Abstract
Despite substantial evidence for benefits of vaccination in mitigating the COVID-19 pandemic, the use of mRNA vaccines faces skepticism built on coincidental health events occurring after vaccination without proven causality. One of the claims regarding excessive amounts of residual DNA from the vaccine manufacturing process has arisen from misinterpretation of improperly conducted analyses. Here, we assessed the quantity, quality and identity of residual DNA in mRNA vaccines based on thoroughly performed and properly interpreted orthogonal methods, including qPCR, fluorometry, capillary electrophoresis and short-read DNA sequencing. Our results show that the quantity of residual DNA in all 15 analysed batches of Comirnaty and Spikevax vaccines is below approved limits and that it consists of small fragments originating from the template used to transcribe mRNA during vaccine production. We demonstrate that reliable mRNA vaccine analysis for DNA impurities requires rigorous application of well-controlled methods that minimise mutual interference of vaccine components.
Similar content being viewed by others
Introduction
mRNA vaccines have represented an unprecedented breakthrough in the management of the COVID-19 pandemic, significantly contributing to reduction of mortality and hospitalisations caused by SARS-CoV-2 coronavirus infection. The primary component of the vaccine is an in vitro transcribed mRNA that encodes the SARS-CoV-2 S protein. This mRNA is encapsulated in lipid nanoparticles (LNPs), which facilitate its delivery into cells. Once inside, mRNA is used for the translation of S protein molecules that subsequently trigger an immune response that protects vaccine recipients from severe manifestations of COVID-191.
In the production of mRNA vaccines, mRNA is in vitro transcribed using a linearised plasmid DNA template, which is then enzymatically hydrolysed into small fragments and removed by a high-performance purification method. In the next step, a thorough check of the mRNA purity is carried out, including verification that the quantity of residual DNA meets the limit accepted by regulatory authorities2,3,4. The purified mRNA is then mixed with lipids in a microfluidic mixer to form LNPs. After further purification and quality control, the final mRNA vaccine product is dispensed into sterilised vials, packaged, stored, and distributed while adhering to the strict cold chain conditions specified by the manufacturer5,6,7.
Given that the quality and purity of the active substance, i.e. mRNA, is thoroughly controlled during the manufacturing process, and that the final vaccine formulation is complex, manufacturers do not recommend quantifying residual DNA in the final vaccine product due to measurement interference with the vaccine’s basic components (mRNA and LNPs). They also explicitly do not recommend quantifying residual DNA in vaccines after their expiration date, as well as in cases of improper storage and handling3. It is important to note that residual DNA is tested for every manufactured batch at the mRNA production stage, and that there is no mechanism for additional residual DNA to be introduced after mRNA production or change/increase after vaccine expiry.
Despite these facts, the public domain contains reports on the evaluation of residual DNA in final COVID-19 mRNA vaccine products. According to the results of analyses published in the form of peer-reviewed scientific publications in reputable scientific journals and the statements by regulatory and control authorities, the amount of residual DNA does not exceed the established limit of 10 ng per vaccine dose2,4,8,9,10.
On the other hand, there are several reports that claim highly excessive amounts of residual DNA based on technically inconsistent analyses, and present unsubstantiated suppositions related to its impact on human health11,12,13,14. Such ideas spread via social media and statements of public figures, where they receive considerable attention. This fuels public concerns and undermines trust in scientific and medical evidence. It then leads to increased doubts about the importance of vaccination and ultimately endangers public health.
In Slovakia, a major controversy was recently sparked by an article published in the HSOA Journal of Angiology & Vascular Surgery15. The paper, accepted just 11 days after the manuscript submission, describes an analysis of expired batches of the Comirnaty and Spikevax vaccines using the multiplex qPCR method including mRNA reverse transcription, which exhibited serious technical deficiencies as detailed further in the Discussion. Based on data misinterpretation, the authors erroneously concluded that the vaccines contain “significant amounts of residual DNA” in “nanogram to microgram quantities of expression vector dsDNA per dose”. They also present alarming hypotheses about the effects of residual DNA on the human body, referencing only three publications of the co-author (in addition to two EMA and FDA Guidelines and three GenBank entries)15.
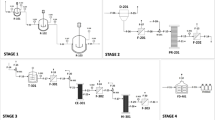
In order to clarify this issue, we aimed to provide an assessment of the quantity, quality and identity of residual DNA in mRNA vaccines that aligns with the standard, rigorous approaches accepted in the scientific community. To meet this aim, we utilised several orthogonal methods with thorough application of controls, biological and technical replicates, and respecting established scientific knowledge about the molecular characteristics of mRNA vaccine components, with an emphasis on minimising their mutual interference. The workflow of the accomplished approaches fulfilling these criteria is illustrated in Fig. 1.
Four orthogonal but complementary approaches were used for the assessment of residual DNA quantity, quality and identity: qPCR, fluorometry, capillary electrophoresis, and sequencing. Essential steps of sample preparation methods with key treatments are depicted for each approach. Details of individual procedures are described in Materials and Methods. Created with Biorender.com elements. PCI phenol-choroform-isoamylalcohol, EtOH - ethanol.
For the purpose of this study, we used batches of Comirnaty and Spikevax vaccines available in Slovakia, obtained from official repositories under the supervision of the State Institute of Drug Control. For direct comparison, we also received vaccine batches identical to those that were subjected to the aforementioned publication15. It is important to note that most of the vaccine batches were expired for an extended period, but their analysis was key for demonstration that proper methodology can provide reliable data also when using such imperfect material.
Results
qPCR assessment of residual DNA
We first decided to utilise quantitative PCR (qPCR). Due to its high sensitivity and specificity, it is accepted by regulatory authorities as a method suitable for testing residual DNA in the active substance. However, DNA quantification can be affected by the efficiency of the qPCR reaction. Errors in primer design and cycling conditions, inaccurate calibration, varying starting amounts of DNA templates, impurities and other deficiencies can lead to significant discrepancies in the resulting concentrations16.
In this study, quantification of residual DNA by qPCR was performed in 15 vaccine batches, 11 of them post expiration date (Table 1, see ‘Methods’). qPCR assays were targeted to 3 regions of the manufacturer-declared plasmid DNA template: the SPIKE region (S protein-coding sequence), the ORI region (origin of replication of the bacterial plasmid), and the KAN region (encoding resistance to kanamycin), see Fig. S1. We used at least two primer sets to amplify each target region separately (Table 2, see ‘Methods’). SPIKE 2 and ORI 2 assay primers were the same as used in ref. 15, to enable direct comparison of the data. SPIKE 1A and ORI 1A primers were optimised versions of the abovementioned primers with regard to melting temperature. Design of additional primers (SPIKE 1B, ORI 1B, KAN 1C and KAN 1S) was based on sequences obtained within this study. Sequence comparison showed 100% identity of the primer-binding regions in SARS-CoV-2 variants used for the development of the analysed vaccines (Original and Omicron).
For each qPCR setting, a calibration curve was generated using dsDNA standards corresponding to the amplified DNA fragments. qPCR assays were performed in parallel in two laboratories, with the involvement of 3 researchers, in accordance with the standard operating procedure for the analysis of medical products outside the manufacturer’s specification.
Calibration curves (Fig. S2) confirmed that the key parameters reached recommended ranges for all designed qPCR assays (−3.1 to −3.6 for the slope, 90– 110% for efficiency, and ≥0.99 for R2). Suboptimal parameters were observed only for the SPIKE 2 assay. Significant improvement of parameters in SPIKE 1A, SPIKE 1B, ORI 1A and ORI 1B assays compared to SPIKE 2 and ORI 2 proved that the slight modifications to the primers used elsewhere15 were beneficial and led to higher qPCR efficiency.
For the qPCR assays, each vaccine sample was treated by Triton X-100 to disrupt LNPs and then directly utilised for analysis. Since the vaccine contains a large amount of SPIKE mRNA, we first wanted to examine its potential interference with the SPIKE qPCR tests. Therefore, we performed a comparative experiment. In the SPIKE 1 and ORI 1A qPCR tests, we used two different concentrations of synthetic standard (100 pg/µl and 1 pg/µl sample) as an input sample. In parallel, for comparison, we also added a regular amount (2 µl) of either undiluted or 1:10 diluted vaccine. If specific PCR inhibition occurred in the presence of the vaccine, it would manifest as higher Ct values in the samples containing undiluted vaccine in the SPIKE 1 test, while the ORI 1A test was intended to serve as a control. As can be seen in Fig. 2, the addition of the vaccine to the qPCR reaction had a negligible effect not only in the case of the ORI 1A test, where it was expected, but also in the case of the SPIKE 1 test. This confirms the legitimacy of our approach of directly using vaccines treated with Triton X-100 as input material for qPCR, without the need for nucleic acid extraction. The results indeed show that not only mRNA, but also the other components of the vaccine do not have a significant negative impact on the course of the qPCR amplification (Fig. 2).
