Abstract
Aging is the predominant risk factor for atherosclerosis—the leading cause of death. Rare smooth muscle cell (SMC) progenitors clonally expand, giving rise to up to ~70% of atherosclerotic plaque cells; however, the effect of age on SMC clonality is not known. Our results indicate that aged bone marrow (BM)-derived cells noncell autonomously induce SMC polyclonality and worsen atherosclerosis. Indeed, in myeloid cells from aged mice and humans, TET2 levels are decreased, which epigenetically silences integrin β3, resulting in increased tumor necrosis factor-α (TNFα) signaling. TNFα signals through TNF receptor 1 on SMCs to promote proliferation, and induces the recruitment and expansion of multiple SMC progenitors into the atherosclerotic plaque. Notably, integrin β3 overexpression in aged BM preserves the dominance of the lineage of a single SMC progenitor and attenuates the plaque burden. Our results demonstrate a molecular mechanism of aged macrophage-induced SMC polyclonality and atherogenesis and suggest novel therapeutic strategies.
Similar content being viewed by others
Main
The revolution of single-cell analysis has unveiled heterogeneity within populations of specific cell types, and clonal expansion of individual cells is increasingly appreciated to underlie a number of diseases beyond cancer, including vascular pathologies, cirrhosis and neurodegeneration1,2,3,4,5,6,7. For instance, multiple smooth muscle cell (SMC) progenitors give rise to the normal arterial wall during development, but a select few progenitors within the wall participate in atherosclerotic plaque formation2,3,4,8. Additionally, in age-related clonal hematopoiesis of indeterminate potential (CHIP), stem cells carrying somatic mutations give rise to dominant leukocyte variants, and CHIP is associated with an increased risk of major atherosclerosis-related diseases, myocardial infarction and ischemic stroke9,10. Recent human studies have indicated a similar progressive accumulation of mutated somatic clones in epithelial cells of diverse organs with normal aging11,12,13,14. Investigations have primarily focused on cell-autonomous mechanisms underlying clonal expansion, and noncell autonomous regulation is not well understood, particularly in aging. Notably, the authors of a recent controversial bioinformatics study challenged the view that increasing somatic mutations with age explain the enhanced incidence of cancer later in life and instead argued that immune system decline is paramount15,16. Herein, we investigate the phenomenon of noncell autonomous regulation of clonality in aging by delineating the role of aged hematopoietic cells in modulating the recruitment and clonal expansion of SMC progenitors in the atherosclerotic plaque.
Inflammatory cells (especially macrophages) and SMCs are key players in atherogenesis. Rare SMC progenitors from the tunica media contribute cells that populate the nascent plaque and clonally expand, giving rise to ~30–70% of the cellularity of an advanced plaque, with cells forming the protective fibrous cap or transitioning into plaque-destabilizing fates2,3,4,17,18,19. Integrins are heterodimeric proteins that link the extracellular and intracellular compartments, and transplant of bone marrow (BM) null for the gene encoding integrin β3 (Itgb3) in atheroprone mice induces multiple SMC progenitors to enter the plaque and clonally expand, exacerbating the disease burden2,20. Importantly, BM transplantation (BMT) from aged mice into young mice worsens atherosclerosis21, but underlying mechanisms and effects on SMC recruitment and clonality are not delineated.
In CHIP, clonally expanded hematopoietic stem cells commonly harbor somatic mutations in epigenetic regulators, such as Tet2 (refs. 9, 10). On an Ldlr-null background, Tet2-deficient BM predisposes mice to develop enhanced Western diet (WD)-induced atherosclerosis10,22. Macrophages recruited to early atherosclerotic plaques proliferate locally during disease progression23, and in atheroprone mice, initiating WD in the aged compared with the young increases the accumulation of macrophages in the aorta21. Importantly, in the context of CHIP in humans or in aged or myeloid cell Tet2-deficient mice, the clonality of myeloid cells in the atherosclerotic plaque itself is elusive. Also, similar to aging, the effect of BM Tet2 deficiency on SMC clonality in the plaque is undefined.
In this Article, we report that the age of the BM is a key factor dictating the clonality of SMC-derived cells in the atherosclerotic plaque: aged BM noncell autonomously induces the recruitment and expansion of multiple SMC progenitors. Mechanistically, decreased ten eleven translocation 2 (TET2) levels in aged monocytes and macrophages epigenetically decrease Itgb3 gene expression, which enhances tumor necrosis factor-α (TNFα)–TNF receptor 1 (TNFR1) signaling. Thus, polyclonality of the SMC lineage and worse atherosclerosis ensue.
Results
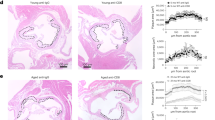
Age dictates the clonality of SMC- or myeloid-derived plaque cells
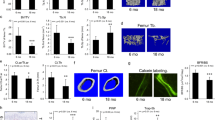
In hypercholesterolemic mice, rare smooth muscle myosin heavy chain-positive cells migrate into each atherosclerotic plaque and clonally expand2,3,4,24. However, the clonality of macrophages in plaques and how aging regulates SMC and macrophage clonality are not elucidated. Young (3-month-old) and aged (18-month-old) mice carrying the multicolor ROSA26R-Rainbow (Rb) Cre reporter and either Myh11-CreERT2 or Csf1r-Mer-iCre-Mer were induced with tamoxifen (1 mg d−1 for 5 or 20 d), injected with recombinant adeno-associated virus (AAV) encoding constitutively active PCSK9 (AAV-Pcsk9) and fed a WD for 16 weeks (Fig. 1a). (The Myh11 gene encodes smooth muscle myosin heavy chain, and colony-stimulating factor-1 receptor (CSF1R) is expressed in committed myeloid progenitors in the BM, as well as in monocytes and macrophages (including tissue-resident macrophages) and a subset of classical dendritic cells25,26). Transverse aortic root sections were stained with hematoxylin and eosin or Oil Red O or for nuclei (4′,6-diamidino-2-phenylindole (DAPI)) and directly imaged for Rb colors (cerulean, membrane cherry and membrane orange). Compared with young mice, plaques in aged mice were larger and had increased lipid content (Fig. 1b–e). Furthermore, the percentage of plaque cells that derived from SMCs (that is, marked by any Rb color) did not change with age, but there was a marked shift toward polyclonality (Fig. 1f–h). Indeed, for young mice, the most prevalent color (indicated by #1) in each plaque comprised 97 ± 1% of marked cells, while the second and third most prevalent colors (#2 and #3) made up the remaining ~3%. For aged mice, the marked cells were substantially more distributed among the Rb colors (50 ± 5, 28 ± 4 and 22 ± 6%, respectively, for colors 1, 2 and 3). Importantly, the distribution of marked cells of the tunica media underlying the plaques was comparable in young and aged mice (Fig. 1i). These results for SMC clonal analysis in the plaque and media of the aortic root were similar for those regions of the brachiocephalic artery (Extended Data Fig. 1a–d). In contrast with SMC tracing, in aortic root plaques of aged mice, CSF1R+ cells gave rise to an approximately threefold higher percentage of cells and were considerably more dominated by a single clone than in young mice (that is, 95 ± 3, 3 ± 1 and 1 ± 1% versus 63 ± 9, 25 ± 5 and 12 ± 3%; Fig. 1j–l). Thus, aging induces polyclonality of SMC-derived cells but dominance by a single clone of the monocyte/macrophage lineage.
Young (3-month-old) and aged (18-month-old) ROSA26RRb/+ mice also carrying either Myh11-CreERT2 or Csf1r-Mer-iCre-Mer were induced with tamoxifen, rested, injected with AAV-Pcsk9 and fed a WD for 16 weeks. Transverse aortic root sections were analyzed. a, Schematic of the experimental plan. d, days; w, weeks; m, months. b–e, For Myh11-CreERT2, ROSA26RRb/+ mice, sections were stained with hematoxylin and eosin (H&E; b; dashed lines demarcate plaque) or Oil Red O (ORO; c) and the areas of the plaque (d) and lipid (e) were quantified. n = 5 young mice and n = 7 aged mice. Triplicate measurements were made for each mouse. f–l, Sections from Myh11-CreER, ROSA26RRb/+ (f–i) and Csf1r-Mer-iCre-Mer, ROSA26RRb/+ mice (j–l) were stained for nuclei (DAPI) and directly imaged for Rb colors (membrane cherry (mCh), membrane orange (mOr) and cerulean (Cer)), then labeled cells were quantified. f,j, Representative sections, with boxed regions displayed as close-ups below. g,k, Quantification of the percentages of DAPI+ plaque cells that were marked by any of the Rb colors. h,l, Of the marked plaque cells, the percentage of cells of each color was quantified for each age group. In a given plaque, color 1 is the color with the greatest number of cells in the plaque (or media), color 2 is the second most common color and color 3 is least frequent color. n = 5 young mice (g–i,k,l) and n = 7 (g–i) or n = 5 (k,l) aged mice. A total of 18 plaques per age group were analyzed, with five sections, a total of ~1,200–1,600 cells and an area of 200 μm analyzed per plaque. In i, of the marked cells in the underlying media, the precentage of cells of each color was quantified. The media underlying 12 plaques per age group was analyzed. All of the data represent means ± s.d. Statistical significance was determined by two-tailed Student’s t-test (d,e,g,k). Scale bars, 100 μm (b,c) or 50 μm (f,j). Lu, lumen; Med, media; Pl, plaque.
BM age is a key determinant of SMC-derived cell clonality
In atheroprone mice, transplant of aged or Itgb3−/− BM worsens WD-induced atherosclerosis, and in the case of Itgb3−/− BMT results in the recruitment and expansion of multiple SMCs in the plaque2,20,21. We next queried whether aged BM is sufficient to induce polyclonality of SMC-derived plaque cells. Young Ldlr−/−, Myh11-CreERT2, ROSA26RRb/+ recipients were induced with tamoxifen and transplanted with BM from young and aged wild-type donors (Fig. 2a). At 4 weeks after BMT, engraftment was confirmed (Fig. 2b) and mice transplanted with aged BM had relative monocytosis and neutrophilia (Extended Data Fig. 1e). After confirming engraftment, recipients were fed a WD for 16 weeks. Aortic root plaques in mice that underwent aged BMT had fewer α-smooth muscle actin-positive (SMA+) cells in the cap and lower collagen content compared with plaques in mice that underwent young BMT (Extended Data Fig. 1f–i). The percentage of aortic root plaque cells derived from SMCs in recipients of aged BM was twice that in young BM transplant recipients (Fig. 2c,d). Furthermore, labeled plaque cells in recipients of young BM were dominated by a single Rb color (95 ± 2%), whereas in recipients of aged BM these cells were more equally distributed (37 ± 3, 34 ± 6 and 29 ± 3%; Fig. 2e). Of note, a distinct mouse model of hypercholesterolemia-induced atherosclerosis yielded similar results (Extended Data Fig. 1j–q). Importantly, the distribution of marked media cells underlying plaques was similar in young and aged BMT groups (Fig. 2f and Extended Data Fig. 1q). As a complementary approach, we transplanted aged Myh11-CreERT2, ROSA26RRb/+ mice with young or aged BM and found that with young BMT the atherosclerotic lesion burden was attenuated and SMC-derived cells made up a smaller percentage of the plaque cells and were dominated by a single clone (Extended Data Fig. 2).
