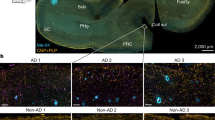
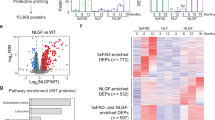
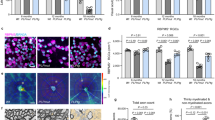
Abstract
Myelin ensheathment is essential for rapid axonal conduction, metabolic support and neuronal plasticity. In Alzheimer’s disease (AD), disruptions in myelin and axonal structures occur, although the underlying mechanisms remain unclear. We implemented proximity labeling subcellular proteomics of the myelin–axon interface in postmortem human brains from AD donors and 15-month-old male and female 5XFAD mice. We uncovered multiple dysregulated signaling pathways and ligand–receptor interactions, including those linked to amyloid-β processing, axonal outgrowth and lipid metabolism. Expansion microscopy confirmed the subcellular localization of top proteomic hits and revealed amyloid-β aggregation within the internodal periaxonal space and paranodal/juxtaparanodal channels. Although overall myelin coverage is preserved, we found reduced paranode density, aberrant myelination and altered paranode positioning around amyloid-plaque-associated dystrophic axons. These findings suggest that the myelin–axon interface is a critical site of protein aggregation and disrupted neuro-glial signaling in AD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Raw proteomics data are provided in Supplementary Table 1. The mass spectrometry proteomics datasets were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD045861. For proteomics sample information see Supplementary Table 2. SwissProt database can be accessed here: https://www.uniprot.org/uniprotkb?query=*&facets=reviewed%3Atrue%2Cmodel_organism%3A9606. GeneOntology.org database can be accessed here: https://geneontology.org/. g:profiler database can be accessed here: https://biit.cs.ut.ee/gprofiler/gost. OmniPath, a comprehensive intercellular database, can be accessed here: https://omnipathdb.org/. CellChat database can be accessed here: http://www.cellchat.org/. CellPhoneDB database can be accessed here: https://www.cellphonedb.org/. CollecTRI can be accessed here: https://github.com/saezlab/CollecTRI. Axogliaosome proteins were cited from ref. 105 (PMID: 20830807). The myelin human proteome was cited from ref. 40 (PMID: 35543322). Bulk brain tissue protein expression was cited from ref. 16 (PMID: 35115731) and ref. 17 (PMID: 32284590). Single-cell RNA expression was cited from ref. 11 (PMID: 31042697), ref. 14 (PMID: 31932797) and ref. 12 (PMID: 33432193).
Code availability
Code for STED imaging data analysis is available at https://github.com/bewersdorflab/Yifei-Lukas-Collab. R codes for the analysis of paranode size distribution can be accessed at the following location: https://github.com/ShawnQin/calcium_trace.
References
Lubetzki, C., Sol-Foulon, N. & Desmazieres, A. Nodes of Ranvier during development and repair in the CNS. Nat. Rev. Neurol. 16, 426–439 (2020).
Nave, K. A. & Werner, H. B. Ensheathment and myelination of axons: evolution of glial functions. Annu. Rev. Neurosci. 44, 197–219 (2021).
Monje, M. Myelin plasticity and nervous system function. Annu. Rev. Neurosci. 41, 61–76 (2018).
Hill, R. A., Li, A. M. & Grutzendler, J. Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat. Neurosci. 21, 683–695 (2018).
McKenzie, I. A. et al. Motor skill learning requires active central myelination. Science 346, 318–322 (2014).
Pan, S. et al. Preservation of a remote fear memory requires new myelin formation. Nat. Neurosci. 23, 487–499 (2020).
Snaidero, N. et al. Myelin replacement triggered by single-cell demyelination in mouse cortex. Nat. Commun. 11, 4901 (2020).
Arancibia-Carcamo, I. L. et al. Node of Ranvier length as a potential regulator of myelinated axon conduction speed. eLife 6, e23329 (2017).
Knowles, J. K. et al. Maladaptive myelination promotes generalized epilepsy progression. Nat. Neurosci. 25, 596–606 (2022).
Chen, W. T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991 e19 (2020).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Leng, K. et al. Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat. Neurosci. 24, 276–287 (2021).
Lau, S. F. et al. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer’s disease. Proc. Natl Acad. Sci. USA 117, 25800–25809 (2020).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Grubman, A. et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019).
Johnson, E. C. B. et al. Large-scale deep multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat. Neurosci. 25, 213–225 (2022).
Johnson, E. C. B. et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 26, 769–780 (2020).
Bai, B. et al. Proteomic landscape of Alzheimer’s disease: novel insights into pathogenesis and biomarker discovery. Mol. Neurodegener. 16, 55 (2021).
Askenazi, M. et al. Compilation of reported protein changes in the brain in Alzheimer’s disease. Nat. Commun. 14, 4466 (2023).
Mayer, M. C. & Meinl, E. Glycoproteins as targets of autoantibodies in CNS inflammation: MOG and more. Ther. Adv. Neurol. Disord. 5, 147–159 (2012).
