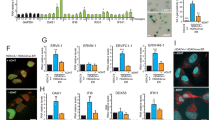
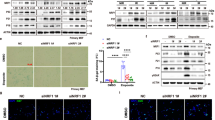
Abstract
Reactivation of endogenous retroviruses (ERVs) has been proposed to be involved in aging. However, the mechanism of reactivation and contribution to aging and age-associated diseases is largely unexplored. In this study, we identified a subclass of ERVs reactivated in senescent cells (termed senescence-associated ERVs (SA-ERVs)). These SA-ERVs can be bidirectional transcriptionally activated by activating transcription factor 3 (ATF3) to generate double-stranded RNAs (dsRNAs), which activate the RIG-I/MDA5–MAVS signaling pathway and trigger a type I interferon (IFN-I) response in senescent fibroblasts. Consistently, we found a concerted increased expression of ATF3 and SA-ERVs and enhanced IFN-I response in several tissues of healthy aged individuals and patients with Hutchinson–Gilford progeria syndrome. Moreover, we observed an accumulation of dsRNAs derived from SA-ERVs and higher levels of IFNβ in blood of aged individuals. Together, these results reveal a previously unknown mechanism for reactivation of SA-ERVs by ATF3 and illustrate SA-ERVs as an important component and hallmark of aging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All datasets have been deposited in the Genome Sequence Archive for Human (GSA-Human) in the National Genomics Data Center70 with accession numbers as indicated: raw RNA-seq data for non-senescent and senescent WI38 cells, HRA005428; raw dsRIP-seq data for senescent WI38 cells, HRA007306; raw WGBS data for non-senescent and senescent WI38 cells, HRA005437; raw ATF3 ChIP-seq data for senescent WI38 cells, HRA007303; raw RNA-seq data for WI38 cells with ATF3 ectopic expression, HRA005436; raw RNA-seq data for whole blood from young and old individuals, HRA005441. Additional public datasets used in this study are listed in Supplementary Table 5. Source data are provided with this paper. All other data supporting the conclusions of the study are available upon reasonable request from the corresponding authors.
References
Cai, Y. et al. The landscape of aging. Sci. China Life Sci. 65, 2354–2454 (2022).
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell 186, 243–278 (2023).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Liang, C. et al. BMAL1 moonlighting as a gatekeeper for LINE1 repression and cellular senescence in primates. Nucleic Acids Res. 50, 3323–3347 (2022).
Gorbunova, V. et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 596, 43–53 (2021).
Feschotte, C. & Gilbert, C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13, 283–296 (2012).
Mao, J., Zhang, Q. & Cong, Y.-S. Human endogenous retroviruses in development and disease. Comput. Struct. Biotechnol. J. 19, 5978–5986 (2021).
Vargiu, L. et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13, 7 (2016).
Johnson, W. E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 17, 355–370 (2019).
Mi, S. et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (2000).
Blaise, S., de Parseval, N., Bénit, L. & Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA 100, 13013–13018 (2003).
Göke, J. et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135–141 (2015).
Burns, K. H. Transposable elements in cancer. Nat. Rev. Cancer 17, 415–424 (2017).
Copley, K. E. & Shorter, J. Repetitive elements in aging and neurodegeneration. Trends Genet. 39, 381–400 (2023).
Mao, J. et al. TERT activates endogenous retroviruses to promote an immunosuppressive tumour microenvironment. EMBO Rep. 23, e52984 (2022).
Liu, X. et al. Resurrection of endogenous retroviruses during aging reinforces senescence. Cell 186, 287–304.e26 (2023).
Battle, A., Brown, C. D., Engelhardt, B. E. & Montgomery, S. B. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Rehwinkel, J. & Gack, M. U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551 (2020).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).
Freund, A., Laberge, R.-M., Demaria, M. & Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23, 2066–2075 (2012).
Hur, S. Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol. 37, 349–375 (2019).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Geis, F. K. & Goff, S. P. Silencing and transcriptional regulation of endogenous retroviruses: an overview. Viruses 12, 884 (2020).
Zhang, Q., Pan, J., Cong, Y. & Mao, J. Transcriptional regulation of endogenous retroviruses and their misregulation in human diseases. Int. J. Mol. Sci. 23, 10112 (2022).
Espinet, E. et al. Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic IFN program linked to a ductal cell of origin. Cancer Discov. 11, 638–659 (2021).
Matteucci, C., Balestrieri, E., Argaw-Denboba, A. & Sinibaldi-Vallebona, P. Human endogenous retroviruses role in cancer cell stemness. Semin. Cancer Biol. 53, 17–30 (2018).
