Abstract
Immune checkpoint inhibitors (ICI) benefit some cancer patients but de novo resistance remains poorly understood. Analyzing transcriptional data from two clinical trial cohorts, GO30140 and IMbrave150, we find B cell lymphoma 9 (BCL9), a Wnt/β-catenin co-factor, associated with resistance. We develop a BCL9-targeting peptide, hsBCL9Z96, which suppresses tumor growth in combination with anti-PD-L1 ab in preclinical hepatocellular carcinoma (HCC) mouse models. Multi-omics analyses implicate targeting BCL9 inhibits BMP4 secretion and downregulates CD24 on tumor cells, reprogramming macrophages toward a tumor-suppressive phenotype and promoting macrophage phagocytosis. This in turn rejuvenates T cell immunity via enhanced macrophage-mediated antigen presentation. Our data extend our understanding of how tumor-derived Wnt/β-catenin signaling impedes the innate and adaptive immune responses in the tumor microenvironment and provide preliminary evidence that targeting BCL9 is a promising preclinical strategy to mitigate ICI resistance in HCC.
Similar content being viewed by others
Introduction
Immune checkpoint inhibitors (ICI) have recently revolutionized the field of clinical oncology, demonstrating remarkable efficacy against a variety of solid tumors1. However, in hepatocellular carcinoma (HCC), which ranks sixth and third in incidence and mortality worldwide, respectively2, the objective response rate (ORR) of single-agent anti-programmed cell death protein 1 (PD-1)/programmed cell death ligand-1 (PD-L1) antibody (ab) remains unsatisfactory. Only ~15–20% of HCCs respond as a result of de novo resistance3. Furthermore, those who do benefit from ICI treatment will inevitably experience disease progression due to the emergence of acquired resistance3,4. Despite extensive research into biomarkers that might predict the efficacy of ICI in HCC, such as the microbiome, tumor mutation burden (TMB), microsatellite instability (MSI), and PD-L1 expression, none has been proven accurate enough to be routinely used in the clinic5,6,7. Therefore, there is an urgent and unmet medical need to identify more effective “mechanistic indicators” that can explore potential targets to enhance therapeutic response for ICI treatment and serve as potential targets to enhance the therapeutic response or mitigate ICI resistance.
The Wnt/β-catenin pathway is a key oncogenic signal in multiple cancers, especially HCC8,9,10. Our previous investigation revealed that a hyperactivated Wnt/β-catenin pathway is critical in promoting hepatocarcinogenesis and represents a core molecular event during HCC development11,12. Furthermore, a desert (immune deficient) phenotype for the tumor immune microenvironment (TIME) was identified in HCC with Wnt/β-catenin pathway dysregulation, highlighting the feasibility of targeting this pathway to mitigate ICI resistance13. Pharmacologically, β-catenin lacks DNA-binding and transcriptional activity, which makes it difficult to target directly14. Additionally, blocking Wnt/β-catenin signaling by various Wnt signaling inhibitors is associated with significant limitations, such as a narrow therapeutic window, weak penetration potential, or intolerable toxicity, which hampers the clinical development of these agents15. Therefore, there remains an urgent need to develop clinically translatable Wnt/β-catenin pathway inhibitors to mitigate ICI resistance and improve the prognosis for patients with unresectable HCC.
A recent study by our group demonstrated that B cell lymphoma 9 (BCL9) is not only a core coactivator for β-catenin-induced transcription factor, but also a critical factor that drives HCC progression and is associated with an immune-excluded TIME12. This suggests that BCL9 might contribute to alternative targeting approaches to mitigate ICI resistance. Indeed, our previous study showed that targeting BCL9 can significantly sensitize colorectal cancer to ICI treatment via selectively inhibiting β-catenin signaling16. However, the composition of the TIME is different between colon cancer and HCC17. Thus, further research is needed to determine whether BCL9 can remodel the TIME as well as whether it can serve as a target to mitigate ICI resistance in patients with HCC.
In this study, we show that tumor-intrinsic BCL9 expression correlates with resistance to ICI treatment in solid tumors. The genetic or pharmacological inhibition of BCL9 can significantly enhance antitumor innate immunity through reprogramming tumor-infiltrating macrophages toward a phenotype with improved phagocytosis and antigen presentation. A highly selective BCL9/β-catenin inhibitor with low toxicity and improved tumor-penetrating capacity, hsBCL9Z96, is developed and used to investigate its function in mitigating ICI resistance in HCC. Preclinical evidence suggests that BCL9-mediated immune reprogramming via specific inhibition can mitigate ICI resistance in HCC.
Results
BCL9 expression correlates with de novo resistance to PD-(L)1-based treatment in HCC
To explore mechanisms for ICI-based immunotherapy resistance in HCC, we conducted an integrative analysis of RNA-sequencing (RNA-seq) data between responders [defined as complete response (CR) or partial response (PR)] and non-responders [defined as stable disease (SD) or progressive disease (PD)] from patients who received atezolizumab (Atezo, anti-PD-L1 ab) + bevacizumab (Bev, anti-VEGF ab), as part of the GO30140 and IMbrave150 cohorts6. Three immunotherapy validation cohorts were also included from our centre: an Atezo + Bev cohort (n = 70), a PD-1 plus tyrosine kinase inhibitors (TKIs) cohort (n = 44), and a patient-derived organotypic tumor spheroids (PDOTS) cohort (n = 77) (Fig. 1A).
A Schematic flowchart of the BCL9 study selection. B Enrichment curves for hallmark Wnt/β-catenin signaling in non-responders versus responders to Atezo + Bev combination therapy in the GO30140 + IMbrave150 cohorts. NES normalized enrichment score, FDR false discovery rate. C Venn diagram representing overlapping genes of the Wnt/β-catenin signaling pathway that were differentially expressed in the transcriptomic data for Atezo + Bev PD vs CR from the GO30140 + IMBrave150 cohorts and CPTAC HCC proteomic data for tumor vs normal tissue. D Waterfall plot showing the patient response to Atezo + Bev from a cohort of 70 patients with advanced HCC treated at Zhongshan hospital (classified according to the RECIST 1.1 criteria). E The ROC curve for BCL9 and EP300 in predicting the response to Atezo + Bev immunotherapy. F The PFS of HCC patients receiving Atezo + Bev with different BCL9 expression levels (n = 70, P value was calculated by the two-sided Log-rank test). G Correlation analysis of BCL9 expression and treatment response in HCC patients receiving Atezo + Bev treatment (n = 70). The association was evaluated using a two-sided chi-square test of independence with SPSS software. H Enrichment of the indicated indicators of a favorable response to immunotherapy, as calculated by the ssGSEA, in the BCL9 low versus BCL9 high samples in the GO30140 + IMBrave150 cohorts. Data are from n = 205 biologically independent patients, unpaired two-tailed t-test. The whiskers represent the minimum and maximum values observed within the range of Q1 − 1.5 × IQR to Q3 + 1.5 × IQR. The box shows the interquartile range (IQR) between the 25th (Q1) and 75th (Q3) percentiles, and the line inside the box indicates the median (50th percentile). I BCL9 IHC staining and representative MRI. J Schematic representation of the HCC patient-derived organotypic tumor spheroid assay. K Viability of PDOTS following anti-PD-1 treatment, and the BCL9 expression levels of corresponding tumor tissues. L The correlation between the PD-1 kill index and BCL9 expression levels (n = 77). A two-sided spearman’s rank correlation test was performed. The shaded band indicates the 95% confidence interval of the regression fit. Schematic in A was created in BioRender. Wu, S. (2025) https://BioRender.com/fo53mwm. Schematic in J was created in BioRender. Wu, S. (2025) https://BioRender.com/l38givb.
When we focused on the intracellular signaling transduction pathway involved in immunotherapy resistance, which lacks systematic analysis in our previous research, it came to our attention that Wnt/β-catenin signaling was the only enriched signaling pathway in non-responders (Figs. 1B and S1A and Supplementary Table 1). Differentially expressed genes analysis between the PD and CR subgroups identified 7 genes involved in Wnt/β-catenin signaling that were significantly upregulated in the PD subgroups, two of which (BCL9 and EP300) were also highly expressed in tumor tissues from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) HCC cohort11 (Fig. 1C).
Further validation of BCL9 and EP300 was conducted against tumor samples from a cohort of 70 patients with advanced HCC in our hospital who received Atezo + Bev treatment. Approximately 32.9%, 47.1%, and 20.0% of patients were identified as having a PR, SD, and PD, respectively, according to the RECIST 1.1 criteria (Fig. 1D). BCL9 expression showed an association with immunotherapy response (area under the curve, AUC = 0.80), higher than EP300 (AUC = 0.69, Fig. 1E). Besides, BCL9 expression correlated more significantly with progression-free survival (PFS) (Figs. 1F and S1B) and response rate (Figs. 1G and S1C) to Atezo + Bev immunotherapy than EP300. To further assess the link between BCL9 and Wnt/β-catenin activity in HCC, we performed immunohistochemistry (IHC) staining of β-catenin from Atezo + Bev-treated patients’ tumor. Tumor with low BCL9 expression showed predominantly membranous localization, while those with high BCL9 expression exhibited mainly nuclear β-catenin (Fig. S1D). Further analysis revealed that BCL9 expression with a median cutoff was negatively correlated with several indicators of a favorable response to ICI, including the inflammatory response, IFN-α response, IFN-γ response, antigen processing and presentation pathway, and cytotoxic T lymphocyte (CTL) (Fig. 1H). The AUC for BCL9 expression demonstrated a stronger correlation than either PD-1 (0.59) or PD-L1 (0.57) expression alone in this cohort (Fig. S1E), suggesting that BCL9 may reflect resistance features beyond these markers. In addition, low expression of BCL9 was also correlated with a better response to ICI in patients with melanoma (Fig. S1F).
Representative cases illustrated the trend; patient 1 (low BCL9) was diagnosed with multifocal HCC, and his tumor was significantly reduced after three cycles of Atezo + Bev treatment (Fig. 1I, left and Fig. S1G, left). In contrast, patient 2 (high BCL9) was diagnosed to progress and have multiple HCCs with metastases in the liver, and he displayed no response toward Atezo + Bev treatment (Fig. 1I, right and Fig. S1G, right). We explored the correlation of BCL9 with the response to ICI in another independent cohort of 44 patients with advanced HCC who received anti-PD-1 ab plus TKIs treatment. High tumoral BCL9 expression showed an association with a reduced response rate than that in patients with low tumoral BCL9 expression in this cohort as well (Fig. S1H). Two typical cases are shown in Fig. S1I.
BCL9 expression at both mRNA and protein levels was assessed using qRT-PCR and IHC, respectively, in the Zhongshan T + A cohort (n = 70) and Zhongshan PD-1-based ICI cohort (n = 44). BCL9 expression was significantly lower in responders compared to non-responders (Fig. S1J, K). In the GO30140 + IMbrave150 cohort, CR exhibited markedly lower BCL9 expression than patients with PD (Fig. S1L).
HCC patient-derived PDOTS were previously used to evaluate the efficacy of ICI in vitro18. We established HCC PDOTS (n = 77) to validate the correlation of the BCL9 expression level with the response to anti-PD-1 ab treatment (Fig. 1J). As expected, the killing index of the anti-PD-1 ab on the HCC PDOTS was negatively correlated with their expression of BCL9 (Fig. 1K, L). Collectively, our data support that BCL9 expression correlates with a poor clinical response to PD-(L)1-based immunotherapy in HCC.
High BCL9 expression is associated with an immune-suppressive microenvironment in HCC
To ascertain the role of BCL9 in the resistance to cancer immunotherapy, we further explored the relationship between the expression of BCL9 and the tumor microenvironments in human HCC. Immunohistochemical staining of patient specimens demonstrated that BCL9 was significantly associated with macrophages, as evidenced by the negative correlation between BCL9 expression and CD68+ macrophage density (Fig. 2A, B). We further evaluated the association between BCL9 expression and relative immune cell abundance estimated algorithmically by ESTIMATE, TIMER, and ssGSEA for a pan-cancer population in the Cancer Genome Atlas (TCGA) dataset19. Compared with the patients with low BCL9 expression, the patients with high BCL9 expression showed relatively low immune cell abundance, including activated CD8+ T cells and macrophages (Fig. 2C). To further clarify whether this pattern reflects immune depletion or active suppression, we analyzed the correlation between BCL9 expression and a panel of immunomodulatory genes across TCGA cancer types. BCL9 expression was positively correlated with immunosuppressive genes (such as VTCN1), and negatively correlated with antigen presentation-related genes (such as HLA-A) and immune effector genes (such as IFNG), which are typically associated with an active immune environment (Fig. S1M).
