Abstract
Ferroptotic waves aggravate kidney ischemia-reperfusion injury and drive delayed graft function (DGF). We demonstrate that elevated glycolysis and lactate production in graft kidney correlate with ferroptosis and functional impairment. A signaling axis composed of the long non-coding RNA IGIP-5, microRNA 670-3p, and lactate dehydrogenase A promotes lactate secretion from injured tubular cells, inducing lactylation and ferroptosis in neighboring cells and triggering ferroptotic waves. Lactylome profiling identifies that nucleophosmin 1 (NPM1), an epigenetic regulator, is lactylated at lysine 257 by the lactyltransferase AARS1. Suppressing NPM1 lactylation—via K257 mutation, AARS1 knockout, or taurochenodeoxycholic acid—upregulates SLC7A11 and inhibits ferroptosis. Mechanistically, lactylation stabilizes NPM1 by reducing MDM2-mediated ubiquitination and strengthens SLC7A11 repression, disrupting cystine metabolism. In mouse allografts, blocking lactate shuttle-mediated NPM1 lactylation prevents ferroptotic propagation and ameliorates graft function. Additionally, we develop an early prediction model for DGF using postoperative urinary lactate concentrations. These findings reveal a metabolic-epigenetic axis driving ferroptotic propagation and propose NPM1 lactylation as a therapeutic target for DGF.
Similar content being viewed by others
Introduction
Kidney transplantation more significantly enhances the quality of life and extends the survival of patients with end-stage renal disease compared to regular dialysis1. However, ischemia-reperfusion injury (IRI) of the graft kidney is inevitable during the organ procurement, storage, and transplantation processes and is a crucial cause of delayed graft function (DGF) and graft failure2. To address the increasing and imbalanced demand for donor organs, donations after circulatory death and extended-criteria donors have been approved, further exacerbating IRI-induced DGF3. Therefore, elucidating the underlying molecular mechanisms and identifying novel therapeutic targets are urgently required to manage DGF.
Metabolic reprogramming controls the fate of tubular epithelial cells (RC) in kidney IRI, making the aberrant metabolism in these cells a viable therapeutic target for preventing kidney IRI. Deletion of kynurenine 3-monooxygenase (KMO), an enzyme highly expressed in the kidney that regulates renal kynurenine metabolism, protects against kidney IRI by reducing neutrophil infiltration4. Inhibition of DNA-binding protein-A (DbpA), a mitochondrial protein deleterious to cell metabolism, protects against kidney IRI by enhancing lipid metabolism and oxygen consumption rates in RC5. Enhancing fatty acid oxidation inhibits pericyte-myofibroblast transition and thereby alleviates kidney IRI6. In addition, glycolysis is a pivotal metabolic pathway for maintaining energy balance and biological functions. Recent evidence reflects that glycolytic reprogramming and the by-product lactate play diverse roles in IRI in various organs and tissues7,8. Deletion of the enzyme responsible for L-2-hydroxyglutarate (L2HG) biosynthesis facilitates a metabolic shift from the pentose phosphate pathway towards glycolysis, which induced oxidative injury and aggravates myocardial injury during low-flow ischemia and ischemia-reperfusion processes9. IRI cardiomyocytes exhibited enhanced glycolysis and suppressed mitochondrial respiration; inhibition of the transcriptional regulator homeobox-containing 1 (Hmbox1) enhanced both metabolic processes and reduced cardiomyocyte apoptosis, protecting against cardiac IRI10. The inhibitor of phosphoenolpyruvate carboxykinase (PCK1) aggravates hepatic IRI by blocking gluconeogenesis (the reverse process of glycolysis) and deepening lactate accumulation11. Neutrophil glycolytic activation facilitates neutrophil extracellular trap formation, exacerbating kidney IRI12. Nevertheless, the regulatory effects of glycolysis on the kidney IRI and associated mechanisms remain unclear.
Lactate-induced lactylation integrates metabolic cues into epigenetic regulation, providing novel insights into the regulatory mechanisms of glycolysis under various physiological and pathological conditions13. Increasing evidence suggests that lactylation plays various roles in IRI. Lipoprotein receptor-related protein-1 (LRP1) suppresses lactate production and ADP-ribosylation factor 1 (ARF1) lactylation protects against brain IRI and ischemic stroke14. The glycolytic enzyme 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) is upregulated in IRI kidneys and prominently potentiates lactate accumulation and histone H4K12 lactylation, thereby promoting chronic kidney disease15. The mechanism through which lactate-derived lactylation regulates graft kidney IRI is not completely understood.
A growing body of evidence has linked ferroptosis to kidney IRI, indicating that ferroptosis may represent a promising diagnostic and therapeutic target for DGF16,17,18. Ferroptotic events, called ferroptotic trigger waves, can extend over long distances and induce large-scale cell death in a non-autonomous manner19,20. Exciting progress in modern research has revealed that dying cell-derived death signals are key propagation mechanisms21,22,23. Platelet-activating factor induces instability of biomembranes and augments synchronized ferroptosis in neighboring cells, thus exacerbating kidney IRI24. Previously, we demonstrated that renal tubular epithelial cells in ischemia-reperfusion injury (IR-RC) secrete small extracellular vesicles and facilitate ferroptotic propagation via the promotion of hexosamine biosynthetic pathway (HBP) metabolic reprogramming and protein O-GlcNAcylation25. However, the roles of other metabolic and epigenetic mechanisms in regulating ferroptotic propagation during kidney IRI remain unclear.
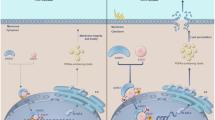
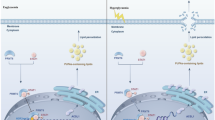
In the present study, we confirmed the pivotal role of lactate-derived lactylation in facilitating ferroptotic waves and exacerbating DGF. Additionally, we found that abnormal activation of the long non-coding RNA IGIP-5/microRNA 670-3p/lactate dehydrogenase A (LDHA) axis induces glycolytic reprogramming and increases lactate production and secretion from IR-RC. Lactate shuttle further augmented Alanyl-tRNA synthetase 1 (AARS1)-mediated nucleophosmin 1 (NPM1) lactylation in adjacent renal tubular epithelial cell (RC) populations, suppressed solute carrier family 7 member 11 (SLC7A11) expression, and induced ferroptosis. These findings imply that the inhibition of lactate shuttle and AARS1-mediated NPM1 lactylation can potentially serve as promising strategies for effective protection against kidney IRI and DGF after kidney transplantation.
Results
The upregulation of glycolysis and generation of byproduct lactate is correlated to the decline in graft kidney function
To determine changes in metabolic patterns during kidney IRI, we established an in situ kidney IRI mouse model (Supplementary Fig. 1a) and delineated the metabolic profiles of injured kidneys. Metabolomic analyses revealed that lactate, a terminal glycolysis metabolite, increased time-dependently during reperfusion (Fig. 1a and Supplementary Fig. 1b), which was validated in mouse models across extended reperfusion periods (Fig. 1b). Lactate dehydrogenase (LDH)-mediated pyruvate conversion drives lactate production. We therefore assessed glycolytic enzyme alterations and observed significant LDHA upregulation during prolonged reperfusion (Fig. 1c, d). Higher lactate concentration was closely related to the decline in renal function (Fig. 1e, f), exhibiting a positive correlation with serum creatinine (SCr) concentration (r = 0.756, p < 0.001, n = 30; Fig. 1g). We further examined the lactate concentration in urine (ULa) of 150 clinical recipients after kidney transplantation (Supplementary Table 1) and detected a considerably higher lactate concentration in recipients exhibiting DGF (DGF recipients; SCr ≥400 μmoL/L on the postoperative day 7) than non-DGF recipients on the postoperative days 2 and 7 (ULa-2 d and ULa-7 d) (Fig. 1h and Supplementary Table 2). In addition, during the recovery of renal function, the ULa declined on the postoperative day 7 in non-DGF patients, whereas, it exhibited no notable change in DGF recipients (Fig. 1i).
a Heatmap illustrated the energy metabolic metabolites in mouse kidney tissues after ischemia and reperfusion for 0 h, 12 h and 24 h (n = 4 group−1). b Lactate assay kit detecting lactate concentration in IRI kidney tissues at different reperfusion stages (n = 6 group−1) and normalized according to the sham group concentrations; one-way ANOVA followed by Tukey’s test. c, d The translational and transcriptional levels of glycolytic enzymes in mouse kidney tissues following ischemia-reperfusion injury (IRI) (n = 6 group−1), and normalized according to sham group levels; one-way ANOVA followed by Tukey’s test. e, f The serum creatinine (SCr) and blood urea nitrogen (BUN) concentration of mice after kidney IRI (n = 6 group−1), and one-way ANOVA followed by Tukey’s test; Boxplot: 25th–75th percentiles; center line: median; whiskers: 1.5 times the interquartile range. g A two-sided Pearson correlation analysis of SCr concentration and lactate concentration (r = 0.756, p < 0.001, n = 30). h, i Urinary lactate concentration of non-DGF (n = 105) and DGF (n = 45) recipients on the postoperative days 2 (ULa-2 d) and 7 (ULa-7 d); unpaired 2-tailed Student’s t test. j A two-sided Pearson correlation analysis of SCr and ULa-2 d concentrations (r = 0.716, p < 0.001, n = 150). k The ROC curve of the DGF prediction model based on ULa-2 d of recipients in the training set (n = 150). l, m Urinary lactate concentration of non-DGF (n = 45) and DGF (n = 19) recipients in the validation set and the ROC curve of the DGF prediction model; unpaired 2-tailed Student’s t test. n Kaplan‒Meier survival curves of the five-year graft survival stratified by the median ULa-2 d level (n = 110); the p value was calculated via the log-rank test. Data are presented as mean ± SD. All experiments were repeated at least three times, yielding similar results. The pvalues are shown for the indicated comparisons. Source data are provided as a Source Data file.
Pearson correlation analyses revealed positive correlations between the SCr and ULa concentrations on postoperative day 2 (r = 0.716, p < 0.001, n = 150; Fig. 1j) and day 7 (r = 0.722, p < 0.001, n = 150; Supplementary Fig. 1c). Serum lactate concentration in recipients on the postoperative day 2 (SLa-2 d) also correlated positively with SCr (r = 0.579, p < 0.001, n = 150; Supplementary Fig. 1d), though this association was weaker than that observed between ULa-2d and SCr. Receiver operating characteristic (ROC) curve analysis demonstrated strong early predictive value for DGF using ULa-2 d (AUC = 0.859, n = 150, Fig. 1k), with performance comparable to the ULa-7d model (AUC = 0.861, n = 150, Supplementary Fig. 1e). The ULa-2 d model was further validated in an independent cohort of 64 recipients (AUC = 0.864, Fig. 1l, m and Supplementary Table 3). While ULa-2 d exhibited comparable predictive accuracy to established DGF biomarkers (urinary neutrophil gelatinase-associated lipocalin (NGAL)−2 d: AUC = 0.857, n = 150, Supplementary Fig. 1f; urinary kidney injury molecule-1 (KIM-1)−2 d: AUC = 0.871, n = 150, Supplementary Fig. 1g), it outperformed conventional predictors including SLa-2 d (AUC = 0.685, n = 150; Supplementary Fig. 1h), SCr-2 d (AUC = 0.730, n = 150; Supplementary Fig. 1i), and 24-h urine volume (AUC = 0.747, n = 150; Supplementary Fig. 1j). Using Firth penalized logistic regression that incorporated established clinical variables associated with graft functional recovery, we identified ULa-2 d, cold ischemia time, 24-h urine volume, and HLA mismatch number as independent risk factors for DGF (Supplementary Table 4). ULa-2 d was the most significant predictor, with high levels conferring a 4.27-fold increase in DGF risk (adjusted odds ratio = 4.27, 95% CI: 1.58–12.14, p = 0.004). Notably, the multivariable model including all four parameters achieved a higher AUC (AUC = 0.879; n = 150) than the model based on ULa-2d (Supplementary Fig. 1k). Moreover, exclusion of ULa-2d from the multivariable model reduced the AUC to 0.788, highlighting its critical role in DGF risk prediction. Additionally, recipients stratified by median ULa-2 d into high- (n = 55) and low-level (n = 55) groups exhibited significantly reduced 5-year graft survival in the high-ULa group (HR = 3.266, p = 0.029, Fig. 1n). These data obtained in patients and mouse models establish lactate as a pivotal mediator of kidney IRI and DGF.