DNA standards were amplified using SPIKE 1 (A) and ORI 1A (B) primers with or without addition of vaccine samples. No increase in Ct values was observed upon addition of vaccine samples suggesting that vaccine mRNA does not interfere with amplification and that qPCR can be effectively performed when DNA extraction is omitted.
Overall, we analysed the presence of DNA in the vaccines using 8 different combinations of primers and targets. The outputs from these qPCR tests of all analysed vaccine batches are summarised in Fig. 3 and individually shown in Figs. S3–S5. Results were initially expressed as the number of copies per µl of sample, and subsequently calculated to the number of copies per vaccine dose. Since regulatory standards are established as the total amount of DNA in nanograms per vaccine dose, and not in copy numbers, we then converted this copy number data to nanograms of DNA per dose, based on the molecular weight of the template plasmid DNA. The results obtained by the analysis of all vaccine batches using all qPCR assays demonstrated no presence of excessive residual DNA.
Quantification of residual plasmid DNA in individual vaccine batches, expressed as nanograms per dose, based on simplex quantitative PCR (qPCR) assays. A DNA fragments derived from the region encoding the SARS-CoV-2 spike protein (SPIKE region). B DNA fragments derived from the region encompassing the bacterial origin of replication (ORI region). C DNA fragments derived from the region encoding kanamycin resistance in Comirnaty (KAN 1C) and Spikevax vaccine (KAN 1S). D Line display of all qPCR results of SPIKE, ORI and KAN assays to show consistent profiles of the batches. Each bar represents the mean value of triplicate reactions using optimised primer/probe sets. The dashed red line indicates the regulatory threshold of 10 ng/dose for residual DNA content. Unexpired vaccine batches are marked with an asterisk.
Quantification of residual DNA and mRNA by fluorometry
When quantifying nucleic acids in vaccine samples, regulatory bodies admit the use of validated orthogonal methods alongside the standard qPCR17,18. One such method is fluorometry, which provides sensitive fluorescence detection of nucleic acid quantities when experimental conditions are properly set and verified. An advantage of this method is its flexibility in sample preparation, allowing the measurement settings for various fluorescent dyes.
However, results of DNA quantification can be significantly affected by the presence of excess RNA, which non-specifically binds DNA-selective fluorescent dyes. In order to perform fluorometric DNA quantification in mRNA vaccines technically correctly, we first evaluated the extent of non-specific signal originating from the mRNA by adding increasing RNA quantities to the sample measured with a kit intended for DNA. In addition, we examined whether the non-specific fluorescent signal originating from RNA can be reduced by the RNase treatment of the sample of residual DNA isolated from the vaccine as described earlier8. We confirmed that elimination of the RNA-related non-specificity is critical for correct results of fluorometric evaluation of residual DNA (Fig. 4). Based on these results, we thoroughly removed RNA prior to fluorometric DNA measurements so that its concentration was maintained below 1 ng/µl.
Samples were quantified using the Qubit 1x dsDNA HS Assay Kit. A Impact of RNA content on measured dsDNA concentration. Quantification of increasing concentrations of a DNase-treated commercial RNA standard (RNA Control Strand; Oxford Nanopore Technologies, Oxford, UK). Each dot represents the mean value of three individual measurements. B Conversion of measured dsDNA concentrations to estimated DNA content per vaccine dose. The values from (A) were converted to reflect the corresponding DNA amount per 300 µl vaccine dose (Comirnaty). The dashed red line indicates the regulatory threshold of 10 ng DNA per dose. C Comparison of the dsDNA signal measured in phenol:chloroform:isoamylalcohol extracts treated with RNase A (+RNase) and without RNase A treatment (−RNase). Each dot represents individual extract from different vials of the batch LK8456, with values corresponding to the average of two measurements. Horizontal lines indicate the mean of three individual extracts with error bars showing standard deviation.
An additional essential prerequisite for the correct fluorometric quantification of residual DNA in mRNA vaccines is DNA extraction that minimises the presence of other vaccine components, including mRNA, while obtaining a DNA-enriched sample in a smaller volume, so that its concentration is within the method’s sensitivity limits. To ensure the reliability and robustness of the DNA quantification results, we isolated residual DNA using two different methods, each including three biological replicates from each batch, subjected to two fluorometric measurements.
As described in Methods, the first DNA isolation method was based on the procedure described earlier with a slight modification8. Vaccine samples were treated with Triton X-100 to disrupt LNPs, nucleic acids were then extracted by phenol-chloroform-isoamyalcohol, followed by chloroform extraction, RNase A treatment, ethanol precipitation and dissolution in ultraclean water of 10-times reduced volume compared to the starting sample volume. Alternatively, DNA was isolated using magnetic beads designed for the isolation of cell-free DNA in biological fluids. The vaccine samples were first treated with Triton X-100, followed by RNase A and proteinase K. Unbound components were removed by a series of washing steps. Finally, DNA was concentrated into a small volume of elution buffer for the subsequent fluorometric analyses.
Results of the fluorometric assessment of residual DNA isolated from all vaccine batches by two alternative methods are summarised in Fig. 5. In accordance with the qPCR analysis, quantity of residual DNA was in all cases below the limits set by the regulatory guidelines. The differences between the data obtained from each extraction method reflect variations in extraction yields between the two methods. Moreover, different vaccine vials were used for each extraction method.
DNA was isolated using two different methods: A extraction with phenol-chloroform-isoamylalcohol, and B the MAGicBead cfDNA Isolation Kit (Zymo Research). The red dashed lines indicate the limit of 10 ng per vaccine dose. Each dot represents an individual vial (biological replicate), with values corresponding to the average of two measurements (technical replicates). Horizontal lines indicate the mean of three biological replicates, with error bars showing the standard deviation. Unexpired vaccine batches are marked with an asterisk.
Since fluorometry is also suitable for quantification of RNA, we used it for assessment of mRNA content in vaccine samples treated by Triton X-100. Results displayed in Fig. 6 showed that all Comirnaty vaccine batches contained RNA quantity above the minimum 90% of the manufacturer-declared amount. Noteworthy, Spikevax vaccines showed lower % of declared mRNA quantity when compared to the Comirnaty batches. However, it should be taken into account that the declared mRNA content in Spikevax Original vaccines (100 µg) and in Spikevax Bivalent vaccines (50 µg) is higher than in Comirnaty vaccines (30 µg). Thus, the absolute mRNA amounts assessed by fluorometry are still higher in Spikevax batches than in Comirnaty ones.
The values are shown as a percentage of the manufacturer-declared mRNA content per vaccine dose (green dashed line). Each dot represents an individual vial (biological replicate), with values corresponding to the average of two measurements (technical replicates). Horizontal lines indicate the mean of three biological replicates, with error bars showing the standard deviation. Unexpired vaccine batches are marked with an asterisk.
Analysis of residual DNA by capillary electrophoresis
Capillary electrophoresis allows for the separation of DNA or RNA fragments, and can determine the size distribution of the fragments, which is important when assessing the quality and potential biological activity of the analysed nucleic acids. Although capillary eletrophoresis is primarily suited for qualitative analyses, we decided to use this approach to either confirm or exclude the potential presence of highly excessive amounts (hundreds to thousands nanograms per dose) that were claimed in some publications12,13,14,15. If that was the case, a strong DNA signal would be clearly visible in electropherograms.
Prior to the analysis of vaccine samples, we assessed the protocols for the vaccine sample preparation and for the precision of separation and quantification by using DNA markers as described in Methods and displayed in Fig. S6. We also examined a defined quantity of DNA fragment of 2000 bp spiked into the physiological saline to visualise the position and height of the peaks in the resulting electropherogram (Fig. S7).
DNA analysis was performed using vaccine samples incubated with Triton X-100 to disrupt LNPs, followed by the RNase A treatment to hydrolyse mRNA and minimise its non-specific signal during the DNA separation. Electropherograms generated by the analysis using DNA kit with a concentration range of 0.5–50 ng/µl input DNA (that would correspond to 150–15,000 ng/vaccine dose) did not show any specific DNA signal in any of the analysed vaccine batches (Fig. 7 and Supplementary Fig. S8). This finding clearly proved no presence of ‘high quantity of residual DNA’ claimed elsewhere12,13,14,15.
Representative electropherograms of FP9632 and 000058A show no specific peaks, indicating the absence of DNA quantities within the detection range of the kit. Two visible peaks correspond to size markers (LM—lower marker of 1 bp, UM—upper marker of 20,000 bp). All other batches exhibited similar profiles of electropherograms (see Supplementary Fig. S8).
