Abstract
Immunotherapy has become a promising treatment for various cancers, including colorectal cancer (CRC). Despite significant progress, identifying immune cell-specific therapeutic targets remains challenging, especially for CD4+ T cells, whose activation influences both anti-tumor and pro-tumor immune responses. This study aims to identify potential immunotherapy targets for CRC by exploring the causal relationships between CD4+ T cell activation-associated genes and CRC through Mendelian randomization (MR) and single-cell RNA sequencing (scRNA-seq). We used transcriptome-wide MR, summary-based MR (SMR), and colocalization analysis, along with validation through multi-omics approaches, to identify 28 dynamic CD4+ T cell-related genes as therapeutic targets. Notably, PARP14 and ORMDL3 emerged as key targets, with strong associations to immune therapy resistance and CRC. This research highlights the critical role of CD4+ T cell activation in CRC progression and identifies novel potential targets for immunotherapy.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths worldwide, with approximately 147,950 new cases and 53,200 deaths reported annually1,2. For patients diagnosed at early stages, surgery remains the primary treatment option3. However, many patients are diagnosed at advanced or metastatic stages, with the 5-year survival rate of approximately 14%4. Immunotherapy has emerged as a promising strategy for CRC treatment in recent years5, with chimeric antigen receptor T-cell (CAR-T) therapy demonstrating efficacy in certain hematologic malignancies. Yet, its success in treating solid tumors like CRC is hindered by the immunosuppressive tumor microenvironment, primarily due to the inhibitory effects of the PD-1/PD-L1 checkpoint pathway on T-cell responses6. Overcoming these limitations to enhance CAR-T therapy in solid tumors remains a significant challenge7. Recent studies have focused on combining immune-related gene targeting with CAR-T therapy, showing potential in enhancing T-cell function. For example, targeting immune molecules such as interferon γ (IFN-γ), interleukin (IL)-6, and IL-12 has been shown to enhance T cell migration and infiltration into CRC cells8.
The tumor microenvironment (TME) is composed of various immune and stromal cells, with much of the current research focusing primarily on CD8+ T due to their well-known cytotoxic effects. However, accumulating evidence has highlighted the crucial role of CD4+ T cells in the tumor immune response9. CD4+ T cells can differentiate into multiple subsets, including Th1, Th2, Th9, Th17, regulatory T cells (Treg), and T follicular helper (TFH) cells10. Among these, Th2 cells are known to enhance angiogenesis and suppress immune cell-mediated tumor cytotoxicity11. Additionally, CD4+ T cells can secrete IFN-γ, which plays a crucial role in promoting the anti-tumor activity of CD8+ T cells12. Despite the recognized importance of T lymphocytes, particularly CD4+ T cells, few studies have systematically explored their potential as immune-mediated targets for CRC prevention or treatment. Thus, understanding the complex interactions between CD4+ T cells and CRC is crucial for identifying promising immune-mediated drug targets for CRC prevention and drug development.
Recent studies have revealed the pivotal role of metabolic reprogramming in cancer progression13. During T cell activation, especially in CD4+ T cell differentiation, significant metabolic shifts occur, which support immune responses. These metabolic changes may contribute to cellular reprogramming, further influencing tumor growth and survival14. While the metabolic alterations driven by immune activation have been recognized as key factors in both cancer progression and immune responses, metabolism-related immune targets for CD4+ T cells remain underexplored. Understanding how CD4+ T cells modulate metabolism within the TME could offer novel insights into how immune responses influence tumorigenesis.
Mendelian randomization (MR) is a method that investigates causal associations between phenotypes based on genetic variations and is increasingly utilized to prioritize drug targets15,16. The primary advantage lies in its ability to minimize confounding factors and reverse causality, compared to conventional epidemiological methods17. Furthermore, previous drug development programs have demonstrated that target-disease pairings identified through MR and colocalization are more likely to result in successful therapeutic approvals18. Genome-wide association study (GWAS) and expression quantitative trait loci (eQTL) have been used in MR analyses to identify disease-associated genes19,20. However, most studies rely on whole blood level data, which lack cell-specific resolution and fail to capture the cellular context accurately. In contrast, single-cell analysis is a revolutionary technology that provides high-resolution insights into cellular mechanisms, enabling a more precise understanding of disease biology21. The integration of single-cell eQTL with GWAS has proven valuable, particularly in identifying of drug targets in cancer22. For instance, Liu et al. performed a causal association analysis between immune cell gene expression and breast cancer, identifying the drug target KCNN4 associated with non-classic monocytes23. Therefore, combining single-cell eQTL with MR not only enhances the understanding of the genetic mechanisms underlying complex diseases but also offers valuable insights for drug development and personalized treatment strategies.
In this study, we integrated dynamic eQTL data with MR, summary-based Mendelian randomization (SMR), and colocalization analyses to comprehensively investigate the role of gene expression in different subsets of activated CD4+ T cells in CRC susceptibility. Specifically, we utilized dynamic single-cell eQTL data collected at distinct time points during CD4+ T cell activation to estimate causal associations between immune-related targets and CRC susceptibility. This MR-based causal inference approach enabled us to unravel the genetic mechanisms linking immune responses to CRC and identify promising immune-mediated therapeutic targets for CRC. Spatiotemporal single-cell RNA sequencing (scRNA-seq) analysis further elucidated immune therapy resistance-associated targets. Additionally, we explored the potential biological functions and unintended effects of targeting the genes identified during CD4+ T cell activation in CRC based on mediation analysis, virtual knockout (KO) experiment, and phenome-wide mendelian randomization (PW-MR). By integrating transcriptome-wide Mendelian randomization and scRNA-seq analysis, our findings deepen the understanding of immune-related pathways in CRC pathogenesis and identify potential immune-mediated therapeutic targets, paving the way for innovative intervention strategies in CRC treatment.
Results
Summary of instrument selection and causal association using dynamic eQTLs across CD4+ T cell activation and non-dynamic eQTLs from DICE and eQTLGen studies
The study design is summarized in Fig. 1. Gene expression eQTL data were extracted from Soskic et al., which included 17 types of CD4+ T cells at five different time points of CD4+ T cell (0 h, low activity-LA, 16 h, 40 h, 5 days) using anti-CD3/anti-CD28 human T-Activator Dynabeads from 119 European individuals. Cis-eQTLs were identified using the tensorQTL (v1.0.3) (ref. 24) R package, which applies linear regression to each SNP–gene pair within a 500-kb window surrounding the transcription start site (TSS) of each gene, as described by the authors25. After applying stringent filtering criteria (p < 5 × 10−8, clumping with r² < 0.001, F-statistics >10, and Steiger direction filtering), a total of 8587 eQTLs from 1440 genes across 46 expression profiles were selected for MR analysis (Supplementary Tables 1 and 2).
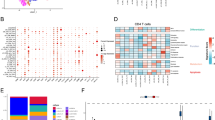
These 8587 eQTLs were used to assess causal association between gene expression and CRC susceptibility. The analysis, using CRC as the outcome (78,473 cases and 107,143 controls of European ancestry)26, identified 216 target-CRC pairs involving 52 genes with cell- and time point-specific causal associations across 32 cell types (pFDR < 0.05) (Fig. 2A, B and Supplementary Table 3). To validate these findings, colocalization and SMR analysis were conducted, further strengthening the causal association. Specifically, 142 target-CRC pairs demonstrated strong colocalization evidence (PP.H4/(PP.H3 + PP.H4) > 0.7) (Supplementary Table 4), and 159 target-CRC pairs met SMR criteria (pSMR < 0.05 and pHEIDI > 0.05), indicating that the observed associations were not confounded by LD (Supplementary Table 5). Integration of these results revealed 115 target-CRC pairs involving 28 genes passed both colocalization and SMR analysis (Fig. 2C and Supplementary Table 6), underscoring the robustness and reliability of the identified causal associations and providing valuable insights into immune-mediated therapeutic targets in CRC.