Chang, K. J. et al. Glial ankyrins facilitate paranodal axoglial junction assembly. Nat. Neurosci. 17, 1673–1681 (2014).
Ho, T. S. et al. A hierarchy of ankyrin-spectrin complexes clusters sodium channels at nodes of Ranvier. Nat. Neurosci. 17, 1664–1672 (2014).
Simons, M. & Nave, K. A. Oligodendrocytes: myelination and axonal support. Cold Spring Harb. Perspect. Biol. 8, a020479 (2015).
Dutta, D. J. et al. Regulation of myelin structure and conduction velocity by perinodal astrocytes. Proc. Natl Acad. Sci. USA 115, 11832–11837 (2018).
Yuan, P. et al. PLD3 affects axonal spheroids and network defects in Alzheimer’s disease. Nature 612, 328–337 (2022).
Safaiyan, S. et al. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat. Neurosci. 19, 995–998 (2016).
Dean, D. C. et al. Association of amyloid pathology with myelin alteration in preclinical Alzheimer disease. JAMA Neurol. 74, 41–49 (2017).
Wang, Q. et al. Quantification of white matter cellularity and damage in preclinical and early symptomatic Alzheimer’s disease. Neuroimage Clin. 22, 101767 (2019).
Nasrabady, S. E. et al. White matter changes in Alzheimer’s disease: a focus on myelin and oligodendrocytes. Acta Neuropathol. Commun. 6, 22 (2018).
Araque Caballero, M. A. et al. White matter diffusion alterations precede symptom onset in autosomal dominant Alzheimer’s disease. Brain 141, 3065–3080 (2018).
Kramer-Albers, E. M. & Werner, H. B. Mechanisms of axonal support by oligodendrocyte-derived extracellular vesicles. Nat. Rev. Neurosci. 24, 474–486 (2023).
Cai, Y. et al. Subcellular proteomics and iPSC modeling uncover reversible mechanisms of axonal pathology in Alzheimer’s disease. Nat. Aging 5, 504–527 (2025).
Hashimoto, N. et al. Proteomic analysis of ganglioside-associated membrane molecules: substantial basis for molecular clustering. Proteomics 12, 3154–3163 (2012).
Bar, D. Z. et al. Biotinylation by antibody recognition—a method for proximity labeling. Nat. Methods 15, 127–133 (2018).
Vallat, J. M. et al. Contactin-associated protein 1 (CNTNAP1) mutations induce characteristic lesions of the paranodal region. J. Neuropathol. Exp. Neurol. 75, 1155–1159 (2016).
Bartsch, U., Kirchhoff, F. & Schachner, M. Immunohistological localization of the adhesion molecules L1, N-CAM, and MAG in the developing and adult optic nerve of mice. J. Comp. Neurol. 284, 451–462 (1989).
Quarles, R. H. Myelin-associated glycoprotein (MAG): past, present and beyond. J. Neurochem. 100, 1431–1448 (2007).
Erb, M. et al. Unraveling the differential expression of the two isoforms of myelin-associated glycoprotein in a mouse expressing GFP-tagged S-MAG specifically regulated and targeted into the different myelin compartments. Mol. Cell. Neurosci. 31, 613–627 (2006).
Li, J. et al. Cell-surface proteomic profiling in the fly brain uncovers wiring regulators. Cell 180, 373–386 e15 (2020).
Gargareta, V. I. et al. Conservation and divergence of myelin proteome and oligodendrocyte transcriptome profiles between humans and mice. eLife 11, e77019 (2022).
Paczkowska, M. et al. Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun. 11, 735 (2020).
Slobodyanyuk, M. et al. Directional integration and pathway enrichment analysis for multi-omics data. Nat. Commun. 15, 5690 (2024).
Tsuchimochi, R. et al. Viral delivery of L1CAM promotes axonal extensions by embryonic cerebral grafts in mouse brain. Stem Cell Rep. 18, 899–914 (2023).
Schafer, M. K. & Frotscher, M. Role of L1CAM for axon sprouting and branching. Cell Tissue Res. 349, 39–48 (2012).
Itoh, K. et al. Disrupted Schwann cell-axon interactions in peripheral nerves of mice with altered L1-integrin interactions. Mol. Cell. Neurosci. 30, 131–136 (2005).
Guo, D. et al. A disintegrin and metalloproteinase 10 (ADAM10) is essential for oligodendrocyte precursor development and myelination in the mouse brain. Mol. Neurobiol. 60, 1675–1689 (2023).
Chen, Y. Y. et al. Targeting of retinal axons requires the metalloproteinase ADAM10. J. Neurosci. 27, 8448–8456 (2007).
Kuhn, P. H. et al. Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. eLife 5, e12748 (2016).
Romi, E. et al. ADAM metalloproteases promote a developmental switch in responsiveness to the axonal repellant Sema3A. Nat. Commun. 5, 4058 (2014).
