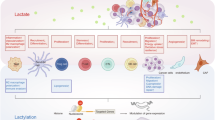
Abstract
Glioblastoma (GBM) is a malignancy with a complex tumour microenvironment (TME) dominated by GBM stem cells (GSCs) and infiltrated by tumour-associated macrophages (TAMs) and exhibits aberrant metabolic pathways. Lactate is a critical glycolytic metabolite that promotes tumour progression; however, the mechanisms of lactate transport and lactylation in the TME of GBM remain elusive. Here we show that lactate is transported from TAMs to GSCs via MCT4–MCT1. TAMs provide lactate to GSCs, promoting GSC proliferation and inducing lactylation of the non-homologous end joining protein KU70 at lysine 317 (K317), which inhibits cGAS–STING signalling and remodels the immunosuppressive TME. Inhibition of lactate transport or targeting the lactylation of KU70, in combination with the immune checkpoint blockade, demonstrates additive therapeutic benefits in immunocompetent xenograft models. This study unveils TAM-derived lactate and lactylation as critical regulators in GSCs to enforce an immunosuppressive microenvironment, opening avenues for developing combinatorial therapy for GBM.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE266884 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE266884). Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD069969 (https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD069969) and PXD070007 (https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD070007). The human GBM data were derived from the TCGA Research Network (http://cancergenome.nih.gov/) and CCGA Research Network (https://www.cgga.org.cn/). All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Louis, D. N., Perry, A. & Wesseling, P. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro. Oncol. 23, 1231–1251 (2021.
Miller, K. D. et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J. Clin. 71, 381–406 (2021).
Ye, Z. et al. Targeting microglial metabolic rewiring synergizes with immune-checkpoint blockade therapy for glioblastoma. Cancer Discov. 13, 974–1001 (2023).
Ostrom, Q. T. et al. CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro. Oncol. 24, v1–v95 (2022).
Tan, A. C., Ashley, D. M. & Lopez, G. Y. Management of glioblastoma: state of the art and future directions. CA Cancer J. Clin. 70, 299–312 (2020).
Lathia, J. D., Mack, S. C., Mulkearns-Hubert, E. E., Valentim, C. L. & Rich, J. N. Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217 (2015).
Prager, B. C., Xie, Q., Bao, S. & Rich, J. N. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell 24, 41–53 (2019).
Lan, X. et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549, 227–232 (2017).
Berg, T. J. & Marques, C. The irradiated brain microenvironment supports glioma stemness and survival via astrocyte-derived transglutaminase 2. Cancer Res. 81, 2101–2115 (2021).
Dapash, M., Hou, D., Castro, B., Lee-Chang, C. & Lesniak, M. S. The interplay between glioblastoma and its microenvironment. Cells 10, 2257 (2021).
Hernandez, A. & Domenech, M. Glioblastoma: relationship between metabolism and immunosuppressive microenvironment. Cells 10, 3529 (2021).
Flint, T. R., Fearon, D. T. & Janowitz, T. Connecting the metabolic and immune responses to cancer. Trends Mol. Med. 23, 451–464 (2017).
Vilbois, S., Xu, Y. & Ho, P. C. Metabolic interplay: tumor macrophages and regulatory T cells. Trends Cancer 10, 242–255 (2024).
Xia, L. et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 20, 28 (2021).
Khan, F. et al. Macrophages and microglia in glioblastoma: heterogeneity, plasticity, and therapy. J. Clin. Invest. 133, e163446 (2023).
Xuan, W., Lesniak, M. S., James, C. D., Heimberger, A. B. & Chen, P. Context-dependent glioblastoma-macrophage/microglia symbiosis and associated mechanisms. Trends Immunol. 42, 280–292 (2021).
Certo, M., Tsai, C. H., Pucino, V., Ho, P. C. & Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 21, 151–161 (2021).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Li, X. et al. Lactate metabolism in human health and disease. Signal Transduct. Target Ther. 7, 305 (2022).