A IHC staining for CD3+ T cells (CD3+), CD4+ T cells (CD4+), CD8+ T cells (CD8+), macrophages (CD68+), and regulatory T cells (FOXP3+) in two representative patients with different BCL9 expression levels treated at Zhongshan Hospital. B Correlation analysis of immune cell infiltration and BCL9 expression in patients from Zhongshan Hospital was performed using two-sided Pearson correlation. The shaded bands indicate the 95% confidence intervals of the regression fits. C Heatmap displaying the correlations of BCL9 expression with immune cell abundances across the TCGA pan-cancer datasets. A two-sided spearman’s rank correlation test was performed for BCL9 and indicated immune cell abundances. D, E Enrichments of the indicated immune cell populations, estimated by TIMER, in BCL9 low versus BCL9 high samples, from the TCGA LIHC cohort (D, data are from n = 186 biologically independent patients) and Zhongshan HCC patients (E, data are from n = 24 biologically independent patients). For each cell type, the comparison between BCL9 low and high groups was performed using an unpaired two-sided student’s t-test. The whiskers represent the minimum and maximum values observed within the range of Q1 - 1.5 × IQR to Q3 + 1.5 × IQR. The box shows the IQR between the 25th (Q1) and 75th (Q3) percentiles, and the line inside the box indicates the median (50th percentile). F Single-cell sequencing analysis (GSE151530) identified differentially expressed CD8 T cell signature scores between patients with high and low BCL9 expression. Data are from n = 46 biologically independent samples and are compared using unpaired two-sided t-tests (Welch’s correction was applied for comparisons with unequal variance for Activated CD8 T cell). G Representative IHC staining of low BCL9 expression in a “hot” tumor, and high BCL9 expression in a “cold” tumor. P peri-tumor, T tumor. H Correlation analysis of BCL9 expression and tumor-immune classification in our Zhongshan cohort (“hot” or “cold” tumors, n = 55). The association was evaluated using a two-sided chi-square test of independence with SPSS software. I, J Tumor growth curves for immunodeficient mice (NCG) and immunocompetent mice (C57BL/6J or BALB/c) implanted with subcutaneous liver cancer cells (Hepa1-6/H22 Bcl9-KO or Hepa1-6/H22 Ctrl-KO cells) that were allowed to grow for 2 weeks (Data are presented as mean ± SD from n = 6 biological replicate). K Quantification of CD8+ T cells in Hepa1-6 Ctrl-KO and Hepa1-6 Bcl9-KO tumors from C57BL/6J mice. (Data are from n = 3 biologically independent mice in each group and presented as mean values +/− SD.).
These patterns were consistently observed across multiple tumor types, suggesting that elevated BCL9 expression may be associated with an immunosuppressive transcriptional profile, beyond reduced immune infiltration alone. Cohorts from both the TCGA LIHC and Zhongshan Hospital consistently confirmed that the infiltrating scores for macrophages, but not other cells, were both elevated in patients with low BCL9 expression (Fig. 2D, E). We also analyzed public single-cell RNA-sequencing (scRNA-seq) data for liver cancer (GSE151530)20, and T cells derived from high BCL9 tumor tissues were frequently found to exhibit an exhausted phenotype, as evidenced by the downregulation of signature score involved in activated CD8 T cell and effector memory CD8 T cell, and the upregulation of signature score for exhausted CD8 T cell (Fig. 2F). Interestingly, only 22.2% (6/27) of patients with low BCL9 expression were considered to have “cold” HCC tumors, while 60.7% (17/28) of patients with high BCL9 expression were considered to have “cold” HCC tumors (Fig. 2G, H). These data suggested a potential immunosuppressive role for tumoral BCL9 in HCC.
Next, we established orthotopic Hepa1-6-bearing and H22-bearing mouse liver cancer models in both immunodeficient NOD clustered regularly interspaced short palindromic repeats (CRISPR) Prkdc II2r Gamma (NCG) mice and immunocompetent C57BL/6J mice to evaluate the extent to which tumor immunity contributes to BCL9-dependent tumor progression. Knocking out BCL9 significantly suppressed tumor growth in both orthotopic models in NCG mice, with tumor growth inhibition rates (TGI) of ~25% (Fig. 2I). However, Bcl9 knockout (KO) led to more substantial tumor growth inhibition of Hepa1-6 tumors implanted in wild-type C57BL/6J mice and H22 tumors growing in wild-type BALB/c mice (both TGI were ~50%), suggesting potential immune-mediated contributions to antitumor effects (Fig. 2J). Consistent with our clinical findings, we observed intra-tumoral infiltration of CD8+ T cells in Bcl9-KO tumors compared to Ctrl-KO (Fig. 2K).
These findings extended the current knowledge regarding the crucial role of BCL9 in regulating the TIME, as well as maintaining an unfavourable “cold” TIME, which suggests that targeting BCL9 merits investigation as a strategy to potentially mitigate ICI resistance and improve the efficacy of PD-(L)1-based immunotherapy for HCC.
Design and characterization of a tumor-penetrating peptide hsBCL9Z96 targeting BCL9
To investigate the feasibility of targeting BCL9 to overcome drug resistance, we developed hsBCL9Z96, a peptide that directly targets BCL9/β-catenin (Fig. 3A). Computational docking of hsBCL9Z96 with β-catenin identified key binding sites in relevant hydrophobic regions (Figs. 3B and S2A). hsBCL9Z96 showed increased potency to disrupt β-catenin/BCL9 binding compared to a BCL9-HD2 linear peptide (Figs. 3C, D and S2B, C). hsBCL9Z96 showed higher activity than Wnt/β-catenin antagonist ICG-001, as evidenced by the LEF/TCF reporter assay (IC50: 0.247 μM vs. 1.272 μM, respectively, Fig. 3E). We then screened other BCL family members and measured the binding capacity of hsBCL9Z96 to these proteins. As shown in Fig. S2D–G, hsBCL9Z96 barely bound other BCL family proteins. Moreover, hsBCL9Z96 demonstrated stronger growth inhibition than hsBCL9CT-24 (a stapled peptide of BCL9/β-catenin previously reported by our team16) (Fig. S2H). Collectively, these in vitro data suggest that hsBCL9Z96 has superior in vitro binding and β-catenin inhibition compared to other synthetic inhibitors.
A Structures of hsBCL9Z96 (right) and hsBCL9CT-24 (left). B Docking of hsBCL9Z96 (yellow) into the β-catenin hydrophobic pocket (blue) as illustrated using GlideXP Maestro Schrodinger. C hsBCL9Z96 inhibited β-catenin/BCL9 binding activity. D Biacore assay of β-catenin with hsBCL9Z96. E LEF/TCF Hepa1-6 reporter assays showed that the IC50 values of hsBCL9Z96 and ICG001 were 0.247 μM and 1.272 μM, respectively. F The cell membrane penetration rates of hsBCL9Z96 and hsBCL9CT-24 in the HCT116 cell line were measured by FACS. Data are presented as mean ± SD from n = 3 biological replicates, with statistical significance determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. G, H The penetration rates of hsBCL9Z96 and hsBCL9CT-24 in liver tissues were measured by in vivo imaging. The bar graphs represent data summarized as the means ± SD from n = 3 independent biological replicates, which were analyzed by one-way ANOVA. I The signal of FITC-positive cells in different regions from the tumor border. Data are presented as mean ± SD from n = 2 independent biological replicates. The identical values for hsBCL9CT-24 at 1 mm and 2 mm depths confirm that no peptide penetration occurred beyond 0.5 mm. J hsBCL9Z96 (orange) forms 4 salt bonds and 10 hydrogen bonds with Caveolin-2 (gray). K The expression of Caveolin-2 on Hepa1-6 cells was measured by immunofluorescence. Caveolin-2 is mainly expressed in HCC cells. The experiment was independently repeated three times with qualitatively identical results. L HeLa cells were treated with FITC-labeled hsBCL9Z96 in the presence or absence of a specific Caveolin-2 inhibitor, MβCD. The main fluorescence signals were measured by flow cytometry (Data are presented as mean ± SD from n = 3 independent biological replicates in each group). M Reporter assay results for non-Wnt signaling cascades, denoting hsBCL9Z96’s lack of off-target effects (IC50 > 10 μM for all).
Due to the complex extracellular matrix, it is difficult for peptides to penetrate deep into solid tumors, leading to reduced efficiency and drug resistance21,22. Flow cytometry indicated a trend toward improved membrane penetration with hsBCL9Z96 compared to hsBCL9CT-24 (Fig. 3F). In addition, in vivo imaging suggested improved tissue penetration of hsBCL9Z96, but not hsBCL9CT-24, into liver tissues (Figs. 3G–I and S2I). Structurally, the hsBCL9Z96 peptide contains a transactivator of transcription (TAT) cell-penetrating signal and palmitic acid modification that improved membrane targeting and cytoplasm transport compared to hsBCL9CT-24. Prior evidence suggests Caveolins, specialized plasma membrane invaginations present in most cell types, may mediate the cell-penetrating capacity of TAT cell-penetrating peptides. Further structural analysis revealed that hsBCL9Z96 forms 4 salt bonds and 10 hydrogen bonds with Caveolin-2 at the pocket outlet and binds to Caveolin-2 at 37 nM (Figs. 3J and S2J, K). We found that Caveolin-2 is highly expressed in HCC cells (Fig. 3K). These observations are consistent with a Caveolin-2-dependent manner. To examine this possibility, we used methyl-beta-cyclodextrin (MβCD), which destroys caveolae, and found that MβCD could largely block the cell-penetrating capacity of hsBCL9Z96 (Figs. 3L and S2L). However, treatment with EIPA and chlorpromazine showed no substantial inhibition of hsBCL9Z96 entry, suggesting that the macropinocytosis and clathrin-mediated pathways are not essential for its cell penetration (Fig. S2M).
To assess potential off-target effects of hsBCL9Z96, we conducted multiple reporter assays and found that hsBCL9Z96’s IC50 values for inhibiting non-Wnt signaling were 22-fold higher than those for Wnt/β-catenin signaling inhibition (Fig. 3M). At an intravenous dose of 5 mg/kg in rats, hsBCL9Z96 displayed low clearance with an extended half-life, suggesting that efficacious exposure levels are achievable at a therapeutic dose (Fig. S2N, O). Thus, hsBCL9Z96 demonstrated tumor-penetrating capacity with a favorable pharmacokinetic profile. Furthermore, no adverse side effects were observed following treatment with up to fourteen daily doses of 100 mg/kg (Fig. S2P–S).
Targeting BCL9 by hsBCL9Z96 mitigates ICI resistance in HCC ex vivo and in vivo
To assess the ability of hsBCL9Z96 to improve the response to ICI, PDOTS from HCC patients who underwent tumor resection (n = 20) were applied for ex vivo tests. These PDOTS were treated ex vivo with PD-L1-based immunotherapy in the presence or absence of hsBCL9Z96 (Fig. 4A). PDOTS with a poor response to anti-PD-L1 ab alone exhibited reduced viability when they were incubated with the combination of anti-PD-L1 ab and hsBCL9Z96 (Fig. 4B). Furthermore, a negative correlation was observed between BCL9 expression and the anti-PD-L1 killing index (Fig. 4C). We then used an oncogenic Myc-driven spontaneous HCC model, which was reported to be an ICI-resistant model23. As expected, these Myc-driven spontaneous tumors exhibited primary resistance to anti-PD-L1 ab (Figs. 4D, E and S3A) with nuclear accumulation of β-catenin (Fig. S3B). Consistent with the PDOTS results, combination therapy with hsBCL9Z96 was associated with reduced tumor burden, as evidenced by fewer tumor nodules in this model (Fig. 4D, E). These therapeutic effects were reproduced in two additional immunocompetent Hepa1-6/H22 orthotopic allograft models (Figs. 4F, G and S3C, D). Hematoxylin and eosin (H&E) staining revealed severe tumor necrosis and reduced metastatic lesions in the lungs following treatment with the combination of anti-PD-L1 ab and hsBCL9Z96 (Figs. 4H, I and S3E–H).
