Abstract
Neural circuits in many brain regions are refined by experience. Sensory circuits support higher plasticity at younger ages during critical periods—times of circuit refinement and maturation—and limit plasticity in adulthood for circuit stability1,2. How astrocytes, a glial subtype, maintain these differing plasticity levels, and whether they stabilize the properties of sensory circuits in adulthood, remain largely unclear. Here we take a comprehensive approach to address these questions and establish astrocytes as key orchestrators of circuit stability. Combining a transcriptomic approach with ex vivo electrophysiology and in vivo imaging, we identify that astrocytes release CCN1 (refs. 3,4) to maintain synapse and circuit stability in the adult visual cortex. Overexpressing CCN1 in astrocytes during the critical period promotes the maturation of inhibitory neurons, limits ocular dominance plasticity and promotes oligodendrocyte differentiation and maturation. Conversely, knocking out astrocyte CCN1 in adults destabilizes binocular circuits and reduces myelination. This establishes CCN1 as an astrocyte-secreted factor that stabilizes neuronal circuits by coordinating the maturation state of multiple cell types, and demonstrates that the composition and properties of sensory circuits require ongoing maintenance in adulthood, and that these maintenance cues are provided by astrocytes.
Similar content being viewed by others
Main
Mammalian neural circuits are highly labile at an early age as the organism learns to adapt to its environment. This is evident in sensory circuits, which typically have higher plasticity at younger ages, a period of circuit refinement, and increased stability and reduced plasticity in adulthood1. This stability in adulthood is necessary to maintain functional neuronal connectivity2. Notably, however, circuit stability in adulthood is reversible. Digesting the extracellular matrix with enzymes, or transplanting juvenile inhibitory neurons or astrocytes, can re-open the window for circuit plasticity5,6,7, demonstrating a requirement for cues to actively stabilize and maintain neural circuits in the adult. Astrocytes, a major glial subtype with important roles in neuronal function8, are poised to have a key role in maintaining circuit stability owing to their stage-specific roles in regulating developmental synapse formation, function, stabilization and plasticity during the critical period7,9,10,11. Whether and how astrocytes restrict plasticity and maintain sensory circuit stability in the adult brain remains unclear.
In this study we establish the astrocyte-secreted protein CCN1 as a factor that promotes circuit stability in mouse visual cortex. CCN1 is a four-domain secreted protein that can bind with many components of the plasma membrane and the extracellular matrix, including heparan sulfate proteoglycans and integrins12. The role of CCN1 in the periphery has been investigated in the context of tumorigenesis, angiogenesis and inflammation3,13,14, although its role in the central nervous system remains largely unknown. Here we show that astrocyte-secreted CCN1 regulates circuit stability by exerting its effect on multiple cell types, including excitatory neurons, inhibitory neurons, oligodendrocytes and microglia.
Astrocyte genes that support stability
To examine how astrocytes regulate circuit stability, we used the mouse visual cortex as our model. The mouse visual cortex has a well-characterized critical period for binocular circuits, which begins at approximately the end of the third postnatal week and lasts for two weeks. By the end of the critical period, essential properties of the visual system are established15,16 and the binocular circuitry is well established and fine-tuned2.
To identify astrocyte factors involved in restricting visual cortex plasticity and maintaining stability, we performed transcriptional profiling of astrocytes during periods of high stability versus high plasticity and after the induction of plasticity, using bulk mRNA sequencing from astrocyte-Ribotag (Rpl22fl/fl; Gfap-cre) mice17,18 (Fig. 1a and Supplementary Table 1). We examined astrocyte transcriptomes during the critical period (postnatal day 28 (P28)), a time of high plasticity, and in adulthood (P120), a time of high stability17,18 (Fig. 1b,c). To induce plasticity, we dark-reared mice from birth until P45 and compared them with normally reared mice at P45 (Fig. 1b). Specifically, dark rearing delays astrocyte maturation and the closure of the critical period, prolonging plasticity19,20,21. Of note, 17 out of 44 genes that are upregulated in dark rearing are also upregulated at P7 versus P28, suggesting delayed maturation of astrocytes (P7 from ref. 18; Fig. 1c and Supplementary Table 1); these genes include Vim, which is known to be upregulated in early development in astrocytes18. As a second model for plasticity induction, we performed two days of monocular deprivation (MD; lid suturing) at P28 to induce ocular dominance plasticity2 and compared the hemisphere contralateral to the deprived eye (most deprived) to the hemisphere ipsilateral to the deprived eye (least deprived; Fig. 1b,c)
a, Experimental setup for bulk transcriptomics of visual cortex using RiboTag (Rpl22fl/fl; Gfap-cre) mice. Created in BioRender. Allen Lab (2025) https://BioRender.com/j3g9rj1. HA, haemagglutinin. b, Experimental conditions. Contra, contralateral to MD; CP, critical period; DR, dark rearing; ipsi, ipsilateral to MD; NR, normal rearing. c, Volcano plots of astrocytic DEGs in different experimental comparisons. Two mice pooled per biological replicate. P7: n = 3; P14: n = 4; P28: n = 5; P120: n = 6; MD contra: n = 3; MD ipsi: n = 3; P45 DR: n = 4; P45 NR: n = 3. Statistics from DESeq2 using HOMER on biological replicates, Benjamini–Hochberg corrections for multiple comparisons; NS, not significant (P ≥ 0.05). d, Proportional Venn diagram of DEGs from c showing overlapping DEGs. e, Left, heat map of fragments per kilobase million (FPKM) of the astrocyte-expressed overlapping genes from d. Right, heat map of log2-transformed fold change (log2FC) of overlapping genes. *, adjusted P value < 0.05. f, FPKM for Ccn1 in different experimental conditions. Developmental data from refs. 17,18. Data are mean ± s.e.m. Symbols indicate biological replicates; *, significant P value. g, Representative z-projection of smFISH in a tiled image of the P120 visual cortex (VC) (left and middle; scale bars, 100 µm) and in different cortical layers (right; scale bars, 5 µm). h, Thresholded Ccn1-positive area within Slc1a3 regions of interest (astrocytes) in different cortical layers in the visual cortex at P7, P14, P28 and P120. Small dots show data for individual astrocytes; circles show mouse averages; and error bars represent s.e.m. n = 3 mice per age. Two-way ANOVA with post hoc Tukey tests on mice (statistics in Supplementary Table 7). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To identify ‘pro-stability’ genes in astrocytes, we examined differentially expressed genes (DEGs) that are upregulated in astrocytes in adulthood compared with the critical period, and downregulated after the plasticity-inducing paradigms (Extended Data Fig. 1a). Pathway analysis identified the involvement of genes in synapses, membrane organization and cell adhesion (Extended Data Fig. 1b–d and Supplementary Table 2). Some DEGs encode known astrocyte synapse-modifying and synapse-associated extracellular matrix factors, including Thbs4, Chrdl1, Ncan and Hapln2 (Extended Data Fig. 2a). To limit the number of potential candidates, we focused on DEGs that were altered in more than one comparison (14 genes; Fig. 1d,e). We identified Ccn1 (also known as Cyr61) as a putative pro-stability factor that is high in adulthood and downregulated after dark rearing and MD (Fig. 1f). Ccn1 encodes the secreted protein CCN1, which contains integrin and heparan sulfate proteoglycan binding sites, and whose predicted functional interaction network highlights its association with integrins (integrin αV, encoded by Itgav), the extracellular matrix and components of the YAP/TAZ Hippo pathway (Extended Data Fig. 2b). We also detected ‘pro-plasticity’ gene candidates, such as Fbln2, that are upregulated during the critical period, downregulated in adulthood, and upregulated after plasticity induction (Fig. 1e).
To confirm that Ccn1 is expressed in astrocytes and is upregulated in adulthood, we used single-molecule fluorescent in situ hybridization (smFISH) to characterize Ccn1 in visual cortex astrocytes across different ages and plasticity paradigms. Ccn1 is expressed by astrocytes in all layers of the visual cortex, and expression increases across development and stays high into adulthood (Fig. 1g,h and Extended Data Fig. 2c). Ccn1 is also expressed by neurons, although at a lower level (Extended Data Fig. 2d). We validated that dark rearing decreases Ccn1 in astrocytes (Extended Data Fig. 2e). Thus, the expression pattern of Ccn1, which is high in adulthood and low after plasticity induction, suggests that CCN1 produced by astrocytes may act a pro-stability factor.
CCN1 restricts remodelling
To test the role of CCN1 as a pro-stability factor, we manipulated astrocyte expression of Ccn1 in vivo in juvenile and adult mice. To test if we could prematurely restrict plasticity, we injected the visual cortex of P14 wild-type mice with AAV2/5-tdTomato (tdT) as a control for viral transduction or AAV2/5-CCN1-HA (CCN1) driven by the minimal Gfap promoter (Fig. 2a). These constructs were specific for astrocytes and had high penetrance (Extended Data Fig. 3a–c). CCN1 overexpression led to production of CCN1 protein, validated in culture and in whole visual cortex lysates, and did not induce astrocyte reactivity in vivo (Extended Data Fig. 3d–i). To probe for large-scale plasticity, we performed monocular enucleation (ME) to induce remodelling of inputs to the visual cortex and measured outcomes with an Arc induction assay. This assay probes for Arc-positive neurons in the binocular zone contralateral to the deprived eye activated by visual stimulation and is thus a proxy for the width of the binocular zone (Fig. 2a,b and Methods). We found that four days of ME during the critical period in tdT-overexpressing (tdT-OE) mice was sufficient to induce binocular zone remodelling, whereas CCN1-overexpressing (CCN1-OE) mice showed much less remodelling (Fig. 2b,c and Extended Data Fig. 4a). These results indicate that CCN1 overexpression restricts plasticity at a time when plasticity is normally high. We then explored whether CCN1 overexpression in adulthood, when plasticity is low and CCN1 expression in astrocytes is already high, would be sufficient to further repress plasticity. We found that CCN1 overexpression in adulthood did not change the size of the binocular zone relative to tdT-OE mice (Extended Data Fig. 4d–f).
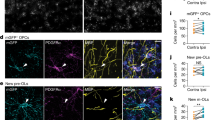
a, Top, experimental setup for overexpression of CCN1 or tdTomato (tdT) in critical period astrocytes (grey). Bottom, Arc induction assay. BZ, binocular zone; WT, wild type. b, Representative image of Arc smFISH. Left, 12 h after ME. Right, 4 days after ME (4 days ME). Scale bars, 1 mm. Dashed yellow lines indicate Arc binocular zone width. c, Quantification of Arc activation width. n = 4 mice per group. d, cKO of Ccn1 in adult mouse astrocytes by intraperitoneal tamoxifen injection. Created in BioRender. Allen Lab (2025) https://BioRender.com/cp7gqpp. e, Representative images of Arc smFISH in Ccn1-cKO visual cortex. Scale bars, 1 mm. f, Arc activation width. n = 6 mice per group. c,f, Two-way ANOVA with post hoc Tukey tests on mouse averages. *, significant P value. Data are mean ± s.e.m. Symbols indicate mouse averages. g, Whole-cell patch clamping of layer 2/3 pyramidal neurons. h, Representative sEPSCs. i, Average inter-event interval. j, Average amplitude. tdT no MD: n = 16 cells, 6 mice; tdT MD: n = 17 cells, 5 mice; CCN1 no MD: n = 17 cells, 7 mice; CCN1 MD: n = 18 cells, 7 mice. k, Representative sIPSCs. l, Average inter-event interval. m, Average amplitude. tdT no MD: n = 17 cells, 7 mice; tdT MD: n = 18 cells, 7 mice; CCN1 no MD: n = 15 cells, 7 mice; CCN1 MD: n = 18 cells, 8 mice. i,j,l,m, Unfilled symbols represent cells and filled symbols represent mouse averages. Data are mean ± s.e.m. Nested one-way ANOVA with post hoc Sidak tests on mouse averages. *, significant P value. n, Representative z-projection of WFA around parvalbumin (PV)-positive cells in critical period visual cortex. Scale bar, 20 µm. o, Integrated WFA density around parvalbumin-expressing cells. Dots represent cells. tdT no MD, n = 204 cells; tdT MD, n = 197 cells; CCN1 no MD, n = 228 cells; CCN1 MD, n = 193 cells. Five mice per group. Kruskal–Wallis test with post hoc Dunn’s tests on cells. *, significant P value. Data are median ± 95% confidence interval. a.u., arbitrary units.
Having established that increasing CCN1 level during the critical period is sufficient to restrict plasticity, we hypothesized that decreasing CCN1 in astrocytes in adulthood would result in increased plasticity. To knock down CCN1 specifically in adult astrocytes, we crossed a Ccn1-floxed mouse with a tamoxifen-inducible Aldh1l1-creERT2 mouse, with tamoxifen treatment at one month of age, to produce wild-type (Ccn1fl/fl;cre−) and astrocyte-specific CCN1 conditional knockout (Ccn1-cKO mice; Ccn1fl/fl;cre+; Fig. 2d and Extended Data Figs. 3j–n and 4c). We performed ME and the Arc induction assay in four-month-old wild-type and Ccn1-cKO mice, a timepoint when plasticity is low (Fig. 2d,e). This revealed increased plasticity after four days of ME in Ccn1-cKO mice (Fig. 2e,f and Extended Data Fig. 4b), demonstrating that CCN1 is actively maintaining visual cortex stability in adulthood. To assess whether reducing astrocyte CCN1 would promote more plasticity during the critical period, a time when plasticity is high, we removed CCN1 from astrocytes in juvenile mice (Extended Data Fig. 4g–k). The Arc induction assay at P28 revealed no differences in the width of the binocular zone after 4 days of ME in cKO mice relative to wild-type mice (Extended Data Fig. 4l,m). Thus, during the critical period when CCN1 level is low, it is not a major regulator of plasticity. Overall, these findings identify a role for astrocyte-secreted CCN1 in restricting plasticity in visual circuits in an age-dependent manner.
CCN1 regulates excitatory synapses
To understand whether CCN1 is affecting circuit stability by altering synapses, we performed ex vivo whole-cell patch clamp recordings of spontaneous excitatory postsynaptic currents (sEPSCs) and spontaneous inhibitory postsynaptic currents (sIPSCs) from layer 2/3 pyramidal neurons in the visual cortex during the critical period (Fig. 2g–m). Previous studies have shown long-term depression of sEPSCs after MD22 and enhancement of inhibition after MD23. We performed recordings in tdT-OE or CCN1-OE P28 mice that underwent 5 days of MD from P23 or no manipulation (Fig. 2g). For sEPSCs, we found an increase in the basal inter-event interval (a decrease in frequency) in CCN1-OE mice relative to tdT-OE mice (Fig. 2h,i and Extended Data Fig. 5a,c,d). We found a trend for a decrease in sEPSC amplitude in tdT-OE mice after MD (Fig. 2j), indicating long-term depression as expected, which was absent in CCN1-OE mice. For sIPSCs (Fig. 2k), we found no differences in inter-event interval or amplitude between tdT-OE or CCN1-OE mice with or without MD (Fig. 2l,m and Extended Data Fig. 5b,e).
We also performed sEPSC recordings from L2/3 pyramidal neurons at a later time point to parallel the experiments in Fig. 2b, with MD being performed at P28 and recordings being done at P33. Similar to the results in Fig. 2i, we observed an increase in the basal sEPSC inter-event interval (a decrease in frequency) in CCN1-OE mice relative to tdT-OE mice, which was reversed with MD at this later age (Extended Data Fig. 5f–j,p). Since we did not detect basal differences at the 12 h timepoint in the Arc induction assay, this suggests that the electrophysiology and Arc induction assays are probing different populations of neurons. These data demonstrate that CCN1 decreases excitatory drive onto pyramidal neurons and that MD reverses some of these changes.
CCN1 promotes maturation of inhibition
Because the maturation of inhibitory neurons occurs during the critical period and is a proposed mechanism underlying its closure, we next examined the role of astrocyte CCN1 in regulating this maturation. Parvalbumin-positive interneurons (PV interneurons) show an increase in EPSC frequency after the critical period24, so we tested whether CCN1-OE could change EPSC frequency in these neurons. We recorded sEPSCs from morphologically and electrophysiologically identified25 fast-spiking interneurons (putative parvalbumin-positive neurons (PV neurons); Extended Data Fig. 5k–o,q) at the end of the critical period, P33, after 5 days of MD or no MD. We found a significant decrease in inter-event interval in CCN1-OE mice relative to tdT-OE mice at baseline, which was reversed with MD (Extended Data Fig. 5l). We also observed an increase in the decay and risetime of sEPSCs in tdT-OE mice, but not CCN1-OE mice, after MD (Extended Data Fig. 5n,o), suggesting an MD-induced change in receptor composition with MD that is absent in CCN1-OE. This demonstrates that the overall effect of CCN1-OE on synaptic networks is to decrease excitatory drive onto excitatory neurons, while increasing excitatory drive onto inhibitory neurons.
The deposition of perineuronal nets (PNNs)—dense extracellular matrices that form predominantly around PV interneurons—also correlates with closure of the critical period and can regulate ocular dominance plasticity5,26. To test whether CCN1-OE mice exhibit premature maturation of PNNs, we performed immunohistochemistry against parvalbumin and labelled PNNs using Wisteria floribunda agglutinin (WFA) in the visual cortex of tdT-OE and CCN1-OE mice at P33, at baseline and after 5 days of MD. CCN1-OE mice had a pronounced increase in WFA signal around PV neurons compared with tdT at baseline (Fig. 2n,o and Extended Data Fig. 6a,b). This effect was reversed with MD, paralleling the reversal of the effects of CCN1 on excitatory drive by MD. Thus, CCN1 promotes the maturity of inhibitory neurons and their PNNs.
PNNs can be remodelled by microglia in the central nervous system27,28; additionally, CCN1 can regulate macrophage and microglial reactivity29,30. To assess whether the observed CCN1-dependent changes in PNN density are mediated by microglia, we examined microglial engulfment of PNNs, finding no basal or MD-induced effects of CCN1 on engulfment at P33 (Extended Data Fig. 6c,d and Methods). We found that CCN1-OE mice, relative to tdT-OE mice, had an increase in IBA1 volume and a decrease in CD68 volume within microglia (Extended Data Fig. 6e–g), suggesting altered microglia morphology, which we examined further by classifying microglia on the basis of morphotypes (Extended Data Fig. 6h,i and Methods). This showed that after MD, CCN1-OE mice have a higher proportion of ramified, resting microglia and a decrease in amoeboid microglia (Extended Data Fig. 6j–n). Thus, we find that CCN1 can regulate microglial state, as we observe a reduction in phagocytotic lysosomes and associated microglial morphotypes in CCN1-OE mice.
