Abstract
Mammalian brains vary in size, structure and function, but the extent to which evolutionarily novel cell types contribute to this variation remains unresolved1,2,3,4. Previous studies suggest that there is a primate-specific population of striatal inhibitory interneurons—the TAC3 interneurons5. However, broader taxonomic and developmental characterization is required to address novelty in cell-type evolution. Here we examine gene expression in inhibitory neurons across 10 mammalian species, spanning 160 million years of divergence from primates. We find that the initial class of newborn TAC3 interneurons specified during development represents an ancestral, medial ganglionic eminence (MGE)-derived striatal population that is also present in pig and ferret cortex. This discovery prompted a re-examination of Glires, including mice, which are thought to lack the TAC3 type5,6. Targeted enrichment of MGE precursors in mice revealed conservation of the TAC3 initial class, camouflaged by reduced expression of Tac2 (the mouse orthologue of TAC3) and a gain of Th expression. Extending our analysis to the adult striatum further supported the homology of primate TAC3 and mouse Th striatal interneurons, and also uncovered a rare Tac2 subpopulation in the mouse ventromedial striatum. This study suggests that initial classes of telencephalic inhibitory neurons are largely conserved, and that during evolution, neuronal types in the mammalian brain change through redistribution and fate refinement, rather than by derivation of novel precursors early in development.
Similar content being viewed by others
Main
Ramón y Cajal first appreciated the abundance and morphological diversity of primate interneurons, linking these elaborations to “the functional superiority of the human brain”7,8. Cortical and striatal inhibitory neurons mainly emerge prenatally from progenitors that line the lateral ventricles of the ventral telencephalon, where spatial and temporal patterning signals influence the specification of inhibitory neuron subtypes9,10, resulting in a newborn neuron initial class. Subsequently, extrinsic cues during neuronal maturation further refine identity11 to a terminal class fate in adults. Unlike in the cortex, most striatal neurons are GABAergic (γ-aminobutyric acid producing), including the principal projection neuron type (spiny projection neurons; SPNs)12. Striatal inhibitory interneuron subtypes, which comprise about 10% of striatal neurons in mice and 15–20% in primates13, are generated mainly from embryonic progenitors that express the transcription factor NKX2-1 and reside in the MGE. In mice, striatal interneurons comprise terminal classes classically denoted by their expression of selective markers and neurochemical pathway genes, including Chat, Pthlh/Pvalb, Sst, Cck and Th (refs. 14,15,16). In addition to these conserved classes, TAC3-expressing striatal interneurons constitute 30% of primate striatal interneurons, but were not observed in the first surveys of mouse and ferret striatum, raising the possibility that they represent a primate-specific population5. Given the conservation of other initial classes of inhibitory neurons and regional patterns of transcription factor expression during development6,17, we sought to examine the developmental specification of this class in a wider taxonomic range and at higher cellular resolution. Notably, we find that initial classes of inhibitory neurons, including TAC3, are shared across placental mammals, with modifications of anatomical allocation and gene expression of the TAC3 class in several taxa. This suggests that evolution of neural cell types in mammals acts on a conserved set of initial developmental classes.
Conservation of initial classes in developing mammals
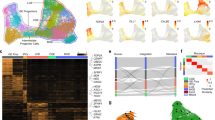
To examine the conservation of initial classes, we surveyed cell-type diversity in placental mammals. We performed single-cell RNA sequencing (scRNA-seq) using 10x Genomics Next GEM Single Cell technology on the cortex and striatum of developing Eutheria (rhesus macaques, rats, mice, rabbits, pigs and ferrets) and developing Marsupialia (sugar gliders and opossums), representing 160 million years of evolution (Fig. 1a and Extended Data Fig. 1a). We reasoned that initial stages of neuronal specification might reveal homologies obscured by fate refinement during later stages of maturation11. To capture cells during the peak stages of interneuron specification, we selected stages comparable with the second trimester of human development, defined using developmental anatomy and the Translating Time tool18,19,20,21 (Fig. 1b). For each species, we applied stringent quality control metrics and performed dimensionality reduction, batch correction and Leiden clustering. To focus our investigation on inhibitory neurons, we isolated this population using the conserved markers DLX1, DLX2, DLX5 and DLX6, and GAD1 and GAD2 (Methods), and performed Leiden clustering on inhibitory cells. We next classified these clusters in each species on the basis of region-specific transcription factors indicating MGE (NKX2-1 and LHX6), lateral ganglionic eminence (LGE; PAX6, MEIS2 and FOXP2) or caudal ganglionic eminence (CGE; NR2F2)9,22 origin, as well as markers of initial classes defined by previous macaque and mouse data6 (Extended Data Figs. 2−5). To identify homologous cell types without disproportionately weighting the results to one species, we downsampled individual species clusters to contain no more than 1,000 cells. We co-embedded these downsampled data using Harmony (Methods) and labelled integrated clusters on the basis of conserved markers (Fig. 1c and Extended Data Fig. 1b). Integrated clusters mostly contained cells from every species and corresponded to cluster labels from independent analysis of each individual species, highlighting conserved marker genes (Fig. 1c,d and Extended Data Fig. 1c–g). Pearson correlation analysis revealed that initial classes were highly correlated across species and segregated according to cell type rather than species (Fig. 1e and Extended Data Fig. 1h), and we observed similar developmental trajectories across species (Extended Data Figs. 1−5). These findings support broad patterns of conservation in inhibitory neuron initial classes across Mammalia.
a, Phylogeny of placental mammals surveyed. Ma, million years ago. b, Stages surveyed compared to human development. PCD, post-conception day. c, Uniform manifold approximation and projection (UMAP) coloured by integrated inhibitory neuron initial classes and individual species (n = 75,000 inhibitory neurons), and key for d,e. d, Heat map of selected markers of inhibitory neuron initial classes plotted by individual species, scaled and normalized. Grey boxes indicate that no cells from that species contribute to the integrated cluster, e, Similarity matrix of Pearson correlations based on principal component analysis (PCA) of initial classes from integrated clusters across species.