Bregin, A. et al. Expression and impact of Lsamp neural adhesion molecule in the serotonergic neurotransmission system. Pharmacol. Biochem. Behav. 198, 173017 (2020).
Thelen, K. et al. Translation of the cell adhesion molecule ALCAM in axonal growth cones – regulation and functional importance. J. Cell Sci. 125, 1003–1014 (2012).
Thelen, K. et al. Depending on its nano-spacing, ALCAM promotes cell attachment and axon growth. PLoS ONE 7, e40493 (2012).
Sharma, K. et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 18, 1819–1831 (2015).
Zhang, J. et al. ACSL4: a double-edged sword target in multiple myeloma, promotes cell proliferation and sensitizes cell to ferroptosis. Carcinogenesis 44, 242–251 (2023).
Zhou, T. et al. A mammalian NudC-like protein essential for dynein stability and cell viability. Proc. Natl Acad. Sci. USA 103, 9039–9044 (2006).
Efremova, M. et al. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Dimitrov, D. et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-seq data. Nat. Commun. 13, 3224 (2022).
Weidemuller, P. et al. Transcription factors: bridge between cell signaling and gene regulation. Proteomics 21, e2000034 (2021).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Dwork, A. J., Schon, E. A. & Herbert, J. Nonidentical distribution of transferrin and ferric iron in human brain. Neuroscience 27, 333–345 (1988).
Gerber, M. R. & Connor, J. R. Do oligodendrocytes mediate iron regulation in the human brain? Ann. Neurol. 26, 95–98 (1989).
Cheli, V. T. et al. Transferrin receptor is necessary for proper oligodendrocyte iron homeostasis and development. J. Neurosci. 43, 3614–3629 (2023).
Li, Y. et al. Transferrin receptor and ferritin-H are developmentally regulated in oligodendrocyte lineage cells. Neural Regen. Res. 8, 6–12 (2013).
Espinosa de los Monteros, A. et al. Transferrin is an essential factor for myelination. Neurochem. Res. 24, 235–248 (1999).
Mietto, B. S. et al. Schwann cells provide iron to axonal mitochondria and its role in nerve regeneration. J. Neurosci. 41, 7300–7313 (2021).
Zhu, X. et al. Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8, e1002853 (2012).
Kaneshiro, N. et al. Lipid flippase dysfunction as a therapeutic target for endosomal anomalies in Alzheimer’s disease. iScience 25, 103869 (2022).
Cali, T. et al. A novel mutation in isoform 3 of the plasma membrane Ca2+ pump impairs cellular Ca2+ homeostasis in a patient with cerebellar ataxia and laminin subunit 1ɑ mutations. J. Biol. Chem. 290, 16132–16141 (2015).
Lecuyer, M. A. et al. Dual role of ALCAM in neuroinflammation and blood-brain barrier homeostasis. Proc. Natl Acad. Sci. USA 114, E524–E533 (2017).
Kunkle, B. W. et al. Novel Alzheimer disease risk loci and pathways in African American individuals using the African Genome Resources Panel: a meta-analysis. JAMA Neurol. 78, 102–113 (2021).
Zhao, L. et al. Alteration of the unfolded protein response modifies neurodegeneration in a mouse model of Marinesco-Sjogren syndrome. Hum. Mol. Genet. 19, 25–35 (2010).
Rao, S. et al. Biological function of HYOU1 in tumors and other diseases. Onco Targets Ther. 14, 1727–1735 (2021).
Zhu, Z. Y. et al. Mitochondrial aldehyde dehydrogenase (ALDH2) rescues cardiac contractile dysfunction in an APP/PS1 murine model of Alzheimer’s disease via inhibition of ACSL4-dependent ferroptosis. Acta Pharmacol. Sin. 43, 39–49 (2022).
Jia, B. et al. ACSL4-mediated ferroptosis and its potential role in central nervous system diseases and injuries. Int. J. Mol. Sci. 24, 10021 (2023).
Hohn, L. et al. Extracellular matrix changes in subcellular brain fractions and cerebrospinal fluid of Alzheimer’s disease patients. Int. J. Mol. Sci. 24, 5532 (2023).
Zhang, Q. et al. Human brain glycoform coregulation network and glycan modification alterations in Alzheimer’s disease. Sci. Adv. 10, eadk6911 (2024).
Chu, T. H. et al. Axonal and myelinic pathology in 5xFAD Alzheimer’s mouse spinal cord. PLoS ONE 12, e0188218 (2017).
Sarkar, D. et al. Revealing nanostructures in brain tissue via protein decrowding by iterative expansion microscopy. Nat. Biomed. Eng. 6, 1057–1073 (2022).
Chapman, T. W. et al. Oligodendrocyte maturation alters the cell death mechanisms that cause demyelination. J. Neurosci. 44, e1794232024 (2024).
Griffiths, I. et al. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science 280, 1610–1613 (1998).
Ogawa, Y. & Rasband, M. N. Proteomic analysis of optic nerve lipid rafts reveals new paranodal proteins. J. Neurosci. Res. 87, 3502–3510 (2009).
