Abstract
Chimeric antigen receptor (CAR) T cell therapy for solid tumours is challenging because of the immunosuppressive tumour microenvironment and a complex manufacturing process. Cellular manufacturing protocols directly impact CAR T cell yield, phenotype and metabolism, which correlates with in vivo potency and persistence. Although metabolic fitness is a critical quality attribute, how T cell metabolic requirements vary throughout the manufacturing process remains unexplored. Here we use optical metabolic imaging (OMI), a non-invasive, label-free method to evaluate single-cell metabolism. Using OMI, we identified the impacts of media composition on CAR T cell metabolism, activation strength and kinetics, and phenotype. We demonstrate that OMI parameters can indicate cell cycle stage and optimal gene transfer conditions for both viral transduction and electroporation-based CRISPR/Cas9. In a CRISPR-edited anti-GD2 CAR T cell model, OMI measurements allow accurate prediction of an oxidative metabolic phenotype that yields higher in vivo potency against neuroblastoma. Our data support OMI as a robust, sensitive analytical tool to optimize manufacturing conditions and monitor cell metabolism for increased CAR T cell yield and metabolic fitness.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study have been deposited and are available via Zenodo at https://doi.org/10.5281/zenodo.15628321 (ref. 68).
Code availability
All relevant analysis codes have been deposited via Zenodo at https://doi.org/10.5281/zenodo.15628321 (ref. 68).
References
Melenhorst, J. J. et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature 602, 503–509 (2022).
Guzman, G., Reed, M. R., Bielamowicz, K., Koss, B. & Rodriguez, A. CAR-T therapies in solid tumors: opportunities and challenges. Curr. Oncol. Rep. 25, 479–489 (2023).
Stefanski, H. E. et al. Higher doses of tisagenlecleucel are associated with improved outcomes: a report from the pediatric real-world CAR consortium. Blood Adv. 7, 541–548 (2023).
Xu, H. et al. Influence of various medium environment to in vitro human T cell culture. In Vitro Cell. Dev. Biol. Anim. 54, 559–566 (2018).
Li, Y. & Kurlander, R. J. Comparison of anti-CD3 and anti-CD28-coated beads with soluble anti-CD3 for expanding human T cells: differing impact on CD8 T cell phenotype and responsiveness to restimulation. J. Transl. Med. 8, 104 (2010).
Fesnak, A. D. The challenge of variability in chimeric antigen receptor T cell manufacturing. Regen. Eng. Transl. Med. 6, 322–329 (2020).
Abou-el-Enein, M. et al. Scalable manufacturing of CAR T cells for cancer immunotherapy. Blood Cancer Discov. 2, 408–422 (2021).
Center for Biologics Evaluation and Research. FDA Investigating Serious Risk of T-cell Malignancy Following BCMA-Directed or CD19-Directed Autologous Chimeric Antigen Receptor (CAR) T cell Immunotherapies (FDA, 2023).
Mueller, K. P. et al. Production and characterization of virus-free, CRISPR-CAR T cells capable of inducing solid tumor regression. J. Immunother. Cancer 10, e004446 (2022).
Hu, Y. et al. Safety and efficacy of CRISPR-based non-viral PD1 locus specifically integrated anti-CD19 CAR-T cells in patients with relapsed or refractory non-Hodgkin’s lymphoma: a first-in-human phase I study. eClinicalMedicine 60, 102010 (2023).
Balke-Want, H. et al. Homology-independent targeted insertion (HITI) enables guided CAR knock-in and efficient clinical scale CAR-T cell manufacturing. Mol. Cancer 22, 100 (2023).
Caruso, H. G. et al. Shortened ex vivo manufacturing time of EGFRvIII-specific chimeric antigen receptor (CAR) T cells reduces immune exhaustion and enhances antiglioma therapeutic function. J. Neurooncol. 145, 429–439 (2019).
Ghassemi, S. et al. Reducing ex vivo culture improves the antileukemic activity of chimeric antigen receptor (CAR) T cells. Cancer Immunol. Res. 6, 1100–1109 (2018).
Shin, J. J. et al. Controlled cycling and quiescence enables efficient HDR in engraftment-enriched adult hematopoietic stem and progenitor cells. Cell Rep. 32, 108093 (2020).
