Abstract
T cells have been identified as correlates of protection in viral infections. However, the level of vaccine-induced T cells needed and the extent to which they alone can control acute viral infection in humans remain uncertain. Here we conducted a double-blind, randomized controlled trial involving vaccination and challenge in 33 adult human volunteers, using the live–attenuated yellow fever (YF17D) and chimeric Japanese encephalitis–YF17D (JE/YF17D) vaccines. Both Orthoflavivirus vaccines share T cell epitopes but have different neutralizing antibody epitopes. The primary objective was to assess the extent to which vaccine-induced T cell responses, independent of neutralizing antibodies, were able to reduce post-challenge viral RNAaemia levels. Secondary objectives included an assessment of surrogate measures of viral control, including post-challenge antibody titres and symptomatic outcomes. YF17D vaccinees had reduced levels of JE/YF17D challenge viraemia, compared with those without previous YF17D vaccination (mean log10(area under the curve genome copies per ml): 2.23 versus 3.22; P = 0.039). Concomitantly, YF17D vaccinees had lower post-JE/YF17D challenge antibody titres that reduced JE virus plaque number by 50%, or PRNT50 (mean log10(PRNT50 titre): 1.87 versus 2.5; P < 0.0001) and symptomatic rates (6% (n = 1/16) versus 53% (n = 9/17), P = 0.007). There were no unexpected safety events. Importantly, after challenge infection, several vaccinees had undetectable viraemia and no seroconversion, even in the absence of neutralizing antibodies. Indeed, high vaccine-induced T cell responses, specifically against the capsid protein, were associated with a level of viral control conventionally interpreted as sterilizing immunity. Our findings reveal the importance of T cells in controlling acute viral infection and suggests a potential correlate of protection against orthoflaviviral infections. ClinicalTrials.gov registration: NCT05568953.
Similar content being viewed by others
Main
Vaccines are undoubtedly one of the most effective public health tools in reducing morbidity and mortality from infectious diseases. Their effectiveness was emphatically demonstrated during the recent COVID-19 pandemic, in which the use of vaccines helped neutralize the sting of the pandemic1. Despite their clear benefit, many untreatable viral diseases that cause substantial health burden still lack effective vaccines. This gap in the vaccine armamentarium is further complicated by the costly, time-consuming and challenging nature of vaccine development2; for every successful vaccine that enters the clinic, multiple others fail to achieve licensure or yield less than desirable outcomes.
An impediment to viral vaccine development has been the lack of clearly established immune correlates of protection (CoP)3. Such CoPs are essential to guide the assessment of vaccine efficacy, allow comparative analysis between different vaccine platforms and even guide public health policies. Conventionally, the assessment of CoPs has focused almost exclusively on antibodies, even though multiple lines of evidence suggest that protection from acute viral infection and disease is mediated not only by humoral (antibody-driven) but also by cellular (T cell-driven) adaptive immune responses4,5,6,7,8. In the context of vaccination, the success of the live–attenuated yellow fever vaccine (YF17D) has been attributed, at least in part, to its ability to induce a broad and polyfunctional T cell response9,10,11,12. The importance of vaccine-induced T cell immunity has been further cemented by our experience with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mRNA vaccines; as vaccine-induced neutralizing antibody titres waned, preservation of vaccine-induced T cell responses, which were less susceptible to escape mutations than neutralizing antibodies, protected against severe COVID-19 even as SARS-CoV-2 evolved13,14.
Despite the emerging body of evidence on cellular immunity being important for protection against acute viral diseases, the extent to which T cell responses are able to also control viral replication and reduce viral loads remains unclear; the reduction of viral burden would not only impact disease outcome but also reduce viral transmission. Mouse studies have previously demonstrated that T cells are able to control viral infection independent of antibodies15,16,17,18,19. However, immune-mediated protection against infection in mouse models has not invariably translated to vaccine efficacy in humans and has occasionally even resulted in immune-enhanced disease20,21.
To understand the extent that cellular immunity protects against human acute viral infections, we conducted a double-blind, randomized clinical trial using two licensed live–attenuated Orthoflavivirus vaccines—YF17D and chimeric Japanese encephalitis–YF17D (JE/YF17D) vaccines, administered either as a vaccination or challenge infection in human volunteers22. As the JE/YF17D vaccine was constructed using the YF17D genomic backbone, vaccination with either would produce homologous T cells; vaccine-induced antibodies, however, would be cross-reactive, and we and others have previously shown that such antibodies often have poor or even no cross-neutralizing activity23,24. Our primary objective was to assess the extent to which vaccine-induced T cell responses, independent of neutralizing antibodies, were able to reduce viral post-challenge RNAaemia levels. Secondary objectives included an assessment of surrogate measures of viral control including post-challenge antibody titres, seroconversion rates and symptomatic outcomes. Thus, our a priori hypothesis was that higher T cell response induced by YF17D vaccination, resulting from previously shown higher viraemia than that induced by JE/YF17D25, would control JE/YF17D infection; the lower T cell response from JE/YF17D vaccination would conversely not alter the YF17D challenge infection outcome22. Indeed, we found that YF17D vaccination, without neutralizing antibodies, was able to control JE/YF17D viral replication. More importantly, we show that high vaccine-induced T cell response to the capsid (C), but not the non-structural (NS) proteins, was associated with viral control. Our findings have bearing on future orthoflaviviral vaccine design and development.
Results
Study cohort
The proteomes of the JE/YF17D and YF17D vaccines are shown schematically in Fig. 1a. We enrolled 34 dengue-seronegative healthy adult volunteers aged 21–45 years and randomized them in a 1:1 ratio to receive either JE/YF17D vaccination on day 0 followed by YF17D heterologous challenge on day 28 (arm 1; n = 17) or YF17D vaccination on day 0 followed by JE/YF17D heterologous challenge on day 28 (arm 2; n = 17) (Fig. 1b). The study was conducted in Singapore where dengue is endemic and Zika virus, another Orthoflavivirus, has also been detected sporadically. A negative finding on an anti-dengue IgG enzyme-linked immunosorbent assay (ELISA) excluded previous immunity against either virus26,27. Baseline demographics such as age, sex, body mass index and race were similar between the two groups (Table 1). In total, 33 out of 34 participants completed the study and were included in the final analysis (Fig. 1c).
a, Pictorial representation of the YF17D and JE/YF17D vaccine constructs. JE/YF17D is constructed by splicing the prM and E genes of the Japanese encephalitis SA 14-14-2 virus (represented in blue) into the C and NS backbone of YF17D (represented in red). b, Overall study design. A total of 34 Orthoflavivirus-naive healthy volunteers were randomized to receive either JE/YF17D vaccination on day 0 followed by YF17D challenge on day 28 (arm 1) or YF17D vaccination on day 0 followed by JE/YF17D challenge on day 28 (arm 2) and followed up until day 56. c, CONSORT diagram showing the flow of study participants through the randomized–controlled trial. D, day. Panel b created by BioRender.com.
Virological and humoral responses after vaccination
We first assessed vaccine infection by measuring plasma viral RNA levels (RNAaemia) using reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR), after either JE/YF17D or YF17D vaccination. Humoral immune response to vaccination was assessed by measuring both neutralizing and binding antibodies.