Fan, M. & Yang, K. Lactate promotes endothelial-to-mesenchymal transition via Snail1 lactylation after myocardial infarction. Sci. Adv. 9, eadc9465 (2023).
Li, Z. et al. Lactate shuttling links histone lactylation to adult hippocampal neurogenesis in mice. Dev. Cell 60, 1182–1198.e1188 (2025).
Qian, Y. et al. MCT4-dependent lactate secretion suppresses antitumor immunity in LKB1-deficient lung adenocarcinoma. Cancer Cell 41, 1363–1380.e1367 (2023).
Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour- derived lactic acid. Nature 513, 559–563 (2014).
Abad, E., Graifer, D. & Lyakhovich, A. DNA damage response and resistance of cancer stem cells. Cancer Lett. 474, 106–117 (2020).
Bao, S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006).
Aleksandrov, R., Hristova, R., Stoynov, S. & Gospodinov, A. The chromatin response to double-strand DNA breaks and their repair. Cells 9, 1853 (2020).
Callen, E. et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153, 1266–1280 (2013).
Xu, G. et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 (2015).
Chen, Y. et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell 187, 294–311.e221 (2024).
Chen, H. et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature 631, 663–669 (2024).
Liu, X. et al. Activation of GPR81 by lactate drives tumour-induced cachexia. Nat. Metab. 6, 708–723 (2024).
Mack, S. C. et al. Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J. Exp. Med. 216, 1071–1090 (2019).
Al Emam, A., Arbon, D., Jeeves, M. & Kysela, B. Ku70 N-terminal lysines acetylation/deacetylation is required for radiation-induced DNA-double strand breaks repair. Neoplasma 65, 708–719 (2018).
Tubbs, A. & Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017).
Tang, M. & Chen, G. SMYD2 inhibition-mediated hypomethylation of Ku70 contributes to impaired nonhomologous end joining repair and antitumor immunity. Sci. Adv. 9, eade6624 (2023).
Li, Z., Sun, C. & Qin, Z. Metabolic reprogramming of cancer-associated fibroblasts and its effect on cancer cell reprogramming. Theranostics 11, 8322–8336 (2021).
Wang, H., Franco, F. & Ho, P. C. Metabolic regulation of Treg in cancer: opportunities for immunotherapy. Trends Cancer 3, 583–592 (2017).
Heuser, C., Renner, K., Kreutz, M. & Gattinoni, L. Targeting lactate metabolism for cancer immunotherapy—a matter of precision. Semin Cancer Biol. 88, 32–45 (2023).
Reinfeld, B. I. & Madden, M. Z. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288 (2021).
Felmlee, M. A., Jones, R. S., Rodriguez-Cruz, V., Follman, K. E. & Morris, M. E. Monocarboxylate transporters (SLC16): function, regulation, and role in health and disease. Pharm. Rev. 72, 466–485 (2020).
Park, S. J. et al. An overview of MCT1 and MCT4 in GBM: small molecule transporters with large implications. Am. J. Cancer Res 8, 1967–1976 (2018).
Floch, R. L. et al. CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc. Natl Acad. Sci. USA 108, 16663–16668 (2011).
Sattler, B. & Kranz, M. Preclinical incorporation dosimetry of [18F]FACH-a novel 18F-labeled MCT1/MCT4 lactate transporter inhibitor for imaging cancer metabolism with PET. Molecules 25, 2024 (2020).
Li, G. et al. Glycometabolic reprogramming-induced XRCC1 lactylation confers therapeutic resistance in ALDH1A3-overexpressing glioblastoma. Cell Metab. 36, 1696–1710.e1610 (2024).
Quinet, A., Tirman, S., Cybulla, E., Meroni, A. & Vindigni, A. To skip or not to skip: choosing repriming to tolerate DNA damage. Mol. Cell 81, 649–658 (2021).
Weinstock, D. M., Richardson, C. A., Elliott, B. & Jasin, M. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst.) 5, 1065–1074 (2006).