CCN1 maintains binocular circuits
The preceding experiments demonstrate sufficiency of CCN1 in inducing maturation of cells in the visual cortex; we next explored whether CCN1 in astrocytes is necessary to maintain visual circuit function in adulthood. To do this we used two-photon in vivo microscopy to examine binocular circuits in adult wild-type and Ccn1-cKO mice. We longitudinally imaged gCaMP6f calcium responses in layer 2/3 pyramidal neurons in the binocular zone of awake mice in response to monocular visual stimulation of both eyes (Fig. 3a,b, Extended Data Fig. 7a–f and Methods).
a, Timeline of experiments. Created in BioRender. Allen Lab (2025) https://BioRender.com/cp7gqpp. b, Two-photon imaging of layer 2/3 pyramidal neurons in the binocular zone. Treadmill drawing from https://doi.org/10.25378/janelia.24691311 (CC BY 4.0). c, Representative image of longitudinal tracking of cell identity. Arrows, cell with changing identity. Scale bar, 100 µm. d–i, Proportions of contralateral-responsive (d), binocular-responsive (f) and ipsilateral-responsive (h) cell types in longitudinally tracked cells, and changes in the number of contralateral-responsive (e), binocular-responsive (g) and ipsilateral-responsive (i) cells following MD. Wild-type responsive cells pre MD: n = 324, wild-type responsive cells post MD: n = 267; Ccn1-cKO responsive cells pre MD: n = 319;Ccn1-cKO responsive cells post MD: n = 203. Four mice per genotype. Chi-square tests on cells with corrections (4 comparisons; α = 0.0125; *, significant P value). j, Alluvial plot of cell-type proportions. B, binocular-responsive; C, contralateral-responsive; I, ipsilateral-responsive; U, unresponsive. k, Histogram of ODI. Inset, hierarchically bootstrapped means. Longitudinally tracked responsive cells. Wild type: n = 229 cells; cKO: n = 163 cells. l, Median ODI by mouse. n = 4 per genotype. Paired t-tests on mice with α = 0.025; *, significant P value. m, Median changes in contralateral and ipsilateral eye responses per pre-MD ODI bin. *P < 0.00625 versus 0% change: wild-type contra, P = 0.002216; wild-type ipsi, P = 8.8 × 10−6; cKO contra, P = 0.000903. Two-sided Wilcoxon rank signed tests with correction (α = 0.00625). Circles, median response changes (%). Inset, bootstrapped mean ± 95% confidence interval; values along axes match those along axes of main plot. n, Summary schematic. o, Top, visual cliff assay. Bottom, representative trajectories. p, Time in cliff across 1 min bins. q, Number of zone transitions. p,q, Two-way ANOVA on mice with post hoc Sidak’s tests against first-minute bin; *, significant P value. Wild type: n = 8 mice; Ccn1-cKO: n = 13 mice. Data are mean ± s.e.m. r, Representative z-projection of WFA around parvalbumin-positive somas. Scale bars, 20 µm. s, Integrated WFA density around parvalbumin-positive cells. Wild type no MD: n = 235 cells; wild type MD: n = 212 cells; cKO no MD: n = 220 cells; cKO MD: n = 198 cells. Dots represent individual cells. Five mice per group. Lines show median ± 95% confidence interval. Kruskal–Wallis tests with post hoc Dunn’s tests on cells; *, significant P value.
To characterize changes in binocular circuitry induced by MD or decreased CCN1 in astrocytes, we performed longitudinal tracking of layer 2/3 pyramidal neurons before and after five days of MD of the eye contralateral to the imaged hemisphere in wild-type and Ccn1-cKO mice (Extended Data Fig. 7d–f and Methods). Imaged neurons were classified on the basis of whether they were contralateral eye-responsive (contralateral-responsive), ipsilateral eye-responsive (ipsilateral-responsive), binocular or unresponsive (Fig. 3c,j and Methods). We found that Ccn1-cKO mice had a higher proportion of contralateral-responsive neurons relative to wild type before MD, and that this proportion decreased with MD of the contralateral eye (Fig. 3d,e and Extended Data Fig. 7i). We also found that Ccn1-cKO mice had a smaller proportion of binocular neurons relative to wild type and an increased binocular cell turnover (Fig. 3f,g and Extended Data Fig. 7j). Paralleling our results with the Arc induction assay, we found that Ccn1-cKO mice showed an increase in the number of ipsilateral-responsive neurons after MD (Fig. 3h,i and Extended Data Fig. 7k). After MD, both wild-type and Ccn1-cKO showed an increase in unresponsive neurons, though this increase was markedly greater in cKO mice (Extended Data Fig. 7g,h,l). This increase in unresponsiveness is similar to what is reported after MD during the critical period in wild-type mice31. Additionally, immature visual circuits have higher proportions of contralateral-responsive cells and lower proportions of binocularly responsive cells32, paralleling the results in adult Ccn1-cKO mice. We performed the same experiment, longitudinally tracking neurons, in naive mice that did not undergo MD. At baseline, we found the alterations in contralateral and binocular proportions in naive Ccn1-cKO versus wild-type mice, with no longitudinal changes (Extended Data Fig. 8a–k). In naive Ccn1-cKO mice, we did not see an increased binocular cell turnover relative to wild-type mice32 (Extended Data Fig. 8h). Together, our findings show that astrocyte CCN1 is necessary for the maturity and stability of binocular visual circuits.
Since ocular dominance of neurons is affected by MD33, we tested whether MD had a differential effect in Ccn1-cKO versus wild-type mice. We calculated ocular dominance index (ODI) of all longitudinally tracked responsive neurons (Methods). We found a robust effect of MD in wild-type mice, with the ODI shifting towards the ipsilateral, non-deprived eye, but no change in Ccn1-cKO mice in either the raw or hierarchically bootstrapped data (Fig. 3k and Extended Data Fig. 7m). The median ODI per mouse also shows a significant shift towards the ipsilateral eye in wild-type but not in Ccn1-cKO mice (Fig. 3l). We plotted the changes in contralateral and ipsilateral eye responses for each longitudinally tracked responsive neuron as a function of its pre-MD ODI, and found that neurons with initially stronger contralateral eye biases experienced a significant decrease in the magnitude of their contralateral eye responses after MD (Fig. 3m). However, only wild-type mice experienced a concomitant increase in ipsilateral response magnitude (Fig. 3m,n). When we examined average response magnitude for all longitudinally responsive cells, regardless of initial ODI, we found no difference at the mouse level between wild-type and cKO mice before or after MD (Extended Data Fig. 7o).
We next explored whether CCN1 could have an effect on additional functional properties of binocular circuits. We found that contralateral neurons and contralateral responses of binocular neurons in Ccn1-cKO mice, compared with wild-type mice, had different preferred orientations and circular variances, but similar tuning to cardinal orientations (Extended Data Fig. 8l,m,p and Methods). Relative to wild-type mice, Ccn1-cKO mice also showed decreased pairwise cellular spiking correlations and cell response reliability during stimulus presentation to the contralateral eye (Extended Data Fig. 8n,o). We did not find any significant modulation of spiking activity by locomotion in either wild-type or Ccn1-cKO mice (Extended Data Fig. 8q,r). We did not find any differences in binocular matching between wild-type and Ccn1-cKO mice (Extended Data Fig. 7n). However, to address whether the reduction in binocular neurons in Ccn1-cKO mice had an effect on binocular vision, we performed the visual cliff assay (Fig. 3o), which probes for depth perception34. We found that Ccn1-cKO mice, relative to wild-type mice, spent more time in the cliff zone (Fig. 3p), less time in the ground (Extended Data Fig. 9a and Methods), and had an increased number of zone transitions (Fig. 3q) with no difference in total distance travelled (Extended Data Fig. 9b). These findings reveal that underlying changes in the binocular circuit in Ccn1-cKO mice have a functional effect on binocular vision.
Together, these data indicate that in the absence of astrocyte CCN1 in adulthood, there is a shift in binocular circuitry, with reduced binocular cells and increased contralateral cells. Additionally, MD in Ccn1-cKO mice leads to a change in the binocular circuitry composition in adults that is absent in the wild type. Whereas we find changes in ODI—as previously reported—in the adult in wild-type mice, namely an increase in the magnitude of non-deprived eye responses35, we do not see these changes in Ccn1-cKO mice, suggesting that wild-type and cKO circuits have different strategies for regulating responsivity after MD (Fig. 3n).
CCN1 regulates PNNs and microglia
Since we found that removal of CCN1 from astrocytes results in immature binocular circuits in the adult, we predicted that reduction of CCN1 in adult astrocytes would result in decreased maturity of inhibitory neurons. To address this, we stained for PNNs with WFA and found that in adult mice, loss of CCN1 in astrocytes results in decreased PNN density around PV interneurons at baseline. PNN density in Ccn1-cKO mice was further reduced after five days of MD, with no change in wild-type mice (Fig. 3r,s and Extended Data Fig. 9c,d). These data suggest that inhibitory neurons are less mature in Ccn1-cKO mice. We then assayed microglial interaction with PNNs in adult wild-type and Ccn1-cKO mice (Extended Data Fig. 9e). We found no differences in PNN engulfment or in CD68 volume across groups (Extended Data Fig. 9f–i). Morphological examination of microglia revealed modest differences between wild-type and cKO mice (Extended Data Fig. 9j–o). Therefore, astrocyte CCN1 may have a larger role in regulating microglial state in younger mice.
ITGAV as a target of CCN1
Having established the role of astrocyte CCN1 in maintaining PNN, binocular circuit and inhibitory neuron maturity, we next set out to identify how CCN1 mediates these effects. In the periphery, CCN1 signals through multiple mechanisms, including by binding to various integrins36. A single point mutation, D125A, renders CCN1 unable to bind to αVβ5 or αVβ3 integrins4 (Fig. 4a). To test the role of these integrins in the effects of CCN1 on plasticity, we overexpressed CCN1(D125A) in juvenile mice using local viral injections into the visual cortex (Fig. 4a–c). We performed the Arc induction assay during the critical period and found no differences between tdT-OE mice and mice overexpressing CCN1(D125A) (CCN1(D125A)-OE mice) after ME (Fig. 4d,e), in contrast to wild-type CCN1, which is repressive of remodelling (Fig. 2). These data indicate that CCN1 binding to αVβ5 or αVβ3 is required for its effect on large-scale remodelling after visual deprivation.
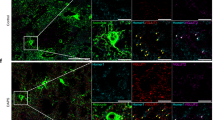
a, Top, CCN1 structure and binding sites. The position of the D125A mutation is indicated. Bottom, timeline of viral injections and experiments. CT, cysteine-knot-containing domain; IGFBP, insulin-like growth factor binding protein domain; HSPGs, heparan sulfate proteoglycans; IHC, immunohistochemistry; TSP-1, thrombospondin type 1 repeat homology domain; VWC, von Willebrand factor type C repeat. b, Representative z-projection of HA tag, SOX9 and NEUN immunofluorescence in a CCN1(D125A)-OE mouse. Scale bar, 100 µm. c, Penetrance and specificity of CCN1(D125A) overexpression. n = 4 mice. d, Representative images of Arc smFISH. Scale bars, 1 mm. Dashed yellow lines indicate Arc binocular zone width. e, Quantification of Arc activation width. n = 4 mice per group. Unpaired t-test on mouse averages. c,e, Symbols show mouse averages. Data are mean ± s.e.m. f, Representative z-projection of WFA and parvalbumin immunofluorescence. Scale bars, 20 µm. g, Integrated WFA density around parvalbumin-positive cells. tdT: n = 147 cells; CCN1: n = 200 cells; CCN1(D125A): n = 284 cells. Four mice per group. Kruskal–Wallis test with post hoc Dunn’s tests on cells; *, significant P value. Dots represent cells. Lines show median ± 95% confidence interval. h, Representative z-projection of aggrecan (ACAN) immunofluorescence. Scale bars, 50 µm. i, Number of ACAN+ cells. n = 4 mice per group. One-way ANOVA with post hoc Tukey’s tests on mouse averages; *, significant P value. Symbols show mouse averages. Data are mean ± s.e.m. j, snRNA-seq experiment. See Supplementary Fig. 2 for fluorescence-activated cell sorting (FACS) gating. Created in BioRender. Allen Lab (2025) https://BioRender.com/o2k2lbh. k, Uniform manifold approximation and projection (UMAP) of MapMyCells cell types from four samples (two replicates for tdT and CCN1). BAM, border-associated macrophage; COP, committed oligodendrocyte precursor; CT, corticothalamic; ET, extratelencephalic; MFOL, myelin-forming oligodendrocyte; MOL, mature oligodendrocyte; NFOL, newly formed oligodendrocyte. Interneurons include Lamp5-, Pvalb-, Sst- and Vip-expressing subtypes. l. Integrin gene expression. Normalized counts. m, AUGUR cell-type prioritization. AUC, area under the receiver operating characteristic curve. n, Excitatory neuron overrepresentation analysis (ORA) Gene Ontology (GO) terms. o, Mean UCell gene signature scores. Kruskal–Wallis test with pairwise Wilcoxon signed rank tests with Holm’s corrections; *, adjusted P value < 0.05.
We then assessed whether mutant CCN1 was able to regulate the density of PNNs using WFA, finding no difference between CCN1-OE and CCN1(D125A)-OE, with the mutant form still increasing PNN density relative to tdT-OE (Fig. 4f,g). Given our finding that CCN1(D125A) was unable to restrict large-scale remodelling, and the link between PNNs and plasticity repression, we investigated this further by specifically analysing aggrecan, a key component of the PNN that regulates plasticity37,38. Aggrecan immunostaining revealed that CCN1-OE mice have an increased number of aggrecan-positive cells in the visual cortex relative to tdT-OE mice, whereas this effect is absent in CCN1(D125A)-OE mice (Fig. 4h,i and Extended Data Fig. 10a). These findings demonstrate that CCN1 regulates multiple components of PNNs through distinct mechanisms, and that integrin binding is probably required for the effects of CCN1 on aggrecan and repression of plasticity.
snRNA-seq identifies cell targets of CCN1
Integrins are ubiquitously expressed among cell types in the brain39; we therefore utilized an unbiased approach to determine which cell types are most affected by CCN1 overexpression in astrocytes. We conducted single-nucleus RNA sequencing (snRNA-seq) of all cell types in the visual cortex of tdT-OE or CCN1-OE mice during the critical period (Fig. 4j, Extended Data Fig. 10b–d and Supplementary Table 3). We used the MapMyCells tool from the Allen Brain Atlas to cluster cells (Fig. 4k and Extended Data Fig. 10e–j), verifying that integrin subunits αV, β5 and β3 are expressed in many different cell types (Fig. 4l). DEG analysis revealed that Ddx5 is a commonly downregulated gene across multiple cell types in the visual cortex of CCN1-OE mice (Extended Data Fig. 11a,b and Supplementary Tables 3 and 4). Ddx5 is a negative regulator of the YAP/TAZ Hippo pathway40, an important pathway in nervous system development that can be regulated by CCN141, indicating direct signalling of astrocyte CCN1 to multiple cell types (Extended Data Fig. 2b).
We then performed AUGUR cell-type prioritization to determine the cell type that is most affected by astrocyte CCN1-OE, and identified excitatory neurons and oligodendrocytes as candidates (Fig. 4m). Many of the top DEGs in excitatory neuron subtypes (L2/3 intra-telencephalic (IT), L4/5 IT and L6 IT) are synapse-associated structural and functional genes (for example, Nrg2, Ctnna3) (Fig. 4n, Extended Data Fig. 11d–f and Supplementary Tables 4 and 5). These findings are in line with the electrophysiological changes that we observed in excitatory synapses onto pyramidal cells. We next examined oligodendrocyte lineage cells, which also express high levels of the αV integrin subunit (Fig. 4l). We performed UCell module signature scoring for myelin and oligodendrocyte-associated pathways and found an increase in the score for myelin sheath formation and cholesterol synthesis in oligodendrocytes in CCN1-OE mice (Fig. 4o and Supplementary Table 6). We also identified myelin-associated DEGs (Mag and Mbp; Extended Data Fig. 11c,g and Supplementary Tables 3 and 4). These data suggest that astrocyte CCN1 may be promoting maturation of oligodendrocytes.