a–f, Young Ldlr−/−, Myh11-CreERT2, ROSA26RRb/+ mice were induced with tamoxifen, irradiated and then transplanted with BM from young or aged mice and fed a WD for 16 weeks. a, Experimental schematic. b, Genomic DNA from the peripheral blood of recipients following BMT from young or aged wild-type (WT) mice was PCR amplified using Ldlr primers. Lanes 7 and 8 are controls from WT and Ldlr−/− mice, respectively. bp, base pairs; MW, molecular weight. c, Transverse aortic root sections were stained with DAPI and directly imaged for Rb colors (mCh, mOr and Cer). The boxed regions are shown as close-ups. d, Quantification of the percentages of DAPI+ plaque cells that were marked by any of the Rb colors. e,f, Of the marked plaque (e) or underlying medial cells (f), the percentage of cells of each color was quantified for each BMT group. n = 5 mice and 15 plaques per BMT group and five sections per plaque, totaling ~1,000–1,500 cells and spanning an area of 200 μm per plaque. g–j, The BM of young and aged WT mice was harvested and differentiated into macrophages. Macrophage-conditioned medium (CM) was added to murine aortic SMCs for migration and proliferation assays. g,h, For the migration assay, confluent SMCs with a central acellular area were cultured in CM for the indicated times. Brightfield images (g) and quantification of the percentage of SMC coverage at 6 and 8 h compared with the uncovered area at 0 h (h) are shown. Dashed lines demarcate uncovered area. n = 6. i,j, For the proliferation assay, SMCs were incubated with CM for 48 h and EdU was added for the last 8 h. SMCs were stained for EdU and nuclei (DAPI) (i), and the percentages of cells expressing EdU were quantified (j). n = 6 mice. All of the data represent means ± s.d. Statistical significance was determined by two-tailed Student’s t-test (d,h,j). Scale bars, 50 μm (c,g) or 25 μm (i).
Macrophages regulate SMC migration and proliferation2,27, which are integral for the recruitment and clonal expansion; thus, we evaluated whether aging influences this regulation. Monocytes were harvested from the BM of young and aged mice and differentiated into macrophages in culture, and aortic SMCs were isolated from young mice. Incubating SMCs with conditioned medium from aged as opposed to young macrophages for 6 or 8 h induced SMC migration without altering proliferation at 8 h (Fig. 2g,h and Extended Data Fig. 1r,s). However, culturing SMCs for 48 h in aged macrophage-conditioned medium enhanced proliferation by around threefold (Fig. 2i,j). Taking the findings from Figs. 1 and 2 and Extended Data Figs. 1 and 2 together, the age of the BM (presumably macrophages) is a decisive factor determining the recruitment and clonality of SMC-derived plaque cells.
Tet2 in BM cells regulates SMC-derived plaque cell clonality
Loss-of-function mutations in a few genes encoding epigenetic regulators, such as TET2, account for most CHIP cases9. Transplant of Tet2-deficient BM to Ldlr−/− mice exacerbates WD-induced atherosclerosis10,22; however, the effect of BM TET2 on SMC clonality is not delineated. Thus, young Ldlr−/−, Myh11-CreERT2, ROSA26RRb/+ recipient mice were induced with tamoxifen, transplanted with Tet2−/− or wild-type BM and fed a WD for 16 weeks (Fig. 3a,b). The percentage of plaque cells that were SMC derived was enhanced with Tet2−/− BMT (Fig. 3c,d). Moreover, labeled cells in plaques of the wild-type BMT group were dominated by a single Rb color (88 ± 3, 8 ± 3 and 4 ± 1%) in contrast with the more even distribution in the Tet2−/− BMT group (48 ± 3, 30 ± 4 and 22 ± 3%), without altering labeled cells in the underlying media (Fig. 3e,f). These effects of Tet2−/− BMT on SMC clonality were confirmed in an alternative model of hypercholesterolemia-induced atherosclerosis (Extended Data Fig. 3a–d). Additionally, similar to studies with aged macrophages, conditioned medium from Tet2−/− macrophages induced SMC migration and proliferation (Fig. 3g–j and Extended Data Fig. 3e,f). Thus, Tet2−/− BM-derived cells (probably macrophages) facilitate the recruitment and polyclonal expansion of multiple preexisting SMCs into the plaque.
a–f, Young Ldlr−/−, Myh11-CreERT2, ROSA26RRb/+ mice were induced with tamoxifen, irradiated and then transplanted with BM from WT or Tet2−/− mice and fed a WD for 16 weeks. a, Experimental schematic. b, Genomic DNA from the peripheral blood of recipient mice after either WT or Tet2−/− BMT was PCR amplified using Tet2 primers. Lanes 7 and 8 are controls from WT and Tet2−/− mice, respectively. c, Transverse aortic root sections were stained with DAPI and imaged for Rb colors. The boxed regions are shown as close-ups. d, Quantification of the percentages of DAPI+ plaque cells that were marked by any Rb color. e,f, Of the marked plaque (e) or underlying medial cells (f), the percentage of cells of each color was quantified for each BMT group. n = 5 mice, 12 plaques per group and five sections per plaque, totaling ~1,000–1,500 cells and spanning an area of 200 μm per plaque. g–j, The BM from Tet2−/− and WT mice was harvested and differentiated into macrophages. Macrophage-conditioned medium (CM) was added to murine aortic SMCs for migration and proliferation assays. g,h, For the migration assay, confluent SMCs with a central area lacking cells were cultured in CM for the indicated times. Brightfield images (g) and quantification of the percentage of SMC coverage at 6 and 8 h compared with the uncovered area at 0 h (h) are shown. Dashed lines demarcate uncovered area. n = 6 mice. i,j, For the proliferation assay, murine aortic SMCs were incubated with CM for 48 h, and EdU was added for the last 8 h. SMCs were stained for EdU and nuclei (DAPI) (i), and the percentages of cells expressing EdU were quantified (j). n = 6. All of the data represent means ± s.d. Statistical significance was determined by two-tailed Student’s t-test (d,h,j). Scale bars, 50 μm (c,g) or 25 μm (i).
Reduced monocyte TET2 links aging and decreased integrin β3
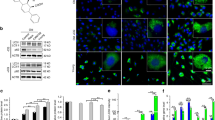
To begin to evaluate how aged BM cells exacerbate atherosclerosis and induce SMC recruitment and expansion in the plaque, we investigated the effects of age on the murine and human monocyte transcriptome. CD3−CD19−Cd11b+Ly6C+ monocytes were isolated from the BM of young and aged mice by flow-activated cell sorting (FACS) (Extended Data Fig. 4a) and subjected to bulk RNA sequencing (RNA-seq) (Supplementary Dataset 1) and pathway analysis. Gene set enrichment analysis indicated that inflammatory and immune pathways were upregulated in aged monocytes (Extended Data Fig. 4b). Ingenuity pathway analysis showed that atherosclerosis is one of the top overrepresented and activated diseases/functions (P = 3.74 × 10−8; z score = 0.65). In addition, 32 differentially expressed gene products significantly overlap with atherosclerosis in the Ingenuity Knowledge Base (Benjamini–Hochberg false discovery rate; P = 1.28 × 10−6; Fig. 4a). Notably, Itgb3 was downregulated in aged monocytes (Fig. 4a) and previous studies have shown that in atheroprone mice Itgb3−/− BMT exacerbates atherosclerosis and induces polyclonal expansion of SMC progenitors in the plaque2,20. However, a link between integrin β3 deficiency in monocytes/macrophages and aging has not previously been established.
a,c–h,j,k, The BM of young and aged mice (a,c–h) or young WT or Tet2−/− mice (j,k) was subjected to FACS to isolate CD3−CD19−Cd11b+Ly6C+ monocytes or differentiated into macrophages. b,i, Peripheral blood CD3−CD19−CD14+ monocytes were isolated from healthy young (25 ± 3-year-old) or older (55 ± 7-year-old) humans. a, Monocyte lysates underwent bulk RNA-seq (n = 3 mice) and pathway analysis. Differentially expressed genes (for example, ITGB3; circled) significantly overlap with atherosclerosis in the Ingenuity Knowledge Base (Benjamini–Hochberg false discovery rate; P = 1.28 × 10−6). Darker and lighter green and red symbols refer to results that were more or less extreme in the dataset, respectively. Darker and lighter orange and blue symbols refer to results with more or less confidence, respectively. b,c, Human (b) and murine (c) monocyte ITGB3 RNA levels were measured by quantitative reverse transcription PCR (qRT-PCR). n = 12 humans per group (b) and n = 5 mice per group (c). d,e, Western blot analysis of integrin β3 and CSF1R from lysates of differentiated macrophages from mice (d) and quantification of the densitometry of these proteins relative to GAPDH and normalized to young mice (e). f,g, Murine monocyte lysates were subjected to Western blot analysis for TET2 and GAPDH (f) and the results of densitometry of TET2 relative to GAPDH were quantified (g). h–j, hMeDIP was performed with antibodies against 5hmC or IgG, followed by qRT-PCR with primers targeting upstream of the ITGB3 transcription start site (chr11:104,608,100–104,608,250 in mice (h and j) and chr17:47,240,300–47,240,400 in humans (i)). *P = 6.2 × 10−8 versus 5hmC, young (h), *P = 4.7 × 10−7 versus 5hmC, young (i) and *P = 3.5 × 10−7 versus WT (j). k, Murine monocyte Itgb3 mRNA levels from WT versus Tet2−/− mice (n = 5 mice). l–n, Apoe−/−, Csf1r-Mer-iCre-Mer, ROSA26RRb/+ mice also carrying Itgb3flox/flox or Itgb3+/+ were induced with tamoxifen, rested and fed a WD for 6 or 9 weeks. l, Transverse aortic root sections were imaged for mCh, mOr and Cer after 9 weeks of WD. Close-ups of the boxed regions are shown to the right. m, Percentage of DAPI+ plaque cells marked by any Rb color. n, Of the marked plaque cells, the percentages of cells of each color per WD duration and genotype group are shown. n = 5 mice and 14 plaques per group and five sections per plaque, with a total of ~1,200–1,400 cells and an area spanning 200 μm per plaque. All of the data represent means ± s.d. Statistical significance was determined by two-tailed Student’s t-test (b,c,e,g,k,m) or multifactor ANOVA with Tukey’s post-hoc test (h–j). Scale bar, 50 μm (l). Ab, antibody.
We further explored this potential link by directly evaluating specific gene product levels in monocytes and macrophages from mice and humans. BM was harvested from young and aged mice and we either isolated monocytes by FACS or differentiated them into macrophages in culture. Additionally, FACS was used to isolate peripheral blood CD3−CD19−CD14+ monocytes from healthy young and older humans (aged 25 ± 3 and 55 ± 7 years, respectively; Supplementary Table 1 and Extended Data Fig. 4c). Quantitative reverse transcription PCR of monocyte lysates showed substantially decreased ITGB3 messenger RNA (mRNA) levels in aged humans and mice (Fig. 4b,c). Furthermore, compared with young macrophages, aged macrophages showed a 75 ± 19% decrease in integrin β3 protein levels but similar levels of CSF1R (Fig. 4d,e).
We then queried whether Itgb3 levels are modulated by TET2, which catalyzes the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine (5hmC)—a key step during DNA demethylation, and thus gene expression. Monocytes from aged mice had decreased TET2 protein levels, and 5hmC levels in proximity to the ITGB3 transcription start site in aged human and murine monocytes were also decreased (Fig. 4f–i). Similarly, monocytes isolated from Tet2−/− mice had reduced 5hmC levels at the Itgb3 proximal promoter and decreased Itgb3 transcript levels (Fig. 4j,k and Extended Data Fig. 4d). Hence, the data suggest that reduced TET2 in aged monocytes leads to decreased ITGB3 levels by attenuating transcription.