RNAaemia was measured in vaccinees on days 4, 7, 10 and 14 after vaccination. Viral RNA was detected in 88% (n = 15/17) and 94% (n = 15/16) of JE/YF17D and YF17D vaccinees, respectively, on at least one sampling timepoint. Among vaccinees with detectable RNAaemia, mean plasma viral RNA levels were higher after YF17D vaccination than after JE/YF17D vaccination (mean log10(area under the curve (AUC) genome copies per ml): 4.17 versus 3.65; P = 0.012) (Fig. 2a). All JE/YF17D and YF17D vaccinees tested negative for viral RNA by day 10 and 14 post-vaccination, respectively (Extended Data Fig. 1a,b).
a, Viral RNA AUC in individuals with detectable RNAaemia after JE/YF17D (n = 15) or YF17D (n = 15) vaccination. b,c, Neutralizing antibody titres (PRNT50) against JE SA 14-14-2 (b) and YF17D (c) 28 days after JE/YF17D (n = 17) or YF17D (n = 16) vaccination. The black dashed line indicates minimum serum dilution of 1:20. PRNT50 titres < 1:20 were conservatively considered to be 1:10. d–f, Anti-JE E antibody levels (d), anti-YF E antibody levels (e) and anti-YF NS1 levels (f) 28 days after JE/YF17D (n = 17) or YF17D (n = 16) vaccination. The P/N ratio was calculated by dividing the absorbance value of the vaccine sera by that of the day 0 (naive) sera. The black dotted lines indicate a positive threshold (P/N > 1). g, Whole blood from JE/YF17D (n = 17) or YF17D (n = 16) vaccinees was stimulated with individual peptide pools covering the entire YF17D C, NS3 and NS5 proteins, and the JE and YF17D E proteins, or DMSO, after which the concentration of secreted IFNγ was measured. h–j, T cell response against YF17D C (h), NS3 (i) and NS5 (j) pre- and post-vaccination in JE/YF17D (n = 17, blue bars; mean) or YF17D (n = 16, red bars; mean) vaccinees. k, Total response was calculated by summing the responses to C, NS3 and NS5 for each individual vaccinee. l,m, T cell response against the heterologous JE E (l) and YF E (m) proteins pre- and post-vaccination in JE/YF17D (n = 17, blue bars; mean) or YF17D (n = 16, red bars; mean) vaccinees. n,o, Frequency of activated (CD38+HLA-DR+) CD4+ and CD8+ T cells on day 14 post-vaccination in a subset of JE/YF17D (n = 12) (n) and YF17D (n = 10) (o) vaccinees. Values plotted are after subtraction of the baseline (day 0 pre-vaccination) activation frequency. In all figures, the dots represent individual vaccinees. The black horizontal bars represent the mean. P values were calculated with a two-tailed paired or unpaired Student’s t-test or two-tailed unpaired Mann–Whitney U test. NS, not significant.
We quantified neutralizing antibody responses in vaccinees using a plaque reduction neutralization test (PRNT). All vaccinees seroconverted by day 28 post-vaccination, developing antibodies to their homologous virus (Fig. 2b,c), although YF17D induced higher levels of neutralizing antibodies, as measured by the antibody titre that reduced virus plaque numbers by 50% (PRNT50) than JE/YF1D (mean log10(PRNT50 titres): 4.06 versus 2.50; P < 0.0001) (Extended Data Fig. 1c). Importantly, neither JE/YF17D nor YF17D vaccination produced cross-neutralizing antibodies against the heterologous virus (Fig. 2b,c).
Aside from producing neutralizing antibodies, vaccination would also produce antibodies against the envelope (E) and NS1 proteins, which may potentially mediate protection and/or disease pathology in orthoflaviviral infection15,28,29,30,31,32. We thus measured antibodies against JE E, YF E and YF NS1 in vaccinees 28 days post-vaccination. Here we used ELISAs with a secondary antibody that detected IgM, IgA and IgG antibodies as all of these classes of antibodies are known to be present at day 28 post-vaccination33. Moreover, both anti-flaviviral IgM34 and IgA 35 antibodies can also contribute to virus clearance.
Antibodies that bound the E protein homologous to the vaccine showed levels that were comparable between the two groups of vaccinees (Extended Data Fig. 1d). It should be noted, however, that the recombinant YF E protein used in this assay was based on an isolate from Brazil, rather than YF17D. By contrast, the JE E protein was homologous to JE/YF17D. YF17D vaccination produced antibodies that bound JE E protein (Fig. 2d), although the converse was not detected (Fig. 2e). Anti-NS1 antibody levels were consistent with what we had observed with the neutralizing antibody responses, in which YF17D vaccination produced higher levels of anti-NS1 antibodies compared with JE/YF17D vaccination (mean positive-to-negative (P/N) ratio: 4.08 versus 2.74; P = 0.015) (Fig. 2f).
In summary, vaccination with YF17D produced higher RNAaemia and 100% seroconversion, higher levels of anti-NS1 antibodies and anti-E antibodies that bound JE E protein but without cross-neutralization.
Cellular immune responses after vaccination
We next assessed the functional antigen-specific T cell responses induced by JE/YF17D and YF17D vaccination, using a whole-blood cytokine release assay (CRA), which measures the concentration of secreted IFNγ in peptide-stimulated whole blood (Fig. 2g)36. Here whole blood was stimulated ex vivo using overlapping peptides covering the entire YF17D C, NS3 and NS5 proteins that are homologous between JE/YF17D and YF17D, as well as the heterologous JE and YF17D E proteins.
Both JE/YF17D and YF17D vaccinees developed antigen-specific T cell responses against the homologous C (Fig. 2h), NS3 (Fig. 2i) and NS5 (Fig. 2j) proteins by day 10 post-vaccination. T cell responses remained high on day 14 and contracted by day 28, although still present at detectable levels. Overall, the magnitude of the T cell responses was higher in YF17D vaccinees than in JE/YF17D vaccinees (Fig. 2h–k), although there was considerable heterogeneity in the magnitude of response within each group. Within individuals, responses between all three peptide pools were significantly correlated, with the strongest correlation observed between the responses against NS3 and NS5, and a slightly weaker albeit still significant correlation between the C and two NS proteins (Extended Data Fig. 2a). Both groups of vaccinees also developed virus-specific T cell responses against their homologous E proteins, with minimal to no responses against the heterologous E (Fig. 2l,m).
To ensure that the CRA findings with orthoflaviviral vaccination correlated with those measured using a more conventional T cell assay, we performed an IFNγ-based enzyme-linked immunospot (IFNγ ELISpot) in 10 vaccinees (n = 5 who received JE/YF17D and n = 5 who received YF17D) on day 14 post-vaccination. Antigen-specific T cell responses measured by IFNγ ELISpot correlated significantly with results of the CRA (Extended Data Fig. 2b).
Finally, we compared the frequency of T cell activation induced by the two vaccines in a subset of vaccinees. Here we measured the frequency of CD4+ and CD8+ T cells expressing the activation markers HLA-DR and CD38, using flow cytometry (Extended Data Fig. 2c). Overall, both JE/YF17D and YF17D induced higher frequencies of activated CD8+ T cells than CD4+ T cells on day 14 post-vaccination (Fig. 2n,o). Unlike the antigen-specific functional T cell responses, however, there were no significant differences in either the CD4+ or CD8+ T cell activation frequency between the two groups of vaccinees (Extended Data Fig. 2d,e).
Collectively, our findings show that YF17D produced greater antigen-specific T cell responses than JE/YF17D.
Outcomes of challenge infection
Next, we compared the outcomes of challenge infection between JE/YF17D and YF17D vaccinees. Here JE/YF17D vaccinees were challenged with YF17D on day 28 post-vaccination, while YF17D vaccinees were challenged with JE/YF17D (Fig. 1b). Given that both JE/YF17D and YF17D vaccination did not induce cross-neutralizing antibodies against each other (Fig. 2b,c), any difference in infection outcomes observed between the two arms would be independent of the effect of neutralizing antibodies.
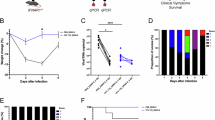
The primary outcome of this trial was the reduction in viral RNA levels (log10(AUC genome copies per ml)) after the challenge infection, compared with viral RNA levels following primary vaccination alone. As hypothesized, mean plasma JE/YF17D RNA levels were lower following challenge infection than following primary vaccination (mean log10(AUC genome copies per ml): 2.23 versus 3.22; P = 0.039) (Fig. 3a). Likewise, as hypothesized, neither mean plasma YF17D RNA levels (mean log10(AUC genome copies per ml): 4.14 versus 3.91; P = 0.33) (Fig. 3b) nor the proportion of vaccinees with RNAaemic infection (94% (n = 15/16) versus 88% (n = 15/17), P > 0.99) (Fig. 3c) was different following the YF17D challenge infection in JE/YF17D vaccinees, compared with those who received YF17D as a primary vaccination.
a, Viral RNA AUC after JE/YF17D vaccination (n = 17) versus JE/YF17D challenge (n = 16). b, Viral RNA AUC after YF17D primary vaccination (n = 16) or YF17D challenge (n = 17). c, Proportion of vaccinees with detectable RNAaemia after vaccination (upper panels) and after challenge (lower panels). d,e, Kinetics of neutralizing antibody titres (PRNT50) against JE SA 14-14-2 28 days after the JE/YF17D challenge (n = 16) (d) and against YF17D after the YF17D challenge (n = 17) (e). The grey arrows indicate the timepoint of the challenge infection. f, Comparison between neutralizing antibody titres (PRNT50) against JE SA 14-14-2 28 days after the primary JE/YF17D vaccination (n = 17) and the JE/YF17D challenge infection (n = 16). g, Comparison between neutralizing antibody titres (PRNT50) against YF17D 28 days after the primary YF17D vaccination (n = 16) and the YF17D challenge infection (n = 17). For d–g, the black dashed lines indicate a minimum serum dilution of 1:20. PRNT50 titres <1:20 were conservatively considered to be 1:10. The pink dashed lines indicate seroconversion defined as a fourfold or greater increase in PRNT50 titres from 1:10 to 1:40. h, Proportion of vaccinees in each arm who experienced systemic symptoms after vaccination (upper panels) and after challenge (lower panels). The dots or circles represent individual vaccinees. The black horizontal bars represent the mean. n represents the number of biological replicates. P values were calculated with a one-tailed unpaired Student’s t-test. NS, not significant.
