Abstract
The cellular and molecular mediators of peri-implant fibrosis—a most common reason for implant failure and for surgical revision after the replacement of a prosthetic joint—remain unclear. Here we show that peri-implant fibrotic tissue in mice and humans is largely composed of a specific population of skeletal cells expressing the leptin receptor (LEPR) and that these cells are necessary and sufficient to generate and maintain peri-implant fibrotic tissue. In a mouse model of tibial implantation and osseointegration that mimics partial knee arthroplasty, genetic ablation of LEPR+ cells prevented peri-implant fibrosis and the implantation of LEPR+ cells from peri-implant fibrotic tissue was sufficient to induce fibrosis in secondary hosts. Conditional deletion of the adhesion G-protein-coupled receptor F5 (ADGRF5) in LEPR+ cells attenuated peri-implant fibrosis while augmenting peri-implant bone formation, and ADGRF5 inhibition by the intra-articular or systemic administration of neutralizing anti-ADGRF5 in the mice prevented and reversed peri-implant fibrosis. Pharmaceutical agents that inhibit the ADGRF5 pathway in LEPR+ cells may be used to prevent and treat peri-implant fibrosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed data generated during the study are available for research purposes from the corresponding author within reasonable request. Transcriptomic data from bulk RNA-seq are available from the Gene Expression Omnibus (GEO) under accession numbers GSE227869 and GSE227128. The genomic data used in Fig. 5n and Supplementary Fig. 7 were extracted from publicly available datasets: GSE108892 (ref. 24), GSE138689 (ref. 23) and GSE136970 (ref. 39). The genomic data used in Supplementary Fig. 6 were extracted from the publicly available dataset GSE147287 (ref. 46). Source data for the figures are provided with this paper.
References
Headon, H., Kasem, A. & Mokbel, K. Capsular contracture after breast augmentation: an update for clinical practice. Arch. Plast. Surg. 42, 532–543 (2015).
Shah, P. K. Inflammation, neointimal hyperplasia, and restenosis: as the leukocytes roll, the arteries thicken. Circulation 107, 2175–2177 (2003).
Apostu, D., Lucaciu, O., Berce, C., Lucaciu, D. & Cosma, D. Current methods of preventing aseptic loosening and improving osseointegration of titanium implants in cementless total hip arthroplasty: a review. J. Int. Med. Res. 46, 2104–2119 (2018).
Wang, T. et al. Institutional experience of in-stent stenosis after pipeline flow diverter implantation: a retrospective analysis of 6 isolated cases out of 118 patients. Medicine 100, e25149 (2021).
Feng, X., Gu, J. & Zhou, Y. Primary total hip arthroplasty failure: aseptic loosening remains the most common cause of revision. Am. J. Transl. Res. 14, 7080–7089 (2022).
Sabah, S. A. et al. No exponential rise in revision knee replacement surgery over the past 15 years: an analysis from the National Joint Registry. Osteoarthr. Cartil. 30, 1670–1679 (2022).
Negm, A. M., Beaupre, L. A., Goplen, C. M., Weeks, C. & Jones, C. A. A scoping review of total hip arthroplasty survival and reoperation rates in patients of 55 years or younger: health services implications for revision surgeries. Arthroplast Today 16, 247–258.e246 (2022).
Brown, M. L., Javidan, P., Early, S. & Bugbee, W. Evolving etiologies and rates of revision total knee arthroplasty: a 10-year institutional report. Arthroplasty 4, 39 (2022).
Tay, M. L., Matthews, B. G., Monk, A. P. & Young, S. W. Disease progression, aseptic loosening and bearing dislocations are the main revision indications after lateral unicompartmental knee arthroplasty: a systematic review. J. ISAKOS 7, 132–141 (2022).
Teeny, S. M., York, S. C., Mesko, J. W. & Rea, R. E. Long-term follow-up care recommendations after total hip and knee arthroplasty: results of the American Association of Hip and Knee Surgeons’ member survey. J. Arthroplast. 18, 954–962 (2003).
Upfill-Brown, A. et al. Epidemiology of revision total knee arthroplasty in the United States, 2012 to 2019. Arthroplast Today 15, 188–195.e186 (2022).
Wooley, P. H. & Schwarz, E. M. Aseptic loosening. Gene Ther. 11, 402–407 (2004).
Drees, P., Eckardt, A., Gay, R. E., Gay, S. & Huber, L. C. Mechanisms of disease: molecular insights into aseptic loosening of orthopedic implants. Nat. Clin. Pr. Rheumatol. 3, 165–171 (2007).
Neale, S. D., Sabokbar, A., Howie, D. W., Murray, D. W. & Athanasou, N. A. Macrophage colony-stimulating factor and interleukin-6 release by periprosthetic cells stimulates osteoclast formation and bone resorption. J. Orthop. Res. 17, 686–694 (1999).
Ingham, E. & Fisher, J. The role of macrophages in osteolysis of total joint replacement. Biomaterials 26, 1271–1286 (2005).
Arora, A. et al. The role of the TH1 and TH2 immune responses in loosening and osteolysis of cemented total hip replacements. J. Biomed. Mater. Res. A 64, 693–697 (2003).
Yang, S. Y. et al. Protective effects of IL-1Ra or vIL-10 gene transfer on a murine model of wear debris-induced osteolysis. Gene Ther. 11, 483–491 (2004).
Apinyankul, R. et al. Isolated versus full component revision in total knee arthroplasty for aseptic loosening. J. Arthroplasty 38, 335–340 (2022).
Yao, J. J. et al. Long-term mortality after revision THA. Clin. Orthop. Relat. Res. 476, 420–426 (2018).
Omatsu, Y. et al. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity 33, 387–399 (2010).
Omatsu, Y., Seike, M., Sugiyama, T., Kume, T. & Nagasawa, T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature 508, 536–540 (2014).
Seike, M., Omatsu, Y., Watanabe, H., Kondoh, G. & Nagasawa, T. Stem cell niche-specific Ebf3 maintains the bone marrow cavity. Genes Dev. 32, 359–372 (2018).
Mo, C. et al. Single-cell transcriptomics of LepR-positive skeletal cells reveals heterogeneous stress-dependent stem and progenitor pools. EMBO J. 41, e108415 (2022).
Tikhonova, A. N. et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019).
Zhong, L. et al. Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. eLife 9, e54695 (2020).
Ishibashi, M. et al. CD200-positive cancer associated fibroblasts augment the sensitivity of epidermal growth factor receptor mutation-positive lung adenocarcinomas to EGFR tyrosine kinase inhibitors. Sci. Rep. 7, 46662 (2017).
Greenbaum, A. et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230 (2013).
Muhl, L. et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun. 11, 3953 (2020).