A The Manhattan plot displays the associations of genetically regulated gene expression in CD4+ T cells and CRC. The Y-axis represents the −log 10 of the pFDR values of the MR estimates. B The volcano plot shows the immune risk and protective targets for CRC. C The bar chart shows the number of MR results with p-values < 0.05 after Benjamini–Hochberg FDR correction, along with the proportion of results passing colocalization and SMR test criteria. D The Venn diagram illustrates the immune cell specificity of the identified targets: 24 targets were recognized exclusively in immune cells, and 15 targets identified solely in dynamic CD4+ T cells. E Nine dynamic CD4+ T cell immune targets were replicated in other immune cells. F The Venn diagram illustrates that the identified targets exhibit activation-specific and time-specific characteristics, with 21 targets (75%) causally associated with CRC only after activation. G Example genes with different causal effects at the activation time point are shown. The effect estimates represent odds ratios and 95% confidence intervals for disease risk per unit change in the related gene expression levels. The error bars indicate 95% confidence intervals.
In the non-dynamic causal association analysis, 104 eQTLs from DICE and 100 eQTLs from eQTLGen databases were used for MR analysis (Supplementary Tables 7–10). After applying a false discovery rate (FDR) threshold of <0.05 to the MR results from DICE, 82 target-CRC pairs involving 15 genes across 15 cell types were identified as having causal associations with CRC. Similarly, MR analysis of eQTLs from eQTLGen identified 11 target-CRC pairs involving 11 genes (Fig. 2C and Supplementary Tables 11 and 12). To ensure the robustness of these findings, colocalization and SMR analyses were also performed. In DICE, 57 target-CRC pairs involving 10 genes demonstrated strong colocalization evidence, and in eQTLGen, 4 target-CRC pairs involving 4 genes also showed similar results (Fig. 2C and Supplementary Tables 13–18).
Cell type- and time-specific causal effect of immune CD4+ T cell gene expression on CRC
Through dynamic and non-dynamic MR analyses, we observed that only 13 of the 28 dynamic CD4+ T cell targets were replicated in the non-dynamic MR analysis (Fig. 2D and Supplementary Table 19). Among these, 10 targets were replicated in the DICE database, and 4 targets were replicated in the eQTLGen database. These findings suggest that 15 genes were unique to CD4+ T cell activation, while 24 genes (15 from dynamic CD4+ T cell and 9 from DICE database) were identified as immune cell-specific targets. For instance, TMEM87B and DCTN5 exhibited strong causal associations with CRC in dynamic CD4+ T cells, but also displayed robust causal links in non-dynamic CD8+ T cells (Fig. 2E).
Among the 28 identified genes, 26 demonstrated causal associations with CRC after T cell activation, while 7 genes exhibited causal associations in the resting state. Notably, T cell activation revealed 21 additional genes with new causal associations to CRC compared to the resting state (Fig. 2F). Interestingly, 2 genes were exclusively associated with CRC in the resting and low activation states, while others showed time-specific associations following activation: 5 genes were associated with CRC exclusively at 16 h, 4 genes at 40 h, and 6 genes at 5 days post-activation. Some genes demonstrated dynamic and time-specific patterns. For example, NDUFA12 showed causal associations with CRC at all five time points, peaking at 40 h, which correlated with an increased CRC risk. Additionally, DCTN5 displayed associations at three time points, peaking at 40 h, but was linked to decreased CRC risk (Fig. 2G). These findings underscore the dynamic nature of gene associations with CRC, highlighting that gene expression and CRC risk vary throughout T cell activation, with distinct risk profiles at different immune response stages.
Identification and mechanistic analysis of key genes in CD4+ T cell as therapeutic targets in CRC for immunotherapy
Using three scRNA-seq datasets (GSE231559, GSE200997, and GSE166555), we identified CD4+ T cells and annotated cell subsets (Supplementary Figs. 1–3). Differential expression gene (DEG) analysis was performed comparing CD4+ T cells from CRC patients and non-tumor controls, applying stringent thresholds (min.pct > 0.1, p < 0.05, and |logFC | > 0.25; Supplementary Table 20). After excluding genes with inconsistent expression directions, two genes were identified as differential genes in three datasets: ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3), Poly (ADP-ribose) and polymerase family, member 14 (PARP14). Additionally, genes like Ribosomal protein L28 (RPL28), Tripartite Motif Containing 4 (TRIM4), NADH: ubiquinone oxidoreductase subunit A12 (NDUFA12) and potassium voltage-gated channel subfamily A member 3 (KCNA3) were identified under less stringent thresholds (|logFC | > 0.1, p < 0.05, Table 1). Importantly, these genes were aligned with the main MR results. For instance, ORMDL3, a risk gene identified in the MR analysis, showed higher expression in CD4+ T cells from CRC patients compared to controls.
Further validation using the TCGA-COADREAD project demonstrated a significant correlation between five targets (ORMDL3, RPL28, NDUFA12, PARP14, KCNA3) and the CD4+ T cell marker CD4 expression (Supplementary Fig. 4A), which were consistent with MR analysis. For instance, ORMDL3 and PARP14 were positively correlated with CD4 expression and associated with an increased risk of CRC. Additionally, both genes exhibited higher mRNA expression in CRC tissues compared to normal tissues (Supplementary Fig. 4B, C). Supporting these findings, the CPTAC database revealed significantly elevated protein expression levels of ORMDL3 and PARP14 in colon cancer samples (Supplementary Fig. 4D, E), which aligned with scRNA-seq results. Based on the above results, the identified targets ORMDL3 and PARP14 were regarded as the key targets.
To investigate the mechanistic roles of key targets in CRC, GSEA was performed using 50 HALLMARK gene sets. The analysis revealed enrichment in immune-related pathways and cancer signaling networks and the differential genes associated with ORMDL3 and PARP14 (Supplementary Table 21). For instance, ORMDL3 was implicated in activating epithelial-mesenchymal transition (EMT), angiogenesis, inflammatory response, and TNFA signaling via the NF-κB pathway in rectal adenocarcinoma (READ). Similarly, PARP14 was associated with EMT, inflammatory response, interferon-gamma response, and TNFA signaling via NF-κB pathway in colon adenocarcinoma (COAD) (Supplementary Fig. 4F, G). Additionally, to further explore the association between the identified targets and immune therapy resistance, we performed spatiotemporal differential expression analysis of CD4+ T cells based on the analytical pipeline outlined in Fig. 3A. Specifically, ORMDL3 and KCNA3 exhibited increased expression in the stable disease (SD) group following immune therapy, while no significant differences were observed in the complete or partial response (CR/PR) group (Fig. 3B–F). Interestingly, PARP14 is elevated in the SD group but decreased in the CR/PR group (Fig. 3C, F and Table 2). Consistent with these overall findings, subtype analysis further revealed that key CD4⁺ T cell targets were associated with immune resistance in MSI/MSS-CRC, with notably higher expression observed in MSI tumors (Fig. S5A–C). Pre-treatment prognostic analysis revealed that PARP14⁺ CD4 T cells and ORMDL3⁺ CD4 T cells were associated with poor patient outcomes (Fig. S6). Compared to clusters with low expression of the three targets, pathways such as TNF-α signaling via NF-κB, IL2/STAT5 signaling, mTORC1 signaling, hypoxia, and inflammatory response were significantly enriched in clusters 3, 7, and 9 (Fig. 3G). Additionally, the expression of immunosuppressive checkpoint genes, such as PDCD1, CTLA4, and LAG3, was also significantly elevated (Supplementary Fig. 7A). Elevated expression of ORMDL3, PARP14, and KCNA3 was associated with increased T cell dysfunction and exhaustion scores, as well as the higher proportion of predicted no-responders to immunotherapy, whereas NDUFA12 exhibited the opposite pattern (Figs. 3H and S7B–D). Collectively, these findings demonstrate that ORMDL3, PARP14, and KCNA3 may represent critical targets for CRC immunotherapy, particularly in addressing immune resistance, offering valuable theoretical insights for future personalized treatment strategies.