Besides RNAaemia, we also assessed challenge infection outcomes using seroconversion rates and antibody titres, and compared them with primary vaccination outcomes; we have previously shown that YF17D viraemia was positively correlated with PRNT50 titres23. Seroconversion was defined as a fourfold or greater rise in PRNT50 titre, using a titre of 10 as baseline. Unlike primary JE/YF17D and YF17D vaccination that resulted in 100% seroconversion rates in vaccinees, 3 of 16 (19%) and 2 of 17 (12%) vaccinees did not seroconvert after the JE/YF17D and YF17D challenge, respectively (Fig. 3d,e). The mean JE PRNT50 titres produced from the JE/YF17D challenge were lower than those produced after primary JE/YF17D vaccination (mean log10(PRNT50 titre): 1.87 versus 2.5; P < 0.0001) (Fig. 3f). By contrast, there was no difference between the YF PRNT50 titres after the YF17D challenge and that after primary YF17D vaccination (mean log10(PRNT50 titre): 4.11 versus 4.06; P = 0.86) (Fig. 3g). In both groups, there was no further boost in the YF and JE neutralizing antibody titres after the JE/YF17D and YF17D challenge, respectively (Extended Data Fig. 3a).
All vaccinees with detectable viral RNA post-challenge had a greater than fourfold rise in PRNT50 titres (Table 2). Remarkably, all 5 participants who showed a less that fourfold rise in PRNT50 titres (n = 3/16 after the JE/YF17D challenge and n = 2/17 after the YF17D challenge) also had undetectable viral RNA post-challenge (Table 2). Indeed, overall antibody titres were significantly lower in both groups of vaccinees with undetectable RNAaemia after the heterologous challenge, compared with those with detectable RNAaemia (Extended Data Fig. 3b,c).
To determine whether the reduced RNAaemic JE/YF17D challenge infection had an impact on clinical outcomes, we compared the rate of systemic symptoms (vaccine-related adverse events) after the challenge infection with those after the primary vaccination. The most commonly reported symptoms after vaccination were lethargy or fatigue (33%, n = 11/33), myalgia (24%, n = 8/33), headache (24%, n = 8/33) and fever (12%, n = 4/33), consistent with what has been previously reported in the literature in individuals receiving JE/YF17D and YF17D vaccination25,37,38,39. While both JE/YF17D and YF17D vaccinees reported comparable rates of symptoms after primary vaccination (29% (n = 5/17) versus 31% (n = 5/16), P > 0.99) (Fig. 3h and Extended Data Table 1), fewer YF17D vaccinees reported symptoms after the JE/YF17D challenge (6% (n = 1/16)), in contrast to the YF17D challenge in JE/YF17D vaccinees (53% (n = 9/17), P = 0.007) (Fig. 3h and Extended Data Table 1).
The primary end-point of the trial was thus met in YF17D vaccinees, in which immunity from YF17D vaccination reduced the extent of JE/YF17D infection, even to the point of undetectable RNAaemia and absence of seroconversion.
Innate immune and binding antibody responses
Given that the YF17D vaccination resulted in viral control after the JE/YF17D challenge, we next conducted a series of post hoc analyses to identify the YF17D vaccination-induced immune response that controlled the JE/YF17D challenge infection.
We first analysed the expression of a panel of immune genes on the day of challenge (day 28), as well as one day after the challenge (day 29). Unsupervised clustering analysis did not reveal any difference in gene expression between vaccinees with and without detectable RNAaemia after the JE/YF17D challenge (Extended Data Fig. 4a,b), suggesting that differences in innate immune response from vaccination, which can be prolonged9,40, were not responsible for viral control. Notably, all six vaccinees who had undetectable RNAaemia post-challenge had demonstrable upregulation of innate immune genes, confirming that they did indeed receive JE/YF17D infection.
Besides innate immunity, we also found no association between vaccination-induced antibody response and RNAaemia. Both pre-challenge anti-YF NS1 antibody levels (Extended Data Fig. 4c) and pre-challenge anti-JE E antibody titres (Extended Data Fig. 4d) did not correlate with post-challenge JE/YF17D RNA levels, suggesting that viral control was not primarily being driven by anti-NS1 or cross-reactive anti-E antibodies.
As expected, there was no association between innate immune responses or binding antibody titres and YF17D RNAaemia in JE/YF17D vaccinees who received YF17D challenge infection (Extended Data Fig. 4e–h).
T cell levels correlate negatively with challenge viraemia
Conversely, the magnitude of the T cell response against C on both day 14 post-vaccination (Fig. 4a) and the day of challenge (Fig. 4b) correlated negatively with post-challenge JE/YF17D viral RNA levels. Likewise, higher T cell responses to C on study days 32 and 35 after the JE/YF17D challenge were also associated with viral control after the JE/YF17D challenge (Fig. 4c,d).
a–p, T cell responses against C (a–d), NS3 (e–h), NS5 (i–l) and the sum of the responses to all three proteins (m–p), on study days 14, 28, 32 and 35, were correlated with the level of viral RNA post-challenge in YF17D vaccinees challenged with JE/YF17D. P values were calculated with two-tailed Spearman’s correlation. Linear regression lines are shown if the correlation was significant (P < 0.05).
There was, however, no association between the T cell responses against NS3 (Fig. 4e–h) and NS5 (Fig. 4i–l), and post-challenge JE/Y17D RNA levels. When the T cell responses to C, NS3 and NS5 were summed (Fig. 4m–p), only the total T cell response before the challenge on day 28 was significantly and negatively correlated with post-challenge JE/YF17D RNA levels (Fig. 4n).
In those who were vaccinated with JE/YF17D and challenged with YF17D, we expectedly did not find any significant correlation between the T cell responses induced by JE/YF17D vaccination and post-challenge YF17D RNA levels (Extended Data Fig. 5).
Aviraemic individuals have higher C-specific T cell levels
Given that higher YF17D-vaccination-induced T cell responses, particularly against C, were associated with reduced viral RNA levels post-JE/YF17D challenge, we next examined post hoc whether the magnitude of T cell responses could differentiate between those with and without detectable post-challenge RNAaemia. Altogether, we identified 6 of 16 vaccinees after the JE/YF17D challenge and 2 of 17 vaccinees after YF17D challenge who had no detectable RNAaemia (Fig. 3c).
Notably, we found that the magnitude of the vaccine-induced T cell response against C on days 14 and 28 (Fig. 5a,b), as well as the post-challenge C-specific T cell response on days 32 and 35 (Fig. 5c,d), was higher in vaccinees with undetectable than detectable RNAaemia post-challenge. There was no difference in the magnitude of the T cell response against NS3 between both groups at any timepoint (Fig. 5e–h). T cell responses against NS5 were higher in vaccinees with undetectable RNAaemia only on day 14 (Fig. 5i) but not at any other timepoint (Fig. 5j–l), with similar findings for the total T cell response against all three proteins (Fig. 5m–p).
a–p, T cell responses against C (a–d), NS3 (e–h), NS5 (i–l) and the sum of the responses to all three proteins (m–p), on days 14, 28, 32 and 35, were compared between vaccinees with and without detectable RNAaemia post-challenge. The open black circles represent vaccinees with detectable RNAaemia post-challenge (n = 25), while the red dots represent those with undetectable RNAaemia (n = 8). The horizontal black bars represent the mean. q,r, ROC curves for comparing vaccinees with and without detectable RNAaemia post-challenge based on the day 14 (q) and day 28 (day of challenge) (r) T cell response against C. P values were calculated with a two-tailed unpaired Mann–Whitney U test. NS, not significant.
