannon indices, revealed lower bacterial diversity and reduced bacterial richness in A. putredinis-colonized mice (Fig. S6A). Beta diversity analysis using PCoA based on Bray‒Curtis distances indicated significant differences in the gut microbiota compositions between A. putredinis-colonized mice and control mice (Fig. S6B).
The intestinal mucus barrier, primarily composed of heavily O-glycosylated mucins, relies exclusively on carbohydrate-active enzymes (CAZymes) for its degradation and remodeling40,41. These CAZymes are essential for preserving barrier integrity and mediating host-microbiota crosstalk, while mucus layer disruption permits translocation of microbial products that can exacerbate cholestatic liver injury via the gut-liver axis42. To investigate how A. putredinis colonization impacts mucus integrity, we surveyed CAZyme gene families in cecal metagenomes using the CAZy database. The results revealed significantly increased levels of 19 carbohydrases in A. putredinis-colonized mice (Fig. 5A). Among them, many host glycans and mucus-targeting enzymes, including glycoside hydrolase (GH) 129, which deconstructs intestinal mucin glycan structures, and carbohydrate-binding module (CBM) 41, which degrades polysaccharide substrates, are significantly elevated (Fig. 5B). To explore whether A. putredinis regulates the composition of carbohydrate-related bacteria, we conducted a screening. As shown in Fig. 5C, the left panel represents bacterial species significantly upregulated after A. putredinis gavage, the middle panel represents bacteria significantly correlated with A. putredinis, and the right panel represents bacteria regulating different carbohydrate enzymes (Tables S3–S5). The shard analysis across these three panels revealed four bacterial species, Gordonibacter urolithinfaciens, Fusicatenibacter saccharivorans, Ruthenibacterium lactatiformans and Holdemania massiliensis, suggesting that A. putredinis is essential for regulating the composition of bacteria associated with mucin degradation (Fig. 5C–E). Measurement of colonic mucosal secretions showed that goblet cell (GC)+ was lower in A. putredinis-colonized mice than control mice (Figs. 5F, G and S6C). Mucin 2 (Muc2) and kruppel-like factor 3 (Klf3) mRNA levels were reduced in A. putredinis-colonized mice, while Muc5ac remained statistically unchanged (Fig. 5H). Colonic histological analysis showed intestinal epithelial damage and inflammatory infiltrates in A. putredinis-colonized mice (Fig. S6D). Moreover, the expression levels of E-cadherin, Zonula occludens-1 (ZO-1), and Claudin-1 were significantly decreased after A. putredinis gavage (Fig. S6E). A. putredinis gavage also resulted in higher serum levels of 4 kDa fluorescein isothiocyanate-dextran (FD4) than control mice (Figs. 5I and S6F–G).
A The volcano plot depicts the fold change and p-value distribution of CAZymes database analysis after shotgun metagenomic sequencing analysis of control and A. putredinis group feces. Red points indicate p < 0.05 and FC of >2; blue points indicate p value < 0.05 and FC of <0.5 (n = 5). B Positive fold-changes in CAZymes database between two group comparisons. Only CAZyme families (x axis) in which >2-fold changes and p < 0.05 (Student’s t test) were listed. C Schematic representation of A. putredinis-regulated carbohydrase-associated bacteria. (Left panel analysis of bacteria differentially upregulated by A. putredinis gavage; center panel, analysis of correlation of bacteria with A. putredinis; right panel, analysis of bacteria expressing differential carbohydrases). D Correlation network of A. putredinis with other bacteria significantly upregulated by A. putredinis and expressing carbohydrases. The red line represents positive correlation and the blue line represents negative correlation. The solid line indicates p < 0.05 and the dotted line indicates p > =0.05. Line thickness denotes the correlation strength. E Relative abundance of indicated bacteria (n = 5). F Alcian blue was performed in colon sections. Scale bar: 100 μm. G Proportion of Alcian blue+ GCs in different colon regions (n = 5). H mRNA levels of Muc2, Muc5ac and Klf3 in colon (n = 6). I Serum level of FD4 (n = 5). J Representative images of liver sections after staining with eubacteria (red), CD45 (white), and DAPI (blue). Scale bar: 100 μm (left) and 20 μm (right). Data are represented as mean ± SEM. #p < 0.05, ##p < 0.01 vs. ANIT’. ANIT alpha-naphthyl isothiocyanate, ANIT’ single ANIT dose, FC fold change, CBM carbohydrate-binding module, GH glycoside hydrolase, GT glycosyltransferase, PL polysaccharide lyase, AA auxiliary activity, CAZymes carbohydrate-active enZYmes, Abs_rho absolute spearman’s rank correlation coefficient, G. urolithinfaciens Gordonibacter urolithinfaciens, F. saccharivorans Fusicatenibacter saccharivorans, R. lactatiformans Ruthenibacterium lactatiformans, H. massiliensis Holdemania massiliensis, GCs goblet cells, Mucmucin, Klf3 kruppel-like factor 3, FD4 4 kDa fluorescein isothiocyanate dextran.
Bacterial translocation to the intestinal submucosal tissue and the liver is encouraged by weakening of the gut barrier43. Fluorescence in situ hybridization (FISH) analysis showed that a higher bacterial load was observed in the submucous in A. putredinis-colonized mice (Fig. S6H–I). To detect the presence of bacteria in the liver, we utilized the EUB338 probe labeled with bacteria and CD45 labeled with immune cells. We found that, in A. putredinis-colonized mice, bacteria were found in the parenchyma outside of immune cells, suggesting that these bacteria may have translocated from the intestine to the liver (Fig. 5J). However, A. putredinis gavage failed to cause a significant increase in hepatic bacterial populations, as determined by bacterial CFU analysis (S6J and K).