Ortinau, L. C. et al. Identification of functionally distinct Mx1+αSMA+ periosteal skeletal stem cells. Cell Stem Cell 25, 784–796.e785 (2019).
Vesprey, A. et al. Tmem100- and Acta2-lineage cells contribute to implant osseointegration in a mouse model. J. Bone Miner. Res. 36, 1000–1011 (2021).
Zhou, B., Yue, R., Murphy, M., Peyer, J. & Morrison, S. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 15, 165–168 (2014).
Shu, H. S. et al. Tracing the skeletal progenitor transition during postnatal bone formation. Cell Stem Cell 28, 2122–2136.e2123 (2021).
Fredriksson, R., Gloriam, D. E., Höglund, P. J., Lagerström, M. C. & Schiöth, H. B. There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem. Biophys. Res. Commun. 301, 725–734 (2003).
Kubo, F. et al. Loss of the adhesion G-protein coupled receptor ADGRF5 in mice induces airway inflammation and the expression of CCL2 in lung endothelial cells. Respir. Res. 20, 11 (2019).
Folts, C. J., Giera, S., Li, T. & Piao, X. Adhesion G-protein-coupled receptors as drug targets for neurological diseases. Trends Pharmacol. Sci. 40, 278–293 (2019).
Purcell, R. H. & Hall, R. A. Adhesion G-protein-coupled receptors as drug targets. Annu. Rev. Pharmacol. Toxicol. 58, 429–449 (2018).
Yang, X. et al. Intermittent parathyroid hormone enhances cancellous osseointegration of a novel murine tibial implant. J. Bone Joint Surg. Am. 97, 1074–1083 (2015).
Kuyl, E.-V. et al. Inhibition of PAD4 mediated neutrophil extracellular traps prevents fibrotic osseointegration failure in a tibial implant murine model: an animal study. Bone Joint J. 103-B, 135–144 (2021).
Matsushita, Y. et al. A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat. Commun. 11, 332 (2020).
Decker, M. et al. Leptin-receptor-expressing bone marrow stromal cells are myofibroblasts in primary myelofibrosis. Nat. Cell Biol. 19, 677–688 (2017).
Baccin, C. et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 22, 38–48 (2020).
Dutta, S. & Sengupta, P. Men and mice: relating their ages. Life Sci. 152, 244–248 (2016).
Tang, X. et al. GPR116, an adhesion G-protein-coupled receptor, promotes breast cancer metastasis via the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 73, 6206–6218 (2013).
Zheng, T. et al. GPR116 overexpression correlates with poor prognosis in gastric cancer. Medicine 100, e28059 (2021).
Georgiadi, A. et al. Orphan GPR116 mediates the insulin sensitizing effects of the hepatokine FNDC4 in adipose tissue. Nat. Commun. 12, 2999 (2021).
Wang, Z. et al. Single-cell RNA sequencing deconvolutes the in vivo heterogeneity of human bone marrow-derived mesenchymal stem cells. Int. J. Biol. Sci. 17, 4192–4206 (2021).
Qiu, X. et al. Single-cell RNA sequencing of human femoral head in vivo. Aging 13, 15595–15619 (2021).
Gao, B. et al. Macrophage-lineage TRAP+ cells recruit periosteum-derived cells for periosteal osteogenesis and regeneration. J. Clin. Invest. 129, 2578–2594 (2019).
Xu, Z. et al. Triple-gene deletion for osteocalcin significantly impairs the alignment of hydroxyapatite crystals and collagen in mice. Front Physiol. 14, 1136561 (2023).
Zhang, J. et al. Roles of SATB2 in osteogenic differentiation and bone regeneration. Tissue Eng. Part A 17, 1767–1776 (2011).
Dillon, S. et al. Ablation of Enpp6 results in transient bone hypomineralization. JBMR Plus 5, e10439 (2021).
Granados-Montiel, J. et al. SERPINA9 and SERPINB2: novel cartilage lineage differentiation markers of human mesenchymal stem cells with kartogenin. Cartilage 12, 102–111 (2021).
Lu, J. et al. Role of paraoxonase-1 in bone anabolic effects of parathyroid hormone in hyperlipidemic mice. Biochem. Biophys. Res. Commun. 431, 19–24 (2013).
Xiang, L., Zheng, J., Zhang, M., Ai, T. & Cai, B. FOXQ1 promotes the osteogenic differentiation of bone mesenchymal stem cells via Wnt/β-catenin signaling by binding with ANXA2. Stem Cell Res. Ther. 11, 403 (2020).
Liu, X. et al. DUSP5 promotes osteogenic differentiation through SCP1/2-dependent phosphorylation of SMAD1. Stem Cells 39, 1395–1409 (2021).
Yuan, B. et al. Aberrant Phex function in osteoblasts and osteocytes alone underlies murine X-linked hypophosphatemia. J. Clin. Invest. 118, 722–734 (2008).
Maranda, V., Gaumond, M. H. & Moffatt, P. The osteogenesis imperfecta type V mutant BRIL/IFITM5 promotes transcriptional activation of MEF2, NFATc, and NR4A in osteoblasts. Int. J. Mol. Sci. 23, 2148 (2022).
Alshbool, F. Z. & Mohan, S. Differential expression of claudin family members during osteoblast and osteoclast differentiation: Cldn-1 is a novel positive regulator of osteoblastogenesis. PLoS ONE 9, e114357 (2014).
Shi, C. et al. Carbonic anhydrase III protects osteocytes from oxidative stress. FASEB J. 32, 440–452 (2018).
Wu, M. et al. Persistent expression of Pax3 in the neural crest causes cleft palate and defective osteogenesis in mice. J. Clin. Invest. 118, 2076–2087 (2008).
Miyamoto, S., Yoshikawa, H. & Nakata, K. Axial mechanical loading to ex vivo mouse long bone regulates endochondral ossification and endosteal mineralization through activation of the BMP-Smad pathway during postnatal growth. Bone Rep. 15, 101088 (2021).
Ha, H. et al. Stimulation by TLR5 modulates osteoclast differentiation through STAT1/IFN-beta. J. Immunol. 180, 1382–1389 (2008).
Xue, Y., Zhao, C. & Liu, T. Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) accelerates osteoclast formation by regulating signal transducer and activator of transcription 3 (STAT3) signalling. Bioengineered 13, 2285–2295 (2022).
Madel, M. B. et al. Specific targeting of inflammatory osteoclastogenesis by the probiotic yeast S. boulardii CNCM I-745 reduces bone loss in osteoporosis. eLife 12, e82037 (2023).
Deng, C. et al. TNFRSF19 inhibits TGFβ signaling through interaction with TGFβ receptor type I to promote tumorigenesis. Cancer Res. 78, 3469–3483 (2018).