A Viability of 20 HCC PDOTS on day 5 following the indicated treatments in a 3-D microfluidic system. B Relative viability of 20 HCC PDOTS on day 5 following the indicated treatments. The bar graph represents data summarized as the means ± SD from n = 20 independent biological replicates in each group, which were analyzed by one-way ANOVA. C The correlation of the PD-L1 ab kill index and BCL9 expression of the 20 HCC PDOTS (the shaded band indicates the 95% confidence interval of the regression fit, and statistical significance was determined by spearman’s correlation test). D, E Representative images and statistical results for Myc transgenic mice that received the indicated treatments (6 mice per group; Vehicle + IgG, 10 mg/kg; hsBCL9Z96, 30 mg/kg; anti-PD-L1 ab, 10 mg/kg; anti-VEGF ab, 10 mg/kg; and combination therapy). Scale bars: 1 cm. The bar graph represents data summarized as the means ± SD from n = 6 independent biological replicates in each group, which were analyzed by one-way ANOVA. F Representative B-ultrasound and growth curves of Hepa1-6 tumors in C57BL/6J mice (Vehicle + IgG, 10 mg/kg, n = 12; hsBCL9Z96, 30 mg/kg, n = 11; anti-PD-L1 ab, 10 mg/kg, n = 11; anti-VEGF ab, 10 mg/kg, n = 11; and combination therapy, n = 11). G Survival curves calculated by the two-sided Log-rank test of C57BL/6J mice bearing Hepa1-6 tumors (n = 10 per group) that received Vehicle + IgG, 10 mg/kg; hsBCL9Z96, 30 mg/kg; anti-PD-L1 ab, 10 mg/kg; anti-VEGF ab, 10 mg/kg; or combination therapy. H, I Representative H&E images of lung metastasis. The incidence of lung metastasis (arrows) is shown in the indicated groups. J The t-distributed stochastic neighbor embedding (t-SNE) plots of tumor-infiltrating immune cells, where each color represents a distinct immune cell subset: MacroMonos, CD4+ T cells, CD8+ T cells, B cells, natural killer (NK) cells, double-negative (DN) T cells, dendritic cells (DC), and granulocytes in the Hepa1-6 tumors as indicated by CyTOF analysis of Hepa1-6 tumors from mice administered the indicated treatments (n = 4). K Quantification of immune cells in Hepa1-6 tumors exposed to the indicated treatments. Data are presented as mean ± SD from n = 4 independent biological replicates. Comparisons were performed using an unpaired two-sided Student’s t-test. L Hepa1-6 tumor volumes in C57BL/6J mice treated as indicated, and macrophages and neutrophils were depleted using anti-CSF1 ab and anti-Ly6G ab, respectively. Data are presented as mean ± SD from n = 6 independent biological replicates, Vehicle + IgG, 10 mg/kg; hsBCL9Z96, 30 mg/kg; anti-PD-L1, 10 mg/kg; anti-CSF1, 10 mg/kg; anti-Ly6G, 10 mg /kg. Statistical significance was determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test.
Cytometry by Time-of-Flight (CyTOF) and whole-transcriptome RNA-sequencing were performed on Hepa1-6 tumors subjected to various treatments to investigate underlying molecular mechanisms (Fig. S3I). FlowSOM clustering identified 8 immune cell subsets, including B cells, CD4+ T cells, CD8+ T cells, dendritic cells (DCs), double-negative T cells (DNTs), granulocytes, macrophages/monocytes (MacroMonos), and undefined cells, based on marker expression (Figs. 4J and S3J). Notably, an increase in MacroMonos proportion was observed following hsBCL9Z96 treatment or its combination with ICI (Figs. 4K and S3K). To validate these findings, flow cytometry and IHC confirmed a consistent increase in macrophage abundance following BCL9 inhibition or combination treatment, corroborating the CyTOF-based profiling (Fig. S3L, M). Importantly, depletion of tumor-associated macrophages (TAMs) using an anti-CSF1 ab attenuated the effects of BCL9 inhibition on tumor immunity, consistent with TAMs, but not other immunocytes, were mainly responsible for the tumoral BCL9-mediated immune modulation in HCC (Fig. 4L). These data indicate that BCL9 inhibition selectively associated with macrophage recruitment and activation.
Collectively, these observations suggest that pharmacological targeting BCL9 by hsBCL9Z96 enhances anti-tumor immunity in HCC through a macrophage-dependent mechanism, thereby potentially improving the efficacy of ICI-based therapies.
Targeting BCL9 reprograms the TIME mainly by reeducating TAMs toward an M1 phenotype via decreasing BMP4 secretion
TAMs, the most abundant innate myeloid infiltrates in the TIME of human solid malignancies24, exhibit a dynamic plasticity, oscillating between a proinflammatory M1 type and an immunosuppressive M2 type. The latter often impedes the efficacy of immunotherapies25. A deconvolution analysis of 24 immune cell types using CIBERSORT suggested increased proportions of M1 macrophages and a reduced proportion of M2 macrophages following hsBCL9Z96 treatment. These effects were further amplified in the hsBCL9Z96 and anti-PD-L1 ab combination treatment group (Fig. 5A). RNA-seq profiling showed trends toward M1 marker upregulation and a downregulation of M2 marker genes following hsBCL9Z96 treatment (Fig. 5B and Supplementary Data 1).
A CIBERSORT analysis for M1 macrophage populations. Statistical significance was determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. Data are presented as mean ± SD from n = 3 biological replicates. The whiskers represent the minimum and maximum values observed within the range of Q1 - 1.5 × IQR to Q3 + 1.5 × IQR. The box shows the IQR between the 25th (Q1) and 75th (Q3) percentiles, and the line inside the box indicates the median (50th percentile). B Heatmap showing the differential expression of genes indicative of protumor M2-like macrophages, antitumor M1-like macrophages, IFN-γ-related pathways (from the Hallmark collection), and T-cell infiltration in the indicated experimental groups. Scale bars represent the normalized FPKM per gene (log2). Representative examples of genes with known roles in the ICI response in patients are labeled. C Enrichment plot for an antitumoral macrophage signature and macrophage activation sets in Hepe1-6 tumors from mice treated with hsBCL9Z96 + anti-PD-L1 ab versus anti-PD-L1 ab alone. D Flow cytometric quantification of M1 macrophages in the Hepa1-6 tumors from each group. Data are presented as the means ± SD from n = 3 biologically independent mice in each group, and were analyzed via one-way ANOVA. E Schematic overview of studies involving BMDMs cocultured with Hepa1-6 cells. F M1 and M2 macrophage markers in BMDMs cocultured with Hepa1-6 cells. G M1 signature scores of BMDMs. Data are presented as mean ± SD from n = 3 biological replicates and were compared by an unpaired two-sided t-test. H M1 macrophages in BMDMs cocultured with Hepa1-6 cells. Data are presented as mean ± SD from n = 3 biological replicates and were compared by an unpaired two-sided t-test. I Overlap of the genes upregulated in non-responders versus responders in the GO30140 + IMbrave150 cohorts, and genes downregulated in mice following hsBCL9Z96 treatment. J Bmp4 in Hepa1-6 cells in the indicated groups. Data are presented as mean ± SD from n = 3 biological replicates and were compared by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. K Bmp4 in Hepa1-6 cells treated with increasing concentrations of hsBCL9Z96. Data are presented as mean ± SD from n = 3 biological replicates, with statistical significance determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. L M1-polarized macrophages after BMP4 ab treatment. Data are presented as mean ± SD from n = 3 biological replicates and were compared by an unpaired two-sided t-test. M, N M1- and M2-polarized macrophages in the indicated groups. Data are presented as mean ± SD from n = 3 biological replicates, with statistical significance determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. Schematic in (E) was created in BioRender. Wu, S. (2025) https://BioRender.com/n1zpx7y.
In line with these observations, the gene expression signatures for the interferon-γ pathway and T-cell infiltration were enriched following hsBCL9Z96 treatment. Combination therapy showed an even greater change in the gene expression signature than single-agent treatment (Fig. 5B). The GSEA also indicated positive enrichment of antitumoral macrophages, macrophage activation, and T-cell activation, along with reduced enrichment of signatures associated with the cell cycle in the hsBCL9Z96 combination group compared to the vehicle control group (Figs. 5C and S4A). Flow cytometry detected similar patterns, revealing an increased proportion of iNOS+CD206− M1 macrophages and a decreased proportion of iNOS−CD206+ M2 macrophages in tumor-bearing mice receiving hsBCL9Z96 or the combination treatment (Figs. 5D and S4B, C). Immunofluorescence staining also showed increased infiltration of F4/80+CD86+ M1 macrophages and a reduction of F4/80+CD163+ M2 macrophages in tumors from mice treated with hsBCL9Z96 or the combination treatment (Fig. S4D, E). Additional single-cell data (GSE151530) consistently supported an inverse correlation between BCL9 and M1 marker genes (CCR7, IRF1, and CCL5) and a positive correlation with M2 markers (CD163, MARCO, and VEGFA)20 (Fig. S4F). Interestingly, treatment with bone marrow-derived macrophages (BMDMs) with hsBCL9Z96 alone did not affect the fractions of M1 and M2 macrophages (Fig. S5A, B). This observation suggests that hsBCL9Z96 plays an indirect role in regulating macrophage polarization.
Supporting our in vivo data, ex vivo 3-D microfluidic PDOTS experiments also showed that supernatant from hsBCL9Z96-treated PDOTS had reduced concentrations of IL-10 and TGF-β1, key immunosuppressive cytokines secreted by M2 macrophages. These cytokines were further reduced in the anti-PD-L1 ab + hsBCL9Z96 combination group (n = 9, Fig. S5C, D). To ascertain the impact of tumor BCL9 on macrophage polarization, Bcl9 was depleted in Hepa1-6 HCC cells using CRISPR-Cas9, and these cells were cocultured with macrophages. Gene profiling and flow cytometry analyses revealed that macrophages cocultured with Bcl9-KO Hepa1-6 cells exhibited an M1-like, rather than an M2-like, phenotype (Fig. 5E–H). We also implanted Bcl9-KO and Ctrl-KO Hepa1-6 cells into C57BL/6J mice and found that Bcl9-KO resulted in increased M1, but decreased M2, macrophages in vivo (Fig. S4G, H). These findings suggest that targeting tumoral BCL9 with hsBCL9Z96 fosters antitumor activity by reeducating immunosuppressive macrophages toward an M1 phenotype.
To unravel how tumoral BCL9 expression regulates macrophage polarization, we assessed the transcriptomes of Hepa1-6 orthotopic tumors and patients’ tumors from the GO30140 + IMbrave150 cohorts. After applying a threshold of > twofold change and a P-value < 0.05, we identified 519 upregulated genes in the non-responders and 215 downregulated genes in hsBCL9Z96-treated mice, respectively. Among the 10 overlapping genes in these groups, BMP4, a secreted protein previously reported to induce macrophage polarization toward an M2 phenotype in bladder cancer cells26, was the most significantly altered gene (Fig. 5I). We thus posited that BMP4 might be a BCL9 target gene that mediates macrophage polarization. A chromatin immunoprecipitation (ChIP)-qPCR analysis further validated that the BCL9 transcription complex bound to the BMP4 promoter in Bcl9-OE Hepa1-6 cells (Fig. S5E).
We found that there was a significant decrease in Bmp4 mRNA expression and secretion in the supernatants from Bcl9-KO Hepa1-6 or Hepa1-6 treated with hsBCL9Z96 compared with the control groups (Figs. 5J, K and S5F, G). Blocking BMP4 in the supernatant of a coculture system with a specific BMP4-neutralizing ab resulted in a higher proportion of CD11b+F4/80+iNOS+ macrophages and a reduced population of CD11b+F4/80+CD206+ macrophages, suggesting that the BMP4 secreted by tumor cells may contribute to the M2 polarization of macrophages (Figs. 5L and S5H). After RAW264.7 cells were exposed to conditioned medium from H22 cells treated with hsBCL9Z96, the expression of M1 macrophage markers (Nos2 and Il-12) was significantly increased, and the expression of M2 macrophage markers (Arg1 and Il-10) was significantly decreased in the cells. Moreover, the addition of recombinant BMP4 protein counteracted the effects of hsBCL9Z96 treatment on the M2 to M1 macrophage polarization of RAW264.7 cells cultured with HCC tumor cells (Fig. S5I, J). Furthermore, the addition of recombinant BMP4 protein to the cells could counteract the effects of BCL9 depletion or hsBCL9Z96 treatment on the proportion of M1 and M2 macrophages, supporting the role of BMP4 in BCL9-induced macrophage polarization (Figs. 5M, N and S5K).
Collectively, these data suggest that BCL9 transcriptionally regulates BMP4 expression, thereby promoting M2 polarization and inhibiting M1 macrophage polarization.
The enhanced phagocytosis and antigen presentation of TAMs after targeting BCL9 occurs via the CD24-SIGLEC-10 axis
A Gene Ontology (GO) enrichment analysis of Hepa1-6 orthotopic tumors after treatment with hsBCL9Z96 or hsBCL9Z96 + anti-PD-L1 ab revealed upregulation of pathways related to macrophage activation, antigen processing and presentation, as well as antitumor T-cell activation (Fig. 6A, B). These data suggest potential activation of antigen processing and presentation of TAMs might be activated via their phagocytic process when the tumoral BCL9 was targeted, since the phagocytic process is crucial for tumor-macrophage interactions and acts as a primary antitumor mechanism for M1 macrophages27.