Lactate secreted from the IR-RC facilitates ferroptotic propagation via inhibiting the cystine/cysteine/GSH metabolism in the normal RC population
Ferroptotic waves exacerbate kidney IRI and DGF. Interestingly, ferroptosis was also augmented (Supplementary Fig. 2a–d) along with the upregulation of glycolysis and lactate synthesis in kidneys with IRI (Supplementary Fig. 2e-f). The inhibition of glycolysis by sodium oxamate significantly decreased ferroptosis in the IRI kidneys and restored the declined renal function, whereas, lactate treatment further induced ferroptosis and deteriorated renal function (Fig. 2a–c). Suppression of ferroptosis via ferrostatin-1, a canonical ferroptosis inhibitor, neutralized lactate-induced kidney injury (Fig. 2d–f). Acknowledging the reported limitations regarding the in vivo stability of ferrostatin-1, we further utilized liproxstatin-1, a ferroptosis inhibitor with superior metabolic stability in vivo26,27. Similar to ferrostatin-1, liproxstatin-1 effectively suppressed ferroptosis and restored lactate-impaired renal function in IRI. Conversely, imidazole ketone erastin (IKE), an inhibitor of cystine–glutamate antiporter system xc–, induced ferroptosis and further exacerbated kidney IRI (Supplementary Fig. 2g-l). Additionally, neither ferrostatin-1 nor liproxstatin-1 altered the expression of glycolytic enzymes or lactate production in the IRI process (Supplementary Fig. 2m-o).
a Flowchart delineating kidney IRI protocol in C57BL/6 mice; sodium oxamate (1 g/kg, OXA), sodium lactate (120 mg/kg, Lac), ferrostatin-1 (5 mg/kg, Fer-1), normal saline (N.S.). SCr (b) and BUN (c) concentration in mice treated with the same experimental setup in a; lipid peroxidation level (d), malondialdehyde (MDA) concentration (e) and iron level (f) in mouse kidney tissues (n = 20 group−1); iron level was normalized according to sham group results; one-way ANOVA followed by Tukey’s test; Boxplot: 25th–75th percentiles; center line: median; whiskers: 1.5 times the interquartile range. g Lactate concentration in conditioned medium of normal and IRI HK-2 cells (NC-CM and IR-CM); normalized according to concentrations in NC-CM (n = 3 group−1); one-way ANOVA followed by Tukey’s test; IR-CMsiMCT4 and IR-CMOXA represent CM from IRI HK-2 cells treated with siMCT4 or OXA. h Schematic representing co-culture models established by normal HK-2 cells and different CM. Lactate concentration (i) and lipid peroxidation (j) in HK-2 cells treated with the same experimental setup in (h) (n = 3 group−1); normalized according to Vehicle group concentrations; one-way ANOVA followed by Tukey’s test. k Flowchart depicting the metabolic flux tracing assay in secreting cells incubated with [13C3]-L-Lactate and importing cells. l, m M + 3 lactate fraction in cell lysate and lactate transport rate (the ratio of m + 3 lactate fraction in importing cells to secreting cells) (n = 3 group−1); one-way ANOVA followed by Tukey’s test. n Metabolite contents in cystine/cysteine/GSH pathway in mouse kidney tissues (n = 6 group−1) and normalized according to sham group concentrations; one-way ANOVA followed by Tukey’s test. o, p Western blotting determining SLC7A11 expression in mouse kidney tissues after different reperfusion phases (n = 6 group−1) and different treatments (n = 20 group−1). Data are presented as mean ± SD. All experiments were repeated at least three times, yielding similar results. The pvalues are shown for the indicated comparisons. Figure 2a, h were created in FigDraw. https://www.figdraw.com; Fig. 2k, n were created in BioRender. Feng, Z. (2025) https://BioRender.com/ye9fs3m and https://BioRender.com/e3ywthw. Source data are provided as a Source Data file.
While determining whether lactate induced ferroptotic waves, initially, we confirmed a large release of lactate from IR-RC (Fig. 2g) and treated normal HK-2 cells using the conditioned medium of IR-RC (IR-CM) and normal RC (NC-CM) (Fig. 2h). We noticed an increase in lactate levels in normal HK-2 cells treated with IR-CM compared to those in the NC-CM treatment group (Fig. 2i). IR-CM considerably induced ferroptosis in normal HK-2 cells, as validated by marked increases in lipid peroxidation and iron levels (Fig. 2j and Supplementary Fig. 3a-b). These increases were substantially attenuated by treatment with CM derived from IR-RC in which monocarboxylate transporter 4 (MCT4) was silenced by siRNA (IR-CMsiMCT4). Additionally, CM derived from IR-RC treated with sodium oxamate (IR-CMOXA) exhibited reduced lactate production, leading to decreased lactate concentrations and ferroptosis levels in normal HK-2 cells. Consistent with the in vivo findings, both ferrostatin-1 and liproxstatin-1 effectively rescued HK-2 cells from IR-CM-induced ferroptotic cell death (Supplementary Fig. 3c–e). Intriguingly, NC-CM supplemented with sodium lactate recaptured the function of IR-CM to induce ferroptosis in normal HK-2 cells; this effect was abolished by AZD3965, a selective monocarboxylate transporter 1 (MCT1) inhibitor blocking lactate import (Supplementary Fig. 3f–h). Notably, pharmacological inhibition of MCT4 (syrosingopine) in IR-RC and MCT1 in normal HK-2 cells effectively blocked intercellular lactate shuttling (Supplementary Fig. 3i–j), consequently suppressing IR-CM-induced ferroptosis in normal HK-2 cells (Supplementary Fig. 3k–m). In vivo validation further revealed that inhibitors of MCT4 and MCT1 suppressed ferroptosis (Supplementary Fig. 3n–p), and protected renal function (Supplementary Fig. 3q–r). Additionally, we employed 13C3-L-lactate isotopic tracing to quantify lactate transport rates (Fig. 2k). The results revealed that lactate transport rate (the m + 3 lactate fraction in importing cells versus secreting cells) was decreased, following MCT4 inhibition in lactate-secreting cells or MCT1 inhibition in lactate-importing cells (Fig. 2l, m and Supplementary Fig. 3s-t). These results indicate that highly glycolytic IR-RC secreted lactate, triggering ferroptotic waves in adjacent normal RC populations. Inhibition of lactate production and MCT4/MCT1-mediated lactate shuttle effectively blocked ferroptosis propagation and ameliorated kidney IRI.
Cystine and cysteine are important anti-ferroptotic metabolites, implicated in glutathione (GSH) synthesis and lipid peroxide detoxification (Supplementary Fig. 4a). Metabolomic analyses indicated that cystine decreased along with the upregulation of ferroptosis in injured kidneys (Fig. 1a and Supplementary Fig. 4b). Furthermore, we detected that metabolites involved in cystine/cysteine/GSH pathway were decreased with a longer reperfusion phase (Fig. 2n), consistent with the alteration of SLC7A11, a key subunit of system xc– cystine–glutamate antiporter (Fig. 2o). Inhibition of glycolysis facilitated SLC7A11 expression and the cystine/cysteine/GSH metabolism in graft kidney; lactate treatment further suppressed this pathway (Fig. 2p and Supplementary Fig. 4c–e). In vitro analyses further demonstrated that IR-CM suppressed SLC7A11 expression and cystine/cysteine/GSH metabolism in normal HK-2 cells, which was substantially abolished by treatment with IR-CMsiMCT4 and IR-CMOXA (Supplementary Fig. 4f-j). However, IR-CM induced no significant changes in other ferroptosis regulators including glutathione peroxidase 4 (GPX4) and acyl-CoA synthetase long-chain family member 4 (ACSL4), with expression remaining unchanged even under lactate pathway inhibition (Supplementary Fig. 4k–m). Consistently, supplementing NC-CM with sodium lactate recapitulated the suppression of SLC7A11 expression and cystine/cysteine/GSH metabolism observed with IR-CM in normal HK-2 cells; this metabolic regulation was abrogated by AZD3965 (Supplementary Fig. 4n–q). These collective findings proposed a possibility that lactate shuttle-triggered ferroptotic waves was due to the inhibition of cystine/cysteine/GSH metabolism in normal HK-2 cells.
LncRNA IGIP-5/miRNA 670-3p/LDHA axis promotes lactate production in the IR-RC and augments ferroptotic waves
Previously, we reported a whole transcriptome sequencing analysis elucidating the whole urine transcription map during kidney transplantation25. We noticed that the lncRNA IGIP-5 was enriched in urine samples acquired from donors and recipients. Intriguingly, lncRNA IGIP-5 was upregulated in IRI kidneys in a time-dependent manner (Fig. 3a), which was consistent with the increases in lactate and LDHA. Knockout of lncRNA IGIP-5 suppressed LDHA expression and lactate production induced by IRI, whereas, a contrasting effect was associated with the overexpression of lncRNA IGIP-5 (Supplementary Fig. 5a and Fig. 3b–d). Additionally, lncRNA IGIP-5 barely affected other metabolic enzymes, such as hexokinase 2 (HK2), phosphofructokinase muscle type (PFKM), and pyruvate kinase (PKM), in the glycolytic pathway (Supplementary Fig. 5b–d and Fig. 3d).
a LncRNA IGIP-5 levels in IRI kidney tissues at different reperfusion stages (n = 6 group−1); normalized according to sham group levels; one-way ANOVA followed by Tukey’s test. Lactate concentration (b) and transcriptional and translational levels of glycolytic enzymes (c, d) in HK-2 cells with lncRNA IGIP-5 overexpression (oeIGIP) and knockout (koIGIP); normalized to sham group levels (n = 3 group−1); one-way ANOVA followed by Tukey’s test. e Relative miRNA levels in wild-type (WT), oeIGIP and koIGIP HK-2 cells; normalized according to WT group levels (n = 3 group−1); one-way ANOVA followed by Tukey’s test. f LDHA transcriptional level in HK-2 cells transfected with miRNA mimics; normalized to Vehicle group levels (n = 3 group−1); one-way ANOVA followed by Tukey’s test. g RIP assays extracted endogenous RNA associated with AGO2 in HK-2 cells transfected with miR 670-3p inhibitor or inhibitor NC; normalized according to IgG group results (n = 3 group−1); one-way ANOVA followed by Tukey’s test. h WT and mutated LDHA sequences containing the predicting miR-670-3p binding site; dual luciferase assays detecting luciferase activity in HEK293T cells co-transfected with pmirGLO reporter plasmids and either mimic NC or miR-670-3p mimic (n = 3 group−1); normalized according to mimic NC group results; unpaired 2-tailed Student’s t test. i, j Transcriptional and translational levels of glycolytic enzymes in HK-2 cells transfected with miRNA 670-3p mimic and inhibitor (n = 3 group−1); normalized according to mimic NC group results; one-way ANOVA followed by Tukey’s test. k–o Lipid peroxidation level, MDA concentration, iron level, GSH content, and SLC7A11 protein expression in normal HK-2 cells cocultured with different CM (n = 3 group−1); IR-CMoeIGIP refers to CM from IGIP5-overexpressing IRI HK-2 cell; iron level and GSH content were normalized to NC-CM group results; one-way ANOVA followed by Tukey’s test. Data are presented as mean ± SD. All experiments were repeated at least three times, yielding similar results. The pvalues are shown for the indicated comparisons. Source data are provided as a Source Data file.