Smirnikhina, S. A., Zaynitdinova, M. I., Sergeeva, V. A. & Lavrov, A. V. Improving homology-directed repair in genome editing experiments by influencing the cell cycle. Int. J. Mol. Sci. 23, 5992 (2022).
Watanabe, N., Mo, F. & McKenna, M. K. Impact of manufacturing procedures on CAR T cell functionality. Front. Immunol. 13, 876339 (2022).
Wang, F., Cheng, F. & Zheng, F. Stem cell like memory T cells: a new paradigm in cancer immunotherapy. Clin. Immunol. 241, 109078 (2022).
López-Cantillo, G., Urueña, C., Camacho, B. A. & Ramírez-Segura, C. CAR-T cell performance: how to improve their persistence? Front. Immunol. 13, 878209 (2022).
Zhou, J. et al. Chimeric antigen receptor T (CAR-T) cells expanded with IL-7/IL-15 mediate superior antitumor effects. Protein Cell 10, 764–769 (2019).
Arner, E. N. & Rathmell, J. C. Metabolic programming and immune suppression in the tumor microenvironment. Cancer Cell 41, 421–433 (2023).
Kao, K.-C., Vilbois, S., Tsai, C.-H. & Ho, P.-C. Metabolic communication in the tumour–immune microenvironment. Nat. Cell Biol. 24, 1574–1583 (2022).
Nanjireddy, P. M., Olejniczak, S. H. & Buxbaum, N. P. Targeting of chimeric antigen receptor T cell metabolism to improve therapeutic outcomes. Front. Immunol. 14, 1121565 (2023).
van Bruggen, J. A. C. et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8+ T cells and impede CAR T-cell efficacy. Blood 134, 44–58 (2019).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013).
Klein Geltink, R. I. et al. Metabolic conditioning of CD8+ effector T cells for adoptive cell therapy. Nat. Metab. 2, 703–716 (2020).
Walsh, A. J. et al. Classification of T-cell activation via autofluorescence lifetime imaging. Nat. Biomed. Eng. 5, 77–88 (2021).
Walsh, A. J. et al. Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res. 73, 6164–6174 (2013).
Chance, B., Legallais, V. & Schoener, B. Metabolically linked changes in fluorescence emission spectra of cortex of rat brain, kidney and adrenal gland. Nature 195, 1073–1075 (1962).
Skala, M. C. et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl Acad. Sci. USA 104, 19494–19499 (2007).
Georgakoudi, I. & Quinn, K. P. Optical imaging using endogenous contrast to assess metabolic state. Annu. Rev. Biomed. Eng. 14, 351–367 (2012).
Quinn, K. P. et al. Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci. Rep. 3, 3432 (2013).
Shah, V. S. et al. Autofluorescence imaging permits label-free cell type assignment and reveals the dynamic formation of airway secretory cell associated antigen passages (SAPs). eLife 12, e84375 (2023).
Neuroblastoma survival rates. (American Cancer Society, 2025); https://www.cancer.org/cancer/types/neuroblastoma/detection-diagnosis-staging/survival-rates.html
Richards, R. M., Sotillo, E. & Majzner, R. G. CAR T cell therapy for neuroblastoma. Front. Immunol. 9, 2380 (2018).
Del Bufalo, F. et al. GD2-CART01 for relapsed or refractory high-risk neuroblastoma. N. Engl. J. Med. 388, 1284–1295 (2023).
MacPherson, S. et al. Clinically relevant T cell expansion media activate distinct metabolic programs uncoupled from cellular function. Mol. Ther. Methods Clin. Dev. 24, 380–393 (2022).
Sudarsanam, H., Buhmann, R. & Henschler, R. Influence of culture conditions on ex vivo expansion of T lymphocytes and their function for therapy: current insights and open questions. Front. Bioeng. Biotechnol. 10, 886637 (2022).
Pearce, E. L. Metabolism in T cell activation and differentiation. Curr. Opin. Immunol. 22, 314–320 (2010).
Wei, J., Raynor, J., Nguyen, T.-L. M. & Chi, H. Nutrient and metabolic sensing in T cell responses. Front. Immunol. 8, 247 (2017).
