Abstract
While epidemiological evidence has long linked obesity with an increased risk of breast cancer, the intricate interactions between adipocytes and cancer cells within the tumor microenvironment remain largely uncharted territory. The use of organotypic three-dimensional (3D) cell cultures that more accurately mimic the spatial architecture of tumors represents an innovative approach to this complex issue. In the present study, we investigated the effects of adipocytes on the proteome of Hs578t breast cancer cells cultured in a 3D microenvironment. Using different treatments, we rigorously optimized the experimental conditions to induce the optimal differentiation of 3T3-L1 fibroblasts into mature adipocytes. Then, we grow the Hs578t cells in a simulated microenvironment using an on-top model for organotypic 3D cultures. Our data showed that cancer cells formed 3D stellate-like architectures when grown over an extracellular matrix proteins-enriched scaffold for 48 h. Proteomic profiling using LC-MS/MS mass spectrometry of Hs578t cells grown in 3D conditions with or without the adipocyte-enriched culture discovered 916 unique proteins. Of these, 605 showed no significant changes in abundance, whereas 87 proteins were significantly upregulated and 224 downregulated after interaction with fat cells (p < 0.05, FC > 2.0). Bioinformatic analysis of upregulated proteins indicated that the most enriched GO terms and molecular functions were related to lipids transport, cell differentiation, hypoxia response, and cell junctions. In addition, several modulated proteins have been previously associated with breast cancer progression. Interestingly, lipid transport proteins, including PITPNM2, ATP2C1, ABCA12, HDLBP, and APOB, showed perturbations in their expression, which were also associated with low overall survival in breast cancer patients. Functional studies showed that the knockdown of apolipoprotein B (APOB) expression in Hs578t cells reduced the size of 3D cellular structures. Moreover, APOB-knocked cells cocultured with adipocytes for 48 h exhibited a significant decrease of intracellular lipids, whereas an increase in the adipocytes was found. Our results indicate that the 3D microenvironment and the adipocytes crosstalk reprogram the proteome of breast cancer cells. These data help us understand the environmental effects in gene expression and contribute to discovering novel tumor proteins with potential intervention in breast cancer therapy.
Similar content being viewed by others
Introduction
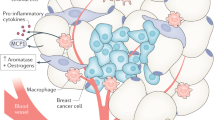
Epidemiologic data indicate that obesity is associated with an increased risk and poor prognosis of breast cancer in postmenopausal women. Recent studies have demonstrated the importance of the tumor microenvironment in the progression of breast cancer, which significantly contributes to the acquisition of cancer hallmarks1. Tumor niches mainly comprise extracellular matrix (ECM) proteins, infiltrating immune cells, fibroblasts, and other stromal cells, including cancer-associated adipocytes2. Remarkably, adipocytes represent the most significant proportion of cells in the mammary gland; therefore, they are considered key players in breast cancer development and progression3,4. Several studies have analyzed the interaction between breast cancer and fat cells in in vitro co-culture models. For instance, it was reported that adipocyte-derived fatty acids drive the proliferation and migration of breast cancer cells5. In addition, the kidnapping of adipocyte-derived fatty acids is facilitated by the upregulation of diverse proteins from tumor cells, which may impact the growth of breast tumors. Among these lipid transporters is the lipoprotein APOB, which has been associated with an increased risk of recurrence and poor prognosis among breast cancer patients6. On the other hand, adipocytes can promote the overexpression of proinflammatory genes related to the NF-κB signaling pathway, which may act as obstacles to therapy, as they are involved in chemotherapy resistance5,7. Recently, it was reported that cancer-associated adipocytes undergo metabolic reprogramming and de-differentiation to myofibroblast- and macrophage-like cells (adipocyte mesenchymal transition), activating ECM remodeling and immune response, contributing to tumor progression8,9,10. However, the complex molecular interactions orchestrated between breast cancer cells and cancer-associated adipocytes on the tumor microenvironment have not been fully elucidated11.
Three-dimensional (3D) cell cultures can better recapitulate the biological and molecular characteristics of in vivo tumor tissues, such as cell heterogeneity, oxygen and nutrient gradients, hypoxia grades, and the activation of oncogenic signaling pathways, which greatly influences the reprogramming of epigenetic12 and gene expression patterns of both non-coding RNAs and mRNAs13. These cell culture models can help decipher the adipocytes and ECM crosstalk, which could induce epithelial to mesenchymal transition (EMT) of cancer cells, promoting metastasis14. Therefore, the implementation of reliable in vitro cancer cell cultures containing components of the EMT may provide information about its impact on the genetic programs leading to breast cancer development and progression, as well as for identifying key proteins that could be translated into the development of novel therapeutic strategies. As the proteins involved in these events remain almost unknown, we propose that a protein profiling of breast cancer cells grown in a 3D environment and co-cultured with adipocytes could be useful in identifying the missing factors. Here, we have developed an in vitro 3D co-culture system to recapitulate, in part, the in vivo interactions between breast cancer cells and adipocytes, not only to understand better the biology of cancer cells and the cellular crosstalk with cellular microenvironment but also to contribute to the discovery of tumor regulated proteins helpful in the development of molecular targeting of proteins for breast cancer therapy.
Results
Differentiation of 3T3-L1 fibroblasts to mature adipocytes
We initiated the study of the functional interactions between adipocytes and triple-negative Hs578t breast cancer cells growing in organotypic 3D cell cultures. To this end, we evaluated the impact of co-culturing adipocytes and Hs578t cells on the proteome of cancer cells using a quantitative proteomics-based approach (Fig. 1).