Subsequently, we explored how lncRNA IGIP-5 regulates LDHA. In a set of miRNAs putatively bound to LDHA identified in the starBase database (https://starbase.sysu.edu.cn), miRWalk database (http://mirwalk.umm.uni-heidelberg.de) and miRDB database (https://mirdb.org), we focused on three metabolism-related miRNAs: miR 670-3p, miR 502-3p and miR 4770 (Supplementary Fig. 5e). Further validation indicated that miR 670-3p is a potential mediator in lncRNA IGIP-5-mediated LDHA regulation because lncRNA IGIP-5 reduced the level of miR 670-3p (Fig. 3e), which plays an inhibitory role in LDHA expression (Fig. 3f and Supplementary Fig. 5f–h). Argonaute 2-RNA immunoprecipitation (AGO2-RIP) assays conducted to validate the interaction between lncRNA IGIP-5 and miR 670-3p revealed that lncRNA IGIP-5 was directly bound to the AGO2-containing miR 670-3p ribonucleoprotein complex; however, this interaction was significantly diminished after the inhibition of miR 670-3p (Fig. 3g). We further validated the direct binding of lncRNA IGIP-5 with miR 670-3p by dual luciferase reporter assay. The results revealed that miR 670-3p mimics prominently decreased the luciferase activities of the reporter plasmid containing the binding sequence of lncRNA IGIP-5, but not the reporter containing mutated sequence (Supplementary Fig. 5i–k). Subsequently, the interaction between LDHA and miR 670-3p was confirmed via dual-luciferase assays (Supplementary Fig. 5l–m); the luciferase activity of the LDHA reporter was significantly reduced by miR 670-3p mimic (Fig. 3h). Additionally, the miR 670-3p mimic reversed lncRNA IGIP-5-induced LDHA expression, whereas, the lncRNA IGIP-5 knockout-mediated suppression of LDHA was recovered by the 670-3p inhibitor (Fig. 3i, j). Other key enzymes in the glycolytic pathway were scarcely regulated by the lncR IGIP-5/miR 670-3p axis (Fig. 3j). These findings indicate that lncRNA IGIP-5 facilitates LDHA expression by inhibiting miR-670-3p via a competitive interaction.
To validate the effects of the lncRNA IGIP-5/miR-670-3p/LDHA axis on ferroptotic propagation during kidney IRI progression, we introduced wild-type lncRNA IGIP-5 and miR-670-3p mimic into IR-RC and treated normal HK-2 cells with CM. Compared to IR-CM, CM derived from lncRNA IGIP-5-overexpressing IR-RC cells (IR-CMoeIGIP) further suppressed SLC7A11 expression and enhanced ferroptotic waves in normal HK-2 cells (Fig. 3k–o and Supplementary Fig. 5n). Conversely, CM from miR-670-3p mimic-transfected IR-RC cells attenuated this effect. However, neither lncRNA IGIP-5 overexpression nor miR-670-3p mimics significantly affected GPX4 or ACSL4 expression (Supplementary Fig. 5o–q).
Lactate-induced NPM1 lactylation at lysine 257 induces ferroptosis in the normal RC by suppressing SLC7A11 expression
Lactylation provides important mechanistic insights into the role of lactate metabolism. We demonstrated a time-dependent increase in global lactylation in IRI kidneys, which was consistent with lactate upregulation (Fig. 4a). Notably, we validated the lactate shuttle-mediated spread of lactylation in the kidney tissue microenvironment; IR-CM considerably augmented global lactylation in normal HK-2 cells (Fig. 4b). The global lactylome analysis conducted to identify lactylated proteins in IR-CM-treated HK-2 cells further elucidated the involvement of lactylation in lactate-induced ferroptosis. A total of 5272 lactylated sites were identified, with the vast majority (5,222 sites, 99.05%) localized to non-histone proteins, and 50 sites (<1%) detected on histones (Supplementary Fig. 6a). Gene Ontology analysis revealed significant enrichment of lactylated proteins in transcription regulation and cellular metabolism (Fig. 4c). We subsequently analyzed the functional enrichment of the top 30 proteins with the highest lactylation levels in the lactylome data. Of these, 15 lactylated proteins were functionally linked to both cellular metabolism and transcriptional regulation (Fig. 4d). Since our previous evidence demonstrated that IR-CM–induced dysregulation of the cystine/cysteine–GSH metabolic axis plays a key role in promoting ferroptosis in HK-2 cells, we specifically focused on highly lactylated proteins involved in this pathway. Notably, NPM1 was identified as a strongly lactylated protein that bridges cystine/cysteine–GSH metabolism and transcriptional regulation (Fig. 4e).
a, b Global lactylation of HK-2 cells cocultured with different CM or lactate for 24 h; IR-CM-12h represents the CM of HK-2 cells experienced ischemia and reperfusion for 12 h (n = 3 group−1). c Bubble plot of significantly enriched GO terms (Biological Process, BP); Fisher’s exact test. d The top 30 proteins with the highest lactylation intensity from the lactylome analysis and the functional enrichment analysis for metabolism and transcriptional regulation. e Venn diagram of lactylated proteins enriched in cystine/cysteine/GSH metabolic pathway and metabolism & transcriptional regulation pathway. IP assays determining endogenous NPM1 lactylation in mouse kidney tissues (f, n = 6 group−1; g, n = 20 group−1) and HK-2 cells stimulated with different CM (h, i, n = 3 group−1). j NPM1 lactylation in endogenous NPM1-knockout (NKO) HK-2 cells reconstituted with full-length NPM1 or its site-directed mutants (n = 3 group−1). k MS analysis identified residue lysine 257 (K257) as the key lactylation site. l KEGG analysis displaying the top 20 enriched pathways of differentially expressed genes (DEGs) from RNA sequencing in NKO-NPM1 WT versus NKO-NPM1 K257R cells (n = 6 versus 6). m Volcano plot highlighting DEGs enriched in ferroptosis pathway (n = 12). n, o Translational and transcriptional levels of ferroptosis-related genes in NKO HK-2 cells expressing either WT or K257R NPM1, following treatment with or without 10 mM sodium lactate (n = 3 group−1); normalized to negative control group results; one-way ANOVA followed by Tukey’s test. p ChIP‒sequencing tracks for SLC7A11 in NKO HK-2 cells reconstituted with WT or K257R NPM1. q ChIP-qPCR was conducted using anti-NPM1 antibody to assess the enrichment of WT or K257R NPM1 at the SLC7A11 promoter in NKO HK-2 cell (n = 3 group−1); one-way ANOVA followed by Tukey’s test. r Lipid peroxidation in HK-2 cells overexpressing WT or K257R NPM (n = 3 group−1). Data are presented as mean ± SD. All experiments were repeated at least three times, yielding similar results. The pvalues are shown for the indicated comparisons. Source data are provided as a Source Data file.
Next, we detected the high expression of NPM1 in HK-2 cells compared to that in other epithelial cells (Supplementary Fig. 6b-c). Notably, we observed a time-dependent increase in NPM1 lactylation in IRI mouse kidneys (Fig. 4 f), which paralleled rising lactate levels and progressive ferroptosis. Glycolytic inhibition with sodium oxamate significantly reduced NPM1 lactylation in kidney grafts, whereas sodium lactate treatment produced the opposite effect (Fig. 4g). Consistent with these findings, sodium lactate-induced elevation of NPM1 lactylation was replicated in HK-2 cells (Supplementary Fig. 6d-e). Importantly, IR-CM treatment considerably elevated endogenous and exogenous NPM1 lactylation in normal HK-2 cells, which was declined when glycolysis was suppressed by oxamate treatment (Fig. 4h and Supplementary Fig. 6f) and knockout of lncRNA IGIP-5 (Fig. 4i and Supplementary Fig. 6g) in IR-RC. We further determined the lactylated sites identified in lactylome analysis (Supplementary Fig. 6h–j); the mutation on lysine 257 (K257) induced a sharp inhibition of NPM1 lactylation, suggesting K257 as a pivotal lactylation site on NPM1 (Fig. 4j, k).
To delineate the transcriptional regulatory function of NPM1-K257 lactylation, we generated NPM1-knockout (NKO) HK-2 cells followed by reconstitution with WT NPM1 (NKO-NPM1 WT) or the K257R mutant (NKO-NPM1 K257R). RNA-sequencing analysis revealed that differentially expressed genes (DEGs) were enriched in ferroptosis and metabolic pathways (Fig. 4l and Supplementary Fig. 6k–l). Notably, among the ferroptosis-associated DEGs, SLC7A11 exhibited the most pronounced downregulation in NKO-NPM1 WT cells (Fig. 4m). Subsequent validation demonstrated that sodium lactate potentiated NPM1-mediated SLC7A11 suppression, whereas the K257R mutation abolished this effect and remained unresponsive to sodium lactate treatment (Fig. 4n, o). However, neither WT nor K257R NPM1 overexpression altered GPX4 or ACSL4 expression, which was consistent with the RNA sequencing data (Supplementary Fig. 6m-n). Chromatin immunoprecipitation sequencing (ChIP-seq) and ChIP-qPCR assays demonstrated NPM1 binding to the SLC7A11 promoter, which was enhanced by sodium lactate (Fig. 4p, q). This binding, however, was significantly reduced by the lactylation-deficient K257R mutation and showed minimal response to sodium lactate treatment. Critically, overexpression of NPM1 activated ferroptosis in HK-2 cells (Fig. 4r) and other epithelial cells (Supplementary Fig. 6o-p), and lactate-stimulated NPM1 lactylation at K257 further promoted ferroptosis. These findings indicated that lactate triggers ferroptotic propagation by inducing NPM1 lactylation and attenuating SLC7A11 expression.
The high lactylation of NPM1 strengthens its protein stability by inhibiting MDM2-mediated ubiquitination in the normal RC
While analyzing the lactylation-regulated NPM1 function, the cycloheximide chase assay revealed that lactate notably prolonged the half-life of wile-type NPM1 (NPM1-WT), which was shortened by oxamate treatment (Fig. 5a, b). However, changes in lactate levels scarcely affected the NPM1-K257 mutant (Fig. 5c, d). These results suggested that lactate-induced lactylation of NPM1 at lysine 257 enhanced protein stability. Subsequently, we demonstrated that enhancement of NPM1 lactylation via sodium lactate directly reduced its interaction with ubiquitin and subsequent proteasomal degradation. Conversely, either lactylation inhibition by sodium oxamate or K257R site-specific mutagenesis - the critical lactylation residue governing this stabilization - enhanced NPM1 ubiquitination (Fig. 5e). To identify the ubiquitin ligase responsible for NPM1 ubiquitination in HK-2 cells, we performed immunoprecipitation followed by mass spectrometry (IP-MS). Murine double minute 2 (MDM2), an E3 ubiquitin ligase, was identified as a key interactor with NPM1 (Fig. 5f). Subsequently, we validated that overexpression of MDM2 facilitated NPM1 ubiquitination in HK-2 cells (Supplementary Fig. 7a). Notably, lactate-stimulated NPM1 lactylation attenuated its interaction with MDM2, but this reduction was reversed by either pharmacological inhibition of lactylation (using sodium oxamate) or the K257R mutation (Fig. 5g). Based on these results, we propose a mechanism: NPM1 lactylation at lysine 257 augments protein stability by suppressing MDM2-mediated ubiquitination.
Protein stability detection and half-life analysis of NPM1 WT (a, b) and K257R (c, d) in HK-2 cells treated with 10 mM sodium lactate, 10 mM sodium oxamate, or vehicle control (n = 3 group−1). e Assessment of ubiquitination for WT and K257R NPM1 in NKO HK-2 cells co-transfected with Flag-tagged ubiquitin (Ub-Flag), following treatment with sodium lactate (10 mM), sodium oxamate (10 mM), or vehicle control (n = 3 group−1). f IP-MS analysis of NPM1-interacting proteins in HK-2 cells treated with IR-CM for 24 h, revealing multiple E3 ubiquitin ligases including MDM. g Co-IP assays detecting the interactions between NPM1-HA and MDM2-His using anti-HA antibody in NKO HK-2 cells treated with 10 mM sodium lactate, 10 mM sodium oxamate, or vehicle control (n = 3 group−1). h, i IP assays determining the effects of diverse CM on the ubiquitination of NPM1-HA using anti-HA antibody in HK-2 cells co-transfected with Ub-Flag and MDM2-His (n = 3 group−1). j, k Translational and transcriptional levels of NPM1 in HK-2 cells treated with different CM (n = 3 group−1); normalized to Vehicle group levels; one-way ANOVA followed by Tukey’s test. ns represents no significant difference. Data are presented as mean ± SD. All experiments were repeated at least three times, yielding similar results. The p-values are shown for the indicated comparisons. Figure 5f was created in FigDraw. Source data are provided as a Source Data file.