Yang, Y. & Sauve, A. A. NAD+ metabolism: bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta 1864, 1787–1800 (2016).
Hu, Y. et al. CRISPR/Cas9-engineered universal CD19/CD22 dual-targeted CAR-T cell therapy for relapsed/refractory B-cell acute lymphoblastic leukemia. Clin. Cancer Res. 27, 2764–2772 (2021).
Khan, A. & Sarkar, E. CRISPR/Cas9 encouraged CAR-T cell immunotherapy reporting efficient and safe clinical results towards cancer. Cancer Treat. Res. Commun. 33, 100641 (2022).
Cappabianca, D. et al. Metabolic priming of GD2 TRAC-CAR T cells during manufacturing promotes memory phenotypes while enhancing persistence. Mol. Ther. Methods Clin. Dev. 32, 101249 (2024).
Xu, X., Gnanaprakasam, J. N. R., Sherman, J. & Wang, R. A metabolism toolbox for CAR T therapy. Front. Oncol. 9, 322 (2019).
Nabe, S. et al. Reinforce the antitumor activity of CD8+ T cells via glutamine restriction. Cancer Sci. 109, 3737–3750 (2018).
Xu, L. et al. PD-1 and TIGIT are highly co-expressed on CD8+ T cells in AML patient bone marrow. Front. Oncol. 11, 686156 (2021).
Banta, K. L. et al. Mechanistic convergence of the TIGIT and PD-1 inhibitory pathways necessitates co-blockade to optimize anti-tumor CD8+ T cell responses. Immunity 55, 512–526.e9 (2022).
Zhao, M.-H., Kim, N.-H. & Cui, X.-S. GlutaMAX prolongs the shelf life of the culture medium for porcine parthenotes. Theriogenology 85, 368–375 (2016).
Kalucka, J. et al. Metabolic control of the cell cycle. Cell Cycle 14, 3379 (2015).
Colombo, S. L. et al. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc. Natl Acad. Sci. USA 108, 21069–21074 (2011).
Heaster, T. M., Walsh, A. J., Zhao, Y., Hiebert, S. W. & Skala, M. C. Optical metabolic imaging identifies tumor cell-cycle status on a single-cell level. J. Biophotonics https://doi.org/10.1002/jbio.201600276 (2018).
Fichter, K. M., Setayesh, T. & Malik, P. Strategies for precise gene edits in mammalian cells. Mol. Ther. Nucleic Acids 32, 536–552 (2023).
Andreadis, S., Fuller, A. O. & Palsson, B. O. Cell cycle dependence of retroviral transduction: an issue of overlapping time scales. Biotechnol. Bioeng. 58, 272–281 (1998).
Verhoeyen, E., Costa, C. & Cosset, F.-L. Lentiviral vector gene transfer into human T cells. In Genetic Modification of Hematopoietic Stem Cells: Methods and Protocols (ed. Baum, C.) 97–114 (Humana Press, 2009).
Yang, D. et al. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 6, 21264 (2016).
Eghbalsaied, S. & Kues, W. A. CRISPR/Cas9-mediated targeted knock-in of large constructs using nocodazole and RNase HII. Sci. Rep. 13, 2690 (2023).
Akbari, B. et al. Metabolic and epigenetic orchestration of (CAR) T cell fate and function. Cancer Lett. 550, 215948 (2022).
Li, W. et al. Cell metabolism-based optimization strategy of CAR-T cell function in cancer therapy. Front. Immunol. 14, 1186383 (2023).
Long, A. H., Orentas, R. J. & Mackall, C. L. Synthetic chimeric antigen receptors (CARs) rapidly induce exhaustion and augmented glycolytic metabolism in human T cells and implicate persistent CD28 signaling as a driver of exhaustion in human T cells. Blood 122, 192 (2013).
Scharping, N. E. et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016).
DePeaux, K. & Delgoffe, G. M. Metabolic barriers to cancer immunotherapy. Nat. Rev. Immunol. 21, 785–797 (2021).
Samimi, K. et al. Autofluorescence lifetime flow cytometry with time-correlated single photon counting. Cytometry A 105, 607–620 (2024).
Samimi, K. et al. Time-domain single photon-excited autofluorescence lifetime for label-free detection of T cell activation. Opt. Lett. 46, 2168–2171 (2021).