A Illustration of the spatiotemporal differential analysis framework. B The t-SNE visualization of CD4⁺ T cell subclusters identified by scRNA-seq. C–F The t-SNE and volcano plots of identified targets in the SD group pre- and post-immunotherapy. Gray dots represent non-differentially expressed genes. SD steady disease, CR complete response, PR partial response. G Biological functions associated with clusters 3, 7, and 9, compared to cell subpopulations with low expression of ORMDL3, PARP14, and KCNA3. H T-cell dysfunction scores predicted using the TIDE database for high and low expression groups of identified targets. Statistical significance is indicated as *p < 0.05, **p < 0.01, ***p < 0.001.
Identification of potential therapeutic drugs for CD4+ T cell-associated immune therapy against CRC
To identify therapeutic drug for CRC targeting these immune-related genes, we classified the identified targets into primary, secondary, and tertiary levels. Among the identified targets, 28 genes passed the main MR analysis, colocalization, and SMR analysis were classified tertiary targets. Five genes were identified as differential genes in CD4+ T cells, classified as secondary targets. Two genes, ORMDL3 and PARP14 were identified as differential genes in the CD4+ T cell and the TCGA project, classifying as primary targets (Fig. 4A).
A Classification of identified targets and multi-omics analysis. ORMDL3 and PARP14 were identified as Level 1 targets, having passed all analyses. KCNA3, RPL28, and NDUFA12, which failed the bulk RNA-seq analysis, were classified as Level 2 targets. The remaining genes, identified exclusively through MR, Coloc, and SMR analyses, were designated as Level 3 targets. Check marks indicate successful completion of the corresponding analysis; Cross marks indicate failure, and combined symbols denote partial fulfillment of analysis conditions. B Dendrogram illustrating the top potential therapeutic compounds targeting ORMDL3 and PARP14 in CRC, along with their mechanisms of action. These target-drug pairs exhibit strong binding affinities, with binding energies below −7 kcal/mol. C–F The molecular docking diagrams of example pairs show hydrogen bond interactions. TA: taurodeoxycholic-acid.
Using the CMap database, we identified 16 compounds with the potential to reverse TME changes induced by ORMDL3 and PARP14 (Table 3). Molecular docking analysis demonstrated that 15 of these compounds exhibited binding energies below −5 kcal/mol, with 8 drugs showing binding energies below −7 kcal/mol, indicating strong target-ligand interactions (Fig. 4B and Table 3). Among the identified compounds, several demonstrated promising binding affinities and therapeutic potential for CRC. Literature analysis confirmed that they offer potential therapeutic benefits through different mechanisms: Seocalcitol, a Vitamin D receptor agonist with well-documented anti-tumor activity in CRC (Fig. 4C)27; Prednisone, a corticosteroid that has shown efficacy in combination with abiraterone acetate for prostate cancer, highlighting its potential for broader oncological applications (Fig. 4D)28; AV-608, an insulinotropic receptor agonist that may enhance anti-CRC effects as an adjunct to chemotherapy (Fig. 4E)29. Haloperidol, a dopamine receptor antagonist, specifically targeting dopamine receptor D2, identified as a novel therapeutic candidate for CRC (Fig. 4F)30. Additionally, the safety considerations of approved or clinically investigated compounds are summarized in Supplementary Table 22. These findings provide a compelling basis for further investigation into the repurposing or development of these compounds as targeted therapies against CRC, leveraging their diverse mechanisms of action to counteract ORMDL3- and PARP14-mediated TME alterations.
Mediation effect of dynamic immune-related targets on CRC outcomes via plasma metabolites
Given that metabolites are closely linked to T cell function and collectively mediate anti-tumor immune responses, a mediation analysis was conducted to explore potential interactions among target genes, CD4+ T cells, metabolites, and CRC. In the causal association analysis between plasma metabolites and CRC, the BH method was applied to control for false positives in multiple hypothesis testing. After excluding results with heterogeneity and pleiotropy, 13 plasma metabolites were identified as having significant causal association with CRC (Fig. 5A). MR-Egger analysis confirmed the absence of pleiotropy, and for results with heterogeneity (p < 0.05), a random-effects model was used to ensure robustness (Supplementary Table 23). Among these, bilirubin degradation product, C17H18N2O4 (2) and Indoleacetoylcarnitine demonstrated causal association with the immune targets ORMDL3 and PARP14, respectively (Fig. 5B). Further investigation using two-step MR revealed that PARP14 mediated CRC progression via Indoleacetoylcarnitine during CD4+ T cell activation, with a mediation proportion of 10.95%, while ORMDL3 mediated CRC progression via C17H18N2O4 (2), with a mediation proportion of 5.81% (Fig. 5C, D). These results suggest that immune targets mediate the progression of colorectal cancer associated with specific metabolites during CD4+ T cell activation, providing potential targets for immune and metabolic intervention strategies.
A Causal association between plasma metabolites and CRC (FDR < 0.05). B Causal associations between primary targets ORMDL3 and PARP14 and plasma metabolites (p < 0.05). C Indoleacetoylcarnitine was identified as mediating 10.95% of the causal relationship between PARP14 and CRC. D The bilirubin degradation product, C17H18N2O4 (2), was identified as mediating 5.81% of the causal association between ORMDL3 (memory CD4+ T cell) and CRC. GCST90200052: 1-(1-enyl-palmitoyl)-2-arachidonoyl-gpc (p-16:0/20:4), GCST90200685: 1-stearoyl-2-arachidonoyl-gpc (18:0/20:4), GCST90200692: 1-palmitoyl-2-arachidonoyl-gpc (16:0/20:4n6), GCST90199899: 1-(1-enyl-palmitoyl)-GPC (p-16:0), GCST90199788: 1-arachidonoyl-gpc (20:4n6), GCST90200219: Cholic acid glucuronide, GCST90199791: 1-arachidonoyl-GPE (20:4n6), GCST90200041: 1-palmitoyl-2-stearoyl-gpc (16:0/18:0), GCST90200203: Indoleacetoylcarnitine, GCST90200702: Bilirubin degradation product, C17H18N2O4 (2), GCST90199754: 7-methylxanthine, GCST90200375: Gamma-glutamylglutamate, GCST90199854: 5alpha-pregnan-3beta,20alpha-diol monosulfate (2). Statistical significance is indicated as *p < 0.05.
Evaluation of the biological functions, pleiotropic effects, and potential adverse effects of targeting ORMDL3 and PARP14
To assess the potential biological functional changes induced by targeting CD4+ T cell therapeutic targets, virtual KO experiments in CD4+ T cell revealed that the KO of ORMDL3 and PARP14 perturbed 75 and 14 genes, respectively (Supplementary Table 24). Functional enrichment analysis using the Enrichr database indicated that the genes significantly perturbed by ORMDL3 and PARP14 were involved in pathways closely associated with CRC progression, including TNF-α signaling via NF-κB, hypoxia, colorectal cancer, and Wnt/β-catenin signaling pathways (Fig. 6A and Supplementary Table 25). Notably, TNF-α signaling via NF-κB and hypoxia were consistently enriched across multiple analyses, including GSEA (Fig. 6B and Supplementary Table 26).