Further analysis of the regulatory program in kidney IRI using normal HK-2 cells treated with CM derived from IR-RC at diverse glycolytic levels revealed that IR-CM significantly reduced the interactions of NPM1 with ubiquitin and MDM2 in normal HK-2 cells, whereas, attenuation of LDHA and glycolysis in IR-RC partially impaired this effect (Fig. 5h). More persuasively, we found that CM derived from lncRNA IGIP-5-overexpressing IR-RC cells (IR-CMoeIGIP) strengthened the suppression of NPM1 ubiquitination in HK-2 cells, whereas, knockout of lncRNA IGIP-5 (IR-CMkoIGIP) exhibited a contrasting effect (Fig. 5i). Consistent with the trend of protein stability and ubiquitination, the transcription of NPM1 in HK-2 cells was not affected by the altered lactate levels (Supplementary Fig. 7b) and grafted kidneys (Supplementary Fig. 7c), which led to an evident change at the translational level (Supplementary Fig. 7d-e). IR-CM-induced lactate shuttling to normal HK-2 cells modulated the protein expression of NPM1 only (Fig. 5j) and did not affect mRNA expression (Fig. 5k). Collectively, these findings present compelling evidence that IR-RC-derived lactate facilitates NPM1 lactylation, and increases protein stability.
Inhibition of AARS1-mediated NPM1 lactylation blocks ferroptotic propagation in the normal RC
Subsequently, we employed short interfering RNAs (siRNAs) to screen for enzymes mediating lactylation. We identified that the siRNA-mediated knockdown of AARS1, a lactate sensor and lactyltransferase, notably inhibited NPM1 lactylation in IR-CM-treated HK-2 cells (Fig. 6a). We confirmed that ectopic expression of AARS1 enhanced both global lactylation and NPM1 lactylation in lactate-treated HK-2 cells, whereas AARS1 knockout exerted the opposite effect (Fig. 6b and Supplementary Fig. 8a). In an in vitro reaction system containing lactate and ATP, we demonstrated that AARS1 catalyzed lactylation of wild-type NPM1, whereas lactylation of the K257R mutant was significantly impaired (Fig. 6c). Furthermore, direct interaction between AARS1 and NPM1 was confirmed in vitro by pull-down assays (Fig. 6d and Supplementary Fig. 8b-c). The endogenous (Fig. 6e) and exogenous (Fig. 6f, g) interactions between AARS1 and NPM1 were also validated in HK-2 cells. IR-CM administration augmented the AARS1-NPM1 interaction, whereas the reduced lactate concentration associated with inhibited glycolysis in IR-RC unequivocally weakened the effect of IR-CM (Fig. 6h). These results imply that AARS1 acts as a lactyltransferase that facilitates NPM1 lactylation in RC. Intriguingly, AARS1 expression in HK-2 cells remained unchanged following treatment with either lactate or IR-CM (Supplementary Fig. 8d-e) and was stably expressed across both normal and diseased human renal tissues (Supplementary Fig. 8f). These findings indicate that elevated lactylation levels in normal RC are primarily driven by increased substrate availability through the lactate shuttle.
a IP of NPM1 lactylation using an anti-NPM1 antibody in IR-CM-treated HK-2 cells following siRNA-mediated knockdown of enzymes (n = 3 group−1). b Detection of NPM1-HA lactylation by anti-HA IP in HK-2 cells with AARS1 knockout (AKO) or overexpression (AOE) following sodium lactate (10 mM) treatment (n = 3 group−1). c In vitro lactylation assay of purified WT and K253R NPM1 proteins incubated with AARS1, lactate (2 mM), and ATP (4 mM) for 2 h at 37 °C, followed by western blot (n = 3 group−1). d GST pull-down assay to detect the direct interaction between GST-NPM1 and His-AARS1 in vitro; arrows represent the indicated proteins (n = 3 group−1). e Co-IP assays of the endogenous NPM1 and AARS1 in HK-2 cells (n = 3 group−1). Co-IP of NPM1-HA and AARS1-His in HK-2 cells using anti-HA (f) and anti-His (g) antibodies (n = 3 group−1). h Co-IP of NPM1-HA and AARS1-His using an anti-HA antibody in HK-2 cells treated with diverse CM (n = 3 group−1). i–l Lipid peroxidation level, MDA concentration, iron level, and GSH content in WT, AOE, and AKO HK-2 cells cocultured with NC-CM or IR-CM (n = 3 group−1); iron level and GSH content were normalized according to negative control group results; one-way ANOVA followed by Tukey’s test. m Western blotting examining NPM1 and SLC7A11 expression in WT, AOE, and AKO HK-2 cells treated with the same experimental setup as described in (i–l) (n = 3 group−1). n Transcriptional level of SLC7A11 in WT, AOE, and AKO HK-2 cells (n = 3 group−1); normalized to negative control group results; one-way ANOVA followed by Tukey’s test. Data are presented as mean ± SD. All experiments were repeated at least three times, yielding similar results. The pvalues are shown for the indicated comparisons. Source data are provided as a Source Data file.
Considering that NPM1 lactylation promoted ferroptosis through SLC7A11 suppression, we explored the effectiveness of AARS1 suppression for interrupting ferroptotic waves. Indeed, AARS1 knockout in normal HK-2 cells markedly attenuated IR-CM-induced ferroptosis, as evidenced by reduced lipid ROS (Fig. 6i), diminished lipid peroxidation (Fig. 6j), decreased iron accumulation (Fig. 6k), and elevated GSH levels (Fig. 6l). Conversely, overexpression of AARS1 significantly downregulated SLC7A11 (Fig. 6m, n), further intensifying ferroptosis in HK-2 cells treated with IR-CM. In addition to its lactyltransferase activity that mediates protein lactylation, AARS1 is a key member of the aminoacyl-tRNA synthetase family involved in protein translation. To determine whether AARS1 regulates IR-CM-induced ferroptosis primarily via its lactyltransferase function, we employed β-alanine, a competitive inhibitor that specifically disrupts lactate binding to AARS1 and thereby suppresses its lactyltransferase function28,29,30,31. Similar to AARS1 depletion, β-alanine suppressed both NPM1 lactylation (Supplementary Fig. 8g) and ferroptosis (Supplementary Fig. 8h) in HK-2 cells induced by IR-CM. Furthermore, we reconstituted AKO cells with either wild-type AARS1 (AARS1-WT) or a lactyltransferase-deficient mutant (AARS1-5M)29,32. AARS1-WT, but not AARS1-5M, restored NPM1 lactylation and ferroptosis. Notably, O-propargyl-puromycin incorporation assays revealed comparable levels of global protein synthesis in AKO cells reconstituted with either AARS1-WT or AARS1-5M (Supplementary Fig. 8i). Despite maintaining global protein synthesis, the AARS1-5M mutant fails to restore NPM1 lactylation and ferroptosis in AKO cells. This result establishes that the suppression of ferroptosis upon AARS1 depletion is specifically attributable to the loss of lactyltransferase activity, independent of its role in protein translation. Therefore, we propose that AARS1 downregulation can block ferroptotic propagation in normal RC mainly through inhibition of NPM1 lactylation.
Suppressing lactate shuttle-induced NPM1 lactylation in the RC blocks ferroptotic waves and ameliorates graft kidney function
The results reflect that lactate shuttle-induced NPM1 lactylation in RC populations triggers ferroptotic propagation during kidney IRI and impairs graft kidney function. We investigated if conditional knockout of lactyltransferase AARS1 (AARS1-CKO) and solute larrier camily 16 member 3 (SLC16A3, the encoding gene of MCT4, SLC16A3-CKO) in the RC could ameliorate the kidney IRI and DGF in allograft kidney transplantation mouse models (Fig. 7a, b). Hematoxylin-eosin (H&E) staining analysis and SCr and blood urea nitrogen (BUN) concentrations revealed that the inhibition of lactate production and shuttling via sodium oxamate treatment and SLC16A3-CKO showed protective effects on graft kidney injury in terms of morphology and function (Fig. 7c–e). AARS1 knockdown in the RC also ameliorated IRI and improved kidney function. Importantly, the regulatory effects of sodium oxamate and AAV9-KSP1.3-shRNA on ferroptosis (Fig. 7f–i) were broadly in line with the morphological and functional alterations in the graft kidney tissues. These findings imply that impairing lactate metabolism, such as production and shuttling, and reduced AARS1-mediated lactylation in RC considerably suppress ferroptotic propagation and IRI in graft kidneys. To further validate the mechanism, we demonstrated that the lactate concentration sharply increased in the graft kidney tissue and urine on the postoperative day 2, whereas, inhibition of LDHA and MCT4 reduced it (Supplementary Fig. 9a-b). In addition, enhanced global lactylation and NPM1 lactylation during kidney transplantation were reversed when lactate and lactyltransferase levels were downregulated in the RC, contributing to NPM1 upregulation (Fig. 7j, k and Supplementary Fig. 9c-d). In contrast, the addition of sodium oxamate and attenuation of MCT4 and AARS1 in RC enhanced SLC7A11 expression and inhibited the cystine/cysteine metabolism in the graft kidney tissues (Fig. 7l, m and Supplementary Fig. 9e-g), which was consistent with a reduction in ferroptosis. More persuasively, the results of IF assays indicated the regional increase of SLC7A11, propagating from the proximal part of blood reperfusion to the distal end, and the regional difference became more significant when lactate shuttle and lactylation were inhibited in graft kidneys (Fig. 7m). These in vivo results suggest that suppression of lactate shuttle-derived NPM1 lactylation blocks ferroptotic waves in the graft kidney tissue and ameliorate graft kidney function.
a Flowchart delineating the procedure of allograft kidney transplantation in WT, Aars1CKO and Slc16a3CKO C57BL/6 mouse. b Schematic representation of the experimental protocols in vivo. For sodium oxamate administration, recipient mice receive intraperitoneal injection of sodium oxamate (1 g/kg, OXA) or normal saline (N.S.) 2 h before kidney transplantation and 1 d after transplantation. c HE staining evaluating kidney morphology and kidney pathological score (n = 6 group−1); scale bar: 50 μm; one-way ANOVA followed by Tukey’s test. d, e SCr and BUN concentrations in mice treated with the same experimental setup as described in a-b (n = 6 group−1); one-way ANOVA followed by Tukey’s test; Boxplot: 25th–75th percentiles; center line: median; whiskers: 1.5 times the interquartile range. Lipid peroxidation level (f), MDA concentration (g), iron level (h), and GSH content (i) in graft kidney tissues (n = 6 group−1); iron level was normalized according to WT-sham group results; one-way ANOVA followed by Tukey’s test. j Western blotting detecting the indicated proteins and global lactylation levels in graft kidney tissues (n = 6 group−1). k IP assays assessing NPM1 lactylation in graft kidney tissues using an anti-NPM1 antibody (n = 6 group−1). l SLC7A11 expression in graft kidney tissues was detected by western blotting (n = 6 group−1). m IF and fluorescence intensity assaying illustrating the expression level and region of SLC7A11 in graft kidney tissue slices (n = 6 group−1); one-way ANOVA followed by Tukey’s test; yellow arrows representing regions from the proximal part of blood reperfusion to the distal end; scale bar: 1000 μm for 1X. Data are presented as mean ± SD. All experiments were repeated at least three times, yielding similar results. The p-values are shown for the indicated comparisons. Figure 7a, b were created in FigDraw. Source Data are provided as a Source data file.