Zhao, B., Rothenberg, E. & Ramsden, D. A. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol. 21, 765–781 (2020).
Woodbine, L., Gennery, A. R. & Jeggo, P. A. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair (Amst.) 16, 84–96 (2014).
Kaneda, M. M. et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature 539, 437–442 (2016).
Qiao, T. et al. Inhibition of LDH-A by oxamate enhances the efficacy of anti-PD-1 treatment in an NSCLC humanized mouse model. Front Oncol. 11, 632364 (2021).
Renner, K. et al. Restricting glycolysis preserves T cell effector functions and augments checkpoint therapy. Cell Rep. 29, 135–150.e139 (2019).
Vander Linden, C. et al. Therapy-induced DNA methylation inactivates MCT1 and renders tumor cells vulnerable to MCT4 inhibition. Cell Rep. 35, 109202 (2021).
Beloueche-Babari, M. & Casals Galobart, T. Monocarboxylate transporter 1 blockade with AZD3965 inhibits lipid biosynthesis and increases tumour immune cell infiltration. Br. J. Cancer 122, 895–903 (2020).
Flavahan, W. A. et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat. Neurosci. 16, 1373–1382 (2013).
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009).
Li, B. et al. Microbiota depletion impairs thermogenesis of brown adipose tissue and browning of white adipose tissue. Cell Rep. 26, 2720–2737.e2725 (2019).
Zhao, F., Kim, W. & Gao, H. ASTE1 promotes shieldin-complex-mediated DNA repair by attenuating end resection. Nat. Cell Biol. 23, 894–904 (2021).
Ying, W., Cheruku, P. S., Bazer, F. W., Safe, S. H. & Zhou, B. Investigation of macrophage polarization using bone marrow derived macrophages. J. Vis. Exp. 23, 50323 (2013).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e324 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Korsunsky, I., Millard, N. & Fan, J. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Aibar, S. & Gonzalez-Blas, C. B. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639–655.e613 (2022).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 82525047 to X.W and 82573312 to Q.Z).
Author information
Authors and Affiliations
Contributions
X.W., D.L., J.N.R., Q.Z. and X.Q. conceived the project and designed the studies. D.L., G.C., C.F.L., Y.J. and L.Z. conducted the experiments. Q.Z. and D.L. wrote the paper. D.G., J.G., Q.L. and H.Y. performed the data analysis and bioinformatics analysis under the supervision of Q.W. and Z.S. Z.Z., J.Z., G.J., Y.C., Q.G.Z., Q.W., F.L., J.S., C. Liu, C. Li, Y.Y. and N.Z. commented on the study. X.W., K.Y., R.C.G. and D.D. revised the manuscript and supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Zhimin Lu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Lactate metabolism and transporter characteristics in GSCs and TAMs.
a, Dot plot of analyze HALLMARK_GLYCOLYSIS pathway in different cell types using scRNA-seq data. b, c, UMAP dimensional reduction of the basis of the expression pattern of lactate transmembrane transport in cell-type clusters. d, Heatmap of expression of 6 genes in lactate transmembrane transport pathway from TCGA-GBM Agilent-4502A database. e, Dot plot of analyze 6 genes in lactate transmembrane transport pathway using scRNA-seq data (HRA004899, GSE182109 and GSE256490). f, UMAP plot showing the expression pattern of SLC16A1 and SLC16A3 using single-cell RNA-seq data (GSE182109 and GSE256490).