A Functional annotation of the top 20 significantly perturbed genes following the virtual KO of ORMDL3 and PARP14. B GSEA analysis of the top 10 of perturbed genes following virtual KO. C The bubble plot illustrates the causal associations between the targets and phenotypes from multiple categories in the FinnGen database (European individuals) (Top10), with a significance level set at FDR < 0.05. Colored points represent FDR < 0.05, while gray points indicate FDR > 0.05. The full list of associations is shown in Supplementary Table S26.
To identify the pleiotropic and potential adverse effects of drugs targeting ORMDL3 and PARP14, we performed PW-MR analysis using FinnGen database (R11), applying BH correction to control for false discovery rates. After excluding phenotypes directly related to intestinal tumors, the analysis included 2417 binary phenotypes (Supplementary Table 27). The results revealed that ORMDL3 was strongly associated with childhood asthma, while PARP14 was associated with psoriasis (Fig. 6C). These findings underscore the therapeutic potential of ORMDL3 and PARP14, highlighting their involvement in critical signaling pathways associated with immune therapy resistance. Also, these results suggest that their impact may extend beyond cancer therapy, providing broader implications for targeting these genes in immune-related diseases.
Discussion
In this study, we aimed to identify immune-related genes causally associated with CRC using a comprehensive multi-omics approach that integrates MR, colocalization, SMR, and HEIDI analyses. By examining the causal associations between 8587 cis-eQTLs from 17 immune cell types during CD4+ T cell activation and CRC, we aimed to elucidate immune therapy targets involved in CRC progression. Unlike traditional observational studies, MR mitigates the interference of reverse causality and confounding factors31,32. The integration of MR with colocalization and SMR analyses further enhances the accuracy of these inferences by leveraging genetic information, minimizing the impact of LD, thereby increasing the credibility of our findings33,34,35. Importantly, the target-disease associations identified through this integrated approach have higher translational potential for clinical approval18. To the best of our knowledge, this is the first study to explore immune-related targets for CRC through a multi-omics framework, incorporating additional scRNA-seq, bulk RNA-seq, immunotherapy prediction, mediation analysis, virtual KO and PW-MR to validate the identified immune targets. The dynamic genes identified during CD4+ T cell activation offer new insights into CRC prevention strategies and potential drug targets.
Recent large-scale genetic studies have identified several druggable protein targets in CRC36,37, but these studies predominantly focused on whole blood level and lacked immune cell-specific analysis, limiting our understanding of how immune cell gene expression impacts CRC. Our study bridges this gap by identifying 28 immune-related genes strongly associated with CRC risk, of which only four were replicated in whole-blood tissue. This finding suggests that immune therapeutic targets for CRC may be distinct from those detectable in whole blood, highlighting the importance of immune cell-specific analysis. While previous research has focused on enhancing the anti-tumor effects of CD8+ T cells38, recent studies emphasize the crucial role of CD4+ T cells in both promoting and inhibiting tumor progression10. For instance, patients with lower CD4+ T cell counts and CD4+/CD8+ ratios may respond better to PD-1 inhibitors in mismatch repair-deficient CRC39. Additionally, TH1-like CD4+ tumor-infiltrating lymphocytes (TILs) have been shown to recruit and enhance the proliferation and cytotoxicity of CD8+ TILs40. These studies highlight the dual roles of CD4+ T cells in the tumor microenvironment, yet immune targets specifically directed at CD4+ T cells remain underexplored. Our study identified 115 target-CRC pairs involving 28 genes in CD4+ T cells, with 21 genes exclusively associated with CRC risk following CD4+ T cell activation, providing valuable therapeutic targets for CRC immunotherapy.
Recent advances in single-cell technologies have revealed dynamic changes in gene expression during tumorigenesis. For instance, NAMPT, BCL2A1, and TREM1 expression levels decrease in peripheral classical monocytes during colon adenocarcinoma progression41. However, observing gene expression changes alone does not necessarily imply causal effects. Our study identified dynamic causal associations between CD4+ T cell gene expression and CRC risk during T cell activation. For instance, the inhibitory effect of NDUFA12 on CRC risk peaked at 40 h, suggesting its potential as a key protein target in CRC treatment.
As level 1 targets, ORMDL3 and PARP14 have been previously linked to immune-related functions. Using a similar approach, ORMDL3 has been associated with cervical cancer risk22. ORMDL3 regulates early signaling events in lymphocyte activation, including store-operated calcium entry (SOCE), a process critical for CD4+ T cells activation42. Interestingly, individuals with asthma risk alleles at the 17q12–21 locus exhibit overexpression of ORMDL3, potentially raising basal calcium levels in naive CD4+ T cells43. Moreover, the overexpression of ORMDL3 may drive CD4+ T cells differentiation toward a Th2 phenotype, promoting chronic inflammation and immune responses linked to allergic asthma44. Our results align with these findings, suggesting that ORMDL3 overexpression contributes to CD4+ T cell activation and inflammation in CRC progression. Interestingly, excessive alcohol consumption has been reported to increase ORMDL3 expression, suggesting that lifestyle factors may influence its regulation45. PARP14, another key target, has been implicated in Th2 differentiation, chronic inflammation, and immune therapy resistance46. Both ORMDL3 and PARP14 were found to correlate with increased CRC risk during CD4+ T cell activation, and both were associated with upregulation of NF-κB signaling and inflammatory response, reinforcing their potential as dynamic immune targets in CRC. Furthermore, our spatiotemporal analysis of CD4+ T cells revealed that ORMDL3, PARP14, and KCNA3 were associated with immune therapy resistance. Specifically, ORMDL3, PARP14 and KCNA3 were significantly upregulated in patients who experienced SD following immunotherapy, and PARP14 was significantly decreased in the CR/PR group. Similarly, TIDE analysis revealed that high expression of ORMDL3, PARP14, and KCNA3 was associated with high T cell dysfunction scores. Notably, the upregulation of CD4⁺ T cell targets in the SD group was positively associated with T cell exhaustion scores and immunosuppressive checkpoint genes. These observations suggest that CD4⁺ T cell targets may modulate immune checkpoint gene expression during immunotherapy, promoting T cell exhaustion and contributing to immunotherapy tolerance. This potential mechanism highlights the role of dynamic CD4⁺ T cell regulation in the development of immunotherapy resistance and underscores the importance of these targets in strategies aimed at overcoming treatment failure in CRC.
Immunotherapy has emerged as a promising strategy in CRC treatment, with a growing body of research focused on identifying actionable targets and developing corresponding therapeutic agents47. However, the clinical application potential of these targets remains insufficiently characterized. In this study, we employed mediation analysis, virtual KO experiments, and PW-MR analysis to elucidate the biological functions and potential side effects of targeting the identified genes, thereby evaluating their therapeutic potential and clinical utility in CRC.
In drug discovery efforts, we identified seocalcitol (a vitamin D receptor agonist) and irbesartan (a renin-angiotensin system inhibitor) as promising candidates for targeting ORMDL3 and PARP14. Seocalcitol has demonstrated growth-inhibitory potential in hepatocellular carcinoma and CRC by modulating the WNT/β-catenin pathway48,49, while irbesartan may inhibit tumor recurrence by blocking the AP-1 transcriptional complex50. Both drugs were found to reverse tumor-promoting microenvironments induced by ORMDL3 and PARP14, highlighting their translational potential in CRC therapy.