Based on genetic evidence that inhibiting NPM1 lactylation ameliorates kidney in mouse allograft models, we sought to identify inhibitors with clinical translational potential. We screened a library of 4,672 FDA-approved or investigational drugs against the hydrophobic region surrounding NPM1 K257 via molecular docking (Source Data file). The top 10% of compounds (467 molecules) ranked by predicted binding affinity were selected for further analysis. These candidates underwent unsupervised K-means clustering based on Molecular ACCess System (MACCS) fingerprints, and the results were visualized using t-SNE, which delineated five distinct molecular clusters (Supplementary Fig. 10a). Projection of binding affinity data onto the t-SNE plot indicated that high-affinity molecules were distributed across all clusters, with comparable mean binding affinities among groups (Supplementary Fig. 10a-b), suggesting that multiple structural classes may interact with the NPM1 K257 region. From each cluster, we selected the top 10 candidates by docking affinity (50 in total) for refined evaluation using the AI-driven Boltz2 model33. Eight compounds showed high binding probability (affinity probability binary >0.5, indicated by checkered bars). Among these, taurochenodeoxycholic acid (TCDCA), a taurine-conjugated bile acid, displayed superior binding affinity and potential clinical suitability for kidney transplant recipients (Supplementary Fig. 10c). Molecular docking (Supplementary Fig. 10d) and surface plasmon resonance (SPR) assays confirmed specific binding between TCDCA and NPM1, with a KD value of 2.09e-5 M (Supplementary Fig. 10e-f). Notably, TCDCA treatment reduced IR-CM-induced NPM1 lactylation and consequently suppressed ferroptosis in HK-2 cells (Supplementary Fig. 10g-h). In cells overexpressing NPM1-WT, TCDCA similarly inhibited NPM1 lactylation and ferroptosis, whereas this effect was substantially reduced in cells expressing the lactylation-deficient NPM1-K257R mutant (Supplementary Fig. 10i-j). Consistent with the in vitro findings, TCDCA administration in a mouse model of in situ kidney IRI improved renal histology and functional parameters (Supplementary Fig. 10k-m). TCDCA-treated mice exhibited lower levels of ferroptosis (Supplementary Fig. 10n), reduced NPM1 lactylation (Supplementary Fig. 10o), and elevated SLC7A11 expression (Supplementary Fig. 10p). Collectively, these results suggest that TCDCA alleviates kidney IRI and improves renal function by inhibiting NPM1 K257 lactylation and blocking ferroptotic propagation, presenting a potential strategy worthy of further preclinical and clinical validation.
Activation of the lncRNA IGIP-5/miRNA 670-3p/LDHA axis in IRI renal tubular epithelial cells promotes glycolytic reprogramming and lactate shuttling in the graft kidney tissue microenvironment. Lactate shuttle enhances AARS1-mediated NPM1 lactylation in adjacent tubular cells, increasing NPM1 protein stability and reinforcing its transcriptional repression of SLC7A11. The subsequent downregulation of SLC7A11 impairs the cystine/cysteine/GSH metabolism, leading to ferroptotic propagation and DGF. Figure 8 was created in GNU Image Manipulation Program (GIMP). https://www.gimp.org/about.
Discussion
The wide propagation of ferroptosis evidently exacerbates graft kidney IRI and DGF after kidney transplantation; however, the associated approaches and mechanisms remain unclear. In this study, we revealed a metabolic and epigenetic mechanism regulating the occurrence and progression of ferroptotic waves. Our results indicate that glycolytic reprogramming in IR-RC promotes lactate production as well as shuttling and induces ferroptosis by strengthening NPM1 lactylation in normal RC. Additionally, we propose an early prediction model for DGF that relies on the postoperative lactate concentration in urine. Mechanistically, we demonstrate that the activated lncRNA IGIP-5/miRNA-670-3p/LDHA axis facilitates glycolysis and increases lactate production and secretion in IR-RC. The lactate shuttle in the kidney tissue microenvironment induces AARS1-mediated NPM1 lactylation in the normal RC, increases NPM1 protein stability and strengthens NPM1-mediated suppression of SLC7A11, thereby inducing ferroptotic waves.
Recent reports indicate that ferroptotic waves cause large-scale cell death in a spatiotemporal manner and intensify the influences of ferroptosis19,21,22,23. The spread of reactive oxygen species (ROS) across cell populations over long distances can regulate ferroptotic waves through a series of ROS amplification-diffusion events19. Nicotinamide adenine dinucleotide phosphate hydrogenated (NADPH) diffuses into dying cells and induces local release of damage-associated molecular patterns and pH changes, rendering the neighboring cells susceptible to ferroptosis34,35. Overholtzer et al. designed ethylene glycol-coated silica nanoparticles that promoted intercellular ferroptotic propagation by trafficking iron36. Moreover, we previously demonstrated the pivotal regulatory roles of IR-RC-derived extracellular nanoparticles in the ferroptotic waves through the regulation of glucose metabolism and O-GlcNAcylation in the normal RC25. The present study elucidates another pivotal metabolic and epigenetic mechanism underlying intercellular ferroptosis promoted by IR-RC-secreted lactate via enhanced protein lactylation. Genetic and pharmacological perturbations associated with lactate production and shuttling in the RC block ferroptotic waves and mitigate graft kidney IRI. Systemic lactate inhibition represents a potential strategy for improving kidney transplant outcomes; however, this approach requires further clinical validation.
Urinary lactate has emerged as a clinically relevant biomarker for kidney outcomes in acute and chronic kidney diseases, including ischemic acute kidney injury, diabetic kidney disease, and chronic kidney failure. In a cohort of patients with diabetic kidney disease, urinary lactate levels correlated significantly with tubular injury markers (kidney injury molecule-1, albuminuria, and Dickkopf-3) and renal functional decline37. Further supporting its biomarker utility, patients with chronic kidney disease exhibit elevated urinary lactate levels compared to healthy controls, with levels inversely correlating with estimated glomerular filtration rate15. Notably, accumulating evidence indicates that the elevated urinary lactate level in kidney IRI and diabetic kidney disease primarily reflect increased lactate production within proximal tubular cells rather than serum lactate excretion38,39,40. This study establishes a ULa-2d-based model with robust predictive performance and specificity for early DGF prediction, outperforming conventional predictors based on SLa-2 d, SCr-2 d and 24-h urine volume. Additionally, elevated ULa-2d levels were associated with significantly inferior 5-year graft survival. Nevertheless, future studies with larger sample sizes are needed to strengthen these conclusions and validate the clinical translational potential of ULa-2d. Furthermore, it should be noted that urinary lactate levels may not fully recapitulate the lactate concentration within the graft tissue microenvironment. Therefore, future studies to establish a direct correlation between urinary and kidney lactate levels in human kidney transplant recipients will significantly strengthen the clinical relevance of these findings.
Recent evidence indicates that lactate metabolism modulates cell susceptibility to ferroptosis41. High SRY-box 8 (SOX8) expression notably decreases the accumulation of lactate rather than earlier glycolytic intermediates, stimulating ferroptosis of hepatocellular carcinoma cells42. Notably, the increasing research focus on lactate-derived histone and non-histone lactylation establishes a novel metabolic and epigenetic link and presents unexpected functional and mechanistic insights into glycolytic metabolism. However, reports of the exact regulatory role of lactylation in ferroptosis, especially in ferroptotic propagation, are rare. A recent study revealed that histone H3K18 lactylation enhances methyltransferase-like 3-mediated N6-methyladenosine (m6A) modification in ACSL4 and induces ferroptosis in sepsis-associated lung injury43. Suppression of tau lactylation decreases ferroptosis and protects against Alzheimer’s disease by regulating mitogen-activated protein kinase signaling pathways44. In this study, we delineated global metabolism and lactylation maps in RC undergoing ferroptotic propagation; we demonstrated that lactate-induced NPM1 lactylation facilitates ferroptosis by inhibiting SLC7A11 expression. Conditional knockdown of the lactyltransferase AARS1 suppresses NPM1 lactylation in RC, limits ferroptotic waves, and ameliorates graft kidney function. The therapeutic potential of targeting this lactyltransferase activity warrants further investigation, particularly considering AARS1’s fundamental role in tRNA aminoacylation and protein synthesis. Additionally, emerging evidence studies establishes the regulatory role of histone and non-histone lactylation in various IRI models14,45,46. Whether non-histone and histone lactylation beyond NPM1 contribute to ferroptosis propagation in graft kidney IRI remains an open question, warranting systematic exploration.
The SLC7A11/GSH/GPX4 axis constitutes a central regulatory hub in ferroptosis, where its suppression aggravates lipid peroxidation and triggers ferroptosis47,48. Reactivation of this axis during ferroptosis inhibition restores cellular antioxidant capacity24,48. In this study, ferroptosis suppression by Fer-1 significantly attenuated lactate-induced renal injury and restored SLC7A11 expression, notably without affecting glycolysis or lactate production. Importantly, SLC7A11 expression is modulated through multiple mechanisms, including p53-mediated SLC7A11 transcriptional repression49, miRNAs-induced SLC7A11 mRNA degradation46, KEAP1/BAP1-dependent ubiquitin-proteasomal degradation of SLC7A11 regulators50, and Gal13/CD44-regulated plasma membrane localization of SLC7A1151. Nevertheless, the precise mechanism driving SLC7A11 upregulation following ferroptosis inhibition in IR-RC remains unresolved and requires further investigation.
NPM1 is a pivotal epigenetic regulator implicated in various biological processes such as embryonic development, tumorigenesis, tissue injury, and repair52,53. However, the effect of NPM1 on ferroptosis in kidney IRI remains unclear. NPM1 mutants restore the depleted glutathione pools in leukemic cells and decrease oxidative stress-associated cell death by regulating cysteine metabolism genes54. The inhibition of aurora kinase A (AURKA) expression was reported to reduce the phosphorylation of NPM1 and trigger ferroptosis in Ewing sarcoma cells55. In this study, we identified that NPM1 is highly lactylated in renal tubular epithelial cells undergoing ferroptosis and that lactylation of NPM1 enhances its protein stability via suppressed MDM2-mediated ubiquitination, which induces the suppression of SLC7A11. However, further research is required to elucidate whether other epigenetic mechanisms involving NPM1 are associated with ferroptotic propagation.
In conclusion, this study revealed that lactate shuttle is a crucial factor regulating the propagation of ferroptotic waves in the graft kidney IRI process. Elevated lactate in early postoperative urine serves as a noninvasive indicator of IRI severity and DGF risk. Mechanistically, AARS1-mediated NPM1 lactylation represents a key epigenetic event associated with lactate shuttle-mediated ferroptotic propagation. Targeting NPM1 lactylation with TCDCA improves graft kidney function. These findings broaden our understanding of ferroptotic waves in kidney IRI, and propose an early prediction model and therapeutic strategies to approach the clinical challenge of DGF.
Methods
Ethics statement
The human research protocol was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (181). Written informed consent was obtained from all participants and/or their relatives following the institutional guidelines. All animal experiments and procedures were approved by the Medicine Policy on the Care and Use of Laboratory Animals of Chongqing Medical University and the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (K276).
Human study
A total of 150 kidney allograft recipients who underwent transplantation at the First Affiliated Hospital of Chongqing Medical University were enrolled in this study (Supplementary Table 1). Donors or recipients under the age of 18 were excluded. Additionally, we excluded recipients with incomplete urine sample collections, and those lacking complete clinical follow-up data. For the lactate assay, clean and fresh urine samples were harvested from the recipients two and seven days after transplantation. According to ethical and safety protocols, the urine samples used in this study were properly disposed of upon completion of all analytical procedures. DGF was defined as a SCr concentration >400 μmol/L on day 7 after transplantation.
Animals
Six-week-old male C57BL/6 mice were purchased from the Animal Center of the Chongqing Medical University (Chongqing, CHN). Mice were randomly assigned to different groups and maintained in specific pathogen-free facilities at the Animal Center of Chongqing Medical University. Mice were housed at a constant temperature of 23 °C with a 12 h/12 h light/dark cycle and provided with standard chow and fresh water ad libitum. Serotype 9 adeno-associated viruses (AAVs) specifically encoding short hairpin RNAs that target Aars1 and Slc16a3 driven by the KSP1.3 promoter in renal tubular epithelial cells (AAV9-KSP1.3-shAars1 and AAV9-KSP1.3-shSlc16a3) were purchased from WZ Biosciences Inc. Jinan, China. The conditional knockout (Aars1CKO and Slc16a3CKO) mouse models were established using caudal intravenous injection of AAVs (2.8×1011 vg mouse−1 of each AAV) and analyzed 14 days after injection. Consequently, eight-week-old male mice were further analyzed. All the mice were fed at a standardized animal facility. Sodium oxamate (1 mg/kg, MCE, CHN), sodium lactate (120 mg/kg, Sigma-Aldrich, CHN), imidazole ketone erastin (40 mg/kg, MCE, China), liproxstatin-1 (10 mg/kg, MCE, China), ferrostatin-1 (5 mg/kg, MCE, China), and TCDCA (10 mg/kg, TargetMol, China) were used to treat mouse as indicated.