CCN1 promotes oligodendrocyte maturation
To more closely examine the role of astrocyte CCN1 in regulating oligodendrocyte differentiation, maturation and myelination, we labelled different stages of oligodendrocyte development in tdT-OE, CCN1-OE or CCN1(D125A)-OE mice during the critical period. We found that CCN1 overexpression decreases the number of oligodendrocyte precursor cells (OPCs), whereas overexpression of CCN1(D125A) does not (Fig. 5a,b). When we examined newly formed oligodendrocytes and new myelin-forming oligodendrocytes, we observed an increase in their numbers in the CCN1-OE mice relative to tdT-OE mice, but not in the CCN1(D125A)-OE mice (Fig. 5c,d). We found no change in the number of mature oligodendrocytes or myelin expression (Fig. 5e–h). Together, these findings demonstrate that CCN1 induces OPC differentiation into myelin-forming oligodendrocytes and that this effect is likely to require CCN1 binding to αVβ5 or αVβ3 integrins.
a,c,e,g, Representative z-projections of oligodendrocyte-associated immunostaining in critical period visual cortex overexpressing tdT, CCN1 or CCN1(D125A). a, NG2 staining of OPCs. Scale bars, 50 µm. b, NG2+ cell density. tdT: n = 5 mice; CCN1: n = 6 mice; CCN1(D125A): n = 6 mice. c, Left, BCAS1 staining shows newly formed oligodendrocytes. Right, BCAS1 and MBP colocalization shows myelin-forming oligodendrocytes in the same sections. Scale bars, 50 µm. d, BCAS1+ (left) and BCAS1+MBP+ cell densities. tdT: n = 5 mice; CCN1: n = 6 mice; CCN1(D125A): n = 6 mice. e, CC1 and OLIG2 staining, showing mature oligodendrocytes. Scale bars, 50 µm. f, CC1+OLIG2+ cell density. tdT: n = 5 mice; CCN1: n = 6 mice; CCN1(D125A): n = 4 mice. b,d,f, One-way ANOVA with post hoc Tukey’s tests on mouse averages; *, significant P value. g, Bottom, myelin basic protein (MBP) staining. Top, illustration of myelin around an axon. Created in BioRender. Allen Lab (2025) https://BioRender.com/1acx0gt. Scale bars, 200 µm. h, MBP integrated density across visual cortex layers. tdT: n = 5 mice; CCN1: n = 4 mice; CCN1(D125A): n = 5 mice. Two-way ANOVA with post hoc Tukey’s tests on mouse averages; *, significant P value. See Supplementary Table 7 for post hoc statistics. i–p, As in a–h but in adult wild-type mice versus Ccn1-cKO mice. j, WT: n = 4 mice; cKO: n = 4 mice. l, WT: n = 5 mice; cKO: n = 5 mice. n, WT: n = 8 mice; cKO: n = 7 mice. p, WT: n = 5 mice; cKO: n = 5 mice. j,l,n, Unpaired t-tests on mouse averages. p, Two-way ANOVA with post hoc Sidak’s tests on mouse averages; *, significant P value. Data are mean ± s.e.m. Symbols represent mouse averages. q, Schematic summarizing mechanisms of CCN1 action.
We next examined whether CCN1 loss from astrocytes in adult mice affects OPCs and oligodendrocytes, and found no differences in the numbers of OPCs or differentiated oligodendrocytes (Fig. 5i–n). We found a significant reduction in myelin as labelled by myelin basic protein in Ccn1-cKO mice, most prominently in the deeper layers of visual cortex (Fig. 5o,p). We did not observe changes in myelin between wild-type and Ccn1-cKO mice in the hippocampus, entorhinal cortex or corpus callosum (Extended Data Fig. 12a,b). Thus, our data identify a role for astrocyte CCN1 in regulating oligodendrocyte differentiation and myelination in the visual cortex.
Discussion
Astrocytes have previously been described as producing stage-specific cues that instruct synapse formation, maturation and elimination8,18. Astrocytes can also modulate synaptic plasticity through secreted factors such as chordin-like 1 and Hevin9,42, clearance of ions and neurotransmitters from the synaptic cleft43,44, and through connexin 30 (ref. 10). In this study, we find that astrocytes also actively induce circuit stability in the adult visual cortex through CCN1, a secreted protein whose expression increases in adulthood and is downregulated when plasticity is induced. Most of the changes that arise after manipulating CCN1 expression in vivo do not require plasticity induction to be revealed, highlighting the role of CCN1 in regulating circuit stability during normal visual experience. We find that CCN1 acts on multiple cell types—excitatory neurons, inhibitory neurons, oligodendrocytes and microglia—underscoring the complex regulation of plasticity and stability by astrocytes (Fig. 5q). Overall, we have identified that CCN1 coordinates the maturational changes that occur in multiple cell types underlying the loss of plasticity in adulthood, highlighting the role of astrocytes as a hub for these effects.
We find that a point mutation that renders CCN1 unable to bind to αVβ5 or αVβ3 integrins occludes the effects of CCN1 on aggrecan, binocular zone remodelling and oligodendrocyte differentiation and maturation. Although this mutation has been characterized to specifically affect signalling through αVβ5 and αVβ3 integrins, and not other integrins or known receptors, we cannot rule out that other unidentified binding partners are also affected by this mutation. We also find that CCN1 reduction in adult astrocytes results in profound deficits in myelination in the visual cortex. This reduction in myelination is likely to be independent of a broad reduction in neuronal activity45, as calcium imaging did not show any reductions in response magnitude in Ccn1-cKO mice. Recent studies have pinpointed oligodendrocyte maturation and myelination as important regulators of critical period plasticity46, and our findings link CCN1 to these results. Together, our findings identify αV integrins as one of the putative signalling partners of astrocyte CCN1 in regulating binocular zone remodelling, aggrecan, and oligodendrocyte differentiation and maturation.
Our study also demonstrates that adult visual circuits, even after the establishment of binocularity47, require ongoing maintenance cues to maintain functional connectivity and visual behaviour, and that astrocytes provide these cues via the secretion of CCN1. Of note, these changes to the binocular visual circuit have an effect on a depth perception-based visual behaviour. As we used the Camk2a promoter to express gCaMP6f in predominantly excitatory neurons, this does not exclude the possibility that some of the imaged neurons were inhibitory cells48. Indeed, future studies could explore effects of CCN1 reduction in astrocytes on inhibitory neuron visual responses.
This study establishes the crucial role of astrocytes in actively stabilizing the connectivity of neuronal circuits. The effect of astrocyte CCN1 on the oligodendrocyte lineage and microglia, in addition to neurons, demonstrates that astrocytes are poised to orchestrate plasticity and circuit stability in the visual cortex by regulating the maturation of many cell types simultaneously. In turn, these findings deepen our understanding of how stability of sensory circuits is actively maintained in the adult brain, through a complex orchestration of multiple cell types by astrocytes. Identifying factors that make the neural environment permissive to plasticity in youth and restrictive in adulthood opens new avenues for therapies for recovery after brain injury. Both Ccn1 (ref. 49) and its transcriptional regulator Srf50 are upregulated after stroke, suggesting that restricting the induction of Ccn1 expression, or its actions, are targets to promote remodelling after injury.
Methods
Animals
All animal experiments were approved by the Salk Institute Institutional Animal Care and Use Committee (IACUC). Rats and mice were typically housed with a standard 12 h:12 h light:dark cycle in the Salk Institute animal facilities, with lights on at 06:00 and lights off at 18:00. Dark-reared mice were housed with a 24 h dark cycle since birth. Mice and rats were provided access to food and water ad libitum. Humidity ranged from 38–62% and temperature from 20 to 22 °C.
Mice
For bulk RNA=sequencing experiments, astrocyte-Ribotag mice were generated by crossing Gfap-cre hemizygous females (B6.Cg-Tg (GFAP-cre)73.12Mvs/J, Jax 012886) to homozygous flox-Rpl22-HA males (B6N.129-Rpl22tm1.1Psam/J, Jax 011029). Male mice hemizygous for cre and heterozygous for flox-Rpl22-HA (Rpl22-HA+;Gfap-cre+) were used for all experiments. Wild-type C57Bl6/J mice were used (Jax 000664) for experiments. For snRNA-seq, male wild-type mice were used. For smFISH experiments (Fig. 1h) validating the bulk RNA sequencing, male mice were used. For adult knockout experiments, Ccn1fl/fl mice were a gift from L. Lau51 and were maintained on a C57Bl6/J background. These mice were crossed to mice expressing tamoxifen-inducible Cre recombinase under an astrocytic promoter for temporal elimination (Aldh1;creERT2, Jax 029655 (ref. 52)). Experimental mice were homozygous for the Ccn1 floxed allele and either cre− or cre+ (wild type or Ccn1-cKO). For adult cKO experiments, mice were injected intraperitoneally with 75 mg kg−1 of tamoxifen (MP Biomedicals 156738) for 5 consecutive days at 1 month of age. For juvenile cKO experiments, mice were injected intraperitoneally once at P3–4 with 100 mg kg−1 of tamoxifen. Mice of both sexes were used and sexes were noted. Sample sizes were chosen on the basis of power analyses (80% power) and literature review. Experimenter was blinded to genotype or manipulation when analysing data.
Rats
Sprague-Dawley rats (Charles Rivers) were used at P1–2 for the preparation of primary cortical astrocyte cultures.
Surgical procedures
Juvenile viral injections
For adeno-associated virus (AAV) injections at P14–15, P11–12, or 3 months of age, C57Bl6/J mice were used. In brief, mice were administered pre-operative carprofen (5 mg kg−1) subcutaneously and anaesthetized using isoflurane. Stereotaxic coordinates for the binocular zone were 2.25 mm lateral and 0.5 mm anterior from lambda. Virus was injected at 3 sites at a depth of 500–600 µm from just below the skull surface. The pipette was kept in the brain for 3 min after each injection to allow the virus to diffuse. TdT, CCN1 and CCN1(D125A) viruses were injected for a total titre of ~2 × 108 viral genomes (vg) per ml. For snRNA-seq and western blotting, bilateral injections of both binocular zones were performed. After injection, mice were sutured and placed back with the dam if pre-weaning.
Monocular enucleation and monocular deprivation
For ME at P28 or 4 months, mice were anaesthetized using isoflurane. The eye was removed using curved forceps and pressure was applied to stop any bleeding. GelFoam was inserted into the eye socket and 2 box sutures (Henry Schein 5616446) were used to close the eyelid. Lidocaine jelly (2.0%, glydo) and erythromycin (0.5%, Bausch + Lomb) was applied to the eyelid. Mice were monitored daily and administered ibuprofen water (0.15 mg ml−1) to minimize any swelling.
For MD at P23, P28, or 4 months, mice were anaesthetized using isoflurane. Eyelashes were trimmed down to the eyelid margins and 4 box sutures using nylon sutures (Ethilon 1647 G) were used to close the eyelid. Lidocaine jelly (2.0%, glydo) and erythromycin (0.5%, Bausch + Lomb) was applied to the eyelid. Mice were monitored daily and administered ibuprofen water (0.15 mg ml−1) to minimize any swelling. Mice were removed from the experiment if the eyelids opened. For suture removal, mice were again placed under isoflurane and sutures were removed. Mice were removed from the experiment if the eye looked damaged or cloudy.
Cranial window implantation and viral injections
For in vivo imaging of ocular dominance plasticity and neuronal response properties, cranial windows were implanted on ~3-month-old Ccn1 wild-type or Ccn1-cKO mice. Mice were injected with buprenorphine SR (1 mg kg−1, subcutaneously), Baytril (10 mg kg−1, intramuscularly) and dexamethasone (2 mg kg−1, intraperitoneally) prior to surgery for anaesthesia, infection prevention and inflammation prevention. Mice were anaesthetized with isoflurane inhalant (3%) and maintained at 1.5–2.5% during the surgery. Mice were mounted on a stereotaxic surgical stage via ear bars and a bite bar. Their body temperature was maintained at 37 °C using a heating pad. The scalp was shaved and the skin was removed and the skull surface was allowed to dry. The skull and scalp margins were covered with a thin layer of Vetbond (Fisher Scientific NC0304169). Avoiding the area above visual cortex, a layer of dental cement (Tetric evoflow A1, Henry Schein 9458634) was applied and a metal head plate was glued to skull. A 3 mm circular piece of skull over the binocular zone (coordinates from lambda: 3 mm lateral, 1.0 mm anterior) was removed with a high speed microdrill with a 0.5 mm burr. Care was taken not to damage the dura. Before window implantation, viral injections of AAV2/1-CAMK1a-gCaMP6f (Addgene #100834-AAV1) were made into the binocular zone. A volume of 150 nl was injected into each of 5 sites at a depth of 300 µm from the pia for a total titre of ~1.2 × 1010 vg. The pipette was kept in the brain after each injection for 3 min to ensure diffusion of the virus before being retracted. A 4 mm diameter coverslip was placed on the dura and sealed to the edges of the skull using Vetbond. Dental cement was then used to further seal the coverslip. Mice were injected subcutaneously with 10 ml kg−1 physiological saline and carprofen (5 mg kg−1,subcutaneously) and placed on a heating pad to recover. Carprofen was administered daily for three days post-surgery. Mice were maintained with Baytril (8.5 mg kg−1 day−1) in their water to prevent infection for 3 days.
Bulk RNA sequencing
Data for experiments from P28 (critical period) and P120 (adult) mice were obtained from Farhy-Tselnicker et al.18 (GEO GSE161398) and Boisvert et al.17 (GEO GSE99791). The samples presented in this study were collected, processed and run at the same time as the samples from Farhy-Tselnicker et al.18. Three P120 biological replicates from Boisvert et al.17 were included and mapped together with the other samples onto the genome. All samples were processed and collected in the same way.
Conditions for analysis
Developmental time course
For experiments comparing mice P28 (critical period) to P120 (adult), collection was performed as described17,18. The visual cortices from two mice (Rpl22-HA+;Gfap-cre+) were pooled for RNA isolation and RNA-sequencing library preparation (P28: n = 5 biological replicates (10 mice, 2 × 5); P120: n = 6 biological replicates (12 mice, 2 × 6)).
Dark rearing
Dark rearing was performed by housing mice in ventilated telemetry cabinets, in complete darkness, and all husbandry and cage changes were done under red light. Mice for dark rearing were born in the dark and remained there until P45, anaesthetized under red light, and perfused with a hood over their head to prevent light from reaching the eyes. For age-matched comparison mice were raised under 12 h light:12 h dark cycle until P45. The visual cortices from 2 mice (Rpl22-HA+; Gfap-cre+) were pooled for RNA isolation and RNA-sequencing library preparation (P45 DR: n = 4 biological replicates (8 mice, 2 × 4); P45: n = 3 biological replicates (6 mice, 2 × 3)).
Monocular deprivation
MD was performed at P26, for 2 days until P28, during the peak of the critical period. The visual cortex contralateral to the deprived eye (major loss of visual input) and ipsilateral to the deprived eye (minor loss of visual input) were collected separately for analysis and comparison. The visual cortices from 2 mice (Rpl22-HA+; Gfap-cre+) were pooled for RNA isolation and RNA-sequencing library preparation (MD contra: n = 3 biological replicates, (contralateral visual cortex from 6 mice, 2 × 3); MD ipsi: n = 3 biological replicates, (ipsilateral visual cortex from 6 mice, 2 × 3)).
Ribotag pulldown and RNA sequencing
Male mice heterozygous for flox-Rpl22-HA (Jax 011029) and hemizygous for Gfap-cre (Jax 012886) (astrocyte-Ribotag) were used to isolated astrocyte mRNA on the basis of a modified Ribotag protocol as described18.
Dissection
All mice were collected between 09:30 and 12:30 on the day of experiment. Dissection was performed as described17,18. In brief, mice were anaesthetized with an intraperitoneal injection of 100 mg kg−1 ketamine (Victor Medical Company) plus 20 mg kg−1 xylazine (Anased) and then transcardially perfused with 10 ml phosphate-buffered saline (PBS) and then 10 ml 1% paraformaldehyde (PFA). Visual cortices were dissected out in 2.5 mM HEPES-KOH (pH 7.4), 4 mM NaHCO3 in Hank’s balanced salt solution with 100 μg ml−1 cycloheximide added day of the dissection. Visual cortices were dissected by cutting at approximately −2.4 mm posterior from Bregma, lateral cuts were made at 1 mm and 3 mm from the midline, and the white matter and any subcortical structures were removed. For each time point or plasticity group, the visual cortices from two mice were pooled.
Ribotag pulldown
A modified Ribotag protocol was performed as described17,18. In brief, brains were homogenized, centrifuged and incubated with HA antibody-conjugated magnetic IgG beads. RNA was purified using the RNeasy plus micro kit (Qiagen 74034) and eluted into water and stored at −80 °C.
RNA-sequencing library generation and sequencing
Library preparation was performed as described17,18. In brief, RNA quality was measured with a TapeStation (Agilent) and Qubit Fluorimeter (ThermoFisher). More than 100 ng of RNA was used to make libraries, and mRNA was extracted with oligo-dT beads to capture polyA tails. cDNA libraries were made with Illumina TruSeq Stranded mRNA Library Preparation Kit (RS-122-2101) by the Salk Institute Razavi Newman Integrative Genomics and Bioinformatics core. Samples were sequenced on an Illumina HiSeq 2500 with single-end 50 base-pairs reads, at 12–60 millions reads per sample.
Processing and analysis
RNA-sequencing mapping, analysis and statistics
Sequencing data mapping, analysis and statistics were done as described17,18. All samples were processed and aligned to the genome at the same time. In brief, raw sequencing data was converted into FASTQ files using CASAVA (v.1.8.2). Alignment to the mm10 genome was performed using STAR aligner (v.2.5.1b). Mapping was performed using the default parameters, and >75% uniquely mapped reads were confirmed with exonic alignment. Raw and normalized (FPKM) gene expression was quantified across all genes using the top-expressed isoform using HOMER (v.4.10). This resulted in 10–55 million uniquely mapped reads in exons. Differential gene expression was carried using DESeq2 (v.1.14.1) using the HOMER getDiffExpression.pl script with default normalization and replicates used to compute within-group dispersion. Significance for differential expression was set using adjusted P <0.05, using Benjamini–Hochberg correction for multiple comparisons adjustments.
Selection of DEGs
DEG analysis was run in the following comparisons: (1) P28 (critical period) versus P120 (adult); (2) P45 DR versus P45 NR; and (3) MD contralateral hemisphere versus ipsilateral hemisphere. Selection of DEGs for subsequent analysis was performed as follows: (1) FPKM >1 in mean of samples of at least one group per comparison; (2) Ribotag pulldown FPKM (astrocyte)/input FPKM (all cells) >0.75 in at least one group per comparison; (3) adjusted P value <0.05; and (4) fold change >|1.25| or log2FC >|0.3219|.
Venn diagrams of overlapping DEGs in different experimental comparisons were generated using R. Heat maps showing the LFC of DEGs were generated in R using the Pheatmap package. The predicted functional interaction network of CCN1 was generated using the STRING database53 (Extended Data Fig. 2b).