Emerging evidence highlights the pivotal role of metabolic reprogramming in shaping the tumor immune microenvironments. Tumor-associated macrophages, for instance, inhibit CD8+ T cell responses by altering arginine metabolism51, while Glut1-dependent metabolic reprogramming supports effector CD4+ T cell activation and effector survival52. Our study revealed that metabolites such as indoleacetoylcarnitine and bilirubin degradation products mediate the associations between PARP14, ORMDL3, and CRC. Notably, these metabolites, derived from tryptophan and bilirubin pathways, play dual roles in cancer progression. For example, tryptophan metabolites like trans-3-indoleacrylic acid promote CRC via the AHR–ALDH1A3 axis53. Although bilirubin has antioxidant properties, elevated serum bilirubin levels have been linked to an increased risk of CRC54. Interestingly, a recent study by Seong-Keun Yoo et al. incorporated bilirubin into a comprehensive metabolite panel and applied this panel to immunotherapy response prediction models, which provides a complementary perspective on the associations we observed between bilirubin degradation products, ORMDL3, and immune therapy tolerance55.
NF-κB signaling plays a central role in tumor immune evasion by fostering the accumulation of immunosuppressive cells and promoting immune-suppressive factor secretion, leading to resistance to immunotherapy56,57. Persistent NF-κB activation impairs immune cell function and reduces anti-tumor efficacy during immunotherapy. In our virtual KO study, the NF-κB signaling pathway emerged as one of the most significantly enriched gene sets, suggesting its potential role in mediating the resistance to tumor immunotherapy induced by ORMDL3 and PARP14.
Furthermore, PW-MR analysis revealed significant associations between ORMDL3 and PARP14 and increased risks of asthma and psoriasis, indicating their potential as targets for therapeutic intervention or drug repositioning. Future development of drugs targeting ORMDL3 or PARP14 in CRC may also provide therapeutic opportunities for these immune-related diseases. This cross-disease relevance underscores the translational potential of ORMDL3 and PARP14 and provides a rationale for further investigation into their broader clinical applications. Notably, our PW-MR analyses did not reveal significant adverse effects, suggesting that interventions targeting ORMDL3 and PARP14 may have a favorable safety profile.
This study has several strengths. First, we analyzed gene expression profiles at different stages of CD4+ T cell activation, revealing immune cell-specific targets for CRC therapy. Second, the integration of multi-omics approaches, including MR, colocalization, SMR, and scRNA-seq, enhanced the robustness of our findings. Third, the use of the CMap drug database facilitated the identification of potential therapeutic agents targeting the immune-related genes identified in our study. Finally, mediation analysis, virtual KO experiments, and PW-MR provided valuable insights into the therapeutic potential and clinical applicability of tumor immunotherapy targets. However, this study has several limitations. The study population was predominantly European, and the lack of immune eQTL data from other populations may introduce biases. Additionally, due to the absence of dynamic data for non-CD4⁺ T cells, the functional roles of the key targets identified within non-CD4 T cell populations remain unclear. Moreover, the relatively small sample size of dynamic immune eQTL data may limit statistical power. Future studies should address these limitations by incorporating diverse populations, larger sample sizes, and experimental validation of dynamic targets across multiple immune cell types.
In conclusion, this study identified 28 putative causal genes associated with CRC, with 24 of these uniquely discovered through immune cell-specific eQTL analyses. Among them, ORMDL3 and PARP14 emerged as primary therapeutic targets for CRC immunotherapy, both linked to immune therapy resistance. Additionally, PARP14 was implicated in mediating CRC risk via the metabolite indoleacetoylcarnitine, while ORMDL3 was associated with CRC progression through the bilirubin degradation product, C17H18N2O4 (2). By conducting cell-type and time-specific causal analyses, along with scRNA-seq analysis, this study provides deeper insights into the dynamic nature of immune gene expression in CRC. In summary, ORMDL3, PARP14, RPL28, KCNA3, and NDUFA12 are highlighted as promising targets for immune-modulating therapeutics, offering novel insights for the strategies against CRC.
Methods
Genetic instrument selection for dynamic expressions of genes in CD4+ T cells
The summarized dynamic CD4+ T cell, immune cell, and blood cis-eQTL data were derived from the study by Soskic et al.25, the Database of Immune Cell Expression (DICE)58, and the eQTLGen Consortium (eQTLGen)59. The data sources and sample information were summarized in Table 4. A total of 46 gene expression profiles from 17 types of CD4+ T cells were identified across five distinct activation states in T cell activation: resting state, low activation, 16-h activation, 40-h activation, and 5-day activation. The CD4+ T cell types included in the analysis were CD4 Naive, TN, TN cycling, TN HSP, TN interferon (IFN), TN nuclear factor κB (NF-κB), TN2, CD4 Memory, heat shock protein (HSP), nTreg, T ER-stress, central memory T cell (TCM), effector memory T cell (TEM), TEM human leukocyte antigen (HLA) positive, effector memory cells re-expressing CD45RA (TEMRA), TM cycling, and TM ER-stress. To perform the MR analysis, the following processing steps were applied to the summary cis-eQTL data: (1) The full summary data were filtered using a p value threshold of < 5 × 10−⁸, ensuring the instrumental variables were strongly associated with exposures60; (2) To mitigate the effect of linkage disequilibrium (LD), the instrumental variables were clumped (r² < 0.001); (3) Instrumental variables with an F-statistic less than 10 were excluded; (4) Instrumental variables within the MHC region (chr6: 25.5–34MB) were excluded. Additionally, Steiger filtering was applied to test the directionality of the eQTL-CRC associations, ensuring that the eQTL influences the outcome through its effect on the exposure61.
Genetic instrument selection for expressions of genes from non-dynamic eQTL datasets
To explore whether the dynamic immune-related targets were also causal associations with CRC in non-dynamic MR analysis, we analyzed immune targets identified through main MR, SMR, and colocalization analyses. The DICE database includes eQTL for gene expression from naive B cells, classical monocytes, non-classical monocytes, CD56dim CD16+ NK cells, CD4+ T cells (memory TREG, naive, activated naive, naive TREG, TFH, TH1, TH2, TH17, TH1/TH17), and CD8+ T cells (naive, activated naive), while eQTLGen includes cis-eQTL from whole blood. The same criteria were applied to filter the non-dynamic eQTLs [p < 5 × 10−⁸, clumping (r2 < 0.001), F > 10, and Steiger test]. The results passing colocalization and SMR analyses were retained for further investigation.
Genetic instrument selection for plasma metabolites
The plasma metabolite data used in this study were derived from individuals of European ancestry, encompassing both individual plasma metabolites and metabolite ratios62. To identify genetic instruments for MR, we selected metabolite quantitative trait loci (mQTL) using stringent criteria consistent with dynamic MR analysis. Specifically, we applied a threshold of p < 5 × 10−⁸ for the association of genetic variants with plasma metabolites, and employed a clumping window of 10,000 kb with an r² < 0.001 to ensure the independence of selected variants. Detailed information regarding the data sources, sample size, and other relevant study parameters can be found in Table 4.
Outcome selection
The CRC GWAS meta-analysis summary statistics, published by Soskic et al., include data from 185,616 individuals of European ancestry, comprising 78,473 cases and 107,143 controls, which is the largest CRC GWAS dataset for European populations to date26 (Table 4). The data used in this study were obtained from publicly available databases, and ethical approval granted by the ethics committee in the original publications. This study adheres to the principles outlined in the Declaration of Helsinki.