Itgb3-deficient CSF1R+ cells are atherogenic and clonally expand
Although transplant of atheroprone mice with Itgb3−/− BM worsens atherosclerosis2,20, the in vivo role of integrin β3 in myeloid cells has not been elucidated. Apoe−/−, Csf1r-Mer-iCre-Mer mice carrying Itgb3flox/flox or Itgb3+/+ were induced with tamoxifen and fed a WD for 16 weeks. Deletion of Itgb3 in CSF1R+ cells exacerbated the aortic root plaque burden, lipid accumulation and acellular necrotic core area (Extended Data Fig. 5a–f).
Next, we studied mice of the same genotypes and additionally with ROSA26RmTmG/+ or ROSA26RRb/+. At 5 d after tamoxifen induction of Apoe−/−, Csf1r-Mer-iCre-Mer, ROSA26RmTmG/+ mice, that were also Itgb3flox/flox or Itgb3+/+, cells expressing green fluorescent protein (GFP) and the monocyte marker Ly6C were isolated from BM by FACS. Peripheral blood was collected as well. Quantitative reverse transcription PCR analysis of Ly6C+GFP+ BM cells showed a 94 ± 4% decrease in Itgb3 levels in mice carrying Itgb3flox/flox (Extended Data Fig. 5g). Deletion of Itgb3 in CSF1R+ cells did not alter circulating levels of total cholesterol, triglycerides or leukocyte subtypes (Extended Data Fig. 5h–j). To evaluate the fate and proliferation of the lineage derived from CSF1R+ cells, following tamoxifen induction, mice of these same genotypes were fed a WD for 16 weeks and then injected with 5-ethynyl-2′ deoxyuridine (EdU) intraperitoneally 12 h before euthanasia. Csf1r-Mer-iCre-Mer-mediated deletion of Itgb3 leads to proliferation and accumulation of cells of this lineage in the plaque as well as an increased percentage of total plaque cells that express the macrophage marker CD68 (Extended Data Fig. 5k–o). Given that CSF1R+ cells clonally expand in the plaques of aged mice (see Fig. 1j–l) and given the link between aging and monocyte/macrophage Itgb3 deficiency (see Fig. 4a–i), we next queried whether CSF1R+ cells deficient in Itgb3 clonally expand. Apoe−/−, Csf1r-Mer-iCre-Mer, ROSA26RRb/+ mice also harboring Itgb3flox/flox or Itgb3+/+ were induced with tamoxifen and then fed a WD for 6 or 9 weeks. For aortic root plaques in mice carrying Itgb3flox/flox, a higher percentage of cells derived from the CSF1R+ lineage and a single Rb color gave rise to ~90% of marked cells at each WD duration, whereas in mice wild type for Itgb3, there was a more equitable distribution of the Rb colors (Fig. 4l–n). Taken together, integrin β3 deficiency in monocytes/macrophages leads to clonal expansion of this population in the atherosclerotic plaque and more severe disease.
Itgb3-deficient SMCs contribute less to the plaque
In addition to myeloid cells, SMCs express high levels of integrin β3, and on an atheroprone background, global Itgb3 nulls have worse atherosclerosis2; however, in this context, the cell-autonomous role of integrin β3 in SMC-derived cells is not defined. Thus, we used Myh11-CreERT2 to delete Itgb3 in SMCs, which attenuates atherosclerosis severity and proliferation and the accumulation of SMC-derived cells without altering the clonality of this lineage in the plaque (Extended Data Fig. 6). To evaluate mechanisms that may link integrin β3 deficiency in SMCs to diminished atherogenesis, we studied signaling pathways implicated in inducing SMC proliferation and migration28,29,30,31. Western blots indicated a 72 ± 7% decrease in phosphorylated AKT in aortic homogenates from Itgb3−/− mice (Extended Data Fig. 7a,b). Alternatively, small interfering RNA targeting of Itgb3 in isolated wild-type aortic SMCs substantially reduced protein levels of phosphorylated AKT, extracellular regulated kinase and p21 activated kinase-1 (Extended Data Fig. 7c,d). Moreover, Itgb3 silencing abrogated platelet-derived growth factor-B-induced dorsal ruffle formation (Extended Data Fig. 7e,f), an early step in cell migration32. In summary, integrin β3-deficient SMCs have decreased proliferation and migration, which probably accounts for their decreased contribution to the atherosclerotic plaque.
Itgb3 −/− monocytes and macrophages are proinflammatory
In contrast with SMCs, integrin β3 deficiency in macrophages noncell autonomously enhances SMC migration and proliferation, and transplant of Itgb3−/− BM induces SMC polyclonal expansion in the atherosclerotic plaque2 (see Extended Data Figs. 5–7). To further interrogate the role of integrin β3 in monocytes and macrophages, we performed single-cell RNA-seq (scRNA-seq) in BM cells from Apoe−/− mice that were also wild type or null for Itgb3. Unsupervised clustering was used to generate t-distributed stochastic neighbor embedding (t-SNE) plots, separating the cells into four major cell type groups: neutrophils, monocytes/macrophages, B cells and T cells (Fig. 5a,b and Extended Data Fig. 8a). We evaluated the expression pattern of classical markers of monocytes/macrophages (Extended Data Fig. 8b–h), and gene set enrichment analysis of these cells on an Apoe−/− background demonstrated that Itgb3 deletion leads to activation of inflammation, including cytokine signaling pathways (for example, nuclear factor kappa B (NF-kB) and TNF; Fig. 5c–e)20. In addition, ingenuity pathway analysis predicted that Itgb3 deletion induces the inflammatory response and monocyte/macrophage migration and proliferation (Extended Data Fig. 8i). Moreover, subcluster analysis of monocytes/macrophages showed diverse subpopulations in both genotypes, with an increased abundance of transcriptionally distinct M1- and M2-like macrophage populations in the Itgb3-null genotype (Fig. 5f,g and Extended Data Fig. 8j). Furthermore, TNF and nuclear factor kappa B signaling pathways were enriched in Itgb3-null subpopulations, most dramatically in M1-like macrophages, compared with the Itgb3+/+ control (Fig. 5h,i and Extended Data Fig. 8k,l).
scRNA-seq was conducted on BM cells isolated from Apoe−/− mice that were also either wild type or null for Itgb3. a, Overlayed t-SNE plots of scRNA-seq of BM cells from mice carrying Itgb3+/+ (blue) or Itgb3−/− (red). b, t-SNE plot combining the cells of both genotypes, showing distinct clusters that were identified as major cell types based on the expression levels of cluster markers (Supplementary Table 3). c–e, Pathway and gene expression analysis of single-cell transcriptomes of the monocyte/macrophage cluster. c, Activated pathways identified by gene set enrichment analysis. Gene ratio indicates the number of activated genes relative to the total number of genes in the corresponding pathway. The dot size represents the number of dysregulated genes in each pathway. The gene set enrichment analysis algorithm, as implemented in the clusterProfiler R package, was used to perform a one-sided test to determine the significance level of pathway enrichment. IL-17, interleukin-17; Padj, adjusted P value. d,e, Dot plot analyses of differentially expressed genes in the TNFα (d) and nuclear factor kappa B (NF-kB) signaling pathways (e). Dot size and the depth of blue shading represent the percentage of cells expressing the gene and the average gene expression, respectively. f,g, t-SNE plots showing the subclustering of monocytes and macrophages from mice carrying Itgb3+/+ or Itgb3−/−. In f, cells are colored by genotype. In g, both genotypes are combined and distinct subclusters of monocyte and macrophage subsets is shown (Supplementary Table 4). h,i, Enrichment scores, defined by differentially expressed genes in TNFα (h) and NF-kB signaling pathways (i), were generated using the Add Module Score function in the Seurat package. n = 3 mice pooled per genotype.
TNFα regulates the clonality of SMC-derived plaque cells
Treatment with an anti-TNFα antibody prolongs the survival of WD-fed Apoe−/−, Itgb3−/− mice20; however, the effects of TNFα inhibition on atherosclerotic burden are controversial33,34 and the effects on SMC clonality are not defined. To this end, Apoe−/−, Csf1r-Mer-iCre-Mer, Itgb3flox/flox mice were induced with tamoxifen and injected intraperitoneally with an anti-TNFα neutralizing antibody or IgG2a isotype control twice per week during 12 weeks of WD feeding (Fig. 6a). TNFα inhibition attenuated plaque area and lipid burden in the aortic root (Fig. 6b–e). For clonal analysis, Apoe−/−, Myh11-CreERT2, ROSA26RRb/+ mice were induced with tamoxifen, transplanted with Apoe−/− BM that was also wild type or null for Itgb3 and started on a 12-week regimen of WD with concomitant twice-weekly injections of anti-TNFα antibody or control IgG2a (Fig. 6f–h). In both the Itgb3 wild-type and Itgb3−/− BMT groups, inhibition of TNFα decreased SMC-derived plaque cells by >50% (Fig. 6i–k). Furthermore, in mice transplanted with the Itgb3−/− BM, anti-TNFα treatment induced the predominance of a single SMC-derived clone (87 ± 6, 10 ± 3 and 3 ± 2%) compared with the more evenly distributed polyclonality with the IgG2a control (40 ± 4, 33 ± 2 and 26 ± 3%) (Fig. 6l). Anti-TNFα treatment did not alter the clonal distribution of SMC-derived cells in either plaques of the Itgb3+/+ BMT group or the underlying media of any BMT group (Fig. 6l,m).
Mice were treated with 12 weeks of WD with concomitant injections of isotype control IgG2a or anti-TNFα antibody (20 mg kg−1) twice per week, then transverse aortic root sections were analyzed. a–e, Before these treatments, Apoe−/−, Csf1r-Mer-iCre-Mer mice also carrying Itgb3flox/flox or wild type for Itgb3 were induced with tamoxifen. a, Experimental schematic. tx, treatment. b,c, Sections were stained with H&E (b) or ORO (c). In b, the dashed lines demarcate lesions. d,e, Lesion area (d) and lipid content (e) were quantified. n = 6 mice. f–m, Before receiving the WD and treatment with control or anti-TNFα antibody, Apoe−/−, Myh11-CreERT2, ROSA26RRb/+ mice were induced with tamoxifen, irradiated and then transplanted with Apoe−/− BM also carrying Itgb3+/+ or Itgb3−/−, as indicated. f,g, Schematics showing the types of BMT received (f) and experimental timeline (g). h, Genomic DNA, prepared from the peripheral blood of recipient mice after BMT or of WT or Itgb3−/− mice, was amplified using primers for Itgb3. i,j, Sections from mice transplanted with BM carrying Itgb3+/+ (i) or Itgb3−/− (j) were stained with DAPI and directly imaged for Rb colors. Close-ups of the boxed regions are shown below. k, The percentages of DAPI+ plaque cells that were marked by any Rb color were quantified. l,m, Of the marked plaque (l) or underlying medial cells (m), the percentages of cells of each color were quantified for each BMT and treatment group. n = 5 mice and 12 plaques per group and 5–6 sections per plaque, with a total of ~1,150–1,650 cells and an area spanning 200 μm per plaque. All of the data represent means ± s.d. Statistical significance was determined by two-tailed Student’s t-test (d,e,k). Scale bars, 200 μm (b,c) or 50 μm (i,j).