Pathway analysis
Gene-set enrichment analysis (GSEA) was performed to determine which predefined sets of genes were significantly enriched across the plasticity paradigms54. Enrichment of gene sets and pathways from the GO, Reactome and KEGG databases was carried out. Selection of DEGs for pathway analysis was performed as described above, but without a log2FC cut-off. Genes were ranked on the basis of descending log2FC in each comparison. A cut-off of adjusted P < 0.05 was used to determine significantly enriched pathways and terms. Simplifying GSEA results for visualization was conducted by only selecting pathways that were differentially expressed in at least two comparisons. Furthermore, we selected the top three downregulated and upregulated (negative and positive normalized enrichment score, respectively) Reactome pathways in each comparison (Extended Data Fig. 1b). ORA was performed to determine GO terms that were enriched in the significant DEGs across the plasticity paradigms (Extended Data Fig. 1c,d). Lists of upregulated and downregulated DEGs based on the same criteria as for the heat maps and Venn diagrams were included and the background list of genes were those that had FPKM >1 in the sample mean of at least one group per comparison. The following criteria were used to select significantly enriched GO terms: adjusted P value <0.05 (Benjamini–Hochberg correction) and q value < 0.01. The clusterProfiler package (v.4.10.0) was used to perform GSEA and ORA55.
Single-nucleus RNA sequencing
Tissue collection
P14 wild-type male mice were bilaterally injected with AAV-tdT or AAV-CCN1-HA. Mice were collected at P28. Mice were anaesthetized with intraperitoneal injection of 100 mg kg−1 ketamine/20 mg kg−1 xylazine mix and then decapitated. The brains were extracted and the injected visual cortices were dissected in ice-cold Dulbecco’s PBS (dPBS). To dissect the entire area expressing AAV, brains were cut from 2.4 mm posterior from bregma; lateral cuts from 1 to 3 mm from the midline were made to dissect the visual cortex. Two mice (four cortices) were pooled for each biological replicate. Two biological replicates were collected per viral group (four samples total).
Nuclei preparation and flow cytometry
Nuclei were extracted from the dissected visual cortices using the protocol in Farhy-Tselnicker et al.18 and nuclei were labelled with Hoechst 33342 solution. Fifty thousand single nuclei were purified using FACS using a BD FACS Aria Fusion with a 70-µm nozzle. Single Hoechst-positive nuclei were gated using fluorescence measured in the BV421 channel, and debris was excluded using forward and side scatter area and width parameters (FSC-A versus FSC-W, and SSC-A versus SSC-W). Nuclei were kept on ice for all the steps. TdT and CCN1 injected samples were processed in parallel on the same day for each repeat.
10X Chromium barcoding, library preparation and sequencing
Single nuclei separation, barcoding and cDNA generation were performed using the Chromium single cell 3′ kit following the manufacturer’s protocol (v.3.1 HT, 10X Genomics, 1000494 Kit and 1000371 Chip). cDNA concentration and quality measurements were performed using an Agilent Tape Station. Library preparation was carried out immediately after cDNA quality control. Libraries were generated as per the 10X instructions using the V3.1 HT kit. Quality and concentration were measured using an Agilent Tape Station and a Qubit Fluorimeter. Sequencing was performed at the University of Calfornia San Diego IGM Genomics Center using a NovaSeqX (Illumina) at 300 million reads per sample, or ~52 K average reads per cell.
Data preprocessing
Sequencing reads were mapped onto the mouse genome (mm10) using CellRanger count v.8.0.1. Median unique molecular identifier (UMI) counts per cell were as follows: tdt_replicate1: 6,428, ccn1_replicate1: 7,626, tdt_replicate2: 2,496, ccn1_replicate2: 2,810. Total genes detected were as follows: tdt_replicate1: 26,053, ccn1_replicate1: 25,318, tdt_replicate2: 24,940, ccn1_replicate2: 24,752. Median genes per cell: tdt_replicate1: 2,502, ccn1_replicate1: 2,720, tdt_replicate2: 1,272, ccn1_replicate2: 1,382. Mean reads per cell: tdt_replicate1: 67,863, ccn1_replicate1: 74,455, tdt_replicate2: 26,638, ccn1_replicate2: 41,400. Biological replicates were aggregated separately as they were sequenced in separate runs (replicate 1 and replicate 2) using CellRanger Aggr v.8.0.1 using normalization default parameters. Subsequent analyses were run in R Studio (R v.4.4.1) using Seurat (v.5.1.0). We discarded nuclei with less than 200 and more than 8,000 detected unique genes, with over 20% of sequencing reads mapped to mitochondrial genes and with over 10,000 detected RNA molecules per nucleus (UMI). Raw counts were normalized and scaled using the SCTransform function from Seurat and the default parameters, and log-normalized using NormalizeData. SCT data (normalized and scaled) were only used for visualization purposes for clustering, whereas normalized counts were used in differential expression analysis and all other analyses and visualizations.
To integrate the samples from the two different sequencing runs, the IntegrateLayers function from Seurat was used, with the Harmony integration method run using the SCT assay. Clustering was performed using the integrated data, with 30 principal component dimensions for reduction. UMAP embedding was used to visualize the data.
Clustering using MapMyCells
A count matrix as an AnnData object was extracted and uploaded to the Allen Brain Atlas MapMyCells website. The reference taxonomy was the 10x Whole Mouse Brain (CCN20230722), with Hierarchical mapping as the mapping algorithm. Classes, subclasses, and supertypes were mapped onto the Seurat object metadata and used to exclude non-visual cortex neuronal cell types.
DEG analyses
DEG analyses were performed using the normalized RNA counts. The Seurat FindMarkers function with Wilcoxon ranked sum tests with Bonferroni corrections for multiple comparisons were used. A min.pct = 0.01 and a log fold change cut-off of 0.0 was used. log2FC and min.pct cut-offs were adjusted for subsequent data visualization in the extended data figures as described in the legends.
Overrepresentation analysis
ORA was carried out using gene lists filtered out for up or downregulated significant DEGs (adjusted P <0.05). The clusterProfiler package (v.4.12.6) was used to perform ORA using GO terms (enrichGO)55.
Gene signatures
UCell Module scores were calculated for each gene using the UCell package (v.2.8.0)56. UCell module scoring is robust to dataset size and composition56, an advantage for examining the heterogeneous oligodendrocyte lineage. Lipid metabolism and myelin-associated gene lists were obtained from Gene Ontology. Wilcoxon signed rank tests were run between the tdT and CCN1 samples for each signature with Holm’s corrections for multiple comparisons.
AUGUR analysis
AUGUR cell-type prioritization analysis57 (v.1.0.3) was performed using the random forest classifier and setting the minimum number of cells to 100 and the subsample size to 100. This excluded some cell types with low abundance in the dataset, such as border-associated macrophages. In brief, AUGUR withholds a subset of labelled cell-type data (‘test’ data) and trains random forest classifiers for each cell type. Predictions are made on the unlabelled test data and accuracy is calculated on the basis of the area under the AUC of the classifier predictions. To minimize the confound introduced by differing number of cells in each cell type, AUGUR selects small subsamples from the dataset and calculates the mean AUC across subsamples.
Cloning
For the overexpression of HA-tagged CCN1, cDNA for the coding sequence of mouse CCN1 (Origene MR221828) was used. This cDNA was MYC-Flag tagged, so we amplified the cDNA with PCR primers (forward: GCGATCGCCATGAGCTCC, reverse: ttaaccggttgcataatccggaacatcatacggataGAGCGGCCGCGTACG) to insert an HA tag at the end of the C terminus in lieu of MYC-Flag. The fragment was then amplified again with primers optimized for InFusion cloning (Takara Bio 638909) (forward: cgactcactataggctagcgccaccATGAGC; reverse: tgtctgctcgaagcggccgcttaaccggttgcataatccggaacatcatacg). The linearized product was run out on a 0.8% agarose TAE gel and extracted. We used a pZac2.1 AAV2 backbone for overexpression. pZac2.1-gfaABC1D-tdTomato (GFAP-tdTomato) from Addgene (#44332) was digested with NheI and NotI to remove the tdTomato. The linearized product was run out on a 0.8% agarose TAE gel and extracted. An InFusion reaction was performed with the linearized PCR-amplified CCN1-HA and the linearized vector. Clones were selected using carbenicillin and sequenced to confirm presence of the inserted CCN1-HA. The cloned CCN1 plasmid was validated in vitro using astrocyte and HEK 293T/Cre cell cultures.
For synthesis of the CCN1(D125A) plasmid, the cloned CCN1 plasmid was sent to Genscript for point mutagenesis. The aspartic acid at position 125 was mutated into an alanine4. The mutated sequence was confirmed by next-generation sequencing.
The GFAP-CCN1-HA and GFAP-CCN1(D125A)-HA plasmids were sequenced and sent to the Salk Institute Gene Transfer, Targeting, and Therapeutics Core and were packaged into an AAV2/5 virus. Obtained titre ranged from 2–4 × 1012 vg ml−1. For the control virus, we obtained the AAV2/5 virus of the GFAP -tdTomato vector from Addgene (#44332-AAV2/5). Titres ranged from 1–4 × 1013 vg ml−1.
Cell culture
For validation of the plasmids, in vitro overexpression was performed in astrocytes purified from Sprague-Dawley P1–2 rats and in human embryonic kidney (HEK) 293T/Cre cells (ATCC CRL-3216, authenticated by ATCC and by morphology). The astrocytes were purified using the McCarthy–de Vellis method58, and were grown in culture media containing DMEM (Life Technologies 11960044), 10% Fetal bovine serum (Life Technologies 10437028), 1% penicillin-streptomycin (Life Technologies 15140122), 1% Glutamax (Life Technologies 35050061), 1% sodium pyruvate (Life Technologies 11360070), 5 μg ml−1 NAC (Sigma A8199), 5 μg ml−1 insulin (Sigma 11882) and 10 µM hydrocortisone (filtered with 0.22-µm filter). Cell culture dishes were coated with poly-D-lysine (Sigma P6407) before splitting cultured astrocytes onto them. Cultured astrocytes were passaged 2–3 times before being transfected.
HEK cells were grown in HEK cell growth media containing DMEM, 10% fetal bovine serum, 1% penicillin-streptomycin, 1% Glutamax, and 1% sodium pyruvate. HEK cells were passaged 4–5 times before being transfected. Cells were not routinely tested for mycoplasma contamination.
Plasmid validation
To validate plasmids via immunocytochemistry (Extended Data Fig. 3d,e), cultured astrocytes and HEK cells were plated in 24-well plates containing glass coverslips. For HEK cells, coverslips were coated with 1:50 CELLstart (ThermoFisher A1014201) diluted in water. For astrocytes, coverslips were coated with poly-D-lysine as described above. HEK cells and astrocytes were transfected the day after plating with 500 ng of plasmid DNA using Lipofectamine 2000 (Invitrogen 11668019) and OptiMEM (Life Technologies 31985-070). After transfection, astrocytes were maintained in astrocyte growth media, while HEK cells were maintained in HEK cell growth media. After 5 days of expression, cells were fixed with 4% PFA. Cells were then permeabilized with 1% BSA and 0.2% Triton X-100. Coverslips were incubated overnight at 4 °C in primary antibodies diluted in 1% BSA. For HEK cells, rabbit anti-HA antibodies (CST 3724), and sheep anti-CCN1 (R&D Systems AF4055) were used at 1:500. Secondaries were used at 1:1,000 for 2 h at room temperature. For astrocytes, mouse anti-GFAP (Millipore 360) and rabbit anti-HA were used. SlowFade Gold with DAPI mounting media (LifeTech S36939) was used. Coverslips were imaged using an Axio Imager.Z2 fluorescent microscope (Zeiss) with an AxioCam HR3 camera (Zeiss) at 20x magnification.
Mouse tissue collection
Tissue for single-molecule fluorescent in situ hybridization (smFISH) in Fig. 1 and Extended Data Fig. 2 was collected at P7, P14, P28, P45 and P120. Tissue for Ccn1-cKO validation was collected at two months of age. Tissue for smFISH against Arc was collected at P33 or 4 months of age. Tissue for immunohistochemistry was collected at approximately 1 month of age and 4 months of age.
smFISH
All mice for smFISH were collected between 13:00 and 17:00. Mice were anaesthetized by intraperitoneal injection of 100 mg kg−1 ketamine (Victor Medical Company)/20 mg kg−1 xylazine (Anased) mix and transcardially perfused with PBS. Brains were removed and embedded in OCT media (Sakura 4583), frozen in dry ice–ethanol slurry solution, and stored at –80 °C until use. Sagittal sections were obtained using a cryostat (Hacker Industries OTF5000) at a slice thickness of 18–20 µm. Sections were mounted on Superfrost Plus slides (Fisher 1255015). smFISH was performed on the same day as sectioning. Three to six mice were used for each experimental group. For each mouse (biological replicate), two or three sections (technical replicates) were imaged and analysed.
Arc induction
All mice for Arc induction were collected from the vivarium right before the end of the dark cycle (lights on at 06:00). Mice were brought back to the laboratory space and exposed to bright light for 30 min. Mice were then anaesthetized by intraperitoneal injection of 100 mg kg−1 ketamine (Victor Medical Company)/20 mg kg−1 xylazine (Anased) mix. Mice were decapitated, brains were extracted, embedded in OCT and flash frozen in dry ice–ethanol slurry mix. Brains were stored at −80 °C until use. Coronal sections were obtained using a cryostat at a slice thickness of 18–20 µm. Sections were mounted on Superfrost Plus slides (Fisher 1255015). smFISH was performed on the same day as sectioning. 3–6 mice were used for each experimental group. For each mouse (biological replicate), 3–6 sections (technical replicates) were imaged and analysed.
Immunohistochemistry
Mice were collected from the vivarium between 13:00 and 17:00 and were anaesthetized by intraperitoneal injection of 100 mg kg−1 ketamine/20 mg kg−1 xylazine mix and transcardially perfused with PBS, then 4% PFA at room temperature. Brains were removed and incubated in 4% PFA overnight at 4 °C, then washed 3 times for 10 min with PBS, and cryoprotected in 30% sucrose for 2–3 days. Brains were then embedded in tissue freezing media (TFM; General Data Healthcare TFM-5), frozen in dry ice–ethanol slurry solution, and stored at –80 °C until use. Brains were sectioned using a cryostat (Hacker Industries OTF5000) in sagittal or coronal orientations depending on experimental needs at a slice thickness of 18–20 µm. Sections were mounted on Superfrost Plus slides (Fisher 1255015). Three to eight mice were used for each experimental group. For each mouse (biological replicate), two or three sections (technical replicates) were imaged and analysed.
Western blot
Mice were collected between 13:00 and 17:00 and were anaesthetized by intraperitoneal injection of 100 mg kg−1 ketamine/20 mg kg−1 xylazine mix and transcardially perfused with dPBS. Brains were removed and bilaterally injected visual cortices were dissected out in ice-cold dPBS. To dissect the entire area expressing AAV, brains were cut from 2.4 mm posterior from bregma; lateral cuts from 1 to 3 mm from the midline were made to dissect the visual cortex. Both cortices were pooled per mouse. RIPA buffer with 1:100 Halt protease and phosphatase inhibitors (ThermoFisher 78430, 78420) was added (300 µl per sample) and homogenates were placed on a rotator for 1 h at 4 °C. Samples were spun down for 20 min at 13,000 RPM at 4 °C and supernatant was collected and frozen for subsequent analysis. Three mice for tdT and six mice for CCN1 were used. For each mouse (biological replicate), two immunoblots (technical replicates) were run and analysed.
Histology
Immunohistochemistry on mouse brain tissue
Slides containing the sections were blocked for 1 h at room temperature in blocking buffer consisting of 1% BSA and 0.2% Triton X-100 diluted in PBS. Primary antibodies were diluted in this blocking buffer and incubated overnight at 4 °C. The next day, slides were washed 3 times for 10 min with PBS and secondary antibodies conjugated to Alexa Fluor were diluted in blocking buffer and applied for 2 h at room temperature. Slides were mounted with the SlowFade Gold with DAPI mounting media, covered with 1.5 glass coverslip (Fisher 12544E), and sealed with clear nail polish. All secondary antibodies were applied at 1:500 dilution.
For validation of the viral vectors, the following antibodies were used: goat anti-SOX9 (R&D Systems af3075, 1:250), rabbit anti-HA (CST 3724, 1:500), mouse anti-NEUN (Millipore MAB377 1:100), mouse anti-GFAP (Millipore 360, 1:500). Secondary antibodies were donkey anti-goat Alexa Fluor 488 (Jackson ImmunoResearch 705-545-147), donkey anti-rabbit Alexa Fluor 568 (ThermoFisher A-10042), donkey anti-mouse Alexa Fluor 647 (Jackson ImmunoResearch 715-605-150), and goat anti-mouse Alexa Fluor 647 (ThermoFisher A-21235).
For PNN deposition experiments, the following antibodies were used: rabbit anti-HA (CST 3724, 1:500), mouse anti-parvalbumin (Millipore Sigma p3088, 1:500). Biotinylated WFA (Vector Laboratories B-1355-2, 1:500) was used at the same time as the primary antibodies to stain for the PNNs. Secondaries were used at 1:500 and included goat anti-rabbit Alexa Fluor 647 (ThermoFisher A-21245), goat anti-mouse Alexa Fluor 488 (ThermoFisher A-11001) and streptavidin conjugated Alexa Fluor 568 (adult mice, ThermoFisher S-11226) or 647 (critical period mice, ThermoFisher S-21374) to stain for PNNs.
For aggrecan staining, the following antibodies were used: rabbit anti-aggrecan (Millipore Sigma AB1031, 1:500), rat anti-HA (Sigma 11867423001, 1:100), and mouse anti-parvalbumin (Millipore Sigma p3088, 1:500). Secondaries were used at 1:500 and were goat anti-mouse Alexa Fluor 647 (ThermoFisher A-21236), goat anti-rat Alexa Fluor Plus 555 (ThermoFisher A48263), and goat anti-rabbit Alexa Fluor 488 (ThermoFisher A-11034).
For microglia morphology and phagocytosis experiments, the following antibodies were used: rabbit anti-IBA1 (Fuji Film Wako 162001, 1:500), rat anti-CD68 (Bio-Rad MCA1957GA, 1:100), and biotinylated WFA (Vector Laboratories B-1355-2, 1:500). Secondaries were used at 1:500 and were goat anti-rabbit Alexa Fluor 488 (ThermoFisher A-11008), goat anti-rat Alexa Fluor 594 (ThermoFisher A-11007), and streptavidin conjugated Alexa Fluor 647 (ThermoFisher S-21374).