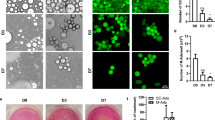
Then, we aim to establish a reproducible method to address the biological questions regarding the influence of adipocytes on the proteome of cancer cells. Using different treatments, we rigorously optimized the experimental conditions to induce the optimal differentiation of 3T3-L1 fibroblasts into mature adipocytes. The used cocktail treatments were: (i) control (basal media), (ii) differentiation medium I (DM-I; DMEM/high glucose, isobutyl-methyl-xanthine 0.5 mM, insulin 1.5 µM, and dexamethasone 0.25 µM supplemented with 10% fetal bovine serum, two mM L-glutamine, 1X non-essential amino acids, one mM sodium pyruvate, 1% penicillin-streptomycin (10,000 units/mL). (iii) DM-I plus insulin 0.75 µM, (iv) DM-I plus rosiglitazone (2.5 µM), and (v) DM-I plus insulin and rosiglitazone during 11 days at 37 ºC. Results showed that all used treatments induced the proper differentiation of fibroblasts to adipocytes, although the differentiation cocktail containing insulin and rosiglitazone had better results as after treatment nearly 94% of 3T3-L1 cells were differentiated into mature adipocytes (Fig. 2A). Optical microscopy analyses of cell cultures indicated that adipocytes showed the typical spherical morphology containing large lipid droplets and peripherally located nuclei. As shown in the representative images in Fig. 2B, on days 3 and 5, after incubation using the differentiation cocktail plus insulin and rosiglitazone, we found initial structural changes in the 3T3-L1 cells, which appeared more prominent than those in the untreated control cells. Interestingly, on day 7, the cells exhibited increased cytoplasmic space. By day 9, we also found a significant accumulation of lipid droplets. Finally, on day 11, most of the 3T3-L1 cells in the culture were differentiated into adipocytes, and a substantial accumulation of lipids was observed within the cells. In agreement with previous reports, the observed changes in cellular morphologies after the treatments were like those described by Xu P and coworkers15. Next, we aim to corroborate the increased intracellular accumulation of neutral lipids using oil-red-O dye, which selectively stains neutral lipids, cholesteryl esters, and lipoproteins of adipocytes. As expected, oil-red-O staining confirmed the increased presence of lipid droplets after the different treatments. The representative images in Fig. 2C and the quantification of stained cellular droplets (Fig. 2D) showed that lipids were more evident in 3T3-L1 cells treated with the differentiation cocktail containing insulin and rosiglitazone. Consequently, we selected this treatment to produce an enriched culture of mature adipocytes for further downstream analysis.
In vitro differentiation of 3T3-L1 fibroblasts to adipocytes. (A) Quantifying the percentage of fibroblasts differentiated into adipocytes after 11 days of incubation with the indicated treatments. Grey and black bars indicate the fibroblasts and mature adipocytes, respectively. (B) Optical microscopy images (40x) showing the morphological changes during the differentiation of 3T3-L1 fibroblasts into adipocytes using the differentiation cocktail containing insulin and rosiglitazone. (C) Optical microscopy images showing the oil-red O staining of adipocytes after 11-day treatment using the differentiation cocktail containing insulin and rosiglitazone. (D) Quantification of oil-red-O staining of mature adipocytes from panel C. ***p < 0.001.
Establishment of a coculture of adipocytes and breast cancer cells grown in 3D microenvironment
To evaluate the effects of adipocytes on the proteome abundance of Hs578t breast cancer cells grown in a simulated microenvironment, an on-top model for organotypic 3D cell culture was implemented using transwell chambers (Fig. 3A). As a scaffold for cancer cell growth, we used Geltrex, an ECM-enriched substrate containing laminin, collagen IV, entactin, and heparin sulfate proteoglycans, as we previously described13. In this system, adipocytes were grown below the semi-solid substrate, whereas Hs578t cancer cells were embedded in Geltrex at the upper chamber (Fig. 3B). Morphological analysis of cultures indicated that Hs578t cells formed stellate-like structures with prominent cell membrane prolongations from 2 to 6 days of incubation over the Geltrex matrix (Fig. 3B). No significant differences in 3D structure formation were found when using different cell densities ranging from 2.8 × 104 to 3.5 × 104 cells. Remarkably, we also detected reduced intracellular lipids accumulation in mature adipocytes after 48 h incubation with breast cancer cells. This suggests that fatty acids stored in adipocytes could be released or degraded in the simulated 3D microenvironment (Fig. 3C and D), although additional experimentation is needed to corroborate this hypothesis.
Coculture of adipocytes with organotypic 3D culture of Hs578t breast cancer cells. (A) On-top 3D cell cultures of Hs578t cells. Optical microscopy images (40x) showing the typical morphology and 3D structures of Hs578 cells grown over ECM-enriched Geltrex substrate over time (0–6 days). The different cell densities used in cultures are indicated on the left. (B) Set up of 3D cell culture of Hs578t cells and adipocytes. (C) Optical microscopy images showing the Oil Red O staining of mature adipocytes (day 11) after incubating with Hs578 cells cultured in 3D for 48 h. Inset indicates a zoom imaging of a single adipocyte. (D) Quantification of lipids using Oil Red O staining in 3T3-L1 fibroblasts, mature adipocytes alone, and adipocytes after 48 h incubation with Hs578t cells. ***p < 0.001. ns = no significant.
Proteomic analysis of organotypic 3D culture of breast cancer cells cocultured with adipocytes
Several cellular effects of cancer-associated adipocytes on cancer cells have recently been reported4,5,6,7,8,9,10; however, the molecular factors involved have not yet been fully identified. Inspired by these studies, we used a proteomic approach to analyze the influence of the adipocyte-enriched culture in the global protein expression profile of Hs578t cells grown in organotypic 3D cultures. We performed differential expression analysis of total proteins extracted from Hs578t breast cancer cells after 48 h incubation with mature adipocytes. After digestion, generated peptides were injected into the mass spectrometer Synapt; the LC-MS/MS analysis was performed, as described in materials and methods. A representative volcano plot of the distribution of modulated proteins is illustrated in Fig. 4A. Proteomic profiling of Hs578t cells co-cultured with adipocytes allows us to identify 916 proteins in the replicates. Of these, 605 showed no significant changes in abundance, whereas 87 and 224 proteins were upregulated and downregulated, respectively, compared to cancer cells grown alone (fold change > 2.0; adjusted p value = 0.05). (Figure 4B and C). Several upregulated (Supplementary Table S1) and downregulated (Supplementary Table S2) proteins are involved in cellular processes related to cancer hallmarks, including cell proliferation, invasion, metastasis, angiogenesis, and resistance to therapy. Interestingly, we found a specific set of upregulated proteins associated with lipids transport, such as PON1, PITPNM2, ATP2C1, RNF213, PLA2G6, ABCA12, ABCA8, HDLBP, ATP8B4, and APOB, in Hs578t breast cancer cells in coculture with adipocytes (Table 1). That makes sense, as our data suggested that cancer cells exert a delipidating effect in adipocytes to obtain triglycerides as an energy source needed to maintain cell proliferation and growth of 3D cellular structures.
Proteomic analysis of 3D organotypic Hs578t cell cultures cocultured with adipocytes. (A) Volcano plot of modulated proteins from Hs578t cells grown in 3D microenvironment with adipocytes during 48 h. (B) Distribution of up and downregulated proteins in Hs578t cancer cells. (C) Venn diagram corresponding to the modulated proteins shown in panel B.