We then used a high sensitivity DNA kit that can detect both DNA fragments in a concentration range of 0.005–0.6 ng/µl and a DNA smear of 0.05–5 ng/µl. Again, no significant DNA signal was detected in any of the analysed vaccine batches. Faint peaks around 60 or 300 bp and weak signal disturbances were visible in some vaccine batches, but similar noise signals were present also in the negative control, suggesting their non-specific origin (Figs. 8 and S9). Altogether, the results of capillary electrophoresis clearly demonstrated the absence of highly excessive amounts of DNA in the analysed vaccine batches.
Representative electro-pherograms of all analysed batches of (A) Comirnaty and (B) Spikevax vaccines show no presence of excessive quantity of residual DNA. Some batches exhibited faint peaks around 60 and/or 300 bp. System-generated quantitative data from the entire electro-pherogram (including noise) as well as biological and technical replicates are shown in the Supplementary information. The control sample included physiological saline treated by Triton X-100 and RNase A alongside the vaccine samples. Unexpired vaccine batches are marked with an asterisk.
Capillary electrophoresis also allows for evaluation of RNA integrity in vaccines by directly detecting the size and relative abundance of RNA fragments using a dedicated RNA kit. The mRNA degradation process can occur during incorrect storage, transport, or handling, especially when the vaccine’s expiration date has passed. We decided to perform the analysis of RNA integrity because most of the available vaccine batches were past their expiration date for an extended period. For this purpose, we used vaccine samples treated by Triton X-100 as described in ‘Methods’.
Results of the mRNA integrity evaluation of all analysed vaccine batches are graphically displayed in Fig. 9. Out of the 15 analysed vaccine samples, all 6 Spikevax batches and 1 Comirnaty batch showed average mRNA integrity below 50%. This was not surprising given their expiration date. Noteworthy, the batches with decreased mRNA integrity exhibited turbid appearance, in contrast to the clear appearance of non-expired batches (Fig. 10). Taking together the results of the fluorometry and the results of capillary electrophoresis, it is possible to conclude that the analysed vaccines contained sufficient amounts of mRNA, but its integrity was not properly preserved particularly in the expired batches.
A Electropherogram of Comirnaty batch FP9632 with well preserved mRNA integrity. B Electropherogram of Comirnaty batch 1L084A with reduced mRNA integrity. C Graph summarising data from all analysed vaccine batches. The dashed line indicates 50% mRNA integrity. Each dot represents an individual vial (biological replicate), with values corresponding to the average of three capillary electrophoresis runs (technical replicates). Horizontal lines indicate the mean of three biological replicates, with error bars showing the standard deviation. Unexpired vaccine batches are marked with an asterisk.
Analysis of residual DNA by short-read sequencing
Finally, the identity of total residual DNA isolated from the available vaccine batches was analysed by the next generation sequencing using a technology of short-read massively parallel sequencing with reversible fluorescently labelled terminators. This technology was chosen as optimal for the detailed characterisation of DNA present in the vaccines due to its specific advantages. It can analyse dsDNA molecules processed into sequence libraries, each molecule can be analysed and counted individually in a separate sequencing cluster, a high number of DNA fragments can be analysed simultaneously in a single run, with reading from both ends of the DNA molecules. Therefore, it is possible to determine the exact size of each DNA molecule during mapping. In this way, the initial state of the DNA (i.e. its integrity) can be determined directly from the sequencing results. Moreover, this technology has a high base-calling quality value, and thus high reliability of the results (reliability of calling each individual nucleotide is at a level of 99.99% or higher for 90% of the analysed sequence).
Isolation of DNA in sufficient quantity and suitable purity from the mRNA vaccine batches was challenging. First, the amounts of residual DNA in vaccines were less than one-tenth of the minimum input amount required for the selected sequencing approach. However, as we have repeatedly confirmed in our published studies, also very low input amounts of the fragmented DNA can be successfully sequenced, even under clinical diagnostic conditions19,20,21. A more significant problem, however, was the presence of a huge excess of mRNA, which interfered with the quantity measurements during the preparation of sequencing libraries. DNA isolation protocol by affinity chromatography was therefore optimised, especially with regard to steps of LNPs disruption and RNase A treatment (for details see ‘Methods’). As a DNA quality criterion, we considered an undetectable amount of RNA (result value ‘too low’) using an RNA high-sensitivity fluorometric assay.
The aim of the analysis was not only to determine which DNA molecules were present in the vaccine batches, but also the state of degradation of these DNA molecules. That forms the basis of fragmentomic analysis, which is of particular importance in the liquid tumour biopsy field22. Therefore, to describe the native state of residual DNA molecules in the mRNA vaccines, it was crucial to skip the further fragmentation step of the DNA sample and thus preserve the original length of the fragments. We therefore chose a ligation-based sequencing library type with the lowest possible DNA input, and omitted the first DNA fragmentation step from the original protocol. Moreover, sequencing libraries were prepared in half volume compared to the original protocol due to low DNA content in the samples. Actually, the average input quantity of DNA isolated from individual vaccine batches was 1.212 ng (range 0.66–1.893 ng) for the Comirnaty samples and 0.794 ng (range 0.801–1.488 ng) for the Spikevax samples.
Sequencing libraries were successfully prepared from all analysed batches of Comirnaty and Spikevax vaccines. The libraries were subjected to quality control using fluorometry and capillary electrophoresis as described in Methods. Fragment profiles of individual libraries are shown in Fig. S10. Values and parameters obtained by quality control of the final libraries are shown in Table 3 below. All prepared libraries met the qualitative requirements for the following sequencing. FastQC analysis23 confirmed consistently high base quality across all sequenced samples, allowing direct use for downstream bioinformatic analysis without additional trimming or filtering.
Bioinformatic assembly of sequencing reads revealed that the DNA content consisted predominantly of degraded plasmid molecules originally used as templates for mRNA synthesis during vaccine production. De novo assembly of overlapping reads produced complete plasmid sequences, indicating the dominant presence of the fragments from plasmid. All reads were then mapped to the assembled contigs, and for each sample, the proportion of plasmid-derived reads was calculated. Averaging these per-sample values showed that plasmid-derived sequences accounted for 96.62% of total reads in Comirnaty vaccines (range: 95.26–98.08%) and 88.71% in Spikevax vaccines (range: 83.52–94.51%), for more information, see Table 3.
Remaining reads in very low relative amounts either mapped to Escherichia coli K12 genome (GenBank accession NC_000913.3; non-pathogenic strain widely used in microbiology research and biotechnologies), bacteriophage discovered in microbiome study (GenBank accession PV093624.1) with predicted specificity to Corynebacter spp. (common commensal and industrial biotechnology organism) or low complexity reads (repeat sequences) with no match to ENA/GenBank nucleotide entry. Given the suboptimal input amount of DNA into the library preparation step, these reads can at least partially be a result of reagent contamination, as described elsewhere24.
Since we used paired-end sequencing, i.e., sequencing from both ends of the fragments, it was possible to determine the precise lengths of these fragments. This analysis revealed that the DNA was heavily degraded, and the median lengths of the sequenced fragments were 154 bp (range 130–181 bp) for the Comirnaty vaccines and 144 bp (range 127–201 bp) for the Spikevax vaccines (Fig. 11). Furthermore, genome coverage was significantly non-uniform across all samples, likely due to uneven residual DNA degradation (Fig. 12).
Spikevax batches 00058A, 200106A, 3005697 and 400012A show larger irregularities in coverage across the respective plasmid genome, compared to Comirnaty batches and Spikevax 200023A and MV1018A batches. All Comirnaty batches have similar coverage patterns especially consistent in non-expired batches.
In summary, we have shown that the residual DNA present in the analysed mRNA vaccine batches is highly degraded into short fragments and originates from the plasmid template, which served as a starting material for vaccine production. Unmodified sequenced reads were deposited in NCBI under Project ID PRJNA1303864.
Discussion
Quality of vaccines is regularly verified before the batch distribution for use by the Official Medicines Control Laboratories (OMCL), working in frame of the Control Authority Batch Release (OCABR) system coordinated by the European Directorate for the Quality of Medicines and HealthCare (EDQM) as described25. Each batch of vaccine is controlled at the OMCL by performing a specific subset of tests from the approved marketing authorisation dossier. The OMCL also reviews the manufacturers’ test results, which are included in the batch protocol. The outcomes of these quality checks are part of internal documentation and are not publicly accessible. This creates a room for various speculations based on scientifically flawed vaccine analyses and/or misinterpreted results11,12,13,14,15. The majority of published reports containing such disinformation are persisting and shared on different platforms in the public domain, where they spread fears and vaccine hesitancy. In contrast, peer-reviewed publications in reputable scientific journals reporting on technically well accomplished vaccine analyses have remained scarce and less effectively disseminated. Therefore, it is of utmost importance to strengthen awareness of the medical relevance of vaccines and transparently inform the general public about the real findings of correctly accomplished mRNA vaccine analyses, together with their fair and evidence-based interpretation.