Bartzokis, G. Age-related myelin breakdown: a developmental model of cognitive decline and Alzheimer’s disease. Neurobiol. Aging 25, 5–18 (2004).
Chen, J. F. et al. Enhancing myelin renewal reverses cognitive dysfunction in a murine model of Alzheimer’s disease. Neuron 109, 2292–2307 e5 (2021).
Depp, C. et al. Myelin dysfunction drives amyloid-β deposition in models of Alzheimer’s disease. Nature 618, 349–357 (2023).
Kedia, S. et al. T cell-mediated microglial activation triggers myelin pathology in a mouse model of amyloidosis. Nat. Neurosci. 27, 1468–1474 (2024).
Auer, F., Vagionitis, S. & Czopka, T. Evidence for myelin sheath remodeling in the CNS revealed by in vivo imaging. Curr. Biol. 28, 549–559 e3 (2018).
Osso, L. A. & Hughes, E. G. Dynamics of mature myelin. Nat. Neurosci. 27, 1449–1461 (2024).
Guo, Q. et al. Amyloid precursor protein revisited: neuron-specific expression and highly stable nature of soluble derivatives. J. Biol. Chem. 287, 2437–2445 (2012).
Skaper, S. D. et al. Oligodendrocytes are a novel source of amyloid peptide generation. Neurochem. Res. 34, 2243–2250 (2009).
Zheng, H. & Koo, E. H. The amyloid precursor protein: beyond amyloid. Mol. Neurodegener. 1, 5 (2006).
Rajani, R. M. et al. Selective suppression of oligodendrocyte-derived amyloid beta rescues neuronal dysfunction in Alzheimer’s disease. PLoS Biol. 22, e3002727 (2024).
Sasmita, A. O. et al. Oligodendrocytes produce amyloid-β and contribute to plaque formation alongside neurons in Alzheimer’s disease model mice. Nat. Neurosci. 27, 1668–1674 (2024).
Jankowsky, J. L. et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: evidence for augmentation of a 42-specific γ secretase. Hum. Mol. Genet 13, 159–170 (2004).
Almeida, R. G. et al. Myelination of neuronal cell bodies when myelin supply exceeds axonal demand. Curr. Biol. 28, 1296–1305 e5 (2018).
Fard, M. K. et al. BCAS1 expression defines a population of early myelinating oligodendrocytes in multiple sclerosis lesions. Sci. Transl. Med. 9, eaam7816 (2017).
Barres, B. A., Lazar, M. A. & Raff, M. C. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development 120, 1097–1108 (1994).
Barres, B. A. et al. Cell death and control of cell survival in the oligodendrocyte lineage. Cell 70, 31–46 (1992).
Garcia-Fresco, G. P. et al. Disruption of axo-glial junctions causes cytoskeletal disorganization and degeneration of Purkinje neuron axons. Proc. Natl Acad. Sci. USA 103, 5137–5142 (2006).
Quintela-Lopez, T. et al. Aβ oligomers promote oligodendrocyte differentiation and maturation via integrin β1 and Fyn kinase signaling. Cell Death Dis. 10, 445 (2019).
Chapman, T. W. et al. Oligodendrocyte death initiates synchronous remyelination to restore cortical myelin patterns in mice. Nat. Neurosci. 26, 555–569 (2023).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Feng, G. et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000).
Harris, M. A. et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261 (2004).
Dhaunchak, A. S. et al. A proteome map of axoglial specializations isolated and purified from human central nervous system. Glia 58, 1949–1960 (2010).
Binder, J. X. et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014, bau012 (2014).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Merico, D. et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5, e13984 (2010).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Kramer, A. et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Saab, A. S. et al. Oligodendroglial NMDA receptors regulate glucose import and axonal energy metabolism. Neuron 91, 119–132 (2016).
Micu, I. et al. The molecular physiology of the axo-myelinic synapse. Exp. Neurol. 276, 41–50 (2016).
Yang, A. C. et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892 (2022).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Gillespie, M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 50, D687–D692 (2022).
Reimand, J. et al. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200 (2007).
Reimand, J. et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14, 482–517 (2019).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
Turei, D. et al. Integrated intra- and intercellular signaling knowledge for multicellular omics analysis. Mol. Syst. Biol. 17, e9923 (2021).
Butler, A. et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Yuan, P. et al. TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron 92, 252–264 (2016).
Schroeder, L. K. et al. Dynamic nanoscale morphology of the ER surveyed by STED microscopy. J. Cell Biol. 218, 83–96 (2019).
Voas, M. G. et al. ɑII-spectrin is essential for assembly of the nodes of Ranvier in myelinated axons. Curr. Biol. 17, 562–568 (2007).
Zhang, C. et al. Membrane domain organization of myelinated axons requires βII spectrin. J. Cell Biol. 203, 437–443 (2013).