Enrichment analysis of modulated proteins in Hs578t in coculture with adipocytes
To obtain a global view of the biological functions associated with modulated proteins in 3D cancer cell cultures incubated with adipocytes, we used the g: profiler database to define the Gene Ontology (GO) functions and pathways enrichment using REACTOME tool (https://reactome.org/). The GO analysis describes the gene functions in terms of three functional groups: cellular component (CC), molecular function (MF), and biological process (BP), where they participate. Results showed that for the upregulated proteins (Fig. 5A), the most enriched GO terms in the biological processes included genes involved in lipid localization and transport, and the most enriched molecular functions were involved in lipid transport activity. The cellular component analysis showed that they mainly involved the cytoplasm. On the other hand, the biological processes enriched for downregulated proteins were mainly associated with organelle and cytoskeleton organization, and molecular functions were involved in protein binding (Fig. 5B). Figure 5C shows robust and highly significant connections between enriched biological functions as a chord graph for upregulated proteins. Lipid transport and lipid localization are depicted as the broadest and most prominent chords. We observed that downregulated proteins have functions related to nucleosome assembly and chromatin organization (Fig. 5D). These data enable us to discern patterns of functional enrichment and highlight the biological functions most prevalent in the dataset under analysis.
Gene ontology (GO) enrichment analysis of modulated proteins in 3D cultures of Hs578t cells co-cultured with adipocytes. (A-B) An advanced bubble plot showed the top 10 GO enrichment significance elements of the upregulated (A) and (B) downregulated proteins ordered by adjusted P-value. Cellular component (CC), molecular function (MF), and biological process (BP). The x-axis label represented the proportion of genes, and the y-axis label represented the GO terms. (C-D) The chord chart showed the distribution of proteins in different GO-enriched functions. The symbols of the genes coding for the proteins were presented on the left side of the graph, with their fold-change values denoted by color scale. Colored connecting lines indicate the involvement of the genes in the GO terms.
Then, we analyzed the biological processes for the 15 upregulated hub proteins (APOB, ABCA9, RNF213, PITPNM2, ABCA8, ATP8B4, CERT1, MYO15B, ANKRD17, ITGA6, ABCA12, TRPM2, FAT3, ATP2C1, RNF17 (Fig. 6). Results showed these hub proteins were mainly associated with lipid transporter activity and localization, cytoskeletal protein binding, and cell organization.
Subsequently, an analysis of signaling pathways was performed using the REACTOME tool to identify routes involving the modulated proteins in 3D cultures of Hs578t cells. Ten and nine pathways were identified for upregulated and downregulated proteins, respectively (Fig. 7). Our data showed that the 87 upregulated proteins are involved in ten signaling pathways, including cell-extracellular matrix interactions, ABC transporters in lipid homeostasis, cell junction organization, plasma lipoprotein clearance, and ABC-family proteins mediated transport signaling pathways. Moreover, the 224 downregulated proteins are involved in signaling pathways such as cell cycle, cellular response to heat stress, cellular responses to stress, and cellular responses to stimuli.
Signaling pathway analysis of regulated proteins in Hs578t cells in coculture with adipocyte-enriched culture. A bubble plot of REACTOME enrichment analysis is shown; on the ordinate axis, the enriched pathways of upregulated proteins (upper panel) and downregulated proteins (below panel) are shown. The circle size represents the number of genes involved in that pathway. The color represents the adjusted p-value.
Protein-protein interaction (PPI) networks
Protein-protein interaction (PPI) network analysis is a valuable tool to understand how proteins interact with each other in the cell. Using the STRING and Cytoscape software, we found interactions between 87 upregulated and 224 downregulated proteins (Fig. 8). These data constructed a complex network using a maximum confidence interaction score (0.9). The most significant biological processes and signaling pathways were selected and marked with colors at the nodes in Fig. 8. Results showed that the 87 upregulated and interacting proteins were involved in functions related to cell cycle and hypoxia regulation, lipid transport, cytoskeleton binding proteins, and cell differentiation (Fig. 8A). On the other hand, the 224 downregulated and interacting proteins were involved in hypoxia regulation, EMT, cadherin binding, and cytoskeletal protein binding activities (Fig. 8B). To increase the robustness of these results, a different approach based on a co-expression network was performed using the central upregulated proteins. The results show a marked enrichment in protein nodes involved in lipid transport and localization (Fig. 8C).
PPI network analysis of regulated proteins in 3D cultures of Hs578t cells in coculture with 3T3-L1 adipocytes. (A) The PPI network of upregulated proteins contains 48 nodes and 39 edges (B) The PPI network of downregulated proteins contains 179 nodes and 627 edges. By STRING visualized in Cytoscape, the color code represents the biological processes in which they are involved. (C) KEGG pathway analysis for upregulated hub proteins.
High expression levels of lipid-transport proteins correlate with low overall survival
Then, we wondered if the changes in lipid transport proteins have clinical implications in breast cancer. Thus, we performed an overall survival analysis using the Start Kaplan-Meier Plotter and protein expression data together with overall survival clinical information from a cohort of breast cancer patients (n = 65), as reported in the kmplot website: https://kmplot.com/analysis/index.php?p=service&cancer=breast_protein. Results showed that the high levels of PITPNM2, ATP2C1, ABCA12, and HDLBP proteins in breast tumors were associated with low overall survival of cancer patients (Fig. 9).
APOB silencing impacts lipid contents in tumor cells and 3D culture formation
As the GO analysis showed that lipids transport and localization were the most enriched biological processes in Hs578t cells cocultured with adipocytes, next we focused on the functional characterization of APOB (Table 1), an essential protein involved in lipids transport and linked to breast cancer progression30. To determine the relevance of APOB in the development of 3D structures, we silenced its expression in Hs578t cancer cells using a specific siRNA, and then they were cocultured with adipocytes for 48 h and imaged. Results showed that the silencing of APOB (Fig. 10A) significantly reduced the size of the Hs578t 3D structures, but not its number, compared to mock and scramble-transfected controls (Fig. 10B-D). Then, we wondered if the diminution in the size of 3D structures was due to a reduced lipid content. Therefore, we decided to quantify the accumulation of intracellular triglycerides using oil-red O staining, as described in the methods section. The data showed that APOB-knocked Hs578t cells cocultured with adipocytes exhibited lower intracellular lipids than mock and scramble-transfected control cells (Fig. 10E-F). In contrast, a significant increase in lipids content was observed in the adipocytes after 48 h of co-culture with APOB-knocked Hs578t cells (Fig. 10G-H).
APOB silencing impacts organotypic 3D culture formation. (A) qRT-PCR assays for control, mock, scramble, and siRNA-APOB transfected cells. (B) Optical microscopy (20X) images show the morphology, number, and size of the 3D structures of Hs578t cells grown over ECM-enriched geltrex substrate after silencing APOB and co-culturing with adipocytes for 48 h. (C-D) Quantification of the number and diameter of 3D cell structures showed in B. (E) Optical microscopy images showing and quantification of lipids using oil-red O of Hs578t cells grown in 3D conditions after silencing of APOB. (F) Quantification of oil-red O in cells showed in panel E. (G) Optical microscopy images showing and quantifying lipids using oil-red O in adipocytes cocultured with Hs578t cells after silencing of APOB. (F) Quantification of oil-red O stain of cells showed in panel G. *p < 0.05, **p < 0.01, ***p < 0.001, ns = no significant.