Samimi, K. et al. Light-sheet autofluorescence lifetime imaging with a single-photon avalanche diode array. J. Biomed. Opt. 28, 066502 (2023).
Gillette, A. A., Pham, D. L. & Skala, M. C. Touch-free optical technologies to streamline the production of T cell therapies. Curr. Opin. Biomed. Eng. 25, 100434 (2023).
Lakowicz, J. R., Szmacinski, H., Nowaczyk, K. & Johnson, M. L. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl Acad. Sci. USA 89, 1271–1275 (1992).
Datta, R., Heaster, T. M., Sharick, J. T., Gillette, A. A. & Skala, M. C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 25, 1–43 (2020).
Pham, D. Label-free metabolic imaging monitors the fitness of chimeric antigen receptor T cells. Zenodo https://doi.org/10.5281/zenodo.15628321 (2025).
Acknowledgements
We thank members of the Capitini, Saha and Skala labs for helpful discussion and comments on the paper, the University of Wisconsin (UW) Carbone Cancer Center Small Animal Imaging and Radiotherapy facility and Flow Cytometry Laboratory (supported by NIH P30 CA014520 and NIH S10 OD025225), M. Brenner (Baylor College of Medicine) for the retroviral 14G2a-OX40-CD28-ζ CAR sequence and C. Mackall (Stanford University) for the retroviral 14G2a-41BB-ζ CAR sequence, the National Cancer Institute for 1A7 antibody for detection of CAR expression, M. Otto (UW–Madison) for the parental CHLA20 cell line, and J. Thomson and J. Zhang (Morgridge Institute for Research) for the AkaLUC-GFP CHLA20 cell line used for in vivo studies. We thank M. Stefely and A. Williams for graphic and paper edits. M.C.S., K. Saha and C.M.C. disclose support for the research described in this study from NIH R01 CA278051, NSF Engineering Research Center (ERC) for Cell Manufacturing Technologies (CMaT) and NSF-EEC 1648035 (C.M.C., K. Saha and M.C.S.). C.M.C. and K. Saha disclose support from St. Baldrick’s Foundation Empowering Pediatric Immunotherapy for Childhood Cancers Team grant, UW–Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation, Hyundai Hope on Wheels (C.M.C. and K. Saha) and Grainger Institute for Engineering at UW–Madison. C.M.C. discloses support for the research in this publication from MACC Fund. K. Saha discloses funding support from NIH R35 GM119644-01.
Author information
Authors and Affiliations
Contributions
D.L.P. and D.C. contributed equally. D.L.P. and C.W. performed the OMI imaging and analysis of T cell samples, and performed the T cell isolation and activation. D.C., A.T. and M.B. isolated and cultured T cells and generated electroporation-based CRISPR-edited CAR T cells. K.P.M. performed viral transductions. M.H.F. performed the mouse study. J.V., A.L. and K. Sylvester performed the ATP production and cellular reducing potential analysis on activated T cells. J.L. performed the metabolomics analysis of two media and the IncuCyte analysis. D.L.P., D.C., M.H.F., K.P.M., C.W., A.T., J.V., A.L., K. Sylvester, J. L. and M.B. performed the experiments and analysed the data. D.L.P. and M.C.S. wrote the paper with inputs from all authors. C.M.C., J.F., K. Saha and M.C.S. supervised the research.
Corresponding authors
Ethics declarations
Competing interests
K. Saha receives honoraria for advisory board membership for Andson Biotech and Notch Therapeutics. C.M.C. receives honoraria for advisory board membership for Bayer, Elephas Biosciences, Nektar Therapeutics, Novartis and WiCell Research Institute. M.C.S., D.L.P., K. Saha and D.C. declare two patents pending based on this work (WO2024229098A3, WO2024229099A1). M.C.S. is an advisory board member for Elephas Biosciences. No other conflicts of interest are reported.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 OMI and cofactor/enzyme activity assessment revealed the impact of culture media on activation kinetics and metabolism of T cells stimulated with aCD3/aCD28 Dynabeads.