As aged myeloid cells have decreased integrin β3 (see Fig. 4a–e) and increased Tnfa transcript levels (Fig. 7a), we similarly assessed the role of TNFα in regulating the SMC clonality of the atherosclerotic plaque following aged BMT. Importantly, aged BMT increases plasma TNFα levels by ~4.5-fold compared with young BMT (Fig. 7b and Extended Data Fig. 9a). Young Ldlr−/−, Myh11-CreERT2, ROSA26RRb/+ mice were induced with tamoxifen, transplanted with BM from aged wild-type mice and then fed a WD for 12 weeks with concomitant twice-weekly anti-TNFα antibody or control IgG2a injections (Fig. 7c,d). Following aged BMT, TNFα inhibition attenuated plaque burden and lipid deposition and increased collagen content and fibrous cap SMA+ cells (Fig. 7e–j and Extended Data Fig. 9b,c). Moreover, in this context, anti-TNFα treatment decreased the contribution of SMC-derived cells to the plaque and promoted the expansion of a single SMC-derived clone compared with IgG2a treatment, without altering clonality in the media (Fig. 7k–n). In addition, incubating aortic SMCs from a young mouse with anti-TNFα antibody-treated conditioned medium from aged macrophages attenuated SMC migration at 8 h and proliferation at 48 h (but not 8 h) compared with exposure to IgG2a-treated conditioned medium (Fig. 7o–q,s and Extended Data Fig. 9d,e). Similarly, RNA silencing of Tnfr1 in isolated murine aortic SMCs attenuated aged macrophage-conditioned medium-induced SMC proliferation (Fig. 7r,s and Extended Data Fig. 9f,g). Additionally, the TNFα–TNFR1 axis induced SMC phenotypic switching: in the presence of soluble cholesterol, TNFα treatment of murine aortic SMCs decreased the expression of the SMC markers Myh11 and Acta2 and induced the expression of the pioneer or intermediate cell marker Lgals3 (refs. 35, 36) and the monocyte/macrophage marker Cd68 (Extended Data Fig. 9h–k). These changes in gene expression were inhibited by pretreatment of SMCs with small interfering RNA targeting Tnfr1 (Extended Data Fig. 9h–k).
a, Tnfa and interleukin-1b (IL-1b) levels of young and aged BM monocytes, as determined by qRT-PCR. n = 5 mice. b, At 4 weeks after young Ldlr−/− mice received young or aged BMT, plasma TNFα levels were quantified. n = 5 mice. c–n, Young Ldlr−/−, Myh11-CreERT2, ROSA26RRb/+ recipients underwent aged WT BMT and then 12 weeks of WD with concomitant anti-TNFα blocking antibody or isotype control IgG2a injections. c,d, Schematics showing the type of BMT received (c) and the experimental timeline (d). e–j, Aortic sections were stained with H&E (e), ORO (f) and Sirius red (g) and lesion area (h), lipid content (i) and collagen content (j) were quantified. n = 5 mice and 8–10 plaques per group, with triplicate measurements taken for each plaque. k,l, Aortic sections were imaged for Rb colors (k) and the percentages of DAPI+ plaque cells marked by any Rb color were quantified (l). m,n, Of marked plaque (m) or underlying medial cells (n), the percentages of cells of each color were quantified. n = 5 mice and ten plaques per group and 5–6 sections per plaque, with a total of ~1,245–1,400 cells and an area spanning 200 μm per plaque. o–s, Young or aged WT BM was differentiated into macrophages. Macrophage-CM was collected and incubated with anti-TNFα antibody or IgG2a for 1 h, and then mouse aortic SMCs were cultured with this CM. o,p, Confluent SMCs with a central acellular area were cultured in CM for 0 or 8 h, followed by brightfield imaging (o) and quantification of the percentage of SMC coverage at 8 h versus the uncovered area at 0 h (p). q–s, SMCs were cultured with young or aged CM after 1 h pre-incubation with anti-TNFα antibody or IgG2a (q) or SMCs were treated with Tnfr1 small interfering RNA (si-Tnfr1) or scrambled RNA (Scr) before young or aged CM exposure (r). SMCs were then cultured with CM for 48 h, and EdU was included for the last 8 h. q,r, SMCs were stained for EdU and DAPI. s, Quantification of the percentages of EdU+ cells. n = 6 mice. *P = 9.3 × 10−6 for IgG and *P = 5.1 x 10−6 for Scr, both versus young. All of the data represent means ± s.d. Statistical significance was determined by two-tailed Student’s t-test (a,b,h–j,l,p) or multifactor ANOVA with Tukey’s post-hoc test (s). Scale bars, 100 µm (e–g), 50 μm (k,o) or 25 μm (q,r).
Finally, given the potential central role of decreased monocyte/macrophage Itgb3 in age-induced increased TNFα and thus, polyclonal SMC progenitor expansion in the plaque, we evaluated the effect of overexpressing Itgb3 in BM cells in aged mice. To this end, BM cells harvested from aged wild-type mice were infected with Itgb3-expressing or control lentivirus and transplanted into aged Myh11-CreERT2, ROSA26RRb/+ mice (Fig. 8a–c). These mice were then induced with AAV-Pcsk9 and fed a WD (Fig. 8b). Itgb3 overexpression in BM-derived monocytes markedly decreased Tnfa mRNA and plasma TNFα but not lipid levels at 4 weeks after BMT (Fig. 8d–f and Extended Data Fig. 9l,m). Furthermore, enhanced Itgb3 expression in aged BM cells decreased plaque burden, increased plaque collagen content and led to the dominance of a single SMC-derived clone in the plaque compared with the control (Fig. 8g–p). Taken together, these studies implicate in aged macrophages an inductive effect of integrin β3 deficiency on TNFα, which signals to SMC progenitors, probably through TNFR1, to induce polyclonal expansion and worse atherosclerosis (Extended Data Fig. 10).
BM cells harvested from aged WT donor mice were infected with empty vector (control) or Itgb3-overexpressing (Itgb3 OE) lentivirus. After 1 d of infection, aged Myh11-CreERT2, ROSA26RRb/+ recipient mice were induced with tamoxifen, irradiated and then transplanted with control and Itgb3 OE lentivirus-treated BM cells, followed by injection with AAV-Pcsk9 and WD feeding for 12 weeks. a,b, Schematics showing the types of BMT received (a) and the experimental timeline (b). c, Genomic DNA, prepared from the peripheral blood of recipient mice before (lanes 1–3) or after Itgb3 OE BMT (lanes 4–6) or of WT (lane 7) or ROSA26RRb/+ mice (lane 8), was PCR amplified with primers for ROSA26RRb/+ alleles. d–f, At 4 weeks after BMT, Itgb3 (d) and Tnfa transcript levels (e) were measured by qRT-PCR in GFP−Ly6C+ BM-derived monocytes, and plasma TNFα levels were measured by enzyme-linked immunosorbent assay (f). g–l, At 4 weeks after BMT, mice were injected with AAV-Pcsk9 and then fed a WD for 12 weeks. Transverse aortic root sections were analyzed by staining with H&E (g), ORO (h) or Sirius red (i) and the lesion area (j), lipid content (k) or collagen content (l) were quantified. n = 6 mice and 10–12 plaques per BMT group, with triplicate measurements taken for each plaque. In g, the dashed lines demarcate lesions. m, Sections were stained with DAPI and directly imaged for Rb colors. The boxed regions are displayed as close-ups below. n, The percentages of DAPI+ plaque cells marked by any Rb color were quantified. o,p, Of the marked plaque (o) or underlying medial cells (p), the percentages of cells of each color were quantified for the control and Itgb3 OE groups. n = 6 mice and 12 plaques per group and 5–6 sections per plaque, with a total of ~1,275–1,350 cells and an area spanning 200 μm per plaque. All of the data represent means ± s.d. Statistical significance was determined by two-tailed Student’s t-test (d–f,j–l,n). Scale bars, 100 µm (g–i) or 50 μm (m).
Discussion
The main finding of this work is that the age of the BM noncell autonomously dictates the clonality of SMCs in the atherosclerotic plaque, regardless of the age of SMCs or other nonhematopoietic cells (Figs. 1 and 2 and Extended Data Figs. 1 and 2). Up to two-thirds of the cellularity of an advanced atherosclerotic plaque in a young mouse derives from clonal expansion of one or two SMC progenitors2,3,4,17,18,24. Herein, we find that in plaques of aged mice SMC expansion is instead polyclonal as a result of aged BM-derived cells—probably macrophages (Figs. 1 and 2). Similar to atherogenesis, neointima formation in ligated carotid arteries of young mice is characterized by oligoclonal expansion of rare SMCs3, and interestingly, SMC senescence does not alter clonality in this model37. The effect of SMC senescence on clonality in atherosclerosis has not been reported.
With regard to the clonality of leukocytes, pivotal incompletely understood CHIP-related questions include: (1) Do leukocyte clones accumulate in atherosclerotic plaques?; (2) Mutations in epigenetic modulators induce expression changes in which downstream genes?; and (3) How do these mutations exacerbate atherosclerosis? Our results show that TET2 and integrin β3 are decreased in monocytes/macrophages of aged mice and/or humans, and aged or Itgb3-deficient monocytes/macrophages clonally expand in plaques (Figs. 1 and 4). Tet2-null BM cells noncell autonomously induce polyclonal expansion of SMCs in the plaque, and conditioned medium from Tet2−/− macrophages induces SMC migration and proliferation (Fig. 3). A regulatory role for ITGB3 promoter methylation has not been reported previously, but herein we find decreased 5hmC on the ITGB3 promoter with aging or Tet2 deletion (Fig. 4). These effects of aging are cell type specific as ITGB3 levels are in fact increased in aged murine liver or human fibroblasts38.
In humans, a polymorphism of the ITGB3 gene (T1565C) is associated with coronary artery disease39. Mice with global Itgb3 deletion or with Itgb3-null BM have polyclonal SMC expansion in the atherosclerotic plaque and worse disease2,20,40. Deletion of Itgb3 in monocytes and macrophages mimics the results with Itgb3−/− BMT in terms of worsening the atherosclerotic burden (Extended Data Fig. 5). In contrast, SMC deletion of Itgb3 attenuates disease and SMC contribution to plaques without altering clonality (Extended Data Fig. 6). Interestingly, aged arteries have increased levels of the anti-efferocytic protein CD47, which was initially co-purified with integrin β3 and regulates ligand binding of integrin αvβ3 (refs. 41,42,43). Anti-CD47 treatment attenuates atherosclerosis and induces a more even distribution of colors of Rb-marked SMC-derived cells (that is, polyclonal expansion)24. Future studies of cell type-specific roles of CD47, its interplay with integrin β3 and downstream signaling in the context of atherosclerosis and aging are warranted.
