Abstract
Long-term nephrocalcinosis leads to kidney injury, fibrosis, and even chronic kidney disease (CKD). Macrophage-to-myofibroblast transition (MMT) has been identified as a new mechanism in CKD, however, the effect of MMT in calcium oxalate (CaOx)-induced kidney fibrosis remains unclear. In this study, abundant MMT cells are identified by immunofluorescence (IF) and flow cytometry in kidney tissues of patients with CaOx-related CKD, a male mouse model, and CaOx-treated macrophages. Clodronate liposome (CLO)-mediated macrophage depletion attenuates fibrosis in male nephrocalcinosis mice. Transcriptomic sequencing reveals that histone methyltransferase (HMTs), EZH2, is highly expressed in nephrocalcinosis. Ezh2 inducible knock-out or inhibition by GSK-126 attenuates MMT and renal fibrosis. Mechanistically, ChIP and transcriptomic sequencing show that EZH2 inhibition reduces the enrichment of H3K27me3 on the Dusp23 gene promoter and elevates Dusp23 expression. The Co-IP and molecular docking analysis shows that DUSP23 mediates the dephosphorylation of pSMAD3 (Ser423/425). Thus, our study found that EZH2 promotes kidney fibrosis by meditating MMT via the DUSP23/SMAD3 pathway in nephrocalcinosis.

Similar content being viewed by others
Introduction
Urolithiasis is a global health concern with high incidence and recurrence rates. The general population incidence ranges from 1.7% to 14.8%1,2. Calcium oxalate (CaOx) is the most common type of nephrocalcinosis. CaOx crystal deposition can induce renal tubular epithelial injury, oxidative stress, inflammation, and various immune cell infiltration3. Incomplete or failed recovery from CaOx crystal-induced kidney injury leads to tubule dysfunction and myofibroblast activation, which ultimately results in interstitium fibrosis and chronic kidney disease (CKD)4. Compared with normal individuals, patients with nephrocalcinosis had approximately 60% increased risk for CKD in 18 years5. It is also reported that nephrocalcinosis was the leading cause of renal failure in 3.2% of dialysis patients6. Therefore, clarifying the mechanism of fibrosis after CaOx crystal-induced injury and delaying the progression of nephrocalcinosis to CKD is essential.
Renal fibrosis originates from the imbalance of synthesis and degradation of the extracellular matrix (ECM), including fibronectin and collagen. Chronic inflammation-induced myofibroblast activation is the main driving force for renal fibrosis7. Recent evidence has shown that macrophages can transition into myofibroblasts, leading to collagen deposition and fibrosis in CKD, defined as a macrophage-to-myofibroblast transition (MMT)8. The MMT cells are characterized by the coexpression of myofibroblast markers (collagen I [Col-I] or α-smooth muscle actin [α-SMA]) and macrophage markers (F4/80 or CD68)9,10. MMT cells were identified in IgA nephropathy, renal allograft rejection patients, and experimental animal models10,11. The count of MMT cells is correlated with collagen deposition and kidney function in CKD patients10. Furthermore, lineage tracing demonstrated that MMT cells accounted for 65–70% of total Col-I+ or α-SMA+ myofibroblast in experimental unilateral ureteral obstruction (UUO) induced kidney fibrosis12. These studies confirm that the MMT process is crucial in chronic inflammation-induced kidney fibrosis. However, the effect of MMT in CaOx crystal-induced kidney fibrosis remains unclear.
Previous studies reported epigenetic modification involves kidney injury and repair13,14,15. Histone methylation is one of the main mechanisms of epigenetic modification, defined as adding a methyl group on core histones by histone methyltransferase (HMTs)13. Enhancer of zeste homolog 2 (EZH2) is an HMT that catalyzes trimethylation of lysine 27 on histone H3 (H3K27me3) and mediates the transcriptional silence of target genes14. It has been reported that EZH2 was upregulated in a mouse model of UUO and CKD patients. EZH2 inhibitor, 3-DZNep, could ameliorate kidney injury and fibrosis by reducing collagen expression in acute kidney injury (AKI) mice14,15. Despite the importance of histone methylation in the kidney, the effect of EZH2 in CaOx crystal-induced kidney injury and fibrosis has yet to be fully clarified.
The present study explored the MMT in CaOx nephrocalcinosis patients and an experimental mouse model. We demonstrated that Ezh2 knockout or pharmacological inhibition reduced MMT cells and ameliorated kidney fibrosis in CaOx nephrocalcinosis mice. Mechanistically, we confirmed that EZH2 promotes kidney fibrosis by meditating MMT via the DUSP23/SMAD3 pathway.
Results
Identification of MMT in patients with nephrocalcinosis-related CKD and nephrocalcinosis mice
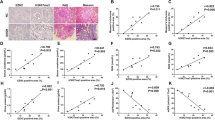
To explore the MMT in nephrocalcinosis patients, we collected kidney specimens from patients with nephrocalcinosis-related CKD. Non-tumoral adjacent kidney tissues were used as a control group. Patient demographics were presented in Supplementary Table 1. Kidney specimens were taken immediately after the nephrectomy. We used immunofluorescence (IF) to identify MMT in kidney tissues by the coexpression of myofibroblast markers (collagen I or α-SMA) and macrophage markers (CD68). In kidney tissues from patients with nephrocalcinosis-related CKD, the histopathological observation showed significant kidney atrophy, renal tubular injury, ECM deposition, and immune cell infiltration (Fig. 1A–C). The coexpressing CD68+ and α-SMA+ cells were not detected in control kidney tissues. In nephrocalcinosis, abundant coexpression of CD68+ macrophages and α-SMA+ myofibroblasts cells was observed in the renal interstitium (Fig. 1D). The CD68+α-SMA+ MMT cells contributed to 32.01% of the total α-SMA+ myofibroblast (Fig. 1E). Moreover, we found that the count of MMT cells correlated with glomerular filtration rate (GFR) of the affected side. MMT cell infiltration was associated with lower glomerular filtration rates (Fig. 1F). Three-color IF exhibited collagen I expression in CD68+α-SMA+ MMT cells in nephrocalcinosis patients (Fig. 1G).
The histopathological images showed significant kidney atrophy, renal tubular injury, ECM deposition, and immune cell infiltration in gross observation (A), HE staining (B), and Sirus rad staining (C) from patients with nephrocalcinosis-related CKD. D Two-color immunofluorescence identifies MMT cells that coexpress macrophage (CD68, green) and myofibroblast (a-SMA, red) markers in nephrocalcinosis patients. Nuclei were stained with DAPI in blue. E Quantifying the number of CD68+ cells, a-SMA+ cells, and CD68+ a-SMA+ cells (experiments with n = 6 biologically independent samples). F Correlation analysis of MMT cell populations and glomerular filtration rate in patients with nephrocalcinosis-related CKD (n = 6). G Three-color immunofluorescence identifies cells coexpressing CD68 (green), a-SMA (red), and collagen I (pink) in patients. Data are presented as the mean ± SEM. *P < 0.05 versus control group. Person correlation and two-tailed Student’s t test were performed.