Furthermore, the oral administration of 50 mg/kg and 100 mg/kg FA markedly increased goblet cell count (Figs. 6A, B and S7A). Histologic examination of colon sections confirmed that inflammation and tissue injury were reduced after FA treatment (Fig. S7B). Furthermore, IF staining confirmed that FA profoundly increased E-cadherin, ZO-1, and Claudin-1 expression (Fig. S7C). The measurement of FD4 leakage following FA treatment revealed a reduction in intestinal barrier disruption (Figs. 6C and S7D). Consistently, FISH analysis and CFU calculations revealed fewer bacteria in the colon and liver tissues of FA- and SA-treated mice compared with ANIT mice (Figs. 6D–F and S7E–F). Collectively, our findings indicate that A. putredinis may induce cholestasis by disrupting the intestinal mucus barrier and tight junction pathways, whereas FA treatment inhibits intestinal mucosal barrier disruption and prevents bacterial invasion into the liver.
A Representative images of Alcian blue staining in different groups (100 μm). B Proportion of Alcian blue+ GCs in different colon regions (n = 5). C FD4 serum levels in the various groups (n = 5). D Bacteria in the liver were stained via FISH with EUB338 (red), CD45 (white), and DAPI (blue). Scale bars: 100 μm. E Liver bacteria culture on Columbia blood agar plates. F Quantification of the bacterial count in the liver (n = 3). Data are represented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ****p < 0.0001. ANIT alpha-naphthyl isothiocyanate, FA ferulic acid, GCs goblet cells, FD44 kDa fluorescein isothiocyanate dextran, CFU colony forming unit.
Clostridium spp. and wogonin promote bile acid excretion
Gut microbiota, especially Clostridium, has been considered a key factor in bile acid metabolism44. This prompted us to further examine the bile acids (BAs) level after mice were gavaged with C. spp. Notably, C. spp. decreased the total hepatic BAs level (Fig. 7A). Liquid Chromatograph-Mass Spectrometer (LC-MS) analysis of serum BAs showed a significant decrease in taurocholic acid (TCA) and tauro-α-muricholic acid (T-α-MCA) in conjugated BAs and a significant increase in deoxycholic acid (DCA), α-muricholic acid (α-MCA) and β-muricholic acid (β-MCA) in unconjugated BAs in C. spp.-colonized mice compared with ANIT mice (Fig. 7B). Furthermore, C. spp. increased the total fecal BAs level, suggesting that C. spp. promotes the excretion of BAs in the intestine (Fig. 7C). In vivo validation showed that BSH activitity was slightly elevated after 20 mg/kg Wog treatment, indicating enhanced bile acid deconjugation activity in bacteria (Fig. 7D). Moreover, compared with ANIT mice, the conjugated BAs TCA and T-α-MCA in serum were apparently decreased, and the unconjugated BAs CA, α-MCA and β-MCA were increased following Wog administration (Fig. 7E). Compared with that of ANIT mice, the total hepatic BA concentration decreased, whereas the total fecal BAs concentration increased after Wog treatment (Fig. 7F, G). Collectively, our data indicate that C. spp. and wogonin promote conjugated bile acid metabolism and excretion.
A Liver total bile acid concentrations in indicated groups. B Serum bile acid concentrations in the ANIT and C. spp. groups. C Fecal total bile acid concentrations were determined in indicated groups. D BSH enzyme activity in feces. E Bile acid concentrations in the serum between ANIT and Wog treatment group. F Liver total bile acid concentrations were determined in indicated groups. G Fecal total bile acid concentrations were determined in indicated groups. Data are represented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01. ANIT alpha-naphthyl isothiocyanate, Wog wogonin, C. spp. Clostridium spp., BAs bile acids, BSH bile salt hydrolase, TDCA taurodeoxycholic acid, TLCA taurolithocholic acid, GLCA glycolithocholic acid, GCA glycocholic acid, TCA taurocholic acid, TUDCA tauroursodeoxycholic acid, T-α-MCA tauro α-Muricholic acid, T-β-MCA tauro β-Muricholic acid, THDCA taurohyodeoxycholic acid, TCDCA taurochenodeoxycholic acid, DCA deoxycholic acid, LCA lithocholic acid, CA cholic acid, UDCA ursodeoxycholic acid, α-MCA α-Muricholic acid, β-MCA β-Muricholic acid, HDCA hyodeoxycholic acid, CDCA chenodeoxycholic acid.
FA combined with Wog and C. spp. can further alleviate cholestatic liver injury in vivo
Considering the preceding findings, we conducted further investigations into the therapeutic efficacy of FA in combination with Wog and C. spp. in the context of cholestatic liver injury. Liver damage indices in the combined treatment group exhibited a marked decrease, according to ALT, AST, ALP, D-BIL and T-BIL levels (Figs. 8A–C and S8A–B). Combination therapy consistently decreased the histological score, collagen volume fraction (%), sirius red positive area (%) and α-SMA positive area (%) (Figs. 8D–G and S8C–F). Overall, these results underscored that FA combined with Wog and C. spp. can more effectively protect the liver against intrahepatic cholestasis compared to individual treatments with FA, Wog or C. spp.