Extended Data Fig. 2 Lactate metabolism and transporter characteristics in GSCs and TAMs.
a-d, Quantitation of BSG and SLC16A1 (a), SOX2 and SLC16A1 (b), AIF1 and SLC16A3 (c), CD163 and SLC16A3 (d) positive cells with analysis of scRNA-seq data. e, Gene expression surface plots illustrate the spatial overlap of SLC16A1 and PROM1, SLC16A3 and CD163 gene expression. f, Kaplan-Meier survival analysis of lactate transmembrane transport signature by the median of different glioma datasets: TCGA GBM LGG and CGGA GBM datasets. g, Kaplan-Meier survival analysis of SLC16A1 combined with SLC16A3 by the median of TCGA GBM LGG and CGGA GBM datasets. h, Representative images of hematoxylin and eosin staining of mouse brains collected on day 40 after transplantation of GL261 in control and Slc16a3-conditionally knockout groups. i, Representative images of hematoxylin and eosin staining of mouse brains collected on day 30 after transplantation of mouse glioma stem cell in control and Slc16a3-conditionally knockout groups. j, L-lactate concentration in GL261-GFP isolated from tumour tissue in control and Slc16a3-conditionally knockout groups (n = 3 biologically independent mice). k, L-lactate concentration in mouse MC9 GSC or TAM isolated from tumour tissue in control and Slc16a3-conditionally knockout groups (n = 3 biologically independent mice). Data are presented as mean ± SD. Statistical significance was determined by a two-tailed log-rank test (f, g) or two-tailed Student’s t-test (j, k).
Extended Data Fig. 3 Lactate transporting between GSCs and TAMs via MCT1–MCT4 supports GSC proliferation.
a, Cell viability detections after different concentrations (0-10 mM) of pyruvate-stimulated GSCs. Results were measured using the CellTiter-Glo assay (n = 6). b, Cell viability detections after different concentrations (0-10 mM) of acetate-stimulated GSCs. Results were measured using the CellTiter-Glo assay (n = 6). c, Cell viability detections after different concentrations (0-10 mM) of β-hydroxybutyrate-stimulated GSCs. Results were measured using the CellTiter-Glo assay (n = 6). d, Cell viability detections of GSCs after being stimulated with the supernatant of TAMs transduced with shCONT, shMCT4-1, shMCT4-2 and rescued by MCT4 (n = 6). e, Representative images of neurospheres of 3028 and MES28 after being stimulated with the supernatant of TAMs transduced with shCONT, shMCT4-1, shMCT4-2 and rescued by MCT4 (n = 6). Scale bar, 100 mm. f, Immunoblot analysis of MCT4 level in TAM transduced with shCONT, shMCT4-1, shMCT4-2 and rescued by MCT4. g, Immunoblot analysis of MCT1 level in 2 GSCs and 2 compared DGCs. h, Fractional abundance of M + 3 lactate in MES28 GSC and MES28 DGC when stimulated with 10 mM 13C3-labelled lactate (n = 3). i, Cell viability detections of DGCs individually or treated with the supernatant of TAMs transduced with control shRNA, shMCT4, or shMCT4 rescued with lactate (n = 6). j, Immunoblot analysis of MCT1 level in 3028 and MES28 GSCs transduced with shCONT, shMCT1-1, shMCT1-2 and rescued by MCT1. k, Oxygen consumption rates of MES28 GSC co-cultured with supernatant of TAM (n = 4). l, Oxygen consumption rates of MES28 GSC expressing non-targeting or MCT1-targeting shRNAs (n = 4). Data are presented as mean ± SD (h, k, l), box plots show the median (centre), the 25th and 75th percentiles (bounds of box), and whiskers indicate the minimum and maximum values (a-d, i). Statistical significance was determined by a two-tailed Student’s t-test.
Extended Data Fig. 4 TAMs-derived lactate induced the lactylation of DNA damage repair protein KU70 K317 leads to GSC proliferation.