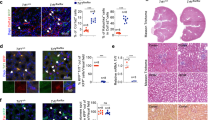
To investigate the MMT in nephrocalcinosis mice, we established a glyoxylic acid (Gly)-induced hyperoxaluria mouse model16(Fig. 2A). The deposited crystals were identified to be CaOx by infrared spectroscopy (Fig. 2B). A significant increase was observed in the level of UREA and Scr in nephrocalcinosis mice (Fig. 2C). The HE, Von Kossa, Masson staining and hydroxyproline assay showed significantly increased renal tubular injury, CaOx crystal, ECM deposition and hydroxyproline content (Fig. 2D, E). Subsequently, transcriptomic sequencing was performed to explore the differentially expressed genes between nephrocalcinosis mice (n = 3) and the control group (n = 3). A total of 3046 differentially expressed genes (DEGs) were identified (Supplementary Fig. 1A, B). We summarized the MMT regulator genes based on literature review8,9,10,11,12 (Supplementary Table 2). And 15 of the 20 MMT regulators were upregulated in nephrocalcinosis (Fig. 2F). Moreover, IF identified abundant F4/80+ α-SMA+ MMT cells in nephrocalcinosis mice, counting for 34.65% of the total α-SMA+ myofibroblast (Fig. 2G, H). Given that endothelial cells are another important source of renal myofibroblasts17, we also utilized IF to detect endothelial-to-mesenchymal transition (EndMT). The results showed that, although EndMT was activated in the kidneys of nephrocalcinosis mice, CD31+ α-SMA+ accounted for only 9.96% of the total α-SMA+ cells (Supplementary Fig. 1C, D). These results indicated that CaOx crystal deposition could induce renal injury and fibrosis. In nephrocalcinosis patients and nephrocalcinosis mice, macrophages are one of the major sources of myofibroblasts.
A Schematic diagram of the experimental procedure, Created in BioRender. Yuqi, X. (2025) https://BioRender.com/z03c224. B The deposited crystals were identified to be CaOx by infrared spectroscopy. C Renal function, body weight and hydroxyproline content were measured on day 12 of modeling (n = 6). D Representative pictures of HE, Von Kossal, and Masson staining (magnification, ×400). E Quantifying kidney tubular injury score, CaOx crystal deposition area, and collagen deposition area (n = 6). F Transcriptomic sequencing in kidney tissues between CaOx nephrocalcinosis mice and the control group (n = 3). G Two-color immunofluorescence identifies MMT cells that coexpress macrophage (CD68, green) and myofibroblast (a-SMA, red) markers in nephrocalcinosis mice. H Quantification of the number of CD68+ cells, a-SMA+ cells, and CD68+ a-SMA+ cells (n = 6). Data are presented as the mean ± SEM. *P < 0.05 versus control group. P values were determined by the two-tailed Student’s t test.
CLO-mediated macrophage depletion attenuates CaOx crystal-induced kidney injury and fibrosis
To further explore the effect of macrophage in CaOx crystal-induced kidney fibrosis, we employed the CLO to deplete macrophages. CLO could enter macrophages through endocytosis and release pro-apoptotic clodronate to target the killing of macrophages18. PBS liposomes (LO) or CLO were injected intraperitoneally three days before Gly treatment18. The body weight, UREA, and Scr levels were measured, and no substantial differences were observed between CLO-treated mice and PBS LO-treated mice (Fig. 3A, B). As expected, CLO treatment significantly reduced the F4/80+ macrophages in the renal interstitium (Fig. 3C). Interestingly, pretreatment with CLO not only improved the renal function but also attenuated the renal tubular injury, ECM deposition, hydroxyproline content and α-SMA expression induced by nephrocalcinosis (Fig. 3C). These findings revealed that macrophage meditated the injury and fibrosis in CaOx nephrocalcinosis.
A Renal function and body weight were measured on day 12 of modeling (n = 6). B Schematic diagram of the experimental procedure, Created in BioRender. Yuqi, X. (2025) https://BioRender.com/d40q387. C Representative pictures of HE, Masson, and immunohistochemical staining (magnification, ×400). D Quantifying hydroxyproline content, F4/80+ cells, collagen deposition area and α-SMA positive area (n = 6). Data are presented as the mean ± SEM. *p < 0.05 versus control group; #p < 0.05 versus CaOx nephrocalcinosis group. P values were determined by the two-tailed Student’s t test.
EZH2 is markedly elevated in CaOx crystal-induced kidney injury and fibrosis
DEGs between nephrocalcinosis mice and the control group were analyzed. GO enrichment analyses revealed that DEGs were significantly enriched in histone binding, methyltransferase activity and SMAD binding between nephrocalcinosis mice and the control group (Fig. 4A). Subsequently, we summarized the HMTs genes based on the literature review19 (Supplementary Table 3). 14 of the 48 histone methylation enzyme genes were upregulated in renal tissues from nephrocalcinosis mice (Fig. 4B). Ezh2 is the most significantly upregulated gene among them. Subsequently, the immunohistochemistry (IHC) showed that the expression of EZH2 was increased in the renal interstitium of nephrocalcinosis patients (Fig. 4C). Single-cell ATAC-seq of AKI mouse kidney indicated that Ezh2 was activated in immune cells during chronic progression of the injury (Fig. 4D). Meanwhile, IF exhibited that EZH2 have increased in F4/80+ macrophages in nephrocalcinosis mice (Fig. 4E, F). Moreover, immunoblotting showed that EZH2 were upregulated in nephrocalcinosis mice and CaOx monohydrate (COM)-treated macrophages in a dose-dependent manner, compared with the control group (Fig. 4G, J).
A GO enrichment analysis of differentially expressed genes between normal and nephrocalcinosis mice. B Heatmap of histone methylation enzyme-related gene expression (n = 3). C Representative pictures of EZH2 immunohistochemistry staining in patients with nephrocalcinosis-related CKD. D The gene activity of Ezh2 in different renal cell types from AKI mouse kidney (from snATAC-Seq database49; http://humphreyslab.com/SingleCell/). E Two-color immunofluorescence identifies EZH2 (red) was increased in F4/80+ (green) macrophages during nephrocalcinosis. F Quantifying the number of F4/80+ and F4/80+ EZH2+ cells (n = 6). Expression of EZH2 was detected by immunoblotting in nephrocalcinosis mice (G, H) and calcium oxalate monohydrate (COM)-treated macrophages (I, J) (n = 3). Data are presented as the mean ± SEM. *p < 0.05 versus control group. P values were determined by the two-tailed Student’s t test.