A Serum ALT in each group (n = 5). B Serum AST in each group (n = 5). C Serum ALP in each group (n = 5). D Representative images of liver sections after staining with H&E (100 μm). E Representative images of liver sections after staining with Masson (100 μm). F Quantification of histological score (n = 3). G Quantification of collagen volume fraction (%) (n = 3). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ANIT alpha-naphthyl isothiocyanate, FA ferulic acid, Wog wogonin, C. spp.Clostridium spp., ALT alanine aminotransferase, AST aspartate transaminase, ALP alkaline phosphatase, H&E hematoxylin and eosin.
Discussion
Cholestasis refers to the buildup of bile in the hepatobiliary system caused by impaired bile formation, secretion, and/or excretion. This condition disrupts the metabolism of bile through the duodenum1. Intestinal microorganisms are vital to the hepatic and intestinal cycling of BAs through their de-conjugation, 7a-dehydroxylation, and other activities that help regulate bile acid composition7.In this study, we conducted research on the specific mechanism and active compounds of the amelioration of cholestatic liver injury by YZH from the point of view of the intestinal microorganisms.
We conducted 16S rRNA sequencing and qPCR analysis and identified significant inhibition of the bacterial genus Alistipes by YZH. CAZyme analysis revealed that A. putredinis significantly upregulated some host glycan and mucus-targeting enzymes and the associated gut microbiota. The gut serves as a competitive niche for microorganisms, where the capacity to break down complex carbohydrates gives specific bacteria a distinct advantage45. When dietary fiber is lacking, certain commensal bacteria exploit host polysaccharides through their enzymatic activities, including GHs, CBMs, carbohydrate esterases (CEs), and proteases46,47. Notably, mucin utilizers like Akkermansia muciniphila, Bacteroides fragilis and Bacteroides thetaiotaomicron bind to and degrade glycans on the mucin, affecting the gut integrity48,49. Further observations revealed that A. putredinis gavage resulted in thinning of mucus and a decrease in the levels of the transcription factors Muc2 and Klf3, which are involved in barrier function. These results point to a potential association between A. putredinis and the degradation of colonic mucins. Further studies will contribute to our understanding of microbiota‒mucin interactions.
Our findings highlight members of the order Clostridiales as key YZH-and Wog responsive taxa with probiotic potential. Not only do Clostridiales species produce bioactive metabolites that reinforce epithelial barrier integrity and promote mucosal immune tolerance44, but also they encode BSH and 7α-dehydroxylase enzymes that deconjugate and transform host-derived bile acids, thereby reshaping both bile acid pool size and signaling properties13. In our antibiotic-depleted mouse model, C. spp. supplementation markedly increased in vivo BSH activity, driving the deconjugation of TCA and T-α-MCA. This enzymatic shift reduced the cytotoxicity of conjugated bile acids and generated secondary bile acids that act as weaker FXR agonists or even antagonists in ileal enterocytes50. Evidence from germ-free and antibiotic-depletion models further supports a causal relationship between microbiota-mediated bile-acid deconjugation51,52,53. Moreover, bile acids can shape gut microbial communities by promoting bile-acid-metabolizing taxa and inhibiting bile-sensitive organisms, establishing a bidirectional feedback loop between host bile-acid metabolism and microbiota54. Together, these findings position Clostridiales not only as promising probiotic effectors but also underscore bile-acid metabolism as a central mediator of microbiota-host crosstalk in cholestasis, offering mechanistic insights and potential therapeutic targets.
Furthermore, our results suggest that C. spp. could upregulate serum levels of DCA and MCA. DCA and MCA are effective regulators of bile acid homeostasis via FXR-mediated feedback55,56. However, MCA is synthesized in rodents via the Cyp2c70-mediated 6β-hydroxylation of chenodeoxycholic acid (CDCA), but these MCAs are essentially absent in adult humans57. This species-specific bile acid profile alters nuclear receptor signaling potentially exaggerating protective effects that human bile acid pools cannot replicate. To overcome these translational limitations, “humanized” mouse models lacking Cyp2c70 have been developed to generate bile acid profiles more similar to humans, thereby enhancing the clinical relevance of preclinical findings58. Despite these interspecies differences, our central observation that modulation of the gut microbiota modifies bile acid composition to ameliorate cholestatic injury remains mechanistically applicable. To enhance the clinical relevance of our findings, future studies should incorporate analyses of serum and fecal bile acid profiles in patients with cholestatic liver diseases.
Recently, the ingestion of several bioactive natural compounds with low bioavailability has been shown to shape a unique pattern of the gut microbiota59,60. Building on our previous studies, we further investigated whether the effective compounds derived from the YZH prescription, which are dependent on A. putredinis and C. spp., have analogous effects on cholestasis. Our research revealed that FA inhibited the growth of A. putredinis and alleviated intestinal dysfunction to ameliorate cholestasis. Moreover, Wog promoted C. spp.-mediated BSH activity and ameliorated cholestatic liver injury by increasing bile acid excretion. Collectively, FA and Wog are potent therapeutic candidates for cholestatic therapy.
Therefore, this study introduces a new research paradigm for discovering naturally active compounds and studying the mechanism of action of TCM compound preparations. Moreover, based on the interpretation of the mechanisms of YZH, C. spp. and A. putredinis, identified as probiotics and pathogenic commensals involved in cholestatic liver injury, were verified (Fig. 9). This study provides a fresh perspective on therapeutic approaches for cholestatic liver injury.