a, Cell viability detections after different concentrations (0-10 mM) of lactate stimulated NSCs (n = 6). b, Fractional abundance of M + 3 lactate in GSCs and NSCs when stimulated with 10 mM 13C3-labelled lactate (n = 3). c, Gene Set Enrichment Analysis (GSEA) scores calculated for RNA sequencing data using Gene Ontology (GO) gene sets showing the correlation between GSCs and NSCs with the Cellular response to CAMP signaling pathway. d, Immunoblot analysis of the pan-lactylation level of 3028 (left) and MES28 (right) after transduction with control shRNA and shGPR81 by stimulating with 1 mM or 10 mM lactate. e, Heatmaps showing lactylated proteins involved in non-homologous end joining, DNA replication, and base excision repair pathways in GSCs. f, KU70 mRNA levels were assayed by qRT-PCR in GSCs with KU70 knockdown using two independent shRNAs. A shCONT was used as the control (n = 3). g, Immunoblot analysis of GSCs γ-H2AX level with the stimulation of 10 mM lactate combined transduced with shCONT, shFEN1, shFEN1 rescued with FEN1-WT or FEN1-K201R. h, Immunoblot analysis of GSCs γ-H2AX level with the stimulation of 10 mM lactate combined transduced with shCONT, shLIG1, or shLIG1 rescued with LIG1-WT or LIG1-K226R. i, Cell viability detections of GSCs under the stimulation of 10 mM lactate combined transduced with shCONT, shFEN1, shFEN1 rescued with FEN1-WT or FEN1-K201R. Results were measured using the CellTiter-Glo assay (n = 6). j, Cell viability detections of GSCs under the stimulation of 10 mM lactate combined transduced with shCONT, shLIG1, or shLIG1 rescued with LIG1-WT or LIG1-K226R. Results were measured using the CellTiter-Glo assay (n = 6). k, Immunoblot analysis of IQGAP3 and IQGAP3 lactylation levels combined transduced with shCONT, shIQGAP3, or shIQGAP3 rescued with IQGAP3-WT or IQGAP3-K981R by stimulating with 10 mM lactate. l, Cell viability detections of GSCs under the stimulation of 10 mM lactate combined transduced with shCONT, shIQGAP3, or shIQGAP3 rescued with IQGAP3-WT or IQGAP3-K981R. Results were measured using the CellTiter-Glo assay (n = 6). Data are presented as mean ± SD (a, b, f), box plots show the median (centre), the 25th and 75th percentiles (bounds of box), and whiskers indicate the minimum and maximum values (i, j, l). Statistical significance was determined by a two-tailed Student’s t-test.
Extended Data Fig. 5 TAMs-derived lactate induced the lactylation of DNA damage repair protein KU70 K317 leads to GSCs proliferation.
a, Quantification of lactylation percentage for KU70 K317 by intensity values of LC/MS. b, Sequence alignment of KU70 in various species. *Lactylation site. c, The synthetic KU70 peptides spanning lactylation K317 and non-modified peptide. d, Dot-blot results validated the efficiency of the KU70 K317lac specific antibodies when detecting modified peptides. e, Immunoblot results showed the KU70 K317lac specific antibodies when detecting the lysate of Hela cell line. f, Coomassie blue staining shown the purified His-KU70 WT and His-KU70 K317R. g, Purified His-KU70 WT or K317R were incubated with or without P300 for 30 min. Lac-CoA was added. Immunoblot analysis were performed. h, Purified His-KU70 WT or K317R were incubated with or without P300 for 30 min. Ac-CoA was added. Immunoblot analysis were performed. i, KU70 K317 lactylation-specific antibody was validated by experiments of the purified His-KU70 WT or K317R incubated with or without the modified peptide (Peptide A). P300 and Lac-CoA were added. j, KU70 K317 lactylation and KU70 K317 acetylation-specific antibody were used to detect the modification level after P300 knockdown in GSCs. k, Cell viability detections of 3028 and MES28 GSCs after deletion of P300 under the stimulation of lactate (n = 6). l, Representative images of neurospheres of 3028 (top) and MES28 (bottom) under the stimulation of 10 mM lactate combined transduced with shCONT, shP300-1 and shP300-2 (n = 6). Scale bar, 100 mm. m, Purified His-KU70 WT or K317R were incubated with or without CBP for 30 min. Lac-CoA (top) or Ac-CoA (bottom) was added. Immunoblot analysis were performed. n, Purified His-KU70 WT or K317R were incubated with or without KAT2B for 30 min. Lac-CoA (top) or Ac-CoA (bottom) was added. Immunoblot analysis were performed. o, KU70 K317 lactylation and KU70 K317 acetylation-specific antibody were used to detect the modification level after CBP knockdown in GSCs. p, KU70 K317 lactylation and KU70 K317 acetylation-specific antibody were used to detect the modification level after KAT2B knockdown in GSCs. The experiments were performed three times independently (d-j, m-p). Box plots show the median (centre), the 25th and 75th percentiles (bounds of box), and whiskers indicate the minimum and maximum values. Statistical significance was determined by a two-tailed Student’s t-test.