Dynamic single-cell eQTL MR analysis (main analysis)
In the dynamic MR analysis, the causal association between cis-eQTL from CD4+ T cells and CRC was assessed by Wald ratio and inverse variance weighted (IVW) methods. Benjamini–Hochberg (BH) false discovery rate (FDR) correction with p-values < 0.05 was applied to select MR results as candidate gene-disease pairs for further investigation. All analyses were conducted using R software (v.4.3.3) with the TwoSampleMR package (v.0.6.6).
Colocalization and SMR analyses of candidate MR signals
To assess whether the main MR results were influenced by LD and confounding factors, we further analyzed the identified targets using colocalization and SMR methods. Colocalization analysis was used to assess the probability that the two traits (exposure and outcome) share the same causal variants, using default parameters. In colocalization analysis, five hypotheses were considered (H0, H1, H2, H3, H4), each representing different scenarios regarding the relationship between the two traits: (1) H0 (No colocalization): no shared causal variant; (2) H1 (colocalization of two distinct variants): the two traits are associated with different variants at a specific locus; (3) H2 (one shared causal variant): the two traits are associated with single variant; (4) H3 (colocalization of two causal variants): two variants in the same region contribute to the traits; (5) H4 (strong colocalization): a single causal variant drives both traits. In this analysis, PP.H3 and PP.H4 were used to assess the probability of shared causal variants, with PP.H4/(PP.H3 + PP.H4) > 0.7 indicating strong colocalization63.
The SMR method was used to investigate associations between gene expression levels and complex traits using summary-level data from GWAS and eQTLs64. The SMR and HEIDI methods were employed whether the effect size of an SNP on the phenotype was mediated by gene expression. In this analysis, genes with pSMR < 0.05 and pHEIDI > 0.05 were considered prioritized targets. The same parameters were applied for non-dynamic MR (FDR < 0.05), colocalization [PP.H4/(PP.H3 + PP.H4) > 0.7], and SMR [pSMR < 0.05 & pHEIDI > 0.05)] analysis.
CD4+ T cell differential gene expression analysis
To investigate whether the identified immune targets were differentially expressed in CD4+ T cells in CRC tissues, we collected single-cell RNA sequencing (scRNA-seq) data from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/), including datasets GSE231559, GSE200997, and GSE166555. scRNA-seq data were normalized using the ‘NormalizeData’ function in Seurat (version 4.1.2). Highly variable genes were identified with ‘FindVariableFeatures’ function, and data were scaled using ‘ScaleData’ function. Principal Component Analysis (PCA) was performed for dimensionality reduction. To correct for batch effects, the Harmony algorithm (RunHarmony) was applied for data integration. Clustering was performed using ‘FindNeighbors’ and ‘FindClusters’ functions (resolution = 0.5). CD3D(+) CD3E(+) clusters were identified as T cells, and CD4(+) clusters were considered as CD4+ T cells. Differentially expressed genes (DEGs) between CD4+ T cells from tumor and non-tumor tissues were identified using ‘FindMarkers’. Genes were considered differentially expressed if they met the following criteria: (1) min.pct >0.1; (2) p < 0.05; (3) average log2fold change (FC) > 0.25.
Gene expression and gene set enrichment analysis (GSEA) in tumor tissue
We used UCSC Xena, a platform that aggregates bulk RNA-seq data from multiple cancer databases, to retrieve the CRC cohort from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression project (GTEx), which includes 514 colon adenocarcinoma (COAD) samples and 177 rectal adenocarcinoma (READ) samples. The universal analysis and visualization of cancer data (UALCAN) database provided protein expression analysis using data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC)65. Additionally, the Gene Expression Profiling Interactive Analysis 2 (GEPIA2) and Tumor Immune Estimation Resource (Timer) database were used to explore gene expression corrections and CD4+ T cell marker.
In order to further elucidate the mechanisms underlying immune targets in cancer, we performed enrichment analysis using the HALLMARK gene sets with the clusterProfiler (v.4.12.2) and enrichplot (v.1.24.2) packages. The HALLMARK gene sets include 50 gene sets derived from diverse biological processes, signaling pathways, and cellular functions that are characteristic of human cancers. These gene sets represent key biological themes such as apoptosis, cell cycle regulation, immune response, and metabolic processes, which are known to play significant roles in tumorigenesis, cancer progression, and metastasis.
Spatiotemporal single-cell sequencing analysis related to immune therapy
To further investigate whether the identified targets are associated with immune therapy resistance, we integrated the most recently published spatiotemporal transcriptomic scRNA-seq data, which included 975,275 high-quality cells from 22 individuals66. The study collected 169 matched samples of blood, tumor tissue, and normal tissue pre- and post-immune therapy, for scRNA-seq analysis. Following immune therapy assessment, the 22 individuals were classified into three groups: non-responders with ‘steady disease’ (SD, N = 3), and responders categorized as ‘complete response’ (CR, N = 12) or ‘partial response’ (PR, N = 7). Using the same analytical pipeline, we performed a secondary analysis on tumor tissue samples from these 22 individuals, conducting differential analysis of CD4+ T cells between SD and CR/PR samples before and after treatment. Based on the method by Zhang et al., CD4⁺ T cells with high gene expression were mapped onto bulk RNA-seq data67. To explore subtype-dependent expression profiles and immunotherapy-related dynamics of CD4⁺ T cell targets in MSI- and MSS-CRC, we performed parallel analyses using bulk and single-cell datasets. Furthermore, based on the Tumor Immune Dysfunction and Exclusion (TIDE) and GEPIA2 databases, we systematically evaluated T cell dysfunction and exhaustion scores in the TCGA-COAD cohort across different levels of target gene expression, and additionally assessed the associations between key CD4⁺ T cell targets and immunosuppressive checkpoint genes in the scRNA-seq cohorts.
Identification of therapeutic targets related to CD4+ T cells
The Connectivity MAP (CMap) database is a valuable tool for drug repurposing, disease mechanism research, and immune-related studies. By analyzing the relationship between drugs and diseases at the gene expression level, CMap aids in identifying new drug candidates, recognizing potential targets, and understanding the mechanisms of drug action68. In this study, the CMap database was used to assess the therapeutic potential of the recognized immune targets and identify associated candidate drugs. Additionally, we employed molecular docking techniques to explore the binding affinity between the drugs and the targets.
Mediation analysis
For immune targets that have a causal association with both CRC and plasma metabolites, we performed a mediation analysis to quantify the effects of the identified immune targets on CRC through metabolites. The mediation proportion was calculated by evaluating both the total effect and indirect effects. The total effect represents the overall impact of the exposure on the outcome, while the indirect effect was estimated using the delta method69.
Virtual knockout experiment of identified targets
To investigate the impact of target gene perturbation, we performed virtual knockout (KO) experiments using the R package scTenifoldKnk70, a machine learning workflow based on scRNA-seq data. This approach allowed us to identify genes perturbed by the virtual KO of target genes. Differentially significant genes (adjusted p < 0.05) were subjected to functional enrichment analysis, while the entire set of genes was analyzed using GSEA to evaluate potential biological functional changes associated with the virtual KO of the targets in CRC.
Phenome-wide Mendelian randomization of identified targets
Phenome-wide Mendelian randomization (PW-MR) is a comprehensive approach that investigates the association between genetic variants and a wide range of phenotypic traits. In this study, we downloaded the binary outcome data from FinnGen R1171. After excluding the intestinal tumor outcomes, the remaining 2417 binary variables were used for PW-MR analysis. This approach enables the identification of potential drug adverse effects, evaluation of pleiotropy, and facilitates the discovery of therapeutic targets, offering insights into the multifaceted effects of drugs.