Ezh2 deletion decreases MMT cells and attenuates CaOx crystal-induced kidney fibrosis
To further investigate the role of EZH2 in MMT and CaOx crystal-induced kidney fibrosis, Ezh2.iKO mice were conducted. Mice with Ezh2 deletion were confirmed by immunoblotting and PCR-based genotyping (Fig. 5, Supplementary Fig. 2A). There were no differences in renal function and ECM deposition between EZH2fl/fl mice and Ezh2.iKO mice. On day 12 of the nephrocalcinosis model, EZH2fl/fl mice exhibited significant renal tubular injury, CaOx crystal, and ECM deposition (Fig. 5A–E). Importantly, Ezh2.iKO attenuated renal injury, ECM deposition, crystal deposition, hydroxyproline content, expression of α-SMA and Collagen I, and MMT cell infiltration caused by CaOx crystal (Fig. 5A–E, Supplementary Fig. 2B). Consistently, immunoblotting showed that the Collagen I, α-SMA and the fibronectin expression was downregulated in Ezh2.iKO mice, compared with that in Ezh2fl/fl mice with nephrocalcinosis (Fig. 5F). Taken together, these data suggested that deletion of Ezh2 decreases MMT cells and attenuates CaOx crystal-induced kidney injury and fibrosis.
A Renal function and body weight were measured on day 12 of modeling (n = 6). Representative pictures of HE staining, Sirus rad staining (B), Von Kossal staining (C), α-SMA and Collagen I immunohistochemistry staining (D) (n = 6). E Two-color immunofluorescence identifies MMT cells (n = 6). F EZH2, Collagen I, α-SMA and fibronectin were expressed by immunoblotting in nephrocalcinosis mice (n = 3). Data are presented as the mean ± SEM. *p < 0.05 versus Ezh2fl/fl group; #p < 0.05 versus CaOx Ezh2fl/fl group. P values were determined by the two-tailed Student’s t test.
Subsequently, macrophages were isolated from kidneys in nephrocalcinosis mice by ficoll-gradient centrifugation and FACS20,21,22 (Fig. 6A, B). Compared with the control group, the expression levels of EZH2 were upregulated in F4/80+ macrophages (Fig. 6C). Moreover, Flow cytometry and immunoblotting showed that Collagen I, α-SMA, and fibronectin were significantly increased in macrophages (Fig. 6D). Consistent with the above results, Ezh2.iKO reversed the expression of pro-fibrotic protein caused by CaOx crystal in macrophages (Fig. 6D). Taken together, these data further confirmed the effect of EZH2 in CaOx crystal-induced MMT in kidney.
A Schematic diagram of macrophage isolation, Created in BioRender. Yuqi, X. (2025) https://BioRender.com/a33h147. B Gating strategy of flow cytometry. The expression of α-SMA in isolated macrophages was detected. C Expression of EZH2 was detected by immunoblotting (n = 3). D Expression of Collagen I and fibronectin was detected by immunoblotting (n = 3). Data are presented as the mean ± SEM. *p < 0.05 versus EZH2fl/fl group; #p < 0.05 versus CaOx EZH2fl/fl group. P values were determined by the two-tailed Student’s t test.
Knockdown of Ezh2 decreases CaOx crystal-induced MMT cells in Vitro
Macrophages were treated with COM for 120 h to mimic in vivo environments. As shown in Fig. 7A, with the increase in concentration (25, 50, 100, 200 μg/ml), the morphology of macrophages changed significantly. Macrophages gradually changed from round cells in a normal state to fusiform shape, which was the most common under 200 μg/ml condition (Fig. 7A). The above studies also confirmed that the expression of EZH2 was most significantly increased under the condition of 200 μg/ml COM (Fig. 4I). Therefore, the subsequent cell experiment used 200 μg/ml COM treatment for 120 h as the cell experiment condition. Then, IF and flow cytometry analysis showed that COM addition significantly induced MMT identified by the coexpression of F4/80 and α-SMA (Fig. 7B, C). Meanwhile, the knockdown of Ezh2 by shRNA significantly decreased the count of MMT cells induced by COM (Fig. 7B, C). Consistently with the in Vivo experiment, immunoblotting showed that collagen I, α-SMA and fibronectin were upregulated in COM-treated macrophages, and knockdown of Ezh2 restored the expression of above fibrosis markers (Fig. 7D).
A Representative pictures of COM-treated macrophages. B Two-color immunofluorescence identifies MMT cells that coexpress macrophage (F4/80, green) and myofibroblast (a-SMA, red) markers in the RAW264.7 cell line (n = 5). C The proportion of MMT cells in the COM-treated macrophages was detected by flow cytometry (n = 3). D EZH2, Collagen I, α-SMA and fibronectin were expressed by immunoblotting in the RAW264.7 cell line (n = 3). Data are presented as the mean ± SEM. *p < 0.05 versus shNC group; #p < 0.05 versus COM-treated shNC group. P values were determined by the two-tailed Student’s t test.
EZH2 modulates MMT by inhibiting the expression of DUSP23 through epigenetic modification
Further, RNA-seq was conducted to investigate the targets via which EZH2 regulates MMT in nephrocalcinosis. A total of 857 differentially expressed genes were identified between wild-type (WT) mice and Ezh2.iKO mice with nephrocalcinosis (Supplementary Fig. 3). We observed that the dual specificity protein phosphatase 23 (Dusp23) was among top 5 increased genes in Ezh2.iKO mice in response to CaOx stimulation (Fig. 8A). Next, IF showed that DUSP23 was downregulated in F4/80+ macrophages of EZH2fl/fl mice with nephrocalcinosis, and upregulated after Ezh2 deletion (Fig. 8B). Mechanistically, a H3K27me3 binding site is predicted on the promoter region of Dusp23 gene by chromatin immunoprecipitation (ChIP)-Seq database, which was further confirmed by ChIP assay and ChIP-PCR that COM treatment significantly enhanced the binding of EZH2 and H3K27me3 on promoter region of Dusp23 in RAW264.7 cells (Fig. 8C). Consistently, we observed a noted upregulated H3K27me3 while downregulated levels of DUSP23 in COM-treated macrophages. Moreover, the knockdown of Ezh2 restored the expression of DUSP23 and reduced the level of H3K27me3 (Fig. 8D). In addition, Dusp23-overexpression RAW264.7 cells were established. The flow cytometry analysis showed that the overexpression of Dusp23 effectively reduced the count of F4/80+α-SMA+ MMT cells induced by COM (Fig. 8E). This indicated that EZH2 targets DUSP23 to regulate MMT via epigenetic modification.