Min, B., Jeon, K., Park, J. S. & Kang, Y.-K. Demethylation and derepression of genomic retroelements in the skeletal muscles of aged mice. Aging Cell 18, e13042 (2019).
Pal, S. & Tyler, J. K. Epigenetics and aging. Sci. Adv. 2, e1600584 (2016).
Kelly, T. K., De Carvalho, D. D. & Jones, P. A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078 (2010).
Zhang, C. et al. ATF3 drives senescence by reconstructing accessible chromatin profiles. Aging Cell 20, e13315 (2021).
Wu, Z. et al. Differential stem cell aging kinetics in Hutchinson–Gilford progeria syndrome and Werner syndrome. Protein Cell 9, 333–350 (2018).
Di Micco, R., Krizhanovsky, V., Baker, D. & d’Adda di Fagagna, F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021).
Balestrieri, E. et al. Transcriptional activity of human endogenous retroviruses in human peripheral blood mononuclear cells. Biomed Res. Int. 2015, 164529 (2015).
Autio, A. et al. Effect of aging on the transcriptomic changes associated with the expression of the HERV-K (HML-2) provirus at 1q22. Immun. Ageing 17, 11 (2020).
Barbot, W., Dupressoir, A., Lazar, V. & Heidmann, T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 30, 2365–2373 (2002).
De Cecco, M. et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging 5, 867–883 (2013).
Li, W. et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 16, 529–531 (2013).
Wood, J. G. et al. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl Acad. Sci. USA 113, 11277–11282 (2016).
Patterson, M. N. et al. Preferential retrotransposition in aging yeast mother cells is correlated with increased genome instability. DNA Repair 34, 18–27 (2015).
Küry, P. et al. Human endogenous retroviruses in neurological diseases. Trends Mol. Med. 24, 379–394 (2018).
Pisano, M. P., Grandi, N. & Tramontano, E. High-throughput sequencing is a crucial tool to investigate the contribution of human endogenous retroviruses (HERVs) to human biology and development. Viruses 12, 633 (2020).
Hai, T., Wolfgang, C. D., Marsee, D. K., Allen, A. E. & Sivaprasad, U. ATF3 and stress responses. Gene Expr. 7, 321–335 (1999).
Inaba, Y. et al. The transcription factor ATF3 switches cell death from apoptosis to necroptosis in hepatic steatosis in male mice. Nat. Commun. 14, 167 (2023).
Wang, L. et al. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc−. Cell Death Differ. 27, 662–675 (2020).
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C. & Santoro, A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018).
Bai, L. et al. Regulation of cellular senescence by the essential caveolar component PTRF/Cavin-1. Cell Res. 21, 1088–1101 (2011).
Chen, J. et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 32, 4812–4820 (2004).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Liu, J. et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 22, 1056–1063 (2020).
Zhang, Q. et al. Human telomerase reverse transcriptase is a novel target of Hippo-YAP pathway. FASEB J. 34, 4178–4188 (2020).
CNCB-NGDC Members and Partners. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 50, D27–D38 (2022).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (U22A20318 and 31730020 to Y.-S.C., 32000512 to J.M. and 31801155 to Q.Z.), the Zhejiang Provincial Department of Science and Technology (2023C03166 to Y.-S.C.) and the Interdisciplinary Research Project of Hangzhou Normal University (2024JCXK06 to Y.-S.C.).
Author information
Authors and Affiliations
Contributions
J.M., Q.Z. and Y.-S.C. conceived of and designed the experiments. J.M., Q.Z. and Y. Zhuang performed most of the experiments and data analyses. Y. Zhang, L.L., J.P., L.X., Y.D. and M.W. helped with experiments. J.M., Q.Z. and Y.-S.C. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Vera Gorbunova and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data
Extended Data Fig. 1 Activation of RIG-I/MDA5-MAVS and IFN-I signaling in old individuals and senescent cells.
a, Differential expression analysis of genes related to viral defense and IFN-I response in adipose tissue, brain, muscle, nerve, skin (not sun exposed), and whole blood between old versus young individuals from GTEx database. b, GSEA enrichment plot (left), enriched biological processes (BP) (middle), and heatmap of genes related to RIG-I/MDA5-MAVS and IFN-I signaling (right) in non-senescent and senescent IMR90 cells through RNA-seq analysis. c, GSEA enrichment plot (left), enriched biological processes (BP) (middle), and heatmap of genes related to RIG-I/MDA5-MAVS and IFN-I signaling (right) in non-senescent and senescent BJ cells through RNA-seq analysis. d, RT-qPCR analysis of genes related to RIG-I/MDA5-MAVS and IFN-I signaling in non-senescent and senescent WI38 or IMR90 cells. Data represent mean ± SEM of three biological replicates. P values were calculated by moderated t-test and adjusted by the Benjamini-Hochberg method in limma (a), accumulative hypergeometric test (b, middle), Wald test with Benjamini-Hochberg adjustment in DESeq2 (b, right), or unpaired two-tailed Student’s t-test (d). NES, normalized enrichment score. non-Sen, non-senescent. Sen, senescent.