This was actually the main purpose of our study, that was aimed at thorough analysis of residual DNA in 9 batches of Comirnaty vaccine and 6 batches of Spikevax vaccine. Eleven of these batches were expired and were previously reported to contain “significant amounts of DNA” including the “DNA of undeclared origin”. That report describes results of improperly designed and poorly performed analysis with unfounded conclusions15. On such a basis, the authors propose an alarming scenario of vaccines’ impact on human health that was presented in public and social media, where it has gained a lot of attention and generated controversy among both the general public and high level decisionmakers.
Analysis presented in that report15 contains a number of critical methodological deficiencies. Principally, the authors used an inappropriate workflow that included DNA/RNA isolation from vaccine samples, followed by reverse transcription of mRNA and finally by multiplex qPCR targeting simultaneously cDNA and residual plasmid DNA. Such a procedure can neither preserve original quantitative ratios of mRNA vs residual DNA nor discriminate between amplification products of SPIKE region from plasmid DNA vs reverse-transcribed cDNA. Moreover, the procedure lacked proper calibration standards and key controls, such as non-template controls to exclude contaminations, controls of nucleic acids isolation efficiency, controls of reverse transcription efficiency, controls of amplification efficiency of individual targets in the simplex qPCR. Thus, from the very beginning and throughout the entire procedure, uneven quantities of input materials were used at every step performed at unknown efficiency15.
However, the major lapse came with data interpretation. Based on qPCR targeted to 3’ region of the sequence encoding the SPIKE protein, the authors of the publication15 claim that vaccines contain “significant amounts of DNA,” specifically: 107 – 108 copies per ml of Spikevax vaccine (corresponding to 5×106 – 5×107 copies per vaccine dose), and 108 – 109 copies per ml of Comirnaty vaccine (corresponding to 3×107 – 3×108 copies per vaccine dose). These copy numbers seem overwhelming to laypersons; however, when converted to the quantity of template DNA in nanograms per dose, the results are well below the limit value of 10 ng per vaccine dose.
The authors of that publication15 also claim that ‘cDNA isolates’ obtained by reverse transcription of purified DNA/RNA were heavily contaminated with the cloning expression vector (‘nanogram to microgram amounts of dsDNA’). They even suggest the presence of template DNA encoding the complete ‘S protein cassette’ (3880 bp), deducing this from the simultaneous amplification of 5’ ORI region and the 3’ SPIKE region in their multiplex qPCR, which is a completely wrong conclusion. The work also contains a consideration of another construct in the given expression vector, which could be ‘admixed Omicron’ ignoring the fact that Omicron subvariants BA.1 and BA.4/BA.5 are declared components of the bivalent batches 200023A, 200106A, MV1018A, and 400012A.
Our analysis of the same vaccine batches completely disproved these claims by performing systematic examination of residual DNA utilising several orthogonal and well-controlled analytical approaches.
Firstly, we performed 8 simplex qPCR assays targeting 3 regions of the template DNA and respecting all key prerequisites of correct methodology. We thoroughly controlled all steps using proper standards for calibration, optimised primers and PCR conditions, all necessary controls, and consistent input material quantities. We also proved that the Triton X-100 treated vaccine can be used for qPCR directly, without preceding DNA purification and that mRNA and other vaccine components do not interfere with the amplification of the target DNA sequences. This was very important since by this means we could completely preserve residual DNA content in the vaccine and avoid yield loss that naturally occurs during the purification process.
As demonstrated in the Results section, our analysis of all vaccine batches, using all qPCR assays, did not show any instance of exceeding the approved limit of 10 ng of DNA per vaccine dose. These results are also consistent with the findings of the Australian ‘Therapeutic Goods Administration’ (TGA). The TGA states on its website that it has analysed 28 final product batches of Comirnaty and Spikevax mRNA vaccines to date and has not confirmed exceeded limits of residual DNA in any batch. Furthermore, our analysis is in line with the TGA observation that Spikevax vaccines from Moderna generally show lower levels of residual DNA compared to Comirnaty vaccines from Pfizer26,27.
Interestingly, the highest copy numbers and, correspondingly, the highest calculated amounts of DNA in nanograms per dose were found using the KAN 1C qPCR (see Fig. 3). The highest average copy number was 9.8 × 108 copies of the fragment, which, when converted to the complete plasmid equivalents, would correspond to 8 ng DNA per dose. The lowest copy numbers and calculated amounts of DNA in the Comirnaty vaccine (0.5–0.7 ng per dose) were found using the SPIKE 1A and SPIKE 2 qPCRs. Tests targeting the ORI region indicated maximum values in the range of 3.3-5.0 ng DNA per dose (Fig. 3).
The differences between the determined amounts of residual DNA using different qPCR tests are, in some cases, quite significant. Given that the parameters of the calibration curves for the individual qPCR tests, with the exception of SPIKE 2, met the recommended ranges, these differences cannot be explained solely by differences in qPCR efficiency. We propose the length of the amplified PCR product to be the most important factor that may influence the observed differences. This is because residual template DNA in the vaccine is not present in the form of complete plasmid DNA molecules, but is randomly cleaved into short fragments. Based on the fragmentomic sequence analysis, the vaccine batches analysed here contain fragments with a median length of 150 bp (see Fig. 11). The longer the amplification product, the greater the probability that the DNA molecule is also cleaved within this region, and therefore fewer such intact, complete fragments are present in the vaccine. This explains why the highest values of residual DNA were found using KAN 1C qPCR, where the amplification product is very short, only 63 bp. On the other hand, the lowest amounts were systematically detected with the SPIKE 1 A and SPIKE 2 tests, which detect fragments of 228–233 bp. This assumption is supported by the data from sequencing, since the profile of >300 bp fragments detected in individual batches (see Fig. 11) roughly corresponds to the profile of batches in qPCR analysis (see Fig. 3).
Importantly, even with the smallest technically feasible amplicon with KAN 1C primers, the calculated amount of template DNA per vaccine dose is below the limit value. At the same time, however, it is important to note that the profile of differences in the number of amplified molecules in different qPCR tests is very consistent between individual batches suggesting that the qPCR method is reliable and suitable for the purpose of residual DNA assessment.
As the second approach, we used Qubit fluorometry, which relies on the use of fluorescent dyes that bind to double-stranded DNA (dsDNA) and, at a specific wavelength, emit fluorescence proportional to the amount of present dsDNA. This method is highly sensitive and allows for accurate quantification of DNA in samples with low concentrations, but it does not provide information about the size or integrity of DNA fragments. However, measurement results can be significantly affected by the presence of a high excess of RNA, which non-specifically binds fluorescent dyes. This fact is underestimated, misinterpreted, or ignored in publications that declare excessive amounts of residual DNA in mRNA vaccines based on fluorometric approaches12,13,14.
To meet the requirements for technical feasibility of DNA quantification using Qubit fluorometry, which is affected by the excess of RNA and works within a specific sensitivity range of DNA detection, we verified concentration-dependent contribution of RNA to non-specific signal in DNA assays and optimised procedures for DNA extraction, which is needed for sample concentration. In agreement with the study8, we found that thorough, carefully controlled treatment with RNase A is crucial for reducing non-specific signal in DNA measurements. Our results showed that even low concentrations of RNA emit a significant fluorescent signal, corresponding to hundredths to tenths of a nanogram of DNA in a 1 μl sample. Although this overestimation may appear negligible in common applications, it can significantly impact the analysis of mRNA vaccines. This is especially true when the measured concentration is converted to a vaccine dose as multiplying low values from the fluorometer can translate even seemingly minor oscillations into a significant difference in the calculated quantity of DNA per dose.
Fluorometric RNA quantification showed that the amount of mRNA in Comirnaty vaccines is at or above the level reported by the manufacturer4. For Spikevax vaccines, four out of six vaccines showed slightly lower values, suggesting their partial degradation (note: all Spikevax vaccines were long past their expiration date). Based on this, we can state that, according to the fluorometric analysis, the ratio of the amount of residual DNA to the amount of mRNA in individual batches was higher than 1:3300 and ranged up to 1:30,000. That is much above the 1:1 ratio declared as acceptable for the correct DNA quantification by Qubit manufacturer and also much above the 10:1 ratio demonstrated as acceptable in the recently published study8. Knowing this, it is apparent that omission or imperfect use of RNase treatment of extracted DNA (short time and/or low concentration) or even complete omission of the DNA extraction from the vaccine is the most plausible explanation of the observations of excessive DNA-like signals in fluorometric measurements proposed by vaccine scepticists12,13,14. This is particularly apparent in the recently published study13, showing the opposing results of qPCR (in few ng/dose) versus fluorometry (in micrograms/dose), where Spikevax vaccines were claimed to contain more residual DNA than Comirnaty vaccines. This is in direct disagreement with our observations, where RNase treatment was essential to obtain valid DNA measurements, the efficiency of the RNase digestion was verified by monitoring RNA content, and both quantitative methods converged on values below the regulatory threshold. We believe that the apparent lack of effect of RNase in the Speicher et al. 13 study reflects methodological differences rather than true biological variability.