A Top 10 differentially expressed genes between Ezh2.iKO mice and WT mice in response to CaOx stimulation (n = 3). B Two-color immunofluorescence identifies DUSP23 (red) was decreased in F4/80+(green) macrophages during nephrocalcinosis and upregulated after Ezh2 deletion. C The H3K27me3 binding site is predicted on the promoter region of the Dusp23 gene by the ChIP-Seq database. ChIP assay confirmed that COM treatment significantly enhanced the binding of EZH2 and H3K27me3 on the predicted promoter region in RAW264.7 cells (n = 3). D DUSP23, H3K27me3, and total H3 were expressed by immunoblotting in the RAW264.7 cell line (n = 3). E The proportion of MMT cells in the Dusp23 overexpression macrophages was detected by flow cytometry (n = 3). F The protein sequences of human and mouse DUSP23 or SMAD3 were analyzed by BLAST. G The molecular docking analysis in humans was conducted to predict the potential amino acid binding sites between DUSP23 and SMAD3. H Co-IP assay in COM-treated RAW264.7 macrophages confirmed the interaction of SMAD3 and DUSP23. I Expression of pSMAD3 (Ser423/425), pSMAD3 (T179), and total SAMD3 was detected by immunoblotting in macrophages (n = 3). J Expression of DUSP23, pSMAD3 (Ser423/425), SMAD3, Collagen I, and α-SMA was detected by immunoblotting in macrophages (n = 3). Data are presented as the mean ± SEM. *p < 0.05 versus control group; #p < 0.05 versus model group. P values were determined by the two-tailed Student’s t test.
DUSP23-meditated dephosphorylation of SMAD3 at Ser423/425 attenuates MMT
Previous studies demonstrate that SMAD3 plays a crucial role in MMT-meditated kidney fibrosis. To further explore the underlying mechanism by which DUSP23 regulates MMT, the interaction between DUSP23 and SMAD3 was investigated. Dusp23 and Smad3 are highly conserved, with 95.3% and 100% amino acid sequence homology between human and mouse orthologs (Fig. 8F). Furthermore, the molecular docking analysis in humans was conducted. The results revealed that proteins DUSP23 and SMAD3 formed a stable protein docking model with high ZDOCK scores (Fig. 8G). Among these binding sites, serine residues Ser423/425 of SMAD3 had the highest score for interacting with DUSP23 (Fig. 8G). The protein-protein interaction interface is presented in Fig. 8G. In mice, the molecular docking analysis gives consistent results (Supplementary Fig. 2D). Further, a Co-IP assay in COM-treated macrophages was carried out. SMAD3 was precipitated by DUSP23, and reverse Co-IP confirmed that SMAD3 could also precipitate DUSP23 in macrophages (Fig. 8H). Moreover, COM-treated RAW264.7 cells showed upregulated phosphorylation of SMAD3 (pSMAD3) at Ser423/425, and knockdown of Ezh2 suppressed pSMAD3 at Ser423/425 (Fig. 8I). Overexpression of Dusp23 effectively inhibited pSMAD3 (Ser423/425) and expression of collagen I and α-SMA in COM-treated macrophages (Fig. 8J). However, the pSMAD3 (T179) was unchanged (Fig. 8I). Together, these results implied that DUSP23 mediated dephosphorylation of SMAD3 at Ser423/425, which can attenuate MMT.
Pharmacological inhibition of EZH2 by GSK-126 ameliorates MMT-meditated kidney fibrosis in nephrocalcinosis
For exploring therapeutic agents, we assessed the effect of a highly selective inhibitor GSK-126, against EZH2 in Vivo and in Vitro. In nephrocalcinosis mice, GSK-126 significantly reduced the levels of Scr, UREA, and F4/80+α-SMA+ MMT cells and attenuated renal injury, ECM deposition, CaOx deposition, and hydroxyproline content (Fig. 9A, D, Supplementary Fig. 2C). And, GSK-126 remarkably decreased CaOx-induced expression of EZH2 and pro-fibrotic proteins (Fig. 9E). In Vitro, IF showed that GSK-126 significantly blocked the fusiform-shaped morphology as well as the MMT process (Fig. 9F). Moreover, the protein expression of EZH2, H3K27me3, pSMAD3 Ser423/425, collagen I, fibronectin, and α-SMA were inhibited by GSK-126 in COM-treated cells. At the same time, the level of DUSP23 was upregulated (Fig. 9G). These findings pharmacologically demonstrated the role of EZH2 in MMT cells and the potential of EZH2 as a therapeutic target in MMT-meditated kidney fibrosis in nephrocalcinosis.
A Renal function and body weight were measured on day 12 of modeling (n = 5). Representative pictures of HE staining, Sirus rad staining (B), and Von Kossal staining (C) (n = 5). D Two-color immunofluorescence identifies MMT cells in nephrocalcinosis mice (n = 5). E EZH2, Collagen I, α-SMA and fibronectin were expressed by immunoblotting in COM-treated macrophages (n = 3). F Two-color immunofluorescence identifies MMT cells in the RAW264.7 cell line (n = 5). G EZH2, Collagen I, α-SMA and fibronectin were expressed by immunoblotting in nephrocalcinosis mice (n = 3). Data are presented as the mean ± SEM. *p < 0.05 versus control group; #p < 0.05 versus CaOx nephrocalcinosis group. P values were determined by the two-tailed Student’s t test.
Discussion
The current study has shown that macrophages are an essential source of myofibroblasts and contribute to CaOx-induced fibrosis in nephrocalcinosis. Moreover, RNA-seq and gene knockout reveal that EZH2 is the critical regulator that promotes kidney fibrosis by meditating MMT. Mechanistically, EZH2 inhibits the expression of Dusp23 by epigenetic modification. Furthermore, DUSP23 mediated dephosphorylation of SMAD3 at Ser423/425, which can attenuate MMT.
Chronic inflammation-induced immune cell infiltration is considered to trigger renal fibrosis23. Macrophages are essential in crystal-induced kidney injury and fibrosis, but their exact role is complex due to different subsets with high functional diversity24,25. Classical and recent studies noted that macrophages could phagocytose and digest crystals present in the interstitial and intraluminal compartments25,26. Moreover, macrophages differentiate into the M1 phenotype and secrete pro-inflammatory cytokines to aggravate renal inflammation in the initial period after injury. During the repair period, M2 macrophages participate in tissue repair through the secretion of anti-inflammatory and pro-fibrotic cytokines, which promote tissue scarring25. The present study demonstrates that macrophage depletion attenuates CaOx crystal-induced kidney injury and fibrosis in the experimental mice model, indicating macrophages’ generally pro-fibrotic role.