Remarkably, not only was aviraemia following the challenge infection associated with no or lower seroconversion, many of these vaccinees showed minimal and, in some cases, complete absence of expansion of antigen-specific T cell responses post-challenge (Extended Data Fig. 6a–h). Even the development of a T cell response against the E protein of the challenge virus was muted (Extended Data Fig. 6a,e), when compared with the T cell responses in vaccinees with detectable RNAaemia post-challenge (Extended Data Fig. 6i–l).
We also examined whether the finding of a higher C-specific T cell response in individuals with undetectable RNAaemia, as measured by IFNγ secretion, could also be observed with other T cell-related cytokines. Here we analysed the concentration of IL-2, granzyme B (GZB), IL-10 and TNF in C, NS3 and NS5 peptide-stimulated whole blood on day 14 post-vaccination, in a subset of 16 vaccinees (all n = 8 with undetectable RNAaemia and n = 8 with detectable RNAaemia post-challenge). Overall, there was significant intra-individual correlation between the levels of these cytokines and IFNγ (Extended Data Fig. 7a–d). Consistent with our findings with secreted IFNγ, vaccinees with undetectable post-challenge RNAaemia also had higher levels of secreted IL-2, GZB and TNF in C-peptide-stimulated whole blood compared with those with RNAaemia (Extended Data Fig. 7e). There was, however, no difference in the levels of these cytokines in NS3- and NS5-stimulated whole blood between individuals with and without detectable RNAaemia (Extended Data Fig. 7f,g), again consistent with our findings with secreted IFNγ.
Finally, we performed a receiver-operating characteristic (ROC) analysis to determine whether we could identify a minimum threshold of T cell response that was needed for viral control to the extent of undetectable RNAaemia. ROC analysis showed that both day 14 and day 28 (day of challenge) C-specific T cell responses were able to clearly distinguish between vaccinees with and without detectable post-challenge RNAaemia (mean AUC = 0.85 (95% CI 0.72–0.99) and mean AUC = 0.81 (95% CI 0.66–0.97), respectively) (Fig. 5q,r). Indeed, based on the ROC, a day 14 post-vaccination C-specific T cell response of 38.8 pg ml−1 of IFNγ in whole blood was able to differentiate between vaccinees with detectable and non-detectable post-challenge RNAaemia, with a sensitivity of 72%, specificity of 87.5% and a likelihood ratio of 5.76.
Discussion
Although the immune response to infection is multifaceted, the extent to which cellular immunity alone can control acute viral infection in humans has remained unclear. Cohort studies on COVID-19 vaccination have found that, until hybrid immunity levels of neutralizing antibody were attained, the spike-specific T cell response was the most important correlate of protection against symptomatic SARS-CoV-2 infection41. In this study, we used two orthoflaviviral vaccines that share a common genomic backbone. This approach allowed us to define experimentally the extent to which T cells control infection in the absence of neutralizing antibodies.
Mouse models have previously shown protection afforded by T cells against lethal orthoflaviviral infection15,16. These studies, however, used immunocompromised mice that do not accurately represent viral infection in immunocompetent humans. Moreover, the mouse studies measured T cells against NS3 and/or NS4b only and would have thus missed nuances in T cells against the C protein.
Although we had hypothesized that T cells, when present at sufficiently high levels after vaccination, would control challenge infection, we did not expect that the control would be to the extent consistent with sterilizing immunity; five of our trial participants had no detectable RNAaemia and no seroconversion. These same five individuals also reported no systemic symptoms following the challenge infection. Indeed, others have previously described the phenomenon of ‘abortive infection’ in the context of SARS-CoV-2, in which exposed but asymptomatic individuals did not seroconvert. However, unlike our findings of minimal to no T cell expansion following challenge infection, the individuals with ‘abortive infection’ showed expansion of antigen-specific T cell responses42. ‘Abortive infection’ in these SARS-Cov-2-exposed individuals was probably mediated by T cells that targeted only conserved epitopes in both human coronaviruses and SARS-CoV-2. As such, the degree of infection control may not have been to the extent achieved in our study, in which, except for the limited heterologous epitopes on the pre-membrane (prM) and E, the T cell epitopes between the vaccine and challenge virus were identical. Indeed, the unexpected absence of seroconversion and lack of a T cell response to JEV after the JE/YF17D challenge led us to terminate the trial after interim analysis, as a lack of immunity from JE/YF17D inoculation made further continuance of the study ethically questionable.
Our findings also have clinical implications on how sequential administration of vaccines with a shared YF17D backbone, both in terms of vaccination order and interval, may interfere with vaccine immunogenicity. Although other studies have concluded that previous YF17D vaccination does not affect immunogenicity of subsequent JE/YF17D (refs. 24,39) or dengue vaccination43, our findings suggest otherwise. Indeed, similar to our study, a previous study demonstrated that the titres of JE neutralizing antibody were significantly lower in JE/YF17D vaccinees who had received YF17D vaccination 1 month before, despite overall seroconversion rates of 90% (ref. 24). The extent to which previous YF17D vaccination affects subsequent vaccine immunogenicity may also be influenced by vaccine viral titres, which can vary from lot to lot.
The finding that vaccine-induced T cell responses against the C, but not the NS3 or NS5, proteins was associated with viral control is intriguing. Upon endocytic cell entry, the E protein is known to fuse with endosomal membrane to reveal the nucleocapsid. The mechanism of nucleocapsid disassembly to release the viral RNA into the cytoplasm for translation and replication remains inconclusive. Tantalizingly, the release of viral RNA from C may be dependent on the ubiquitylation of C (ref. 44). Although proteasomal function is not required for RNA release from C, ubiquitylated C could nonetheless be processed through the proteosomal pathway and presented on MHC class I molecules45. T cell response against C could thus be initiated even before expression and processing of the viral proteome.
Despite the findings on antiviral potency C-specific T cell responses, our findings do not conclude that NS3- and NS5-specific T cells are redundant. C-specific T cell responses could have masked the effects of NS3- and NS5-specific T cells in controlling infection from being revealed. Further experimental studies on the extent to which NS3- and NS5-specific T cells control infection will be needed. Nonetheless, the usefulness of the C-specific T cell response has substantial bearing on orthoflaviviral vaccine design; incorporation of the homologous capsid into orthoflaviviral vaccines, such as those against dengue, would be desirable for protection. Among all the Orthoflavivirus proteins, C is the least genetically conserved46, with limited C-specific T cell cross reactivity between the different Orthoflavivirus species47.
Our study has several limitations. YF17D is known to replicate to higher viraemia levels than JE/YF17D vaccination25,39. While the extent of infection shapes adaptive immune response to vaccination48, the immunity threshold needed to control YF17D infection may be higher than that needed to control JE/YF17D infection; further statistical modelling may be helpful to define the exact contribution of the T cell responses in such scenarios. We have also not measured T cell responses to the entire proteome of YF17D. Our choice of measuring T cell response to C, NS3 and NS5 proteins was because these proteins are known to be immunodominant YF proteins49,50. Attenuated viruses were used as in the challenge infection, which was done at 28 days after vaccination. It is unlikely that memory T cell responses would have been able to fully develop in this short timeframe51. Hence, although we have shown experimental evidence of T cell control of virus infection without neutralizing antibodies, the threshold of virus-specific T cells needed for long-term protection against wild-type viruses will need to be determined separately. Finally, our study was statistically powered to test the hypothesis that YF17D-specific T cells produced from vaccination would control JE/YF17D challenge infection. We found no correlation between anti-NS1 and anti-E cross-reactive antibodies, which have shown protective effects in animal models15,28,29,30,31,32, and post-challenge RNAaemia. This lack of correlation may be due to statistical factors rather than a lack of functional protection.
Collectively, our results suggest that, without neutralizing antibodies, high vaccine-induced T cell responses, especially to C, are able to control viral infection to the point of undetectable viraemia and absence of seroconversion. In addition, the C-specific T cell response that is induced 14 days after vaccination could serve as an important CoP against orthoflaviviral infection.