As the third approach, we employed capillary electrophoresis using the 5200 Fragment Analyser System, which was not used for the analysis of residual DNA in mRNA vaccines so far. Therefore, it was important to optimise and justify the methods specifically with respect to vaccine compositions. Sample preparation procedure as well as process of electrophoretic separation was thoroughly tested to ensure technically correct methodology. In any of the analysed vaccine batches we did not detect any specific signal indicating highly excessive amounts of residual DNA, which is consistent with the results of Qubit fluorometry. If residual DNA levels were as high as reported in some studies12,13,14, these quantities would be readily visible by capillary electrophoresis in the form of distinct peaks or pronounced smears. The absence of such signals in our analysis, even using highly sensitive protocols, directly refutes these claims and supports the conclusion that DNA impurities remain well below the regulatory threshold.
Finally, we performed sequencing analysis using short-read massively parallel sequencing, formerly referred to as next generation sequencing. Using ligation-based library preparation with the omission of fragmentation step we obtained detailed, highly accurate information on present individual DNA molecules including precise physical sizing with single base precision as well as information on relative quantities of different DNA entities. Using the state-of-art bioinformatic de novo assembly method, from the overlapping sequence fragments we could consistently recover complete sequences of plasmid vectors, which served as templates in the vaccine production process. Importantly, subsequent mapping of all reads revealed that 83.52 – 98.08% of the DNA sequences were derived from plasmids used as templates during in vitro transcription of mRNA, rather than from exogenous sources or host genomes. These findings confirm that the detected DNA is a known and expected by-product of the manufacturing process, not an evidence of unintended contamination.
To further characterise the nature of the residual DNA, we applied a fragmentomics-based approach by omitting the enzymatic fragmentation step during NGS library preparation and using paired-end sequencing. This allowed us to preserve and analyse the native fragment size distribution. We found that the residual DNA was highly fragmented, with a median fragment length of approximately 150 base pairs. This degree of degradation is consistent with prior regulatory assumptions that residual DNA in biologics is typically non-functional and not competent for replication or integration. The observed fragment length falls below the threshold associated with potential biological activity, providing additional reassurance regarding its biological inertness, especially when production of mRNA vaccines does not include eukaryotic cell substrates3.
In summary, this study was initiated as part of a national effort to address growing public concerns and misinformation regarding the presence of residual DNA in mRNA-based COVID-19 vaccines. Claims suggesting the presence of high levels of potentially harmful DNA have been widely disseminated by anti-vaccine groups, fuelling vaccine hesitancy and undermining public confidence11,12,13,14,15. To tackle this infodemic with independently verified data, we performed a comprehensive analysis of 15 mRNA vaccine batches, including 11 expired batches, using four orthogonal molecular methods: quantitative PCR, fluorometry, fragment analysis, and sequencing analysis (short-read massively parallel).
Across all samples and quantitative analytical methods, residual DNA content was consistently below 10 ng per dose, complying with regulatory thresholds established by EMA and FDA. This consistent finding across different methodologies highlights the robustness of the data and supports the conclusion that residual DNA levels in these vaccines are minimal and well-controlled. Moreover, the inclusion of expired batches further supports the chemical stability and regulatory compliance of mRNA vaccines over time with respect to DNA content. The low DNA quantity, plasmid origin, and high fragmentation collectively suggest negligible risk to human health.
It is worth noting that simultaneous employment of independent molecular approaches was key to obtaining reliable evidence for the compliance of vaccine batches with regulatory requirements, as each individual approach has its inherent advantages as well as limitations.
qPCR can be applied on vaccine samples treated only for LNP disruption without prior DNA purification step thereby preserving actual DNA content. Moreover, it allows for highly sensitive and specific amplification and relative quantification of selected DNA regions upon proper design of primers, probes, controls and amplification conditions. However, high specificity is also a limitation as this approach itself cannot detect presence of unexpected or unknown DNA and particularly in case of vaccine analysis, amplification of larger qPCR products is more affected by random degradation of residual DNA when compared to smaller amplicons, as demonstrated in this study.
Fluorometry can reliably quantify total residual DNA, but its proper application in mRNA vaccine analysis requires DNA purification and concentration steps, including LNPs disruption and minimisation of interference caused by huge excess of mRNA present in vaccine as the main active substance. DNA purification is obviously associated with its partial loss during the procedure. Depending on the method, DNA isolation may result in 60-95% yield and this fact should be taken into account when considering final residual DNA quantities. On the other hand, fluorometry is fully appropriate for quantification of vaccine mRNA. Limitation of this approach in the vaccine evaluation context is that it does not allow for assessment of the quality and identity of nucleic acids. However, it has to be mentioned that fluorometry is fully suitable and preferable for a broad spectrum of applications when analysing biological fluids or matrixes as these do not contain huge imbalances in DNA:RNA ratio.
DNA as well as RNA quality can be assessed by capillary electrophoresis on a fragment analyser. Using this approach, care should be taken to comply with the sensitivity range using specific detection kits and requirements for sample purity as excessive impurities can obscure specific signals. In addition, it is necessary that the suitability of the method for use in biotherapeutic analysis is demonstrated experimentally as different factors can impact its sensitivity. This approach can also provide information on relative quantities of nucleic acids.
Finally, sequencing is the only approach that can uncover the full identity of the residual DNA to a very deep detail. Moreover, upon proper selection of the methodology and bioinformatic tools described in this study, it can provide precise information on the size distribution and relative quantitative representation of residual DNA fragments. However, accomplishment of this approach requires specific equipment and expertise.
Taking into account all these aspects, we recommend that a combination of independent but complementary approaches is the suitable and reliable attitude to vaccine analysis, particularly when accomplished out of the official batch control system.
Overall, our findings provide independent confirmation of the regulatory compliance of mRNA vaccines with respect to residual DNA content. In addition to supporting ongoing public vaccination programs, this study also demonstrates the value of transparent, science-based investigations in addressing misinformation and strengthening vaccine confidence. Future research may explore the dynamics of DNA degradation over extended storage or evaluate alternative purification technologies, though our findings indicate no current need for concern under existing manufacturing practices.
Methods
Vaccine samples
A total of 15 vaccine batches (at least 10 vials each) were received for the study from official repositories in Slovakia under supervision of State Institute for Drug Control with approval of the Ministry of Health of the Slovak Republic (Table 1). Factory-sealed vaccine vials were stored according to the manufacturer’s specifications (Comirnaty: −90 to −60 °C; Spikevax: −50 to −15 °C). Prior to analysis, vaccines were thawed at room temperature and subsequently kept on ice. Where applicable, vaccine samples were diluted with sterile saline solution to an equal working RNA concentration of 0.1 μg/μl.
Quantification of residual DNA by qPCR
Quantitative real-time PCR (qPCR) was used to quantify residual plasmid DNA in mRNA vaccine formulations. Assays targeted three distinct regions of the DNA template used for in vitro transcription: (i) the SARS-CoV-2 spike protein gene (SPIKE), (ii) the bacterial origin of replication (ORI), and (iii) the kanamycin resistance gene (KAN). All reactions were performed in triplicate, independently in two laboratories, and validated using synthetic standards.
Vaccine samples were processed by detergent lysis. Briefly, 100 µl of vaccine sample was treated with Triton X-100 to a final concentration of 1% (v/v), incubated at room temperature for 15 min to disrupt LNPs and release nucleic acids, then transferred to ice. Aliquots were either used immediately for qPCR or stored at −20 °C. Two microliters of processed lysate were used per qPCR reaction.
Eight TaqMan-based qPCR assays were used (Table 2). For SPIKE and ORI regions, both published primer/probe sets15 and in-house optimised versions were employed. The KAN assay was designed specifically for Comirnaty, as Spikevax plasmid sequences showed substantial divergence in the kanamycin resistance region. Final concentrations were 300 nM for primers and 200 nM for probes.
Synthetic dsDNA fragments (IDT gBlocks) corresponding to each qPCR target were used to construct standard curves. Seven-point 10-fold serial dilutions (100 pg to 0.1 fg) were prepared in triplicate. Copy numbers per reaction were calculated from molecular weights. Calibration curves were used to assess assay slope, amplification efficiency, and linearity (R2). Optimised in-house assays exhibited efficiency between 90 and 110% and R2 ≥0.99. The SPIKE2 assay, adopted from ref. 13, showed lower efficiency (89.4%) but was retained for comparison (Fig. S2).
qPCR reactions were performed using a QuantStudio 5 Real-Time PCR System (Applied Biosystems) with TaqMan Fast Advanced Master Mix (Applied Biosystems). Thermal cycling conditions were: 50 °C for 2 min, 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Each run included a no-template control (NTC) and standard dilutions. Fluorescence thresholds were set automatically, and copy number quantification was based on the average Ct values from triplicates. To convert the number of DNA copies into mass concentration expressed in nanograms, we used the following formula: mass (ng) = (copies × MW (g/mol))/NA × 10⁹, where copies is the number of qRT-PCR-measured DNA molecules, MW is the molecular weight of the fragment in grams per mole (based on its length in base pairs), and NA is Avogadro’s number (6.022 × 10²³ mol⁻¹).