The most significantly upregulated functional class of genes in Tet2-null BM-derived macrophages consists of cytokines, chemokines and their receptors10, and herein, scRNA-seq of Itgb3-null BM-derived monocytes/macrophages showed enhanced chemokine and cytokine signaling, and in particular TNF signaling (Fig. 5). Plasma monocyte counts and TNFα levels increase with aging21,44 and are thus associated with polyclonality of SMC-derived plaque cells (Fig. 1). Importantly, monocytosis and the TNFα pathway have been implicated as protagonists in atherosclerosis and myocardial infarction45,46,47. Two single-nucleotide polymorphisms of the TNFR1 gene are associated with coronary artery disease in aged adults48. Furthermore, carotid arteries from aged and young Tnfr1−/− mice were grafted into Apoe−/− mice and, most interestingly, arteries from aged Tnfr1−/− mice are protected from age-induced atherosclerosis48. We now report that pharmacological inhibition of TNFα attenuates atherosclerosis, limiting the recruitment and clonal expansion of SMCs with Itgb3−/− or aged BM, and that Tnfr1 knockdown in SMCs or TNFα inhibition decreases SMC proliferation and/or migration induced by aged macrophage-conditioned medium (Figs. 6 and 7). Of note, rheumatoid arthritis is associated with increased atherosclerosis-related disease, especially among the aged49, and we suggest that some of the cardiovascular benefit of anti-TNFα drugs in this context50,51 may result from limiting the clonal expansion of SMC progenitors. In contrast, Itgb3 overexpression in the BM of aged mice via BMT decreases plasma TNFα and thus prevents the clonal expansion of multiple SMC-derived progenitors in the plaque and attenuates plaque burden (Fig. 8). It is intriguing to consider this result in light of a recent study showing the efficacy of autologous transplantation of genetically modified hematopoietic stem and progenitor cells as a treatment for patients with sickle cell disease52.
Aging is the predominant risk factor for atherosclerosis. We show that aged macrophages express reduced levels of TET2, inhibiting Itgb3 expression, and that decreased integrin β3 in macrophages enhances TNFα levels, which induces polyclonal expansion of SMCs in the atherosclerotic plaque and worsens disease. Thus, our studies put forth deficient regulation of SMC clonal expansion by aged BM-derived macrophages as a critical underlying factor in atherogenesis. Looking forward, this concept of impaired regulation of clonal expansion by aged BM (perhaps via myeloid cell Itgb3 deficiency) warrants intense investigation in other contexts, such as normal aging of the esophagus, endometrium, skin and bronchi, as well as in cirrhosis and neurodegeneration5,6,11,12,13,14.
Methods
Animals
All of the mouse experiments were approved by the Institutional Animal Care and Use Committee (protocol 11381) at Yale University and performed in accordance with the relevant ethical guidelines. Wild-type (C57BL/6), Ldlr−/−, Apoe−/−, Csf1r-Mer-iCre-Mer, ROSA26RmTmG/mTmG, Itgb3−/−, Itgb3flox/flox and Tet2−/− mice were purchased from The Jackson Laboratory53,54,55,56,57. Mice carrying Myh11-CreERT2 or the multicolor Rb Cre reporter ROSA26RRb have been described8,58,59. Experiments with Myh11-CreERT2 mice were restricted to males as this transgene is incorporated on the Y chromosome59. Otherwise, studies utilized both male and female adult mice. Young (3-month-old) or aged (18-month-old) mice were used for most of the experiments. Mice were housed in rooms with 12 h light/12 h dark cycles, ambient temperature of 21-22°C and 38–50% humidity.
Induction of atherogenesis and administration of AAV-Pcsk9
For the atherosclerosis studies, we used mice null for Ldlr or Apoe and/or injected with recombinant AAV encoding constitutively active PCSK9 (rAAV8.ApoEHCR-hAAT.D377Y-mPCSK9.bGH (AAV-Pcsk9); Penn Vector Core, University of Pennsylvania). For the AAV-Pcsk9 studies, 1 week after either tamoxifen treatment or, if relevant, 4 weeks after BMT, a single retro-orbital injection containing 1.0 × 1011 genome copies was administered. After 4 weeks, the mice were fed a WD (40% fat by calories and 1.25% cholesterol by weight; D12108C; Research Diets) for up to 16 weeks. For mice not injected with AAV-Pcsk9, a WD was initiated 4 weeks after BMT.
BMT
At 1 week after tamoxifen treatment, mice that were to receive a BM transplant were first lethally irradiated with two doses of 550 rads (5.5 Gy) from an X-RAD 320 unit (Precision X-ray), administered 4 h apart. BM was collected from femurs and tibias of donor mice by flushing with sterile Opti-MEM medium (Thermo Fisher Scientific). Each recipient mouse was injected retro-orbitally with 2 × 106 BM cells. At 4 weeks after BMT, peripheral blood was collected by retro-orbital venous plexus puncture and genomic DNA was extracted with a QIAamp DNA Blood Mini Kit (51104; Qiagen) for PCR analysis of BM reconstitution. Primer sequences are provided in Supplementary Table 2. Mice were then switched to a WD for up to 16 weeks and euthanized.
Immunohistochemistry
The aortic root was cryosectioned in the transverse axis and 10 µm sections were incubated with blocking solution (0.1% Triton X-100 in phosphate-buffered saline (PBS) supplemented with 5% goat serum) and then with primary antibodies diluted in blocking solution overnight at 4 °C. On the next day, sections were washed with 0.1% Triton X-100 in PBS and then incubated with secondary antibodies for 1 h. The primary antibodies used were anti-GFP (1:500; ab13970; Abcam), anti-CD68 (1:200; MCA1957; Bio-Rad) and directly conjugated Cy3 anti-SMA (1:500; C6198; Sigma–Aldrich). The secondary antibodies were conjugated to either FITC or goat anti-chicken Alexa Fluor 488 or goat anti-rat Alexa Fluor 647 fluorophores (1:500; Invitrogen). Nuclei were visualized by DAPI staining (1:1.000; D9542; Sigma–Aldrich). Note that membrane-localized tdTomato (mT) fluorescence in the aorta of ROSA26RmTmG/+ mice was very weak and essentially undetectable compared with the strong fluorescence in the red channel of staining with the anti-SMA antibody.
Fate mapping, clonal analysis and proliferation in atherogenesis
Mice carrying an inducible Cre recombinase and a Cre reporter (ROSA26RmTmG/+ or ROSA26RRb/+) were induced with tamoxifen (1 mg d−1) for 5 or 20 d in the case of Myh11-CreERT2 or Csf1r-Mer-iCre-Mer, respectively and then rested for 1 week. Following BMT and/or injection with AAV-Pcsk9, mice were fed a WD for up to 16 weeks and euthanized. For the in vivo proliferation studies, 2.5 mg EdU (A10044; Thermo Fisher Scientific) was injected intraperitoneally 12 h before euthanasia. A Click-iT EdU Alexa Fluor 647 Imaging Kit (C10340; Thermo Fisher Scientific) was used per the manufacturer’s instructions to detect EdU incorporation into proliferating cells. Serial sections through the aortic root were stained for nuclei (DAPI). In the case of mice carrying the ROSA26RmTmG/+ reporter, sections were also stained for GFP and SMA and either EdU or CD68. For ROSA26RRb/+ mice, sections were directly imaged using fluorescence filters for cerulean, membrane orange and membrane cherry, and marked cells in the media and plaque were quantified by scoring for the expression of these fluorophores. In all quantitative cellular studies, total cells were determined by counting DAPI+ nuclei. In select studies, cap cells were scored as SMA+ or SMA−, with the cap defined as the region of plaque within 30 µm of the lumen boundary, as previously classified18.
Anti-TNFα treatment and plasma TNFα measurement
Mice underwent BMT, were rested for 4 weeks and were then fed a WD for 12 weeks. During WD feeding, mice were intraperitoneally injected twice per week with mouse monoclonal anti-TNFα blocking antibody or an IgG2a isotype matched control (Janssen Research and Development) at a dose of 20 mg kg−1 body weight. For experiments with cultured cells, medium conditioned by young or aged BM-derived macrophages was pretreated with 20 µg ml−1 anti-TNFα antibody or IgG2a for 1 h at 37 °C before initiating SMC migration or proliferation assays. Plasma TNFα was quantified by enzyme-linked immunosorbent assay per the manufacturer’s instructions (BMS607-2INST; Thermo Fisher Scientific).
Cell isolation from murine BM
BM cells were harvested by flushing the femurs and tibias of mice in PBS with 2% fetal bovine serum (FBS) on ice. Erythrocytes and dead cells were removed from BM cells by density gradient centrifugation using Lympholyte (CL5031; Cedarlane). After centrifugation at 1,300g for 20 min, the remaining BM cells were collected from the interface and washed in Ca2+/Mg2+-free Hanks’ Balanced Salt Solution. These cells were dissociated to a single-cell suspension by filtering through a 70-µm nylon mesh and then subjected to scRNA-seq, monocyte isolation by FACS or differentiation to macrophages. For monocyte isolation, a suspension of mononuclear cells was stained with Alexa 700 anti-CD3 (1:100; 100215; BioLegend), PE-Cy7 anti-CD19 (1:100; 115520; BioLegend), FITC anti-CD11b (1:100; 553310; BD Pharmingen) and APC anti-Ly6c antibodies (1:100; 128015; BioLegend) in PBS with 0.5% FBS for 30 min. CD11b+Ly6c+ monocytes were isolated with a BD FACSAria ΙΙ cell sorter. Flow cytometry data were analyzed using FlowJo version 10.5.0.
Macrophage differentiation
Similar to our previous approach2, BM-derived mononuclear cells were resuspended in Iscove’s Dulbecco’s Modified Eagle Medium (DMEM) with 20% FBS, 20% L-929 cell-conditioned medium and 25 μg ml−1 fungizone and plated at a density of 1.8 × 106 cells per ml on uncoated plates. After 7 d of culture, nonadherent cells were washed away. Adherent cells were cultured in RPMI and 10% FBS for 12 h and then centrifuged at 1,300g for 5 min. The supernatant (that is, macrophage-conditioned medium) and pellet (that is, macrophages) were used immediately or stored at −80 °C.
Human peripheral blood monocyte isolation
All of the procedures involving human participants were approved by the Institutional Review Boards of Yale University (IRB number 1005006865), and we complied with all of the relevant ethical regulations. Written informed consent was obtained from all participants before inclusion in the study. Human peripheral blood mononuclear cells were isolated by Ficoll (GE Healthcare) density gradient centrifugation and then stained with PE anti-CD3 (1:100; 300308; BioLegend), PE-Cy7 anti-CD19 (1:100; 115520; BD Pharmingen) and FITC anti-CD14 (1:100; 325604; BioLegend). Human CD14+CD3−CD19− monocytes were sorted in a BD FACSAria ΙΙ cell sorter.
Aortic SMC isolation
Aortic SMCs were isolated by modification of a previously described protocol60. Briefly, aortas from the root to the iliac bifurcation were harvested from adult wild-type mice and opened longitudinally. Harvested aortas were enzymatically digested with 175 U ml−1 collagenase and 1.25 U ml−1 elastase (LS004176 and LS002279, respectively; Worthington) in PBS at 37 °C in a shaking incubator for 30 min. The adventitia was manually peeled off and the endothelium was removed by gentle scraping. Aortas were sequentially washed with 1% penicillin/streptomycin in PBS and then with 100% FBS. Washed aortas were cut into small pieces and cultured in plastic dishes in DMEM supplemented with 20% FBS and 1% penicillin/streptomycin. After 3 d, the medium was replaced by fresh medium that was the same except with 10 instead of 20% FBS. SMCs that migrated out of the aortic pieces and adhered to the dish were trypsinized and expanded, and passaged SMCs were used until the fifth passage.