Discussion
Breast cancer is one of the most common fatal malignancies and the leading cause of cancer-related death in women worldwide31. Within the molecular subtypes of breast cancer, triple-negative breast cancer is the most aggressive subtype, showing a poor prognosis due, in part, to a high probability of distant metastasis as well as a lack of specific targets for targeted therapies32. These facts highlighted the need to discover novel therapeutic targets for personalized therapies in triple-negative breast cancers. Obesity is a well-known risk factor for breast cancer in menopausal women. Recent primary research has demonstrated that key interactions of adipocytes with ECM could promote migration, invasion, and metastasis of cancer cells, suggesting that cancer-associated adipocytes could be an adequate target for cellular therapies. Here, we performed a proteomic analysis of breast cancer cells for proteins regulated by the presence of adipocytes in a simulated 3D microenvironment, which exhibits advantages compared to traditional two-dimensional monolayer cultures of cancer cells. Organotypic 3D cultures of cancer cells mimic several characteristics occurring in vivo tumors, including (i) cell-cell and cell-ECM interactions, (ii) natural cellular structure preserved, (iii) high reproducibility, (iv) gradients in nutrients and oxygen availability, (v) reprogramming of the metabolism and transcriptional programs; and (vi) more accurate drugs response, among others. Thus, to recreate a proper microenvironment for the interactions between cancer cells and adipocytes, the Hs578t cells were grown in an ECM-enriched environment, aiming to establish a reproducible method to address the biological questions. In these conditions, we have reported that reprogramming of transcriptional programs of mRNAs, microRNAs, and lncRNAs may occur10,11,12. More importantly, these variations in transcriptional profiles are close to the genetic patterns occurring in tumors in vivo. Although contaminating proteins potentially contained in Geltrex originating from a primary source could not completely be excluded, we sticked to protocols published previously10,11,12.
Taking advantage of the 3D cell cultures, we performed a proteomics-based analysis. This allowed us to identify a comprehensive list of modulated proteins in tumor cells grown in a 3D microenvironment in response to the presence of adipocytes, most of them with potential oncogenic roles. We focused on their study as the most enriched GO terms and molecular functions were related to lipids localization and transport. That makes sense, as we suggested that cancer cells exert a delipidation effect in adipocytes, obtaining triglycerides as an energy source to maintain cell proliferation and the growth of 3D cellular structures. However, additional functional experiments on these proteins are needed to corroborate these assumptions. The set of upregulated proteins involved in lipids-transport includes PON1, PITPNM2, ATP2C1, RNF213, PLA2G6, ABCA12, ABCA8, HDLBP, ATP8B4, and APOB. The functions of these proteins are various, and they are related to lipids. For instance, directly binding to phospholipids, the PON1 protein mediates enzymatic protection of low-density lipoproteins against oxidative modification. In breast cancer, PON1-L55M genetic polymorphism might be a genetic risk factor for the risk of breast cancer33. In breast cancer patients treated with doxorubicin with or without trastuzumab, high levels of PON1 were associated with increased cardiac dysfunction risk34. On the other hand, PITPNM2 catalyzes the transfer of phosphatidylinositol and phosphatidylcholine between membranes. In glioma stem cells, PITPNM2 is involved in two pathways controlled by ELF4 transcriptional factor affecting lipid homeostasis and proliferation35. Importantly, the ABCA12, PITPNM2, ATP2C1, and HDLBP proteins showed a clinical value as their overexpression correlates with poor overall survival in breast cancer patients. Finally, APOB transports lipids within the cells and human tissues36. The polymorphisms 12,669 GA and 7673 CT in the APOB gene have been associated with an increased risk of breast cancer37. In addition, ApoA1 and ApoB are risk factors for intraocular metastasis in patients with breast cancer27. Nonetheless, very little information about the role of APOB in breast cancer progression has been reported. For instance, an interesting report showed the upregulation of lipid metabolism- and fatty acid transport-related genes in the susceptible breast epithelium and adipose tissues from normal breast, respectively. These genes include fatty acid uptake/transport (CD36 and AQP7), lipolysis (LIPE), and lipid peroxidation (AKR1C1), suggesting that in the early stages of carcinogenesis, the activation of lipid metabolism and transport could influence the microenvironment for cell proliferation, and survival38. On the other hand, a report from Ben Hassen and coworkers indicates that forced expression of apolipoprotein A1 (APOA1) in estrogen-positive MCF-7 breast cancer cell lines activates cell proliferation and tumor growth, whereas in the triple-negative MDA-MB-231 reduces proliferation and migration, indicating that the APOA1 function depends on the subtype of breast cancer39. Here, we show for the first time that APOB knockdown inhibited the development of 3D cell structures of triple-negative breast cancer cells, suggesting the existence of complex interactions orchestrated between breast cancer cells and cancer-associated adipocytes in the tumor microenvironment. These findings highlight the critical role of lipid transport proteins in cancer progression and suggest that they may represent attractive therapeutic targets. Finally, the limitations of our study, which remain as perspectives for ongoing research, include: (i) lack of data for deregulated proteins in the diverse breast cancer subtypes, (ii) the validation of regulated proteins using human-derived adipocytes, and (iii) lack of data about the potential secreted factors by adipocytes mediating the proteomic response of cancer cells. Future investigations will provide additional clues into the complex interaction between adipocytes and cancer cells.
Conclusions
In conclusion, our results suggested that the growth of Hs578t cancer cells in 3D conditions and cocultured with an adipocyte-enriched culture reshapes the proteome of cancer cells, inducing the upregulation of lipid-transport proteins. Moreover, the knockdown of APOB protein suggested that it is essential to maintain the growth of cancer cells in 3D architectures. Our results contribute to discovering proteins modulated in the microenvironment that could be translated into novel breast cancer therapy strategies.
Materials and methods
Cell cultures
Mouse 3T3-L1 fibroblasts obtained from the American Type Culture Collection (ATCC-CL-173) were grown in growth medium (GM) containing Dulbecco’s modified Eagle’s medium (DMEM, ATCC-30-2002) supplemented with 10% fetal bovine serum, two mM of L-glutamine, 1X nonessential amino acids (Sigma M7145), one mM of sodium pyruvate (Gibco 11360), and penicillin-streptomycin 1% (10,000 unit/mL; Gibco 30-2300) as suggested the manufacturer. Triple-negative Hs578t (ATCC, HTB-126) human breast carcinoma cell line was also obtained from the ATCC and maintained in DMEM supplemented with 10% fetal bovine serum and penicillin-streptomycin (10,000 unit/mL; Gibco 30-2300) at 37 °C in a humidified incubator with 5% CO2 atmosphere, as suggested the manufacturer.