Methods
Clinical trial and study participant details
Study conduct and approvals
The study was approved by the SingHealth Centralised Institutional Review Board (reference: 2021/2738) and registered on ClinicalTrials.gov (NCT05568953). The full study protocol has been previously published22. The study sponsor was the Singapore General Hospital, Singapore, in collaboration with Duke-NUS Medical School, Singapore. The study was conducted at the SingHealth Investigational Medicine Unit and carried out in accordance with the principles of the Singapore Good Clinical Practice guidelines and in compliance with the Helsinki Declaration. Participants were compensated for their time and inconvenience. Study enrolment, follow-up and data collection were conducted between 30 March 2023 and 31 October 2023 at the SingHealth Investigational Medicine Unit, Singapore. Data were collected using REDCap (v13.1.30).
Study participants
Healthy adult volunteers aged 21–45 years with a negative serum anti-dengue IgG ELISA at screening were enrolled into the study, after providing written informed consent. Other inclusion criteria included a satisfactory baseline medical assessment and stable health status, willingness to use adequate and reliable contraceptive measures or practice abstinence for 10 days after vaccination, and a negative urine pregnancy test (for female volunteers of child-bearing potential) at screening and day of vaccination. Key exclusion criteria included a history of cardiovascular, respiratory, hepatic, renal, gastrointestinal, neuropsychiatric, haematological, endocrine or immunosuppressive disorders that would be a risk factor for study vaccine administration; a positive serum anti-dengue IgG ELISA; previous receipt of any yellow fever or Japanese encephalitis vaccines; known allergy to JE/YF17D or YF17D vaccines; known allergy to egg; thymus gland disease; acute infection in the preceding 7 days or presence of a temperature ≥38.0 °C on the day of the first vaccination; pregnant or breastfeeding women; receipt of anti-inflammatory drugs (such as non-steroidal anti-inflammatory drugs or systemic steroids) in the past 7 days; and receipt of any licensed vaccine in the past 30 days before the first study vaccine dose.
Study design
Participants were randomized into one of two arms in a 1:1 ratio. Both the investigators and study participants were blinded to the allocation. Participants in arm 1 received JE/YF17D vaccine (IMOJEV, Sanofi Pasteur) on day 0, followed by YF17D (STAMARIL, Sanofi Pasteur) challenge on day 28. Participants in arm 2 received YF17D vaccine on day 0 followed by JE/YF17D challenge on day 28. To ensure consistency in the antigen load delivered, all vaccines administered were from the same manufacturing lot. Participants were followed up for 56 days from the first vaccination, and blood samples were collected at multiple timepoints (both pre- and post-vaccination) for viral RNA quantification, T cell and serological analysis, and transcriptomic analysis. Participants were also monitored for vaccine-related symptoms after the vaccination and challenge, and these symptoms were recorded. A detailed study schedule, including study visit and biological sampling timepoints, can be found in the published study protocol22.
Vaccine administration
Both JE/YF17D and YF17D vaccinations were administered via subcutaneous injection into the deltoid region of the left or right arm. The second vaccination (heterologous challenge, day 28) was delivered into the ipsilateral arm as the first vaccination (day 0).
PBMC isolation
Peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient centrifugation using Ficoll-Paque. Isolated PBMCs were either studied directly or cryopreserved and stored in liquid nitrogen until used in the T cell assays.
Peptide pools
Fifteen-mer peptides that overlapped by 10 amino acids and covering each of the following YF17D proteins (C, E, NS3 and NS5) and the E protein of JE SA 14-14-2 were synthesized (GenScript).
Whole-blood CRA
A total of 320 μl of whole blood drawn on the same day was mixed with 80 μl RPMI and stimulated with peptide pools at 2 μg ml−1 or a DMSO control, following the same protocol previously described for SARS-CoV-2 (ref. 36). After 16 h of culture, the culture supernatant (plasma) was collected and stored at −80 °C until quantification of cytokines. Cytokine concentrations in the plasma were quantified using an Ella machine with microfluidic multiplex cartridges measuring IFNγ, IL-2, GZB, IL-10 and TNF following the manufacturer’s instructions (ProteinSimple). The level of cytokines present in the plasma of DMSO controls was subtracted from the corresponding peptide-pool-stimulated samples. A cutoff of 10× the lower limit of detection (1.7 pg ml−1) was used to determine a positive response.
IFNγ ELISpot assay
ELISpot plates (Millipore) were coated with human IFNγ antibody (1-D1K, Mabtech; 5 μg ml−1) overnight at 4 °C. Then, 200,000–400,000 PBMCs were seeded per well and stimulated for 18 h with peptide pools of the individual proteins (2 μg ml−1). Subsequently, the plates were developed with human biotinylated IFNγ detection antibody (7-B6-1, Mabtech; 1:2,000), followed by incubation with streptavidin-AP (Mabtech) and KPL BCIP/NBT Phosphatase Substrate (SeraCare). Spot-forming units (SFU) were quantified with ImmunoSpot. To quantify positive peptide-specific responses, 2× the mean spots of the unstimulated wells were subtracted from the peptide-stimulated wells, and the results expressed as SFU/106 PBMCs. Results were excluded if negative control wells had >30 SFC/106 PBMC or positive control wells (phorbol 12-myristate 13-acetate and ionomycin) were negative.
Activation-induced marker assay
Fresh PBMCs were stained using the yellow LIVE/DEAD fixable dead cell stain kit (Invitrogen) and anti-CD3 (clone SK7; 2:50 dilution), anti-CD4 (clone SK3; 3:50 dilution) and anti-CD8 (clone SK1; 3:50 dilution) antibodies. For analysis of the activation status, cells were additionally stained with surface markers anti-HLA-DR (clone L243; 1:20 dilution) and anti-CD38 (clone HIT2; 10:50 dilution). All samples were acquired on a CytoFLEX S (Beckman Coulter) and analysed with FlowJo software v10.10 (BD). The gating strategy is shown in Extended Data Fig. 2c.
Dengue IgG ELISA
Dengue serostatus was determined using the Abbott Panbio Dengue IgG Indirect ELISA kit according to the manufacturer’s instructions. Panbio units >11 indicate the presence of dengue-virus-specific IgG antibodies; the specificity of this ELISA for anti-dengue virus antibodies is 100%, according to the product brochure. Cross-reactive anti-Zika virus antibodies can also be detected on this ELISA with specificity above 85% (ref. 52).
PRNT
Neutralizing antibody titres induced by YF17D and JE/YF17D vaccination were measured by PRNT as described previously23,53. Briefly, 40 pfu of YF17D or JE SA 14-14-2 virus were reacted with serial twofold-diluted serum samples and incubated at 37 °C. This mixture was then added to a monolayer of baby hamster kidney cells and incubated for 1 h. The mixture was aspirated and cells were overlaid with maintenance medium with 1% carboxymethylcellulose. The viruses and cells were then incubated for 3–5 days, following which the cells were washed, fixed with 20% formaldehyde and stained with 1% crystal violet. The number of plaques was then counted and the PRNT50 titres was calculated using a sigmoid dose response curve and reported as reciprocal values.
Anti-NS1 serology
Ninety-six-well Maxisorp plates (Thermo Scientific) were coated with 1 μg ml−1 of purified recombinant YF NS1 proteins (Native Antigen Company) in 0.1 M sodium carbonate (pH 9.5) overnight at 4 °C. The plates were then washed thrice with washing buffer (PBS + 0.1% Tween-20 (PBS-T)) and blocked with blocking buffer (5% bovine serum albumin in PBS-T) for 1.5 h at room temperature with gentle shaking. Next, heat-inactivated sera from day 0 and 28 of primary vaccination were diluted 1:40 in blocking buffer, added into the respective wells and further incubated for 2 h at room temperature with gentle shaking. The plates were then washed thrice with washing buffer, using an automated washer, to remove any unspecific binding of sera components to the well. To detect IgM, IgG and IgA antibodies, secondary anti-human IgM, IgG and IgA HRP conjugated (Dako) were then added at 1:1,000 dilutions in blocking buffer and further incubated for 1 h at room temperature with gentle shaking. After 3 more times of washing, 3,3',5,5'-tetramethylbenzidine (TMB) substrate (KPL) was added into each well, and the reaction was stopped after 25 min with stopping solution (1 M H2SO4). Absorbance at 450 nm with 595 nm correction in the Varioskan Lux microplate reader (Thermo Scientific) was then recorded. After wavelength correction, the absorbance values were further subjected to blank subtraction using signal values read in the empty wells. Antibody levels were determined using the P/N ratio, which is the standard for test result interpretations in the diagnostic lab in the US Centers for Disease Control and Prevention54. This ratio was calculated by dividing the average absorbance of the technical replicate of the sample sera by the average absorbance of the technical replicate of the day 0 sera. The day 0 sera were used as the naive control as all study participants had a negative dengue IgG ELISA at enrolment; neither JE nor YF is endemic in Singapore.