Proliferation and migration assays in cell culture
For the proliferation studies, wild-type aortic SMCs were cultured in DMEM, 10% FBS and 1% penicillin/streptomycin until they reached ~60% confluency. Cells were then serum starved in DMEM overnight, followed by incubation with macrophage-conditioned medium for 8 or 48 h. During the last 8 h of this incubation, EdU (10 µM) from the Click-iT EdU Alexa Fluor 647 Imaging Kit (Thermo Fisher Scientific) was added to the conditioned medium. Subsequently, cells were fixed with 4% PFA for 30 min, permeabilized in 0.5% Triton X-100 in PBS and stained for EdU and nuclei (DAPI).
The migration assay used cell culture inserts (Ibidi) composed of two chambers flanking a central insert that prevents cell growth. Wild-type aortic SMCs were added to both chambers and allowed to attach and grow to confluency. SMCs were serum starved overnight and washed with PBS before removal of the insert and then cultured in macrophage-conditioned medium for 6 and 8 h. The cell coverage of the area that was blocked by the insert was measured immediately after insert removal and 6 and 8 h later.
Hydroxymethyl DNA immunoprecipitation
Hydroxymethyl DNA immunoprecipitation (hMeDIP) was carried out using a hMeDIP kit (C02010031; Diagenode) as previously published61. Briefly, genomic DNA from murine or human monocytes was isolated using an AllPrep DNA/RNA/miRNA Universal Kit (80224; Qiagen). DNA (1 µg) was sheared with a sonicator and DNA fragments (200–500 base pairs) were immunoprecipitated with a mouse monoclonal anti-5hmC antibody (2.5 μg per immunoprecipitation) following the protocol from Diagenode. The DNA–antibody mixture was incubated with magnetic beads overnight at 4 °C and then DNA was isolated. Multiple primers were designed to scan the proximal promoter region of ITGB3 in murine and human monocytes. Open chromatin regions were identified using the University of California, Santa Cruz genome browser (https://genome.ucsc.edu/). The forward and reverse primers spanning the proximal promoter region upstream of the transcription start site of mouse Itgb3 were 5′-AGGATGCGAGCGCAGTG-3′ and 5′-CGCACCTCTGCTTCTCAGT-3′, respectively. The corresponding PCR amplification product was located at chr11:104,608,100–104,608,250. For human ITGB3, the forward and reverse primers spanning the promoter region upstream of the transcription start site were 5′-GAAGTGGTCAGGACCTGGAA-3′ and 5′- TTCTGTGCCACTAGCCTGAG-3′. The location of this PCR amplification product was chr17:47,240,300–47,240,400. Hydroxymethylated DNA and input fractions were analyzed with hMeDIP-qPCR using ITGB3 primers, per the manufacturer’s protocol, to confirm enrichment of the hydroxymethylated gene.
Mouse Itgb3 cloning and lentivirus production
Mouse Itgb3 was cloned into the pLVX-EF1α-IRES-Puro vector (Takara Bio) per a previously described cloning strategy62. Briefly, total RNA was isolated from mouse aorta using a PureLink RNA Mini Kit (Invitrogen) and complementary DNA (cDNA) was synthesized with a ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs) using 1 µg RNA. PCR was performed using 1 µl cDNA template with AccuPrime Taq HIFI Polymerase (Invitrogen) in a 50 µl reaction volume using Itgb3-specific cloning primers to incorporate EcoRI and BamHI flanking sites. The primers used were Itgb3-EcoR1 forward (5′-AGCAGAATTCATGCGAGCGCAGTGG-3′) and Itgb3-BamHI reverse (5′-AGCAGGATCCTTAAGTCCCCCGGTAGGTG-3′). The Itgb3 amplicons were cleaned with the Qiagen QIAquick PCR Purification Kit, digested with EcoRI and BamH1 along with the pLVX-EF1α-IRES-Puro vector at 37 °C for 1 h, gel purified with the Qiagen QIAquick Gel Extraction Kit and ligated overnight at 14 °C using T4 ligase. Positive clones were verified by sequencing. A Lenti-X Expression System (Takara Bio) was utilized to generate lentivirus, per the manufacturer’s instructions. Briefly, 5 × 106 Lenti-X 293T cells were cotransfected with Lenti-X Packaging Single Shots (VSV-G) and 7 µg pLVX-EF1α-IRES-Puro vector with or without the Itgb3 insert. Lentiviral supernatants were collected 48 h after transfection and concentrated by adding one volume of Lenti-X Concentrator (Takara Bio) to three volumes of lentivirus-containing supernatant and incubating at 4 °C overnight. The next day, lentiviral pellet was obtained by centrifuging the mixture at 1,500g for 45 min, resuspended in 1 ml sterile PBS, aliquoted and stored at −80 °C. Before transplantation, BM cells purified by Ficoll density gradient centrifugation were transduced with empty vector (control) or Itgb3-overexpressing lentivirus in X-VIVO 15 medium (BioWhittaker) supplemented with stem cell factor (100 ng ml−1), thrombopoietin (50 ng ml−1), Flt3 ligand (50 ng ml−1) and interleukin-3 (20 ng ml−1) on to plates coated with retronectin (Takara Bio).
Imaging
Images were acquired with Nikon microscopes (Eclipse 80i upright fluorescent or Eclipse TS100 inverted) or Leica SP8 confocal microscope. For image processing, analysis and cell counting, Adobe Photoshop and ImageJ software were used.
Statistical analysis and reproducibility
No statistical methods were used to predetermine sample sizes, but sample sizes were similar to those reported in previous publications1,2. Mice were randomly assigned to experimental groups. Data collection and analysis were conducted in a manner that was blinded to the experimental conditions. Data distribution was assumed to be normal but was not formally tested. No data were excluded from the analyses. The two-tailed Student’s t-test and one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test were used to analyze the data (GraphPad Prism version 8.0.0). The data were derived from biological replicates and are presented as averages ± s.d. A P value of ≤0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data associated with this study are presented within the paper and its Supplementary Information files. The bulk and scRNA-seq data generated in this study are available from the Gene Expression Omnibus (accession code GSE206431). Trimmed bulk RNA-seq reads were aligned to the reference mouse genome mm10 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/). Reagents and materials associated with this study are available from the corresponding author D.M.G. Source data are provided with this paper.
References
Sheikh, A. Q., Misra, A., Rosas, I. O., Adams, R. H. & Greif, D. M. Smooth muscle cell progenitors are primed to muscularize in pulmonary hypertension. Sci. Transl. Med. 7, 308ra159 (2015).
Misra, A. et al. Integrin beta3 regulates clonality and fate of smooth muscle-derived atherosclerotic plaque cells. Nat. Commun. 9, 2073 (2018).
Chappell, J. et al. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ. Res. 119, 1313–1323 (2016).
Jacobsen, K. et al. Diverse cellular architecture of atherosclerotic plaque derives from clonal expansion of a few medial SMCs. JCI Insight 2, e95890 (2017).
Tay, T. L. et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20, 793–803 (2017).
Brunner, S. F. et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature 574, 538–542 (2019).
Dobnikar, L. et al. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat. Commun. 9, 4567 (2018).
Greif, D. M. et al. Radial construction of an arterial wall. Dev. Cell 23, 482–493 (2012).
Jaiswal, S. & Libby, P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat. Rev. Cardiol. 17, 137–144 (2020).
Jaiswal, S. et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017).
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018).
Moore, L. et al. The mutational landscape of normal human endometrial epithelium. Nature 580, 640–646 (2020).
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Yoshida, K. et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 578, 266–272 (2020).
Palmer, S., Albergante, L., Blackburn, C. C. & Newman, T. J. Reply to Jimenez-Alonso et al., Schooling and Zhao, and Mortazavi: further discussion on the immunological model of carcinogenesis. Proc. Natl Acad. Sci. USA 115, E4319–E4321 (2018).
Palmer, S., Albergante, L., Blackburn, C. C. & Newman, T. J. Thymic involution and rising disease incidence with age. Proc. Natl Acad. Sci. USA 115, 1883–1888 (2018).
Basatemur, G. L., Jorgensen, H. F., Clarke, M. C. H., Bennett, M. R. & Mallat, Z. Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol. 16, 727–744 (2019).
Shankman, L. S. et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 21, 628–637 (2015).
Wirka, R. C. et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 25, 1280–1289 (2019).
Schneider, J. G., Zhu, Y., Coleman, T. & Semenkovich, C. F. Macrophage β3 integrin suppresses hyperlipidemia-induced inflammation by modulating TNFα expression. Arterioscler. Thromb. Vasc. Biol. 27, 2699–2706 (2007).
Du, W. et al. Age-associated vascular inflammation promotes monocytosis during atherogenesis. Aging Cell 15, 766–777 (2016).
Fuster, J. J. et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017).
Robbins, C. S. et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 19, 1166–1172 (2013).
Wang, Y. et al. Clonally expanding smooth muscle cells promote atherosclerosis by escaping efferocytosis and activating the complement cascade. Proc. Natl Acad. Sci. USA 117, 15818–15826 (2020).
Grabert, K. et al. A transgenic line that reports CSF1R protein expression provides a definitive marker for the mouse mononuclear phagocyte system. J. Immunol. 205, 3154–3166 (2020).
Summers, K. M., Bush, S. J. & Hume, D. A. Network analysis of transcriptomic diversity amongst resident tissue macrophages and dendritic cells in the mouse mononuclear phagocyte system. PLoS Biol. 18, e3000859 (2020).
Ntokou, A. et al. Macrophage-derived PDGF-B induces muscularization in murine and human pulmonary hypertension. JCI Insight 6, e139067 (2021).
Nelson, P. R., Yamamura, S., Mureebe, L., Itoh, H. & Kent, K. C. Smooth muscle cell migration and proliferation are mediated by distinct phases of activation of the intracellular messenger mitogen-activated protein kinase. J. Vasc. Surg. 27, 117–125 (1998).
Stabile, E. et al. Akt controls vascular smooth muscle cell proliferation in vitro and in vivo by delaying G1/S exit. Circ. Res. 93, 1059–1065 (2003).
Gerthoffer, W. T. Mechanisms of vascular smooth muscle cell migration. Circ. Res. 100, 607–621 (2007).
Fernández-Hernando, C., József, L., Jenkins, D., Di Lorenzo, A. & Sessa, W. C. Absence of Akt1 reduces vascular smooth muscle cell migration and survival and induces features of plaque vulnerability and cardiac dysfunction during atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 29, 2033–2040 (2009).
Gu, Z., Noss, E. H., Hsu, V. W. & Brenner, M. B. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J. Cell Biol. 193, 61–70 (2011).
Branen, L. et al. Inhibition of tumor necrosis factor-α reduces atherosclerosis in apolipoprotein E knockout mice. Arterioscler. Thromb. Vasc. Biol. 24, 2137–2142 (2004).
Oberoi, R. et al. Anti-tumor necrosis factor-α therapy increases plaque burden in a mouse model of experimental atherosclerosis. Atherosclerosis 277, 80–89 (2018).
Alencar, G. F. et al. Stem cell pluripotency genes Klf4 and Oct4 regulate complex SMC phenotypic changes critical in late-stage atherosclerotic lesion pathogenesis. Circulation 142, 2045–2059 (2020).
Pan, H. et al. Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation 142, 2060–2075 (2020).
Uryga, A. K. et al. Telomere damage promotes vascular smooth muscle cell senescence and immune cell recruitment after vessel injury. Commun. Biol. 4, 611 (2021).
Rapisarda, V. et al. Integrin beta 3 regulates cellular senescence by activating the TGF-β pathway. Cell Rep. 18, 2480–2493 (2017).