Mouse model of allograft kidney transplantation
Syngeneic kidney transplantation was performed as described previously25,56. Donor mice were anesthetized using an intraperitoneal injection of pentobarbital (50 mg/kg of the body weight). The right-side kidney of the donor mouse was excised and stored in 4 °C cold Ringer lactate solution for 6 h. During transplantation, recipient mice were similarly anesthetized and underwent right-side nephrectomy, followed by the orthotopic graft kidney vascular anastomosis performed using 10–0 silk suture within 30 min. The end of the ureter was fixed to the lateral wall of the bladder. The contralateral kidney of the recipient was removed and the abdomen was sutured using a 4–0 silk suture. The recipient mice were kept on a thermostatic pad to maintain their body temperature and intraperitoneal moisture was maintained by continually instilling warm normal saline (NS; Beyotime, CHN). The recipient mice were euthanized two days after transplantation and blood and graft kidney samples were harvested following the guidelines promulgated by the American Veterinary Medicine Association.
In situ kidney IRI model
An in situ kidney warm ischemia-reperfusion model was constructed as previously described25,56,57,58. The mice were anesthetized and subjected to a left-sided nephrectomy. To induce warm ischemia, the right renal pedicle was occluded for 35 min using a microvascular clamp and released to induce reperfusion injury. Mice in the sham group underwent left-sided nephrectomy only. Blood and kidney samples were collected after different reperfusion periods.
Cell culture and treatment
The human proximal tubular cell line (HK-2) and human embryonic kidney cell line (HEK293T) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HK-2 was cultured using Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, BioInd, Israel), 100 μg/mL streptomycin (Beyotime, CHN), and 100 U/mL penicillin (Beyotime, CHN) at 37°C in a humidified atmosphere maintaining 5% CO2; HEK293T was cultured using DMEM (Gibco, USA) under the same condition. The cell lines were tested for Mycoplasma contamination. Sodium oxamate (10 mM, MCE, CHN), sodium lactate (10 mM, Sigma-Aldrich, CHN), ferrostatin-1 (5 μM, MCE, China), and TCDCA (100 μM, TargetMol, China) were used to treat the cells as indicated.
The in vitro IRI model was established through oxygen-glucose deprivation following a previously described method. A glucose-free medium (Gibco, USA) supplemented with antimycin A (5 μM, Sigma-Aldrich, CHN) and 2-deoxyglucose (5 mM, Sigma-Aldrich, CHN) were used to treat HK-2 cells for one hour and simulate the ischemic process. The culture medium was replaced with a complete medium to restore the reperfusion phase.
An in vitro co-culture model was established by treating normal HK-2 cells for 24 h with conditioned medium derived from HK-2 cells. IR-CM was collected from wild-type HK-2 cells (2.5 × 106 cells) subjected to 1 h of oxygen–glucose deprivation followed by 24 h of reperfusion in complete medium consisting of DMEM/F12 (Gibco, USA) supplemented with 10% FBS (BioInd, Israel). Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 during reperfusion. NC-CM was obtained from the same number of wild-type HK-2 cells cultured under identical medium for 24 h, but without exposure to oxygen–glucose deprivation or reperfusion.
Generation of stable cell lines with lentivirus and CRISPR-Cas9 systems
For generating stably overexpressed cell lines, the pLVX-AcGFP1-N1 lentiviral vectors containing the full-length cDNA fragments of lncRNA IGIP-5, NPM1, or AARS1 were transfected into HK-2 cells, followed by the selection using puromycin (5 μg/mL) for two weeks. An empty vector was used as the negative control (NC). Additionally, individual guide sequences targeting lncRNA-IGIP-5 and AARS1 were cloned into the pSpCas9 BB-2A-Puro vector, and the CRISPR-Cas9 system was used to establish a cell line knockout of IGIP (koIGIP) or AARS1 (sgAARS1), following a previously reported method59. The oligonucleotide sequences used in this study are listed in Supplementary Table 5.
Oligonucleotides, plasmids, siRNA, and cell transfection
The wild-type and mutant cDNA of NPM1, AARS1, ubiquitin, and MDM2 were separately cloned into the pcDNA 3.1 or pEGFP-N1 plasmid with different tags as indicated. MiRNA mimics and inhibitors (NC, miR-670-3p, miR-502-3p, and miR-4770 mimic) and small interfering RNAs targeting MCT1 (siR-MCT1) and MCT4 (siR-MCT4) were synthesized by Tsingke Biotechnology Co., Ltd., China. Plasmids and oligonucleotides were transfected using Lipofectamine 3000 transfection reagent (Invitrogen, USA).
Metabolomic profiling
Mouse kidney tissues were washed and suspended in 500 μL of a pre-cooled methanol-acetonitrile mixture (MeOH/ACN = 1:1, V/V, Merck, GER) to extract metabolites and remove proteins. After centrifugation at 14,000 × g for 20 min at 4 °C, 300 μL of the supernatant was incubated at –20 °C for 1 h and dried via rotary evaporation. Subsequently, the sample was redissolved into 100 μL of 50% ACN solvent (ACN/H2O = 1:1, V/V) and centrifugated at 14,000 × g for 10 min at 4 °C. The supernatant was analyzed via LC-MS chromatography-mass spectrometry. An LC-ESI-MS/MS system (UPLC, ExionLC AD; MS, QTRAP 6500+system; https://sciex.com) was used for the detection of metabolites and targeted metabolomic profiling.
Identification of proteins with lactylation by LC-MS/MS analysis
The HK-2 cells were snap-frozen in liquid nitrogen and supplemented with urea lysis buffer for protein extraction followed by quantification. Next, equal amounts of protein were mixed dropwise with trichloroacetic acid (Sigma-Aldrich, CHN) at a final concentration of 20% and precipitated at 4 °C for 2 h. The cell pellet was washed twice with pre-cooled acetone (Sigma-Aldrich) and resuspended in 200 mM triethylammonium bicarbonate buffer (TEAB, Sigma-Aldrich, CHN). The supernatant was incubated with trypsin (trypsin: protein = 1:50, w/v; Promega) and digested overnight. After the treatment with 5 mM dithiothreitol (Sigma-Aldrich, CHN) for 1 h at 37 °C, 11 mM iodoacetamide (Sigma-Aldrich, CHN) was added to the supernatant followed by incubation for 30 min at room temperature.
The prepared peptides were suspended in NP-40 buffer (Beyotime, CHN) and incubated overnight with the pre-washed anti-L-lactyllysine antibody beads (PTM-1404, PTM Bio, CHN) at 4 °C. Enriched lactylated peptides bound to the beads were eluted twice with 0.1% TCA and purified using C18 ZipTips (Sigma-Aldrich, St. Louis, MO, USA). Lactylated peptides were identified using a Bruker timsTOF Pro mass spectrometer equipped with a NanoElute ultra-high-performance system (Bruker, GER).
The resulting MS/MS data were processed using MaxQuant search engine (v.1.6.15.0). Tandem mass spectra were searched against the Homo_sapiens_9606_SP_20220107.fasta (20376 entries) concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme. The mass tolerance for precursor ions was set as 20 ppm in first search and 20 ppm in main search, and the mass tolerance for fragment ions was set as 20 ppm. Carbamidomethyl on cysteine was specified as fixed modification, and acetylation on protein N-terminal, oxidation on methionine and lactylation on lysine were specified as variable modifications. FDR was adjusted to <1%.
In vitro lactylation assay
An in vitro lactylation assay was performed as described previously28. Purified AARS1 proteins were incubated with NPM1 proteins (WT or K257R mutant) in a reaction mix containing 50 mM HEPES (pH 7.5), 30 mM KCl, 5 mM MgCl2, 2 mM lactate, and 4 mM ATP. After incubating at 37 °C for 2 h, the reactions were stopped for subsequent SDS-PAGE and western blotting assays.
Recombinant protein purification
Bacterial expression constructs (pET28a and pGEX-6P-1) containing the indicated genes (AARS1, NPM1-WT, or NPM1-K257R) were transformed into Escherichia coli BL21 (DE3), which were induced by 0.2 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 20 °C with 160 rpm rotation for 20 h. Collected bacterial pellet was resuspended in 35 mL of lysis buffer (20 mM Tris-HCl, pH = 7.5, 500 mM NaCl, 5% glycerol, 20 μg/mL lysozyme, 1 mM PMSF, 1 mM DTT, and 1 mM EDTA) and subjected to ultrasonic cell disruption via an ultrasonic homogenizer. The lysate was clarified by ultracentrifugation at 15,000 × g for 30 min at 4 °C. The supernatants were incubated with glutathione or Nickel beads for affinity chromatography according to standard procedures.
Pull-down assay
Recombinant GST-NPM1 (200 μg) and GST control protein (200 μg) were separately incubated with Glutathione MagBeads for 2 h at room temperature in an incubation buffer, followed by incubation with purified prey protein (500 μg) in a binding buffer (20 mM HEPES, pH 7.5, 100 mM NaCl) overnight at 4 °C. After washing three times with ice-cold washing buffer, bound complexes were eluted in 100 μL of elution buffer by boiling at 95 °C for 10 min. The eluates were then separated by centrifugation at 12,000 × g for 5 min at 4 °C, and supernatants were collected for subsequent SDS-PAGE analysis.
ChIP assay
NKO-NPM1 WT and NKO-NPM1 K257R HK-2 cells were cross-linked with 1% formaldehyde for 10 min at room temperature to stabilize DNA-protein complexes. The reaction was quenched with 0.125 M glycine for 5 min. Cells were then lysed in ice-cold lysis buffer (10 mM HEPES pH 7.5, 0.1 mM EDTA, 0.5% NP-40) supplemented with protease inhibitor cocktail. Nuclei were pelleted by centrifugation at 2000 × g for 10 min at 4 °C, and chromatin was fragmented to an average size of 100–500 bp using sonication. Fragmented chromatin was immunoprecipitated overnight at 4 °C with anti-NPM1 antibody or rabbit IgG, followed by incubation with protein G-agarose beads. After washing, bound complexes were eluted and reverse-crosslinked. DNA from both input and IP samples was purified by phenol-chloroform extraction. Sequencing libraries were constructed using the VAHTS Universal DNA Library Prep Kit for Illumina V3 (Vazyme, China) and subjected to sequencing on an Illumina Novaseq™ X Plus platform. For ChIP-qPCR validation, aliquots of immunoprecipitated DNA fragments were analyzed by quantitative PCR using target-specific primers (Supplementary Table 5).
Transcriptome sequencing
Total RNA was extracted using Trizol reagent (ThermoFisher, USA) following the manufacturer’s procedure. Subsequently, mRNA was purified from total RNA (2 μg) using mRNA Caprure Beads2.0 (Yeasen, CHN) with two rounds of purification. RNA fragments were reverse-transcribed to synthesize first-strand cDNA by reverse transcriptase, which were next used to synthesise second-stranded DNAs with E.coli DNA polymerase I, RNase H and dUTP Solution (Yeasen, CHN). Dual-index adapters were ligated to the fragments, followed by amplification with PCR. PCR amplification products were purified by Hieff NGS DNA Selection Beads (Yeasen, CHN) and subjuected to ssequencing on an Illumina Novaseq™ X Plus platform.