Susuki, K. et al. Glial βII spectrin contributes to paranode formation and maintenance. J. Neurosci. 38, 6063–6075 (2018).
Colakoglu, G. et al. Contactin-1 regulates myelination and nodal/paranodal domain organization in the central nervous system. Proc. Natl Acad. Sci. USA 111, E394–E403 (2014).
Boyle, M. E. et al. Contactin orchestrates assembly of the septate-like junctions at the paranode in myelinated peripheral nerve. Neuron 30, 385–397 (2001).
Rasband, M. N. et al. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J. Neurosci. 19, 7516–7528 (1999).
Sherman, D. L. et al. Neurofascins are required to establish axonal domains for saltatory conduction. Neuron 48, 737–742 (2005).
Rasband, M. N., Taylor, C. M. & Bansal, R. Paranodal transverse bands are required for maintenance but not initiation of Nav1.6 sodium channel clustering in CNS optic nerve axons. Glia 44, 173–182 (2003).
Cousin, M. A. et al. Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome. Nat. Genet. 53, 1006–1021 (2021).
Dolapchieva, S., Eggers, R. & Kuhnel, W. Expression of neural cell adhesion molecule (NCAM) in the peripheral nerve fibers demonstrated by postembedding immunogold method on ultrathin sections. A preliminary study. Ann. Anat. 184, 137–140 (2002).
Liu, C. H. et al. Nodal β spectrins are required to maintain Na+ channel clustering and axon integrity. eLife 9, e52378 (2020).
Brunner, C. et al. Differential ultrastructural localization of myelin basic protein, myelin/oligodendroglial glycoprotein, and 2′,3′-cyclic nucleotide 3′-phosphodiesterase in the CNS of adult rats. J. Neurochem. 52, 296–304 (1989).
Roth, A. D. et al. Septin 7: actin cross-organization is required for axonal association of Schwann cells. Biol. Res. 46, 243–249 (2013).
Pavlou, O. et al. Analysis of interactions of the adhesion molecule TAG-1 and its domains with other immunoglobulin superfamily members. Mol. Cell. Neurosci. 20, 367–381 (2002).
Kozar-Gillan, N. et al. LGI3/2-ADAM23 interactions cluster Kv1 channels in myelinated axons to regulate refractory period. J. Cell Biol. 222, e202211031 (2023).
Patzig, J. et al. Septin/anillin filaments scaffold central nervous system myelin to accelerate nerve conduction. eLife 5, e17119 (2016).
Sukhanov, N. et al. Differential contribution of Cadm1-Cadm3 cell adhesion molecules to peripheral myelinated axons. J. Neurosci. 41, 1393–1400 (2021).
Martens, A. K. et al. Targeted inactivation of the Septin2 and Septin9 genes in myelinating Schwann cells of mice. Cytoskeleton (Hoboken) 80, 290–302 (2023).
Yoshimura, T. et al. GSK-3β regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 120, 137–149 (2005).
Oliver, D. et al. Integrins have cell-type-specific roles in the development of motor neuron connectivity. J. Dev. Biol. 7, 17 (2019).
Apostolo, N. et al. Synapse type-specific proteomic dissection identifies IgSF8 as a hippocampal CA3 microcircuit organizer. Nat. Commun. 11, 5171 (2020).
Ray, A. & Treloar, H. B. IgSF8: a developmentally and functionally regulated cell adhesion molecule in olfactory sensory neuron axons and synapses. Mol. Cell. Neurosci. 50, 238–249 (2012).
Vardar, G. et al. Distinct functions of syntaxin-1 in neuronal maintenance, synaptic vesicle docking, and fusion in mouse neurons. J. Neurosci. 36, 7911–7924 (2016).
De Rossi, P. et al. Neuronal BIN1 regulates presynaptic neurotransmitter release and memory consolidation. Cell Rep. 30, 3520–3535 e7 (2020).
Barrecheguren, P. J. et al. SNARE proteins play a role in motor axon guidance in vertebrates and invertebrates. Dev. Neurobiol. 77, 963–974 (2017).
Parcerisas, A. et al. The hidden side of NCAM family: NCAM2, a key cytoskeleton organization molecule regulating multiple neural functions. Int. J. Mol. Sci. 22, 10021 (2021).
King, R. H. et al. Ndrg1 in development and maintenance of the myelin sheath. Neurobiol. Dis. 42, 368–380 (2011).
Marechal, D. et al. N-myc downstream regulated family member 1 (NDRG1) is enriched in myelinating oligodendrocytes and impacts myelin degradation in response to demyelination. Glia 70, 321–336 (2022).
Higurashi, M. et al. Localized role of CRMP1 and CRMP2 in neurite outgrowth and growth cone steering. Dev. Neurobiol. 72, 1528–1540 (2012).
Ishimoto, T. et al. Mice lacking BCAS1, a novel myelin-associated protein, display hypomyelination, schizophrenia-like abnormal behaviors, and upregulation of inflammatory genes in the brain. Glia 65, 727–739 (2017).