Anti-envelope serology
Ninety-six-well Maxisorp plates (Thermo Scientific) were separately coated with either 1 μg ml−1 of purified recombinant YF Envelope (Native Antigen Company) or 1 μg ml−1 of purified recombinant JE Envelope (Native Antigen Company) in 0.1 M sodium carbonate (pH 9.5) overnight at 4 °C. The plates were then washed thrice with washing buffer (PBS-T) and blocked with blocking buffer (1% casein in PBS; Thermo Scientific) for 1.5 h at room temperature with gentle shaking. Next, the heat-inactivated vaccinee sera from day 0 and 28 of primary vaccination were diluted 1:10 in blocking buffer, added into the respective wells and further incubated for 2 h at room temperature with gentle shaking. The plates were then washed thrice with washing buffer, using an automated washer, to remove any unspecific binding of sera components to the well. To detect IgM, IgG and IgA antibodies, secondary anti-human IgM, IgG and IgA HRP conjugated (Dako) were then added at 1:1,000 dilution in blocking buffer and further incubated for 1 h at room temperature with gentle shaking. After three more times of washing, TMB substrate (KPL) was added into each well and the reaction was stopped after 7 min with stopping solution (1 M HCl). Absorbance at 450 nm with 595 nm correction in the Varioskan Lux microplate reader (Thermo Scientific) was then recorded. After wavelength correction, the absorbance values were further subjected to blank subtraction using signal values read in the empty wells. Antibody levels were determined using the P/N ratio, which is the standard for test result interpretations in the diagnostic laboratory of the US Centers for Disease Control and Prevention54. This ratio was calculated by dividing the average absorbance of the technical replicate of the sample sera by the average absorbance of the technical replicate of the day 0 sera. The day 0 sera were used as the naive control as all study participants had a negative dengue IgG ELISA at enrolment; neither JE nor YF is endemic in Singapore.
Viral RT-qPCR
Viral RNAaemia was measured using RT-qPCR directed against the NS5 gene of YF17D (ref. 10). Viral RNA was extracted from 200 µl of plasma samples using the Roche MagNA Pure 24 Total NA Isolation Kit, and the elution volume was 50 µl. The YF17D qPCR was conducted using the Superscript III Platinum One-Step qRT-PCR Kit (Invitrogen). Briefly, 5 µl of extracted RNA was added to 20 µl master mix containing 10 μM of YF17D-specific primers and probe as shown below:
YF17D forward: GAACAGTGATCAGGAACCCTCTCT
YF17D reverse: GGATGTTTGGTTCACAGTAAATGTG
YFV-17D probe: 5′ HEX–CTACGTGTC/ZEN/TGGAGCCCGCAGCAAT–IABkFQ 3′
The human RNase P gene55 was amplified as an internal control for successful nucleic acid extraction using the same master mix containing 10 μM of RNase P specific primers and probe as shown below:
RNase P forward: AGA TTT GGA CCT GCG AGC G
RNase P reverse: GAG CGG CTG TCT CCA CAA GT
RNase P probe: 5′ HEX–TTCTGACCT/ZEN/GAAGGCTCTGCGCG–IABkFQ 3′
Reverse transcription was performed at 50 °C for 15 min and then at 95 °C for 2 min, followed by 45 cycles of PCR amplification in a Roche LC96 RT-PCR System at an annealing temperature of 54 °C. Samples were tested in technical duplicates for YF17D and singlicates for RNase P. RNA extracted from pooled human serum matrix (Seracare) was used as an extraction control; a positive signal for RNase P from the extraction control showed the integrity of the RNA extraction. An in vitro-transcribed RNA of the YF17D NS5 gene was used to construct a standard curve ranging from 106 copies µl−1 to 1 copy µl−1, which was used to quantitate vaccine RNAaemia in plasma samples. Raw cycle threshold (Cq) values were subsequently converted into log10(genome copies per ml) for reporting purposes.
nCounter profiling of gene expression
Nanostring profiling of host response was performed using the nCounter Human Immunology v2 Panel, comprising 594 genes (the full gene list can be found at https://nanostring.com/resources/ncounter-human-immunology-v2-panel-gene-list/). RNA from whole blood was isolated from Tempus blood RNA tubes according to the manufacturer’s protocol (Tempus Spin RNA Isolation Kit, Thermo Fisher Scientific). 50 ng of RNA was hybridized to reporter and capture probe sets of the nCounter Human Immunology v2 panel (Nanostring Technologies) at 65 °C for 24 h. Hybridized samples were loaded on the nCounter cartridge, and post-hybridization steps and scanning were performed on the nCounter Sprint Profiler. RCC files were analysed using nSolver analysis software (v4.0) as per the manufacturer’s protocols. Negative and positive controls included in probe sets were used for background thresholding, and normalizing samples for differences in hybridization or sample input, respectively.
Partek Genomics Suite (v7.21) was used to analyse the transcriptomic differences between individuals with and without detectable post-challenge RNAaemia. Heatmaps were constructed using the R package (v4.3.2) pheatmap (v1.0.12).
Statistical analysis
A sample size of 28 per arm (assuming a 10% drop-out rate) was initially estimated to provide 80% power at a 5% type 1 error rate, to detect an effect size of 0.8 standard deviation in mean RNA levels on a log10 scale, between primary vaccination and challenge infection. However, a decision was made to terminate enrolment after the first 34 participants, as 5 participants did not seroconvert after JE/YF17D or YF17D challenge infection, indicating that a proportion of study participants would not be fully protected from a future JE or YF infection, despite being vaccinated.
The primary end-point of the study was the reduction in viral RNA levels (as measured by log10(RNA AUC genome copies per ml)) after the challenge infection compared with after the primary vaccination alone. This was analysed using a one-tailed Student’s t-test, as the a priori hypothesis was that high vaccine-induced T cell responses would reduce RNAaemia after heterologous challenge infection. All other analyses of continuous variables were performed using a two-tailed Student’s t-test for parametric outcomes or a Mann–Whitney U test for non-parametric outcomes. Differences in the proportion of symptomatic participants between the two arms were analysed using Fisher’s exact test. Post hoc correlation analyses were performed using Spearman’s correlation. Statistical analyses were performed using GraphPad Prism v9.0; details are provided in the figure legends. In all instances, n refers to the number of participants analysed.
Material availability statement
Requests for unique biological materials should be made via email to the corresponding authors.
Inclusion and ethics statement
All collaborators of this study who have fulfilled the criteria for authorship required by Nature Portfolio journals have been included as authors, as their participation was essential for the design and implementation of the study. All researchers involved in this study reside and work full time in Singapore, where the study was conducted. The work includes findings that are locally relevant. The research was not severely restricted or prohibited in the setting of the researchers and does not result in the stigmatization, incrimination or discrimination of the participants. The study was approved by the local institutional ethics committee, carried out in accordance with the principles of the Singapore Good Clinical Practice guidelines and in compliance with the Helsinki Declaration. Local and regional research relevant to our study was taken into account in citations.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings in this study are available in the Article. Source data are provided with this paper.
References
Watson, O. J. et al. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 22, 1293–1302 (2022).
Maslow, J. N. Challenges and solutions in the development of vaccines against emerging and neglected infectious diseases. Hum. Vaccin. Immunother. 15, 2230–2234 (2019).
Plotkin, S. A. Recent updates on correlates of vaccine-induced protection. Front. Immunol. 13, 1081107 (2023).
Braciale, T. J. & Hahn, Y. S. Immunity to viruses. Immunol. Rev. 255, 5–12 (2013).
Hope, J. L. & Bradley, L. M. Lessons in antiviral immunity. Science 371, 464–465 (2021).
Koutsakos, M. et al. SARS-CoV-2 breakthrough infection induces rapid memory and de novo T cell responses. Immunity 56, 879–92.e4 (2023).
Painter, M. M. et al. Prior vaccination promotes early activation of memory T cells and enhances immune responses during SARS-CoV-2 breakthrough infection. Nat. Immunol. 24, 1711–1724 (2023).
Kalimuddin, S. et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med 2, 682–688.e4 (2021).