For oligodendrocyte experiments, sections were counterstained with DAPI at 1:10,000 to enable easy cell counting. For all juvenile experiments, to confirm the presence of CCN1 or CCN1(D125A), an antibody against HA was used. To label oligodendrocyte progenitor cells (OPCs), rabbit anti-NG2 (1:200, Sigma Aldrich AB5320) and rat anti-HA (1:100, Sigma 11867423001) for juvenile experiments were used. Secondary antibodies were goat anti-rabbit Alexa Fluor 488 (ThermoFisher A-11034) and goat anti-rat Alexa Fluor Plus 555 (ThermoFisher A48263).
To label newly differentiated oligodendrocytes and myelin, mouse anti-BCAS1 (1:300, Santa Cruz SC-136342), rat anti-myelin basic protein (MBP, 1:200, Millipore MAB386), and rabbit anti-HA (1:500, CST 3724S) for juvenile experiments were used. For juvenile experiments, secondaries were goat anti-mouse Alexa Fluor Plus 488 (ThermoFisher A32723), goat anti-rat Alexa Fluor Plus 647 (ThermoFisher A48265), and goat anti-rabbit Alexa Fluor 555 (ThermoFisher A21429). For adult experiments, secondaries were goat anti-mouse Alexa Fluor 555 (ThermoFisher A21424) and goat anti-rat Alexa Fluor Plus 647 (ThermoFisher A48265).
To label mature oligodendrocytes, rabbit anti-OLIG2 (1:400, Millipore AB9610), mouse anti-APC (1:100, Millipore OP80), and rat anti-HA for juvenile experiments were used. For juvenile experiments, secondaries were goat anti-mouse Alexa Fluor Plus 488 (ThermoFisher A32723), goat anti-rat Alexa Fluor Plus 555 (ThermoFisher A48263), and goat anti-rabbit Alexa Fluor 647 (ThermoFisher A21245). For adult experiments, secondaries were goat anti-mouse Alexa Fluor 488 (ThermoFisher A-11001) and goat anti-rabbit Alexa Fluor 555 (ThermoFisher A21429).
For all immunohistochemistry, two or three sections (technical replicates) per mouse were imaged and averaged.
smFISH
All smFISH experiments were done as described18 except on fresh- frozen tissue as described in ‘Mouse tissue collection’. For tissue from P7 mice, slides were incubated with protease plus for 15 min; for P14–P120, protease 3 or 4 10–20 min. Probes used were: Arc (Biotechne 316911-C1), Ccn1 (Biotechne 429001-C1), Tubb3 (Biotechne 423391-C3) and Slc1a3 (Biotechne 430781-C2).
For experiments done in Fig. 2 and Extended Data Fig. 3k,m, the RNAScope V1 Multiplex assay was performed (Biotechne 320851). For the experiments done in Extended Data Fig. 4d–m, the RNAScope V2 Multiplex assay was run (Biotechne 323100) according to manufacturer instructions. The tissue baking step was eliminated and the hydrogen peroxide step was performed at room temperature for 10 min.
For Arc smFISH, three to six sections (technical replicates) were imaged and averaged per mouse (biological replicate). For Ccn1 smFISH, two to three sections (technical replicates) were imaged and averaged per mouse (biological replicate).
Western blot
Protein concentrations of the whole visual cortex lysates were quantified using the Bradford Protein Assay (Bio-Rad 5000006) on a plate reader (Tecan Infinite 200 PRO). Samples were diluted at least 40× to ensure accurate protein measurements and BSA was used as a standard. Reducing sample buffer (ThermoFisher 39000) was added to 20 µg of total proteins and samples were denatured at 55 °C for 45 min. Samples were then loaded into a 12-well Bolt 4–12% Bis-Tris gradient gel (ThermoFisher NW04122), using a PageRuler Prestained Protein Ladder (ThermoFisher 26626). Gels were run at 100 V for 1.5 h using a Mini gel tank (ThermoFisher A25977) and Bio-Rad PowerPac using MOPS running buffer (ThermoFisher NP000102). Transfer was done in Mini Trans-Blot Gel tank (Bio-Rad 1703930), using Tris-glycine transfer buffer (ThermoFisher 28363) with 20% methanol onto a PVDF membrane (Millipore Immobilin-FL IPFL00010). Membranes were blocked at room temperature for 1 h using 1% casein in Tris buffered saline (TBS; Bio-Rad 1610782). Primary antibodies were incubated overnight at 4 °C. Following primary antibody incubation, membranes were washed 3 times for 10 min in TBS (ChemCruz SC-362186) with 1% Tween (TBS-T). Secondary antibodies were used at 1:10,000 for 1 h at room temperature. Membranes were washed with 3 times for 10 min TBS-T and then placed in TBS until they were imaged.
For CCN1 overexpression validation, sheep anti-CCN1 (1:650, R&D AF4055) and mouse anti-pan actin (1:5000, Sigma Aldrich A1978) were used. Secondaries were donkey anti- sheep Alexa Fluor 680 (ThermoFisher A21102) and donkey anti-mouse Alexa Fluor 800 (ThermoFisher A32789). CCN1 and actin were probed for in the same blot, with CCN1 imaged in the 700 nm channel and actin in the 800 nm channel. The expected molecular mass of CCN1 is 37 kDa and the expected molecular mass of actin is 42 kDa.
Imaging and analysis
Membranes were imaged on a Licor Odyssey Imager Clx at a resolution of 84 μm (300 dpi). Automatic exposure was used. Analysis was conducted using ImageStudio and background lane fluorescence was used for normalization. CCN1 band intensities were additionally normalized using the actin loading control within each blot. The CCN1 group was also normalized to the tdT group for each membrane.
Histology imaging and analysis
Epifluorescence microscopy
Imaging was performed using an Axio Imager.Z2 fluorescent microscope (Zeiss) with the apotome module (Apotome2) and AxioCam HR3 camera (Zeiss) at 10× or 20× magnification, depending on the experiment. Tile images that contained the entire primary visual cortex (from pial surface to white matter tract) were acquired. Number of tiles were maintained consistent within each experiment. Images were 14 bit. All tiles had 10% overlap. For critical period experiments looking at WFA expression, all sections were confirmed to have viral vector expression.
For the Arc smFISH (Figs. 2b,e and 4d and Extended Data Fig. 4e,l), images were acquired at a single plane at 10× magnification, and the entire section was tiled. Arc activation width was measured along layer 4 (ref. 59) using the distance tool in Zen Blue.
For smFISH in Extended Data Figs. 3k and 4h for the Ccn1-cKO validation, images were acquired at 20×, with 3 tiles of the entire visual cortex taken, and a z-stack width of 10–12 μm.
For the immunohistochemistry for viral vector validation in Fig. 5 and Extended Data Fig. 2, images were acquired at 20× with 6 tiles taken of the visual cortex at 10% overlap and a z-stack width of 10–12 μm. For WFA and parvalbumin staining, images were acquired at 20× with 3 tiles taken of the visual cortex at 10% overlap and a z-stack width of 10–12 μm. For NG2, CC1 and OLIG2 staining in Fig. 5, images were also acquired at 20× with 3 tiles taken of the cortex at 10% overlap and a z-stack width of 10–12 μm.
In all cases, when comparing wild type and cKO or different viruses per given experiment, slides were imaged on the same day using the same acquisition settings, most notably camera exposure time was kept the same.
Confocal and super-resolution microscopy
smFISH images used for layer-specific developmental profile of Ccn1 expression were acquired using a Zeiss LSM 700 confocal scanning microscope. Images were acquired at 8-bit depth, 1,024 × 1,024 resolution using a 20× objective with a pixel dwell time of 0.79 µs. The scaling per pixel was 0.31 µm × 0.31 µm and 2 frames were averaged per plane. z-stacks at 1 µm steps were taken and the visual cortex was tiled at 10% overlap. Number of tiles remained consistent between experiments.
For microglia engulfment analysis, a Zeiss LSM 880 with Airyscan module was used. The images were acquired in super-resolution mode with a Fast Airyscan module using an oil-immersion 63× objective with a numerical aperture of 1.46 at a pixel dwell time of 0.60 µs. The scaling per pixel was 0.041 µm × 0.041 µm. z-stacks with 0.110 µm steps were acquired using a piezo, typically 150–200 planes per image. For the critical period mice expressing tdTomato, the 563 laser was used to photobleach the tdTomato signal prior to imaging the engulfment. To confirm that critical period mice injected with CCN1–HA, we confirmed viral vector expression with half the sections on each slide that were stained with rabbit anti-HA. The WFA channel (647) was imaged with an excitation beamsplitter of 488/561/633 and an emission filter set of 570–620 bandpass and 645 longpass. The CD68 channel (594) was imaged with an excitation beamsplitter of 488/594 and an emission filter set of 420–480 bandpass and 495–620 bandpass. The IBA1 channel (488) was imaged with an excitation beamsplitter of 488/561/633 and an emission filter set of 420–480 bandpass and 495–550 bandpass. Laser power for acquisition was kept the same across experiments. Images were processed by Airyscan in automatic mode.
For microglia morphology analysis, the same microscope was used as for the engulfment assay but in normal confocal mode. Images were acquired at 16-bit depth using a 20× objective with a pixel dwell time of 2.05 µs. The scaling per pixel was 0.21 µm × 0.21 µm. z-stacks at 2 µm steps were taken and the visual cortex was tiled at 15% overlap. Number of tiles remained consistent between experiments.
For analysis of newly differentiated oligodendrocytes (BCAS1-labelled oligodendrocytes) and myelin (MBP levels), a Zeiss LSM 900 was used. Images were acquired at 16-bit depth, 2,392 × 2,456 resolution using a 20× objective with a pixel dwell time of 0.41 µs. The scaling per pixel was 0.124 µm × 0.124 µm. z-stacks at 1 µm steps were taken and the visual cortex was tiled at 10% overlap. Four times two tiles were acquired (total eight tiles).
Image analysis
All image analysis of the smFISH in Fig. 1 and the Ccn1-cKO validation in Extended Data Figs. 3k and 4h was performed in ImageJ using a custom macro developed in the laboratory. The Slc1a3 signal was thresholded and used to segment astrocyte regions of interest (ROI). The Ccn1 probe signal was also thresholded and the total area within each astrocyte ROI was quantified. Thresholds were kept the same within experiments (for example, images acquired on the same day).
For quantification of the immunohistochemistry validating the viral vectors, the cell counter plug-in in ImageJ was used to count the number of transduced cells. For quantifying astrocyte GFAP level in Extended Data Fig. 3h,i, GFAP signal was thresholded equally in each pair of mice (tdT versus CCN1) and total area was quantified and compared.
To quantify the PNN around parvalbumin-expressing (PV) cells, a custom ROI detection pipeline was developed in CellProfiler. The PV channel intensity was rescaled in order to use the full intensity range to increase the brightness of the image for ROI selection. Both channels had a gaussian filter with a sigma of 1 applied. In order to reduce image heterogeneity and optimize ROI detection, the lower quartile of pixel intensity values were subtracted from the WFA and PV channels. ROIs were manually curated and added or removed. Finalized WFA and PV ROIs were then overlaid with the original, unprocessed images and the integrated density of WFA or PV per ROI was obtained. To separately quantify only WFA that surrounded PV cells, ROIs were overlaid and excluded if overlap was absent. The overlapping ROIs were overlaid with the original, unprocessed images and the integrated density per ROI was obtained. Values were exported to a csv file.
For oligodendrocyte and oligodendrocyte progenitor cells (OPCs), the cell counter plug-in in ImageJ was used to count the number of positive cells labelled by the different markers.
For analysis of microglia engulfment, images were analysed using pyclesperanto in the Napari viewer (https://github.com/clEsperanto/napari_pyclesperanto_assistant). In brief, the WFA, IBA1 and CD68 channels were independently segmented. A gaussian filter followed by a gamma correction at 1.5 were applied to the WFA signal prior to thresholding using the Otsu method. The IBA1 and CD68 signals were thresholded using the Otsu method. However, for critical period mice, for the CD68 signal the manual thresholding value was set to 900 instead of using the Otsu method due to differences in the CD68 signal in critical period aged mice. The overlapping regions (WFA + CD68 + IBA1 and CD68 + IBA1) were generated using the segmentations and saved as tiff files. To extract volumes of the segmentations, the tiff files were analysed in ImageJ using the 3D Object Counter plug-in and using the same thresholding for all images (Extended Data Figs. 6 and 9).
Microglia morphological classification
For microglia morphology analysis, images were processed in a custom ImageJ macro and analysed using a custom CellProfiler pipeline. In ImageJ, the contrast for each image was normalized so that at least 70% of the pixels are saturated, and the image was despeckled to remove noise. The pia and white matter were excluded from analysis to standardize the microglia morphology in each image. Then, in CellProfiler, the EnhanceOrSuppressFeatures module was used to suppress feature sizes of ten to segment soma from processes. The soma was identified as the primary object via the adaptive two-class Otsu thresholding method (threshold smoothing scale: 1, threshold correction factor: 1, size of adaptive window: 100). Soma area was measured using the MeasureObjectSizeShape module. The non-suppressed image underwent a gaussian filter (sigma 1) and thresholded using the global robust background method (averaging method: mean, variance method: standard deviation, no. of deviations: 0.8, threshold smoothing scale: 1, threshold correction factor: 0.9). From the thresholded image, secondary objects (processes) were identified via propagation from the overlaid primary objects (soma) using the adaptive two-class Otsu thresholding method (threshold smoothing scale: 0.3, threshold correction factor: 1, size of adaptive window: 10, regularization factor: 1). Microglia that had processes touching the borders of the image were excluded from analysis to remove artificial decreases in measurements. The MorphologicalSkeleton module was used to generate skeletons within the processes, and the MeasureObjectSkeleton module quantified number of trunks, non-truck branches, and total skeleton length of each microglia. Average branch length was calculated by dividing the total skeleton length by the sum of the number of trunk and non-trunk branches. All measurements were converted to microns.
For microglial morphology clustering analysis, Python sci-kit learn was used to scale and perform dimensionality reduction on all the extracted morphology features using principal component analysis (PCA). The number of components was chosen as the minimum number that explained 80% of the variance. Then, k-means clustering was performed and the elbow method was used to minimize the within cluster sum of squares. The number of chosen clusters was four for both critical period and adult mice, which is in line with the literature60,61,62,63. To plot the cluster heat maps, we used the scaled mean of each feature; we identified each cluster as amoeboid, rod-like, ramified (resting) or hyper-ramified on the basis of descriptions from the literature. The mean proportion per cluster for each mouse was calculated and a linear mixed effects model with a beta distribution was built in R using the glmmTMB and DHARMa libraries64. Post hoc tests looking at pairwise comparisons of clusters across viral groups or genotypes and manipulations were performed with P value adjustments for multiple comparisons.
Electrophysiology
Acute slice preparation
Coronal slices of the binocular zone of the visual cortex were prepared from P27–28 or P33–34 wild-type mice, that had been injected at P14 with GFAP–TdTomato or CCN1–HA, following described protocols65,66. Animals were deeply anaesthetized by injection with Avertin and decapitated. The brain was removed, hemi-sected, and cut into 300 µm coronal sections using a Leica VT1000s vibratome. The brain dissection was performed in cold, sucrose-based dissection solution consisting of (in mM): 2.5 KCl, 7.0 MgCl2, 1.25 Na2HPO4, 11 glucose, 234 sucrose, 0.50 CaCl2, and 24 NaHCO3 and equilibrated with carbogen (95% O2/5% CO2). Slices were then placed in a recovery chamber containing artificial cerebrospinal fluid (aCSF) consisting of (in mM): 126 NaCl, 26 NaHCO3, 1.25 Na2HPO4, 2.5 KCl, 2 CaCl2, 1 MgCl2, 25 glucose, and saturated with carbogen. Slices recovered for 30 min at 34 °C and were then maintained at room temperature until recordings were performed for 4–6 h after slicing.
Electrophysiology
Slices were placed in a recording chamber and perfused with a recirculating bath of carbogen-saturated aCSF maintained at 31 °C. Whole-cell patch clamp recordings were obtained from neurons in layer 2/3 of the injected binocular zone that were visualized using IR-DIC on a Scientifica microscope. Open pipette resistances were 2–5 MΩ (borosilicate glass pipette; Harvard Apparatus 30-0057). Recordings were performed using a Multiclamp 700B amplifier (Molecular Devices). All recordings were sampled at 10 kHz. For measuring the cell membrane properties, data were filtered at 10 kHz and measurements for analysis were taken 5 min after patching onto the cell. Recordings were discarded if the series resistances were >25 MΩ or changed >25% during the entire recording. For each mouse, 1–5 cells were recorded.
For experiments recording sEPSCs and sIPSCs at P27–28, sEPSCs were recorded in voltage clamp holding the cell at −60 mV and sIPSCs were recorded at +0 mV. Five minutes after breaking into the cell, cell membrane properties were measured as described above at −60 mV. sEPSCs were then recorded for 5 min with a filter of 2 kHz and a gain of 10 to allow the detection of small events and then the cell membrane properties were recorded again as described above. The holding voltage was then set to +0 mV to record sIPSCs for 5 min, filtered at 2 kHz and with a gain of 10. After recording sIPSCs, the holding voltage was set back to −60 mV to measure the membrane properties again. For each cell, sEPSCs or sIPSCs were excluded if series resistances changed >25% before and after recording or if cell seal was inadequate. Thus, for some cells, both sEPSCs and sIPSCs were analysed but for others only sEPSCs or sIPSCs were analysed. A caesium methanesulfonate internal was used (in mM): 100 caesium methanesulfonate, 20 KCl, 10 HEPES, 4 Mg-ATP, 0.3 Na-GTP, 10 Na-phosphocreatine, 3 QX 314 Chloride (Tocris 2313). Osmolarity and pH of the internal solutions were adjusted to 290–310 mOsm and pH 7.3–7.4 with double-distilled water and with CsOH.