Extended Data Fig. 2 RNA-seq and ATAC-seq analysis for ERVs in non-senescent and senescent WI38, IMR90, and BJ cells.
a, Scatterplot representing differences in ERVs expression in non-senescent and senescent BJ cells by RNA-seq analysis. Red dots indicate ERV elements with significantly increased expression, while blue dots indicate ERV elements with significantly decreased expression. b, Regression analysis of the correlation for the log2FoldChange (senescent versus non-senescent) of ERV elements expression between WI38 and BJ cells. R is Pearson correlation coefficient. P value was calculated using a two-sided Pearson’s correlation test. c, The profiles for genomic regions of ERV1, ERVK, ERVL, and ERVL-MaLR subfamilies by ATAC-seq analysis in BJ cells. d, Heatmap showing expression by RNA-seq analysis (left) and chromatin accessibility by ATAC-seq analysis (right) of ERV elements in non-senescent and senescent BJ cells. e, Venn diagram showing the overlap of significantly activated ERV elements in senescent WI38, IMR90, and BJ cells. f, log2FoldChange differences of chromatin accessibility (left) and expression (right) of LTR39, MER61C, MER61-int, MSTC, and MST-int, between senescent and non-senescent BJ cells, revealed by ATAC-seq and RNA-seq respectively. g, Signal distributions of ATAC-seq and RNA-seq at the representative genomic loci of LTR39, MER61C, MER61-int, MSTC, and MST-int, in non-senescent and senescent BJ cells. P values were calculated by Wald test in DESeq2 (a and f). non-Sen, non-senescent. Sen, senescent.
Extended Data Fig. 3 Expression of SA-ERVs is gradually upregulated with passaging in normal fibroblasts, but remains unchanged in hTERT-immortalized cells.
a, The expression patterns of LTR39, MER61C, MER61-int, MSTC, and MST-int during passaging in WI38 and HFF cells by RNA-seq analysis. b, The expression of SA-ERVs (left), and genes related to RIG-I/MDA5-MAVS and IFN-I signaling (right), in PD46 and PD109 hTERT-immortalized WI38 cells by RNA-seq analysis. c, The profiles of ATAC-seq signals in genomic regions of ERV1, ERVK, ERVL, and ERVL-MaLR subfamilies in PD46 and PD109 hTERT-immortalized WI38 cells. Data represent mean ± SEM of three biological replicates (a and b). PD, population doublings.
Extended Data Fig. 4 Treatments with DNMTi DAC activate RIG-I/MDA5-MAVS and IFN-I signaling in human non-senescent fibroblasts.
a, log2FoldChange differences of CpG methylation levels of genomic LTR39, MER61C, MER61-int, MSTC, and MST-int loci between senescent versus non-senescent WI38 or IMR90 cells by WGBS. b, Heatmap of CpG methylation levels in the genomic regions of ERV1, ERVK, ERVL, and ERVL-MaLR subfamilies in human primary dermal fibroblasts (HDFs) with DAC treatment and control by RRBS analysis. c, Scatterplot representing differences in ERVs expression between HDFs with DAC treatment versus control by RNA-seq analysis. Red dots indicate ERV elements with significantly increased expression, while blue dots indicate ERV elements with significantly decreased expression. d, GSEA enrichment plots between HDFs with DAC treatment versus control. e, Heatmap showing expression of genes related to RIG-I/MDA5-MAVS and IFN-I signaling in HDFs with DAC treatment and control. f, Scatterplot representing differences in ERVs expression between human gingival fibroblasts (HGFs) with DAC treatment versus control by RNA-seq analysis. Red dots indicate ERV elements with significantly increased expression, while blue dots indicate ERV elements with significantly decreased expression. g, Expression analysis of sense (upper) and antisense (lower) transcripts of the SA-ERVs in HGFs between DAC treatment and control, using strand-specific RNA-seq. Data represent mean ± SEM. n = 5 biological replicates per group. h, GSEA enrichment plots between HGFs with DAC treatment versus control. i, Heatmap showing expression of genes related to RIG-I/MDA5-MAVS and IFN-I signaling in HGFs with DAC treatment and control. P values were calculated by Wald test in DESeq2 (c, e, f, and i), or paired two-tailed Student’s t-test (g).