Bogatyreva, K. B. et al. Association of the ITGB3 gene T1565C polymorphism with the development of atherosclerosis and in-stent restenosis in patients with stable coronary artery disease. Res. Results Biomed. 4, 3–9 (2018).
Weng, S. et al. β3 integrin deficiency promotes atherosclerosis and pulmonary inflammation in high-fat-fed, hyperlipidemic mice. Proc. Natl Acad. Sci. USA 100, 6730–6735 (2003).
Ghimire, K. et al. CD47 promotes age-associated deterioration in angiogenesis, blood flow and glucose homeostasis. Cells 9, 1695 (2020).
Lindberg, F. P., Gresham, H. D., Schwarz, E. & Brown, E. J. Molecular cloning of integrin-associated protein: an immunoglobulin family member with multiple membrane-spanning domains implicated in alpha v beta 3-dependent ligand binding. J. Cell Biol. 123, 485–496 (1993).
Brown, E., Hooper, L., Ho, T. & Gresham, H. Integrin-associated protein: a 50-kD plasma membrane antigen physically and functionally associated with integrins. J. Cell Biol. 111, 2785–2794 (1990).
Paolisso, G. et al. Advancing age and insulin resistance: role of plasma tumor necrosis factor-α. Am. J. Physiol. 275, E294–E299 (1998).
Bruunsgaard, H., Skinhoj, P., Pedersen, A. N., Schroll, M. & Pedersen, B. K. Ageing, tumour necrosis factor-alpha (TNF-α) and atherosclerosis. Clin. Exp. Immunol. 121, 255–260 (2000).
Ridker, P. M. et al. Elevation of tumor necrosis factor-α and increased risk of recurrent coronary events after myocardial infarction. Circulation 101, 2149–2153 (2000).
Hilgendorf, I., Swirski, F. K. & Robbins, C. S. Monocyte fate in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 35, 272–279 (2015).
Zhang, L. et al. Aging-related atherosclerosis is exacerbated by arterial expression of tumor necrosis factor receptor-1: evidence from mouse models and human association studies. Hum. Mol. Genet. 19, 2754–2766 (2010).
Crowson, C. S. et al. Brief report: accelerated aging influences cardiovascular disease risk in rheumatoid arthritis. Arthritis Rheum. 65, 2562–2566 (2013).
Dixon, W. G. et al. Reduction in the incidence of myocardial infarction in patients with rheumatoid arthritis who respond to anti-tumor necrosis factor α therapy: results from the British Society for Rheumatology Biologics Register. Arthritis Rheum. 56, 2905–2912 (2007).
Westlake, S. L. et al. Tumour necrosis factor antagonists and the risk of cardiovascular disease in patients with rheumatoid arthritis: a systematic literature review. Rheumatology (Oxford) 50, 518–531 (2011).
Kanter, J. et al. Biologic and clinical efficacy of lentiglobin for sickle cell disease. N. Engl. J. Med. 386, 617–628 (2022).
Qian, B. Z. et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 (2011).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Hodivala-Dilke, K. M. et al. β3-integrin-deficient mice are a model for Glanzmann thrombasthenia showing placental defects and reduced survival. J. Clin. Invest. 103, 229–238 (1999).
Morgan, E. A. et al. Dissection of platelet and myeloid cell defects by conditional targeting of the β3-integrin subunit. FASEB J. 24, 1117–1127 (2010).
Ko, M. et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl Acad. Sci. USA 108, 14566–14571 (2011).
Rinkevich, Y., Lindau, P., Ueno, H., Longaker, M. T. & Weissman, I. L. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476, 409–413 (2011).
Wirth, A. et al. G12-G13–LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 14, 64–68 (2008).
Karnik, S. K. et al. A critical role for elastin signaling in vascular morphogenesis and disease. Development 130, 411–423 (2003).
Liu, R. et al. Ten-eleven translocation-2 (TET2) is a master regulator of smooth muscle cell plasticity. Circulation 128, 2047–2057 (2013).
Dave, J. M. et al. Hic-5 mediates the initiation of endothelial sprouting by regulating a key surface metalloproteinase. J. Cell Sci. 129, 743–756 (2016).
Acknowledgements
We thank members of the D.M.G. laboratory and J. Testani for input. Schematics created with BioRender.com (Figs. 1, 2a, 3a, 6a,f, 7c, 8a and Extended Data Figs. 2a, 10) I.K. was supported by a Postdoctoral Fellowship (18POST34030015) and Career Development Award (938472) from the American Heart Association. Funding was also provided by the National Institutes of Health (R35HL150766, R01HL125815, R01HL142674, R21AG062202 and 1R21NS123469 to D.M.G.) and American Heart Association (Established Investigator Award 19EIA34660321 to D.M.G.).
Author information
Authors and Affiliations
Contributions
I.K., J.M.D., R.C., R.Q., N.R., K.A.M., C.F.-H. and D.M.G. conceived of and designed the experiments. I.K., X.Z., J.M.D., R.C., R.R.C., A.N., E.G.-V., B.A. and N.R. performed them. R.Q. and Y.K. conducted the scRNA-seq analysis. R.G.-M. helped with the bulk RNA-seq analysis. J.H. provided infrastructure for access to human blood. I.K. and D.M.G. analyzed the results, prepared the figures and wrote the manuscript. All authors reviewed and provided input on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Alan Tall, Gary Owens and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Aged BMT promotes polyclonal SMC progenitor expansion.
a–d, Young and aged Myh11-CreERT2, ROSA26R(Rb/+) mice were injected with AAV-Pcsk9 and fed a WD for 16 weeks. Brachiocephalic artery sections were imaged for Rb colors (a). Percent of DAPI+ plaque cells marked by any Rb color was quantified (b). For marked plaque (c) or underlying media (d) cells, the percent of cells of each color was quantified for each age group. In a given plaque, color 1 has greatest number of cells, color 2 is second most common color and color 3 is least frequent. In c, n = 5 (young) and 7 (aged) mice and 10 plaques per age group, 5 sections with a total of ~1100–1350 cells and spanning 200 μm per plaque. In d, n = 5 (young) and 7 (aged) mice and media underlying 14 plaques analyzed per group. e-i, Young Ldlr(−/−), Myh11-CreERT2, ROSA26R(Rb/+) mice underwent young or aged BMT. In e, after 4 weeks CD feeding, peripheral leukocyte counts were analyzed. n = 6 mice. In f,g, following 16 weeks WD feeding, transverse aortic root sections were stained with SMA or Sirius red, with dashed lines demarcating cap. Percent of cap cells that are SMA+ and percent of plaque area stained with Sirius red (h,i). n = 6 mice. j–q, Young Apoe(−/−), Myh11-CreERT2, ROSA26R(Rb/+) mice underwent young or aged BMT, AAV-Pcsk9 injection and 16 weeks WD feeding. Aortic sections were stained with H&E and Oil Red O (j,k), with quantification of lesion area (l; demarcated by dashed lines in j), lipid content (m), necrotic core area (n). Sections were imaged for Rb colors (o), and of the marked plaque (p) or medial cells (q), the percent of cells of each color was quantified. n = 5 mice, 12 plaques per group, 5–6 sections with a total of ~1000–1150 cells and spanning 200 μm per plaque. r,s, Murine aortic SMCs were cultured for 8 h with EdU in medium pre-conditioned by BM-derived macrophages of young vs. aged mice. In r, SMCs were stained for EdU and DAPI, and in s, percent of DAPI+ nuclei expressing EdU was quantified. n = 6 mice. Lu, lumen; Med, tunica media; Pl; plaque. All data are means ± SD. Two-tailed Student’s t-test. Scale bars, 100 µm (j,k), 50 µm (a,f,g,o), 25 µm (r).
Extended Data Fig. 2 Aged BM induces polyclonal expansion of SMMHC+ progenitors in the plaque.
Aged Myh11-CreERT2, ROSA26R(Rb/+) mice were induced with tamoxifen, irradiated, transplanted with BM from young or aged mice, injected with AAV-Pcsk9 and fed a WD for 16 weeks. Transverse aortic root sections were analyzed. a, Schematic of experiments. b–e, Sections were stained with H&E (dashed lines demarcate lesion; b) and Oil Red O (c) to determine lesion area (d) and lipid content (e). n = 5 and 7 mice per young and aged BMT group, respectively. Triplicate measurements from each mouse. f–i, Sections were stained with DAPI and directly imaged for Rb colors (mCh, mOr, Cer). In f, representative sections are shown with close-ups of boxed regions displayed on the right. In g, percent of DAPI+ plaque cells that were marked by any of the Rb colors was quantified. Of the marked plaque (h) or underlying medial cells (i), percent of cells of each color was quantified for each BMT group. In a given plaque (or media), color 1 is color with the greatest number of cells in plaque (or media), color 2 is second most common color and color 3 is least frequent color. n = 6 and 8 mice per young and aged BMT group, respectively, 14 plaques per BMT group, 5 sections with a total of ~1000–1200 cells and spanning 200 μm per plaque. Lu, lumen; Med, media; Pl, plaque. All data are means ± SD, and two-tailed Student’s t-test was used. Scale bars, 100 μm (b,c), 50 μm (f).
Extended Data Fig. 3 Tet2(−/−) BMT promotes recruitment and expansion of multiple SMC progenitors in the plaque.
Apoe(−/−), Myh11-CreERT2, ROSA26R(Rb/+) mice were induced with tamoxifen, irradiated and transplanted with Tet2(+/+) or Tet2(−/−) BM, injected with AAV-Pcsk9 to reduce hepatic LDLR receptor level and fed a WD for 16 weeks. a, Transverse aortic root sections were stained with DAPI and directly imaged for Rb colors (mCh, mOr, Cer). b, Percent of DAPI+ plaque cells that were marked by one of the Rb colors was quantified. n = 5 mice per group. c,d, Of the marked plaque (c) or medial cells (d), the percent of cells of each color was quantified for each BMT group. In a given plaque, color 1 is the most dominant color of cells in a plaque, color 2 is the second most common color and color 3 is least frequent. n = 5 mice per group, 9 plaques per genotype, 5 sections with a total of ~950–1100 cells and spanning 200 μm per plaque were analyzed. e,f, Murine aortic SMCs were cultured for 8 h in the presence of EdU in a medium conditioned by BM-derived macrophages of Tet2(+/+) and Tet2(−/−) mice. In e, SMCs were stained for EdU and nuclei (DAPI), and in f, the percent of DAPI+ nuclei that also express EdU was quantified. n = 6 mice. Lu, lumen; Med, tunica media; Pl; plaque. All data are means ± SD. Two-tailed Student’s t-test. Scale bar, 50 µm (a), 25 µm (e).
Extended Data Fig. 4 Isolation of mouse BM-derived and human peripheral blood monocytes and pathway analysis of young vs old monocytes.
a,c,d, Dot plots showing gating schemes of monocyte isolation by FACS. In a, BM from the femurs of young and aged mice was harvested and subjected to FACS to isolate CD3−CD19−Cd11b+ Ly6C+ monocytes (n = 9 mice per group). In c, human peripheral blood monocytes were harvested from young and aged subjects, and CD14+ monocytes were sorted (n = 12 humans per age group). In d, BM from Tet2(+/+) and Tet2(−/−) mice was harvested and subjected to FACS to isolate CD3−CD19−Cd11b+Ly6C+ monocytes (n = 9 mice per group). b, Lysates of monocytes isolated from young or aged mice underwent bulk RNA-seq (n = 3 mice per age group), and the transcriptome was subjected to gene set enrichment analysis (GSEA). Activated pathways are shown with gene ratio indicating the number of activated genes relative to the total number of genes in the corresponding pathways and size of the dot representing number of activated genes. The GSEA algorithm as implemented in clusterProfiler R package was used to perform a one-sided test to determine the significance level of pathway enrichment.