Nakamura, S. et al. Suppression of autophagic activity by Rubicon is a signature of aging. Nat. Commun. 10, 847 (2019).
Yi, X. et al. TNF-polarized macrophages produce insulin-like 6 peptide to stimulate bone formation in rheumatoid arthritis in mice. J. Bone Miner. Res. 36, 2426–2439 (2021).
Sun, Y. et al. TGF-β1 and TIMP-4 regulate atrial fibrosis in atrial fibrillation secondary to rheumatic heart disease. Mol. Cell. Biochem. 406, 131–138 (2015).
Ni, C. et al. FMO2 (flavin containing monooxygenase 2) prevents cardiac fibrosis via CYP2J3-SMURF2 axis. Circ. Res. 131, 189–206 (2022).
Pang, X. et al. Targeting integrin pathways: mechanisms and advances in therapy. Signal Transduct. Target Ther. 8, 1 (2023).
Toriumi, K. et al. LRRC15 expression indicates high level of stemness regulated by TWIST1 in mesenchymal stem cells. iScience 26, 106946 (2023).
Wang, G., Wu, H., Liang, P., He, X. & Liu, D. Fus knockdown inhibits the profibrogenic effect of cardiac fibroblasts induced by angiotensin II through targeting Pax3 thereby regulating TGF-β1/Smad pathway. Bioengineered 12, 1415–1425 (2021).
Cribb, A. E. et al. Assessment of arylamine N-acetyltransferase (NAT1) activity in mononuclear leukocytes of cystic fibrosis patients. Br. J. Clin. Pharm. 39, 85–89 (1995).
Di Fazio, P. et al. Toll-like receptor 5 tunes hepatic and pancreatic stellate cells activation. BMJ Open Gastroenterol. 10, e001148 (2023).
Shirai, K. et al. Serum amyloid P component and pro-platelet basic protein in extracellular vesicles or serum are novel markers of liver fibrosis in chronic hepatitis C patients. PLoS ONE 17, e0271020 (2022).
Nhieu, J., Lin, Y. L. & Wei, L. N. CRABP1 in non-canonical activities of retinoic acid in health and diseases. Nutrients 14, 1528 (2022).
Königshoff, M. et al. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J. Clin. Invest. 119, 772–787 (2009).
Minoura, I. et al. Reversal of axonal growth defects in an extraocular fibrosis model by engineering the kinesin-microtubule interface. Nat. Commun. 7, 10058 (2016).
Yi, Y. et al. Regulation of atrial fibrosis by the bone. Hypertension 73, 379–389 (2019).
Yang, K. et al. Transforming growth factor-β induced protein regulates pulmonary fibrosis via the G-protein signaling modulator 2/Snail axis. Peptides 155, 170842 (2022).
Song, P. et al. Effect of the Wnt1/β-catenin signalling pathway on human embryonic pulmonary fibroblasts. Mol. Med. Rep. 10, 1030–1036 (2014).
Collins, C. A. et al. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS ONE 4, e4475 (2009).
Yan, Y. & Jiang, Y. RACK1 affects glioma cell growth and differentiation through the CNTN2-mediated RTK/Ras/MAPK pathway. Int. J. Mol. Med. 37, 251–257 (2016).
Liu, Z., Zhao, M., Yokoyama, K. K. & Li, T. Molecular cloning of a cDNA for rat TM4SF4, a homolog of human il-TMP (TM4SF4), and enhanced expression of the corresponding gene in regenerating rat liver. Biochim. Biophys. Acta 1518, 183–189 (2001).
Tiang, J. M., Butcher, N. J. & Minchin, R. F. Small molecule inhibition of arylamine N-acetyltransferase type I inhibits proliferation and invasiveness of MDA-MB-231 breast cancer cells. Biochem. Biophys. Res. Commun. 393, 95–100 (2010).
Stoupa, A. et al. TUBB1 mutations cause thyroid dysgenesis associated with abnormal platelet physiology. EMBO Mol. Med. 10, e9569 (2018).
Xiao, B. et al. Glutamate ionotropic receptor kainate type subunit 3 (GRIK3) promotes epithelial-mesenchymal transition in breast cancer cells by regulating SPDEF/CDH1 signaling. Mol. Carcinog. 58, 1314–1323 (2019).
Wang, Z., Liu, N., Shi, S., Liu, S. & Lin, H. The role of PIWIL4, an argonaute family protein, in breast cancer. J. Biol. Chem. 291, 10646–10658 (2016).
Li, W. & Gu, M. NUDT11 rs5945572 polymorphism and prostate cancer risk: a meta-analysis. Int J. Clin. Exp. Med. 8, 3474–3481 (2015).
Chen, G. et al. EphA1 receptor silencing by small interfering RNA has antiangiogenic and antitumor efficacy in hepatocellular carcinoma. Oncol. Rep. 23, 563–570 (2010).
Charfi, C., Levros, L. C., Jr. Edouard, E. & Rassart, E. Characterization and identification of PARM-1 as a new potential oncogene. Mol. Cancer 12, 84 (2013).
Pidugu, V. K. et al. IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene 38, 3232–3247 (2019).
Bolitho, C., Hahn, M. A., Baxter, R. C. & Marsh, D. J. The chemokine CXCL1 induces proliferation in epithelial ovarian cancer cells by transactivation of the epidermal growth factor receptor. Endocr. Relat. Cancer 17, 929–940 (2010).
Maruyama, T. et al. Downregulation of carbonic anhydrase IX promotes Col10a1 expression in chondrocytes. PLoS ONE 8, e56984 (2013).
Niu, G. et al. Overexpression of CPXM2 predicts an unfavorable prognosis and promotes the proliferation and migration of gastric cancer. Oncol. Rep. 42, 1283–1294 (2019).
Leblanc, N., Harquail, J., Crapoulet, N., Ouellette, R. J. & Robichaud, G. A. Pax-5 inhibits breast cancer proliferation through MiR-215 up-regulation. Anticancer Res. 38, 5013–5026 (2018).
Wang, P. et al. The role of Sprouty1 in the proliferation, differentiation and apoptosis of epidermal keratinocytes. Cell Prolif. 51, e12477 (2018).
Mongan, M. et al. Loss of MAP3K1 enhances proliferation and apoptosis during retinal development. Development 138, 4001–4012 (2011).
Xu, J. X. et al. Down-regulation of ALDOB during metabolic reprogramming mediates malignant behavior in hepatocellular carcinoma and insensitivity to postoperative adjuvant transarterial chemoembolization. Clin. Sci. 137, 303–316 (2023).
Li, X. & Yu, Q. PON1 hypermethylation is associated with progression of renal cell carcinoma. J. Cell. Mol. Med. 23, 6646–6657 (2019).