IP assay
For immunoprecipitation assays, NPM1-HA, NPM1-mutant-HA, Ub-Flag, and/or MDM2-His were co-transfected into HK-2 cells using Lipofectamine 3000. Cell samples were lysed using IP buffer (Beyotime, China) supplemented with protease and phosphatase inhibitors (Bimake, China). The cell lysates were resuspended and immunoprecipitated by incubating overnight with 2.5 μg of species-specific immunoglobulin G (IgG, CST, USA) or an equivalent amount of target antibodies at 4 °C. Subsequently, the immunoprecipitated complexes were captured by incubating with prewashed protein A/G magnetic beads (MCE, China) at 4 °C for 8 h. The immunoprecipitated proteins were eluted and analyzed by western blotting. To reduce the masking of IgG bands (heavy and light chains), anti-mouse and anti-rabbit IgG Veriblot secondary antibodies (Abcam, UK) were used to detect immunoprecipitated proteins at a dilution of 1:5000.
RIP assay
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) was used to conduct the RIP assay following a previously described method25. Initially, processed HK-2 cells were mechanically disrupted and lysed using ice-cold RIP lysis buffer. The lysates were incubated separately with 5 μg of IgG and AGO2 antibody (Abcam, USA) overnight at 4°C. Protein A/G magnetic beads were introduced to capture the co-precipitated RNA-AGO2 complexes. Proteinase K was added to the eluate to digest proteins and extract RNA. The coprecipitated RNA was quantified through quantitative real-time polymerase chain reaction (qRT-PCR).
Luciferase reporter assay
Luciferase reporter assay was performed following a previously described method59. The wild-type 3′-UTR fragment of human LDHA containing the predicted miR-670-3p binding site, was cloned downstream of the firefly luciferase gene in the pmirGLO vector (designated pmirGLO-LDHA-3′UTR WT). Similarly, a mutated sequence targeting the miR-670-3p binding site was constructed in the same vector (pmirGLO-LDHA-3’UTR MUT). To assess the regulatory role of miR-670-3p in the LDHA 3′-UTR, HEK293T cells were seeded in 96-well plates at 2 × 10⁴ cells/well and co-transfected with either pmirGLO-LDHA-3′UTR WT or pmirGLO-LDHA-3’UTR MUT reporter plasmid, along with miR-670-3p mimics or mimic negative control (NC). Similarly, HEK293T cells were co-transfected with either the pmirGLO-LncR-IGIP-5-WT or pmirGLO-LncR-IGIP-5-MUT reporter plasmid, together with miR-670-3p mimics or mimic NC, respectively. After 24 h, firefly and Renilla luciferase activities were quantified using the Dual-Luciferase Reporter Assay System (Promega, USA). The Renilla luciferase activity was normalized to firefly luciferase activity.
NPM1 protein stability assay
HK-2 cells transfected with plasmids containing NPM1 full-length (NPM1-HA) or lysine 257 mutant (NPM1-K257-HA) sequence were incubated with 40 μM cycloheximide (MCE, China) to block protein synthesis. Cells were harvested and lysed after the addition of sodium oxamate or sodium lactate for 0, 6, 12, and 24 h. Protein expression was quantified using western blot analysis.
Western blotting
Total protein from the cells and kidney tissues was extracted using RIPA buffer supplemented with 1% protease inhibitor (Beyotime, CHN). Equal amounts of protein were separated by SDS-PAGE and transferred onto PVDF membranes (Millipore, USA). Nonspecific proteins were blocked using tris-buffered saline (TBS; Beyotime, CHN) supplemented with 5% skim milk, followed by incubation with the primary antibody at 4 °C overnight. After washing three times with TBST (TBS supplemented with Tween), the membranes were incubated with secondary antibodies against mouse or rabbit, which were conjugated with HRP, for 1 h at room temperature. Protein blots were visualized using the VILBER FUSION FX5 system and analyzed using ImageJ software (NIH, USA).
Quantitative real-time PCR
Total RNA was extracted from pretreated cells using TRIzol (ThermoFisher, USA) and reverse-transcribed to generate cDNA using a high-capacity ABScript III RT Master Mix for qPCR with gDNA (ABclonal, China). Quantitative real-time PCR (qRT-PCR) analysis was conducted on an ABI-7500 system using the SYBR Green reaction mix (ABclonal, China). The gene expression level was estimated considering β-actin the internal control, which was further normalized according to the expression level in the control group. The primer sequences are listed in Supplementary Table 5.
Immunofluorescence (IF)
The kidney tissue slices were stained using anti-NPM1, anti-SLC7A11 and anti-LTL antibodies. Goat anti-rabbit Alexa Fluor 488 or anti-mouse Alexa Fluor 594 secondary antibodies were used to visualize the signal. For nuclear staining, the slides were rinsed and mounted with an antifade reagent containing DAPI. High-resolution images were captured using a confocal laser microscope (Leica Microsystems AG).
Lactate assay
Lactate concentrations in the urine, kidney tissues, HK-2 cells, and cell culture medium were measured using lactate assay kit (Abbkine, CHN) following the instructions provided by the manufacturer. Initially, samples were homogenized with lactate assay buffer in an ice bath and centrifuged at 12,000 × g for 5 min at 4 °C. The production of lactate was measured according to the instructions of the lactic acid assay kit. The absorbance was measured using a microplate reader (TECAN, USA).
Renal morphology and function assessment
Renal morphology was analyzed through hematoxylin and eosin (H&E) staining assay. The renal tubular injury was assessed based on tubular epithelial cell swelling, tubular atrophy and dilatation, loss of brush borders, vacuolization, cast formation, and desquamation. Ten areas randomly selected from the outer medulla of the kidney were photographed at ×400 magnification using a microscope. The severity of damage was classified based on the percentage of the injured area, using pathological scoring ranging from 0 to 5 (0: no injury; 1: injured area <10%; 2: 10–25%, 3: 25–50%, 4: 50–75%; and 5: >75%). Renal function was determined by BUN and SCr concentrations.
Cell viability
Cell viability was measured using Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). HK-2 cells were seeded into 96-well plates (5 × 103 cells/well). Next, the cells were treated with different conditioned media. The treated cells were incubated with CCK-8 reagent at 10% concentration for 2 h, and the absorbance was measured at 450 nm using a microplate reader.
Lipid peroxidation assay
Lipid peroxidation was assessed using the C11-BODIPY 581/591 probe. Cell suspensions were incubated with C11-BODIPY 581/591 (5 μM final concentration) at 37 °C for 30 min. After washing, cells were resuspended in PBS and analyzed by flow cytometry. Data analysis was performed using FlowJo software.
Malondialdehyde (MDA) concentration was determined using a lipid peroxidation assay kit (Abcam, UK) following the instructions provided by the manufacturer. Standards and samples were incubated with TBA reagent for 1 h at 95 °C and quickly cooled to room temperature. Next, the resulting mixture was transferred to a 96-well plate and absorbance was measured at a wavelength of 532 nm using a microplate reader.
Iron assay
Iron levels were measured using a commercially available iron assay kit (ElabScience). HK-2 cells and kidney tissues were homogenized in pre-cooled iron assay buffer and centrifuged at 16,000 × g for 10 min (4 °C). Subsequently, 10 μL of supernatant was transferred to a 96-well plate and adjusted to 100 μL using iron assay buffer. Next, 5 μL of iron reducer was added to the mixture and incubated at 25 °C for 30 min. Following incubation with the iron probe in the dark, the absorbance was measured at 593 nm using a microplate reader.
Glutathione assay
Glutathione (GSH) concentration was quantified using a Glutathione Assay Kit (Elabscience, CHN). For this assay, HK-2 cells and kidney tissues were treated with a 5% 5-sulfosalicylic acid solution. Next, 10 μL of supernatant was mixed with the reaction mix and incubated at room temperature for 5 min. GSH concentration was quantified by measuring the absorbance at 405 nm.
Computational and AI-driven screening of NPM1-K257-targeted compounds
The full-length sequence of NPM1 was retrieved from the UniProt database, and its three-dimensional structure was obtained from the AlphaFold Database (https://alphafold.ebi.ac.uk). The structures of 4,672 FDA-approved or investigational small molecules (TargetMol) were initially prepared with RDKit program (https://www.rdkit.org), followed by geometry optimization using the MMFF94s force field to correct molecular strains. The protein structure was prepared with AutoDock Tools (version 1.5.6) by adding hydrogen atoms, assigning Gasteiger charges, and merging nonpolar hydrogens. Molecular docking was performed using AutoDock Vina software (version 1.1.2) in a semi-flexible mode, with the docking grid centered on the K257 region of NPM1 (center: x = 23.6, y = –30.3, z = 34.1; grid box size: 30 × 30 × 30 Å3).
The top 10% of compounds (467 molecules) ranked by binding affinity were selected for subsequent analysis. MACCS structural fingerprints were computed using RDKit program, and unsupervised K-means clustering was applied based on molecular similarity. The optimal number of clusters (K = 5) was determined using the elbow method. The chemical space was visualized using t-SNE, with binding affinity values projected onto the plot—darker shades indicating stronger binding (lower binding energy). From each cluster, the top 10 molecules by binding affinity (50 in total) were further evaluated using the AI-based Boltz2 model. Binding affinity scores (<0 indicating strong binding) and affinity probability binary values (> 0.5 indicating high binding probability) were used to prioritize candidates.
Surface plasmon resonance assay
SPR experiments were performed on a Biacore T200 instrument (Cytiva, Sweden) using a Series S Sensor Chip CM5 (Lot #10344853), as described previously60. The running buffer (1× PBS containing 0.05% Tween-20 and 1% DMSO) was degassed before use. The CM5 sensor chip was activated with a 1:1 mixture of 0.4 M EDC and 0.1 M NHS (Amine Coupling Kit, Cytiva) at a flow rate of 10 μL/min. Purified NPM1 protein, diluted in 10 mM sodium acetate (pH 4.0), was immobilized on the sensor surface to a level of approximately 8000 response units (RU). Blocking was performed using ethanolamine at 10 μL/min. TCDCA was dissolved in PBST (1% DMSO) and serially diluted for concentration analysis. Binding kinetics were assessed at 25 °C using a multi-cycle method with a flow rate of 30 μL/min, 90 s association phase, and 120 s dissociation phase. Prior to sample analysis, three start-up cycles and blank injections were performed to stabilize the baseline and correct for background signals. All sensorgrams were double-referenced, and kinetic parameters (kon and koff) and equilibrium dissociation constants (KD) were derived by global fitting to a 1:1 Langmuir binding model using Biacore Evaluation Software (Cytiva).
Statistics and reproducibility
Firth penalized logistic regression was performed using the logistf package in R to identify independent risk factors for DGF and to mitigate small-sample bias and the issue of separation. ROC analysis was performed using the pROC package in R to compare the predictive performance of the different DGF models, and the AUC was calculated for each. SPSS (version 21.0; IBM, USA) and GraphPad Prism 8.0 (GraphPad Software, USA) were used to analyze data and the Pearson correlation. Data are presented as mean ± standard deviation (SD). Comparisons between two groups were conducted using unpaired two-tailed Student’s ttests and multiple-group comparisons were conducted using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Sample independence, normal distribution, and variance equality were assumed to be satisfied before the tests. Results were validated through at least three biological replicates. Statistical significance was defined as p < 0.05.
Data availability
The lactylome data generated in this study has been deposited in ProteomeXchange via the PRIDE database under accession code PXD058534. The RNA-seq and ChIP-seq data are available in the GEO database under accession codes GSE306588 and GSE306531, respectively. The raw metabonomics and metabolic flux data are available in CNSA database under accession codes CNP0008378 and CNP0008376, respectively. Source data are provided with this paper.
References
Miller, G. et al. Kidney transplantation outcome predictions (KTOP): A risk prediction tool for kidney transplants from brain-dead deceased donors based on a large European cohort. Eur. Urol. 83, 173–179 (2023).
Lindeman, J. H. et al. Results of an explorative clinical evaluation suggest immediate and persistent post-reperfusion metabolic paralysis drives kidney ischemia reperfusion injury. Kidney Int. 98, 1476–1488 (2020).
Smith, M., Dominguez-Gil, B., Greer, D. M., Manara, A. R. & Souter, M. J. Organ donation after circulatory death: Current status and future potential. Intensive Care Med. 45, 310–321 (2019).