In cholestasis, dysbiosis drives hepatic inflammation and barrier disruption. Ferulic acid reverses Alistipes putredinis–mediated intestinal barrier damage, preventing bacterial translocation to the liver. Wogonin, together with Clostridium spp., enhances bile salt hydrolase activity and promotes bile acid excretion. These combined actions restore gut–liver homeostasis, reduce hepatocellular injury, and attenuate inflammatory responses.
Methods
Reagents
Ferulic acid (CAS No. 1135-24-6, B20007) was supplied by Yuanye (Shanghai, China). Wogonin (CAS No. 632-85-9, H-029) was supplied by Herb purity (Chengdu, China). Alpha-nephthyl isothiocyanate (ANIT) (CAS No. 551-06-4, N4525) was supplied by Sigma‒Aldrich (St. Louis, USA).α-SMA (19245), ZO-1 (13663S) and CD45 (13917S) antibodies were acquired from Cell Signaling Technology (Beverly, USA). Antibodies against E-cadherin (SC8426), MUC2 (SC515032) were acquired from Santa Cruz (CA, USA). Antibody against Claudin-1 (DF6919) was acquired from Affinity Biosciences (Changzhou, China). Antibody against EUB338 (FB-001) was obtained from Focofish (Guangzhou, China).
Human sample collection
Human faecal samples were collected at Nanjing Children’s Hospital using germ-free collection tubes. All samples were immediately transferred on dry ice. Detailed information for all human samples is provided in Table S1. The study had been approved by the Medical Ethics Committee of the Children’s Hospital of Nanjing Medical University (202205012-1).
Animal study and experimental design
C57BL/6 J mice (male, 6‒8 weeks) were obtained from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). All experimental protocols were approved by the Animal Care and Use Committee of the Nanjing University of Chinese Medicine (Nanjing, China) and conformed to the Guidelines for the Care and Use of Laboratory Animals (202306A072). During the experimental period, the room temperature and relative humidity were set to 22 ± 2 °C and 50 ± 5%, respectively. Mice were acclimatized to a 12-h light/dark cycle with food and water available ad libitum.The mice were anesthetized with 1.5–2.5% inhaled isoflurane using a gas anesthesia machine to ensure minimal discomfort. Upon reaching a surgical plane of deep anesthesia, the animals were humanely euthanized via cervical dislocation, in strict accordance with institutional ethical guidelines and approved animal care protocols to minimize suffering.
To evaluate the effects of traditional Chinese medicine, bacteria, and natural compounds on cholestatic liver injury, mice were divided into three main experimental cohorts. In the YZH-treated cohort, animals were randomly assigned to control, ANIT, and YZH-treated groups (1.35, 2.7, or 5.4 g/kg; n = 10 per group). In the bacteria-treated cohort, mice were randomized into four groups (n = 10 per group): ANIT (two-hit model), ANIT’ (single-hit model), A. putredinis-treated, and C. spp.-treated. In the natural compound-treated cohort, animals were assigned to control, ANIT, and treatment groups with either wogonin (20 or 40 mg/kg) or ferulic acid (50 or 100 mg/kg; n = 10 per group).
To model cholestatic liver injury, mice in the therapeutic study received either a single or repeated (two-hit) intraperitoneal injection of α-naphthylisothiocyanate (ANIT, 62.5 mg/kg in olive oil). The two-hit model (day 1 and day 13) induced sustained cholestatic injury, resembling chronic intrahepatic cholestasis and triggering compensatory changes in hepatobiliary transporters34,35. This approach was used to assess the protective effects of YZH, wogonin, ferulic acid, and C. spp. In contrast, the one-hit model (day 1 only) generated acute biliary injury without subsequent transporter adaptation, allowing more accurate evaluation of the pathogenic role of A. putredinis. Control animals received equal volumes of olive oil.
YZH, wogonin and ferulic acid were administered via oral gavage once daily for 14 consecutive days, while mice in the control and ANIT group received an equal volume of saline. Mice in the bacterial treatment groups were gavaged with 2 × 10⁸ CFU of A. putredinis or C. spp. suspended in 0.2 mL of PBS every three days for two weeks, whereas the ANIT and ANIT’ groups received an equal volume of PBS. To deplete endogenous gut microbiota, an antibiotic cocktail consisting of ampicillin (1 mg/mL), vancomycin (0.5 mg/mL), neomycin (0.5 mg/mL) and metronidazole (1 mg/mL) (Rhawn, Shanghai, China) was administered orally once daily for five days.
The BDL procedure was performed as previously described61. Briefly, mice were anesthetized with 1.5–2.5% inhaled isoflurane. Following laparotomy, the bile duct was double-ligated using non-absorbable surgical sutures. In sham-operated controls, the bile duct was not ligated. The incision was disinfected with an alcohol swab, and 0.5 mL of 0.9% saline was applied at the incision site to facilitate post-operative recovery and improve survival. Mice were maintained at 37.5 °C until fully recovered. During the 9-day observation period, mice received bacterial gavage every three days or daily administration of ferulic acid.