Extended Data Fig. 6 Lactylation of KU70 K317 enhances the NHEJ function and maintains stemness of GSC.
a, KU70 K317 lactylation and acetylation level detection after stimulated with different concentration of lactate. b, Representative pictures of comet assay under the stimulation of lactate combined transduced with shCONT, shKU70 and rescued using KU70 WT and KU70 K317R in GSCs. Scale bars, 100μm. c, Apoptosis assay under the stimulation of lactate combined transduced with shCONT, shKU70 and rescued using KU70 WT and KU70 K317R in GSCs. d, Immunoblot analysis under the stimulation of lactate combined transduced with shCONT, shKU70, or shKU70 rescued with KU70 WT or KU70 K317R. e, Senescence-associated β-galactosidase assay under the stimulation of lactate combined transduced with shCONT, shKU70 and rescued using KU70 WT and KU70 K317R in GSCs. The experiments were performed three times independently (a, b, d).
Extended Data Fig. 7 Lactylation of KU70 K317 enhances the NHEJ function and maintains stemness of GSC.
a, b, Two independent shRNAs targeting MCT1 decreased the self-renewal of GSCs compared with shCONT, as measured by sphere number quantification (n = 6) (a) and the extreme limiting dilution assays (b) in 3028 and MES28. c, d, Sphere number quantification (n = 6) (c) and the extreme limiting dilution assays (d) performed in 3028 and MES28 with the deletion of KU70 and rescued by KU70 WT and KU70 K317R. e, Cell viability of six patient-derived GSCs (MES28, 3028, 2907, GSC23, RKI and 3264) after 48 h treatment of vehicle control (DMSO) and various concentrations of NHEJ inhibitor NU7441 combined with the stimulation of lactate (n = 6). f, GST pull-down assay performed to detect the interaction between KU80 and KU70 WT or KU70 K317R in vitro. g, Immunoblot results showing the interaction between KU70 and KU80 with anti-KU70 antibody in 3028 and MES28 GSCs with or without P300-Flag (1287-1663AA) and the 10 mM lactate stimulated. h, Immunoblot analysis of GSCs γ-H2AX level and the interaction level between KU70 and KU80 under the stimulation of 10 mM lactate combined transduced with shCONT, shKU70 and rescued by KU70 (WT) or KU70 (E330A) or KU70 (K317R) or KU70 (K317R E330A) after KU70 knockdown. i, Effects of shCONT, shKU70 and rescued by KU70 (WT) or KU70 (E330A) or KU70 (K317R) or KU70 (K317R E330A) after KU70 knockdown on the efficiency of NHEJ in MES28 GSC with the stimulation of lactate (n = 3). j, Immunoblot analysis after KU70 K317-Peptide blockade in GSCs. The experiments were performed three times independently (f, g, j). Data are presented as mean ± SD (e, i), box plots show the median (centre), the 25th and 75th percentiles (bounds of box), and whiskers indicate the minimum and maximum values (a, c). Statistical significance was determined by a two-tailed Student’s t-test (a, c, i) and a likelihood ratio test (LRT) comparing the single-hit model to alternative models (two-sided). P-values were computed based on the χ² distribution of the likelihood ratio statistic (b, d).