See also Figs. S6 and 7. A, B GO analysis showed enrichment of pathways involved in macrophage activation, antigen processing and presentation, and antitumor T-cell activation pathways in Hepa1-6 cells after hsBCL9Z96 treatment. GO analysis was performed using a two-sided hypergeometric test with Benjamini–Hochberg FDR adjustment for multiple comparisons; terms with an adjusted P value < 0.05 were considered significant. C Putative ligand–receptor (L–R) interactions between tumor cells and macrophages were inferred using CellPhoneDB. Statistical significance was assessed using a two-sided permutation test. D Correlation of BCL9 and ligand expression. A two-sided Spearman’s rank correlation test was performed for BCL9 and indicated genes. E Effects of Bcl9-KO on CD24 expression in HCC cells as determined by FACS. Data are presented as mean ± SD from n = 3 biological replicates and were compared by an unpaired two-sided t-test. F Effects of treatments with hsBCL9Z96 on the CD24 expression as determined by FACS. Data are presented as mean ± SD from n = 3 biological replicates, with statistical significance determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. G ChIP-qPCR analysis using BCL9 ab for the promoters of CD24 in Hepa1-6 cells. Data are presented as mean ± SD from n = 3 biological replicates and were compared by paired two-sided t-test. H ChIP-qPCR analysis using TCF4 ab for the promoters of CD24 in Hepa1-6 cells. Data are presented as mean ± SD from n = 3 biological replicates, with statistical significance determined by paired two-sided t-test. I Effects of hsBCL9Z96 treatment on CD24 expression in HCC cells as determined by qRT‒PCR assays. Data are presented as mean ± SD from n = 3 biological replicates and were compared by an unpaired two-sided t-test. J, K The phagocytic capacity of macrophages treated with the indicated agents in vivo using mIHC assays. Data are presented as mean ± SD from n = 3 biological replicates, with statistical significance determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. L The surface H-2Kb OVA complex in macrophages cocultured with OVA257–264-loaded Hepa1-6 cells with or without Bcl9-KO. Data are presented as mean ± SD from n = 4 biological replicates and were compared by an unpaired two-sided t-test. M The surface H-2Kb OVA complex in macrophages was cocultured with OVA257–264-loaded Hepa1-6 cells with or without hsBCL9Z96.Data are presented as mean ± SD from n = 5 biological replicates and were compared by an unpaired two-sided t-test. N Macrophages cocultured with the OVA257–264-loaded Hepa1-6 cells with the indicated treatment. Data are presented as mean ± SD from n = 3 biological replicates, with statistical significance determined by two-sided one-way ANOVA followed by Tukey’s multiple comparisons test. O Quantification of granzyme B production in OT-I cells. Data are presented as mean ± SD from n = 3 biological replicates and were compared by an unpaired two-sided t-test. P Representative mIHC staining on TMAs. CD68/CD86 and CD68/CD163 images were acquired from the same microscopic fields, with different fluorescence channels displayed. Scale bars, 200 μm (overview images) and 10 μm (magnified and inset images). Q Correlations between the tumoral BCL9 and CD24, M1/M2 phenotypes, antigen-processing potentials of macrophages, and infiltrating active effector T-cell proportions. R Evaluation of the associations between tumoral BCL9 and the CTL score, MHC score, and PD-1 score in multiple clinical studies. S Schematic diagram of the proposed model showing how hsBCL9Z96 enhances immunotherapy in HCC. Schematic in (S) was created in BioRender. Wu, S. (2025) https://BioRender.com/nxpkml6.
Employing single-cell computational analyses with an integrated HCC scRNA-seq data joint with GSE125449 and GSE14022828,29, we confirmed that BCL9 is primarily expressed in malignant cells (Fig. S6A), and ligand-receptor (L-R) analysis showed enrichment of the interactions of the top 10 L-R pairs between tumor cells and M1 macrophages (Fig. 6C). To further investigate the potential mechanism underlying how the macrophage-dependent anti-tumor effects are mediated by targeting BCL9, the correlation between BCL9 and the top 10 receptors were analyzed in four independent HCC datasets, and a correlation between BCL9 and CD24 was observed in these datasets, including TCGA LIHC (R = 0.350, P < 0.001), GSE22058 (R = 0.566, P < 0.001), GSE43619 (R = 0.487, P < 0.001) and GSE19977 (R = 0.327, P < 0.001). These findings suggest that this pair may play a pivotal role in mediating interactions between tumor cells and macrophages (Fig. 6D). A positive correlation between BCL9 and CD24 was confirmed at both the mRNA and protein levels in clinical samples of HCC (Fig. S6B, C). Moreover, the depletion of Bcl9 in Hepa1-6 cells led to decreased Cd24a mRNA expression and a reduced fraction of CD24+ tumor cells (Figs. 6E and S6D). Similarly, hsBCL9Z96 diminished the CD24+ cell fraction and Cd24a mRNA in a concentration-dependent manner (Figs. 6F and S6E, F). ChIP-qPCR and RT-qPCR also confirmed that BCL9 has a role in Cd24a transcription (Fig. 6G–I).
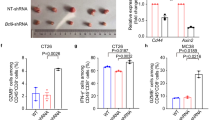
Since CD24 is the ligand for SIGLEC-10, their interaction acts as a “don’t eat me” signal that enables tumor cells to evade macrophage-mediated phagocytosis30. We therefore hypothesized that targeting BCL9 might promote phagocytosis by disrupting tumoral CD24. Supporting this, BMDMs cocultured with Bcl9-KO Hepa1-6 demonstrated enhanced phagocytic capacity (Fig. S6G). Public HCC scRNA-seq datasets and multiplex immunohistochemistry (mIHC) confirmed SIGLEC-10 is expressed in M1 macrophages28 (Fig. S6H, I). When Hepa1-6 were cocultured with BMDMs treated with hsBCL9Z96, the CD24 overexpression in HCC cells restored the phagocytic phenotype of the macrophages, indicating that CD24 appears as a downstream effector of BCL9 in regulating macrophage phagocytosis (Fig. S6J). Additionally, hsBCL9Z96 or an anti-CD24 neutralizing ab educated macrophage polarization in tumor-BMDMs coculture and enhanced phagocytic activity of macrophages (Fig. S6K). mIHC also confirmed that hsBCL9Z96 exposure resulted in a significant increase in inclusion bodies derived from HCC cells (in white, labeled with HNF4α31) within M1 macrophages (in green and red, labeled with F4/80 and CD86) (Fig. 6J, K), indicating that hsBCL9Z96 activated M1 macrophages to engulf tumor cells in vivo.
The antigen-presenting capacity of M1-like macrophages is crucial to immunotherapy32,33,34,35,36,37,38. Given that antigen-presenting cells (APC) initiate innate antitumor immunity39,40, we explored whether inhibiting tumoral BCL9-mediated macrophage phagocytosis could enhance the antigen presentation process. We noted a diminished antigen processing and presentation score in the high BCL9 group, and a negative correlation between BCL9 and antigen processing and presentation was identified in the GO30140 + IMbrave150 cohorts (Fig. S6L). Responders to Atezo + Bev exhibited higher antigen processing and presentation scores (Fig. S6M), and tumors with lower BCL9 levels in the TCGA LIHC also had higher antigen processing and presentation scores (Fig. S6N).
Notably, BCL9 inhibition in mice with Hepa1-6 orthotopic tumors was associated with upregulated expression of MHC-I antigen presentation-related genes, including Tap1, Tap2, B2m, and Psmb9 (Fig. S6O), and GSEA also demonstrated the enrichment of antigen processing and presentation following hsBCL9Z96 treatment (Fig. S6P). This suggests that tumoral BCL9 may influence antigen presentation. To examine the effects of tumor BCL9 in modulating the functions of APC, we cultured BMDMs with Hepa-1-6 cells incubated with surface OVA-loaded H2-Kb complex, and found that the surface OVA-loaded H2-Kb complex was increased on BMDMs when they cocultured with Bcl9-KO Hepa1-6 compared with Ctrl-KO Hepa1-6 (Fig. 6L) or when hsBCL9Z96 was added to the cultures (Fig. 6M). Conversely, this enhancement was reduced in macrophages cocultured with CD24-overexpressing tumor cells, which could be prevented by hsBCL9Z96 (Fig. 6N). CD8+ T cells cocultured with Bcl9-KO Hepa1-6 cells and macrophages showed trends toward increased GZMB secretion and proliferation (Figs. 6O and S6Q, R). Additionally, mIHC staining confirmed that tumoral BCL9 could enhance the infiltration of M1 macrophages, active antigen-processing macrophages (defined as CD68+MHC-I+cells), and cytotoxic GZMB+CD8+ T-cells, while decreasing the infiltration of M2 macrophages (Figs. 6P, Q and S7A–F).
In addition, a TCGA pan-cancer analysis revealed that there were correlations between BCL9 expression and macrophage infiltration, antigen presentation, and phagocytosis (Fig. S7G–J), and negative relationships between BCL9 expression and CTL infiltration, the MHC score, and the PD-1 score, which were validated in multiple clinical studies of ICI (Fig. 6R).
Thus, targeting tumoral BCL9 enhanced phagocytosis and antigen presentation by TAMs by decreasing the CD24 expression in tumor cells. Targeting BCL9 may represent a promising strategy to combat the resistance to immunotherapy in multiple cancer types, including HCC.
Discussion
Although the association between Wnt/β-catenin signaling and resistance to immunotherapy is well-established in HCC41, the mechanism(s) by which this signaling contributes to ICI treatment resistance are not fully understood. In this study, we identified BCL9 as a major regulator of Wnt/β-catenin signaling activation-induced immunotherapy resistance in HCC and identified pathways where TAM phenotype shifts may occur toward a tumor-suppressing M1 macrophage phenotype via the BCL9/BMP4 axis, and where TAMs’ phagocytosis and antigen presentation were increased through the BCL9/CD24/SIGLEC-10 axis to mitigate ICI resistance (Fig. 6S). Our findings highlight BCL9 as a targetable key player linking macrophage-mediated innate immunity and T-cell-mediated adaptive immunity and extend our understanding of how the innate immune system connects with the adaptive immune response through Wnt/β-catenin signaling to reverse ICI resistance in HCC. Given that 43% of HCC patients acquire Wnt/β-catenin signaling alterations42, our study lays a solid foundation toward understanding the link between Wnt/β-catenin signaling abnormalities and resistance to immunotherapy. While BCL9 expression correlates with ICI response in our cohorts, its predictive value as a standalone biomarker is limited by HCC heterogeneity. Future studies should validate these findings in larger multicenter cohorts and explore combinatorial biomarker approaches.
Multiple factors, including PD-L1 expression and MSI, are known to affect the efficacy, but are insufficient to predict the outcomes of treatment. The absence of reliable biomarkers prevents the precise selection of patients for ICI treatment, which results in poorer patient response rates. Here, we reported that BCL9, a co-transcription factor of β-catenin in the Wnt/β-catenin signaling pathway, serves as a correlated factor for responses of solid tumors to ICI. Our findings, which identified a key resistance-driving pathway (Wnt/β-catenin signaling) and a potential target (BCL9) from high-throughput screening of data from first-line clinical trials of immunotherapy, may have significant clinical implications. Our preclinical data further demonstrated that treatment targeting BCL9 in combination with ICI agents could generate tumor growth inhibition, overcoming ICI resistance with a favorable safety profile.
Macrophages are a vital component of adaptive immunity and are one of the most abundant infiltrates within the TIME of HCC43,44. Similar to DCs, another type of APC that exhibits poor infiltration and dysfunction in HCC45,46, macrophages have the capacity for antigen cross-presentation to initiate an adaptive immune response47,48. However, TAMs often lose their antigen-presenting function and are then considered tumor-promoting49. However, several clinical trials aiming to deplete macrophages have failed to achieve satisfactory outcomes50. Reinvigorating the antigen-presentation capacity of the macrophages already present at the tumor site and rekindling their antitumor potential could represent an alternative approach in macrophage-targeting immunotherapy. The central mechanism underlying the macrophage antigen-presentation process is phagocytosis, which is generally impeded in HCC43. Our mechanistic investigations uncovered the role for tumoral BCL9 in regulating phagocytosis and the antigen processing activity of TAMs. Targeting BCL9 could induce the M2 to M1 polarization of infiltrated TAMs via decreasing BMP4 secretion and can promote M1 phagocytosis by reducing tumoral CD24 expression. This subsequently enhanced the immune response mediated by CD8+ T cells through M1 macrophage-mediated antigen presentation via inhibiting the CD24-SIGLEC-10 axis. In addition, as TAMs are the most abundant immune cells in the TIME of human solid malignancies, reinvigorating their phagocytosis and antigen-processing function might provide a therapeutic target for the development of more precise and effective treatments for liver cancer and other solid tumors.