Ethical statement
A portion of the data used in this study was obtained from publicly available sources. The authors of the original GWAS and GEO datasets had obtained all necessary ethical approvals, and all participants provided informed consent. These approvals were granted based on adherence to the ethical principles outlined in the Declaration of Helsinki.
Data availability
The data used in this study were obtained from publicly available databases. The GWAS summary data for immune cells eQTL are available at Zendo (https://zenodo.org/records/6006796) and DICE (https://dice-database.org/downloads#info_anchor) database25,58. The GWAS summary data for whole blood eQTL can be accessed at https://eqtlgen.org/59. The GWAS summary data for CRC are available at the GWAS catalog (https://www.ebi.ac.uk/gwas/studies/GCST90255675)26. The GWAS summary data for plasma metabolites mQTL are available at the GWAS catalog (https://www.ebi.ac.uk/gwas/studies/GCST90199621-GCST90201020)62. The summary data utilized in PW-MR analysis were sourced from the FinnGen database (https://www.finngen.fi/en/access_results)71. The single-cell RNA-seq data for human colon tumor tissue and adjacent normal tissues can be accessed at GEO, via GSE231559, GSE200997, and GSE16655 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE231559, GSE200997, and GSE166555). The bulk RNA-seq data for human colon and rectum tumor tissue and adjacent normal tissues are available at the UCSC Xena (https://xenabrowser.net/datapages/). The scRNA-seq data used for immunotherapy analysis can be accessed at GEO: GSE236581. The T cell dysfunction assessment scores were derived from the TIDE database (http://tide.dfci.harvard.edu/login/). Potential drug candidates were identified using the CMap database (https://clue.io/). The scRNA-seq data used in the virtual KO experiments were sourced from the GEO database at GSE231559 and GSE200997 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE231559, GSE200997). The bulk RNA-seq data of MSI- and MSS-subtype colorectal cancer can be accessed from the GEO dataset GSE271172. The Enrichr database was used for the enrichment analysis of significantly disturbed genes (https://maayanlab.cloud/Enrichr/). Kaplan–Meier survival analyses were performed using the Kaplan–Meier plotter database (https://kmplot.com/analysis).
References
Grothey, A., Fakih, M. & Tabernero, J. Management of BRAF-mutant metastatic colorectal cancer: a review of treatment options and evidence-based guidelines. Ann. Oncol. 32, 959–967 (2021).
Wang, S. et al. Photothermal therapy mediated by gold nanocages composed of anti-PDL1 and galunisertib for improved synergistic immunotherapy in colorectal cancer. Acta Biomater. 134, 621–632 (2021).
Manjelievskaia, J. et al. Chemotherapy use and survival among young and middle-aged patients with colon cancer. JAMA Surg. 152, 452–459 (2017).
Liu, N. et al. Traditional Chinese medicine combined with chemotherapy and cetuximab or bevacizumab for metastatic colorectal cancer: a randomized, double-blind, placebo-controlled clinical trial. Front. Pharm. 11, 478 (2020).
Zhang, Y. & Zhang, Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol. Immunol. 17, 807–821 (2020).
Chen, J. et al. Target delivery of a PD-1-TREM2 scFv by CAR-T cells enhances anti-tumor efficacy in colorectal cancer. Mol. Cancer 22, 131 (2023).
Kamrani, A. et al. New immunotherapy approaches for colorectal cancer: focusing on CAR-T cell, BiTE, and oncolytic viruses. Cell Commun. Signal 22, 56 (2024).
Chen, N. et al. Chimeric antigen receptor T cells targeting CD19 and GCC in metastatic colorectal cancer: a nonrandomized clinical trial. JAMA Oncol. 10, 1532–1536 (2024).
Tay, R. E., Richardson, E. K. & Toh, H. C. Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 28, 5–17 (2021).
Speiser, D. E., Chijioke, O., Schaeuble, K. & Münz, C. CD4(+) T cells in cancer. Nat. Cancer 4, 317–329 (2023).
Shang, Q. et al. Polysaccharides regulate Th1/Th2 balance: a new strategy for tumor immunotherapy. Biomed. Pharmacother. 170, 115976 (2024).
Laidlaw, B. J., Craft, J. E. & Kaech, S. M. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat. Rev. Immunol. 16, 102–111 (2016).
Faubert, B., Solmonson, A. & DeBerardinis, R. J. Metabolic reprogramming and cancer progression. Science 368, eaaw5473 (2020).
Wilfahrt, D. & Delgoffe, G. M. Metabolic waypoints during T cell differentiation. Nat. Immunol. 25, 206–217 (2024).
Chong, M. et al. Novel drug targets for ischemic stroke identified through Mendelian randomization analysis of the blood proteome. Circulation 140, 819–830 (2019).
Zhang, Y. et al. Evaluating the causal effect of circulating proteome on the risk of osteoarthritis-related traits. Ann. Rheum. Dis. 82, 1606–1617 (2023).
Wootton, R. E. et al. Bidirectional effects between loneliness, smoking and alcohol use: evidence from a Mendelian randomization study. Addiction 116, 400–406 (2021).
Zheng, J. et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 52, 1122–1131 (2020).
Jiang, W. et al. Potiential therapeutic targets for sarcopenia identified by Mendelian randomisation. Age Ageing 52, afad024 (2023).
Mao, R., Li, J. & Xiao, W. Identification of prospective aging drug targets via Mendelian randomization analysis. Aging Cell 23, e14171 (2024).
Qi, C. et al. Cell-type eQTL deconvolution of bronchial epithelium through integration of single-cell and bulk RNA-seq. Allergy 77, 3663–3666 (2022).
Li, N. et al. Revealing genes associated with cervical cancer in distinct immune cells: a comprehensive Mendelian randomization analysis. Int. J. Cancer 155, 149–158 (2024).
Liu, J. et al. Uncovering immune cell-associated genes in breast cancer: based on summary data-based Mendelian randomized analysis and colocalization study. Breast Cancer Res. 26, 172 (2024).
Taylor-Weiner, A. et al. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 20, 228 (2019).
Soskic, B. et al. Immune disease risk variants regulate gene expression dynamics during CD4(+) T cell activation. Nat. Genet. 54, 817–826 (2022).
Fernandez-Rozadilla, C. et al. Deciphering colorectal cancer genetics through multi-omic analysis of 100,204 cases and 154,587 controls of European and east Asian ancestries. Nat. Genet. 55, 89–99 (2023).
Pálmer, H. G. et al. The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nat. Med. 10, 917–919 (2004).
Lambert, E. et al. Treatment of patients with newly diagnosed metastatic hormone sensitive prostate cancer (mHSPC) in Belgium: a real world data analysis. Acta Clin. Belg. 77, 897–905 (2022).
Shi, Y. et al. A novel mechanism of endoplasmic reticulum stress- and c-Myc-degradation-mediated therapeutic benefits of antineurokinin-1 receptor drugs in colorectal cancer. Adv. Sci. 8, e2101936 (2021).
Joo, S. H. & Chun, K. S. Therapeutic strategies for colorectal cancer: antitumor efficacy of dopamine D2 receptor antagonists. Toxicol. Res. 40, 533–540 (2024).
Bao, Y. et al. Autoimmune diseases and cardiovascular risk: Mendelian randomization analysis for the impact of 19 autoimmune diseases on 14 cardiovascular conditions. J. Transl. Autoimmun. 9, 100259 (2024).
Sanderson, E. et al. Mendelian randomization. Nat. Rev. Methods Primers 2, 6 (2022).