Adipocytes differentiation
Adipocyte differentiation was conducted according to a previously reported protocol15.
Firstly, the mouse 3T3-L1 cells were seeded at a density of 5 × 10⁴ cells per well in six-well plates containing growth medium (GM) and incubated at 37 °C with 5% CO₂ for 48 h until the cells reached 80% confluence (day 0). Then, the differentiation of 3T3-L1 fibroblasts into adipocytes was performed in the six-well plates as follows: GM was then replaced with Differentiation Medium I (DM-I; DMEM/high glucose, insulin 1.5 µM, isobutyl-methyl-xanthine 0.5 mM, and dexamethasone 0.25 µM supplemented with 10% fetal bovine serum, two mM L-glutamine, 1X non-essential amino acids, one mM sodium pyruvate, 1% penicillin-streptomycin (10,000 units/mL). Then, the cells were incubated at 37 °C with 5% CO₂ for three days to initiate adipocyte differentiation. Subsequently, DM-I was replaced with Differentiation Medium II (DM-II; DMEM/high glucose, insulin 0.75 µM, supplemented with 10% fetal bovine serum, two mM L-glutamine, 1X non-essential amino acids, one mM sodium pyruvate, 1% penicillin-streptomycin). The cells were then incubated at 37 °C with 5% CO₂ for two days to promote adipocyte maturation and differentiation. After this period, cells were maintained in a maintenance medium (MM) containing DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (10,000 units/mL; Gibco 30-2300) in an incubator at 37 °C with 5% CO2. The medium was refreshed every two days until the morphological changes associated with adipocyte differentiation were observed under a microscope. Differentiation was confirmed using oil-red-O staining for microscopic visualization of lipid droplets in mature adipocytes. We try additional treatments to maximize and obtain a complete differentiation of 3T3-L1 cells16,17. The treatments were as follows: (i) control 3T3-L1 cells grown in GM medium, (ii) 3T3-L1 cells grown in DM-I (3 days), then the media was replaced by DM-II (2 days), and finally DM-II replaced by MM medium, (iii) 3T3-L1 cells grown in DM-I (3-days), then the media was replaced by DM-II (2 days), and finally, DM-II replaced by MM medium supplemented with insulin (0.75 µM) which was refreshed every two days for maintenance, (iv) 3T3-L1 cells grown in DM-I supplemented with rosiglitazone (2.5 µM) for 3-days, then the media was replaced by DM-II (2 days), and finally DM-II replaced by MM medium which was refreshed every two days for maintenance, (v) 3T3-L1 cells grown in DM-I supplemented with rosiglitazone (2.5 µM) for 3-days, then the media was replaced by DM-II (2 days), and finally, DM-II replaced by MM medium supplemented with insulin (0.75 µM) which was refreshed every two days for maintenance. These treatments were applied for 11 days at 37 °C in an incubator with 5% CO₂. Then, the morphological changes associated with adipocyte differentiation were assessed by optical microscopy. Differentiation was confirmed using oil-red-O staining for microscopic visualization of lipid droplets in mature adipocytes.
Oil-red O staining in 2d and 3D cultures
The 3T3-L1 adipocytes were fixed with 3.7% formaldehyde for 30 min, washed three times with PBS 1X, and stained with oil-red O dye (ORO, Sigma O 0625), 60% isopropanol/PBS solution (1.2 mg of dye/mL isopropanol-PBS 1X solution) for 10 min. Then, cells were washed with 70% ethanol/PBS 1X to remove excess dye, and stained lipid droplets were documented by optical microscopy. Dye droplets were recovered with isopropanol and quantified by spectrophotometry at 510 nm. Data were expressed as the mean ± SD from three independent experiments performed in triplicate and normalized against cells without treatment. For 3D cultures, the cells were washed with phosphate-buffered saline (PBS, pH 7.4, Sigma-Aldrich) and adhered to a culture plate coated with poly-L-lysine. Subsequently, they were permeabilized with 100X Triton (0.05%) and fixed with 4% paraformaldehyde for 20–30 min at room temperature. The cells were washed with 1X PBS and incubated with oil-red O dissolved in a solution of 60% isopropanol and 1X PBS for 20 min at room temperature. Following this, they were washed with 500 µl of a solution containing 70% ethanol and 1X PBS. Images were captured using an optical microscope. The dye retained within the cells was extracted with pure isopropanol and quantified at 510 nm using a microplate reader.
Co-cultures of adipocytes with breast cancer cells grown in 3D cultures
The interaction of adipocytes with organotypic 3D cell cultures was performed in a transwell chamber of 8.0 μm pore size, 24-well with 12 polyester membrane inserts (Corning Life Sciences, Lowell, MA, USA). Adipocytes were induced to differentiate in the lower part of the 24-well transwell system for five days, as described before. Then, the Hs578t cells were cultured in 3D in the upper part of the transwell chamber as follows. As a scaffold for cancer cell growth, we used Geltrex which is an ECM-enriched substrate containing laminin, collagen IV, entactin, and heparin sulfate proteoglycans. Briefly, the upper part was coated with 20 µl of Geltrex without LDEV, reduced growth factor, and incubated at 37 °C for 30 min. Next, 3.2 × 104 cells were resuspended in a final volume of 50 µl and incubated at 37 °C for 30 min. Finally, 50 µl of 5% Geltrex medium was added, and 3D cell cultures were maintained for six days, with DMEM medium changes every two days. Both mature adipocytes and Hs578t cancer cells were grown in DMEM supplemented with 10% fetal bovine serum and penicillin-streptomycin (10,000 unit/mL; Gibco 30-2300) at 37 °C in a humidified incubator with 5% CO2 atmosphere.
RT-PCR assays
Total RNA was obtained using TRIzol reagent (Invitrogen). For real-time PCR, relative gene expression was determined using SuperScript™ III Platinum™ SYBR™ Green One-Step qRT-PCR Kit (Thermo Fisher Scientific) and 1 µg of total RNA as a template. The primers used were APOB Forward: 5´-GGGATTGCCAGACTTCCACA-3´ and APOB Reverse: 5´-TAGGAGGCGGACCAGTTGTA-3´. Polymerase chain reaction conditions were as follows: 95 °C for 10 min, 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. Tests were normalized using GAPDH as internal control. Experiments were performed three times in triplicate, and results were expressed as mean S.D. p < 0.05 was considered statistically significant.