Serum biochemical analysis
Blood samples were collected and subjected to centrifugation at 3000 × g for 10 min. The supernatant was carefully separated and used for analysis. Serum levels of aspartate transaminase (AST) (C010-2-1), alanine aminotransferase (ALT) (C009-2-1), alkaline phosphatase (ALP) (A059-2-2), direct bilirubin (D-BIL) (C019-2-1) and total bilirubin (T-BIL) (C019-1-1) were determined via commercial kits obtained from the Jiancheng Institute of Biotechnology (Nanjing, China).
Measurement of FITC-dextran leakage
The mice were fasted for 6 h and then gavaged with 50 mg/mL FITC-dextran (Sigma). The serum collected 4 h later was analyzed for fluorescence (Perkin Elmer) (Excitation: 480 nm, Emission: 520 nm).
Histological analysis
Mouse liver tissue was fixed in 4% paraformaldehyde (PFA), while colon tissue was fixed in Carnoy’s fixative, dehydrated, and embedded in paraffin. Hematoxylin‒eosin (H&E) staining (Leagene, DH0006) was performed on these tissue sections to assess their histological features. To assess pathological alterations, a trained and blinded pathologist inspected the sections. For intestinal tissues, Alcian blue staining (Leagene, 0041) was also applied following the manufacturer’s instructions. All the stained sections were mounted onto glass slides, observed under a light microscope, and imaged using Mantra 1.0.1 (PerkinElmer, MA, USA).
RT‒qPCR analysis
To isolate total RNA, liver and intestinal tissues were homogenized and lysed in TRIzol reagent (Thermo Fisher Scientific, MA, USA). A NanoDrop spectrophotometer was used to measure the concentration and purity of RNA. 500 ng of total RNA was converted to cDNA using the HiScript II QRT SuperMix (Yeasen, China) in accordance with the manufacturer’s recommendations. Quantitative real-time PCR (qPCR) was subsequently conducted on an ABI 7500 Real-Time PCR System (Applied Biosystems, CA, USA). The specific primers used are listed in Table S2.
Liver bacterial culture
For this investigation, an anaerobic apparatus (Electrotek) was used to cultivate anaerobic bacteria, adapting the intestinal CFU culture method. Liver tissues (30–50 mg) were collected and homogenized in sterile PBS for the purpose of analyzing the bacterial burden in vivo. Columbia blood agar plates were covered with the tissue homogenates that were produced. The amount of bacteria present in the tissues was measured following a 48-h incubation period.
Fecal DNA extraction and quantification of 16S rDNA
The QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) was used to extract all of the fecal DNA. Bacteria-specific 16S primers (F: 5′-ACTCCTACGGGAGGCAGCAGT-3′ and R: 5′-GTATTACCGCGGCTGCTGGCAC-3′) and A. putredinis primers (F: 5′-GTAGTTGCGGTAGGCGGAAT-3′ and R: 5′-GTAAGCTGCCTTCGCAATCG-3′) and C. spp. primers (F: 5′- GCAGGTTCTCCTACGGCTAC-3′ and R: 5′- CCCGTCACACCATGAGAGTT-3′) were used to measure the absolute abundance of bacteria 16S rRNA and A. putredinis and C. spp. gene copies, respectively. Relative abundance was calculated by normalizing absolute copy numbers of A. putredinis and C. spp. 16S rRNA genes to total bacterial 16S rRNA gene copy numbers.All qPCR reactions were carried using ChamQ SYBR qPCR Master Mix (Vazyme #R331-02) and ABI 7500 Real-Time PCR System (Applied Biosystems, CA, USA). The primers are listed in Table S2.
Fecal bile salt hydrolase (BSH) enzyme activity analysis and liver and fecal total bile acid determination
The assessment of BSH activity was adapted from the method established by Mullish. For the BSH activity assay across different experimental groups, the reaction mixture consisted of 0.02 mM sodium phosphate buffer, 1 mM taurodeoxycholic acid and 500 μg of fecal protein. The total bile acid concentration was quantified via the Mouse Total Bile Acids Assay Kit (#80470, Crystal Chem).
Fecal bacteria culture
Fresh stool collected from mice at harvest was resuspended in 5% sterile defibrillated sheep blood (TX0030, Solarbio,) ATCC medium 260 (M2216, Elite Media) supplemented with 7.5 μg/ml vancomycin (R051682, Rhawn) and 100 μg/ml kanamycin (R107148, Rhawn) and homogenized for 5 min, followed by dilution on tryptone soy agar plates (HB4114, Hopebio). The fecal commensal bacteria were cultured anaerobically in anaerobic chamber (AW500SG/TG, Electrotek, UK) at 37 °C for 48 h. Each clone was transferred to liquid medium with 2 ml of brain heart infusion (BHI) (HB8297-5, Hopebio) and incubated anaerobically at 37 °C for 48 h.
Screening of fecal bacteria by indole detection
To detect indole production, we used Ehrlich’s reagent (DM0051, Leagene), whose active ingredient is p-dimethylaminobenzaldehyde (PDL). PDL reacts with indole to form a red-colored rose indole. The reaction solution containing 400 μL of Ehrlich’s reagent and 1 mL of bacterial culture supernatant in a 1.5 mL tube. After gentle mixing, tubes were incubated at room temperature for 5 min, and the development of a red coloration was recorded as a positive indole reaction and bacteria showing positive indole reaction were sub-cultured in BHI broths for further analysis.
Identification of bacteria with responsive to indole
To confirm the identity of bacteria candidates, polymerase chain reaction–based amplification of bacterial V3-V4 region of the 16S rRNA gene followed by sequencing was performed. The primers for amplification were 27 F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492 R (5′- TACGGCTACCTTGTACGACTT -3′).