Phenol-chloroform-isoamylalcohol extraction
Phenol-chloroform-isoamyl alcohol extraction is a traditional and widely used method in molecular biology for nucleic acid purification. For each analyzed batch, three separate vials were used as biological replicates for DNA purification using this method. To disrupt the LNPs and release the encapsulated nucleic acids, 30 μl of Triton X-100 (10%; Sigma Aldrich, St. Louis, MO, USA; final concentration 1%, v/v) was added to 300 μl of the vaccine sample. After a 10-min incubation at room temperature on a rotary mixer, an equal volume of phenol:chloroform:isoamyl alcohol mixture (25:24:1, pH ~8; Sigma–Aldrich, St. Louis, MO, USA) was added. The sample was mixed vigorously by vortexing for 60 s, followed by a 2-min incubation at room temperature and centrifugation at 16,000 × g for 10 min at 4 °C. The upper (aqueous) phase was carefully transferred to a new sterile microcentrifuge tube, and an equal volume of chloroform (Sigma-Aldrich, St. Louis, MO, USA) was added. After brief mixing (5 s) and centrifugation (5 min, 16,000 × g, 4 °C), the upper phase was recovered. For enzymatic degradation of mRNA, RNase A (10 mg/ml; Millipore, Darmstadt, Germany) was added to the sample to achieve a final concentration of 0.3 μg/μl. The sample was incubated for 2 h at 37 °C in a water bath and then cooled on ice.
For nucleic acid precipitation, 1/10 volume of sodium acetate (3 M, pH 5.2; Sigma-Aldrich, St. Louis, MO, USA) and glycogen (20 mg/ml; Thermo Fisher Scientific, Waltham, MA, USA; final concentration 100 ng/μl) were added to the sample. After mixing, 2.5 volumes of pre-chilled absolute ethanol (Sigma–Aldrich, St. Louis, MO, USA) were added. The sample was gently mixed by inverting several times and incubated for 1 h at −80 °C. Following incubation, the sample was centrifuged (15 min, 16,000 × g, 4 °C), the supernatant was discarded, and the pellet was washed with 500 μl of 70% ethanol. After washing and thorough removal of residual ethanol, the tube was left open at room temperature for 5–10 min to allow the pellet to air-dry. Finally, the pellet was resuspended in 30 μl of nuclease-free water, representing a 10-fold concentration of the original input volume. To validate the expected extraction yield, a GeneRuler Low Range DNA Ladder containing fragments of 25–700 bp (Thermo Fisher Scientific, Waltham, MA, USA) of known concentration was spiked into the vaccine sample prior to extraction. The recovery efficiency of this extraction method ranged from 60 to 80%.
DNA extraction using MAGicBeads cfDNA Isolation Kit
The MAGicBead™ cfDNA Isolation Kit (Zymo Research, Irvine, California, USA) is designed for the extraction of cell-free circulating DNA from biological fluids, including blood plasma, saliva, urine, and others. For each analyzed vaccine batch, three separate vials were used as biological replicates for DNA extraction using this method. Initially, 200 μl of the vaccine sample was transferred into a sterile 1.5 ml microcentrifuge tube. Samples were first treated with Triton X-100 (10%, Sigma–Aldrich, St. Louis, MO, USA) for 10 min at room temperature to disrupt the LNPs and release the encapsulated nucleic acids. Subsequently, enzymatic degradation of mRNA was performed by adding RNase A (10 mg/ml; Millipore, Darmstadt, Germany; final concentration 0.3 μg/μl), followed by incubation for 2 h at 37 °C in a water bath. The subsequent steps were performed following the manufacturer’s protocol. The digestion step was carried out according to the manufacturer’s recommendations for non-plasma sample types (Appendix A) with the following digestion conditions: 55 °C 15 min, then 37 °C 15 min. The DNA was eluted in a final volume of 20 μl, representing a 10-fold concentration of the original input volume. To validate the expected extraction yield, DNA (GeneRuler Low Range DNA Ladder containing fragments of 25–700 bp, Thermo Fisher Scientific, Waltham, MA, USA) of known concentration was spiked into the vaccine sample prior to extraction. The recovery efficiency of this extraction method ranged from 90 to 95%.
Fluorometric quantification of nucleic acids
Quantity of nucleic acids (both DNA and RNA) in vaccine samples and extracts was determined using Qubit™ Flex fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). For mRNA quantification in vaccine samples, the Qubit™ RNA BR Assay Kit was used, with a quantification range of 0.5–1200 ng/μl. For DNA quantification in extracts, the Qubit™ 1X dsDNA HS Assay Kit was used, which is designed for highly sensitive detection of double-stranded DNA in the concentration range of 0.005-120 ng/μl. For the quantification of residual RNA in DNA extracts, the Qubit™ RNA HS Assay kit was used, with a quantification range of 0.2–200 ng/μl. Measurements were performed according to the manufacturer’s instructions. Sample input volumes were 10 μl for mRNA in vaccine samples, 4 μl for DNA in extracts, and 5 μl for residual RNA in extracts. These volumes were chosen to achieve an appropriate quantification range for each assay, based on the expected sample concentrations. Depending on the experiment, each sample was measured in two or three replicates.
To estimate the total amount of DNA or mRNA per vaccine dose (300 μl for Comirnaty and 500 μl for Spikevax), the measured nucleic acid concentrations (in ng/μl) were converted accordingly. For extracted samples, the final 10-fold concentration of the eluate relative to the original vaccine volume was taken into account. Calculations also accounted for the volume of reagents (Triton X-100 and RNase A) added to the vaccine sample. The final values reflect the total DNA or mRNA content per vaccine dose, adjusted for dilution introduced during sample processing.
Fragment analysis by capillary electrophoresis
Capillary electrophoresis-based fragment analysis of nucleic acids was performed using the Agilent Fragment Analyzer 5200 system (Agilent Technologies, Santa Clara, CA, USA) with manufacturer-supplied reagent kits. To detect residual DNA in vaccine samples, we used the Agilent DNF-930 dsDNA 930 Reagent Kit intended for analysis of DNA fragments between 75 and 20,000 bp over a concentration range of 0.5–50 ng/μl, and the Agilent DNF-464 HS Large Fragment 50 kb Kit designed for highly sensitive analysis of DNA fragments ranging from 75 to 48,500 bp, with a concentration range of 5–600 pg/μl per fragment (50 pg/μl to 5 ng/μl per sample).
Prior to the analysis, vaccine samples were treated with Triton X-100 (10%; Sigma Aldrich, St. Louis, MO, USA; final concentration 1%, v/v) for 10 min at room temperature to disrupt the LNPs and release the encapsulated nucleic acids. Following Triton X-100 treatment, RNase A (10 mg/ml; Millipore, Darmstadt, Germany) was added to each sample to a final concentration of 0.3 μg/μl, and the samples were incubated at 37 °C for 2 h to achieve enzymatic degradation of mRNA. Treated vaccine samples were directly loaded into the sample plate wells pre-filled with dilution marker or 0.1× TE buffer according to the manufacturer’s protocol for each kit. For concentrations less than 10 ng/μl, 4 μl of sample was diluted with 20 μl of 0.1× TE buffer, for concentrations above 10 ng/μl, 2 μl of sample was diluted with 22 μl of 1× TE buffer. All vaccine batches were analysed in three biological replicates (individual vials) and three technical replicates (electrophoretic runs). The electrophoretic separation was performed using the corresponding separation method under the following conditions: capillary effective length 33 cm, prerun 8 kV for 30 s, marker 3.0 kV for 5 s, sample injection 3.0 kV for 5 s, separation 8 kV for 30 min (DNF-930-33 – dsDNA 75-20000 bp), or capillary effective length 33 cm, prerun 8 kV for 30 s, sample injection 3.0 kV for 5 s, separation 5 kV for 55 min (DNF-464-33 – HS Large Fragment 50Kb).