For experiments recording sEPSCs from pyramidal and fast-spiking cells at P33–34, aCSF containing 40 µM bicuculline methochloride (Tocris 0131) was washed in. sEPSCs were recorded at −70 mV for 5 min and were filtered at 2 kHz and acquired at a gain of 10 to allow the detection of small events. Fast-spiking cells were confirmed as fast-spiking with little-to-no spike-frequency adaptation by injecting a 100 ms current step and noting the frequency and AP width in current-clamp mode. Additionally, fast-spiking neurons had a low input resistance (<150 MΩ) and multipolar morphology25. In these experiments, a potassium gluconate internal consisting of (in mM) was used: 115.0 potassium gluconate, 20.0 KCl, 10.0 phosphocreatine disodium salt, 10.0 HEPES acid, 0.2 EGTA, 4.0 Mg-ATP, and 0.3 Na-GTP. Osmolarity and pH of the internal solutions were adjusted to 290–310 mOsm and pH 7.3-7.4 with double-distilled water and with KOH.
Data analysis
Off-line data analyses were performed using MiniAnalysis (Synaptosoft) and MATLAB. sEPSC and sIPSC recordings were filtered post hoc with a 1 kHz low-pass Bessel 8-pole filter in Clampfit (Molecular Devices). For all experiments, 1–5 cells were analysed per mouse.
Visual cliff assay
The visual cliff experiments34,67,68 were conducted in a quiet room during the 12-h light cycle. The apparatus consisted of an open field behaviour box with clear walls and bottom. The arena was 40 cm × 40 cm and placed on the edge of a 1-m-tall table with half the box extended out over the table. A black and white checkboard mat was placed on the bench top and dropped down to the floor and extended out to create the ‘cliff’. A lamp was placed directly over the box to provide 30 lux of light but oriented to prevent any reflections on the plexiglass bottom. A camera was placed directly overhead pointed down at the arena and connected to AnyMaze tracking software. Mice were brought to the room 1 h prior to testing for habituation. The mouse’s movements were tracked during the duration of the entire 5 min trial. Measurements were binned by 1 min intervals. The arena was cleaned with Virkon followed by RO water between each mouse.
In vivo two-photon imaging
Two-photon calcium imaging
Recordings and habituation were performed during the 12 h light cycle. After recovery from viral injection and cranial window implantation, mice were habituated to handling and head fixation on a linear treadmill (LabMaker) over the course of one week. Imaging was performed during the day cycle of the mouse. We used a custom-built two-photon laser scanning microscope from Neurolabware equipped with a pulsed femtosecond Ti:Sapphire laser (Chameleon Vision II, Coherent) controlled by Scanbox acquisition software. The laser was tuned to 920 nm for imaging gCaMP6f and focused through a 16× water-immersion objective (Nikon, 0.8 numerical aperture). Images were acquired at a frequency of 15.5 Hz at a depth of 150–300 µm below the pial surface for layer 2/3. Images were 512 × 796 pixels (500 × 600 µm). A rotary encoder was used to track the running speed of the mice on the linear treadmill. A camera fitted with a 740 nm longpass filter was used to track pupil diameter during imaging (Mako U-015B). Both the encoder and the pupil camera were triggered at the scanning frame rate. To measure the responses of neurons to presentation of visual stimuli to either eye independently, an opaque eye path was placed in front of the other, non-imaged eye.
Binocular zone confirmation
Imaged areas that were stereotaxically identified during cranial window implantation (see above) as the binocular zone were confirmed as such by performing retinotopic mapping. A BENQ LCD 27-inch monitor (60 Hz refresh rate) was used for visual stimulus presentation, was placed 20 cm from the eyes, and covered approximately 113° in azimuth and 60° in elevation. Stimuli were designed in PsychoPy 2.22 and custom code was written to enable communication between the stimulus computer and the imaging computer. Stimulus presentation onset was tracked using a photodiode (Thorlabs, FDS1010) that was fed to an Arduino UNO rev. 3 and generated a transistor-transistor logic (TTL) pulse that was sampled by the imaging computer and time-stamped with the imaged frame. To confirm that the imaged area was indeed binocular, small checkerboard stimuli were presented pseudo-randomly at 15 neighbouring positions at a rate of 3 Hz. Stimuli were presented to either eye independently and randomly and regions were considered binocular if the peak responses in both eyes were evoked by stimuli presented in the central, upper visual field (−20° to +20° azimuth relative to the midline)33 (Extended Data Fig. 7a–c). Regions with no or weak ipsilateral responses were also not considered binocular.
Visual stimulus presentation for ocular dominance mapping
Drifting sinusoidal gratings were presented to each eye independently and randomly. The stimuli consisted of 16 directions (from 0° to 337.5°) and 2 spatial frequencies (0.03 and 0.13 cycles per degree) at 80% contrast. They were generated in PsychoPy 2.22. The full stimulus set was presented 5 times in one experiment in pseudo-random sequence. Gratings drifted at 1 Hz for 2 s, followed by a 4 s grey screen. Stimulus presentation onset was tracked using a photodiode and TTL pulses were generated and sampled by the imaging microscope to synchronize stimulation and imaging data.
Analysis
Image processing
Scanbox.sbx files were converted to tiff format and motion-corrected and segmented using Suite2p in Python (https://github.com/MouseLand/suite2p). The files from the contralateral and ipsilateral eye stimulation were aligned together to ensure the same cells were segmented. Files from different days were aligned and segmented separately. Rigid and non-rigid registration were run. After automated cell detection, the registered binary was manually checked and additional ROIs were drawn if necessary.
Identification of visually responsive cells
The F for the entire fluorescent trace of each cell was calculated by subtracting 0.7 × Fneuropil from Fcell. ΔF/F was calculated as (F − F0)/F0, where F0 is the baseline fluorescence. F0 was calculated as the 25th percentile of the fluorescence signal in a 30 s sliding window69,70. ΔFstimulus/F was calculated as the ΔF/F during the stimulus window and ΔFoff/F was calculated as the ΔF/F during the grey screen presentation. To compute whether a neuron was significantly visually responsive, we performed a Wilcoxon signed ranks test comparing ΔFstimulus/F and ΔFoff/F with a Bonferroni-corrected α = 0.05/number of unique trials (32 trials; α = 0.00156) to correct for multiple comparisons. Cells were considered visually responsive if they passed significance threshold for at least 25% of the trials of the single stimulus condition35,71. Cells were considered monocular if they had significant visual response to one or more stimulus conditions presented to either the contralateral or ipsilateral eye. Cells were considered binocular if they had a significant visual response to one or more stimulus conditions presented to each eye.
Ocular dominance index
The ODI of individual neurons was calculated as the ratio between the difference and the sum of the mean ΔFstimulus-peak/F in response to the ipsilateral or contralateral eye experimentally determined preferred drifting direction: ODI = (Rc − Ri)/(Rc + Ri), where Rc is the contralateral eye response to its preferred direction, and Ri is the ipsilateral eye response to its preferred direction. An ODI of −1 or +1 indicates ipsilateral or contralateral eye dominance, respectively. The stimulus-triggered response, ΔFstimulus-peak/F, was calculated by subtracting the mean ΔF/F for the 16 frames prior to stimulus presentation from the mean ΔF/F 7 to 31 frames (0.5 s to 2 s) after stimulus presentation (averaged 4 frames around the peak). Only neurons that were longitudinally tracked and responsive across imaging sessions were used for ODI calculations. For Fig. 3m, we used data only from longitudinally tracked neurons and binned the cells on the basis of their pre-MD ODI. We made 8 bins and calculated the median change in the binned cells’ contralateral eye responses and ipsilateral eye responses.
Orientation tuning
The preferred orientation of a neuron was calculated as:
On is the peak neuronal response to the 18 different orientations (0° to 170° spaced every 18°). θ is the orientation.
Global orientation selectivity was calculated as:
Cardinal proportions were calculated as the proportion of neurons having an orientation preference to 0° or 90°, ± 11.25°.
Binocular matching for binocular neurons was quantified as the absolute difference in preferred orientation of the contralateral and the ipsilateral eyes. \(\Delta {\rm{Orientation}}=|{{\rm{Ori}}}_{{\rm{contra}}}-{{\rm{Ori}}}_{{\rm{ipsi}}}|\).
If ΔOrientation > 90°, then the actual value is 180°- ΔOrientation.
Spiking correlations
Using Suite2p, the deconvolved spike train was extracted from each cell. An unconstrained non-negative deconvolution using exponential kernels was used. The kernel decay timescale was set to 1.0 and a gaussian filter with a smoothing constant of 10 was applied to the neuropil subtracted fluorescence trace. The neuropil subtraction coefficient was set to 0.7. The spiking activity of each pair of significantly responsive, longitudinally tracked neurons was calculated by concatenating the spiking activity for all trial-on periods and performing a pairwise Pearson’s correlation. This was done independently for contralateral and ipsilateral eye responses.
Response reliability
For all responsive, longitudinally tracked neurons, the response reliability was calculated by the coefficient of variation (cv = σ/μ) of the neuron across all trials of each unique stimulus condition. σ is the standard deviation of the neuron’s responses to all trials of a specific stimulus and μ is the mean of the trial responses.
Longitudinal imaging
To locate the same imaging plane for longitudinal imaging, we used the images acquired on previous days as reference, such as the vascular map of the brain surface acquired with the PCO camera and epifluorescence and the mean motion-corrected two-photon fluorescence images. The angle of the objective was kept the same. Fine adjustment of imaging depth and x,y location was performed using the Scanbox built-in plug-in searchref. In brief, a z-stack was automatically acquired and projected onto the mean motion-correct image from the prior imaging. The plug-in computes (using fast Fourier transform) the optimal translation of the microscope and moves the scope to best align the images (https://scanbox.org/2019/07/18/scanbox-searches-for-a-population/).
Longitudinal cell tracking
To track cells across multiple imaging sessions across different days, roiMatchPub.m, a MATLAB package written by A. Ranson was used (https://github.com/ransona/ROIMatchPub). In brief, the package uses a control point-based affine geometric transformation to correct for the plane rotation. The transformation is then applied to the image from the second imaging time point (‘post MD’). The overlap of ROIs between the mask file from the reference image and the transformed mask file from the second imaging time point is calculated. Overlapping ROIs were considered as longitudinally tracked after visual inspection and verification (Extended Data Fig. 7d,e). The average percentage of cells that were segmented before MD that were longitudinally tracked and also segmented post MD was 46 ± 6% (Extended Data Fig. 7f). For naive mice, the average percentage of cells that were longitudinally tracked from day 1 to day 6 of imaging was 26 ± 3.0% for wild-type mice and 35.9 ± 9.2% for Ccn1-cKO mice (Extended Data Fig. 8j). The difference in number of neurons that were longitudinally tracked between the two genotypes, combined with a lower number of mice in the wild-type naive condition, could account for the difference in the unresponsive number of neurons for all segmented cells versus longitudinally tracked unresponsive neurons (Extended Data Fig. 8d,j,k).
For experiments looking at cell proportions (Fig. 3d–j and Extended Data Figs. 7g–l and 8a–h), we used cells that were longitudinally tracked but not necessarily responsive, as we were also interested in cells that were unresponsive and became responsive after MD or vice versa. Contralateral, ipsilateral and binocular proportions were reported as a proportion of those cells that were responsive either pre MD or post MD32,47,72. For experiments looking at ODI, spiking correlations, locomotion, or tuning properties (Fig. 3k–n and Extended Data Figs. 7m,n and 8l–q), we looked at cells that were longitudinally tracked as responsive pre and post MD.
Bootstrapping
For bootstrapping analysis in Fig. 3k, we performed a hierarchical bootstrapping method73 with two levels: the animal level and the cells level. Sampling was performed 10,000 times for the dataset. A sample was taken with replacement at the first level (mice) with the sample size being equal to the number of mice. Then, a sample was taken with replacement at the second level, the neurons imaged. We chose a sample size of 100 which was slightly larger than the maximum number of neurons imaged per mouse. We reported the median across the 10,000 samples and directly calculated a P value (Pboot) that represents the probability that the mean ODI of the pre-MD group is larger than that of the post-MD group. The Pboot was used to determine statistically significant differences for the pre-MD and post-MD samples. For the bootstrapping analysis in Fig. 3m, inset, we used the bootci function in MATLAB for 1,000 samples for each of the pre-MD ODI bins. We calculated the mean for these 1,000 samples and a 95% confidence interval.
Locomotion analysis
Spiking data for each trial were extracted as described above. Locomotion data was obtained from the Scanbox quadrature file. Locomotion was converted to speed during each trial. Correlations between speed and spiking for all trials per responsive, longitudinally tracked cell were calculated using pairwise Spearman correlations to account for any non-normality in the locomotion data.
Statistical analysis and reproducibility
For most experiments, analyses were performed in GraphPad Prism (v.8.4.3 and v.10), with P values calculated to four decimal points. For WFA and PV quantifications, two-photon in vivo imaging data, and microglial morphology analyses were run in MATLAB (MathWorks) and Python. Significance was set at α = 0.05. Data were tested for normality, and two-tailed parametric or non-parametric tests were run as appropriate. Tests were corrected for multiple comparisons using either the Tukey or Sidak method for two-way ANOVA. For nested ANOVA for the electrophysiology, Tukey’s correction for multiple comparisons was used. For non-parametric multiple comparisons, the Dunn method was applied. Adjusted P values are shown. For the majority of statistical tests presented, the adjusted P values corrected for multiple comparisons are shown. For chi-square tests of proportions done in MATLAB, a Bonferroni correction for multiple comparisons was used to calculate the corrected α threshold (corrected α = 0.05/number of comparisons). For chi-square tests in Fig. 3 and Extended Data Figs. 7g and 8a–h, unadjusted P values are shown, with significance indicated based on Bonferroni-corrected α. For Wilcoxon Signed Rank tests of locomotion modulation the unadjusted P value is shown, with significance indicated based on Bonferroni-corrected α. For Kolmogorov–Smirnov tests, the unadjusted P value is shown, with significance indicated based on Bonferroni-corrected α. For microglia morphology, bulk RNA-sequencing and snRNA-seq analyses, R studio was also used. All statistical analyses excluding the sequencing are in Supplementary Table 7.
For data comparing mouse averages, biological replicates are an average of technical replicates, with the exception of the visual cliff assay, which was run once per mouse. Each experiment was repeated a minimum of two times. No data were excluded from analyses unless viral injection was off-target or if electrophysiology recordings had series resistances that were >25 MΩ or changed > 25% during the entire recording.
Data presentation
Figure legends indicate whether mean and s.e.m. or median and the 95% confidence interval are shown. P values are shown to 2 significant figures, except in the case of P < 0.0001 or P > 0.9999. Box plots show median and upper and lower quartiles. Violin plots represent all the data points, with medium smoothing. Bar graphs represent mean, with error bars showing s.e.m. Dot plots show all data points, with line at median and error bars showing 95% confidence intervals.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Bulk RNA-sequencing data have been deposited at GEO: GSE161398 (P7, P14, P28, P120 Ribo-seq), GSE99791 (P120 Ribo-seq), and GSE259341 (P45 NR, P45 DR, P28 contra and ipsi MD Ribo-seq). snRNA-seq data are deposited at GSE298814. Two-photon data processed with suite2p is available at https://doi.org/10.5281/zenodo.15785197 (ref. 74). Other data available on reasonable request from the authors. Source data are provided with this paper.
Code availability
All code and analysis pipelines, including the in vivo calcium imaging analysis, transcriptomics, microglia morphology and classification, CellProfiler WFA quantification and spiking correlations analysis, are available at https://doi.org/10.5281/zenodo.15785196 (ref. 75).
References
Hensch, T. K. Critical period plasticity in local cortical circuits. Nat. Rev. Neurosci. 6, 877–888 (2005).
Espinosa, J. S. & Stryker, M. P. Development and plasticity of the primary visual cortex. Neuron 75, 230–249 (2012).
Chen, C.-C., Mo, F.-E. & Lau, L. F. The angiogenic factor Cyr61 activates a genetic program for wound healing in human skin fibroblasts. J. Biol. Chem. 276, 47329–47337 (2001).
Chen, N., Leu, S. J., Todorovic, V., Lam, S. C. & Lau, L. F. Identification of a novel integrin αvβ3 binding site in CCN1 (CYR61) critical for pro-angiogenic activities in vascular endothelial cells. J. Biol. Chem. 279, 44166–44176 (2004).
Pizzorusso, T. et al. Reactivation of ocular dominance plasticity in the adult visual cortex. Science 298, 1248–1251 (2002).
Davis, M. F. et al. Inhibitory neuron transplantation into adult visual cortex creates a new critical period that rescues impaired vision. Neuron 86, 1055–1066 (2015).
Muller, C. M. & Best, J. Ocular dominance plasticity in adult cat visual cortex after transplantation of cultured astrocytes. Nature 642, 427–430 (1989).
Farhy-Tselnicker, I. & Allen, N. J Astrocytes, neurons, synapses: a tripartite view on cortical circuit development. Discov. Neurosci. 13, 7 (2018).
Blanco-Suarez, E., Liu, T. F., Kopelevich, A. & Allen, N. J. Astrocyte-secreted chordin-like 1 drives synapse maturation and limits plasticity by increasing synaptic GluA2 AMPA receptors. Neuron 100, 1116–1132.e13 (2018).
Ribot, J. et al. Astrocytes close the mouse critical period for visual plasticity. Science 373, 77–81 (2021).
Ackerman, S. D., Perez-Catalan, N. A., Freeman, M. R. & Doe, C. Q. Astrocytes close a motor circuit critical period. Nature 592, 414–420 (2021).
Lau, L. F. Cell surface receptors for CCN proteins. J. Cell Commun. Signal. 10, 121–127 (2016).
Kim, K.-H., Chen, C.-C., Monzon, R. I. & Lau, L. F. Matricellular protein CCN1 promotes regression of liver fibrosis through induction of cellular senescence in hepatic myofibroblasts. Mol. Cell. Biol. 33, 2078–2090 (2013).
Sin, W. C., Bechberger, J. F., Rushlow, W. J. & Naus, C. C. Dose-dependent differential upregulation of CCN1/Cyr61 and CCN3/NOV by the gap junction protein Connexin43 in glioma cells. J. Cell. Biochem. 103, 1772–1782 (2008).
Hooks, B. M. & Chen, C. Circuitry underlying experience-dependent plasticity in the mouse visual system. Neuron 106, 21–36 (2020).
Tagawa, Y., Kanold, P. O., Majdan, M. & Shatz, C. J. Multiple periods of functional ocular dominance plasticity in mouse visual cortex. Nat. Neurosci. 8, 380–388 (2005).