Bacterial strains and culture conditions
C. spp. (BNCC 195293) were purchased from BeNa Culture Collection (BNCC). They were cultured in reinforced Clostridium medium (RCM) at 37 °C under aerobic conditions. A. putredinis were cultured in ATCC Medium 0260 in a 37 °C anaerobic incubator.
16S rRNA gene sequencing
Total genomic DNA was extracted from fecal samples using the E.Z.N.A.® Stool DNA Kit (Omega Bio-Tek, USA), following the manufacturer’s protocol. DNA concentration and purity were evaluated using 1% agarose gel electrophoresis to ensure sample integrity. The variable V3–V4 regions of the 16S rRNA gene were amplified using barcoded universal primers 341 F (5′-CCTACGGGNGGCWGCAG-3′) and 805 R (5′-GACTACHVGGGTATCTAATCC-3′). PCR reactions were carried out using TransStart FastPfu DNA Polymerase (TransGen, China) to ensure high-fidelity amplification. Amplicons were purified using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific, USA), quantified with Qubit (Invitrogen, USA), and pooled in equimolar concentrations. Sequencing libraries were constructed, and quality control was performed using the Agilent 2100 Bioanalyzer and the Library Quantification Kit (Kapa Biosystems). Sequencing was conducted on an Illumina MiSeq platform to generate high-quality paired-end reads (2 × 250 bp). Alpha diversity (Observed OTUs and Simpson index) and nonmetric multidimensional scaling (NMDS) of weighted UniFrac distance were calculated using QIIME2 and visualized with R (v3.5.2). Differential taxa (LDAå 2) were identified using a three-part LEfSe workflow. The Kruskal–Wallis rank-sum test was applied to detect significant differences in taxon abundance across groups. The Wilcoxon rank-sum test assessed whether sub-taxa of significantly different taxa exhibited consistent abundance trends. LDA was then used to evaluate effect sizes and rank taxa by their contribution to group separation.
Metagenomic sequencing and analysis
Total microbial DNA were extracted via HiPure Bacterial DNA Kits (Magen, Guangzhou, China). Qubit (Thermo Fisher Scientific, Waltham, MA) and Nanodrop (Thermo Fisher Scientific, Waltham, MA) instruments were used to determine the quality of the DNA. Using pair-end technology (PE 150), genome sequencing was carried out on an Illumina NovaSeq 6000 sequencer. FASTP (version 0.18.0) was used to filter the raw data from the Illumina platform. In order to create sample-derived assemblies, clean reads from each sample were assembled separately via MEGAHIT (version 1.1.2), with steps over a k-mer range of 21–99. We annotated the constructed sequences using a number of complementary techniques. By matching the deposited unigenes in several protein databases, DIAMOND (version 0.9.24) was used to annotate the unigenes. Kaiju (version 1.6.3) was utilized to create a taxonomy profile based on clean reads. The R vegan program was employed to compute the PCoA of Bray-curtis distances, and the R ggplot2 tool was used to plot the results. The R ggplot2 software was used to graph the violin plot and box plot. Using the R project Vegan package, statistical analyses of the Welch’s t-test, ANOVA (analysis of variance), and Adonis and Anosim test were computed.
Immunofluorescence staining
Colon and liver tissues were sectioned into 4 μm slices and rehydrated by immersion in xylene and graded ethanol solutions. Antigen retrieval was performed using a sodium citrate buffer. To reduce nonspecific binding, the tissue sections were blocked with 5% bovine serum albumin solution and subsequently treated with the designated antibodies. Finally, the stained samples were visualized via a Leica fluorescence microscope.
Fluorescence in situ hybridization (FISH)
The EUB338 16S rRNA gene probe (5’-GCTGCCTCCCGTAGGAGT-3’) was employed to identify the presence of bacteria in the tissues of the liver and colon. A Leica fluorescence microscope was used for fluorescence microscopy.
Serum bile acid metabolomics analysis
100 μl serum was mixed with 100 μl of CA-d4 and 100 μl of GCA-d5. Subsequently, 1000 μl of methanol solution was added, and the mixture was vortexed at 2000 rpm for 5 min. Liquid‒liquid extraction was performed at 4 °C for 1 h. The sample was then centrifuged at 12,000 rpm for 5 min, and 1000 μl of the supernatant was vacuum-dried.
Next, 200 μl of 50% methanol solution was added to the dried sample, which was subsequently vortexed at 2000 rpm for 5 min and centrifuged again at 12,000 rpm for 5 min. A 100 μl aliquot of the resulting supernatant was analyzed via UPLC‒MS‒MS (Sciex, Netherlands). Targeted metabolite analysis of serum BAs was conducted at the Nanjing Jiangbei New Area Biopharmaceutical Public Service Platform.
Statistical analysis
Data were represented as the mean ± SEM and p < 0.05 was regarded as statistically significant. Sample size (n) was listed in each figure legend. Data were performed by unpaired, two-tailed Student’s t test, Manne–Whitney test, one-way ANOVA, and two-way ANOVA (GraphPad, RRID:SCR_000306).
Data availability
Data of metagenomic sequencing and 16S rRNA sequencing have been deposited at the National Genomics Data Center (https://ngdc.cncb.ac.cn/) (accession numbers: PRJCA023311) or the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra)(accession numbers: PRJNA1070795).