One of the major reasons for the poor response of solid tumors to immunotherapy is the highly complex underlying TIME51. The microenvironment of HCC consists of dynamic cancer cells, intricate cytokine networks, extracellular matrix components, immune cell subsets, and various other components52. Thus, the difficulty for systemic regimens to penetrate the microenvironment and interact with the target explains the failure of many clinical trials of developed regimens. In this study, we designed and synthesized hsBCL9Z96, a BCL9 inhibitor with a cell-penetrating signal. Our data indicated that hsBCL9Z96 infiltrated deep into the tumor to trigger an immune reaction and promote antitumor immunity while simultaneously exhibiting desirable specificity for its target. Therefore, hsBCL9Z96 showed favorable inhibitory properties targeting the Wnt/β-catenin signaling pathway that overcomes many critical drawbacks of existing drugs. As expected, pharmacological inhibition of BCL9 with hsBCL9Z96 led to tumor growth inhibition in multiple in vivo HCC models, including transgenic spontaneous HCC models. Targeting BCL9 might be particularly useful against HCC with a “cold” TIME because it can remodel the TIME to have more favorable immune responsiveness. Thus, we herein developed a candidate therapeutic approach to improve the treatment of HCC.
Although our study indicates that pharmacological inhibition of BCL9 may be therapeutically valuable in HCC when combined with anti-PD-L1 therapy, several limitations warrant consideration. The predictive performance of BCL9 (AUC = 0.80) remains below thresholds for clinical implementation, compounded by the observation that approximately 50% of non-responders exhibited low BCL9 expression—a discrepancy potentially attributable to molecular heterogeneity within HCC. Mechanistic insights into the BCL9-BMP4-CD24 axis, while supported by preclinical models, require further validation in human HCC, where tumor spatial architecture and microenvironmental complexity may alter pathway dynamics. Translational development of hsBCL9Z96 necessitates rigorous evaluation of target engagement in human tumors, potential toxicity profiles in primates, and penetration efficiency in fibrotic microenvironments. Future studies should validate BCL9-based stratification in multi-institutional cohorts, develop integrated biomarker panels incorporating TMB and PD-L1, and assess effects on myeloid-derived suppressor cells.
In summary, BCL9 acts as a key regulator of Wnt/β-catenin signaling activation-induced immunotherapy resistance. A regulatory network was identified which involves to a tumor-suppressing phenotype in HCC. We also identified associations suggesting tumor-infiltrating macrophages showed trends toward a tumor-suppressing phenotype with enhanced phagocytosis and antigen presentation through the BMP4/CD24/SIGLEC-10 axis. Targeting BCL9 could remodel the TIME to reinvigorate innate and adaptive immunity to enhance the response to ICI in HCC. These findings build upon the current understanding of the mechanisms underlying the resistance of HCC (and other cancers) to immunotherapies and provide a rationale for the further development of BCL9 inhibitors. Therefore, strategies that target BCL9 hold promise to improve the efficacy of ICI-based treatments in a broad spectrum of cancers.
Methods
Patients and specimens
This study complied with all relevant ethical regulations. The study was approved by the Institutional Ethics Committee of Zhongshan Hospital, Fudan University with approved No. B2022-063R. This study was conducted in accordance with the Declaration of Helsinki. Sex was not considered in this study of public human RNA-seq data, as BCL9 is autosomal, and no reports indicated that BCL9 function is sex-biased and the study was not designed to investigate sex-based differences but rather tumor-intrinsic pathways. Written informed consent was obtained from all patients prior to tumor collection. No participants received compensation for this study, in accordance with our institutional guidelines and the informed consent protocol. Tissue microarrays (TMA) were obtained from Zhongshan Hospital, Fudan University. Biopsy punctures of unresectable HCC patients who received Atezolizumab (atezo) plus Bevacizumab (Bev) or anti-PD-1 plus TKIs were obtained from Zhongshan Hospital (Supplementary Table 2), Fudan University. Patients who accepted the Atezo + Bev treatment received 1200 mg of Atezo plus 15 mg per kilogram of body weight of Bev intravenously every 3 weeks. Patients who accepted the anti-PD-1 plus TKIs treatment received 200 mg of anti-PD-1 ab every 3 weeks intravenously plus the oral TKIs. No grade 3 or 4 treatment-related adverse events occurred during treatment. The treatment response for patients was assessed using the RECIST 1.153. Radiographic assessment of the response was performed by independent radiology review. The ZS T + A cohort consisted of 70 patients (57 male, 13 female) with a median age of 54 years (range, 30–77 years). The ZS PD-1 plus TKI cohort consisted of 44 patients (37 male, 7 female) with a median age of 53 years (range, 33–74 years). The ZS PDOTS cohort consisted of 77 patients (69 male, 8 female) with a median age of 62 years (range, 35–69 years). Detailed clinicopathological characteristics are provided in Supplementary Table 2. Postoperative patient surveillance was conducted. All patients were routinely followed up at an interval of 2–3 months during the first two years after surgery and then every 3–4 months after. All patients were routinely monitored by serum alpha-fetoprotein measurements, liver function tests, abdominal ultrasonography, and chest radiography at an interval of 1 to 6 months, depending on the postoperative time. Enhanced Magnetic Resonance Imaging (MRI) or computed tomography (CT) of the abdomen was performed every six months. If intrahepatic recurrence and/or distal metastasis were clinically suspected based on symptoms or unexplained elevations in tumor marker levels, MRI, CT, or bone scans were performed immediately. The Recurrence-Free Survival was defined as the interval between resection and intrahepatic recurrence or extrahepatic metastasis. The overall survival (OS) was defined as the interval between resection and death by any cause or the last observation date, and the data for surviving patients were censored at the last follow-up period.
Cell lines and cell culture
The Hepa1-6 and H22 cell lines were kindly provided by Dr. Xun Huang at Lingang Laboratory, Shanghai, China. The Bcl9-targeting sgRNA sequences (5′-AGGCGTCTAACATTCAGTAGAGG-3′ and 5′-GTTCCTTGTCATGAGCCTGGGGG-3′) were cloned into the CRISPR-U plasmid (Ubigene), and Hepa1-6 cells were transfected by electroporation. Following puromycin selection and single-cell cloning, stable Bcl9-KO Hepa1-6 cell lines were established and validated by PCR and Sanger sequencing. The HepG2 cell line was purchased from the Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, Shanghai, China). RAW264.7, HEK293, and HeLa cells were kindly provided by the Stem Cell Bank, Chinese Academy of Sciences. B16F10 cells were kindly provided by the Laboratory of Pengyuan Yang at Fudan University, Shanghai, China. CellSensor CRE-bla Jurkat cells were purchased from Thermo Fisher Scientific. Cell lines were identified by short tandem repeat authentication. RAW264.7, Hepa1-6, HEK293, HeLa, B16F10, and HepG2 cells were cultured in DMEM. H22 cells and CellSensor CRE-bla Jurkat cells were cultured in RPMI 1640 medium. All media were supplemented with 10% FBS (Gibco, 10099-141) and penicillin-streptomycin, and cells were cultured at 37 °C in a 5% CO2 incubator. Mouse BMDMs were obtained by flushing the femurs and tibias of 6- to 8-week-old healthy C57BL/6 mice. Bone marrow cells were then cultured for 6 days in RPMI 1640 complete medium containing 20 ng/mL carrier-free recombinant mouse M-CSF (BioLegend, 576404) prior to use in experiments. All cell cultures were tested for identity using STR and tested for Mycoplasma by qRT-PCR (monthly testing).
Animal studies
Animal studies followed the ARRIVE guidelines54. All animal care and experimental protocols performed were approved by the Institutional Animal Care and Use Committee (IACUC) of Zhongshan Hospital, Fudan University. Our IACUC permitted a maximal tumor size of 20 mm, which was not exceeded in our study. All animal experiments were conducted using male mice only; therefore, data disaggregation by sex was not applicable. BALB/c mice, C57BL/6J mice, and NOD SCID mice were purchased from Shanghai SLAC Laboratory Animal Ltd., Co. Mice were housed under specific pathogen-free conditions in the Experimental Animal Center of Zhongshan Hospital. Mice were maintained at 12-h/12-h dark–light cycle, approximately 25 °C and approximately 60% humidity and given food and water ad libitum. The mice were assigned to indicated group by random allocation. During daily monitoring, mice were checked for any effects of tumor growth and treatment on normal behavior, such as mobility, food and water consumption, body weight, and for other effects. Mice were not fasted before assessments were carried out; All experiments were optimized to minimize mouse suffering and to reduce the number of mice used. For euthanasia at the end of the treatment period, mice were humanely killed using CO2 asphyxiation followed by cervical dislocation or by intraperitoneal injection of a pentobarbital-phenytoin solution. All efforts were made to minimize suffering. Following euthanasia, tumors were dissected, photographed, and weighed. The tumor volume was calculated using the formula length (mm) × width2 (mm) × 0.5. Mice were kept in the same cage according to the same experimental group, and the experiment operators were aware of the groups of animals with different treatment.
For in vivo studies, the following antibodies were used: a VEGF ab (Bevacizumab, Selleck, #A2006), isotype control for the VEGF ab (BioXCell, #BE0297), PD-L1 ab (BioXCell, clone 10 F.9G2, #BE0101), and isotype control for the PD-L1 ab (BioXCell, clone LTF-2, #BE0090).
An orthotopic liver tumor model was generated. For this model, 5-week-old male C57BL/6J or BALB/c mice were used. A total of 5 × 106 Hepa1-6 or H22 cells in 100 μL DMEM were first subcutaneously injected into the right flank of new 5-week-old male C57BL/6J or BALB/c mice. After two weeks, the subcutaneous tumors were dissected into small pieces (1 mm3) under aseptic conditions and then implanted into the right liver lobe of new mice. When Hepa1-6 or H22 orthotopic tumor growth reached an estimated 100 mm2, the mice were randomized into five groups according to our previous experimental protocol55: isotype control group (n = 12), hsBCL9Z96 group (n = 11), PD-L1 ab group (n = 11), VEGF ab + PD-L1 ab group (n = 11), and hsBCL9Z96 + PD-L1 ab group (n = 11). The VEGF ab or isotype control was injected intraperitoneally at a dosage of 5 mg/kg twice a week. hsBCL9Z96 was injected intraperitoneally at a dose of 40 mg/kg daily. The PD-L1 ab or isotype control was injected via the caudal vein for the first time at a dose of 10 mg/kg and subsequently administered by intraperitoneal injection at a dose of 5 mg/kg three times per week. The doses and medications used for combination therapy were the same as those used for the corresponding monotherapies. The above five groups were treated for 21 days. Ultrasound tumor imaging was performed with a Preclinical Vevo 3100 micro-ultrasound imaging platform (Visual Sonics) every three days according to the group of different treatment, and images were analyzed using VevoLAB software (Visual Sonics) to determine the tumor length, width, and volume.
A c-Myc-driven spontaneous HCC model was generated by crossing mice expressing Cre recombinase under the control of the albumin promoter (AlbCre/Cre mice) with mice harboring LoxP alleles of c-Myc (MycMut/Mut mice). At the age of 6 weeks, male mice were selected for in vivo experiments (n = 6 per group). The mice used were purchased from the Shanghai Model Organisms Center.
Immunochemistry
Tumor microarrays were subjected to IHC staining for BCL9, CD3, CD8, CD68, and FOXP3. The antibodies used are shown in the Supplementary Table 3. The quantification of BCL9 staining was performed using the mean optical density values of BCL9 staining obtained in the IHC assay. The quantification of CD3, CD8, CD68, and FOXP3 IHC used the positive cell count per field.
Multiplex immunofluorescence staining
mIHC was performed by staining 4-μm-thick formalin-fixed, paraffin-embedded whole tissue sections with standard, primary antibodies sequentially paired with a TSA 7-color kit (abs50015-100T, Absinbio, Shanghai). Then samples were stained with DAPI. For example, deparaffinized slides were incubated with an anti-CD68 ab (CST76437, Cell Signaling Technology) for 30 min and then treated with an anti-rabbit/mouse horseradish peroxidase-conjugated secondary ab (abs50015-02, Absinbio, Shanghai) for 10 min. Then, labelling was developed for a strictly observed 10 min, using TSA 520 per the manufacturer’s instructions. Slides were washed in TBST buffer and then transferred to preheated citrate solution (90 °C) before being heat-treated using a microwave set at 20% of maximum power for 15 min. Slides were cooled in the same solution to room temperature. Between all steps, the slides were washed with Tris buffer. The same process was repeated for the following antibodies/fluorescent dyes, in order: anti-CD86/TSA 570 and anti-CD163/TSA 620. Each slide was then treated with 2 drops of DAPI (abs47047616, Absinbio, Shanghai), washed in distilled water, and manually cover slipped. Slides were air-dried, and pictures were taken with a Pannoramic MIDI II (3D HISTECH). Images were analyzed using Indica Halo software.