Proteomic analysis
Sample preparation for mass spectrometry
All proteomic procedures were performed as previously reported18. 50 µg of protein contained in Tris-TAPS-EDTA (TNTE) buffer for each condition (biological replicate, n = 3) was precipitated using MeOH/Chloroform, and resulting pellets were enzymatically digested using iST Sample Preparation Kit® (PreOmics GmbH, Munich, GER) according to manufacturer instructions. Briefly, 50 µL of Lyse reagent was added to protein pellets and placed in a heating block for 10 min, 95 °C at 1000 rpm. Then, samples were sonicated using BioRuptor Pico® (Diagenode, Liège, BEL). Protein samples were digested using 50 µL of a Lys-C/Trypsin mix and heating at 37 °C for 2 h. Resulting peptides were cleaned in an iST cartridge using a buffer to eliminate hydrophobic (Wash buffer 11) and hydrophilic molecules (Wash buffer 2); afterward, peptides were eluted using Elute reagent and subsequently evaporated to dryness in a SpeedVac (Thermo Fisher Scientific, Waltham, MS, USA ); finally, peptides were resuspended with LC-Load reagent and stored at -80 °C until LC-MS analysis.
Label-free quantification by mass spectrometry
Peptides were injected into the mass spectrometer Synapt G2-Si (Waters, Milford, MA) in MSE mode to calculate the area under the curve (AUC) of the total ion chromatogram (TIC) to normalize the injection before LC-MS analysis and therefore, inject a comparable sample quantity for both conditions1. Afterward, peptides in each sample were loaded and separated on an HSS T3 C18 column; 75 μm × 150 mm, 100 Å pore size, 1.8 μm particle size; using a UPLC ACQUITY M-Class with mobile phase A (0.1% formic acid in H2O) and mobile phase B (0.1% formic acid in acetonitrile) under the following gradient: 0 min 7% B, 121.49 min 40% B, 123.15 to 126.46 min 85% B, 129 to 130 min 7% B, at a flow of 400 nL·min − 1 and 45 °C. The spectra data were acquired in the mass spectrometer using nanoelectrospray ionization (nanoESI) and ion mobility separation (IMS) using a data-independent acquisition (DIA) approach through High-Definition Multiplexed MS/MS mode (HDMSE) mode. On the tune page, for the ionization source, parameters were set with the following values: 2.75 kV on the capillary emitter, 30 V on the sampling cone, 30 V on the source offset, 70 °C for the source temperature, 0.5 bar for the nanoflow gas and 150 L·h − 1 for the purge gas flow. Two chromatograms (low and high energy chromatograms) were acquired in positive mode in a range of m/z 50 − 2000 with a scan time of 500 ms. No collision energy was applied to obtain the low-energy chromatogram, while for the high-energy chromatograms, the precursor ions were fragmented in the transfer cell using a collision energy ramp from 19 to 55 eV.
Data analysis
The MS and MS/MS measurements contained in the generated *.raw files were analyzed and relatively quantified using Progenesis QI for Proteomics software v4.2 (Waters, Milford, MA) against a using a target decoy strategy against a Homo sapiens *.fasta database (obtained from UniProt, UP000005640, 77027 protein sequences, last modification on 29th January 2021), which was concatenated with the same *.fasta file in the reverse sense. Parameters used for the protein identification were trypsin as an enzyme and one missed cleavage allowed; carbamidomethyl (C) as a fixed modification and oxidation (M), amidation (C-terminal), deamidation (Q, N) and phosphorylation (S, T, Y) as variable modifications; default peptide and fragment tolerance (maximum normal distribution of 10 and 20 ppm respectively) and false discovery rate ≤ 4%. Synapt G2-Si was calibrated with [Glu1]-fibrinopeptide fragments through the precursor ion [M + 2 H]2 + = 785.84261 fragmentation of 32 eV with a result less than 1.2 ppm across all MS/MS measurements. Data generated from Progenesis software were exported to *.xlsx files to calculate statistical parameters. All proteins considered differentially expressed display a ratio of ± 1 (expressed as a base two logarithm); these proteins had at least ± 2 absolute fold change. The ratio was calculated based on the average MS signal response of the three most intense tryptic peptides (Top3)2 of each protein in the cocultured sample by the Top 3 of each protein in the control sample.
Enrichment analysis
The g: Profiler database (https://biit.cs.ut.ee/gprofiler/gost) was used to perform gene ontology (GO) analysis of differentially expressed proteins in 3D cultures of the Hs578t cell line in coculture with adipocytes19. The species was restricted to Homo sapiens, and the adjusted p-value (from the Benjamini-Hochberg method) of 0.05 was considered statistically significant18. GO terms included the following three criteria: molecular function (MF), cellular component (CC), and biological process (BP). Enriched GO terms were presented as enrichment scores. Signaling pathway analysis was performed using the REACTOME database (https://reactome.org/) to determine the involvement of differentially expressed proteins20.
Protein-protein interaction (PPI) analysis
The analysis and protein-protein interaction (PPI) networks were predicted using the STRING database (http://www.string-db.org) with protein accessions as input and a confidence level threshold of 0.4 (medium confidence). The interaction networks were visualized with Cytoscape v3.2.0 software (http://www.cytoscape.org)21.
Kaplan Meier analysis
Overall survival analysis using Kaplan Meier plotter for lipids-transport proteins in breast cancer patients was evaluated as previously described22. We used the Start KM plotter for breast cancer tool, which uses protein data expression and clinical information from a cohort of breast cancer patients (n = 65). The dataset was obtained from the study by Tang et al.23. The tool calculated a Kaplan-Meier survival plot comparing the two patient cohorts, the hazard ratio with 95% confidence intervals, and the log-rank P value, as described in the URL: https://kmplot.com/analysis/index.php?p=service&cancer=breast_protein.
Silencing of APOB expression using siRNAs
The Hs578t breast cancer cells (12,000) were cultured in 8-well dish plates until a confluence of 60% was reached. Two independent mixtures were made for each well to carry out the transfections. The first one contained lipofectamine 2000 (Invitrogen) mixed with Opti-MEM (Gibco, Waltham, MA, USA) and was gently homogenized and incubated again for 15 min at room temperature. In another tube, the specific siRNA for APOB (AM16708, Invitrogen) or scramble negative control (AM17110, Invitrogen), both at a concentration of 30 nM, were mixed with Opti-MEM medium and incubated for 15 min at room temperature. After that, the mixture of lipofectamine and the siRNA for APOB and scramble were incubated for 20 min to promote the formation of complexes. The mixture was added to each well, mixed gently by rocking the plate for 2 min, and fresh Opti-MEM medium (250 ml) was added. Transfected cells were incubated at 37 °C for four h. Finally, the medium was removed, DMEM with 10% Glutamax medium (Invitrogen) was added, and cells were incubated at 37 °C for 48 h. After the co-culture period, 3D structures of Hs578t cells were imaged using optical microscopy at 40X magnification. The morphology, number, and size of these structures were analyzed. The quantification of the number and diameter of the spheroids was performed using ImageJ software. Experiments were performed three times in triplicate, and results were expressed as mean (SD). p < 0.05 was considered statistically significant.