Meschkat, M. et al. White matter integrity in mice requires continuous myelin synthesis at the inner tongue. Nat. Commun. 13, 1163 (2022).
Thomas, R. et al. LGI1 is a Nogo receptor 1 ligand that antagonizes myelin-based growth inhibition. J. Neurosci. 30, 6607–6612 (2010).
Balaji, V. et al. Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems. Autophagy 14, 2139–2154 (2018).
Aparicio, G. I. et al. Endogenous glycoprotein GPM6a is involved in neurite outgrowth in rat dorsal root ganglion neurons. Biomolecules 13, 594 (2023).
Schneider, F. et al. Mutual functional dependence of cyclase-associated protein 1 (CAP1) and cofilin1 in neuronal actin dynamics and growth cone function. Prog. Neurobiol. 202, 102050 (2021).
Thakurela, S. et al. The transcriptome of mouse central nervous system myelin. Sci. Rep. 6, 25828 (2016).
Acknowledgements
This project was supported by National Institutes of Health (NIH) grants no. RF1AG058257, no. R01NS115544 and no. R01NS111961 (J.G.), a Cure Alzheimer’s Fund Research Grant (J.G.), a Yale/NIDA Neuroproteomics Center Pilot Project Grant 2019 (Y.C.), a BrightFocus Foundation Postdoctoral Fellowship Program in Alzheimer’s Disease Research (grant no. A2021003F) (Y.C.), a Yale ADRC Research Scholar Award (Y.C.), an Alzheimer’s Association Research Fellowship (grant no. 23AARF-1020552) (Y.C.), an EMBL Corporate Partnership Programme Fellowship (Y.C.) and Yale ADRC grant no. P30 AG066508 (A.C.N). I.P.-d.-S. and E.P. were supported by EMBL-EBI Core funding. EMBL IT Support is acknowledged for provision of computer and data storage servers. M.S. and J.R. were supported by the Investigator Award to J.R. from Ontario Institute for Cancer Research (OICR), an NSERC Discovery Grant no. RGPIN-2023-04646 to J.R. and a CIHR Catalyst Grant no. DV1-197665 to J.R. Funding to OICR is provided by the Government of Ontario. We thank the Yale/NIDA Neuroproteomics Center (grant no. P30 DA018343) for providing experimental design advice, technical support and funding opportunities. We thank F. Collin from the Keck MS & Proteomics Resource at the Yale School of Medicine for processing the LC–MS/MS experiments. We also thank the Keck MS & Proteomics Resource at the Yale School of Medicine for providing the necessary mass spectrometers and the accompanying biotechnology tools funded in part by the Yale School of Medicine and by the Office of The Director, NIH (grants no. S10OD02365101A1, no. S10OD019967 and no. S10OD018034). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank the Yale ADRC (grant no. P30 AG066508) and D. Davis and the University of Miami Brain Endowment Bank, an NIH NeuroBioBank, for providing human postmortem AD and control brain tissues. We thank S. Qin (Flatiron Institute) for performing the quantile–quantile plot data analysis. We thank the staff at the Center for Cellular and Molecular Imaging, Electron Microscopy Facility at Yale Medical School for assistance with the electron microscopy experiments. We thank M. Rasband (Baylor College of Medicine) and R. Hill (Dartmouth College) for critical comments and suggestions on this project. Schematic figures were created with BioRender.com (Figs. 1a,p, 2a, 3a,b,d,g,h, 4e, 5a,e–g, 6a, 7a,i and 8a,m).
Author information
Authors and Affiliations
Contributions
Y.C. and J.G. conceived and designed the study. Y.C. performed proximity labeling proteomics and western blotting and proteomics data analysis. J.K. and T.T.L. performed LC–MS/MS and data analysis. A.C.N. supervised the proteomics experiments and analysis. M.S., Y.C. and J.R. performed the integrative pathway enrichment analysis. I.P.-d.-S., Y.C. and E.P. performed the cell–cell communication analysis. A.H. performed pathological evaluation of human brain specimens and provided the tissues. Y.C., P.T., A.B. and T.H. performed immunofluorescence and confocal imaging. P.T., T.H. and P.Y. performed myelin analysis. F.C., Y.C., T.H. and L.T. performed expansion microscopy and imaging. L.A.F., Y.C. and T.H. performed STED imaging, image analysis and quantification. T.H., P.T., A.B. and R.W. performed quantifications. Y.C. performed electron microscopy sample preparation. Y.C., T.H., P.T., A.B. and R.W. performed statistical analysis. Y.C. and J.G. prepared the manuscript. J.G. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Junmin Peng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Expression of myelin marker proteins in AD versus controls from snRNAseq and bulk proteomics studies.
Fold changes in myelin marker protein expression in human AD frontal cortex (including prefrontal cortex and frontal gyrus) were extracted from recent snRNA-seq and bulk proteomics datasets11,14,16,17. RNA and protein expression levels in oligodendrocyte clusters are shown in the three left columns and two right columns, respectively. Abbreviation: n.s. = not significant, sig. = significant.