Chen, W. et al. A small molecule inhibitor of VSIG-8 prevents its binding to VISTA. Invest. N. Drugs 40, 690–699 (2022).
Wang, B. et al. Carbonic anhydrase IV inhibits cell proliferation in gastric cancer by regulating the cell cycle. Oncol. Lett. 20, 4 (2020).
Richter, A. M., Walesch, S. K., Wurl, P., Taubert, H. & Dammann, R. H. The tumor suppressor RASSF10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis 1, e18 (2012).
Merighi, S. et al. A3 adenosine receptor activation inhibits cell proliferation via phosphatidylinositol 3-kinase/Akt-dependent inhibition of the extracellular signal-regulated kinase 1/2 phosphorylation in A375 human melanoma cells. J. Biol. Chem. 280, 19516–19526 (2005).
Li, Y. & Guo, L. The versatile role of Serpina3c in physiological and pathological processes: a review of recent studies. Front. Endocrinol. 14, 1189007 (2023).
Zhao, X. et al. SPIB acts as a tumor suppressor by activating the NFκB and JNK signaling pathways through MAP4K1 in colorectal cancer cells. Cell Signal. 88, 110148 (2021).
Stoeckle, C. et al. RhoH is a negative regulator of eosinophilopoiesis. Cell Death Differ. 23, 1961–1972 (2016).
Choi, J. et al. Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2. Nat. Cell Biol. 20, 586–596 (2018).
Luo, N. et al. Prognostic role of M6A-associated immune genes and cluster-related tumor microenvironment analysis: a multi-omics practice in stomach adenocarcinoma. Front. Cell Dev. Biol. 10, 935135 (2022).
Zhang, Y., Godavarthi, J. D., Williams-Villalobo, A., Polk, S. & Matin, A. The role of DND1 in cancers. Cancers 13, 3679 (2021).
Avraham, S. et al. Structural and functional studies of the intracellular tyrosine kinase MATK gene and its translated product. J. Biol. Chem. 270, 1833–1842 (1995).
Liu, S. et al. PCDH17 increases the sensitivity of colorectal cancer to 5-fluorouracil treatment by inducing apoptosis and autophagic cell death. Signal Transduct. Target Ther. 4, 53 (2019).
Ghafouri-Fard, S. et al. A review on the role of HAND2-AS1 in cancer. Clin. Exp. Med. 23, 3179–3188 (2023).
Ni, R. et al. MiR-216a-3p inhibits the proliferation and invasion of fibroblast-like synoviocytes by targeting dual-specificity phosphatase 5. Int J. Rheum. Dis. 26, 699–709 (2023).
Kechele, D. O. et al. Orphan Gpr182 suppresses ERK-mediated intestinal proliferation during regeneration and adenoma formation. J. Clin. Invest. 127, 593–607 (2017).
Yue, Q. et al. Nitric oxide hinders club cell proliferation through Gdpd2 during allergic airway inflammation. FEBS Open Bio. 13, 1041–1055 (2023).
Yang, W. et al. Claudin-10 overexpression suppresses human clear cell renal cell carcinoma growth and metastasis by regulating ATP5O and causing mitochondrial dysfunction. Int J. Biol. Sci. 18, 2329–2344 (2022).
Castro, C. N. et al. NCKAP1L defects lead to a novel syndrome combining immunodeficiency, lymphoproliferation, and hyperinflammation. J. Exp. Med. 217, e20192275 (2020).
Heino, S. et al. Lef1 restricts ectopic crypt formation and tumor cell growth in intestinal adenomas. Sci. Adv. 7, eabj0512 (2021).
Salazar-Olivo, L. A. et al. SerpinA3g participates in the antiadipogenesis and insulin-resistance induced by tumor necrosis factor-alpha in 3T3-F442A cells. Cytokine 69, 180–188 (2014).
Parikh, A. et al. Malignant cell-specific CXCL14 promotes tumor lymphocyte infiltration in oral cavity squamous cell carcinoma. J. Immunother. Cancer 8, e001048 (2020).
Delmonte, O. M. et al. SASH3 variants cause a novel form of X-linked combined immunodeficiency with immune dysregulation. Blood 138, 1019–1033 (2021).
Wang, J. et al. SNP-mediated lncRNA-ENTPD3-AS1 upregulation suppresses renal cell carcinoma via miR-155/HIF-1alpha signaling. Cell Death Dis. 12, 672 (2021).
Xu, X. et al. Molecular mechanism of MYL4 regulation of skeletal muscle development in pigs. Genes 14, 1267 (2023).
Jiao, S., Li, N., Cai, S., Guo, H. & Wen, Y. Inhibition of CYFIP2 promotes gastric cancer cell proliferation and chemoresistance to 5-fluorouracil through activation of the Akt signaling pathway. Oncol. Lett. 13, 2133–2140 (2017).
Zhou, J., Jiang, J., Wang, S. & Xia, X. DKK1 inhibits proliferation and migration in human retinal pigment epithelial cells via the Wnt/beta-catenin signaling pathway. Exp. Ther. Med. 12, 859–863 (2016).
Xu, F. et al. Comprehensive analyses identify RIPOR2 as a genomic instability-associated immune prognostic biomarker in cervical cancer. Front. Immunol. 13, 930488 (2022).
Zhang, Q. et al. Glycoprotein M6A suppresses lung adenocarcinoma progression via inhibition of the PI3K/AKT pathway. J. Oncol. 2022, 4601501 (2022).
Balinas-Gavira, C. et al. Frequent mutations in the amino-terminal domain of BCL7A impair its tumor suppressor role in DLBCL. Leukemia 34, 2722–2735 (2020).
Mao, H. et al. HMGCS2 serves as a potential biomarker for inhibition of renal clear cell carcinoma growth. Sci. Rep. 13, 14629 (2023).
Pan, S. et al. Decreased expression of ARHGAP15 promotes the development of colorectal cancer through PTEN/AKT/FOXO1 axis. Cell Death Dis. 9, 673 (2018).
Chen, X., Lv, Q., Ma, J. & Liu, Y. PLCgamma2 promotes apoptosis while inhibits proliferation in rat hepatocytes through PKCD/JNK MAPK and PKCD/p38 MAPK signalling. Cell Prolif. 51, e12437 (2018).
Wang, Y., Li, S. J., Wu, X., Che, Y. & Li, Q. Clinicopathological and biological significance of human voltage-gated proton channel Hv1 protein overexpression in breast cancer. J. Biol. Chem. 287, 13877–13888 (2012).
Vallath, S. et al. CADM1 inhibits squamous cell carcinoma progression by reducing STAT3 activity. Sci. Rep. 6, 24006 (2016).