Pulendran, B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat. Rev. Immunol. 9, 741–747 (2009).
Miller, J. D. et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity 28, 710–722 (2008).
Akondy, R. S. et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J. Immunol. 183, 7919–7930 (2009).
James, E. A. et al. Yellow fever vaccination elicits broad functional CD4+ T cell responses that recognize structural and nonstructural proteins. J. Virol. 87, 12794–12804 (2013).
Vardhana, S., Baldo, L., Morice, W. G. 2nd & Wherry, E. J. Understanding T cell responses to COVID-19 is essential for informing public health strategies. Sci. Immunol. 7, eabo1303 (2022).
Bertoletti, A., Le Bert, N. & Tan, A. T. SARS-CoV-2-specific T cells in the changing landscape of the COVID-19 pandemic. Immunity 55, 1764–1778 (2022).
Mishra, N. et al. A chimeric Japanese encephalitis vaccine protects against lethal yellow fever virus infection without inducing neutralizing antibodies. mBio 11, e02494-19 (2020).
Kum, D. B. et al. A chimeric yellow fever–Zika virus vaccine candidate fully protects against yellow fever virus infection in mice. Emerg. Microbes Infect. 9, 520–533 (2020).
Fumagalli, V. et al. Antibody-independent protection against heterologous SARS-CoV-2 challenge conferred by prior infection or vaccination. Nat. Immunol. 25, 633–643 (2024).
Yauch, L. E. et al. A protective role for dengue virus-specific CD8+ T cells. J. Immunol. 182, 4865–4873 (2009).
Jain, N. et al. CD8 T cells protect adult naive mice from JEV-induced morbidity via lytic function. PLoS Negl. Trop. Dis. 11, e0005329 (2017).
Kim, H. W. et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89, 422–434 (1969).
Nossal, G. J. Inactivated measles vaccine and the risk of adverse events. Bull. World Health Organ. 78, 224–225 (2000).
Kalimuddin, S. et al. An experimental medicine decipher of a minimum correlate of cellular immunity: study protocol for a double-blind randomized controlled trial. Front. Immunol. 14, 1135979 (2023).
Chan, K. R. et al. Cross-reactive antibodies enhance live attenuated virus infection for increased immunogenicity. Nat. Microbiol. 1, 16164 (2016).
Nasveld, P. E. et al. Concomitant or sequential administration of live attenuated Japanese encephalitis chimeric virus vaccine and yellow fever 17D vaccine: randomized double-blind phase II evaluation of safety and immunogenicity. Hum. Vaccin. 6, 906–914 (2010).
Monath, T. P. et al. Clinical proof of principle for ChimeriVax: recombinant live, attenuated vaccines against flavivirus infections. Vaccine 20, 1004–1018 (2002).
Priyamvada, L. et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl Acad. Sci. USA 113, 7852–7857 (2016).
Katzelnick, L. C. et al. Dengue and Zika virus infections in children elicit cross-reactive protective and enhancing antibodies that persist long term. Sci. Transl. Med. 13, eabg9478 (2021).
Schlesinger, J. J., Foltzer, M. & Chapman, S. The Fc portion of antibody to yellow fever virus NS1 is a determinant of protection against YF encephalitis in mice. Virology 192, 132–141 (1993).
Schlesinger, J. J., Brandriss, M. W., Cropp, C. B. & Monath, T. P. Protection against yellow fever in monkeys by immunization with yellow fever virus nonstructural protein NS1. J. Virol. 60, 1153–1155 (1986).
Modhiran, N. et al. A broadly protective antibody that targets the flavivirus NS1 protein. Science 371, 190–194 (2021).
Biering, S. B. et al. Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction. Science 371, 194–200 (2021).
Rathore, A. P. S. & St John, A. L. Cross-reactive immunity among flaviviruses. Front. Immunol. 11, 334 (2020).
Waickman, A. T. et al. Low-dose dengue virus 3 human challenge model: a phase 1 open-label study. Nat. Microbiol 9, 1356–1367 (2024).
Singh, T. et al. A Zika virus-specific IgM elicited in pregnancy exhibits ultrapotent neutralization. Cell 185, 4826–40.e17 (2022).
Wegman, A. D. et al. DENV-specific IgA contributes protective and non-pathologic function during antibody-dependent enhancement of DENV infection. PLoS Pathog. 19, e1011616 (2023).
Tan, A. T. et al. Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. J. Clin. Invest. 131, e152379 (2021).
Chan, C. Y. et al. Early molecular correlates of adverse events following yellow fever vaccination. JCI Insight 2, e96031 (2017).
Monath, T. P. et al. Comparative safety and immunogenicity of two yellow fever 17D vaccines (ARILVAX and YF-VAX) in a phase III multicenter, double-blind clinical trial. Am. J. Trop. Med. Hyg. 66, 533–541 (2002).
Monath, T. P. et al. Chimeric live, attenuated vaccine against Japanese encephalitis (ChimeriVax-JE): phase 2 clinical trials for safety and immunogenicity, effect of vaccine dose and schedule, and memory response to challenge with inactivated Japanese encephalitis antigen. J. Infect. Dis. 188, 1213–1230 (2003).
Querec, T. D. et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10, 116–125 (2009).
Zhong, Y. et al. Correlates of protection against symptomatic SARS-CoV-2 in vaccinated children. Nat. Med. 30, 1373–1383 (2024).
Swadling, L. et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 601, 110–117 (2022).
Guirakhoo, F. et al. Live attenuated chimeric yellow fever dengue type 2 (ChimeriVax-DEN2) vaccine: phase I clinical trial for safety and immunogenicity: effect of yellow fever pre-immunity in induction of cross neutralizing antibody responses to all 4 dengue serotypes. Hum. Vaccin. 2, 60–67 (2006).
Byk, L. A. et al. Dengue virus genome uncoating requires ubiquitination. mBio 7, e00804–e00816 (2016).
Pishesha, N., Harmand, T. J. & Ploegh, H. L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 22, 751–764 (2022).
Jones, C. T. et al. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 77, 7143–7149 (2003).
Grifoni, A. et al. T cell responses induced by attenuated flavivirus vaccination are specific and show limited cross-reactivity with other flavivirus species. J. Virol. 94, e00089-20 (2020).
Akondy, R. S. et al. Initial viral load determines the magnitude of the human CD8 T cell response to yellow fever vaccination. Proc. Natl Acad. Sci. USA 112, 3050–3055 (2015).
Stryhn, A. et al. A systematic, unbiased mapping of CD8+ and CD4+ T cell epitopes in yellow fever vaccinees. Front. Immunol. 11, 1836 (2020).
Mateus, J. et al. Identification of novel yellow fever class II epitopes in YF-17D vaccinees. Viruses 12, 1300 (2020).
Kaech, S. M., Wherry, E. J. & Ahmed, R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2, 251–262 (2002).
Medialdea-Carrera, R. et al. A systematic evaluation of IgM and IgG antibody assay accuracy in diagnosing acute Zika virus infection in Brazil: lessons relevant to emerging infections. J. Clin. Microbiol. 59, e0289320 (2021).
Low, J. G. et al. Phase 1 trial of a therapeutic anti-yellow fever virus human antibody. N. Engl. J. Med. 383, 452–459 (2020).
Chao, D. Y., Galula, J. U., Shen, W. F., Davis, B. S. & Chang, G. J. Nonstructural protein 1-specific immunoglobulin M and G antibody capture enzyme-linked immunosorbent assays in diagnosis of flaviviral infections in humans. J. Clin. Microbiol. 53, 557–566 (2015).
Kudo, E. et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. PLoS Biol. 18, e3000867 (2020).
Acknowledgements
We would like to acknowledge all study coordinators and staff of the SingHealth Investigational Medicine Unit and the Singapore General Hospital Clinical Trials Research Centre for their dedication to the study, as well as all study volunteers. We would also like to thank A. Tan and N. Le Bert for their helpful advice. This study was funded by the National Research Foundation, Singapore (award number: NRF-CRP25-2020-0003). S.K. receives salary support from the NMRC Research Training Fellowship (award number: MOH-000617-00); E.E.O. and A.B. receive salary support from the Singapore Translational Research Award (award number: MOH-001271-00 and MOH-001633-00, respectively), all administered by the National Medical Research Council of Singapore.