Extended Data Fig. 2 AI-guided myelin quantification in Cnp-EGFP-5XFAD and Cnp-EGFP-WT mice.
a. Myelin in Cnp-EGFP-5XFAD mice and Cnp-EGFP-WT mice was quantified using AI-guided segmentation of immunofluorescence confocal images. b. Representative images showing tiled confocal images of brains from Cnp-EGFP-WT and Cnp-EGFP-5XFAD mice. Myelin (grey) was labeled by anti-GFP, and lysosomes/dystrophic neurites (red) were labeled by anti-Lamp1. Brain regions used for quantification are marked by yellow boxes. Low zoom and high zoom representative images with myelin (anti-GFP), AI-generated masks and objects. Scale bars: full-tiled images = 500 μm; low zoom myelin images = 100 μm; high zoom images = 5 μm. c. Quantification of myelin volume and mean fluorescence intensity in regions of interest for Cnp-EGFP-5XFAD mice (n = 4) and Cnp-EGFP-WT mice (n = 5); (Mann Whitney test, two-tailed). Abbreviations: RSPd = Retrosplenial area, dorsal part; PTLp = Posterior parietal association areas; DG = dentate gyrus. Error bars indicate SEM.
Extended Data Fig. 3 Super-resolution STED microscopy demonstrates spatial precision of proximity labeling.
a. Bead imaging illustrates the resolution difference between confocal microscopy (250 nm) and STED microscopy (50 nm). Scale bar = 250 nm. b-c. Representative confocal and STED images of proximity labeling in human postmortem brains showing (B) paranodes (anti-CASPR, magenta) and (C) internodes (anti-MAG, magenta), with biotinylated proteins detected by streptavidin (green); scale bars: (B) 500 nm, (C) 5 μm. d. A representative line plot shows the radius measurements of signals from the secondary antibody channel (magenta) and the streptavidin channel (green). e. A dot plot showing the radius ratio between the secondary antibody (magenta) and streptavidin (green). The average radius ratio is 1.07 (SD= 0.1); the orange line indicates the median, with the lower and upper edges representing the 25th (Q1) and 75th (Q3) percentiles. Whiskers extend to the minimum and maximum values within 1.5 times the interquartile range, and outliers (pink circles) are plotted individually (and excluded from analysis). Each dot represents a myelin segment (n = 45).
Extended Data Fig. 4 Correlation analysis of proteomics samples in humans and mice.
Correlation analysis for biological replicates of (a) MAG-labeled and (b) CASPR-labeled samples, along with the no-antibody labeled proteomic controls in (A-B) human brains and (C) in mice. Pearson correlation coefficient (R2) values are provided in each comparison box.
Extended Data Fig. 5 Caspr-labeled paranode-enriched proteomes in 5XFAD mice.
A. Schematic illustrating the experimental design. B. Partial volcano plot of proteomic hits in paranode-enriched samples from 5XFAD mice. Known paranode-related hits are shown in red and other hits in green. The gene names of the top 10 proteomic hits and known paranodal proteins are indicated. C. Venn diagram showing the number of paranode-enriched proteomic hits shared between AD humans and 5XFAD mice. D. Gene Ontology cellular component (GO-CC) analysis of the shared hits between AD humans and mice, displaying the top 8 GO-CC terms. Quantifications in panels B and D were performed two-sided.
Extended Data Fig. 6 Top 100 hits in the CASPR-labeled paranode-enriched proteome and MAG-labeled myelin-axon interface proteome in unaffected human brains.
(a) The top 100 proteomic hits identified in the paranode-enriched proteome. Proteins known to be expressed in the paranode are marked with an asterisk (‘*’): SPTAN1123, SPTBN1124,125, CNTN1126,127, CNTNAP135, ANK321,128, NFASC129, and SCN2A130. Proteins known to be expressed in the node of Ranvier and juxtaparanode are marked with a hash (‘#’): TNR1, ACTN4131, SPTBN41, VCAN1, NCAM1132, CNTNAP2129, SPTB133, CNTN21, HAPLN21, NCAN1, EPB41L21. (b) The top 100 proteomic hits identified in the myelin-axon-interface proteome. Proteins known to be expressed at the myelin-axon interface are marked with an asterisk (‘*’): CNP134, CNTN1126,127, CNTNAP135, NFASC129, NCAM1132, MOG134, MAG36,37,38, SEPTIN7135, NRCAM136, CNTN236,37,38, PLP120, CNTNAP21, LGI3137, SEPTIN8138, CADM420,139, ADAM22137, SEPTIN2138,140, SEPTIN4138. Proteins known to be related to myelin or axon are marked with a hash (‘#’): DPYSL2141, INA1142, IGSF8143,144, STX1B145, BIN1146, SNAP25147, NCAM2148, NDRG1149,150, CRMP1151, BCAS1152, KCNAB2153, LGI1154, MAPT155, GPM6A156, CAP1157, PLEKHB1158.