Boisvert, M. M., Erikson, G. A., Shokhirev, M. N. & Allen, N. J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269–285 (2018).
Farhy-Tselnicker, I. et al. Activity-dependent modulation of synapse-regulating genes in astrocytes. eLife 10, e70514 (2021).
Müller, C. M. Dark-rearing retards the maturation of astrocytes in restricted layers of cat visual cortex. Glia 3, 487–494 (1990).
Mower, G. D. The effect of dark rearing on the time course of the critical period in cat visual cortex. Dev. Brain Res. 58, 151–158 (1991).
Stogsdill, J. A. et al. Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197 (2017).
Lambo, M. E. & Turrigiano, G. G. Synaptic and intrinsic homeostatic mechanisms cooperate to increase L2/3 pyramidal neuron excitability during a late phase of critical period plasticity. J. Neurosci. 33, 8810–8819 (2013).
Kannan, M., Gross, G. G., Arnold, D. B. & Higley, M. J. Visual deprivation during the critical period enhances layer 2/3 GABAergic inhibition in mouse V1. J. Neurosci. 36, 5914–5919 (2016).
Miyamae, T., Chen, K., Lewis, D. A. & Gonzalez-Burgos, G. Distinct physiological maturation of parvalbumin-positive neuron subtypes in mouse prefrontal cortex. J. Neurosci. 37, 4883–4902 (2017).
Sancho, L. & Bloodgood, B. L. Functional distinctions between spine and dendritic synapses made onto parvalbumin-positive interneurons in mouse cortex. Cell Rep. 24, 2075–2087 (2018).
Ye, Q. & Miao, Q.-L Experience-dependent development of perineuronal nets and chondroitin sulfate proteoglycan receptors in mouse visual cortex. Matrix Biol. 32, 352–363 (2013).
Venturino, A. et al. Microglia enable mature perineuronal nets disassembly upon anesthetic ketamine exposure or 60-Hz light entrainment in the healthy brain. Cell Rep. 36, 109313 (2021).
Nguyen, P. T. et al. Microglial remodeling of the extracellular matrix promotes synapse plasticity. Cell 182, 388–403.e15 (2020).
Bai, T., Chen, C.-C. & Lau, L. F. Matricellular protein CCN1 activates a proinflammatory genetic program in murine macrophages. J. Immunol. 184, 3223–3232 (2010).
Yan, L. et al. Single and compound knock-outs of microRNA (miRNA)-155 and its angiogenic gene target CCN1 in mice alter vascular and neovascular growth in the retina via resident microglia. J. Biol. Chem. 290, 23264–23281 (2015).
Brown, T. C. & McGee, A. W. Monocular deprivation during the critical period alters neuronal tuning and the composition of visual circuitry. PLoS Biol. 21, e3002096 (2023).
Tan, L., Tring, E., Ringach, D. L., Zipursky, S. L. & Trachtenberg, J. T. Vision changes the cellular composition of binocular circuitry during the critical period. Neuron 108, 735–747.e6 (2020).
Mrsic-Flogel, T. D. et al. Homeostatic regulation of eye-specific responses in visual cortex during ocular dominance plasticity. Neuron 54, 961–972 (2007).
De Jesús-Cortés, H. et al. Using the visual cliff and pole descent assays to detect binocular disruption in mice. Preprint at bioRxiv https://doi.org/10.1101/2023.05.29.542767 (2024).
Rose, T., Jaepel, J., Hübener, M. & Bonhoeffer, T. Cell-specific restoration of stimulus preference after monocular deprivation in the visual cortex. Science 352, 1319–1322 (2016).
Lau, L. F. CCN1/CYR61: the very model of a modern matricellular protein. Cell. Mol. Life Sci.68, 3149–3163 (2011).
Rowlands, D. et al. Aggrecan directs extracellular matrix-mediated neuronal plasticity. J. Neurosci. 38, 10102–10113 (2018).
Ueno, H. et al. Expression of aggrecan components in perineuronal nets in the mouse cerebral cortex. IBRO Rep. 4, 22–37 (2018).
Cui, Y., Rolova, T. & Fagerholm, S. C. The role of integrins in brain health and neurodegenerative diseases. Eur. J. Cell Biol. 103, 151441 (2024).
Park, S. et al. Cerebral cavernous malformation 1 determines YAP/TAZ signaling-dependent metastatic hallmarks of prostate cancer cells. Cancers 13, 1125 (2021).
Chaqour, B. CCN–Hippo YAP signaling in vision and its role in neuronal, glial and vascular cell function and behavior. J. Cell. Commun. Signal. 17, 255–262 (2023).
Singh, S. K. et al. Astrocytes assemble thalamocortical synapses by bridging NRX1α and NL1 via hevin. Cell 164, 183–196 (2016).
Sibille, J., Pannasch, U. & Rouach, N. Astroglial potassium clearance contributes to short-term plasticity of synaptically evoked currents at the tripartite synapse. J. Physiol. 592, 87–102 (2014).
Pita-Almenar, J. D., Zou, S., Colbert, C. M. & Eskin, A. Relationship between increase in astrocytic GLT-1 glutamate transport and late-LTP. Learn. Mem. 19, 615–626 (2012).
de Faria, O., Gonsalvez, D. G., Nicholson, M. & Xiao, J. Activity-dependent central nervous system myelination throughout life. J. Neurochem. 148, 447–461 (2019).
Xin, W. et al. Oligodendrocytes and myelin limit neuronal plasticity in visual cortex. Nature 633, 856–863 (2024).
Tan, L., Ringach, D. L., Zipursky, S. L. & Trachtenberg, J. T. Vision is required for the formation of binocular neurons prior to the classical critical period. Curr. Biol. 31, 4305–4313.e5 (2021).
Veres, J. M., Andrasi, T., Nagy-Pal, P. & Hajos, N. CaMKIIα promoter-controlled circuit manipulations target both pyramidal cells and inhibitory interneurons in cortical networks. eNeuro https://doi.org/10.1523/ENEURO.0070-23.2023 (2023).
Guo, K. et al. Single-cell RNA sequencing with combined use of bulk RNA sequencing to reveal cell heterogeneity and molecular changes at acute stage of ischemic stroke in mouse cortex penumbra area. Front. Cell Dev. Biol. 9, 624711 (2021).
Hanna, M. et al. Mechanical regulation of the proangiogenic factor CCN1/CYR61 gene requires the combined activities of MRTF-A and CREB-binding protein histone acetyltransferase. J. Biol. Chem. 284, 23125–23136 (2009).
Monzon, R. I., Kim, K.-H. & Lau, L. F. in Methods in Molecular Biology (ed. Takigawa, M.) 361–376 (Springer, 2016).
Srinivasan, R. et al. New transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo. Neuron 92, 1181–1195 (2016).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Andreatta, M. & Carmona, S. J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021).
Squair, J. W., Skinnider, M. A., Gautier, M., Foster, L. J. & Courtine, G. Prioritization of cell types responsive to biological perturbations in single-cell data with Augur. Nat. Protoc. 16, 3836–3873 (2021).
McCarthy, K. D. & de Vellis, J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 85, 890–902 (1980).
Djurisic, M. et al. PirB regulates a structural substrate for cortical plasticity. Proc. Natl Acad. Sci. USA 110, 20771–20776 (2013).
Choi, S. et al. Automated characterisation of microglia in ageing mice using image processing and supervised machine learning algorithms. Sci. Rep. 12, 1806 (2022).
Fernández-Arjona, M. D. M., Grondona, J. M., Granados-Durán, P., Fernández-Llebrez, P. & López-Ávalos, M. D. Microglia morphological categorization in a rat model of neuroinflammation by hierarchical cluster and principal components analysis. Front. Cell. Neurosci. 11, 235 (2017).
Vidal-Itriago, A. et al. Microglia morphophysiological diversity and its implications for the CNS. Front. Immunol. 13, 997786 (2022).
Reddaway, J., Richardson, P. E., Bevan, R. J., Stoneman, J. & Palombo, M. Microglial morphometric analysis: so many options, so little consistency. Front. Neuroinformatics 17, 1211188 (2023).
Kim, J., Pavlidis, P. & Ciernia, A. V. Development of a high-throughput pipeline to characterize microglia morphological states at a single-cell resolution. eNeuro https://doi.org/10.1523/ENEURO.0014-24.2024 (2024).
Kuhlman, S. J., Lu, J., Lazarus, M. S. & Huang, Z. J. Maturation of GABAergic inhibition promotes strengthening of temporally coherent inputs among convergent pathways. PLoS Comput. Biol. https://doi.org/10.1371/journal.pcbi.1000797 (2010).
Xue, M., Atallah, B. V. & Scanziani, M. Equalizing excitation-inhibition ratios across visual cortical neurons. Nature 511, 596–600 (2014).
Boone, H. C. et al. Natural binocular depth discrimination behavior in mice explained by visual cortical activity. Curr. Biol. 31, 2191–2198.e3 (2021).
Fox, M. W. The visual cliff test for the study of visual depth perception in the mouse. Anim. Behav. 13, 232–233 (1965).
Schoenfeld, G., Carta, S., Rupprecht, P., Ayaz, A. & Helmchen, F. In vivo calcium imaging of CA3 pyramidal neuron populations in adult mouse hippocampus. eNeuro https://doi.org/10.1523/ENEURO.0023-21.2021 (2021).
Li, R. et al. Two-photon functional imaging of the auditory cortex in behaving mice: from neural networks to single spines. Front. Neural Circuits 12, 00033 (2018).
Salinas, K. J., Huh, C. Y. L., Zeitoun, J. H. & Gandhi, S. P. Functional differentiation of mouse visual cortical areas depends upon early binocular experience. J. Neurosci. 41, 1470–1488 (2021).
Tan, L., Ringach, D. L. & Trachtenberg, J. T. The development of receptive field tuning properties in mouse binocular primary visual cortex. J. Neurosci. 42, 3546–3556 (2022).
Saravanan, V., Berman, G. J & Sober, S. J Application of the hierarchical bootstrap to multi-level data in neuroscience. Neuron. Behav. Data Anal. Theor. 3, 1–25 (2020).
Sancho Fernandez, L. Astrocyte CCN1 stabilizes neural circuits in the adult visual cortex [Data set]. Zenodo https://doi.org/10.5281/zenodo.15785197 (2025).
Sancho, L. & Allen, N. J. Data from: Astrocyte CCN1 stabilizes neural circuits in the adult brain. Zenodo https://doi.org/10.5281/zenodo.15785196 (2025).
Acknowledgements
This work has been supported by US National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke (NINDS) 1R01NS105742, NIH NINDS DP11NS142587, the L.I.F.E. Foundation, a Pew Scholars Award, the Chan Zuckerberg Initiative, and the Roger Guillemin Chair to N.J.A. L.S. was supported by NIH National Eye Institute National Research Service Award 5F32EY033629 and a Salk Pioneer Fund Award. J.B. was supported by NINDS 1F99NS134205. M.C. was supported by a National Academies of Sciences, Engineering, and Medicine Ford Foundation Predoctoral Fellowship and NINDS 1F99NS1139511. L.L.-B. was supported by a Hewitt-Eckhart Postdoctoral Fellowship. L.T. was supported by the Shurl and Kay Curci Foundation. This work was supported by the following Salk Institute core facilities: Waitt Advanced Biophotonics Core, GT3 Core, Razavi Newman Integrative Genomics and Bioinformatics Core, Flow Cytometry Core, and In Vivo Scientific Services (NIH National Cancer Institute Cancer Center Support Grant P30 CA01495, NIH National Institute on Aging San Diego Nathan Shock Center P30 AG068635, NIH National Eye Institute R24NS092943, NIH National Institute on Aging Liver Cancer P01 AG073084-04, the Howard and Maryam Newman Family Foundation and the Helmsley Trust). Single-nucleus sequencing was conducted at the IGM Genomics Center at University of California, San Diego, La Jolla, CA. We thank the members of the Allen laboratory, E. Callaway, B. Emery and M. V. Sofroniew for discussion and feedback on the manuscript.
Author information
Authors and Affiliations
Contributions
N.J.A. and L.S. conceived of the project and designed the experiments. L.S. performed the experiments, designed the analysis pipelines and conducted the data analysis. M.M.B. performed the bulk RNA-sequencing experiments. J.B. performed the analysis and generated plots for the bulk RNA sequencing. M.C. and L.L.-B. performed the snRNA-seq. T.E., E.W. and L.T. contributed to immunostaining and image analysis for PNNs, microglia and oligodendrocytes, respectively. L.S. and N.J.A. interpreted the results and wrote the paper with comments from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Marc Freeman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Bulk ribo-Seq of visual cortex astrocytes.
a. Upset plot showing up- and down-regulated genes across the different comparisons (MD con vs ipsi, DR vs NR, CP vs Adult). MD, monocular deprivation. Con, contralateral eye to deprivation. Ipsi, ipsilateral eye to deprivation. CP, critical period. Adult, P120. DR, dark rearing. NR, normal rearing. 2 mice pooled per biological replicate. P28, n = 5. P120, n = 6. MD con/ ipsi, n = 3. P45 DR, n = 4. P45 NR, n = 3. Statistics done with DESeq2 using HOMER on biological replicates, Benjamini-Hochberg’s corrections for multiple comparisons. p-adjusted <0.05. b. Gene-set enrichment analysis (GSEA) results using the Reactome Database. NES is normalized enrichment score. Cutoff of adjusted p < 0.05 with a Benjamini-Hochberg correction. Pathways were significant in at least two comparisons. c, d. Overrepresentation analysis (ORA) to determine Gene Ontology (GO) terms enriched in DEGs across the comparisons. BP: Biological Process, CC: Cellular Component, MF: Molecular Function. Cutoff of adjusted p < 0.05 with a Benjamini-Hochberg correction.
Extended Data Fig. 2 Identifying CCN1 as a pro-stability astrocyte factor.
a. Heatmaps showing FPKM values and Log2FC of astrocyte-expressed synaptogenic, synapse elimination, and extracellular matrix genes. Stars, significance. P-adjusted <0.05. b. Predicted functional protein network of CCN1 using STRING database. c. Tiled sagittal brain section of an adult mouse smFISH for Ccn1. Scale bar, 1 mm. d. Representative z-projection of P120 smFISH in a tile image of the visual cortex (VC). smFISH against Slc1a3, Tubb3, and Ccn1. Same image from Fig. 1g. e. Ccn1 relative area per astrocyte after dark-rearing to P45 (DR) or normal rearing to P45 (NR) in the VC. Two-way ANOVA with post-hoc Tukey tests on mouse averages. Small dots, individual astrocytes. Large circles and bars, mouse averages ± SEM. N = 3 mice/ group.
Extended Data Fig. 3 In vivo manipulation of Ccn1 expression in astrocytes.
a. Experimental set-up for critical period overexpression (o/e) of CCN1 or tdTomato (tdT) viral vectors in astrocytes. BZ, binocular zone. b. Representative z-projection of P28 VC expressing tdT (left) or CCN1 (right). Scale bar, 50 µm. c. Left, penetrance of viral vectors quantified as % SOX9+ cells also expressing tdT/HA-tag. Right, specificity quantified as % total cells expressing tdT or CCN1 that are co-stained with SOX9, NEUN or no co-staining (other). tdT, n = 6 mice. CCN1, n = 5 mice. Unpaired T-test on mouse averages. d. Immunocytochemistry (ICC) of cultured astrocytes transfected with pAAV-GFAP-CCN1-HA. Cyan, HA-tag. Scale bar, 100 µm. e. ICC of HEK cells transfected with pAAV-GFAP-CCN1-HA and stained for CCN1 protein and the HA-tag. Scale bar, 100 µm. f. Cropped representative image of immunoblot against CCN1 (top) and β-actin (bottom). All uncropped immunoblots are shown in Supplementary Fig. 1. β-actin loading control and CCN1 immunoblotted on the same gel. g. Quantification of immunoblotted CCN1 band at ~42 kDa. tdT, n = 3 mice. CCN1, n = 6 mice. Unpaired T-test on mouse averages. h. Representative z-projection of VC GFAP in tdT and CCN1 o/e P28 mice. Scale bar, 200 µm. i. GFAP quantified as % total area of section. N = 5 mice/ group. Unpaired T-test on mouse averages. j. Schematic of adult conditional knockout validation experiments. Created in BioRender. Allen Lab (2025) https://BioRender.com/cp7gqpp. k. Representative z-projection of 2 month VC smFISH against Slc1a3 and Ccn1. Scale bar, 100 µm. l. Left, quantification of Ccn1 threshold area per astrocyte. Unpaired T-test done on mouse averages. Right, Ccn1 threshold area per neuron (labeled with Tubb3, bottom). Mann-Whitney U-test on mouse averages. m. Representative image of Ccn1 smFISH in layer 6 VC of adult WT and CCN1 cKO mice. Scale bar, 50 µm. Z-projection. n. Ccn1 threshold area per Tubb3+ cell in layer 6 of VC. Unpaired T-test on mouse averages. l,n. N = 6 mice/ genotype. For all bar graphs, data shown as mean ± SEM with symbols denoting mouse averages.