Microfluidic 3D culture and combined treatment assay
The microfluidic 3D culture study was approved by the Institutional Ethics Committee of Zhongshan Hospital, Fudan University with approval No. B2021-689R. Resected primary tumors from patients with HCC were mechanically minced and dissociated into 1 mm3 fragments, diluted in Advanced DMEM/F-12 (Thermo Fisher Scientific, 12634010) with 0.1% Collagenase, Type 4 (Worthington, LS004188), 0.05% Hyaluronidase (Sigma, H3506), 0.01% deoxyribonuclease (Sigma, DN25), and shaken on a horizontal platform for 30 min at 37 °C. Spheroids (40–100 μm) were resuspended in type I rat tail collagen (Corning, 354236), and the spheroid-collagen mixture was injected into the center gel channel of a 3D microfluidic chamber (9 µL mixture for each channel). After incubation for 30 min at 37 °C in a 5% CO2 incubator, basic medium (advanced DMEM/F12 supplemented with 10% fetal bovine serum (FBS), 500 U/mL IL-2, and 1× CD3/CD28 T cell activator) with or without the indicated drugs and/or abs (10 μM hsBCL9Z96, 500 µg/mL PD-L1 ab, and 10 µg/mL VEGF ab) was added to the media port. The medium was refreshed every three days. After 5 days of incubation, the medium was removed, 10 µL AO/PI reagent was injected into each media port, and the chamber was incubated in the dark at room temperature, followed by removal of the AO/PI reagent and incubation with DAPI for 10 min at room temperature. Images were captured with an OLYMPUS fluorescence microscope, and areas of live cells or dead cells were quantified using ImageJ software. The kill index was calculated as the (Live NC – Live Treatment)/Live NC. The Live NC is the live area ratio in the negative control group [Live NC = Green Area/(Green Area + Red Area)], and the Live Treatment refers to the live area in the indicated treatment group [Live Treatment = Green Area/(Green Area + Red Area)]. Each PDOTS had three replicates, and the average of the three kill index was the final kill index used for the calculation.
Preparation of HCC-conditioned medium (CM) and coculture with macrophages
To obtain Hepa1-6 CM and H22 CM, cells were seeded 1 day before treatment with hsBCL9Z96 (10 μM) for 48 h at 37 °C in a 5% CO2 incubator, with DMSO used as the vehicle control. To obtain Ctrl-KO Hepa1-6 CM and Bcl9-KO Hepa1-6 CM, Hepa1-6 Scramble and Bcl9-KO cells were cultured at 106 cells/well in 6-well flat-bottom culture plates containing complete culture media for 48 h. The supernatants were collected and filtered through a 0.22-μm-diameter sterile syringe filter. H22 CM or hsBCL9Z96-treated H22 CM was added to cultures of RAW264.7 cells (M0) for 48 h. Hepa1-6 CM, Ctrl-KO Hepa1-6 CM, hsBCL9Z96-treated Hepa1-6 cell CM, or Bcl9-KO Hepa1-6 CM was added to cultures of BMDMs (M0) for 48 h. To assess the role of tumor cell-derived soluble factors in macrophage polarization, 50 ng/mL mouse recombinant BMP4 (PeproTech, 315-27) was added to the coculture system. In some experiments, the soluble BMP4 present in the coculture system was neutralized by a rat anti-mouse BMP-4 monoclonal ab (R&D Systems, MAB50201). A rat IgG1 isotype control (MAB005, R&D Systems) was used as a negative control. After incubation for 48 h, the macrophages were harvested and assessed by flow cytometry.
Flow cytometry-based evaluation of phagocytosis
The in vitro phagocytosis assay was performed by coculturing CFSE-labeled Hepa1-6 cells and BMDMs at a ratio of 200,000 Bcl9-KO or Ctrl-KO Hepa1-6 cells to 100,000 BMDMs for 2 h in ultralow-attachment 48-well plates (Corning) in serum-free RPMI 1640 medium. To explore the effects of hsBCL9Z96 treatment on macrophage-mediated phagocytosis, Hepa1-6 cells were pretreated with hsBCL9Z96 (10 μM) or DMSO vehicle control for 2 h before the coculture and harvested with 0.25% trypsin-EDTA (Meilunbio, MB4376). Then, the Hepa1-6 cells were fluorescently labeled with 0.5 μM CFSE (Invitrogen, C34554) according to the manufacturer’s instructions, followed by coculture with BMDMs. Phagocytosis was quantified by measuring the percentage of F4/80+CFSE+ cells among the total CD11b+F4/80+ macrophage population.
In vitro antigen presentation analysis
To detect surface OVA257–264-peptide presentation on H-2Kb (MHC-I), macrophages (BMDMs) were cocultured with OVA257–264-loaded Hepa1-6 cells for 48 h followed by staining with PE-conjugated mouse anti-OVA257-264 (SIINFEKL) peptide bound to anti-H-2Kb antibodies (eBioscience, 12-5743-81), PE-Cyanine7-conjugated anti-mouse CD11b (eBioscience, 25-0112-82), and APC-conjugated anti-mouse F4/80 (eBioscience, 17-4801-80). The cells were analyzed by fluorescence-activated cell sorting (FACS). To perform RNA-seq, macrophages in the coculture system were magnetically purified using the EasySep Mouse F4/80 Positive Selection Kit (StemCell, 100-0659). Total RNA was extracted using the RNeasy Micro Kit (Qiagen, 74004) according to the manufacturer’s protocol.
T-cell proliferation and activation assay
For the in vitro T-cell proliferation assay, 104 macrophages were cocultured overnight with equal numbers of OVA257–264-loaded Hepa1-6 cells in serum-free RPMI 1640 medium. The next day, spleens were isolated from OT-I transgenic mice and CD8+ T cells were magnetically enriched using the EasySep Mouse CD8+ T Cell Isolation Kit (StemCell,19853) and labeled with 0.5 mM CFSE. Then, 5 × 104 CD8+ T cells and 20% FBS were added to the cultures. After a 3-day incubation, the cells were harvested and analyzed for CFSE dilution and evaluation of GZMB expression by FACS.
Quantitative real-time PCR
Total RNA was isolated from cultured cells or tumor tissue using the TRIzol reagent (Ambion, 15596-026) and was reverse transcribed into cDNA using the Hifair II 1st Strand cDNA Synthesis Kit (Yeasen, 11141ES60) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using SYBR Green Master Mix (Yeasen, 11184ES08) according to the manufacturer’s instructions. A housekeeping gene, β-actin, was used as an internal control. The primer sequences are listed in the Supplementary Table 4.
RNA-seq and analysis
The TRIzol method was adopted to extract total RNA according to the manufacturer’s protocol. A NanoDrop 2000 spectrophotometer (Thermo Scientific) was applied to assess the RNA purity and for quantification, and an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) was used to assess the RNA integrity. Sequencing libraries were generated using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations.
Prepared libraries were sequenced on an Illumina HiSeq X Ten platform. Raw sequence reads were first aligned to the mouse UniGene transcriptome with Bowtie (v1.0.0) to estimate the insert fragment size and the standard deviation needed by TopHat2 to align the reads to the genome. Then, TopHat2 was used to align the reads to a reference mouse genome (GRCm38) with the aligning parameter bowtie1 and Ensembl-annotated transcripts (version 77) set as a guide reference. Uniquely mapped reads were used to quantify gene expression, and differential gene expression was analyzed by Cuffdiff, a subpackage of Cufflinks (v2.1.1) with the Ensembl-annotated genes (version 77). The abundances of transcripts (including those of mRNAs, pseudogenes, noncoding RNAs, and other predicted RNAs) were calculated and normalized in fragments per kilobase of transcript per million mapped reads (FPKM), as described above, from the raw RNA-seq data and were used for GSEA (Broad Institute).
To estimate the cellular composition of bulk RNA-seq data for mouse samples, we used CIBERSORT to deconvolute bulk RNA-seq data. CIBERSORT can estimate relative cell abundances in a mixed cell population. The CIBERSORT gene signature matrix, which contains 512 genes and distinguishes 24 mouse cell types, was downloaded from ImmuCC. For the bulk RNA-seq data for TCGA LIHC samples, we used ssGSEA to estimate the cellular composition. The R codes were uploaded.
CyTOF sample preparation and data analysis
Freshly harvested tumors were digested with the Mouse Tumor Dissociation Kit (Miltenyi Biotec) according to the manufacturer’s instructions. Briefly, dissociated tumor samples were filtered through a 70-μm strainer and centrifuged with 36% Percoll reagent to remove cell debris and enrich immune cells. ACK lysis buffer was then utilized to lyse erythrocytes. The cells were resuspended in 100 μL Living Cell Staining Mix (250 nM cisplatin in PBS) and incubated for 5 min on ice. The samples were blocked and stained for 30 min with a surface Ab panel developed in-house (see Supplementary Table 5), followed by fixation overnight in DNA intercalator solution (250 nM 191/193-iridium).
To stain intracellular markers, permeabilization buffer (eBioscience, San Diego, California, USA) was utilized following the manufacturer’s instructions. Cells were then washed, resuspended in ddH2O, and stored at 4 °C before data were acquired on a Helios instrument (Fluidigm).
Mass cytometry data were randomized and normalized before processing. A doublet filtering scheme was used on mass-tagged barcodes for differentiating samples. Single cells were manually gated to exclude calibration beads, debris, dead cells, and doublets. Next, live CD45+ cells were gated and exported for further downstream analysis. Separate viSNE plots and heatmaps were developed for all CD45+ cells to specifically characterize the T cell clusters. X-shift was applied to the T cell compartment defined in the first X-shift analysis. A dimensionality reduction analysis was performed with the viSNE tool, clustering analysis with X-shift.
Single-cell transcriptomic analysis
The HCC scRNA-seq data in the NCBI GEO GSE125449 and GSE140228 datasets were downloaded. We downloaded both the raw count matrices and corresponding cell annotations. Conos (https://github.com/kharchenkolab/conos) was used to integrate multiple scRNA-seq datasets. Each individual dataset was first normalized using the basicP2proc function in pagoda2 with default parameters. Different samples were then aligned using Conos with default parameter settings (PCA space with 30 components, angular distance, mNN matching, k = 15, and k.self = 5), and UMAP embedding was estimated using default parameter settings. Leiden clustering (Conos: findCommunities) was used to determine joint cell clusters across the entire collection of datasets. For the macrophage phenotypes, the signature scores were estimated as average expression values of the genes in a given gene set, which were included in Supplementary Data 1. Validation HCC scRNA-seq data GSE151530 was obtained from GEO, including the gene expression matrix and corresponding cell annotations. Subsequent analyses were performed using the Seurat R package with default parameters.
Ligand-receptor analysis
Ligand-receptor interactions were inferred using an approach similar to that Efremova M. et al.56. We collected 1263 well-annotated ligand-receptor pairs from CellPhoneDB. We first screened each of the ligands and receptors based on their expression within each cell type, requiring that the gene be expressed in more than 10% of the cells. Next, we calculated the average expression of ligand-receptor pairs across cell type pairs in normalized scRNA-seq data. The product of average ligand expression in cell type A and average receptor expression in cell type B was used to measure ligand-receptor pair expression. To evaluate the robustness and statistical significance of ligand-receptor pairs, we calculated a null distribution for average ligand-receptor pairs by shuffling cell identities in the aggregated data and recalculating the average ligand-receptor pair expression across 1000 permutations of randomized cell identities. The P value represents the number of randomized pairs exceeding the observed data.
Enzyme-linked immunosorbent assay (ELISA)-based measurement of soluble factors
The supernatants of Hepa1-6 cells or HCC PDOTS culture medium were collected. The concentration of BMP4 secreted by Hepa1-6 cells was measured with the mouse bone morphogenetic protein 4 (BMP-4) ELISA Kit (Cusabio, CSB-E04512m) according to the manufacturer’s instructions. The concentrations of TGFβ and IL10 in the HCC PDOTS culture medium were measured with a Human/Mouse/Rat TGF-β ELISA Kit (MultiSciences, EK981-96) and Human IL-10 ELISA Kit (MultiSciences, EK110-96) according to the manufacturer’s instructions.