Extended Data Fig. 5 Analysis for the functional characteristics of ATF3.
a, Motif analysis based on the sequences of SA-ERVs. P value was calculated by binomial test in HOMER. b, Scatterplot representing the enrichment of ATF3 on genomic ERV elements. Red dots indicate ERV elements with ATF3 binding. There are 357 ERV elements potentially bound by ATF3. c, Analysis for enrichment of ATF3 on the promoters of IFIH1, RIGI, IFNB1, OAS1, and CCL2 in senescent WI38 cells by ChIP-seq. β-actin promoter as negative control. d, ChIP-qPCR analysis for ATF3 targets on the promoters of IFIH1, RIGI, IFNB1, OAS1, and CCL2 in senescent WI38 cells. LTR39 and β-actin promoter were used as positive and negative controls, respectively. Data represent mean ± SEM of three biological replicates. P values were calculated by two-way ANOVA with Sidak’s multiple comparisons test. e, Scatterplot representing differences of genes expression in WI38 cells expressing ATF3 versus control by RNA-seq analysis. Red dots indicate significantly upregulated genes, while blue dots indicate significantly downregulated genes. P values were calculated by Wald test with Benjamini-Hochberg adjustment in DESeq2. f, Enriched biological processes (BP) for WI38 cells expressing ATF3 versus control. GO was analyzed for downregulated genes in WI38 cells expressing ATF3 compared to control. P values were calculated by the accumulative hypergeometric test.
Extended Data Fig. 6 ERVs and IFN-I signaling are activated in aged mice.
a, RT-qPCR analysis of mouse ERVs in kidney or lung from young and aged mice. b, The profiles for genomic regions of ERV1, ERVK, ERVL, and ERVL-MaLR subfamilies by ATAC-seq analysis for mouse kidney. c, Enrichment of dsRNA over ssRNA calculated by normalizing the ΔCt between RNase A treated and non-treated of mouse ERVs (dsRNA) against β-actin (ssRNA). SINEB1 and Rplp0 served as positive and negative controls, respectively. d, ELISA analysis of dsRNA in total RNA extracted from kidney or lung of young and aged mice. e, Western blot analysis of ATF3, RIG-I, MDA5, and MAVS in kidney or lung from young and aged mice. β-actin served as loading control. f, RT-qPCR analysis of genes related to RIG-I/MDA5-MAVS and IFN-I signaling in kidney or lung from young and aged mice. g, GSEA enrichment plots showing “KEGG: RIG-I-like receptor signaling pathway” enriched signature between aged versus young mice. NES, normalized enrichment score. h, log2FoldChange expression differences of genes related to RIG-I/MDA5-MAVS and IFN-I signaling in kidney or lung between young and aged mice by RNA-seq analysis. Data represent mean ± SEM. n = 5 samples per group (a, c, d, and f). P values were calculated by unpaired two-tailed Student’s t-test (a, c, d, and f), or Wald test in DESeq2 (h).
Extended Data Fig. 7 Schematic representation of key findings.
Schematic model showing that SA-ERVs activated by ATF3 form dsRNAs through bidirectional transcription, drive IFN-I response via RIG-I/MDA5-MAVS signaling pathway in senescent cells, thereby contributing to SASP and other senescence-associated phenotypes. non-Sen, non-senescent. Sen, senescent.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mao, J., Zhang, Q., Zhuang, Y. et al. Reactivation of senescence-associated endogenous retroviruses by ATF3 drives interferon signaling in aging. Nat Aging 4, 1794–1812 (2024). https://doi.org/10.1038/s43587-024-00745-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-024-00745-6
This article is cited by
-
CDK9 Inhibition with enitociclib reveals influence on HERV and LINE RNA abundances in whole blood, T-, and B-Cell lines
BMC Medical Genomics (2026)
-
Ectopic co-expression of canonical and LINE1 and THE1A-exonizing IL23R transcripts in sarcoid myopathy
Journal of Human Genetics (2026)
-
DNA fragmentation factor B suppresses interferon to enable cancer persister cell regrowth
Nature Cell Biology (2025)
-
Transcriptional and post-transcriptional regulation of transposable elements and their roles in development and disease
Nature Reviews Molecular Cell Biology (2025)
-
Roles of Immunity and Endogenous Retroelements in the Pathogenesis of Rheumatoid Arthritis and Treatment Strategies
Functional & Integrative Genomics (2025)