Extended Data Fig. 5 Deletion of Itgb3 in CSF1R+ cells exacerbates atherosclerosis and induces proliferation and accumulation of this cell lineage in the plaque.
a–j, Apoe(−/−), Csf1r-Mer-iCre-Mer mice also carrying Itgb3(fl°x/flox) or wild type for Itgb3 were induced with tamoxifen. In a–f, mice were then fed a WD for 16 weeks, and transverse aortic root sections were stained with H&E (dashed lines demarcate lesion and necrotic core in a,e, respectively) and Oil Red O (b). Quantification of lesion area (c), lipid content (d) and necrotic core area (f). n = 5 mice and 10–12 plaques per genotype (triplicate measurements per plaque). BM-derived GFP+ myeloid cells (Ly6C+) were isolated by FACS after 5 days of tamoxifen induction and subjected to qRT-PCR for Itgb3 (g). Itgb3 mRNA is relative to Gapdh and normalized to control. n = 6 mice per group, and qRT-PCR was done in triplicate. Four weeks following tamoxifen induction, peripheral blood total cholesterol and triglycerides (after a 16 h fast; n = 5–7 mice per group; h,i) and leukocyte counts (j) were determined. n = 5 and 7 mice in Itgb3(+/+) and Itgb3(flox/flox) groups, respectively. k–o, Apoe(−/−), Csf1r-Mer-iCre-Mer, ROSA26R(mTmG/+) mice also carrying Itgb3(+/+) or Itgb3(flox/flox) were induced with tamoxifen and fed a WD for 16 weeks. In k,l, after 16 weeks of WD feeding, EdU was injected intraperitoneally 12 h prior to euthanasia. Transverse aortic root sections were stained for markers of fate (GFP), SMCs (SMA), nuclei (DAPI) and either proliferation (EdU) in k or CD68 in m. Boxed regions in k are shown as close-ups below with arrowheads indicating GFP+EdU+ cells. In l, the percentage of GFP+ plaque cells that are EdU+ is shown. In, n,o, percentage of plaque cells that are GFP+ or CD68+ was quantified. n = 5 and 7 mice in Itgb3(+/+) and Itgb3(flox/flox) groups, respectively, 10 plaques per genotype, 4 sections with a total of ~1000–1250 cells and spanning 150 μm per plaque (l,n,o). All data are means ± SD, and two-tailed Student’s t-test was used. Lu, lumen; Med, media; Pl, plaque. Scale bars, 100 μm (a,b,e), 50 μm (k,m).
Extended Data Fig. 6 SMC-specific Itgb3 deletion attenuates atherosclerosis and SMC contribution to the plaque without altering plasma lipids.
Apoe(−/−), Myh11-CreERT2 mice also carrying Itgb3(flox/flox) or Itgb3(+/+) and either ROSA26R(mTmG/+) (a–k) or ROSA26R(Rb/+) (l–n) were induced with tamoxifen. a–e and i–n, Mice were rested, subjected to WD for 16 weeks, and aortic root transverse sections were analyzed. In a–e, sections were stained with H&E [dashed lines demarcating lesion; (a)] and Oil Red O (b), and lesion area (c), lipid content (d) and necrotic core area (e) were quantified. n = 7 mice (c–e) in Itgb3(+/+) and n = 8 (c,d), n = 7 (e) mice in Itgb3(flox/flox) groups. In i, EdU was injected 12 h prior to euthanasia. Sections were stained for SMA, GFP (fate marker), EdU and nuclei (DAPI). Close-ups of boxed regions are shown. Arrowheads indicate GFP+EdU+ cells. Percent of total plaque cells that are GFP+ and percent of total plaque GFP+ cells that are EdU+ are shown in j,k, respectively. n = 5 mice and 10 plaques per genotypes, 5–6 sections with a total of ~1200–1500 cells and spanning 200 μm per plaque. In l, sections were imaged for Rb colors with close-ups of boxed regions. Of the marked plaque (m) or underlying medial cells (n), the percent of cells of each color was quantified. Colors #1–3 are defined as in previous figures. n = 5 mice and 12 plaques per genotype, 6 sections with a total of ~1000–1300 cells and spanning 250 μm per plaque. Lu, lumen; Med, media; Pl, plaque. f, After 5 days of tamoxifen induction, GFP+ SMCs were isolated by FACS and subjected to qRT-PCR for Itgb3. Itgb3 mRNA is represented relative to Gapdh and normalized to Itgb3(+/+) group. n = 6 mice per group, and qRT-PCR was done in triplicate. g,h, Four weeks following tamoxifen induction, plasma total cholesterol and triglycerides were measured after fasting 16 h; n = 5 and 7 mice in Itgb3(+/+) and Itgb3(flox/flox) groups, respectively. All data are means ± SD, and two-tailed Student’s t-test was used. Scale bars, 100 μm (a,b), 50 μm (i,l).
Extended Data Fig. 7 Integrin β3 reduction in aortic SMCs attenuates phosphorylation of AKT and ERK and PDGF-B-induced dorsal ruffles.
a,b, Aortic lysates of wild type (WT) or Itgb3(−/−) mice were subjected to Western blot for integrin β3, GAPDH and phosphorylated (p-) and total AKT (a) with densitometry of each protein relative to GAPDH and normalized to WT (b). n = 6 mice per genotype. c–f, Mouse aortic SMCs were isolated and treated with Itgb3 siRNA or scrambled (Scr) RNA. Cell lysates were subjected to Western blot for integrin β3, GAPDH as well as phosphorylated and total AKT, PAK1 and ERK (c) with densitometry of each protein relative to GAPDH and normalized to Scr (d). In e, following siRNA treatment, SMCs were serum starved overnight, treated with PDGF-B (10 ng/mL) for the indicated times and stained with FITC-phalloidin. Arrowheads indicate dorsal ruffles with boxed regions shown as close-ups below. Scale bar, 25 μm. Quantification of dorsal ruffles is shown in (f). n = 6 experiments. All data are means ± SD, and two-tailed Student’s t-test was used.
Extended Data Fig. 8 scRNA-seq analysis of Apoe(−/−), Itgb3(−/−) and Apoe(−/−), Itgb3(+/+) BM cells, focusing on monocyte/macrophage cluster.
BM was isolated from Apoe(−/−) mice carrying Itgb3(−/−) or Itgb3(+/+) and subjected to scRNA-seq. a, Proportion of leukocyte sub-types in BM cells from scRNA-seq data. b-h, t-SNE plots of pooled scRNA-seq data from BM of both genotypes showing the expression of monocyte/macrophage markers. i, IPA predicts the effects of differentially expressed genes on the inflammatory responses and monocyte/macrophage migration or proliferation. n = 3 mice pooled per genotype. j, Differential abundance analysis of sub-clustered monocytes/macrophages show abundance of Apoe(−/−), Itgb3(−/−) cells in circled regions. k, l, Dot plot analysis of monocytes/macrophage sub-cluster showing differentially expressed genes in TNFα and NF-kB signaling pathways, respectively. Dot size represents the fraction of cells expressing the gene, and shade of blue color represents average gene expression.
Extended Data Fig. 9 Aged BM, TNFα and SMC biology.
a–c, Young Ldlr(−/−), Myh11-CreERT2, ROSA26R(Rb/+) mice underwent young or aged BMT. In a, DNA isolated from peripheral blood of recipients before (lanes 1–3) or after (lanes 4–6) aged BMT or of WT (lane 7) or ROSA26R(Rb/+) (lane 8) mice was amplified using primers for ROSA26R Rainbow (Rb) and WT alleles. In b,c, BMT recipients were fed a WD feeding for 12 weeks with concomitant twice weekly anti-TNFα antibody or control IgG2a injections. Transverse aortic root sections were stained with SMA and DAPI, and the percent of SMA+ cells in the cap area was assessed (cap area demarcated by dashed lines in b). n = 6 mice per group. Lu, lumen; Med, media; Pl, plaque. d,e, BM of young or aged wild type mice was harvested and differentiated into macrophages and isolated mouse aortic SMCs were cultured with macrophage conditioned medium. Anti-TNFα blocking antibody or control IgG was added to conditioned medium, 1 h prior to incubation with SMCs. SMCs were incubated with conditioned medium with EdU for 8 h. SMCs were stained for EdU and DAPI, and percent of EdU+ cells was quantified. n = 3 mice per group, each experiment done in triplicate. f,g, Mouse aortic SMCs were isolated and treated with Tnfr1 siRNA or scrambled (Scr) RNA. Cell lysates were subjected to Western blot for TNFR1 and GAPDH with densitometry relative to GAPDH and normalized to Scr. n = 3 mice per group, each experiment done in triplicate. h–k, Tnfr1-silenced SMCs were exposed to methyl-β-cyclodextrin complexes and TNFα for 3 days, and then mRNA levels were assessed by qRT-PCR. n = 3 experiments, each experiment done in triplicate. l,m, BM harvested from aged wild type mice was infected with Itgb3-expressing or control lentivirus and transplanted into aged Myh11-CreERT2, ROSA26R(Rb/+) mice. Four weeks after BMT, fasting total cholesterol and triglycerides levels were assessed in blood plasma. n = 4 mice in control and n = 6 in Itgb3 o/e groups. All data are means ± SD. Two-tailed Student’s t-test. Scale bars, 50 μm (b), 25 μm (d).
Extended Data Fig. 10 Age of BM-derived monocytes/macrophages determines clonality of SMC lineage in the atherosclerotic plaque.
Atherogenesis is depicted in a young (a) or aged (b) host. Aged monocytes/macrophages have decreased levels of the epigenetic regulator TET2, leading to reduction of the 5-hydroxymethylcytosine (5hmC) mark on the Itgb3 promoter. The resulting low integrin β3 levels in aged monocytes/macrophages induces high TNFα levels, facilitating recruitment and expansion of multiple SMC progenitors (polyclonality) in the atherosclerotic plaque and worse disease burden. In contrast, the young control is characterized by mono/oligoclonal SMC expansion in a smaller plaque.
Supplementary information
Supplementary Information
Supplementary Tables 1–5 and Supplementary Methods.
Supplementary Data
Bulk RNA-seq of BM-derived monocytes isolated from aged mice relative to young mice.
Source data
Source Data Fig. 4
Unprocessed Western blots.
Source Data Extended Data Fig. 7
Unprocessed Western blots.
Source Data Extended Data Fig. 9
Unprocessed Western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kabir, I., Zhang, X., Dave, J.M. et al. The age of bone marrow dictates the clonality of smooth muscle-derived cells in atherosclerotic plaques. Nat Aging 3, 64–81 (2023). https://doi.org/10.1038/s43587-022-00342-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-022-00342-5
This article is cited by
-
Roadmap for alleviating the manifestations of ageing in the cardiovascular system
Nature Reviews Cardiology (2025)
-
Chronic inflammation and vascular cell plasticity in atherosclerosis
Nature Cardiovascular Research (2024)
-
Systemic and local regulation of hematopoietic homeostasis in health and disease
Nature Cardiovascular Research (2024)
-
Hallmarks of cardiovascular ageing
Nature Reviews Cardiology (2023)
-
Old bones control smooth muscle clones
Nature Aging (2023)