Accumulating evidence has shown that MMT cells participate in the development and progression of multiple organ fibrosis, including kidney10, liver27, subretinal28, and pulmonary fibrosis29. The mainstream detection methods for MMT include IF, immunoblotting, flow cytometry, lineage tracing, and scRNA-seq. Firstly, Meng et al.12 and Wang et al.11 identified MMT cells by confocal microscopy and flow cytometry in the kidneys of CKD patients and UUO mice. Moreover, the lineage tracing studies using LysM-Cre/Rosa26-tdTomato transgenic mice demonstrated that MMT cells originated from bone marrow-derived macrophages (BMDMs)11,12. Then, Wang et al.10 identified MMT cells in human and experimental chronic renal allograft rejection. In Vitro, as described by Tang et al.30. TGFβ1 treatment for 5–7 days promoted the transition of macrophages into collagen-producing α-SMA+ myofibroblasts in BMDMs. Moreover, by employing scRNA-seq, Tang et al.31 reported the occurrence of MMT in the Lewis lung carcinoma model, which promoted cancer-associated fibroblast formation and disease progression. In this study, we identified abundant MMT cells by two or three-color IF and flow cytometry in kidney tissues of patients with CaOx-related CKD, nephrocalcinosis mouse model, and CaOx-treated macrophages. The RNA-seq revealed the up-regulation of MMT regulators in nephrocalcinosis mice. The most compelling finding is that macrophages isolated from kidneys in nephrocalcinosis mice were also identified as MMT cells. Moreover, MMT cell infiltration was associated with lower glomerular filtration rates in nephrocalcinosis patients. These data confirm that MMT cells are essential in CaOx crystal-induced renal fibrosis.
Mechanistically, the regulatory gene networks in the MMT process received considerable attention. Extensive research has shown that ΤGFβ1–SMAD3 signaling is the centric regulatory pathway of the MMT process31. In general, TGFβ1 facilitates the recruitment and phosphorylation of SMAD332. The activated SMAD3 then mediate the transcription of fibrosis-related genes, such as collagen I and fibronectin32. Deletion of SMAD3 in bone marrow cells reduced the number of F4/80+αSMA+ MMT cells and protected against renal fibrosis in experimental UUO and chronic renal allograft rejection mice10,12. In Vitro, TGFβ1-treated Smad3−/− BMDMs similarly presented fewer MMT cells30. In addition, many genes are involved in SMAD3-mediated MMT process. Tang et al.30,33 found tyrosine kinase Src and neural transcription factor Pou4f1 served as a direct SMAD3 target gene by scRNA-seq and ChIP assay in MMT process. Furthermore, Chen et al.34 demonstrated that P2Y12 is upregulated in macrophages with the MMT phenotype and induced by ΤGFβ1 via an SMAD3-dependent mechanism. In the current study, we revealed that Ezh2 was highly expressed in kidneys with CaOx deposition, especially in macrophages. Further study demonstrated that Ezh2.iKO or pharmacological inhibition by GSK-126 attenuated MMT and renal fibrosis in Vivo and in Vitro. Crystal deposition in the kidney is a complex process often accompanied by renal interstitial fibrosis35. Recent studies have shown that collagen deposition can further promote kidney stone formation through collagen mineralization and the expansion of crystal plaques3. Consequently, Ezh2 deletion or inhibition not only reduced collagen deposition but also prevented further crystal deposition from the early stages.
Subsequently, RNA-seq and ChIP assay were conducted, and the Dusp23 was considered the direct target gene of EZH2. DUSP23 belongs to the dual specificity protein phosphatase family, which regulates proteins’ enzymatic activity, stability, and spatiotemporal distribution36. Studies have shown that the inhibition of DUSP9, DUSP12, and DUSP26 leads to dysregulation of phosphorylation and dephosphorylation and promotes fibrosis, including renal, cardiac, and hepatic fibrosis36,37,38. DUSP23 is a protein phosphatase and mediates dephosphorylation of phosphorylated proteins on Tyr and Ser residues, such as p-ERK139. Chen et al.40 found that DUSP23 mediated dephosphorylation of glial cells missing homolog 1 (GCM1), which maintains the stability of GCM1. In this work, DUSP23 was downregulated in F4/80+ macrophages of nephrocalcinosis mice. Moreover, Dusp23 overexpression could alleviate SMAD3-mediated MMT. In addition, Co-IP, molecular docking analysis and immunoblotting showed that DUSP23 could bind with SMAD3 directly and mediate dephosphorylation of pSMAD3 (Ser423/425).
Taken together, we identify that MMT contributes to CaOx-induced fibrosis in nephrocalcinosis. EZH2 is the critical regulator that promotes kidney fibrosis by meditating MMT. Mechanistically, we confirmed that EZH2 promotes MMT via the DUSP23/SMAD3 pathway. Thus, the current study represented a potential therapeutic target for MMT-meditated kidney fibrosis in nephrocalcinosis.
Methods
Study approval
The clinical specimen collection was approved by the Ethics Committee of the Renmin Hospital of Wuhan University (approval number: WDRY2021-KS047). All ethical regulations relevant to human research participants were followed. The informed consent was obtained from the patients. We have complied with all relevant ethical regulations for animal use. The experimental animal protocol was approved by the Laboratory Animal Welfare & Ethics Committee of the Renmin Hospital of Wuhan University (approval number: WDRM-20200604).
Human nephrocalcinosis-related CKD samples
Kidney specimens and clinical data were collected from patients with nephrocalcinosis-related CKD (n = 6) and non-tumoral adjacent kidney tissues (n = 6) in Renmin Hospital of Wuhan University between November 2021 and November 2022 (details in Supplementary Table 1). Clinically indicated nephrocalcinosis-related CKD was defined if the sustained estimated GFR of <60 ML/min/1.73 m2 for >3 months with nephrocalcinosis. Exclusion criteria were as follows: the presence of CKD risk factors such as diabetic nephropathy, hypertensive nephropathy, glomerulonephritis, and severe hydronephros. The renal tissue specimen is collected immediately after surgery, rinsed with normal saline, and fixed in 4% paraformaldehyde. After 24 hours, it is embedded in paraffin and subjected to histological staining within 1 week.