Author information
Authors and Affiliations
Contributions
E.E.O., S.K., J.G.L. and A.B. conceived of and/or designed the study. S.K., Y.F.Z.C., C.Y.Y.C., D.H.L.N. and J.X.Y.S. ran the clinical trial. C.Y.L.T., S.K.H., K.K., A.C., H.-C.T., A.S., A.Q.N., N.Z.H., V.C., Y.S.L. and J.X.Y. conducted the assays. S.K., C.Y.L.T., A.S., A.Q.N., K.R.C. and E.Z.O. performed the analysis. E.E.O. obtained funding for the project. S.K. wrote the first draft of the paper. All authors reviewed and approved of the paper.
Corresponding authors
Ethics declarations
Competing interests
S.K. and J.G.L. have served in advisory capacities for Takeda on dengue. A.B. is co-founder of T Cell Diagnostics, a company developing virus-specific T cell assays. E.E.O. has served in various advisory capacities for Sanofi Pasteur, Takeda, MSD, Janssen Pharmaceuticals, Novartis and Arcturus Therapeutics on dengue and vaccines. The other authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Kai Dallmeier, Andrew Fiore-Gartl, Alba Grifoni and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 RNAemia and antibody responses after vaccination.
a, Kinetics of RNAemia (genome copies/ml) after JE/YF17D vaccination. Individual vaccinees are connected by blue-dashed line. b, Kinetics of RNAemia (genome copies/ml) after YF17D vaccination. Individual vaccinees are connected by red-dashed line. c, Neutralizing antibody titers (PRNT50) against JE SA 14-14-2 and YF17D 28 days after JE/YF17D (n = 17) or YF17D (n = 16) vaccination, respectively. d, Anti-JE E antibody levels (P/N ratio) and anti-YF E antibody levels (P/N ratio) 28 days after JE/YF17D (n = 17) or YF17D (n = 16) vaccination, respectively. P/N ratio was calculated by dividing the absorbance value of the vaccine sera by that of the day 0 (naïve) sera. Dots represent individual vaccinees. Black horizontal bar represents mean. P values were calculated with two-tailed unpaired Student’s t-test or two-tailed unpaired Mann-Whitney U test. ns, not significant.
Extended Data Fig. 2 Cellular immune responses after vaccination.
a, Two-tailed Spearman’s correlation of IFN-γ concentration in whole-blood stimulated with capsid, NS3 or NS5 (314 samples from 33 participants). Dotted lines denote the 95% confidence interval. b, Two-tailed Spearman’s correlation of IFN-γ concentration in peptide-stimulated whole blood (C, NS3 or NS5) and the corresponding frequency of reactive T cells in peptide-stimulated cryopreserved PBMCs quantified by IFN-γ ELISPot (30 samples from 10 participants). Dotted lines denote the 95% confidence interval. c, Gating strategy of activation induced marker flow cytometry assay. d, Absolute frequency of activated CD4+ (left panel) and CD8+ (right panel) T cells pre- and post-JE/YF17D (n = 12, blue bars; mean) or YF17D (n = 10, red bars; mean) vaccination. e, Comparison of frequency of activated CD4+ (left panel) and CD8 + T cells (right panel) on day 14 after JE/YF17D (n = 12) and YF17D (n = 10) vaccination. Values plotted are those after subtraction of baseline (day 0 pre-vaccination) activation frequency. For (d,e), dots or circles represent individual participants. Black horizontal bar represents mean. P values were calculated with two-tailed unpaired Student’s t-test. ns, not significant.
Extended Data Fig. 3 Antibody responses after JE/YF17D or YF17D challenge.
a, Kinetics of neutralizing antibody titers (PRNT50) against YF17D after JE/YF17D challenge (left panel) and against JE SA 14-14-2 after YF17D challenge (right panel). b, Comparison of neutralizing antibody titers (PRNT50) against JE SA 14-14-2 28 days after JE/YF17D challenge infection in participants with (n = 10) or without (n = 6) detectable RNAemia. c, Comparison of neutralizing antibody titers (PRNT50) against YF17D 28 days after YF17D challenge infection in participants with (n = 15) or without (n = 2) detectable RNAemia. Black-dotted line indicates minimum serum dilution of 1:20. PRNT50 titers <1:20 were conservatively considered to be 1:10. Seroconversion defined as fourfold or greater increase in PRNT50 titers from 1:10 to 1:40, represented by pink dashed-line. Circles represent individual participants. P values were calculated with two-tailed paired or unpaired Student’s t-test. ns, not significant.
Extended Data Fig. 4 Association between innate immune response or binding antibodies and post-challenge viremia.
a,b, Heat map showing gene expression profiles of YF17D vaccinees on day 28 (day of JE/YF17D challenge) (a) and (b) day 29 (one day post-JE/YF17D challenge) from the nCounter human immunology panel. Z-scores of log2 counts are displayed. c,d, Correlation between anti-YF NS1 (c) and anti-JE E (d) antibody levels on day 28 post-YF17D vaccination (day of JE/YF17D challenge), and viral RNA levels post-JE/YF17D challenge. e,f, Heat map showing gene expression profiles of JE/YF17D vaccinees on day 28 (day of YF17D challenge) (e) and day 29 (one day post-YF17D challenge) (f) from the nCounter human immunology panel. Z-scores of raw log2 counts are displayed. g,h, Correlation between anti-YF NS1 (g) and anti-YF E (h) antibody levels on day 28 post-JE/YF17D vaccination (day of YF17D challenge), and viral RNA levels post-YF17D challenge. P values calculated with two-tailed Spearman’s correlation.
Extended Data Fig. 5 Correlation between T-cell responses and post-YF17D challenge viremia.
T cell responses against capsid (a-d), NS3 (e-h), NS5 (i-l), and the sum of the response to all three proteins (m-p), on study days 14, 28, 32 and 35 were correlated with the level of viral RNA post-challenge in JE/YF17D vaccinees challenged with YF17D. P values calculated with two-tailed Spearman’s correlation.
Extended Data Fig. 6 Kinetics of T cell response after heterologous challenge infection.
Log2 fold-change of magnitude of T cell response against E homologous to the challenge virus (top row), C (second row), NS3 (third row) and NS5 (last row), in vaccinees on days 4, 7, 10, 14 and 28 post-challenge, compared to baseline levels on day of challenge. Dotted line indicates no change from baseline. a-d, Vaccinees with no detectable RNAemia and absence of seroconversion post-challenge (n = 5). e-h, Vaccinees with no detectable viremia but who seroconverted post-challenge (n = 3). i-l, Vaccinees with detectable RNAemia post-challenge (n = 25). Each line with connected dots represents an individual participant.
Extended Data Fig. 7 Multi-cytokine antigen-specific T cell responses in vaccinees.
Correlation analysis of concentration of IFN-γ in C peptide-stimulated whole blood and the corresponding concentrations of IL-2 (a), granzyme-B (b), IL-10 (c) and TNF-α (d) (n = 16). Two-tailed Spearman’s correlation. Dotted lines denote the 95% confidence interval. e-g, Concentration of secreted cytokines in C (e), NS3 (f) and NS5 (g) stimulated whole-blood of vaccinees with (n = 8, blue bars; mean) and without (n = 8, red bars; mean) detectable RNAemia post-challenge. Circles represent individual vaccinees. P values calculated with two-tailed unpaired Mann-Whitney U test. ns, not significant.
Supplementary information
Source data
Source Data Figs. 2–5 and Extended Data Figs. 1–5 and 7
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kalimuddin, S., Tham, C.Y.L., Chan, Y.F.Z. et al. Vaccine-induced T cell responses control Orthoflavivirus challenge infection without neutralizing antibodies in humans. Nat Microbiol 10, 374–387 (2025). https://doi.org/10.1038/s41564-024-01903-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-024-01903-7
This article is cited by
-
Multi-antigen DNA vaccine targeting non-structural proteins confers robust T Cell-mediated protection against Zika virus
npj Vaccines (2026)
-
Endothelial leak and knowledge gaps: yellow fever virus non-structural protein 1 and the antibodies that bind it
Virology Journal (2025)
-
Cross-genotype immunogenicity and antibody-dependent enhancement of KD-382 dengue vaccine in flavivirus-naïve adults
npj Vaccines (2025)
-
Towards evidence-based immunology
Nature Immunology (2025)
-
Efficacy of emergency maternal MVA-ZIKV vaccination in a rapid challenge model of lethal Zika infection
npj Vaccines (2025)