Extended Data Fig. 8 Lactylation of KU70 K317 inhibits cGAS-STING signaling and maintains the immunosuppressive TME.
a, Gene Set Enrichment Analysis (GSEA) scores calculated for RNA sequencing data using Gene Ontology (GO) gene sets showing MCT1 expression correlates with a transcriptional signature of Interferon Alpha production signaling (left) and Interferon Beta production signaling (right) in MES28 GSC. b, GSEA scores calculated for RNA sequencing using GO gene sets showing KU70 expression correlates with type I interferon production signaling in MES28 GSC. c, GSEA scores calculated for RNA sequencing using GO gene sets showing KU70 K317lac level correlates with type I interferon production signaling in MES28 GSC. d, e, GSEA using GO gene sets showing MCT1 (d) and KU70 (e) expression correlate with immune response related signals in TCGA GBM dataset. f, Immunoblot analysis of stemness marker level of 3028 (left) and MES28 (right) after transduced with control shRNA, shcGAS and shMCT1 by stimulating with lactate. g, Diagram of the NHEJ and HR reporter assay. Effects of Mct1 knockdown on the efficiency of NHEJ and HR in mouse glioma cell line GL261 under the stimulation of lactate (n = 3). h, ELISA experiments detecting the levels of type I interferon (IFNα and IFNβ) while GL261 expressing shCont and shMct1 (n = 3). i, The gating strategy of GSCs in flow cytometric analysis. j, Flow cytometry plots and quantification of Sox2+ Prom1+ GSCs in GFP+ cells as indicated (n = 3 biologically independent mice). Data are presented as mean ± SD. Statistical significance was determined by a two-tailed Student’s t-test.
Extended Data Fig. 9 Lactylation of KU70 K317 inhibits cGAS-STING signaling and maintains the immunosuppressive TME.
a, Flow cytometry percentage of macrophage, microglia, MDSC, Dendritic cell, CD8 + T cell, NK cell and CD4 + T cell from CD45+ cells and Treg from CD4 + T cells in Lyz2-Cre with Slc16a3wt/wt and Lyz2-Cre with Slc16a3fl/fl C57BL/6 J mice groups bearing 5 × 104 mouse glioma stem cell MC9 (n = 3 biologically independent mice). b, In vivo bioluminescent imaging of tumour growth was performed in immunocompetent mice bearing 5 × 104 mouse glioma stem cell MC9 and 5 × 104 BMDMs using KU70 K317-Peptide or KU70 K317R-Peptide on days 8, 15, 35. c, Representative images of hematoxylin and eosin staining of mouse brains collected on day 25 after transplantation of MC9 and BMDMs treated with KU70 K317-Peptide or KU70 K317R-Peptide. Scale bar, 2 mm. Each image is representative of at least three similar experiments. d, Kaplan-Meier survival curves of immunocompetent mice after being treated with KU70 K317-Peptide or KU70 K317R-Peptide (n = 10). e, Representative images showing immunohistochemically stained with KU70 K317lac in as indicated brain sections of the indicated mice on day 25 after treated with KU70 K317-Peptide or KU70 K317R-Peptide. Scale bar, 50 μm. f, Flow cytometry analysis of CD8 + T cells and granzyme B + CD8 + T cells in BMDMs and treated MC9-derived tumours at day 25 after injection (n = 3 biologically independent mice). g, Flow cytometry analysis of exhaustion markers in CD8 + T cells in BMDMs and treated MC9-derived tumours at day 25 after injection (n = 3 biologically independent mice). h, Expression of PD-1 on CD8 + T cells after the cytotoxic experiments with or without the syrosingopine treatment. i, Kaplan-Meier survival analysis of lactate transmembrane transport signature combined with PD-1 expression level by the median of TCGA GBM LGG and CGGA GBM datasets. j, After co-cultured with the supernatant of PBMC-derived macrophages, cytotoxicity experiments of 3028 and MES28 GSCs with activated CD8 + T cells. The ratios between CD8 + T cells and GSCs are indicated (n = 3). k, ELISA experiments detecting the levels of IFNγ in the supernatant of co-cultured system of activated T cells and GSCs (n = 3). Data are presented as mean ± SD. Statistical significance was determined by a two-tailed Student’s t-test (a, f, g, j, k) or two-tailed log-rank test (d, i).