Animal experiments
To avoid embryo death due to Ezh2 deletion41, we generated the tamoxifen-inducible knock out (iKO) transgenic mice. Ezh2fl/fl mice42 (Ezh2tm2Sho,Cre-negative, C57BL/6 J background, JAX:022616) were obtained from Pro. Xi Wang (Department of Immunology, School of Basic Medical Sciences; Advanced Innovation Center for Human Brain Protection, Beijing Key Laboratory for Cancer Invasion and Metastasis, Department of Oncology, Capital Medical University, Beijing, China). CAG-creER mice (stock no.:004682) were purchased from the Jackson Laboratory and used in our previous study43,44,45. CAG-CreER mice and Ezh2fl/fl mice were crossed over two generations, and PCR-based tail genotyping was performed to obtain the CAG-creEREZH2fl/fl (Ezh2.iKO) mice. Ezh2.iKO mice were conducted at six weeks of age with 75 mg/kg tamoxifen intraperitoneal injection (T5642, Sigma-Aldrich, formulated in corn oil) for five days. The primers for genotyping are as follows: Forward 5’-CCCATGTTTAAGGGCATAGTGACATG-3’, Reverse 5’-ATGTGCAGGTCAGTCAGCAACTTCAG-3’. The mice were acclimatized with free access to water and normal chow. Male mice were selected for model construction in order to improve the success rate of the model.
Male C57BL/6 J mice (weight 24–26 g, 6–8 weeks old, n = 6, each group, obtained from the Animal Experiment Center of the Renmin Hospital of Wuhan University) were treated with 120 mg/kg of glyoxylic acid (G10601, Sigma-Aldrich, formulated in saline) intraperitoneal injection once a day for 12 days to construct the CaOx nephrocalcinosis model16. For macrophage deletion, clodronate liposomes (40337ES10, Yeasen, China) or PBS liposomes were intraperitoneally injected once three days before modeling with 200 μL each mouse18. For Ezh2 inhibition, GSK-126 (500580, Sigma-Aldrich, formulated in 10% DMSO and 90% corn oil) administered intraperitoneally injection (1 mg/kg) on days −1, 1, 3, 5, 7, 9, 11 of modeling. After euthanasia by CO2 inhalation, mice kidneys were harvested. The renal samples were rinsed with saline and longitudinally bisected. One portion is rapidly frozen and stored at −80 °C for molecular biological analysis, while the other is fixed in paraformaldehyde for histological examination.
Macrophage isolation and flow cytometry
Macrophages were isolated from kidneys in Ezh2fl/fl mice and Ezh2.iKO mice by ficoll-gradient centrifugation and fluorescence-activated cell sorting (FACS)20,21,22. Briefly, kidney tissues were minced and sieved with 70-μm cell filter. Cellular suspension was added to a 10 ml tube with ficoll (1.084 g/ml) and centrifuged at 400 × g for 30 min. Cells at the ficoll interface were collected. Then, cells were incubated at 4 °C for 30 min with anti-F4/80-APC antibody (E-AB-F0995E, Elabscience). After intracellular fixation and permeabilization (554722, BD Bioscience), cells were incubated for 30 min with anti-α-SMA-PE antibody (NBP2-34522PE, Novusbio). Then, FACS sorting was conducted using a Beckman CytoFLEX with the gating strategy presented in Fig. 6B. F4/80+ macrophages were harvested for further detection. For flow cytometry in RAW264.7 cells, cells were incubated at 4 °C for 30 min with anti-F4/80-BV421 antibody (565411, BD Bioscience), and the following experimental methods are the same as above.
Cell culture and treatment
Mice macrophage cell line (RAW264.7) was purchased from Procell Life Science&Technology Co., Ltd. RAW264.7 cells were cultured in 1640 medium (R8758, Sigma-Aldrich) added with 10% fetal bovine serum and 1% antibiotics. CaOx monohydrate (COM, C124191, Aladdin) was added for 120 h with the concentration of 25, 50, 100, 200 μg/ml. The lentivirus was obtained from OBiO Technology (China). The Ezh2 shRNA sequence was 5’-CGGCTCCTCTAACCATGTTTA-3’. The negative sequence of the control was 5’-CCTAAGGTTAAGTCGCCCTCG-3’.The lentiviral expression vector was applied for Dusp23 (NM_026725.3) gene overexpression.
Histological staining
Kidney tissues of patient and mice were fixed with 4% paraformaldehyde, imbedded in paraffin and cut into 5 μm slices for the histology examination. Hematoxylin and eosin (HE), Von Kossal, Masson and Sirius red staining were performed as standard protocols16. Tubular injury was assessed by scoring (0–4) based on the percentage of injured tubular cells in HE staining. The crystal deposition area and renal fibrosis area were quantified using ImageJ software.
Hydroxyproline assay
Quantification of collagen deposition in kidney was evaluated by measuring hydroxyproline concentration46,47. The hydroxyproline assay was conducted according to the manufacturer’s instructions (A030-2-1, Nanjing Jiancheng Bioengineering Institute). Kidney tissues (60 mg) were hydrolyzed at 95 °C for 30 minutes. Subsequently, the pH of the solution was adjusted to 6.0-6.8. The reaction reagents were then added sequentially, and the mixture was heated to 60 °C for 15 minutes. The supernatant was collected, and absorbance was measured at 550 nm.
IF and immunohistochemistry staining
Kidney tissue slices and RAW264.7 cells were permeabilized with 0.2% Triton X-100 after 4% paraformaldehyde fixation. After that, samples were incubated with the primary antibodies (Supplementary Table 4) at 4 °C overnight. Then, slides were incubated with FITC- or CY3- or CY5-conjugated secondary antibodies for 60 mins and examined by fluorescence microscopy(Olympus). Images were quantified by measuring positive stained cells between groups in ImageJ software.
Immunoblotting
Proteins were extracted and quantified from kidney tissues, isolated macrophages, and RAW264.7 cells by RIPA buffer and BCA assay. In general, equivalent proteins were separated on SDS-PAGE and transferred to PVDF membrane. After blocked with 5% milk, membranes were incubated with primary antibodies (Supplementary Table 4). The membranes were incubated with secondary antibodies. Chemiluminescence was performed and integrated densities were measured by ImageJ software.
Chromatin Immunoprecipitation
The RAW264.7 cells were treated with 0 μg/ml or 200 μg/ml COM for 120 hours. Cells were collected and cross-linked (1% formaldehyde), after which cells were lysed and sonicated in SDS buffer. Then, the 10% supernatant was collected as input and lysate was incubated with EZH2 (#5246, CST), H3K27me3 (A2363, ABclonal) or lgG (#2729, CST) overnight at 4 °C. Lysates were incubated with protein A/G magnetic beads (P2078, Beyotime) for 2 h at 4 °C. Subsequently, DNA was decross-linked (NaCl 0.2 M, 65 °C, 4 h) and purified. Collected DNA was determined by PCR and qPCR using primers that targeting the predicted binding site on the promoter region of DUSP23 sequence: forward 5’-CGTGATTATTGCAGCGGGGA-3’ and reverse 5’-ATGAAGAGACCAGGCCGTTAG-3’. Enrichment was evaluated by normalizing to input.