Extended Data Fig. 4 Context-dependent effects of manipulating Ccn1 expression in astrocytes.
a. Cumulative probability distribution of all sections Arc induction assay in critical period mice after 12 h of monocular enucleation (12 h) or 4 days of monocular enucleation (4D ME). N = 24 sections per group. b. As in a but in 4 month WT and cKO mice. WT 12 h, n = 30 sections. WT 4D ME, n = 34 sections. cKO 12 h, n = 27 sections. cKO 4D ME, n = 32 sections. a,b. Kolmogorov-Smirnov tests (α = 0. 0125) on sections. c. Arc induction assay in 4 month old Aldh1l1-creERT2 mice, not crossed to Ccn1fl/fl mice. Two-way ANOVA with post-hoc Tukey tests on mouse averages. Symbols, mouse averages. Cre- 12 h, n = 4 mice. Cre+ 12 h, n = 4 mice. Cre- 4D ME, n = 3 mice. Cre+ 4D ME, n = 4 mice. d. Experimental timeline for tdT or CCN1 o/e in adult WT mice. e. Representative image of Arc smFISH in the BZ after 4 days of monocular enucleation (ME). Left, tdT. Right, CCN1. Scale bar, 1 mm. Dashed yellow lines, Arc BZ width. f. Quantification of Arc activation width in layer 4 of hemisphere contralateral to enucleated eye. Unpaired T-tests on mouse averages. N = 3 mice/ group. Symbols, mouse averages. g. Schematic of experimental set-up for juvenile cKO of CCN1 in astrocytes. Created in BioRender. Allen Lab (2025) https://BioRender.com/cp7gqpp. h. Representative z-projection of 1 month VC smFISH against Slc1a3 and Ccn1. Scale bar, 50 µm. i. Quantification of Ccn1 threshold area per astrocyte. WT n = 4 mice, cKO n = 3 mice. j. Representative z-projection of 1 month VC smFISH against Tubb3 and Ccn1. Scale bar, 50 µm. k. Ccn1 threshold area per neuron (labeled with Tubb3, bottom). WT n = 4 mice, cKO n = 3 mice. l. Representative image of Arc smFISH in the juvenile BZ after 4 days of monocular enucleation (ME). Left, WT. Right, CCN1 cKO. Scale bar, 1 mm. Dashed yellow lines, Arc BZ width. m. Quantification of Arc activation width. N = 3 mice/ genotype. i,k,m. Unpaired T-tests on mouse averages. Symbols, mouse averages. For all bar graphs, data shown as mean ± SEM with symbols denoting mouse averages.
Extended Data Fig. 5 Astrocyte CCN1 regulates excitation onto pyramidal and fast-spiking neurons.
a. Decay (left) and 10–90% risetime (right) for sEPSC recordings in Fig. 2h at P28. tdT no MD: n = 16 cells, 6 mice. tdT MD: n = 17 cells, 5 mice. CCN1 no MD: n = 17 cells, 7 mice. CCN1 MD: n = 18 cells, 7 mice. b. As in a. but for sIPSC. tdT no MD: n = 17 cells, 7 mice. tdT MD: n = 18 cells, 7 mice. CCN1 no MD: n = 15 cells, 7 mice. CCN1 MD: n = 18 cells, 8 mice. c. Membrane resistance (left, MΩ) and capacitance (right, pF) at P28. tdT no MD: n = 16 cells, 6 mice. tdT MD: n = 16 cells, 5 mice. CCN1 no MD: n = 17 cells, 7 mice. CCN1 MD: n = 18 cells, 7 mice. d. Cumulative probability distributions of sEPSC amplitudes (left) and IEIs (right). e. Cumulative probability distributions of sIPSC amplitudes (left) and IEIs (right). d,e. Kolmogorov-Smirnov tests (α = 0.0125). f-q. Whole-cell patch clamp recordings done at P33 after no MD or 5 days of MD starting at P28. f. Representative spontaneous excitatory postsynaptic currents (sEPSCs) from each of the four conditions. g. Interevent interval (ms). h. Amplitude (pA). i. Decay (ms). j. 10- 90% risetime (ms). tdT no MD: n = 17 cells, 8 mice. tdT MD: n = 16 cells, 7 mice. CCN1 no MD: n = 13 cells, 7 mice. CCN1 MD: n = 16 cells, 8 mice. k. Representative sEPSCs as in f from fast-spiking neurons. l. Interevent interval (ms). m. Amplitude (pA). n. Decay. o. 10- 90% risetime. tdT no MD: n = 16 cells, 10 mice. tdT MD: n = 14 cells, 11 mice. CCN1 no MD: n = 12 cells, 7 mice. CCN1 MD: n = 10 cells, 6 mice. p. Pyramidal cells. Left, membrane resistance (MΩ). Right, capacitance (pF). q. Fast-spiking cells. Left, membrane resistance. Right, capacitance. Symbols, cells. Larger symbols, mouse averages. Thick black line, mean ± SEM. For all tests except in d and e, nested one-way ANOVAs with post-hoc Sidak tests on mouse averages.
Extended Data Fig. 6 Astrocyte CCN1 regulates microglial reactivity.
a. Representative single channel z-projections of WFA and PV staining. Same images as in Fig. 2n. Scale bar, 10 µm. b. WFA integrated density in all WFA+ cells, not just PV cells. N = 5 mice/ group. tdT no MD n = 577 cells. tdT MD n = 572 cells. CCN1 no MD n = 491 cells. CCN1 MD n = 538 cells. c. Representative z-projection of IBA1, WFA, and CD68 staining in critical period VC. Scale bar, 10 µm. d. Volume of WFA within CD68+ puncta within IBA1 volume. e. IBA1 volume per imaged microglia. f. Representative z-projection of IBA1and CD68 staining in critical period VC. Scale bar, 10 µm. g. Volume of CD68+ puncta within IBA1 volume. N = 39-40 microglia/group from 4 mice/group. b-g. All statistics are Kruskal Wallis with post-hoc Dunn’s tests on cells. h. Representative z-projection and skeletons of amoeboid microglia, rod-like, ramified, and hyper-ramified, left to right. i. Representative z-projection of IBA1 staining from critical period VC, top. Representative cell body and process tracing (skeleton), bottom. Scale bar, 10 µm. tdT no MD, n = 747 microglia from 4 mice. tdT MD, n = 876 microglia from 4 mice. CCN1 no MD, n = 496 microglia from 3 mice. CCN1 MD, n = 379 microglia from 3 mice. j. Proportion of microglial morphotypes- amoeboid, rod-like, ramified, and hyper-ramified. Two-way ANOVA with post-hoc Tukey tests on mouse averages. Only significant tests are shown. Symbols, mouse averages. Box plots, median with upper and lower quartiles. Whiskers, full range of data. k. Form factor (4*π*Area/Perimeter2). l. Total skeleton length. m. Mean branch length. n. Soma eccentricity. k-n. All statistics are Kruskal Wallis with post-hoc Dunn’s tests on cells. Violin plots show median, upper and lower quartiles. For all dot plots, each dot is an individual cell, line at median ± 95% CI.
Extended Data Fig. 7 Validation of in vivo two-photon calcium imaging of binocular zone neurons.
a. Retinotopic mapping of the BZ with small checkerboard boxes. b. Representative retinotopy in degrees of visual field in example mouse “L022”. Top, horizontal retinotopy, “azimuth”. Bottom, vertical retinotopy, “elevation”. Left, contralateral eye responses. Right, ipsilateral eye responses. Scale bar, 100 µm. c. Example mouse “L022”. Histograms of number of cells with peak responses to a particular azimuth (horizontal) or elevation (vertical). d. Validation of longitudinal tracking. Example mouse “L022”. Mean images pre, left, and post, right, MD. e. Left, fused image. Right, white denotes overlapping ROIs. f. Percent of longitudinally tracked cells across WT and CCN1 cKO mice. g. Unresponsive cells as a proportion of total cells. Corrected chi-square tests (4 comparisons; α = 0.0125). Stars, significance. h. Change in unresponsive cell proportion after MD, data and statistics from g. Stars, significant change relative to pre MD. i. C2I, contralateral to ipsilateral. C2B, contralateral to binocular. C2U, contralateral to unresponsive. j. B2M, binocular to monocular. M2B, monocular to binocular. Stable B, stable binocular. k. C2I, contralateral to ipsilateral. B2I, binocular to ipsilateral. U2I, unresponsive to ipsilateral. l. I2U, ipsilateral to unresponsive. C2U, contralateral to unresponsive. B2U, binocular to unresponsive. i-l. Corrected chi-square tests (3 comparisons; α = 0.01667). Stars, significance. WT total longitudinally tracked cells n = 453. CCN1 cKO total longitudinally tracked cells n = 511. m. ODI of longitudinally imaged responsive neurons only. WT n = 229 cells. cKO. n = 163 cells. n. Binocular matching as difference in preferred orientation for contralateral and ipsilateral responses for longitudinally tracked binocular neurons. WT n = 73 cells. cKO n = 37 cells. 4 mice/ genotype. m and n. Kruskal-Wallis tests with post-hoc Dunn’s tests on cells. For all dot plots, dots are individual cells. Lines and bars, median ± 95% CI. o. Mouse average peak ΔF/F for contralateral and ipsilateral. Two-way ANOVAs with paired uncorrected Fisher’s LSD tests on mouse averages. 4 comparisons; α = 0.0125. Starred p-values, significance. Bar graphs, mean ± SEM.
Extended Data Fig. 8 Binocular zone responses are altered in adult CCN1 cKO mice.
a-i. Longitudinally tracked neurons in naïve WT and CCN1 cKO mice. a-c. Contralateral, binocular, ipsilateral cells as a percentage of responsive cells. WT responsive cells day 1, n = 102. WT responsive cells day 6, n = 74. CCN1 cKO responsive cells day 1 n = 75. CCN1 cKO responsive cells day 6 n = 71. d. Unresponsive cells as a proportion of total cells. WT total cells n = 134. CCN1 cKO total cells n = 155. e. C2I, contralateral to ipsilateral. C2B, contralateral to binocular. C2U, contralateral to unresponsive. f. C2I, contralateral to ipsilateral. B2I, binocular to ipsilateral. U2I, unresponsive to ipsilateral. g. I2U, ipsilateral to unresponsive. C2U, contralateral to unresponsive. B2U, binocular to unresponsive. h. B2M, binocular to monocular. M2B, monocular to binocular. Stable B, stable binocular. a-h. Corrected chi-square tests of proportions. Stars, significance (a-d, α = 0.0125. e-h, α = 0.01667). i. Ocular dominance index for longitudinally tracked responsive cells. WT, n = 64 cells. cKO, n = 52 cells. Kruskal-Wallis test with post-hoc Dunn’s tests on cells. j. Percent of longitudinally tracked neurons in WT and cKO naïve mice. k. Pie chart of cell proportions for all cells, not just longitudinally tracked. a-k. Data from 2 WT mice, 3 CCN1 cKO mice. l-p. Longitudinally tracked responsive neurons from mice that underwent MD. l-m. Longitudinally tracked contralateral and binocular neurons in WT and CCN1 cKO adult mice. l. Orientation preferences. Kolmogorov-Smirnov tests on cells (α = 0.0125). Brackets, p-values. m. Global orientation tuning measured by circular variance. l-m. Contralateral neurons: WT n = 60, cKO n = 51. Binocular neurons: WT n = 73, cKO n = 37. n. Response reliability of all responsive, longitudinally tracked neurons. Left, contralateral eye responses. Right, ipsilateral eye responses. WT n = 229 neurons, cKO n = 163 neurons. o. Spiking correlations of all responsive, longitudinally tracked neurons. WT n = 9736 pairwise correlations, cKO n = 4241 pairwise correlations. m-o. Kruskal-Wallis tests with post-hoc Dunn’s tests on cells or correlation coefficients. p. Relative proportions of contralateral, ipsilateral, and binocular neurons with cardinal orientation preferences. Chi-square tests on cells. For binocular neurons, contralateral and ipsilateral response cardinal proportions were averaged. WT mice: contralateral neurons n = 60, binocular neurons n = 73, ipsilateral neurons n = 4. CCN1 cKO mice: contralateral neurons n = 51, binocular neurons n = 37, ipsilateral neurons n = 3. q. Histogram of locomotion during trial presentation in mice. r. Modulation by locomotion. Pair-wise Spearman’s coefficient of correlation between spiking activity and speed on treadmill. One sample Wilcoxon signed rank test on cells done against Spearman’s coefficient = 0. Corrected α = 0.0125 (4 comparisons). For all dot plots, dots represent individual cells. Lines and bars, median ± 95% CI.
Extended Data Fig. 9 Impaired depth perception and decreased PNN density in adult CCN1 cKO mice.
a. Exploration index in 1 min bins. Two-way ANOVA with post-hoc Tukey tests. b. Total distance traveled by mice during full 5 min of trial. Unpaired T-test. Symbols, mice. a,b. WT, n = 8. CCN1 cKO, n = 13. c. Representative single channel z-projections of WFA and PV staining. Same images as in Fig. 3r. Scale bar, 10 µm. d. WFA integrated density quantified per WFA+ cell. WT no MD n = 493 cells. WT MD n = 506 cells. cKO no MD n = 474 cells. cKO MD n = 446 cells. 5 mice/group. e. Representative z-projection of IBA1, WFA, and CD68 staining in adult VC. Scale bar, 10 µm. f. WFA colocalized to CD68+ puncta within IBA1 volumes. g. IBA1 volume within each field of view (FOV). h. Representative z-projection of IBA1and CD68 staining in adult VC. Scale bar, 10 µm. i. Volume of CD68+ puncta within IBA1 volume. N = 39-40 microglia/group from 4 mice/group. d-i. Kruskal Wallis tests with post-hoc Dunn’s tests on cells. j. Representative z-projection of IBA1 staining from adult VC, top. Representative cell body and process tracing (skeleton), bottom. Scale bar, 10 µm. WT no MD, n = 883 microglia, 4 mice. WT MD, n = 612 microglia, 4 mice. cKO no MD, n = 667 microglia, 4 mice. cKO MD, n = 543 microglia, 3 mice. k. Proportion of microglial morphotypes. Black symbols, mouse averages. Box plots, median with upper/lower quartiles. Whiskers, full range of data. Two-way ANOVA with post-hoc Tukey’s tests on mouse averages. l. Form factor. m. Total skeleton length. n. Mean branch length. o. Soma eccentricity. l-o. Kruskal Wallis test on cells with post-hoc Dunn’s tests. Bar graphs, mean ± SEM. Violin plots, median, upper and lower quartiles. For all dot plots, dots represent individual cells. Lines and bars, median ± 95% CI.
Extended Data Fig. 10 Validation of single-cell transcriptomics of visual cortex cell types.
a. Left, aggrecan (ACAN) integrated density per cell. Right, Proportion of PV+ cells expressing ACAN. One-way ANOVA with Tukey’s post-hoc tests on mouse averages. N = 4 mice/ group. Symbols, mouse averages. Bar graphs, mean ± SEM. b. Post-filtering genes per cell for all cell types. c. Post-filtering UMIs per cell across cell types. d. Post-filtering mitochondrial gene % per cell. b-d. dots, individual cells. e. Violin plots of canonical cell markers in different cell types. Normalized counts. f. UMAP after harmony integration of two biological replicates. g. UMAP of cell types with counts in different biological replicates, rep1 and rep2, and experimental groups, tdT and CCN1 o/e. h. UMAP of MapMyCells Subclass Annotations. i. Number of DEGs in each cell type for CCN1 vs tdT. min_pct > 0.01 and p-adjusted <0.05 using Seurat FindMarkers function with Wilcoxon Ranked Sum tests and Bonferroni corrections for multiple comparisons. j. Relative differences in cell proportions for CCN1 vs tdT using scProportion. Dot, mean of bootstrapped differences. Line, 95% CI. FDR set at 0.05.
Extended Data Fig. 11 Identifying targets of CCN1 using single-cell transcriptomics.
a. Violin plot of expression of Ddx5 across cell types. P-adjust shown from DEG analysis. Seurat FindMarkers function with Wilcoxon Ranked Sum tests and Bonferroni corrections for multiple comparisons. |Fold change| > 1.25 in all significant comparisons in CCN1 vs tdT. Dots, cells. b. Violin plot of Ddx5 expression in selected cell types with biological replicates separated. c. As in b. but for Mbp and Mag. b,c. Boxes inside violin plots, median and upper/lower quartiles. Dots, outliers (>1.5*inter-quartile distance). d. Over-representation analysis (ORA). GO terms (cellular components) of upregulated genes in excitatory neurons. e. ORA of Reactome pathway of upregulated genes in excitatory neurons. d,e. P-adjusted <0.05. f. Top DEGs in L2/3, L4/5, and L6 IT. IT is Allen Brain Atlas nomenclature for intra-telencephalic. min.pct >0.10. P-adjusted <0.05. Log2FC > |0.10 |. g. Top DEGs in oligodendrocytes. Seurat FindMarkers function with Wilcoxon Ranked Sum tests and Bonferroni corrections for multiple comparisons. min.pct >0.10. P-adjusted <0.05. Log2FC > |0.10|. Normalized count data.
Extended Data Fig. 12 Myelin basic protein in different brain regions of adult CCN1 cKO mice.
a. Representative z-projections of MBP in different brain regions. Left, hippocampus. Scale bar 200 µm. Middle, entorhinal cortex (EC). Scale bar 100 µm. Right, corpus callosum (CC) below VC. Scale bar 50 µm. b. Quantification of MBP integrated density. Unpaired T-tests on mouse averages. N = 5 mice/genotype. Symbols, mouse averages. Bar graphs, mean ± SEM.
Supplementary information
Supplementary Figures
Supplementary Figs. 1 and 2. Supplementary Fig. 1 includes the uncropped western blots from Extended Data Fig. 3 and Supplementary Fig. 2 includes an example plot for the single-nuclei gating strategy for FACS for snRNA-seq.
Supplementary Table 1
Data from the bulk RNA (Ribo-seq) sequencing experiments. Developmental data from Farhy-Tselnicker et al. 2021 and Boisvert et al. 2018. Ccn1 is named Cyr61. Related to Fig. 1 and Extended Data Fig. 2a.
Supplementary Table 2
Gene-set enrichment analysis (GSEA) data and overrepresentation analysis (ORA) for Ribo-seq experiment. Related to Extended Data Fig. 1.
Supplementary Table 3
DEGs for different cell types from the snRNA-seq. Related to Fig. 4j and Extended Data Figs. 10b–j and 11a–c.
Supplementary Table 4
DEGs in excitatory neuron and oligodendrocyte lineage subclusters from snRNA-seq data. Each tab is a different cluster. Related to Extended Data Fig. 11f,g.
Supplementary Table 5
ORA for snRNA-seq using GO terms that were enriched in the significant DEGs across the plasticity paradigms. Related to Fig. 4n and Extended Data Fig. 11d,e.
Supplementary Table 6
UCell module signature summary scores with statistics for snRNA-seq. Related to Fig. 4o.
Supplementary Table 7
Statistical tests and summary statistics from studies.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sancho, L., Boisvert, M.M., Eddy, T. et al. Astrocyte CCN1 stabilizes neural circuits in the adult brain. Nature 649, 948–958 (2026). https://doi.org/10.1038/s41586-025-09770-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09770-w