Flow cytometry
At the end of the study, tumors were dissected from HCC rodent models, cut into pieces, and digested in RPMI 1640 medium containing 1 mg/mL collagenase IV (Sigma, C5138) and 1 mg/mL DNase I (Meilunbio, MB3069). Single-cell suspensions were prepared and stained for flow cytometric analysis. For surface-marker analyses, cells were stained with fluorochrome-conjugated antibodies against surface markers. For assays involving intracellular or nuclear markers, cells were first stained for surface markers, then fixed and permeabilized using an intracellular fixation & and permeabilization buffer set (88-8824-00, eBioscience) according to the manufacturer’s protocol, and then were stained with intracellular antibodies. The complete list of antibodies is provided in Supplementary Table 3.
In in vitro studies, Hepa1-6 cells were stained with Brilliant Violet 421-conjugated anti-mouse CD24 (BioLegend, 101826) ab. BMDMs were digested in 0.25% trypsin-EDTA (Meilunbio, MB4376) and stained with PE-Cyanine7-conjugated anti-mouse CD11b (eBioscience, 25-0112-82), APC-conjugated anti-mouse F4/80 (eBioscience, 17-4801-80), PE-conjugated anti-mouse CD206 (eBioscience, 12-2061-82) and Alexa Fluor 488-conjugated anti-mouse iNOS (eBioscience, 53-5920-82) antibodies. The LIVE/DEAD Fixable Yellow Dead Cell Stain Kit (Invitrogen, L34967) was used to gate living cells. Samples were analyzed using a FACSAria II cell sorter (BD Biosciences) or CytoFlex S flow cytometer (Beckman). The data reported in this paper were analyzed using FlowJo V10 software.
To identify macrophages, the following gating strategies were used: (I) selection of live single-cell leukocytes [SSC–A, CD45+], (II) selection of macrophages [CD11b+F4/80+], and (III) selection of M1 macrophages [iNOS+CD206−] and M2 macrophages [iNOS−CD206+]. We also selected for activated GZMB+CD8+ cells.
ALPHA assays
ALPHA screening (Perkin Elmer) was performed according to a standard protocol, with modifications to account for the optimized readout. The maximum average signal in the ALPHA assay was 31,618.9, with a standard deviation of 2,872.1. The minimum average signal was 526.4, with a standard deviation of 68.6, and the Z-factor was 0.7. Biotinylated BCL9-HD2 and hsBCL9Z96 were conjugated with streptavidin-coated donor beads via a PEG linker, which was customized to allow a larger binding area between BCL9 peptides and β-catenin, while β-catenin was conjugated with protein A-coated acceptor beads using an anti-β-catenin Ab (Abcam ab32572). Following cross-titrations, 100 nM BCL9-HD2B (biotinylated BCL9-HD2 peptide), which lacked the PEG tag in comparison to BCL9-HD2A, and 100 nM β-catenin were used in further studies. ALPHA assay readouts were measured on an EnVision Multimode Plate Reader and Enspire Multimode Plate Reader (Perkin Elmer). All assays were performed in duplicate and repeated at least twice with freshly prepared reagent solutions.
Biacore assay
Beta-catenin (Feng et al. SA, 2019) was immobilized on a CM5 chip (GE Health) using a Biacore T200. A sensogram was obtained by using serially diluted concentrations of hsBCL9Z96. The surface plasmon resonance sensograms had association time intervals of 45 s and dissociation time intervals of 60 s. Data were analyzed using Biacore Evaluation Software.
TCF/LEF signal reporter assay
To measure the TCF/LEF signal reporter activity, 1 × 105 Hepa1-6 cells were added to 96-well plates and transfected with Lipofectamine 2000 (Invitrogen, 11668-019)-loaded LEF/TCF plasmids (QIAGEN, 7601316241). Lipofectamine 2000 was mixed with LEF/TCF plasmids at 25 °C for 20 min before being added into 96-well plates. Cells were cultured at 37 °C in a 5% CO2 incubator and treated with different concentrations of hsBCL9Z96 or ICG-001 for 24 h. The luciferase reporter activity was measured using the Firefly & Renilla Luciferase Reporter Assay Kit (meilunbio, MA0518).
Other reporter assays
The following signaling pathways and cell lines were analyzed in other assays: the cAMP/PKA pathway was tested in CRE-bla Jurkat cells (Life Technologies) following forskolin stimulation, the heat shock response was examined in HepG2 cells after 17-(allylamino)-17-demethoxygeldanamycin (17-AAG) stimulation, the MAPK/ERK pathway activity in luc2P/SRE/Hygro HEK293 cells was examined using EGF stimulation, the interferon regulation pathway in HeLa cells was tested following IFN-γ stimulation, the STAT3 pathway in HEK293 cells was examined after IL-6 stimulation, and the NF-κB pathway was assessed in luc2P/NF-κB-RE/Hygro HEK293 cells with hTNF stimulation.
cAMP/PKA-CRE-bla Jurkat cells-inhibitor screen
CRE-bla Jurkat cells were thawed and prepared as described in the manufacturer’s protocol. A 4 μL aliquot of 10× serial dilutions of H-89 (control inhibitor; starting concentration: 10,000 nM) or a test compound was added to a tissue culture-treated assay plate. Then, 32 μL of cell suspension was added to each well, and the plate was preincubated at 37 °C/5% CO2 in a humidified incubator with compound and control inhibitor titrations for 30 min. Next, 4 μL forskolin (10×, control activator) at the predetermined 80% maximal effective concentration (EC80) was added to wells containing the control inhibitor or compounds. The plate was incubated for 5 h at 37 °C/5% CO2 in a humidified incubator before 8 μL substrate loading solution (1 μM) was added to each well, followed by incubation for 2 h at RT. The plate was read on a fluorescence plate reader.
Heat shock response–HepG2 cells–inhibitor screen
HepG2 cells (1 × 104 cells per well) were plated in a solid-white 96-well plate (Corning Cat. # 3917) and incubated for 24 h in a 37 °C, 5% CO2 incubator. Vector constructs were diluted at a 10:1 mass ratio to 10 ng total DNA/μL in Opti-MEM I. Then, FuGENE HD (E2311) was added at a 4.5:1 lipid:DNA ratio. Samples were mixed by pipetting and incubated at RT for 20 min. A 10 μL volume of transfection complex was added to each well (100 ng DNA/well) and the plate was incubated for 18 h in a 37 °C/5% CO2 incubator. The culture medium was removed from the cells and replaced with 72 μL DMEM + 0.5% charcoal-stripped FBS per well. Then, 8 μL 17-AAG (1 μM) was added to each well and the plate was incubated for another 6 h in a 37 °C/5% CO2 incubator. The plates were removed from the 37 °C/5% CO2 incubator and allowed to cool to RT for approximately 15 min, and then luminescence was measured.
MAPK/ERK-luc2P/SRE/Hygro HEK293 cells-inhibitor screen
HEK293 cells (1 × 104–1.5 × 104 cells per well) were plated in a solid-white 96-well plate (Corning Cat# 3917) and incubated for 24 h in a 37 °C/5% CO2 incubator. The pGL4.33 [luc2P/SRE/Hygro] vector (Promega, E134A) was diluted at a 10:1 mass ratio to 10 ng total DNA/μL in Opti-MEM I. FuGENE HD (E2311) was added at a 4.5:1 lipid:DNA ratio. The samples were mixed by pipetting and incubated at RT for 20 min. Then, 10 μL of transfection complex was added per well (100 ng DNA/well) and the plate was incubated in a 37 °C/5% CO2 incubator. After a 4–6-h incubation, the medium was changed to DMEM/0.5% FBS (100 µl per well) to start serum starvation. Then, 50 µL of the medium was removed, and 50 µL 2× induction solution (40% FBS plus 20 ng/mL EGF in DMEM) was added to the cells to be induced, or 50 µL 2× control solution (DMEM) was added to the control/noninduced cells. The cells were incubated for 6 h in a 37 °C/5% CO2 incubator. The plates were then removed from the 37 °C/5% CO2 incubator and allowed to cool to RT for approximately 15 min, and luminescence was measured.
Interferon regulation pathway–HeLa cells–inhibitor screen
HeLa cells (1 × 104 per well) were plated in a solid-white 96-well plate (Corning Cat# 3917) and incubated for 24 h in a 37 °C/5% CO2 incubator. A 10 μL aliquot of transfection complex (pDD006, pFirefly, polyethylenimine) was added to the cells, which were incubated for another 4 h in a 37 °C/5% CO2 incubator. The culture medium was then removed from the cells and replaced with complete medium (DMEM + 10% FBS). The plate was incubated for 4 h in a 37 °C/5% CO2 incubator. Then, the culture medium was removed from the cells, and IFN-γ (final concentration; 10 ng/mL) or test compound(s) was added to appropriate wells of the plate. The plate was then incubated for 16 h in a 37 °C/5% CO2 incubator. The plates were removed from the 37 °C/5% CO2 incubator and allowed to cool to RT for approximately 15 min prior to luminescence detection.
STAT3 pathway–HEK293 cells–inhibitor screen
A total of 2.5 × 104 HEK293 cells per well were plated in a solid-white 96-well plate (Corning Cat. # 3917) and incubated for 24 h in a 37 °C/5% CO2 incubator. A 20 μL aliquot of transfection complex (pDD021, pFirefly, polyethylenimine) was added to the cells, which were incubated for 4 h in a 37 °C/5% CO2 incubator. At that time, the culture medium was removed from the cells and replaced with a complete medium (DMEM + 10% FBS). The plate was then incubated for 24 h in a 37 °C/5% CO2 incubator. Then, the culture medium was removed from the cells, and IL-6 (final concentration of 10 ng/ml) or test compound(s) was added to appropriate wells of the plate. The plate was then incubated for 24 h in a 37 °C/5% CO2 incubator. The plates were removed from the 37 °C/5% CO2 incubator and allowed to cool to RT for approximately 15 min, and then luminescence was measured.
NF-κB pathway–luc2P/NF-κB-RE/Hygro HEK293 cells
A total of 1.5 × 104 HEK293 cells per well were plated in a solid-white 96-well plate (Corning Cat. # 3917) and incubated for 24 h in a 37 °C/5% CO2 incubator. Each well of the 96-well plate to be transfected received 0.1 µg pGL4.32 [luc2P/NF-κB-RE/Hygro] plasmid DNA (Promega, E849A). The plate was then incubated for another 24 h in a 37 °C/5% CO2 incubator. The medium was removed from the wells, and 100 µl 1× induction solution (20 ng/mL TNFα in DMEM/FBS) was added to the cells to be induced, or 100 µl 1× control solution (DMEM/FBS) was added to the control noninduced cells. The cells were incubated for 5 h in a 37 °C/5% CO2 incubator. The plates were then removed from the incubator and allowed to cool to RT for approximately 15 min prior to the measurement of luminescence.
PK profiling
PK analysis of hsBCL9Z96 was conducted in 3 male Sprague–Dawley rats (weighing 220–250 × g) (Shanghai SLAC Laboratory Animal Co. Ltd.) in accordance with a standard protocol (Viva Biotech) and ethical regulations (IACUC). Rats were administered hsBCL9Z96 by intravenous injection via the tail vein. Positive control vehicles were prepared with 10% DMSO and 90% deionized H2O to generate a mixed solvent, with peptides tested at nominal concentrations of 0.5 mg/mL (incubated at RT before administration). Blood samples (300 mL) were collected periodically from the retro-orbital vein at 0.083, 0.25, 0.5, 1, 2, 4, 8, 24, 48, and 72 h after dosing, followed by plasma separation for subsequent bioanalysis.
Cell viability assay
Cell viability was assessed using the cell counting kit-8. Tumor cells (100 µl; 10,000 cells/well) were seeded in a solid-white 96-well plate (Corning Cat# 3917) and incubated for 24 h in a 37 °C/5% CO2 incubator. Then the cells were treated with different concentrations of hsBCL9CT-24 or hsBCL9Z96 for 24 h at 37 °C under 5% CO2. Subsequently, 10 µl of CCK8 solution (MA0218-2, Meilunbio) was added into the medium and the plate was incubated at 37 °C for 1 h away from light. The absorbance (450 nm) was measured using a Synergy H1 microplate reader (BioTek).
Cell membrane penetration assay
To determine the cell membrane penetration ratio of hsBCL9Z96, HCT116 cells were treated with hsBCL9CT-24-FITC or hsBCL9Z96-FITC in vitro for 24 h, and the proportion of FITC+ live cells was analyzed by flow cytometry.
The penetration rates of hsBCL9Z96 and hsBCL9CT-24 in Hepa1-6 tumor tissues were measured by in vivo imaging. Briefly, the tumor samples were fixed with 4% paraformaldehyde, then pruned, dehydrated, embedded, sliced, stained with DAPI (Absinbio, abs47047616), and sealed in strict accordance with the SOP procedure of the pathology department. Finally, samples qualified for microscopic examination were obtained. Pictures were taken with a Pannoramic MIDI II (3D HISTECH). Images were analyzed using Indica Labs software (U.S.A.).