Statistical analysis
Experiments were performed three times, and the results were presented as mean ± S.D. One-way analysis of variance (ANOVA) followed by Tukey’s test was used to compare the differences between means. p < 0.05 was considered statistically significant.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
References
Protani, M., Coory, M. & Martin, J. H. Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res. Treat. 123(3), 627–635. https://doi.org/10.1007/s10549-010-0990-0 (2010).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell. 144(5), 646–674. https://doi.org/10.1016/j.cell.2011.02.013 (2011).
Deepak, K. et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple-negative breast cancer. Pharmacol. Res. 153, 104683. https://doi.org/10.1016/j.phrs.2020.104683 (2020).
Rybinska, I., Mangano, N., Tagliabue, E. & Triulzi, T. Cancer-Associated adipocytes in breast Cancer: causes and consequences. Int. J. Mol. Sci. 22(7), 3775. https://doi.org/10.3390/ijms22073775 (2021).
Balaban, S. et al. Adipocyte lipolysis links obesity to breast cancer growth: adipocyte-derived fatty acids drive breast cancer cell proliferation and migration. Cancer Metabolism. (5), 1. https://doi.org/10.1186/s40170-016-0163-7 (2017).
Harborg, S. et al. Circulating lipids and breast cancer prognosis in the Malmö diet and cancer study. Breast Cancer Res. Treat. 191(3), 611–621. https://doi.org/10.1007/s10549-021-06462-7 (2022).
Nickel, A. et al. Adipocytes induce distinct gene expression profiles in mammary tumor cells and enhance inflammatory signaling in invasive breast cancer cells. Sci. Rep. 8(1), 9482. https://doi.org/10.1038/s41598-018-27210-w (2018).
Dirat, B. et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 71(7), 2455–2465. https://doi.org/10.1158/0008-5472.CAN-10-3323 (2011).
Choi, J., Cha, Y. J. & Koo, J. S. Adipocyte biology in breast cancer: from silent bystander to active facilitator. Prog. Lipid Res. 69, 11–20. https://doi.org/10.1016/j.plipres.2017.11.002 (2018).
Zhu, Q. et al. Adipocyte mesenchymal transition contributes to mammary tumor progression. Cell. Rep. 40(11), 111362. https://doi.org/10.1016/j.celrep.2022.111362 (2022).
Zhao, C. et al. Cancer-associated adipocytes: emerging supporters in breast cancer. J. Experimental Clin. cancer Research: CR. 39(1), 156. https://doi.org/10.1186/s13046-020-01666-z (2020).
Heredia-Mendez, A. J., Sánchez-Sánchez, G. & López-Camarillo, C. Reprogramming of the Genome-Wide DNA Methylation Landscape in Three-Dimensional Cancer Cell Cultures. Cancers, 15(7), 1991. (2023). https://doi.org/10.3390/cancers15071991
Nuñez-Olvera, S. I. et al. Breast Cancer cells reprogram the oncogenic lncRNAs/mRNAs coexpression networks in three-Dimensional Microenvironment. Cells. 11 (21), 3458. https://doi.org/10.3390/cells11213458 (2022).
Lee, Y., Jung, W. H. & Koo, J. S. Adipocytes can induce epithelial-mesenchymal transition in breast cancer cells. Breast Cancer Res. Treat. 153(2), 323–335. https://doi.org/10.1007/s10549-015-3550-9 (2015). Epub 2015 Aug 19. PMID: 26285644.
Xu, P. et al. Mature adipocytes observed to undergo reproliferation and polyploidy. FEBS Open. Bio. 7, 652–658. https://doi.org/10.1002/2211-5463.12207 (2017).
Mandujano-Lázaro, G. et al. A short S-Equol exposure has a long-term inhibitory effect on adipogenesis in mouse 3T3-L1 cells. Appl. Sci. 11, 9657. https://doi.org/10.3390/app11209657 (2021).
Zebisch, K., Voigt, V., Wabitsch, M. & Brandsch, M. Protocol for effectively differentiating 3T3-L1 cells to adipocytes. Anal. Biochem. 425 (1), 88–90. https://doi.org/10.1016/j.ab.2012.03.005 (2012). Epub 2012 Mar 13. PMID: 22425542.
Ríos-Castro, E., Souza, G.H.M.F., Delgadillo-Álvarez, D.M., Ramírez-Reyes, L., Torres-Huerta, A.L., Velasco-Suárez, A., Cruz-Cruz,C., Hernández-Hernández, J.M., Tapia-Ramírez, J. Quantitative proteomic analysis of MARC-145 cells infected with a Mexican Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) strain using a label-free based DIA approach. J. Am. Soc. Mass. Spectrom. 31(6), 1302–1312. https://doi.org/10.1021/jasms.0c00134 (2020).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res., 47(W1), W191–W198. https://doi.org/10.1093/nar/gkz369 (2019).
Gillespie, M. et al. The Reactome pathway knowledgebase 2022. Nucleic Acids Res. 50 (D1), D687–D692. https://doi.org/10.1093/nar/gkab1028 (2022).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Ősz, Á., Lánczky, A. & Győrffy, B. Survival analysis in breast cancer using proteomic data from four independent datasets. Sci. Rep. 11 (1), 16787. https://doi.org/10.1038/s41598-021-96340-5 (2021).
Tang, W. et al. Integrated proteotranscriptomics of breast cancer reveals globally increased protein-mRNA concordance associated with subtypes and survival. Genome Med. 10(1), 94. https://doi.org/10.1186/s13073-018-0602-x (2018). PMID: 30501643; PMCID: PMC6276229.
Dang, D., Prasad, H. & Rao, R. Secretory pathway Ca2+ -ATPases promote in vitro microcalcifications in breast cancer cells. Mol. Carcinog. 56(11), 2474–2485. https://doi.org/10.1002/mc.22695 (2017).
Banh, R. S., Iorio, C., Marcotte, R., Xu, Y., Cojocari, D., Rahman, A. A., Pawling, J., Zhang, W., Sinha, A., Rose, C. M., Isasa, M., Zhang, S., Wu, R., Virtanen, C., Hitomi, T., Habu, T., Sidhu, S. S., Koizumi, A., Wilkins, S. E., Kislinger, T., … Neel, B. G. PTP1B controls non-mitochondrial oxygen consumption by regulating RNF213 to promote tumour survival during hypoxia. Nature cell biology, 18(7), 803–813. https://doi.org/10.1038/ncb3376 (2016).