The Agilent DNF-471 RNA Kit designed for total RNA analysis with fragment sizes ranging from 200 to 6000 nucleotides and concentrations between 5 and 500 ng/μl was used to assess the integrity of mRNA in vaccine samples. Following the manufacturer’s recommendations, vaccine samples were heat-treated at 70 °C for 2 min to disrupt the LNPs and release the encapsulated mRNA. Treated vaccine samples were directly loaded into to the sample plate wells pre-filled with dilution marker according to the manufacturer’s protocol. All vaccine batches were analysed in three biological replicates (individual vials) and three technical replicates (electrophoretic runs). The electrophoretic separation was performed using the corresponding separation method under the following conditions: capillary effective length 33 cm, prerun 8 kV for 30 s, sample injection 5.0 kV for 4 s, separation 8 kV for 45 min (DNF-471-33-SS Total RNA 15 nt). Integrity of mRNA was calculated by as the ratio of the main peak area representing the full-length mRNA of expected size to the total peak area of all RNA fragments, expressed as a percentage. Visualisation and analysis of data obtained from all capillary electrophoresis runs, including calculation of mRNA integrity were performed using ProSize Data Analysis Software (Agilent Technologies, Santa Clara, CA, USA).
Analysis by short-read massively parallel sequencing
Fluorescently labelled reversible terminator chemistry based short-read massively parallel sequencing using NextSeq 2000 system (Illumina, San Diego, CA, USA) and P3 XLEAP-SBS sequencing kits with 200 cycles was performed for all analysed vaccine samples. For DNA extraction of samples intended for sequencing, silica membrane based QIAamp DNA mini kit (Qiagen, Hilden, Germany) was used with separately optimised protocols for Comirnaty and Spikevax batch samples.
DNA isolation from Comirnaty vaccines was performed using 300 μl of vaccine (equivalent to one vaccine dose). Samples were first heated at 95 °C for 10 min to release nucleic acids from LNPs, then cooled on ice. Subsequently, 60 μl of RNase A was added, followed by vortexing and brief centrifugation (15 s). Lysis buffer AL (300 μl) and proteinase K (30 μl) were added, vortexed, briefly centrifuged, and incubated in a thermoblock for 90 min at 37 °C, followed by 10 min at 56 °C.
After incubation, 300 μl of absolute ethanol was added. The mixture was vortexed for 15 s, briefly centrifuged, and applied to a QIAamp spin column in three successive loads. After DNA binding, the column was washed in three steps: 500 μl of AW1 buffer (first wash), 500 μl of AW2 buffer (second wash), and 500 μl of absolute ethanol (third wash). All centrifugation steps were performed at 6000 × g for 1 min unless otherwise specified. To remove residual ethanol, the column was transferred to a new collection tube and centrifuged for 3 min at 14,000 × g. It was then placed into a sterile 1.5 ml microtube and incubated at 56 °C for 2 min to fully dry the membrane.
DNA was eluted with either 200 μl of MPW (millipore water) (for non-expired batches LN0605, LK8842, LK8456, LM9076) or 60 μl (for expired batches FP9632, 1F1051A, 1L084A, 1F1059A, PCB0020). After a 10-min incubation at room temperature, the column was centrifuged for 1 min at 14,000 × g. The eluate (purified DNA) was used for subsequent analyses.
DNA isolation from Spikevax vaccines
A volume of 500 μl of Spikevax vaccine (equivalent to one dose) was used. For effective RNA degradation with RNase A, samples from batches 000058A and 3005697 were diluted 1:3 in MPW, while booster vaccines (400012A, 200106A, MV1018A, 200023A) were diluted 1:1, based on their labelled mRNA content (100 μg or 50 μg, respectively). As with the Comirnaty vaccines, the samples were first heated at 95 °C for 10 min to release nucleic acids from LNPs, then cooled on ice. Subsequently, 100 μl of RNase A was added, followed by vortexing and brief centrifugation (15 s). Lysis buffer AL (500 μl) and proteinase K (50 μl) were added, vortexed, briefly centrifuged, and incubated in a thermoblock for 90 min at 37 °C, followed by 10 min at 56 °C.
After incubation, 500 μl of absolute ethanol was added, vortexed (15 s), and briefly centrifuged, and applied to a QIAamp spin column in three successive loads. Following three-steps column wash was performed as described in the previous protocol. Finally, DNA was eluted with 60 μl MPW. After 10 min at room temperature, columns were centrifuged at 14,000 × g for 1 min, and the eluate was used for downstream analysis.
DNA and RNA quantification
The concentration of nucleic acids (DNA and RNA) in the extracted samples was determined using the Qubit™ 3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) with Qubit™ assay tubes and corresponding detection kits. For DNA, the Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used, providing high sensitivity in the range of 0.005–120 ng/μl. For RNA, the Qubit™ RNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used, with a detection range of 0.2–200 ng/μl. Measurements were performed according to the manufacturer’s instructions, including calibration with kit standards. DNA quantity in each sample was measured from 5 μl of extract in technical triplicate. The same volume was entered into the device to allow automated calculation of nucleic acid concentration in ng/μl.
As extractions were performed from vaccine volumes equivalent to a single dose, DNA/RNA content per dose was calculated by adjusting for: (1) elution volume, (2) input sample dilution prior to extraction, and (3) any dilution applied before fluorometric measurement.
Library Preparation Protocol
For the library preparation, Illumina Truseq DNA NANO Library Prep kit was used (Illumina, San Diego, CA, USA) according to the manufacturer’s protocols, ommitting DNA fragmentation step, with the subsequent step of size selection replaced by a complete capture of fragments using magnetic beads. For low-input DNA, a half-volume preparation was chosen to optimise the process. In detail, a 30 µl aliquot of isolated DNA was transferred to a DNA low-binding PCR plate (LoBind, Eppendorf). To each sample, 20 µl of End Repair Mix 2 was added to blunt the DNA fragment ends. The mixture was gently mixed by pipetting and incubated at 30 °C for 30 minutes, followed by cooling to 4 °C. Subsequently, 200 µl of Sample Purification Beads (SPB; 4× the sample volume) were added to capture all DNA fragments. After pipette mixing and a 5-min incubation at room temperature, the plate was placed on a magnetic stand for 10 min. The supernatant was removed, and the pellet was washed twice with 200 µl of freshly prepared 80% ethanol. Residual ethanol was removed by air-drying at room temperature.
The pellet was resuspended in 10 µl of Resuspension Buffer (RSB), mixed thoroughly, incubated for 2 min at room temperature, and placed on a magnetic stand for 5 min. From the eluate, 8.75 µl was transferred to a new PCR well. Adenylation of the 3’ ends (addition of a single adenine nucleotide) was performed by adding 6.25 µl of A-Tailing Mix to the eluate. The mixture was incubated at 37 °C for 30 min, then at 70 °C for 5 min, and finally cooled to 4 °C for 5 min. Adaptor ligation was performed by adding 1.25 µl of Indexed Adaptor (unique for each sample), 1.25 µl of Ligation Mix 2, and 1.25 µl of RSB. The mixture was incubated at 30 °C for 10 min and then cooled to 4 °C. Ligation was stopped by adding 2.5 µl of Stop Ligation Buffer. Unligated adaptors and adaptor dimers were removed by a two-step bead cleanup using SPB. In the first step, 21.25 µl of SPB was added, mixed, and incubated for 5 min at room temperature. The beads were separated on a magnetic stand (5 min), the supernatant was discarded, and the pellet was washed twice with 200 µl of 80% ethanol. After ethanol removal, 26.25 µl of RSB was added, the sample was mixed, incubated for 2 min, and separated again. From the eluate, 25 µl was transferred to a new PCR well.
In the second cleanup step, 25 µl of SPB was added, mixed, and incubated for 5 min at room temperature. Following magnetic separation, the supernatant was removed, and the pellet was washed twice with 200 µl of 80% ethanol. After drying, 13.25 µl of RSB was added, the sample was mixed, incubated for 2 min, and separated again. A final volume of 12.5 µl was transferred to a new PCR well.
PCR enrichment of the library was carried out by adding 10 µl of Enhanced PCR Mix (EPM) and 2.5 µl of PCR Primer Cocktail (PPC). The thermal cycling conditions were as follows: Initial denaturation at 95 °C for 3 min, 10 cycles of denaturation at 98 °C for 20 s, annealing at 60 °C for 15 s, extension at 72 °C for 30 s, final extension at 72 °C for 5 min, hold at 4 °C.
Final library cleanup involved adding 25 µl of SPB to the PCR product. After pipette mixing and a 5-min incubation at room temperature, magnetic separation was performed, the supernatant was removed, and the pellet was washed twice with 200 µl of 80% ethanol. After ethanol removal and drying, 13 µl of RSB was added. The sample was mixed, incubated for 2 min at room temperature, separated again, and 12.5 µl of eluate was transferred to a new PCR well.
Final sequencing libraries were quality-checked using fluorometric quantification (Qubit 3.0, Qubit DNA HS Assay) and fragment size analysis (Agilent Bioanalyzer 2100, High Sensitivity DNA Kit). Library molarity (nM) was calculated using the formula:
(Concentration [ng/µl]/Average fragment length [bp] × 660) × 106
Libraries not meeting the required 2 nM threshold were re-amplified with an additional three PCR cycles, followed by SPB cleanup and repeated quality control. Libraries that still did not meet the concentration threshold were concentrated by vacuum evaporation of the eluate using a Concentrator Plus (Eppendorf AG, Hamburg, Germany).