Extended Data Fig. 7 Integrative pathway enrichment analysis of paranode and myelin-axon interface proteomes using the ActivePathways method.
(a-b) The Enrichment Map depicts a network of pathways (FDR < 0.05) where edges connect pathways sharing many genes. Node size reflects the number of genes in each pathway, and node color indicates the dataset contribution (combined AD and control). Theme labels were curated based on the main pathways represented in each subnetwork. Only subnetworks with at least four pathways connected by edges are shown. Grey nodes indicate combined evidence of pathway enrichment in which the respective pathways were detected in the integrative analysis but not detected in either the AD or Control proteomes alone.
Extended Data Fig. 8 Cell-cell communication analysis revealing ligand-receptor interaction at the myelin-axon interface.
a-b. (A) Cell clustering and (B) cell type annotation of snRNAseq data from AD human frontal cortex (Braak stage 6) and controls (Braak stage 0). c. Enrichment analysis shows that myelin-axon interface proteomics (MAG or CASPR-labeled) are highly enriched in neurons and oligodendrocytes, but not in other cell types (related to Fig. 5b). Each row depicts contingency tables for each hypergeometric test (from top to bottom: p-values: 0.0005275, 0.0007784, 0.009328, 0.0001295). In these rows, values with two decimal places indicate residuals and the size of the circles; positive residuals denote that the observed values were more frequent than expected, while negative residuals indicate lower-than-expected frequencies. Quantifications were performed two-sided. d-e. Violin plots showing RNA expression levesl of ligand-receptor pairs in (D) control human postmortem brains (Braak stage 0) and (E) AD human postmortem brains (Braak stage 6).
Extended Data Fig. 9 Paranode-enriched and myelin-axon interface Alzheimer’s disease proteomes reveal unique subcellular changes not observed by bulk proteomics or single cell RNA transcriptomics.
Heatmaps display (a) Paranode Alzheimer’s-associated proteomes (PAPs) and (b) Myelin-axon interface Alzheimer’s proteomes (MAPs). Heatmap denotes log10 (spectral counts). Comparison between PAPs or MAPs and bulk proteomics data (middle panel, Johnson et al.16), or single nuclei RNA sequencing transcriptomics (right panels, Mathys et al.11) were performed. Both bulk proteomics and snRNAseq data were obtained from their original studies. Neuronal cell types (yellow box) and oligodendrocyte/OPC (green box) were highlighted in the snRNAseq data. Abbreviations: FC = fold change; DEG = differentially expressed genes. (A and B) Quantifications of subcellular proteomic data derived from this study were performed two-sided.
Extended Data Fig. 10 Diagram of myelin-axon disruption in AD.
a. Diagram illustrating how amyloid toxicity to axons and myelin (#1 and #2) may lead to axonal spheroid formation (a), myelin paranode/juxtaparanode disruption (b), protein perturbation at the myelin-axon interface (c) and amyloid accumulation at the interface (d). Together, these events may create a vicious cycle of dysregulated myelin-axon crosstalk and degeneration (#3). b. Diagram outlining potential signaling pathways that contribute to myelin-axon disruption, based on findings from myelin-axon interface proteomics and imaging validations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15, titles of Supplementary Tables 1–6 and titles and captions of Supplementary Videos 1 and 2.
Supplementary Table 1
Raw and analyzed proteomics results of paranode-enriched and myelin–axon interface proteomes in humans and mice.
Supplementary Table 2
Proteomic sample information (also available at ProteomeXchange).
Supplementary Table 3
Subcellular location of proteomic hits.
Supplementary Table 4
Full lists of pathway enrichment analysis.
Supplementary Table 5
Full lists of cell–cell communication analysis in neuron-oligodendrocytes and the ligand–receptor pairs identified in subcellular proteomics.
Supplementary Table 6
Full lists of predicted downstream signaling processes induced by the ligand–receptor pairs identified at the myelin–axon interface.
Supplementary Video 1
Representative large tiling and magnified images of AI-guided immunofluorescence and imaging annotation of myelin, axons and paranodes in postmortem human brains of AD and age-matched controls, as well as Cnp-EGFP mice with or without 5XFAD background. The masks generated by AI annotation are in yellow, and the objects are in blue. Scale bar is shown in real-time during the video at the lower left corner.
Supplementary Video 2
Representative amyloid fiber accumulation at the myelin–axon interface in 5XFAD mice. Amyloid fibers (4G8 labeled, red) were observed along the axon (labeled by NFH, green), and dense amyloid coils were observed at the paranode and juxtaparanode regions (CASPR and Kv7.3 labeled, gray).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cai, Y., Pinheiro-de-Sousa, I., Slobodyanyuk, M. et al. Myelin–axon interface vulnerability in Alzheimer’s disease revealed by subcellular proteomics and imaging of human and mouse brain. Nat Neurosci 28, 1418–1435 (2025). https://doi.org/10.1038/s41593-025-01973-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-025-01973-8