(a–e) Liquid chromatography-mass spectrometry analysis revealed significantly different composition in ImmunoCult XF and TexMACs media. Concentration of (a) glucose, (b) glutamine, (c) glutamax, (d) pyruvate, and (e) arginine quantified in ImmunoCult XF and TexMACs media. n = 3 replicates, two-sided unpaired t test. (f) Representative NAD(P)H τm images from T cells activated with αCD3/αCD28 Dynabeads at 1:1 and 2:1 bead-to-T cell ratio in ImmunoCult XF and TexMACs media. αCD3/αCD28 Dynabeads were removed immediately prior to imaging. (g, h) T cells activated in ImmunoCult XF and TexMACs media demonstrated different activation kinetics. NAD(P)H α1 quantification of T cells from two healthy donors activated with αCD3/αCD28 Dynabeads in (g) ImmunoCult XF and (h) TexMACs media at 24-, 48-, and 72-hours post activation. For (g) n = 449, 437, and 432 activated T cells from 3 donors in ImmunoCults media at 24, 48, and 72 hours. For (h), n = 475, 446, and 410 activated T cells from 3 donors in TexMACs media at 24, 48, and 72 hours. For (g, h) two-way ANOVA with Tukey’s posthoc test for multiple comparisons. (i, j) UMAP based on Euclidean distances of 11 OMI parameters (NAD(P)H τm, τ1, τ2, α1, intensity, FAD τm, τ1, τ2, α1, intensity, redox ratio) and cell size of T cells from two healthy donors activated with αCD3/αCD28 Dynabeads, color coded based on (i) culture media and (j) bead-to-T cell ratio. n = 2649 cells. (k) NAD/NADH ratio, and fold increase in (l) NADH concentration, (m) ATP production, (n) LDH, o) GAPDH, and p) IDH activity measured from T cells activated with αCD3/αCD28 Dynabeads (at 1:1 and 3:1 bead-to-T cell ratio), StemCell, or TransAct antibodies for 72 hours in ImmunoCult XF or TexMACs media. The fold increase was calculated as compared to quiescent cells. n = 3 replicates/condition, two-way ANOVA with Sidak’s posthoc test for multiple comparisons. Scale bar is 50 μm. Bars are mean ± SD. * p < 0.0001.
Extended Data Fig. 2 T cells progressed through cell cycle and proliferated upon activation.
(a) Experimental design and timeline. CD3 T cells were isolated from 3 healthy donors and activated with StemCell αCD2/αCD3/αCD28 in ImmunoCult XF media (Imm) or TransAct αCD3/αCD28 in TexMACS media (Tex). Cells were divided into 7 groups with duration of activation ranging from 0 hours (quiescent T cells) to 72 hours prior to electroporation to introduce the anti-GD2 CAR transgene. When activated T cells were collected for electroporation, their metabolic features were characterized using OMI. Meanwhile, their cell cycle stage and proliferation capacity were assessed using flow cytometry of Hoechst 33342 and Ki-67 staining. These activated T cells were then electroporated to incorporate the anti-GD2 CAR transgene and expanded in corresponding culture media. Genome editing efficiency was determined after 7 days of expansion with GD2 CAR antibody using flow cytometry. (b, c) T cells progressed through cell cycle upon activation. Density plots of Hoechst MFI in T cells activated with (b) Imm and (c) Tex methods. For each donor, Hoechst MFI was normalized to the average of 12-hour-activated group. n = 393,999 cells and 370,802 cells for Imm and Tex activation methods, respectively. (d) Correlation between NAD(P)H α1 and cell cycle stage (% cells in S/G2/M phase) at electroporation. Each dot represents one sample average, color coded based on the method of activation (Imm or Tex) and duration of activation (0–72 hours). n = 42 samples, Pearson R analysis. (e, f) T cell proliferation (% Ki-67+) following activation with (e) Imm method or (f) Tex method. n = 3 donors, Brown-Forsythe and Welch ANOVA test with Dunnett’s post hoc test for multiple comparisons. Bars are mean ± SD. Color lines connected donor-match averages.
Extended Data Fig. 3 T cell characteristics at the time of CAR gene transfer correlated with transgene incorporation efficiency.