Co-immunoprecipitation (co-IP)
The RAW264.7 cells were treated with 200 μg/ml COM for 120 hours. Proteins were extracted and quantified from RAW264.7 cells. Cell lysis solutions were incubated with anti-SMAD3 (#9523, CST) or anti-DUSP23 (108245, Genetex) overnight at 4 °C. The IgG was used as a negative control. Then, protein A-agarose (P2175, Beyotime) was used for precipitation for 2 h. After that, the immunocomplexes were eluted by boiling. The supernatant was subjected to immunoblotting.
Transcriptomic sequencing and bioinformatics analysis
The kidney tissues from normal mice, CaOx nephrocalcinosis mice, and CaOx nephrocalcinosis Ezh2.iKO mice were randomly selected for transcriptome sequencing (n = 3, each group). Transcriptome sequencing was performed by BGI Co., Ltd. Differential expression genes were determined by using the “limma” package48 in the R Software with the criteria of |log2FC | >0.75 and adjust. The “ggplot2” package and normalized data were used to draw heatmaps and volcano plots for visualization.
Single-nucleus Assay for Transposase-Accessible Chromatin (snATAC-Seq) data was obtained using the Kidney Interactive Transcriptomics database49 (http://humphreyslab.com/SingleCell/). In brief, access the aforementioned website, select the Mouse IRI scATAC-seq database, and enter the target gene name. Use the platform’s tools to visualize the scATAC-seq data. To predict the possible binding site of H3K27me3 to the Dusp23 promoter region, ChIP-Seq data of mouse kidney tissues were obtained from Cistrome DB database50,51(http://cistrome.org/db/#/).
Protein BLAST and molecular docking analysis
The protein sequences of human and mouse DUSP23 or SMAD3 were analyzed by Basic Local Alignment Search Tool (BLAST) using UniProt platform. PDB database (https://www.rcsb.org/) and Alpha Fold database (https://alphafold.com/) were used to download the protein structure files. The accession numbers are as following: DUSP23 (4erc.1.A, human); DUSP23 (Q6NT99, mouse); SMAD3 (AF-P84022-F1, human); SMAD3 (Q8BUN5, mouse). Then, the molecular docking of DUSP23 and SMAD3 was performed in ZDOCK52 (https://zdock.umassmed.edu/). The possible phosphorylation sites of SMAD3 were selected to calculate the ZDOCK Score. PyMOL software was used to visualize the analysis results.
Statistics and reproducibility
Data are presented as the mean ± SEM. Statistical analysis was conducted using the FlowJo 10.8 (BD Bioscience), ImageJ 1.46r (NIH), PyMOL 2.6 (DeLano Scientific LLC), GraphPad Prism 9.0 (GraphPad Software Inc), and SPSS 20.0 software (IBM). Student’s t test was conducted to compare differences between groups. Person correlation was performed. A P value of <0.05 was considered statistically significant. The sample size was based on previous studies. All experiments were performed with biological replicates at least three times.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
References
Zeng, G. et al. Prevalence of kidney stones in China: an ultrasonography based cross-sectional study. BJU Int. 120, 109–116 (2017).
Thongprayoon, C., Krambeck, A. E. & Rule, A. D. Determining the true burden of kidney stone disease. Nat. Rev. Nephrol. 16, 736–746 (2020).
Khan, S. R., Canales, B. K. & Dominguez-Gutierrez, P. R. Randall’s plaque and calcium oxalate stone formation: role for immunity and inflammation. Nat. Rev. Nephrol. 17, 417–433 (2021).
Venkatachalam, M. A., Weinberg, J. M., Kriz, W. & Bidani, A. K. Failed tubule recovery, AKI-CKD transition, and kidney disease progression. J. Am. Soc. Nephrol. 26, 1765–1776 (2015).
Rule, A. D. et al. Kidney stones and the risk for chronic kidney disease. Clin. J. Am. Soc. Nephrol. 4, 804–811 (2009).
Jungers, P., Joly, D., Barbey, F., Choukroun, G. & Daudon, M. Nephrolithiasis-induced ESRD: frequency, causes and prevention. Nephrol. Ther. 1, 301–310 (2005).
Yuan, Q., Tang, B. & Zhang, C. Signaling pathways of chronic kidney diseases, implications for therapeutics. Signal Transduct. Target. Ther. 7, 182 (2022).
Tang, P. M.-K., Nikolic-Paterson, D. J. & Lan, H.-Y. Macrophages: versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 15, 144–158 (2019).
Nikolic-Paterson, D. J., Wang, S. & Lan, H. Y. Macrophages promote renal fibrosis through direct and indirect mechanisms. Kidney Int. Suppl. 4, 34–38 (2014).
Wang, Y.-Y. et al. Macrophage-to-myofibroblast transition contributes to interstitial fibrosis in chronic renal allograft injury. J. Am. Soc. Nephrol. 28, 2053–2067 (2017).
Wang, S. et al. TGF-β/Smad3 signalling regulates the transition of bone marrow-derived macrophages into myofibroblasts during tissue fibrosis. Oncotarget 7, 8809–8822 (2016).
Meng, X.-M. et al. Inflammatory macrophages can transdifferentiate into myofibroblasts during renal fibrosis. Cell Death Dis. 7, e2495 (2016).
Guo, C., Dong, G., Liang, X. & Dong, Z. Epigenetic regulation in AKI and kidney repair: mechanisms and therapeutic implications. Nat. Rev. Nephrol. 15, 220–239 (2019).
Zhou, X. et al. Enhancer of zeste homolog 2 inhibition attenuates renal fibrosis by maintaining Smad7 and phosphatase and tensin homolog expression. J. Am. Soc. Nephrol. 27, 2092–2108 (2016).
Mimura, I. et al. Genome-wide analysis revealed that DZNep reduces tubulointerstitial fibrosis via down-regulation of pro-fibrotic genes. Sci. Rep. 8, 3779 (2018).
Xia, Y. et al. Construction and analysis of immune infiltration-related ceRNA network for kidney stones. Front. Genet. 12, 774155 (2021).
Lovisa, S. et al. Endothelial-to-mesenchymal transition compromises vascular integrity to induce Myc-mediated metabolic reprogramming in kidney fibrosis. Sci. Signal. 13, eaaz2597 (2020).
Belliere, J. et al. Specific macrophage subtypes influence the progression of rhabdomyolysis-induced kidney injury. J. Am. Soc. Nephrol. 26, 1363–1377 (2015).