Bioinformatics processing of sequenced reads
Quality of sequenced reads was assessed using FastQC (v 0.12.1)23. Given the high quality of the raw data, no read trimming or filtering was applied prior to analysis.
De novo assembly into plasmid contigs was performed using Unicycler (v 0.5.1)28. For most samples, assemblies were generated from the first 100,000 reads to optimise computational performance. For two Spikevax bivalent Original/Omicron BA.4/BA.5 batches (400012A and MV1018A), the assembly from 1,000,000 reads of sample MV1018A was used for subsequent analyses. To account for the circular nature of plasmids, each assembled contig (in linear form) was extended by an additional 100 bp (maximum read length) from its 5′ end to the 3′ terminus, ensuring proper mapping of reads spanning the artificial break point. Each sample was thus assigned a single contig, which served as the reference for read mapping with Bowtie2 (v 2.5.4)29. Mapping results were assessed with Qualimap (v 2.3)30, extracting key metrics including the total number of reads, number of mapped reads, reference coverage, and fragment length distribution.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information file. Unmodified short-read sequences were deposited in NCBI under Project ID PRJNA1303864 and are accessible via https://www.ebi.ac.uk/ena/browser/view/PRJNA1303864, or https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1303864/.
References
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines – A new era in vaccinology. Nat. Rev. Drug. Discov. 17, 261–279 (2018).
Guidance for Industry – Characterization and Qualification of Cell Substrates and Other Biological Materials Used in the Production of Viral Vaccines for Infectious Disease Indications, U.S. DHHS, FDA, CBER, February 2010.
Paul-Ehrlich-Institut 2023. Information for Healthcare professionals. Testing of COVID-19 mRNA vaccines. https://www.pei.de/SharedDocs/Downloads/EN/newsroom-en/notification/231222-testing-mrna-vaccinas-dna-contamination.pdf?__blob=publicationFile&v=3.
European Commission, Directorate-General for Health and Food Safety: ADVANCE PURCHASE AGREEMENT („APA”) for the development, production, priority-purchasing options and supply of a successful COVID-19 vaccine for EU Member States. SANTE/2020/C3/043 -S1I2.838335 (2020).
Chaudhary, N., Weissman, D. & Whitehead, K. A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug. Discov. 20, 817–838 (2021).
Schoenmaker, L. et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 601, 120586 (2021).
Crommelin, D. J. A., Anchordoquy, T. J., Volkin, D. B., Jiskoot, W. & Mastrobattista, E. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 110, 997–1001 (2021).
Kaiser, S., Kaiser, S., Reis, J. & Marschalek, R. Quantification of objective concentrations of DNA impurities in mRNA vaccines. Vaccine 55, 127022 (2025).
Leban, M. et al. Determination of linearized pDNA template in mRNA production process using HPLC. Anal. Bioanal. Chem. 416, 2389–2398 (2024).
Vieth, S., Waibler, Z. & Schumann, G. G. Opting for the appropriate methodology for quantification of residual DNA impurities in mRNA vaccines. Vaccine 56, 127186 (2025).
Kämmerer, U., Schulz, V. & Steger, K. BioNTech RNA-Based COVID-19 Injections Contain Large Amounts Of Residual DNA Including An SV40 Promoter/Enhancer Sequence. https://publichealthpolicyjournal.com/biontech-rna-based-covid-19-injections-contain-large-amounts-of-residual-dna-including-an-sv40-promoter-enhancer-sequence/ (2024).
König, B. & Kirchner, J. O. Methodological considerations regarding the quantification of DNA impurities in the COVID-19 mRNA vaccine comirnaty®. Methods Protoc. 7, 41 (2024).
Speicher, D. J., Rose, J. & McKernan, K. Quantification of residual plasmid DNA and SV40 promoter-enhancer sequences in Pfizer/BioNTech and Moderna modRNA COVID-19 vaccines from Ontario, Canada. Autoimmunity 58, 2551517 (2025).
McKernan, K., Helbert, Y., Kane, L. T. & McLaughlin, S. Sequencing of bivalent Moderna and Pfizer mRNA vaccines reveals nanogram to microgram quantities of expression vector dsDNA per dose. Preprint at https://doi.org/10.31219/osf.io/b9t7m (2023).
Fleming, R. M., Kotlar, P. & Pekova, S. Quantitative analysis of nucleic acid content in spikevax (Moderna) and BNT162b2 (Pfizer) COVID-19 vaccine lots. HSOA J. Angiol. Vasc. Surg. 10, 124 (2025).
Bustin, S. A. et al. MIQE 2.0: revision of the minimum information for publication of quantitative real-time PCR experiments guidelines. Clin. Chem. 71, 634–651 (2025).
EUROPEAN PHARMACOPOEIA 01/2020:20635 2.6.35. Quantification and characterisation of host-cell DNA. © Council of Europe, 67075 Strasbourg Cedex, France - 2019.
17 EMA/CHMP/ICH/82072/2006. Committee for Medicinal Products for Human Use, ICH Q2(R2) Guideline on validation of analytical procedures. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-quality-non-clinical-clinical-requirements-investigational-advanced-therapy-medicinal-products-clinical-trials_en.pdf.
Minarik, G. et al. Utilization of benchtop next generation sequencing platforms ion torrent PGM and MiSeq in noninvasive prenatal testing for chromosome 21 trisomy and testing of impact of in silico and physical size selection on its analytical performance. PLoS ONE 10, e0144811 (2015).
Kucharik, M. et al. Non-invasive prenatal testing (NIPT) by low coverage genomic sequencing: Detection limits of screened chromosomal microdeletions. PLoS ONE 15, e0238245 (2020).
Holesova, Z. et al. Understanding genetic variability: exploring large-scale copy number variants through non-invasive prenatal testing in European populations. BMC Genom. 25, 366 (2024).
Bao, H., Yang, S., Chen, X. & Zhou, Q. Early detection of multiple cancer types using multidimensional cell-free DNA fragmentomics. Nat. Med. https://doi.org/10.1038/s41591-025-03735-2 (2025).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
Asplund, M. et al. Contaminating viral sequences in high-throughput sequencing viromics: A linkage study of 700 sequencing libraries. Clin. Microbiol. Infect. 25, 1277–1285 (2019).
Milne, C. et al. Independent control of COVID-19 vaccines by EU official control authority batch Release: challenges, strengths and successes. npj Vaccines 8, 22 (2023).
Therapeutic Goods Administration (TGA), Australian Government, Department of Health and Aged Care. Summary report of residual DNA and endotoxin on COVID-19 mRNA vaccines conducted by TGA Laboratories. https://www.tga.gov.au/resources/publication/tga-laboratory-testing-reports/summary-report-residual-dna-and-endotoxin-covid-19-mrna-vaccines-conducted-tga-laboratories (2024).
Therapeutic Goods Administration (TGA), Australian Government, Department of Health and Aged Care. Batch release assessment of COVID-19 vaccines. https://www.tga.gov.au/products/covid-19/covid-19-vaccines/batch-release-assessment-covid-19-vaccines (2025).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
García-Alcalde, F. et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679 (2012).
Acknowledgements
This work was funded by the Government of the Slovak Republic via the Ministry of Health of the Slovak Republic, and supported by the Slovak Academy of Sciences. We would like to thank the State Institute for Drug Control and Ministry of Health of the Slovak Republic for arranging the provision of vaccine samples from official repositories at UNIPHARMA and IMUNA PHARM. We are grateful to Roman Dorcik, Director of the State Institute for Drug Control, for his kind advises and support.The funder played no role in the study design, data collection, analysis and interpretation, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
S.P., J.K., B.K., and T.Sz. designed the study, A.A., T.Se., L.P., M.B., V.Z., D.R., M.Me., M.Mi., V.G., and B.C. performed the experiments and/or data analysis, J.B. accomplished bioinformatic analysis of sequencing data, S.P. prepared the original manuscript, J.K., B.K., T.Sz., A.A., T. Se., L.P., and J.B. contributed to the writing, reviewing and editing the manuscript. S.P. and J.K. were involved in funding acquisition and project management. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Achs, A., Sedlackova, T., Predajna, L. et al. Systematic analysis of COVID-19 mRNA vaccines using four orthogonal approaches demonstrates no excessive DNA impurities. npj Vaccines 10, 259 (2025). https://doi.org/10.1038/s41541-025-01304-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41541-025-01304-9
This article is cited by
-
Process and analytical strategies for the safe production of mRNA vaccines and therapeutics
Molecular Biology Reports (2026)