Takahashi, K. et al. Lysophosphatidic acid (LPA) signaling via LPA4 and LPA6 negatively regulates cell motile activities of colon cancer cells. Biochem. Biophys. Res. Commun. 483, 652–657 (2017).
Wang, Y., Lim, R. & Nie, G. HtrA4 may play a major role in inhibiting endothelial repair in pregnancy complication preeclampsia. Sci. Rep. 9, 2728 (2019).
Ma, Y., Han, C. C., Huang, Q., Sun, W. Y. & Wei, W. GRK2 overexpression inhibits IGF1-induced proliferation and migration of human hepatocellular carcinoma cells by downregulating EGR1. Oncol. Rep. 35, 3068–3074 (2016).
Han, Y. et al. RASSF4 inhibits cell proliferation and increases drug sensitivity in colorectal cancer through YAP/Bcl-2 pathway. J. Cell. Mol. Med. 26, 3538–3547 (2022).
Dionne, G. et al. Mechanotransduction by PCDH15 relies on a novel cis-dimeric architecture. Neuron 99, 480–492.e485 (2018).
Vikhorev, P. G. et al. Titin-truncating mutations associated with dilated cardiomyopathy alter length-dependent activation and its modulation via phosphorylation. Cardiovasc. Res. 118, 241–253 (2022).
Barry, A. K., Wang, N. & Leckband, D. E. Local VE-cadherin mechanotransduction triggers long-ranged remodeling of endothelial monolayers. J. Cell Sci. 128, 1341–1351 (2015).
Stassen, O. M. J. A., Ristori, T. & Sahlgren, C. M. Notch in mechanotransduction—from molecular mechanosensitivity to tissue mechanostasis. J. Cell Sci. 133, jcs250738 (2020).
Wilde, C., Mitgau, J., Suchý, T., Schöneberg, T. & Liebscher, I. Translating the force-mechano-sensing GPCRs. Am. J. Physiol. Cell Physiol. 322, C1047–C1060 (2022).
Diaz-Horta, O. et al. Ripor2 is involved in auditory hair cell stereociliary bundle structure and orientation. J. Mol. Med. 96, 1227–1238 (2018).
Chen, Y. C. et al. ATOH1/RFX1/RFX3 transcription factors facilitate the differentiation and characterisation of inner ear hair cell-like cells from patient-specific induced pluripotent stem cells harbouring A8344G mutation of mitochondrial DNA. Cell Death Dis. 9, 437 (2018).
Lam, M. & Calvo, F. Regulation of mechanotransduction: emerging roles for septins. Cytoskeleton 76, 115–122 (2019).
Dela Paz, N. G., Melchior, B. & Frangos, J. A. Shear stress induces G-protein-coupled receptor (GPCR)-independent heterotrimeric G-protein activation in endothelial cells. Am. J. Physiol. Cell Physiol. 312, C428–C437 (2017).
Friess, M. C. et al. Mechanosensitive ACKR4 scavenges CCR7 chemokines to facilitate T cell de-adhesion and passive transport by flow in inflamed afferent lymphatics. Cell Rep. 38, 110334 (2022).
Ambrosi, T. H. et al. Distinct skeletal stem cell types orchestrate long bone skeletogenesis. eLife 10, e66063 (2021).
Debnath, S. et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature 562, 133–139 (2018).
Jeffery, E. C., Mann, T. L. A., Pool, J. A., Zhao, Z. & Morrison, S. J. Bone marrow and periosteal skeletal stem/progenitor cells make distinct contributions to bone maintenance and repair. Cell Stem Cell 29, 1547–1561.e1546 (2022).
Di Carlo, S. E. & Peduto, L. The perivascular origin of pathological fibroblasts. J. Clin. Invest. 128, 54–63 (2018).
El Agha, E. et al. Mesenchymal stem cells in fibrotic disease. Cell Stem Cell 21, 166–177 (2017).
Carlson, S., Trial, J., Soeller, C. & Entman, M. L. Cardiac mesenchymal stem cells contribute to scar formation after myocardial infarction. Cardiovasc. Res. 91, 99–107 (2011).
Kramann, R. et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66 (2015).
Xie, T. et al. Transcription factor TBX4 regulates myofibroblast accumulation and lung fibrosis. J. Clin. Invest. 126, 3626 (2016).
Kanisicak, O. et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun. 7, 12260 (2016).
Stoveken, H. M., Hajduczok, A. G., Xu, L. & Tall, G. G. Adhesion G-protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl Acad. Sci. USA 112, 6194–6199 (2015).
Bridges, J. P. et al. Regulation of pulmonary surfactant by the adhesion GPCR GPR116/ADGRF5 requires a tethered agonist-mediated activation mechanism. eLife 11, e69061 (2022).
Kienzle, A. et al. High rates of aseptic loosening after revision total knee arthroplasty for periprosthetic joint infection. JB JS Open Access 5, e20.00026 (2020).
de Steiger, R. N., Lewis, P. L., Harris, I., Lorimer, M. F. & Graves, S. E. What is the outcome of the first revision procedure of primary THA for osteoarthritis? A study from the Australian Orthopaedic Association National Joint Replacement Registry. Clin. Orthop. Relat. Res. 480, 1952–1970 (2022).
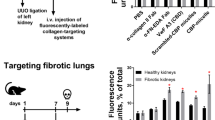
Li, F., Xu, X., Geng, J., Wan, X. & Dai, H. The autocrine CXCR4/CXCL12 axis contributes to lung fibrosis through modulation of lung fibroblast activity. Exp. Ther. Med. 19, 1844–1854 (2020).
Parvizi, J. et al. New definition for periprosthetic joint infection: from the Workgroup of the Musculoskeletal Infection Society. Clin. Orthop. Relat. Res. 469, 2992–2994 (2011).
Chan, C. K. et al. Identification and specification of the mouse skeletal stem cell. Cell 160, 285–298 (2015).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Bouxsein, M. L. et al. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 25, 1468–1486 (2010).
Müller, R. & Rüegsegger, P. Three-dimensional finite element modelling of non-invasively assessed trabecular bone structures. Med. Eng. Phys. 17, 126–133 (1995).
Hildebrand, T. & Ruegsegger, P. A new method for the model-independent assessment of thickness in three-dimensional images. J. Microsc. 185, 67–75 (1997).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Acknowledgements
This project was funded by the OREF under awards 994088 and 892405, a Hospital for Special Surgery Surgeon-in-Chief Grant and a Complex Joint Reconstruction Center grant given to V.J.S. X.Y. is supported by grant UL1 TR000457 from the Clinical and Translational Science Center at Weill Cornell Medicine, the Feldstein Medical Foundation and grant W81XWH-21-1-0900 from the Department of Defense. M.B.G. is supported by a Pershing Square Sohn Cancer Research Alliance Award and R01AR075585. The content is solely the responsibility of the authors and does not represent the official views of the sources of research support. We thank E. Monica for the assistance on the article figure editing and formatting. We thank E. Kuyl for the assistance on patient sample procurement. We thank the Flow Cytometry Core, Genomics Resources Core, Optical Microscopy Core and Citigroup Biomedical Imaging Core at Weill Cornell Medicine for their technical support.