Bobin-Dubigeon, C. et al. Paraoxonase 1 (PON1) as a marker of short-term death in breast cancer recurrence. Clin. Biochem. 45(16–17), 1503–1505 (2012).
Liu, J. X. et al. Apolipoprotein A1 and B as risk factors for development of intraocular metastasis in patients with breast cancer. Cancer Manage. Res.11, 2881–2888. https://doi.org/10.2147/CMAR.S191352 (2019).
Zhou, Y. & Luo, G. Apolipoproteins, as the carrier proteins for lipids, are involved in the development of breast cancer. Clinical & translational oncology: official. Publication Federation Span. Oncol. Soc. Natl. Cancer Inst. Mexico. 22(11), 1952–1962. https://doi.org/10.1007/s12094-020-02354-2 (2020).
Chen, J., Qian, X., He, Y., Han, X. & Pan, Y. Novel key genes in triple-negative breast cancer identified by weighted gene co-expression network analysis. J. Cell. Biochem. 120(10), 16900–16912. https://doi.org/10.1002/jcb.28948 (2019).
Zhou, Y. & Luo, G. Apolipoproteins, as the carrier proteins for lipids, are involved in the development of breast cancer. Clin. Transl. Oncol. 22(11), 1952–1962. https://doi.org/10.1007/s12094-020-02354-2 (2020).
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. Cancer J. Clin. 71(3), 209–249. https://doi.org/10.3322/caac.21660 (2021).
Denkert, C., Liedtke, C., Tutt, A. & von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet (London England). 389(10087), 2430–2442. https://doi.org/10.1016/S0140-6736(16)32454-0 (2017).
Karimian, M. A common genetic variation in paraoxonase 1 and risk of breast cancer: a literature review, meta-analysis, and in silico analysis. Nucleosides, nucleotides & nucleic acids, 1–16. Advance online publication. (2022). https://doi.org/10.1080/15257770.2022.2107216
Thompson, E. W. et al. Paraoxonase-1 activity in breast Cancer patients treated with Doxorubicin with or without Trastuzumab. JACC Basic. Translational Sci. 7(1), 1–10. https://doi.org/10.1016/j.jacbts.2021.10.010 (2021).
Kosti, A. et al. F. ELF4 is a critical component of a miRNA-transcription factor network and is a bridge regulator of glioblastoma receptor signaling and lipid dynamics. Neuro-oncology, noac179. Advance online publication. (2022). https://doi.org/10.1093/neuonc/noac179
Bodde, M. C. et al. Apolipoproteins A1, B, and apoB/apoA1 ratio are associated with first ST-segment elevation myocardial infarction but not with recurrent events during long-term follow-up. Clin. Res. Cardiology: Official J. German Cardiac Soc. 108(5), 520–538. https://doi.org/10.1007/s00392-018-1381-5 (2019).
Liu, X. et al. Associations of polymorphisms of rs693 and rs1042031 in apolipoprotein B gene with risk of breast cancer in Chinese. Jpn J. Clin. Oncol. 43(4), 362–368. https://doi.org/10.1093/jjco/hyt018 (2013).
Marino, N. et al. Upregulation of lipid metabolism genes in the breast prior to cancer diagnosis. NPJ Breast Cancer. 6:50. (2020). https://doi.org/10.1038/s41523-020-00191-8. Erratum in: NPJ Breast Cancer. 2024;10(1):47. doi: 10.1038/s41523-024-00649-z.
Ben Hassen, C. et al. Apolipoprotein-mediated regulation of lipid metabolism induces distinctive effects in different types of breast cancer cells. Breast Cancer Res. 22(1), 38. https://doi.org/10.1186/s13058-020-01276-9 (2020). PMID: 32321558; PMCID: PMC7178965.
Acknowledgements
The authors acknowledge Instituto Nacional de Enfermedades Respiratorias “Ismael Cosio Villegas” for supporting the publication fee of this manuscript. We also thank Universidad Autónoma de la Ciudad de México for its support. Karla Tovar-Hernández and Yarely M. Salinas-Vera received fellowships from Consejo Nacional de Humanidades Ciencias y Tecnologías (CONAHCyT). Alejandra P. García-Hernández received a postdoctoral fellowship from CONAHCyT.
Funding
CONAHCYT, México, funded this research. Programa Presupuestario F003 Grant 51207/2020 to Dr. César López Camarillo.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.L.C, L.A.M, E.I.S, A.C.R, E.R.C and J.T.R; Data curation: K.T.H, Y.M.S.V, E.C.R, J.T.R, A.P.G.H, and A.V.S; Formal analysis: K.T.H, Y.M.S.V, E.C.R, J.T.R, A.V.S, G.M.L, A.P.H.G, and C.L.C; Funding acquisition: A.C.R, C.L.C; Investigation: K.T.H, Y.M.S.V, E.C.R, E.I.S, J.T.R, A.V.S, G.M.L, A.P.H.G, and C.L.C; Methodology: K.T.H, Y.M.S.V, E.C.R, J.T.R, A.V.S, G.M.L, and A.P.H.G; Project administration: C.L.C; Resources: L.A.M., G.M.L, A.C.T.C, E.I.S, H.V.M, A.C.R and C.L.C; Supervision: L.A.M, A.P.G.H, and C.L.C; Writing – original draft: L.A.M, A.C.T.C; C.L.C; Writing – review & editing: C.L.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Authorship contribution
Conceptualization: C.L.C, L.A.M, E.I.S, A.C.R, E.R.C and J.T.R; Data curation: K.T.H, Y.M.S.V, E.C.R, J.T.R, A.P.G.H, and A.V.S; Formal analysis: K.T.H, Y.M.S.V, E.C.R, J.T.R, A.V.S, G.M.L, A.P.H.G, and C.L.C; Funding acquisition: A.C.R, C.L.C; Investigation: K.T.H, Y.M.S.V, E.C.R, E.I.S, J.T.R, A.V.S, G.M.L, A.P.H.G, and C.L.C; Methodology: K.T.H, Y.M.S.V, E.C.R, J.T.R, A.V.S, G.M.L, and A.P.H.G; Project administration: C.L.C; Resources: L.A.M., G.M.L, A.C.T.C, E.I.S, H.V.M, A.C.R and C.L.C; Supervision: L.A.M, A.P.G.H, and C.L.C; Writing – original draft: L.A.M, A.C.T.C; C.L.C; Writing – review & editing: C.L.C.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tovar-Hernández, K., Salinas-Vera, Y.M., Carlos-Reyes, Á. et al. Adipocytes reprogram the proteome of breast cancer cells in organotypic three-dimensional cell cultures. Sci Rep 14, 27029 (2024). https://doi.org/10.1038/s41598-024-76053-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-76053-1