Zheng, X. et al. Kynurenine 3-monooxygenase is a critical regulator of renal ischemia-reperfusion injury. Exp. Mol. Med. 51, 1–14 (2019).
Reichardt, C. et al. DNA-binding protein-A promotes kidney ischemia/reperfusion injury and participates in mitochondrial function. Kidney Int. 106, 241–257 (2024).
Xu, C. et al. Regulation of pericyte metabolic reprogramming restricts the AKI to CKD transition. Metab.: Clin. Exp. 145, 155592 (2023).
She H. et al. Dexmedetomidine ameliorates myocardial ischemia-reperfusion injury by inhibiting MDH2 lactylation via regulating metabolic reprogramming. Advanced science (Weinheim, Baden-Wurttemberg, Germany), e2409499 (2024).
Oeing, C. U. et al. MTORC1-regulated metabolism controlled by TSC2 limits cardiac reperfusion injury. Circ. Res. 128, 639–651 (2021).
He, H. et al. L-2-hydroxyglutarate protects against cardiac injury via metabolic remodeling. Circ. Res. 131, 562–579 (2022).
Bei, Y. et al. Inhibition of Hmbox1 promotes cardiomyocyte survival and glucose metabolism through Gck activation in ischemia/reperfusion injury. Circulation 150, 848–866 (2024).
Yu, S. et al. m 6 A-mediated gluconeogenic enzyme PCK1 upregulation protects against hepatic ischemia-reperfusion injury. Hepatology (Baltimore, Md.) 81, 94–110 (2023).
Zhuang, S. et al. Targeting P2RX1 alleviates renal ischemia/reperfusion injury by preserving mitochondrial dynamics. Pharmacol. Res. 170, 105712 (2021).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Zhou, J. et al. Astrocytic LRP1 enables mitochondria transfer to neurons and mitigates brain ischemic stroke by suppressing ARF1 lactylation. Cell Metab. 36, 2054–2068.e2014 (2024).
Wang, Y. et al. The glycolytic enzyme PFKFB3 drives kidney fibrosis through promoting histone lactylation-mediated NF-κB family activation. Kidney Int. 106, 226–240 (2024).
Li, Y. et al. 7-Dehydrocholesterol dictates ferroptosis sensitivity. Nature 626, 411–418 (2024).
Chu, L. K. et al. Autophagy of OTUD5 destabilizes GPX4 to confer ferroptosis-dependent kidney injury. Nat. Commun. 14, 8393 (2023).
Li, R. et al. FTO deficiency in older livers exacerbates ferroptosis during ischaemia/reperfusion injury by upregulating ACSL4 and TFRC. Nat. Commun. 15, 4760 (2024).
Co, H. K. C., Wu, C. C., Lee, Y. C. & Chen, S. H. Emergence of large-scale cell death through ferroptotic trigger waves. Nature 631, 654–662 (2024).
Zhou, X. et al. Combinational analysis of metabolomic and O-GlcNAcylation omics reveals the HBP metabolic regulation of chemoresistance via GFPT1/NR3C1 O-GlcNAcylation/GPX4 axis. Res. (Wash., D.C.) 8, 0809 (2025).
Bayır, H., Dixon, S. J., Tyurina, Y. Y., Kellum, J. A. & Kagan, V. E. Ferroptotic mechanisms and therapeutic targeting of iron metabolism and lipid peroxidation in the kidney. Nat. Rev. Nephrol. 19, 315–336 (2023).
Fiore, A. et al. Kynurenine importation by SLC7A11 propagates anti-ferroptotic signaling. Mol. cell 82, 920–932.e927 (2022).
Riegman, M. et al. Ferroptosis occurs through an osmotic mechanism and propagates independently of cell rupture. Nat. Cell Biol. 22, 1042–1048 (2020).
Zhang, Q. et al. PAFAH2 suppresses synchronized ferroptosis to ameliorate acute kidney injury. Nat. Chem. Biol. 20, 835–846 (2024).
Li, X. et al. Small extracellular vesicles delivering lncRNA WAC-AS1 aggravate renal allograft ischemia‒reperfusion injury by inducing ferroptosis propagation. Cell Death Differ. 30, 2167–2186 (2023).
Devisscher, L. et al. Discovery of novel, drug-like ferroptosis inhibitors with in vivo efficacy. J. Med. Chem. 61, 10126–10140 (2018).
Farmer, L. A. et al. Intrinsic and extrinsic limitations to the design and optimization of inhibitors of lipid peroxidation and associated cell death. J. Am. Chem. Soc. 144, 14706–14721 (2022).
Zong, Z. et al. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell 187, 2375–2392.e2333 (2024).
Hong, J. et al. AARS1-mediated lactylation of H3K18 and STAT1 promotes ferroptosis in diabetic nephropathy. Cell Death Differ. https://doi.org/10.1038/s41418-025-01587-4 (2025).
Lin, J. et al. NUDT21 lactylation reprograms alternative polyadenylation to promote cuproptosis resistance. Cell Discov. 11, 52 (2025).
Xing, Z. et al. High-glucose-associated YTHDC1 lactylation reduces the sensitivity of bladder cancer to enfortumab vedotin therapy. Cell Rep. 44, 115545 (2025).
Ju, J. et al. The alanyl-tRNA synthetase AARS1 moonlights as a lactyltransferase to promote YAP signaling in gastric cancer. J. Clin. Investig. 134, e174587 (2024).
Passaro, S. et al. Boltz-2: Towards Accurate and Efficient Binding Affinity Prediction. bioRxiv: the preprint server for biology https://doi.org/10.1101/2025.06.14.659707 (2025).
Martin-Sanchez, D. et al. TWEAK and RIPK1 mediate a second wave of cell death during AKI. Proc. Natl. Acad. Sci. USA 115, 4182–4187 (2018).
Vanden Berghe, T., Linkermann, A., Jouan-Lanhouet, S., Walczak, H. & Vandenabeele, P. Regulated necrosis: The expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. cell Biol. 15, 135–147 (2014).
Kim, S. E. et al. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol. 11, 977–985 (2016).
Azushima, K. et al. Abnormal lactate metabolism is linked to albuminuria and kidney injury in diabetic nephropathy. Kidney Int. 104, 1135–1149 (2023).
Lan, R. et al. Mitochondrial Pathology and Glycolytic Shift during Proximal Tubule Atrophy after Ischemic AKI. J. Am. Soc. Nephrology: JASN 27, 3356–3367 (2016).
Darshi, M. et al. Glycolytic lactate in diabetic kidney disease. JCI Insight 9, e168825 (2024).
Zhang, X. et al. Lactate drives epithelial-mesenchymal transition in diabetic kidney disease via the H3K14la/KLF5 pathway. Redox Biol. 75, 103246 (2024).
Friedmann Angeli, J. P., Krysko, D. V. & Conrad, M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer 19, 405–414 (2019).
Yang, X. et al. Excessive SOX8 reprograms energy and iron metabolism to prime hepatocellular carcinoma for ferroptosis. Redox Biol. 69, 103002 (2024).
Wu, D., Spencer, C. B., Ortoga, L., Zhang, H. & Miao, C. Histone lactylation-regulated METTL3 promotes ferroptosis via m6A-modification on ACSL4 in sepsis-associated lung injury. Redox Biol. 74, 103194 (2024).
An, X. et al. The effect of tau K677 lactylation on ferritinophagy and ferroptosis in Alzheimer’s disease. Free Radic. Biol. Med. 224, 685–706 (2024).
Yu, W. et al. HSPA12A maintains aerobic glycolytic homeostasis and Histone3 lactylation in cardiomyocytes to attenuate myocardial ischemia/reperfusion injury. JCI Insight 9, e169125 (2024).
Zhang, Y. et al. CircTTC13 promotes sorafenib resistance in hepatocellular carcinoma through the inhibition of ferroptosis by targeting the miR-513a-5p/SLC7A11 axis. Mol. Cancer 24, 32 (2025).
Jin, C. et al. Piezo1-mediated ferroptosis delays wound healing in aging mice by regulating the transcriptional activity of SLC7A11 through activating transcription factor 3. Research. (Wash., D.C.) 8, 0718 (2025).
Ye, Y. et al. Repression of the antiporter SLC7A11/glutathione/glutathione peroxidase 4 axis drives ferroptosis of vascular smooth muscle cells to facilitate vascular calcification. Kidney Int. 102, 1259–1275 (2022).
Jiang, L. et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015).
Zhang, Y. et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 20, 1181–1192 (2018).
Zhang, H. L. et al. Galectin-13 reduces membrane localization of SLC7A11 for ferroptosis propagation. Nat. Chem. Biol. 21, 1531–1543 (2025).
Zhou, F. & Müller-Tidow, C. NPM1 functions in epitranscriptomics. Nat. Genet. 51, 1436–1437 (2019).
Wang, X. et al. NPM1 inhibits tumoral antigen presentation to promote immune evasion and tumor progression. J. Hematol. Oncol. 17, 97 (2024).
Pardieu, B. et al. Cystine uptake inhibition potentiates front-line therapies in acute myeloid leukemia. Leukemia 36, 1585–1595 (2022).
Chen, H. et al. AURKA inhibition induces Ewing’s sarcoma apoptosis and ferroptosis through NPM1/YAP1 axis. Cell Death Dis. 15, 99 (2024).
Li, X. et al. Roles of Akirin1 in early prediction and treatment of graft kidney ischemia‒reperfusion injury. Smart Med. 3, e20230043 (2024).
Wang, C. et al. BMSCs protect against renal ischemia-reperfusion injury by secreting exosomes loaded with miR-199a-5p that target BIP to inhibit endoplasmic reticulum stress at the very early reperfusion stages. FASEB J. 33, 5440–5456 (2019).
Li, X. et al. Negative feedback loop of autophagy and endoplasmic reticulum stress in rapamycin protection against renal ischemia-reperfusion injury during initial reperfusion phase. FASEB J. 32, 6002–6018 (2018).
Li, X. et al. Bladder cancer-derived small extracellular vesicles promote tumor angiogenesis by inducing HBP-related metabolic reprogramming and SerRS O-GlcNAcylation in endothelial cells. Adv. Sci. (Weinh., Baden. -Wurtt., Ger.) 9, e2202993 (2022).
Li, X. et al. Irinotecan alleviates chemoresistance to anthracyclines through the inhibition of AARS1-mediated BLM lactylation and homologous recombination repair. Signal Transduct. Target. Ther. 10, 214 (2025).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82403698 to Xinyuan Li; 82303655 to Hubin Yin; 82372881 to Weiyang He; 82472779 to Xin Gou), the Postdoctoral Fellowship Program of CPSF grant (GZC20233348 to Xinyuan Li), Chongqing Natural Science Foundation (CSTB2024NSCQ-MSX1030 to Xinyuan Li), Chongqing Medical Youth Top Talent Project (YXQN202401 to Xinyuan Li), Chongqing Postdoctoral Special Funding Project (2023CQBSHTB3054 to Xinyuan Li). We thank Jiaxiu Liu from Chongqing Medical University (College of Artificial Intelligence Medicine) for guidance on statistical analysis. We are also grateful to Professor Hao Yu from Sun Yat-sen Memorial Hospital for helpful discussions.
Author information
Authors and Affiliations
Contributions
X.L., T.L., W.H., and J.L. directed the study. X.L., W.Z., and X.G. designed experiments; Haitao Y., Hubin Y., Y.M., and X.B. performed experiments and analyzed data. X.L. and Haitao Y. wrote the manuscript. C.Z., Z.F., and H.C. assisted in animal experiments and bioinformatic analysis. M.L. and S.L. provided clinical assistance. Hubin Y., Haitao Y., Y.M., and X.B. revised the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Xiaoming Ding, Hina Bhart, Lingyi Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yu, H., Yin, H., Mei, Y. et al. Nucleophosmin 1 lactylation in graft kidney induces ferroptotic trigger waves that exacerbate delayed graft function. Nat Commun 17, 277 (2026). https://doi.org/10.1038/s41467-025-66991-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66991-3