Author information
Authors and Affiliations
Contributions
V.J.S. and A.O. designed, conducted and analysed the majority of experiments. X.Y., V.J.S., A.O. and M.P.G.B. conceived the project. X.Y. and M.P.G.B. supervised the project. X.Y. performed all mouse surgeries. V.J.S. and A.O. performed μCT scans and analysis and RNA-seq data analysis. V.J.S., J.L. and A.O. maintained and genotyped all mice. J.M. supervised or conducted flow cytometry. V.J.S., A.O., Q.L., M.H., Y.N., Q.L., A.T. and J.L. assisted with surgeries and performed tissue collection, immunofluorescence and histology analysis. M.H. and J.L. helped with human sample processing. M.P.G.B. provided access to human samples and supervised human studies. U.M.A., M.B.G. and L.B.I. helped with the study design and data interpretation. V.J.S., A.O., M.H. and X.Y. prepared the article. All authors read and approved the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
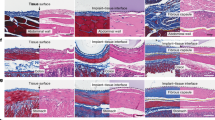
Extended Data Fig. 1 LEPR, ACTA2, and ADGRF5 are expressed by peri-implant fibrotic tissue from multiple patients underwent revision surgery for aseptic loosening.
a,c, Expression of LEPR (red), ACTA2(green), ADGRF5 (cyan) by peri-implant fibrotic tissue from two different patients suffering from peri-implant fibrosis around prior total hip arthroplasty. Far left, overlap between DAPI and LEPR. Second from left, overlap between ACTA2 and DAPI. Third from left, overlap between ADGRF5 and DAPI. Third from right, overlap between LEPR and ACTA2. Second from right, overlap between LEPR and ADGRF5. Far right, overlap between LEPR, ACTA2, and ADGRF5. Scale bar, 250 μm. b,d, Expression of LEPR (red), ACTA2 (green), ADGRF5 (cyan) by peri-implant fibrotic tissue from two different patients suffering from peri-implant fibrosis around prior total knee arthroplasty. Far left, overlap between DAPI and LEPR. Second from left, overlap between ACTA2 and DAPI. Third from left, overlap between ADGRF5 and DAPI. Third from right, overlap between LEPR and ACTA2. Second from right, overlap between LEPR and ADGRF5. Far right, overlap between LEPR, ACTA2, and ADGRF5. Scale bar, 250 μm. Images in a–d are representative of 4 biologically independent experiments.
Extended Data Fig. 2 COL3A1, S100A4, SM22A, ACTA2, FBLN2, and SDC4 are expressed by peri-implant fibrotic tissue in both human and mice.
Expression of COL3A1 (a, green), S100A4 (c, green), SM22A (e, green), ACTA2-RFP (g, green), FBLN2 (i, green), SDC4 (k, green) in murine peri-implant fibrotic tissue and its overlap with LepR– tdTomato cells (red). Scale bar, 50 μm. Expression of COL3A1 (b, green), S100A4 (d, green), SM22A (f, green), ACTA2 (h, green), FBLN2 (j, green), SDC4 (l, green) in human peri-implant fibrotic tissue and its overlap with LEPR+ cells (red). Scale bar, 50 μm. Images in a–l are representative of at least 3 independent biological replicates.
Extended Data Fig. 3 A subset of LEPR+ cells in the peri-implant fibrotic tissue expresses Cxcl12-GFP.
a, Expression of LepR-tdTomato and Cxcl12-GFP in the peri-implant fibrotic tissue of LepRcre;Rosa26tdTomato; Cxcl12GFP mice underwent fibrous-integrated surgery at postoperative day 14. A subset of LEPR+ cells (red) co-localize with cells expressing Cxcl12 (green). Scale bar, 500 μm. b-e, Enlarged view of the outlined blue box of figure in panel a. Scale bar, 50 μm. Images in a-e are representative of at least 3 independent experiments. f, Expression of LepR-tdTomato and Cxcl12-GFP in the peri-implant osseous tissue of LepRcre;Rosa26tdTomato; Cxcl12GFP mice underwent osseointegrated surgery at postoperative day 14. A subset of LEPR+ cells (red) in the perivascular area co-localizes with cells expressing Cxcl12 (green). Scale bar, 500 μm. g–j, Enlarged view of the outlined blue box of figure in panel f. Scale bar, 50 μm. Images in f-j are representative of at least 3 independent experiments. k–l, Expression of CXCL12-GFP by Lin-LepR-tdTomato+ (k) or osseointegrated surgery (l). m-n, Immunofluorescence quantification of expression of and Cxcl12-GFP in LepRcre;Rosa26tdTomato; Cxcl12GFP mice underwent fibrous-integrated or osseointegrated surgery at postoperative day 14. Data are mean ± s.d. n = 11 for both osseointegrated and fibrous-integrated model. n, Immunofluorescence quantification of co-localization between of LepR-tdTomato and Cxcl12-GFP in LepRcre;Rosa26tdTomato; Cxcl12GFP mice underwent fibrous-integrated or osseointegrated surgery at postoperative day 14. Unpaired, two-tailed Student’s t-test. Data are mean ± s.d. n = 11 for both osseointegrated and fibrous-integrated model. Each dot in m and p corresponds to biologically independent replicates.
Extended Data Fig. 4 Small percentage of Lin– 6C3– CD90–CD200+ CD105– (mSSC) and Lin– 6C3–CD90– CD200var CD105+ (BCSP) are Lin– LepR– tdTomato+.