(a) Experimental setup for viral transduction experiment. T cells were isolated from peripheral blood of two healthy donors and activated with Imm method. Activated CD3 T cells underwent electroporation with Cas9 ribonucleoproteins targeting the human TRAC locus to knockout the T cell receptor (TRAC KO) 2 days prior to transduction with retrovirus to express anti-GD2 CAR receptor using two constructs (OX40-CD28-CAR or 41BB-CAR) as previously described9. OMI was performed immediately before transduction. Transduction efficiency was quantified as %CAR+ after 7 days of expansion. (b) Representative NAD(P)H τm images of TCR-intact and TRAC KO T cells at transduction. OMI parameters including (c) NAD(P)H τm, (d) NAD(P)H α1, and (e) cytoplasm size of cells with intact T cell receptor (TCR-intact) and TRAC KO T cells at transduction. n = 480 TCR intact and 374 TRAC KO cells from 2 independent donors, two-sided Mann-Whitney test. (f) Transduction efficiency (% CAR) of TCR-intact and TRAC KO cells at the end of the manufacturing process. n = 17 samples from 2 donors and 2 anti-GD2 CAR constructs, unpaired two-sided T test. (g) Experimental timeline of electroporation experiment. (h–k) Correlation between cell characteristics (h) NAD(P)H α1, (i) normalized redox ratio, (j) %S/G2/M and (k) %Ki-67+ at electroporation and genome editing outcome (% CAR). Each dot represents one sample average, color coded based on the method of activation (Imm or Tex) and duration of activation (0–72 hours). n = 36 samples, Pearson R analysis. Quiescent T cells with 0-hour activation duration did not undergo genome editing. Bars are mean ± SD. Color lines connected donor-match averages. * p < 0.0001.
Extended Data Fig. 4 CAR T cells expanded in Imm and Tex media displayed distinct metabolic and phenotypic features.
(a) Experimental timeline. (b) Quantification of FAD mean lifetime (FAD τm) of CAR T cells expanded in either ImmunoCult XF or TexMACS media. n = 666 and 751 CAR T cells from 3 independent donors expanded in ImmunoCult XF and TexMACs media supplemented with various cytokine combinations, two-sided Mann-Whitney test. (c–e) OMI metabolic profiles of CAR+ T cells differed based on expansion media. (c) Representative image of CAR+ T cells identified based on PerCP conjugated GD-2 CAR antibody (red). (d) UMAP based on 13 OMI parameters of CAR+ T cells expanded in Imm or Tex media. (e) ROC curves and AUCs of three models based on OMI parameters (Table 1) to classify CAR+ T cells by expansion media (Imm vs. Tex); n = 494 cells for training (80%), n = 123 cells for testing (20%). (f) Quantification of NAD(P)H τm from CAR T cells expanded in ImmunoCult XF media supplemented with 500 U/mL IL-2 compared to other cytokine combinations, n = 123, 83, 96, and 129 CAR T cells expanded in IL-2, IL-7, IL-17, and IL-7 + IL-15(high) supplemented media from 2 donors, ANOVA with Kruskal-Wallis test. MFIs of (g) CCR7 and (h) CD62L of CAR+ T cells expanded in either Imm or Tex media supplemented with different cytokine cocktails, normalized by fluorescence-minus-one (FMO) controls. (n = 87,317 cells from 3 independent donors). UMAP based on Euclidean distances of MFIs of 6 surface markers (CD27, CD45RO, CD62L, CD28, CD45RA and CCR7) of CAR+ T cells expanded in Imm and Tex media, color coded based on (i) culture media or (j) supplemented cytokines. Dots are CAR+ T cells. n = 87,317 cells from 3 independent donors. Bars are mean ± SD. * p < 0.0001.
Extended Data Fig. 5 CAR T cell phenotypes and metabolic profile before in vivo treatment course.
(a) Experimental timeline. (b) Phenotypes of CAR+ T cells expanded in Imm, Tex→Imm, and Tex conditions based on expression of 7 surface markers. (c) Extracellular flux analysis of baseline oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) from CD3 T cells that were unactivated (quiescent) or activated and expanded in Imm or Tex→Imm condition. n = 9–18 samples/condition. Brown-Forsythe and Welch ANOVA tests with Dunnett’s T3 post hoc test for multiple comparisons. Bars are mean ± SD. * p < 0.0001.