Extended Data Fig. 10 Inhibition of lactate transporters synergizes with immune checkpoint inhibitor to prolong survival in glioblastoma orthotopic xenograft models.
a, Flow cytometry analysis of CD8 + T cells and granzyme B + CD8 + T cells in BMDMs and treated GL261-derived tumours at day 25 after injection (n = 3 biologically independent mice). b, Immunoblot results show tumour-promoting macrophage markers and apoptosis markers after MCT4 konckdown in TAMs (left) and BMDMs (right). c, Immunoblot analysis showing the Ku70 and cgas level in GL261 after Ku70 and cgas knockdown. d, In vivo bioluminescent imaging of tumour growth was performed in C57BL/6 J mice bearing 5 × 104 BMDMs and 5 × 104 GL261 transduced with shCont, shKu70, shcgas or combined with shKu70 and shcgas on days 8, 15, 35. e, Kaplan-Meier survival curves of immunocompetent mice bearing GL261 and BMDMs transduced with shCont, shKu70, shcgas or combined with shKu70 and shcgas derived intracranial tumours (n = 5). f, Representative images of hematoxylin and eosin staining of mouse brains collected on day 25 after transplantation of BMDMs and Gl261 transduced with shCont, shKu70, shcgas or combined with shKu70 and shcgas. Scale bar, 2 mm. Each image is representative of at least three similar experiments. g, In vivo bioluminescent imaging of tumour growth was performed in immunocompetent mice bearing GL261 and BMDMs using combination of the KU70 K317-Peptide and PD-1 antibody on days 8, 15, 35. h, Representative images of hematoxylin and eosin staining of mouse brains collected on day 25 after transplantation of GL261 and BMDMs treated with KU70 K317-Peptide and PD-1 antibody. Scale bar, 2 mm. Each image is representative of at least three similar experiments. i, Kaplan-Meier survival curves of immunocompetent mice after being treated with KU70 K317-Peptide and PD-1 antibody (n = 5). j, Schematic diagram illustrating that targeting lactate transporter MCT1 and MCT4 or KU70 K317 lactylation combined with anti-PD-1 remodels the immune suppressive microenvironment. k, Schematic diagram illustrating that TAMs-derived lactate remodels glioblastoma immune microenvironment through regulation of NHEJ repair by KU70 K317 lactylation. Data are presented as mean ± SD. Statistical significance was determined by a two-tailed Student’s t-test (a) or two-tailed log-rank test (e, i). Panels j and l created with BioRender.com.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and associated legends and Flow cytometry gating strategies.
Supplementary Data 1
Source data for Supplementary Figs. 1 and 2.
Supplementary Data 2
Unprocessed western blots for Supplementary Figs. 1 and 2.
Supplementary Table 1
Genotyping of conditional knockout mice.
Supplementary Table 2
DNA oligos used in this study.
Supplementary Table 3
Exact P value in Figs. 1–6, Extended Data Figs. 1–10 and Supplementary Figs. 1 and 2.
Source data
Source Data Figs. 1–6
Statistical source data for Figs. 1–6.
Source Data Figs. 3–5 and Extended Data Figs. 3–8 and 10
Unprocessed western blots for Figs. 1–6 and Extended Data Figs. 1–10.
Source Data Extended Data Figs. 2–5 and 7–10
Statistical source data for Extended Data Figs. 1–10.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, D., Cui, G., Yang, K. et al. Inhibiting macrophage-derived lactate transport restores cGAS–STING signalling and enhances antitumour immunity in glioblastoma. Nat Cell Biol (2026). https://doi.org/10.1038/s41556-025-01839-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41556-025-01839-y
This article is cited by
-
Immune evasion by macrophage-derived lactate
Nature Cell Biology (2026)