a, Schematic of flow cytometry comparison experiment between LepRcre;Rosa26tdtomato mice underwent fibrous-integrated or osseointegrated implantation surgery at postoperative day 14. b-d, Schematic representation of the strategy used for FACS analysis of Lin– 6C3– CD90– CD200+ CD105– (mSSC) and Lin-6C3-CD90-CD200varCD105+ (BCSP)of mice underwent fibrous integrated surgery (c) or osseointegrated surgery (d). e–f, There is no difference in abundance of mSSC and BCSP in the peri-implant area between fibrous- and osseointegrated mice. Data are mean ± s.d. Unpaired, two-tailed Student’s t-test. n = 8 for fibrous-integrated model and n = 9 for osseointegrated model. g, TdTomato+ subset of Lin– 6C3– CD90– CD200+ CD105– are more abundant in fibrous-integrated peri-implant area than in osseointegrated peri-implant area. Data are mean ± s.d. Unpaired, two-tailed Student’s t-test. n = 8 for fibrous-integrated model and n = 9 for osseointegrated mode. h, There is no statistically significant difference in the abundance of tdTomato+ subset of Lin-6C3– CD90– CD200var CD105+ in fibrous-integrated peri-implant and in osseointegrated peri-implant area. Unpaired, two-tailed Student’s t-test. Data are mean ± s.d. n = 8 for fibrous-integrated model and n = 9 for osseointegrated model. i, LepR– tdTomato+ cells in fibrous-integrated interface were immunostained for CD200 demonstrating presence of LepR-TdTomato+ CD200+ at the bone-fibrous tissue interface. Scale bar, 500 μm. Middle column and right column enlarged view of the outlined yellow box. Scale bar, 50 μm. j, LepR– tdTomato+ cells in osseointegrated interface were immunostained for CD200 demonstrating presence of LepR– TdTomato+ CD200+ at the perivascular area. Scale bar, 500 μm. Middle column and right column, enlarged view of the outlined yellow box. Scale bar, 50 μm. Each dot in e–h corresponds to biologically independent replicates. Images in b–d, i–j are representative of at least 3 independent experiments.
Extended Data Fig. 5 A subset of LEPR+ cells in the peri-implant.
a, Expression of LepR-zsGreen and Acta2-RFP in the peri-implant fibrotic tissue of LepRcre;Rosa26Zsgreen; Acta2mRFP mice underwent fibrous-integrated surgery at postoperative day 14. A subset of LEPR+ cells (green) co-localize with cells expressing Acta2 (red). Scale bar, 500 μm. b-e, Enlarged view of the outlined yellow box of the figure in panel a. Scale bar, 50 μm. Images in a-e are representative of at least 3 independent experiments. f, Expression of LepR-zsGreen and Acta2-RFP in the peri-implant osseous tissue of LepRcre;Rosa26Zsgreen; Acta2mRFP mice underwent osseointegrated surgery at postoperative day 14. A subset of LEPR+ cells (green) in the perivascular area co-localize with cells expressing Acta2 (red). Scale bar, 500 μm. Images in a are representative of at least 3 independent experiments. g–j, Enlarged view of the outlined yellow box of figure in panel f. Scale bar, 50 μm. Images in f–j are representative of at least 3 independent experiments. k–l, Representation of the strategy used for FACS analysis of expression of LepR-zsGreen and Acta2-RFP in Lin-6C3-CD90-CD200+ CD105– fibrous integrated surgery (k) or osseointegrated surgery (l). LepRcre;Rosa26Zsgreen; Acta2mRFP (red), LepRcre;Rosa26Zsgreen (blue). m-n, Immunofluorescence quantification of expression of LepR-zsGreen and Acta2-RFP in LepRcre;Rosa26Zsgreen; Acta2mRFP mice underwent fibrous-integrated or osseointegrated surgery at postoperative day 14. Data are mean ± s.d. n = 5 for both osseointegrated and fibrous-integrated model. Unpaired, two-tailed Student’s t-test. o, Immunofluorescence quantification of co-localization between LepR-zsGreen and Acta2-RFP in LepRcre;Rosa26Zsgreen; Acta2mRFP mice underwent fibrous-integrated or osseointegrated surgery at postoperative day 14. Data are mean ± s.d. n = 5 for both osseointegrated and fibrous-integrated model; Unpaired, tailed Student’s t-test. p, Flow cytometry quantification of relative abundance of Lin-LepR-Zsgreen+ Acta2-RFP+ 6C3–CD90– CD200+ CD105 in LepRcre;Rosa26Zsgreen; Acta2mRFP mice underwent fibrous-integrated or osseointegrated surgery at postoperative day 14. Data are mean ± s.d. n = 5 for both osseointegrated and fibrous-integrated model; Unpaired, two-tailed Student’s t-test. Each dot in m-p corresponds to biologically independent replicates. Images in k-l are representative of at least 3 independent experiments.
Extended Data Fig. 6 Peri-implant fibrotic tissue originate from osteogenic LEPR+ cells.
a–c, Immunofluorescent imaging of proximal tibia of LepRcre;Rosa26tdtomato, BGLAP-GFP that underwent fibrous-integration surgery at post-operative day 3 (a), 7 (b), and 14 (c). Scale bar, 50 μm. d–f, Immunofluorescent imaging of ALPL-stained proximal tibia of LepRcre;Rosa26tdtomato, BGLAPGFP that underwent fibrous-integration surgery at post-operative day 3 (d), 7 (e), and 14 (f). Scale bar, 50 μm. g, Violin plots showing the expression density levels of Alpl and Bglap showed that they are selectively expressed by osteogenic (OLC1, OLC2) and chondrogenic (CLC) cells. h, Immunofluorescence quantification of co-localization between LepR-tdTomato and BGLAP-GFP in LepRcre;Rosa26tdtomato, BGLAPGFP mice underwent fibrous-integrated surgery at postoperative day 3, 7, and 14. Data are mean ± s.d. n = 6 for both osseointegrated and fibrous-integrated model; Unpaired, tailed Student’s t-test. i, Immunofluorescence quantification of co-localization between LepR-tdTomato and ALPL antibody staining in LepRcre;Rosa26tdtomato, BGLAPGFP mice underwent fibrous-integrated surgery at postoperative day 3, 7, and 14. Data are mean ± s.d. n = 6 for both osseointegrated and fibrous-integrated model; Unpaired, tailed Student’s t-test. j, Immunofluorescence quantification of tdTomato+ cells from CXCL12creERT2;Rosa26tdtomato, AdipoqcreERT2;Rosa26tdtomato and LepRcre;Rosa26tdtomato that underwent fibrous-integration surgery. N = 7 per group. Data are mean ± s.d.; two-tailed unpaired t-test. k-m, Immunofluorescent imaging of CXCL12creERT2;Rosa26tdtomato (k), AdipoqcreERT2;Rosa26tdtomato (l), and LepRcre;Rosa26tdtomato (m) that underwent fibrous-integration surgery at post-operative day 14. Each dot in h-j corresponds to biologically independent replicates. Images in a–f, k–m are representative of at least 3 independent experiments.