Extended Data Fig. 6 Expansion conditions shaped T cell expansion capacity and metabolism.
(a, b) Imm and Tex-Imm T cells exhibited high expansion capacity. Fold expansion of T cells from two healthy donors under various expansion conditions (a) throughout and (b) on day 8 of the expansion process n = 4-6 replicates/condition across two donors, two way ANOVA test with Tukey’s posthoc test for multiple comparisons. (c) ATP production by T cells under several expansion conditions. (d–m) Media transition from TexMACs to ImmunoCult XF (Tex-Imm) shifted T cell metabolism towards balanced oxidative phosphorylation while maintaining ATP production. (d) Representative NAD(P)H τm images of T cells on day 9. (e) Quantification of NAD(P)H τm and (f) normalized NAD(P)H intensity of expanded T cells from two healthy donors on day 9. n = 361-580 cells/conditions across 2 donors, color coded by donors. (g, h) Metabolic cofactors, (i–m) dehydrogenases activity measured from expanded T cells of two healthy donors on day 9. n = 6 replicates across 2 donors. Brown-Forsythe and Welch ANOVA test with Dunnett’s T3 post hoc test for multiple comparisons. Bars are mean ± SD. * p < 0.0001.
Extended Data Fig. 7 In vivo treatment response and phenotyping analysis of CAR T cells post in vivo treatment.
(a) Experimental timeline. (b) Fold change in tumor flux (measured with IVIS imaging) in NSG tumor-bearing mice after CAR T cell treatment. Dash line represents no change in tumor flux (fold change = 1). (c) CAR T treatment outcomes on day 17 post treatment. (d–g) Expression of stem central memory (d, e) and exhaustion phenotypes (f, g) in CAR T cells isolated from treated NSG mouse spleen at day 21 post-treatment. (d, f) Representative flow histograms from one individual mouse in each treatment group. (e, g) Quantification of stem central memory (CD62 + CCR7+) and exhausted (PD-1 + TIGIT+) populations in treated mouse spleen. n = 12 mice, color coded by CAR T treatment groups. Two-sided non-parametric Mann-Whitney test. Bars are mean ± SD.
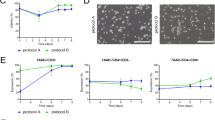
Extended Data Fig. 8 Tex→Imm CAR T cells demonstrated higher in vitro cytotoxicity than Imm CAR T cells against two cancer models at low effector-to-target ratios.
(a) Representative NAD(P)H τm images of donor-matched CAR T cells expanded in Imm and Tex-Imm conditions. (b, c) Quantification of NAD(P)H α1 and NAD(P)H τm from Imm and Tex-Imm CAR T cells at the end of manufacturing. (d–g) GFP + GD2 + M21 melanoma and (h–k) GFP + GD2 + MG63 osteosarcoma were cocultured with donor-matched GD2-CAR Imm and CAR Tex-Imm T cells at various effector-to-target ratios (1:1, 1:2, 1:5, and 1:10) for 72 hours. (d, e) Normalized GFP intensity and percentage of GFP+ cells in the coculture of CAR T cells and M21 melanoma throughout 72 hours and (f, g) at 72-hour timepoint. (h, i) Normalized GFP intensity and percentage of GFP+ cells in the coculture of CAR T cells and MG63 osteosarcoma throughout 72 hours and (j, k) at 72-hour timepoint. For (d, e) and (h, i) n = 2 well/condition x 72 time points, Friedman test (one-way repeated measures analysis of variance by ranks) with Dunn’s posthoc test for multiple comparisons. For (f, g) and (j, k), n = 2 replicates/condition, unpaired t-test. Bars are mean ± SD. * p < 0.0001. Scale bar is 50 μm.
Extended Data Fig. 9
Gating strategy for analysis of spectral immunophenotyping flow cytometry of CAR T cells post-manufacturing (a) or isolated from treated NSG mouse spleen post-treatment (b).
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pham, D.L., Cappabianca, D., Forsberg, M.H. et al. Label-free metabolic imaging monitors the fitness of chimeric antigen receptor T cells. Nat. Biomed. Eng (2025). https://doi.org/10.1038/s41551-025-01504-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41551-025-01504-7