References
Jansen, P. L. M. et al. The ascending pathophysiology of cholestatic liver disease. Hepatology 65, 722–738 (2017).
Zollner, G. & Trauner, M. Mechanisms of cholestasis. Clin. Liver Dis. 12, 1–26 (2008).
Schaap, F. G., Trauner, M. & Jansen, P. L. M. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 11, 55–67 (2014).
Dyson, J. K., Beuers, U., Jones, D. E. J., Lohse, A. W. & Hudson, M. Primary sclerosing cholangitis. Lancet 391, 2547–2559 (2018).
Sahoo, S. M. & Mahapatra, S. J. Intrahepatic cholestasis of pregnancy: are we expecting too much from ursodeoxycholic acid?. Lancet Gastroenterol. Hepatol. 6, 886 (2021).
Chappell, L. C. et al. Ursodeoxycholic acid versus placebo in women with intrahepatic cholestasis of pregnancy (PITCHES): a randomised controlled trial. Lancet Lond. Engl. 394, 849–860 (2019).
Tripathi, A. et al. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411 (2018).
Hsu, C. L. & Schnabl, B. The gut–liver axis and gut microbiota in health and liver disease. Nat. Rev. Microbiol. 21, 719–733 (2023).
Hartmann, P. et al. Modulation of the intestinal bile acid–FXR–FGF15 axis improves alcoholic liver disease in mice. Hepatol. Baltim. Md 67, 2150–2166 (2018).
Collins, S. L., Stine, J. G., Bisanz, J. E., Okafor, C. D. & Patterson, A. D. Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 21, 236–247 (2023).
Fuchs, C. D. & Trauner, M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 19, 432–450 (2022).
Funabashi, M. et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570 (2020).
Fleishman, J. S. & Kumar, S. Bile acid metabolism and signaling in health and disease: molecular mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 9, 1–51 (2024).
Ma, C. et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018).
Cheng, P. et al. Capsaicin shapes gut microbiota and pre-metastatic niche to facilitate cancer metastasis to liver. Pharmacol. Res. 188, 106643 (2023).
Lei, Y. et al. Disulfiram ameliorates nonalcoholic steatohepatitis by modulating the gut microbiota and bile acid metabolism. Nat. Commun. 13, 6862 (2022).
Wang, K. et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 26, 222–235.e5 (2019).
Fan, X. et al. Herbal formula BaWeiBaiDuSan alleviates polymicrobial sepsis-induced liver injury via increasing the gut microbiota Lactobacillus johnsonii and regulating macrophage anti-inflammatory activity in mice. Acta Pharm. Sin. B 13, 1164–1179 (2023).
Wu, T.-R. et al. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut 68, 248–262 (2019).
Liu, Y. et al. Probiotic Lactobacillus rhamnosus GG prevents liver fibrosis through inhibiting hepatic bile acid synthesis and enhancing bile acid excretion in mice. Hepatology 71, 2050–2066 (2020).
Wu, L. et al. Lactobacillus acidophilus ameliorates cholestatic liver injury through inhibiting bile acid synthesis and promoting bile acid excretion. Gut Microbes 16, 2390176 (2024).
Yao, Q. et al. Yin Zhi Huang, a traditional Chinese herbal formula, ameliorates diet-induced obesity and hepatic steatosis by activating the AMPK/SREBP-1 and the AMPK/ACC/CPT1A pathways. Ann. Transl. Med. 8, 231 (2020).
Wu, R.-H. et al. Yinzhihuang oral liquid combined with phototherapy for neonatal jaundice: a systematic review and meta-analysis of randomized clinical trials. BMC Complement. Altern. Med. 18, 228 (2018).
Tan, Y. et al. Integrated serum pharmacochemistry, 16S rRNA sequencing and metabolomics to reveal the material basis and mechanism of Yinzhihuang granule against non-alcoholic fatty liver disease. J. Ethnopharmacol. 310, 116418 (2023).
Li, D. et al. Ferulic acid: a review of its pharmacology, pharmacokinetics and derivatives. Life Sci. 284, 119921 (2021).
Huynh, D. L., Ngau, T. H., Nguyen, N. H., Tran, G.-B. & Nguyen, C. T. Potential therapeutic and pharmacological effects of Wogonin: an updated review. Mol. Biol. Rep. 47, 9779–9789 (2020).
Banik, K. et al. Wogonin and its analogs for the prevention and treatment of cancer: a systematic review. Phytother. Res. 36, 1854–1883 (2022).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Sun, D. et al. Angiogenin maintains gut microbe homeostasis by balancing α-Proteobacteria and Lachnospiraceae. Gut 70, 666–676 (2021).
Zhang, M. et al. Microbiota-derived urocanic acid triggered by tyrosine kinase inhibitors potentiates cancer immunotherapy efficacy. Cell Host Microbe 33, 915–931.e9 (2025).
Guerin, E. et al. Isolation and characterisation of ΦcrAss002, a crAss-like phage from the human gut that infects Bacteroides xylanisolvens. Microbiome 9, 89 (2021).
Parker, B. J., Wearsch, P. A., Veloo, A. C. M. & Rodriguez-Palacios, A. The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 11, 906 (2020).
Lamb, A. C., Federico-Perez, R. A. & Xue, Z. Product in indole detection by Ehrlich’s reagent. Anal. Biochem. 484, 21–23 (2015).