Extended Data Fig. 7 Peri-implant fibrosis is persistent up to at least 16 weeks in murine model of peri-implant fibrosis.
a, Micro-computed tomography (μCT) of proximal tibia of LepRcre;Rosa26tdtomato underwent fibrous-integrated surgery at post-operative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Scale bar, 500 μm. b, Hematoxylin and eosin staining of proximal tibia of LepRcre;Rosa26tdtomato underwent fibrous-integrated surgery at post-operative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Scale bar, 500 μm. c, Immunofluorescent imaging proximal tibia of LepRcre;Rosa26tdtomato underwent fibrous-integrated surgery at post-operative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Scale bar, 500 μm. Bottom row, enlarged view of the outlined yellow box at each time-point. Scale bar, 50 μm. d, Bone volume/total volume (BV/TV)) of mice at post-operative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Data are mean ± s.d. n = 6. e, Histology quantification of peri-implant fibrosis (% Fibrosis) and peri-implant bone (% bone) of mice at at post-operative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Data are mean ± s.d. n = 6. f, Immunofluorescent quantification of peri-implant fibrosis (% Fibrosis) and peri-implant bone (% bone) of mice at at post-operative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Data are mean ± s.d. n = 6. Each dot in d-f corresponds to biologically independent replicates. Images in a–c are representative of at least 3 independent experiments.
Extended Data Fig. 8 Analysis of peri-implant bone of mice underwent osseointegrated surgery at postoperative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks.
a, Micro-computed tomography (μCT) of proximal tibia of LepRcre;Rosa26tdtomato underwent osseointegrated surgery at post-operative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Scale bar, 500 μm. b, Haematoxylin and eosin staining of proximal tibia of LepRcre;Rosa26tdtomato underwent osseointegrated surgery at postoperative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Scale bar, 500 μm. c, Immunofluorescent imaging proximal tibia of LepRcre;Rosa26tdtomato underwent osseointegrated surgery at postoperative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Scale bar, 500 μm. Bottom row, enlarged view of the outlined yellow box at each time-point. Scale bar, 50 μm. d, Bone volume/total volume (BV/TV) of mice at postoperative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Data are mean ± s.d. n = 6. e, Histology quantification of peri-implant fibrosis (% Fibrosis) and peri-implant bone (% bone) of mice at postoperative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Data are mean ± s.d. n = 6; 3 independent experiments. f, Immunofluorescent quantification of peri-implant fibrosis (% Fibrosis) and peri-implant bone (% bone) of mice at postoperative day 3, day 7, 2 weeks, 4 weeks, 8 weeks, and 16 weeks. Data are mean ± s.d. n = 6. Each dot in d-f corresponds to biologically independent replicates. Images in a-c are representative of at least 3 independent experiments.
Extended Data Fig. 9 Human mSSC (Lin– PDPN+ CD146– CD164+ CD73+) are more abundant in bone than in fibrous membranes, but Lin– LEPR+ are more abundant in fibrous membranes than in bone.
a, Schematic representation of the strategy used for FACS analysis to obtain Lin- population. b–c, Schematic representation of the strategy used for FACS analysis to obtain LEPR expressing a subset of Lin– PDPN+ CD146– CD164+ CD73+ from bone (b) or from fibrous tissue (c). d, Flow cytometry quantification of human fibrous tissue and bone demonstrates human mSSC (Lin– PDPN+ CD146– CD164+ CD73+) are more abundant in bone than in fibrous membrane. Data are mean ± s.d. n = 10 patients for fibrous membrane and n = 10 for bone patients. Unpaired, two-tailed Student’s t-test. e, Flow cytometry quantification of human fibrous tissue and bone demonstrates the non-significant difference in osteolineage progenitor (Lin–PDPN+ CD146+) between fibrous membrane and bone. Data are mean ± s.d. n = 10 for fibrous membrane and n = 10 for bone. Unpaired, two-tailed Student’s t-test. f, Flow cytometry quantification of human fibrous tissue and bone demonstrates significantly higher amount of Lin– LEPR+ PDPN+ CD146– in the fibrous membrane than in bone. Data are mean ± s.d. n = 10 patients for fibrous membrane and n = 10 patients for bone. Unpaired, two-tailed Student’s t-test. Each dot in d-f corresponds to biologically independent replicates. Images in a-c¬ are representative of at least 3 independent experiments.
Extended Data Fig. 10 Administration of neutralizing antibody against reduces LepR– tdTomato+ cells but does not affect Lin– 6C3– CD90– CD200+ CD105– (mSSC) subpopulation.
a, Immunofluorescent imaging of CD200-antibody stained. proximal tibia of LepRcre;Rosa26tdtomato receiving either anti-ADGRF5 or isotype control as prophylaxis. Scale bar, 500 μm. Right column, enlarged view of the outlined yellow box. Scale bar, 50 μm. b, Immunofluorescent imaging of CD200-antibody-stained proximal tibia of LepRcre;Rosa26tdtomato receiving either anti-ADGRF5 or isotype control as treatment starting from postoperative day 14. Scale bar, 500 μm. Right column, enlarged view of the outlined yellow box. Scale bar, 50 μm. c,f FACS analysis of TdTomato+ subset of Lin-6C3– CD90–CD200+ CD105– (TdTomato+ mSSC) in mice group receiving anti-ADGRF5 as compared to isotype control, either as prophylaxis (c) or as treatment (f). Anti-ADGRF5 (n = 5), isotype control (n = 5). Unpaired, two-tailed Student’s t-test. Data are mean ± s.d. d,g, FACS analysis of Lin– 6C3– CD90– CD200+ CD105– (mSSC) between mice receiving daily anti-ADGRF5 or isotype control either as prophylaxis (d) or as treatment (f). Anti-ADGRF5 (n = 5), isotype control (n = 5). ns= not significant. Unpaired, two-tailed Student’s t-test. treatment (f). Anti-ADGRF5 (n = 5), isotype control (n = 5). Unpaired, two-tailed Student’s t-test. Data are mean ± s.d. e, Host bone-implant failure load of LepRcre;Rosa26tdtomato receiving either daily anti-ADGRF5 or isotype control as prophylaxis at postoperative day 28. n = 5 for mice receiving anti-ADGRF5 and n = 5 for mice receiving isotype control. Unpaired, two-tailed Student’s t-test. Data are mean ± s.d. h, Host bone-implant failure load of LepRcre;Rosa26tdtomato receiving either daily anti-ADGRF5 or isotype control as treatment starting from postoperative day 14 and evaluated at postoperative day 28. n = 5 for mice receiving anti-ADGRF5 and n = 5 for mice receiving isotype control. Unpaired, two-tailed Student’s t-test. Data are mean ± s.d. Images in a-b are representative of at least 3 independent biological replicates. Each dot in c–e and f–h corresponds to biologically independent replicates.
Supplementary information
Source data
Source Data Figs. 1–8 and Extended Data Figs. 1–10
Source data for all figures.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suhardi, V.J., Oktarina, A., Hammad, M. et al. Prevention and treatment of peri-implant fibrosis by functionally inhibiting skeletal cells expressing the leptin receptor. Nat. Biomed. Eng 8, 1285–1307 (2024). https://doi.org/10.1038/s41551-024-01238-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41551-024-01238-y