Greer, E. L. & Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 (2012).
Alfaro, C. et al. Tumor-produced interleukin-8 attracts human myeloid-derived suppressor cells and elicits extrusion of neutrophil extracellular traps (NETs). Clin. Cancer Res. 22, 3924–3936 (2016).
Ribas, J. L. C. et al. Effects of anti-inflammatory drugs in primary kidney cell culture of a freshwater fish. Fish. Shellfish Immunol. 40, 296–303 (2014).
Wen, Y. et al. KLF4 in macrophages attenuates TNFα-mediated kidney injury and fibrosis. J. Am. Soc. Nephrol. 30, 1925–1938 (2019).
Dhana, E., Ludwig-Portugall, I. & Kurts, C. Role of immune cells in crystal-induced kidney fibrosis. Matrix Biol. 68–69, 280–292 (2018).
Hammerich, L. & Tacke, F. Hepatic inflammatory responses in liver fibrosis. Nat. Rev. Gastroenterol. Hepatol. 20, 633–646 (2023).
Mulay, S. R. & Anders, H.-J. Crystal nephropathies: mechanisms of crystal-induced kidney injury. Nat. Rev. Nephrol. 13, 226–240 (2017).
He, J. et al. Renal macrophages monitor and remove particles from urine to prevent tubule obstruction. Immunity 57, 106–123.e7 (2024).
Xia, S. et al. Role of macrophage-to-myofibroblast transition in chronic liver injury and liver fibrosis. Eur. J. Med. Res. 28, 502 (2023).
Little, K. et al. Macrophage to myofibroblast transition contributes to subretinal fibrosis secondary to neovascular age-related macular degeneration. J. Neuroinflamm. 17, 355 (2020).
Geng, F. et al. Quercetin alleviates pulmonary fibrosis in silicotic mice by inhibiting macrophage transition and TGF-β-Smad2/3 pathway. Curr. Issues Mol. Biol. 45, 3087–3101 (2023).
Tang, P. M.-K. et al. The proto-oncogene tyrosine protein kinase Src is essential for macrophage-myofibroblast transition during renal scarring. Kidney Int. 93, 173–187 (2018).
Tang, P. C.-T. et al. Smad3 promotes cancer-associated fibroblasts generation via macrophage-myofibroblast transition. Adv. Sci.9, e2101235 (2022).
Meng, X.-M., Nikolic-Paterson, D. J. & Lan, H. Y. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338 (2016).
Tang, P. M.-K. et al. Neural transcription factor Pou4f1 promotes renal fibrosis via macrophage-myofibroblast transition. Proc. Natl. Acad. Sci. USA 117, 20741–20752 (2020).
Chen, J. et al. P2Y12 inhibitor clopidogrel inhibits renal fibrosis by blocking macrophage-to-myofibroblast transition. Mol. Ther. 30, 3017–3033 (2022).
Evan, A., Lingeman, J., Coe, F. L. & Worcester, E. Randall’s plaque: pathogenesis and role in calcium oxalate nephrolithiasis. Kidney Int. 69, 1313–1318 (2006).
Huang, F., Sheng, X.-X. & Zhang, H.-J. DUSP26 regulates podocyte oxidative stress and fibrosis in a mouse model with diabetic nephropathy through the mediation of ROS. Biochem. Biophys. Res. Commun. 515, 410–416 (2019).
Li, W.-M. et al. Dual specific phosphatase 12 ameliorates cardiac hypertrophy in response to pressure overload. Clin. Sci.131, 141–154 (2017).
Ye, P. et al. Dual-specificity phosphatase 9 protects against nonalcoholic fatty liver disease in mice through ASK1 suppression. Hepatology 69, 76–93 (2019).
Wu, Q. et al. Molecular cloning and characterization of a novel dual-specificity phosphatase 23 gene from human fetal brain. Int. J. Biochem. Cell Biol. 36, 1542–1553 (2004).
Lin, F.-Y. et al. Dual-specificity phosphatase 23 mediates GCM1 dephosphorylation and activation. Nucleic Acids Res. 39, 848–861 (2011).
Shen, X. et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 (2008).
Simon, C. et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 26, 651–656 (2012).
Yang, Y. et al. Interactions between macrophages and cyst-lining epithelial cells promote kidney cyst growth in Pkd1-deficient mice. J. Am. Soc. Nephrol. 29, 2310–2325 (2018).
Sun, Y. et al. Activation of P-TEFb by cAMP-PKA signaling in autosomal dominant polycystic kidney disease. Sci. Adv. 5, eaaw3593 (2019).
Li, B. et al. Histone H3K27 methyltransferase EZH2 regulates apoptotic and inflammatory responses in sepsis-induced AKI. Theranostics 13, 1860–1875 (2023).
Krones, E. et al. NorUrsodeoxycholic acid ameliorates cholemic nephropathy in bile duct ligated mice. J. Hepatol. 67, 110–119 (2017).
Fickert, P. et al. Bile acids trigger cholemic nephropathy in common bile-duct-ligated mice. Hepatology 58, 2056–2069 (2013).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Muto, Y. et al. Epigenetic reprogramming driving successful and failed repair in acute kidney injury. Sci. Adv. 10, eado2849 (2024).
Zheng, R. et al. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 47, D729–D735 (2019).
Mei, S. et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 45, D658–D662 (2017).
Pierce, B. G. et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 (2014).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China 82370765 (to F.C.), 82400895 (to Y.X.), 82170775 (to F.C.), and 82270802 (to X.Z.). The authors thank Prof. Mingxia Fan and the staff from the Animal Experiment Center at the Renmin Hospital of Wuhan University for their technical support. The graphic abstract and featured image were created in BioRender. Yuqi, X. (2025) https://BioRender.com/w62v979.
Author information
Authors and Affiliations
Contributions
F.C., X.Z., Y.X., and Z.Y. designed the study. Y.X., Z.Y., B.L., X.Y., T.Y., L.L., B.S. performed the main experiments and collected the data. W.Y., T.R., J.Z., J.N., X.L., S.M., and Z.M. participated in some experiments and analyzed the data. Y.X., Z.Y. wrote the manuscript. F.C. and X.Z. revised the manuscript and approved the manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Chandraiah Godugu, Guoping Zheng, and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary handling editors Rosie Bunton-Stasyshyn & David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xia, Y., Ye, Z., Li, B. et al. EZH2-mediated macrophage-to-myofibroblast transition contributes to calcium oxalate crystal-induced kidney fibrosis. Commun Biol 8, 286 (2025). https://doi.org/10.1038/s42003-025-07735-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07735-3