Zuber, V. et al. Combining evidence from Mendelian randomization and colocalization: review and comparison of approaches. Am. J. Hum. Genet. 109, 767–782 (2022).
Baird, D. A. et al. Identifying drug targets for neurological and psychiatric disease via genetics and the brain transcriptome. PLoS Genet. 17, e1009224 (2021).
Lei, T. et al. Genetic influence of the brain on muscle structure: a Mendelian randomization study of sarcopenia. J. Cachexia Sarcopenia Muscle 16, e13647 (2024).
Nounu, A. et al. A combined proteomics and Mendelian randomization approach to investigate the effects of aspirin-targeted proteins on colorectal cancer. Cancer Epidemiol. Biomark. Prev. 30, 564–575 (2021).
Sun, J. et al. Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome. Genome Med. 15, 75 (2023).
Chu, X. et al. Integrative single-cell analysis of human colorectal cancer reveals patient stratification with distinct immune evasion mechanisms. Nat. Cancer 5, 1409–1426 (2024).
Cheng, Y. K. et al. Association of peripheral blood biomarkers with response to anti-PD-1 immunotherapy for patients with deficient mismatch repair metastatic colorectal cancer: a multicenter cohort study. Front. Immunol. 13, 809971 (2022).
Montauti, E., Oh, D. Y. & Fong, L. CD4(+) T cells in antitumor immunity. Trends Cancer 10, 969–985 (2024).
Xu, J. et al. Single-cell sequencing reveals alterations in the peripheral blood mononuclear cell landscape and monocyte status during colorectal adenocarcinoma formation. Clin. Transl. Med. 14, e1609 (2024).
Carreras-Sureda, A. et al. ORMDL3 modulates store-operated calcium entry and lymphocyte activation. Hum. Mol. Genet. 22, 519–530 (2013).
Schmiedel, B. J. et al. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat. Commun. 7, 13426 (2016).
Schedel, M. et al. Polymorphisms related to ORMDL3 are associated with asthma susceptibility, alterations in transcriptional regulation of ORMDL3, and changes in TH2 cytokine levels. J. Allergy Clin. Immunol. 136, 893–903.e814 (2015).
Brown, R. D. R. et al. Overexpression of ORMDL3 confers sexual dimorphism in diet-induced non-alcoholic steatohepatitis. Mol. Metab. 79, 101851 (2024).
Wong, C. W. et al. PARP14 inhibition restores PD-1 immune checkpoint inhibitor response following IFNγ-driven acquired resistance in preclinical cancer models. Nat. Commun. 14, 5983 (2023).
Pan, Y. et al. IRIS: discovery of cancer immunotherapy targets arising from pre-mRNA alternative splicing. Proc. Natl. Acad. Sci. USA 120, e2221116120 (2023).
Dalhoff, K. et al. A phase II study of the vitamin D analogue Seocalcitol in patients with inoperable hepatocellular carcinoma. Br. J. Cancer 89, 252–257 (2003).
Yu, J. et al. Vitamin D receptor prevents tumour development by regulating the Wnt/β-catenin signalling pathway in human colorectal cancer. BMC Cancer 23, 336 (2023).
Jones, M. R. et al. Response to angiotensin blockade with irbesartan in a patient with metastatic colorectal cancer. Ann. Oncol. 27, 801–806 (2016).
Tharp, K. M. et al. Tumor-associated macrophages restrict CD8(+) T cell function through collagen deposition and metabolic reprogramming of the breast cancer microenvironment. Nat. Cancer 5, 1045–1062 (2024).
Macintyre, A. N. et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 20, 61–72 (2014).
Cui, W. et al. Gut microbial metabolite facilitates colorectal cancer development via ferroptosis inhibition. Nat. Cell Biol. 26, 124–137 (2024).
Seyed Khoei, N. et al. Circulating bilirubin levels and risk of colorectal cancer: serological and Mendelian randomization analyses. BMC Med. 18, 229 (2020).
Yoo, S. K. et al. Prediction of checkpoint inhibitor immunotherapy efficacy for cancer using routine blood tests and clinical data. Nat. Med. 31, 869–880 (2025).
Guo, Q. et al. NF-κB in biology and targeted therapy: new insights and translational implications. Signal Transduct. Target Ther. 9, 53 (2024).
Khan, A., Zhang, Y., Ma, N., Shi, J. & Hou, Y. NF-κB role on tumor proliferation, migration, invasion and immune escape. Cancer Gene Ther. 31, 1599–1610 (2024).
Schmiedel, B. J. et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715.e1716 (2018).
Võsa, U. et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 53, 1300–1310 (2021).
Perrot, N. et al. A trans-omic Mendelian randomization study of parental lifespan uncovers novel aging biology and therapeutic candidates for chronic diseases. Aging Cell 20, e13497 (2021).
Hemani, G., Tilling, K. & Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 13, e1007081 (2017).
Chen, Y. et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 55, 44–53 (2023).
Wu, X. et al. Transcriptome-wide Mendelian randomization during CD4(+) T cell activation reveals immune-related drug targets for cardiometabolic diseases. Nat. Commun. 15, 9302 (2024).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Chandrashekar, D. S. et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia 25, 18–27 (2022).
Chen, Y. et al. Spatiotemporal single-cell analysis decodes cellular dynamics underlying different responses to immunotherapy in colorectal cancer. Cancer Cell 42, 1268–1285.e1267 (2024).
Zhang, F. et al. Inhibiting PLA2G7 reverses the immunosuppressive function of intratumoral macrophages and augments immunotherapy response in hepatocellular carcinoma. J. Immunother. Cancer 12, e008094 (2024).
Musa, A. et al. A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 19, 506–523 (2018).
MacKinnon, D. P., Fairchild, A. J. & Fritz, M. S. Mediation analysis. Annu. Rev. Psychol. 58, 593–614 (2007).
Osorio, D. et al. scTenifoldKnk: an efficient virtual knockout tool for gene function predictions via single-cell gene regulatory network perturbation. Patterns 3, 100434 (2022).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Acknowledgements
We would like to thank all the participants from the dynamic immune research, CRC GWAS, FinnGen cohort, and GWAS catalog, as well as the institutions and their staff for providing the data.This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2024A1515013108), the National Natural Science Foundation of China (82573391, 82473172, 82203243, 82293680, 82293681, 82321004, 82430108, 81973341, 82103185), the Science and Technology Projects in Guangzhou (202102070001), the fundamental research funds for the central universities (21623109).
Author information
Authors and Affiliations
Contributions
Conceptualization: K.C. and Q.M.Z.; Formal analysis: K.C., Q.M.Z., X.Y.Q. and Y.Y.; Methodology: Q.M.Z., Y.Y., X.Y.Q. and J.X.S.; Investigation: Y.Y., M.S.L., Y.Y., L.L.L., Y.T.S., S.H.W., X.S.Z. and K.C.; Visualization: K.C., Q.Q. and X.F.Z.; Funding acquisition: Q.Q., W.C.Y. and B.F.Q.; Project administration: Q.Q. and W.C.Y.; Supervision: Q.Q., W.C.Y. and B.F.Q.; Writing—original draft: K.C., Q.M.Z. and Y.Y.; Writing—review and editing: Q.Q. and W.C.Y. All authors have approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cui, K., Zou, Q., Qu, X. et al. Transcriptome-wide Mendelian randomization and single-cell analysis during CD4+ T cell activation deciphers immunotherapeutic targets for colorectal cancer. npj Precis. Onc. 10, 32 (2026). https://doi.org/10.1038/s41698-025-01236-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-01236-6