Drug safety assessment
To perform a preliminary toxicology study of hsBCL9Z96, SD rats (n = 3) were treated with vehicle control (25% glucose) or hsBCL9Z96 (25, 50, and 100 mg/kg) via intravenous (i.v.) injection, QD over 14 days. The body weight and food intake were recorded. Serum chemistry and hematology studies were performed. In another study, Balb/c mice (n = 3) were treated with vehicle control (25% glucose) or hsBCL9Z96 at 10 mg/kg via i.v. injection, QD over 14 days. The numbers of different blood cell types and parameters were analyzed, including white blood cells, lymphocytes (Lym), monocytes (Mon), neutrophils (Neu), red blood cells, hematocrit (HCT), hemoglobin (HGB), platelets (PLT), and the mean platelet volume (MPV).
Statistical analysis
GraphPad software (version 9.0) and R statistical software (version 4.2.0) were used for most data analyses. Significance was determined by Student’s t-test when comparing two groups. Survival and recurrence curves were generated by the Kaplan–Meier method, and the differences between groups were evaluated by the log-rank test. A two-tailed P < 0.05 was considered to be statistically significant. Data are presented as the means ± SD unless otherwise indicated.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data generated in this study have been deposited in the Genome Sequence Archive (GSA) under project accession number CRA031201 and CRA031323. Publicly available datasets are available under accession codes: GSE125449 (GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125449), GSE140228 (GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140228), GSE151530 (GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151530), GSE22058 (GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22058), GSE43619 (GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43619), GSE19977 (GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19977), and TCGA cancer data (TCGA database: https://www.cancer.gov/tcga). Clinical and RNA-seq data of the GO30140 and IMbrave150 cohorts were downloaded from European Genome-Phenome Archive under accession no. EGAS00001005503. Proteomic data of HCC cohorts were obtained from CPTAC under accession No. PDC000198. All newly generated materials are available from the corresponding author upon request. Requests will be processed under a Materials Transfer Agreement to ensure appropriate use and to address any intellectual property constraints. The remaining data are available within the article, Supplementary Information, Source Data file or from the corresponding author on request. Source data are provided with this paper.
Code availability
Software used for single-omic and multi-omic analyses were described and referenced in the respective Method Details subsections with default parameters and listed in the Supplementary Table 3. No additional customer code was generated in this article
References
Kubli, S. P. et al. Beyond immune checkpoint blockade: emerging immunological strategies. Nat. Rev. Drug Discov. 20, 899–919 (2021).
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Yau, T. et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): a randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 23, 77–90 (2022).
Kelley, R. K. et al. Safety, efficacy, and pharmacodynamics of tremelimumab plus durvalumab for patients with unresectable hepatocellular carcinoma: randomized expansion of a phase I/II study. J. Clin. Oncol. 39, 2991–3001 (2021).
Morad, G. et al. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184, 5309–5337 (2021).
Zhu, A. X. et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat. Med. 28, 1599–1611 (2022).
Aun, R. S. & Sandeep, M. Impact of gut microbiome on immunotherapy resistance in hepatocellular carcinoma: a review article. Hepatoma Res. 10, 48 (2024).
Xu, C. et al. β-Catenin signaling in hepatocellular carcinoma. J. Clin. Invest 132, e154515 (2022).
Perugorria, M. J. et al. Wnt-β-catenin signalling in liver development, health and disease. Nat. Rev. Gastroenterol. Hepatol. 16, 121–136 (2019).
Zhan, T., Rindtorff, N. & Boutros, M. Wnt signaling in cancer. Oncogene 36, 1461–1473 (2017).
Gao, Q. et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell 179, 561–577 (2019).
Zhou, S. L. et al. Whole-genome sequencing reveals the evolutionary trajectory of HBV-related hepatocellular carcinoma early recurrence. Signal Transduct. Target Ther. 7, 24 (2022).
Sun, Y. et al. Integrated multi-omics profiling to dissect the spatiotemporal evolution of metastatic hepatocellular carcinoma. Cancer Cell 42, 135–156.e17 (2024).
Wang, Z., Li, Z. & Ji, H. Direct targeting of β-catenin in the Wnt signaling pathway: Current progress and perspectives. Med. Res. Rev. 41, 2109–2129 (2021).
Jung, Y. S. & Park, J. I. Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond β-catenin and the destruction complex. Exp. Mol. Med. 52, 183–191 (2020).
Feng, M. et al. Pharmacological inhibition of β-catenin/BCL9 interaction overcomes resistance to immune checkpoint blockades by modulating T cells. Sci. Adv. 5, eaau5240 (2019).
Liu, Y. et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell 40, 424–437.e5 (2022).
Song, F. et al. Anlotinib potentiates anti-PD1 immunotherapy via transferrin receptor-dependent CD8(+) T-cell infiltration in hepatocellular carcinoma. Clin. Transl. Med. 14, e1738 (2024).
Hoadley, K. A. et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018).
Ma, L. et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 75, 1397–1408 (2021).
Xie, J. et al. Cell-penetrating peptides in diagnosis and treatment of human diseases: from preclinical research to clinical application. Front. Pharm. 11, 697 (2020).
Li, Z. et al. Applications of surface modification technologies in nanomedicine for deep tumor penetration. Adv. Sci. 8, 2002589 (2020).
Chiu, D. K. et al. Hepatocellular carcinoma cells up-regulate PVRL1, stabilizing PVR and inhibiting the cytotoxic T-cell response via TIGIT to mediate tumor resistance to PD1 inhibitors in mice. Gastroenterology 159, 609–623 (2020).
Christofides, A. et al. The complex role of tumor-infiltrating macrophages. Nat. Immunol. 23, 1148–1156 (2022).
Duan, Z. & Luo, Y. Targeting macrophages in cancer immunotherapy. Signal Transduct. Target Ther. 6, 127 (2021).
Martínez, V. G. et al. BMP4 induces M2 macrophage polarization and favors tumor progression in bladder cancer. Clin. Cancer Res. 23, 7388–7399 (2017).
Lecoultre, M., Dutoit, V. & Walker, P. R. Phagocytic function of tumor-associated macrophages as a key determinant of tumor progression control: a review. J. Immunother. Cancer 8, e001408 (2020).
Ma, L. et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell 36, 418–430.e6 (2019).
Zhang, Q. et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179, 829–845.e20 (2019).
Barkal, A. A. et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572, 392–396 (2019).
Seehawer, M. et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 562, 69–75 (2018).
Biswas, S. K. & Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11, 889–896 (2010).
Anfray, C. et al. Current strategies to target tumor-associated-macrophages to improve anti-tumor immune responses. Cells 9, 46 (2019).
Wang, C. et al. Interaction of glioma-associated microglia/macrophages and anti-PD1 immunotherapy. Cancer Immunol. Immunother. 72, 1685–1698 (2023).
Zhang, X. et al. Cell microparticles loaded with tumor antigen and resiquimod reprogram tumor-associated macrophages and promote stem-like CD8(+) T cells to boost anti-PD-1 therapy. Nat. Commun. 14, 5653 (2023).
Wang, S. et al. Macrophage-tumor chimeric exosomes accumulate in lymph node and tumor to activate the immune response and the tumor microenvironment. Sci. Transl. Med. 13, eabb6981 (2021).
Lampreht Tratar, U. et al. Gene electrotransfer of plasmid-encoding IL-12 recruits the M1 macrophages and antigen-presenting cells inducing the eradication of aggressive B16F10 murine melanoma. Med. Inflamm. 2017, 5285890 (2017).
David, T. et al. Anti-cathepsin D immunotherapy triggers both innate and adaptive anti-tumour immunity in breast cancer. Br. J. Pharmacol. https://doi.org/10.1111/bph.16291 (2023).
Feng, M. et al. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat. Rev. Cancer 19, 568–586 (2019).
Lin, H. et al. Stanniocalcin 1 is a phagocytosis checkpoint driving tumor immune resistance. Cancer Cell 39, 480–493.e6 (2021).
Ruiz de Galarreta, M. et al. β-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 9, 1124–1141 (2019).
Sanchez-Vega, F. et al. Oncogenic signaling pathways in the Cancer Genome Atlas. Cell 173, 321–337.e10 (2018).
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382 (2019).
Lee, M. S. & Bensinger, S. J. Reprogramming cholesterol metabolism in macrophages and its role in host defense against cholesterol-dependent cytolysins. Cell Mol. Immunol. 19, 327–336 (2022).
Sun, Y. et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 184, 404–421.e16 (2021).
Sachdeva, M., Chawla, Y. K. & Arora, S. K. Immunology of hepatocellular carcinoma. World J. Hepatol. 7, 2080–2090 (2015).
Zhou, D. et al. Tumor-associated macrophages in hepatocellular carcinoma: friend or foe?. Gut Liver 15, 500–516 (2021).
Muntjewerff, E. M., Meesters, L. D. & van den Bogaart, G. Antigen cross-presentation by macrophages. Front Immunol. 11, 1276 (2020).
Lurje, I., Hammerich, L. & Tacke, F. Dendritic cell and T cell crosstalk in liver fibrogenesis and hepatocarcinogenesis: implications for prevention and therapy of liver cancer. Int. J. Mol. Sci. 21, 7378 (2020).
Mantovani, A. et al. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 21, 799–820 (2022).
Giraldo, N. A. et al. The clinical role of the TME in solid cancer. Br. J. Cancer 120, 45–53 (2019).
Fu, Y. et al. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 38, 396 (2019).
Fereshteh, Y. et al. Measures for response assessment in HCC treatment. Hepatoma Res. 10, 27 (2024).
Percie du Sert, N. et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 18, e3000410 (2020).
Hu, B. et al. IFNα potentiates anti-PD-1 efficacy by remodeling glucose metabolism in the hepatocellular carcinoma microenvironment. Cancer Discov. 12, 1718–1741 (2022).
Efremova, M. et al. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Acknowledgements
This research was supported by the following funding awards: National Natural Science Foundation of China: 82341027 (to X.R.Y.), 82072715 (to X.R.Y.), 81802364 (to B.H.), 81772551 (to J.F.), 81872895 (to D.Z.), 82073881 (to D.Z.), 82073272 (to Y.X.), 81830102 (to J.Z.), 81772578 (to J.Z.), 82473284 (to B.H.), and 82303161 (to J.L.S.). National Key R&D Program of China: 2019YFC1315800 (to J.Z.), 2019YFC1315802 (to J.Z.). Shanghai Municipal Health Commission Collaborative Innovation Cluster Project: 2019CXJQ02 (to J.F.). Eastern Talent Program Leading project (to X.R.Y.). Projects from the Shanghai Science and Technology Commission: 21140900300 (to X.R.Y.) and 22140900600 (to B.H.). Project from Shanghai hospital development center: SHDC2023CRD025 (to X.R.Y.). Projects from Science Foundation of Zhongshan Hospital, Fudan University: 2021ZSCX28 (to X.R.Y.) and 2020ZSLC31 (to X.R.Y.).
Author information
Authors and Affiliations
Contributions
X.R.Y., B.H., D.Z., J.F., and A.X.Z. designed the study, interpreted the data, and wrote the manuscript. B.H., S.Y.W., Y.Y.Z., J.L.S., and C.Y.W. conducted most of the experiments. Y.L.W., Y.Y.N., F.S., X.H., Z.C., T.H., L.A.S., Y.X., C.H., S.J.Q., and J.Z. helped design experiments, coordinated the studies, and contributed key reagents. X.H., T.H., and C.H. assisted technically in molecular biology and functional analyses performed in vitro. X.R.Y., B.H., D.Z., J.F., and A.X.Z. contributed to study supervision.
Corresponding authors
Ethics declarations
Competing interests
A.X.Z. received consulting fees from Bayer, Eisai, Eli Lilly, Exelixis, F. Hoffmann–La Roche, Merck, Gilead, Sanofi Aventis, and Sirtex. The other authors (S.Y.W., Y.Y.Z., J.L.S., C.Y.W., Y.L.W., Y.Y.N., F.S., X.H., Z.C., T.H., L.A.S., Y.X., C.H., S.J.Q., J.Z., J.F., D.Z., B.H., and X.R.Y.) declare no competing financial or non-financial interests.
Peer review
Peer review information
Nature Communications thanks Jamila FAIVRE and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, SY., Zhu, YY., Sun, JL. et al. Targeting tumor-intrinsic BCL9 reverses immunotherapy resistance by eliciting macrophage-mediated phagocytosis and antigen presentation. Nat Commun 16, 10039 (2025). https://doi.org/10.1038/s41467-025-65945-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65945-z