Kosters, A., Felix, J. C., Desai, M. S. & Karpen, S. J. Impaired bile acid handling and aggravated liver injury in mice expressing a hepatocyte-specific RXRα variant lacking the DNA-binding domain. J. Hepatol. 60, 362–369 (2014).
Wagner, M., Zollner, G. & Trauner, M. Nuclear receptor regulation of the adaptive response of bile acid transporters in cholestasis. Semin. Liver Dis. 30, 160–177 (2010).
Luo, X. et al. Yinzhihuang formula modulates the microbe‒gut‒liver axis and bile acid excretion to attenuate cholestatic liver injury. Phytomedicine 139, 156495 (2025).
Chen, F. et al. Could the gut microbiota reconcile the oral bioavailability conundrum of traditional herbs?. J. Ethnopharmacol. 179, 253–264 (2016).
Zhang, F. et al. Bioavailability based on the gut microbiota: a new perspective. Microbiol. Mol. Biol. Rev. MMBR 84, e00072–19 (2020).
Qu, Y., Zhang, Z., Lu, Y., Zheng, D. & Wei, Y. Network pharmacology reveals the molecular mechanism of cuyuxunxi prescription in promoting wound healing in patients with anal fistula. Evid. Based Complement. Altern. Med. 2019, 3865121 (2019).
Paone, P. & Cani, P. D. Mucus barrier, mucins and gut microbiota: the expected slimy partners?. Gut 69, 2232–2243 (2020).
Breugelmans, T. et al. The role of mucins in gastrointestinal barrier function during health and disease. Lancet Gastroenterol. Hepatol. 7, 455–471 (2022).
Blesl, A. & Stadlbauer, V. The gut-liver axis in cholestatic liver diseases. Nutrients 13, 1018 (2021).
Plaza-Díaz, J. et al. The gut barrier, intestinal microbiota, and liver disease: molecular mechanisms and strategies to manage. Int. J. Mol. Sci. 21, 8351 (2020).
Guo, P., Zhang, K., Ma, X. & He, P. Clostridium species as probiotics: potentials and challenges. J. Anim. Sci. Biotechnol. 11, 24 (2020).
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P. & Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306 (2012).
Martens, E. C., Neumann, M. & Desai, M. S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 16, 457–470 (2018).
Luis, A. S. & Hansson, G. C. Intestinal mucus and their glycans: a habitat for thriving microbiota. Cell Host Microbe 31, 1087–1100 (2023).
Desai, M. S. et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016).
Pudlo, N. A. et al. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6, e01282–15 (2015).
Chiang, J. Y. L. & Ferrell, J. M. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol. Cell. Endocrinol. 548, 111618 (2022).
Sayin, S. I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Yang, M. et al. Antibiotic-induced gut microbiota dysbiosis modulates host transcriptome and m6A epitranscriptome via bile acid metabolism. Adv. Sci. 11, 2307981 (2024).
Zarrinpar, A. et al. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 9, 2872 (2018).
Wahlström, A., Sayin, S. I., Marschall, H.-U. & Bäckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016).
Wang, H., Chen, J., Hollister, K., Sowers, L. C. & Forman, B. M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3, 543–553 (1999).
Hu, X., Bonde, Y., Eggertsen, G. & Rudling, M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J. Intern. Med. 275, 27–38 (2014).
Zheng, D. et al. Comparative profiling of serum, urine, and feces bile acids in humans, rats, and mice. Commun. Biol. 7, 1–13 (2024).
Gillard, J. & Leclercq, I. A. Importance of the gut microbiota in mice with a ‘humanized’ bile acid pool. Clin. Sci. Lond. Engl. 138, 61–64 (2024).
Zhou, W. et al. The polysaccharides from the fruits of Lycium barbarum L. confer anti-diabetic effect by regulating gut microbiota and intestinal barrier. Carbohydr. Polym. 291, 119626 (2022).
Zhang, Y. et al. Stigmasterol attenuates hepatic steatosis in rats by strengthening the intestinal barrier and improving bile acid metabolism. Npj Sci. Food 6, 38 (2022).
Miyoshi, H., Rust, C., Roberts, P. J., Burgart, L. J. & Gores, G. J. Hepatocyte apoptosis after bile duct ligation in the mouse involves Fas. Gastroenterology 117, 669–677 (1999).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82474375, 82374322, 82004124, 82104184), the Open Project of State Key Laboratory of Natural Medicines (No. SKLNMKF202301), Fundamental Research Funds for the Central Universities (2632023TD03), the Open Project of Chinese Materia Medica First-Class Discipline of Nanjing University of Chinese Medicine (ZYXJC2024-002) and Jiangsu College graduate research and innovation projects (KYCX22_2040). We would like to thank Nanjing Jiangbei New Area Bio pharmaceutical Public Service Platform for the quantification of Bile Acids.
Author information
Authors and Affiliations
Contributions
Study conceptualization, supervision, and editing: Z.W., Y.L. and Y.S.; all experiments and analyses were performed by X.L., P.C., M.L., X.W., F.W. and L. Z.; writing, review, and visualization: X.L., T.M. and R.T. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, X., Mao, T., Wang, X. et al. Synergistic effects of ferulic acid and wogonin on cholestatic liver injury via gut microbiota modulation. npj Biofilms Microbiomes 11, 231 (2025). https://doi.org/10.1038/s41522-025-00862-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-025-00862-z