The MGE_CRABP1/TAC3 class, previously observed exclusively in primates, gives rise to TAC3 neurons6, whereas the closely related MGE_CRABP1/MAF initial class is conserved in primates and rodents, and in mice is thought to give rise to Pvalb, Pthlh and Th striatal interneurons15. Notably, co-embedding revealed that cells isolated from pigs, ferrets, opossums and sugar gliders contribute to both MGE_CRABP1/MAF (5,924 cells) and MGE_CRABP1/TAC3 (2,880 cells) integrated clusters (Fig. 2a,b). Within the integrated MGE_CRABP1/TAC3 cluster, we observed enriched expression of known markers6, including TRHDE, STXBP6 and TAC3 (Fig. 2c and Extended Data Fig. 1b). In the MGE_CRABP1/MAF integrated cluster, we also observed enriched expression of known markers, including MAF, MAFB and COL19A1 (Fig. 1d). Cells within each initial class showed strong correlations across species, supporting the overall conservation of both MGE_CRABP1 classes (Fig. 1e).
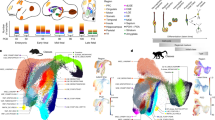
a, Laurasiatherian and marsupial species studied. b, UMAP coloured by MGE_CRABP1 initial classes (MGE_CRABP1/MAF, n = 5,924 cells; MGE_CRABP1/TAC3, n = 2,880). Inset illustrates laurasiatherian and marsupial species that contribute to the MGE_CRABP1 initial class clusters. c, Dot plot of marker-gene expression in the MGE_CRABP1/TAC3 cluster, scaled and normalized. d, RNAscope in developing pigs at E62 (whole-hemisphere image acquired at 10×), with markers LHX6, CRABP1 and TAC3. Boxed regions highlight the MGE_CRABP1/TAC3 class in the striatum (e) and cortex (f). Scale bar, 1,000 μm. e,f, High-magnification (100×) maximum intensity projection of striatum (e) and cortex (f), with arrowheads indicating the MGE_CRABP1/TAC3 initial class. Scale bars, 25 μm. g, RNAscope in developing ferrets at P1 (whole-section image acquired at 20×), with markers LHX6, CRABP1 and TAC3. Boxed region highlights the MGE_CRABP1/TAC3 class in the striatum (see h). Scale bar, 1,000 μm. h, High-magnification (100×) maximum intensity projection, with arrowheads indicating the MGE_CRABP1/TAC3 class. Scale bar, 25 μm. i, RNAscope in the P22 ferret brain (whole-section image acquired at 10×), with markers LHX6, CRABP1 and TAC3. Boxed regions highlight striatum (j) and cortex (k). Scale bar, 1,000 μm. j,k, High-magnification (100×) maximum intensity projection of striatum (j) and cortex (k), with arrowheads indicating the MGE_CRABP1/TAC3 class. Scale bars, 25 μm.
Despite the overall homology of MGE_CRABP1 classes, we observed changes in marker-gene expression—consistent with studies in primate and rodent motor cortex23—including genes that could influence signalling pathways. For example, CRABP1, encoding a retinoic-acid-binding protein, was not detected in either the MGE_CRABP1/MAF or the MGE_CRABP1/TAC3 initial class in both marsupial species, perhaps indicating the modification of these initial classes early in the diversification of placental mammals. Similarly, CER1, encoding a BMP antagonist, was enriched in primate TAC3 neurons (Fig. 2c). TAC3, encoding the neuropeptide originally used to name the subpopulation, was also detected in COL19A1-, MAF- and MAFB-expressing cells in ferrets, designated by integrated clustering (Extended Data Fig. 4p). Functional studies will be required to ascertain whether marker-gene turnover, including signalling-pathway genes, reflects neutral drift or contributes to lineage-specific functional specializations among homologous cell types.
Analyses in marmosets, macaques and humans suggest that cells derived from the MGE_CRABP1/TAC3 initial class are present in the primate striatum but not in the neocortex5,6,24. To confirm the spatial location of the TAC3 population in Laurasiatheria, we performed RNA in situ hybridization (RNAscope) (Fig. 2d–k and Extended Data Fig. 6) and observed a LHX6-, CRABP1- and TAC3-positive population in the striatum of developing pigs and ferrets, as well in adult porpoises (Fig. 2d,e,g–j and Extended Data Fig. 6a), supporting the conservation of the MGE_CRABP1/TAC3 initial class in Laurasiatheria. Of note, we also observed LHX6-, CRABP1- and TAC3-expressing cells migrating towards and within the cortex of pigs as early as embryonic day (E)50 (Fig. 2f and Extended Data Fig. 6b–o). We did not observe an LHX6-, CRABP1- and TAC3-positive population in or migrating towards the cortex of ferret at postnatal day (P)1, but we did observe this population in the cortex at later time points (Fig. 2g–k and Extended Data Fig. 6p–r). Finally, we performed RNAscope in adult pigs and ferrets and confirmed that these populations persist past development in both the striatum and cortex (Extended Data Fig. 6s–x).
The cortical and striatal populations in pigs and ferrets overall showed similar gene expression, but we observed a small number of differentially expressed genes that could contribute to migratory or functional differences within this cell type between brain structures (Extended Data Fig. 4f–h,q). Differential gene-expression analysis using DEseq2 revealed that cortical MGE_CRABP1/TAC3 cells in pigs were enriched for the transcription factor DACH1, the cell-adhesion-linked gene TENM3 and the diazepam binding inhibitor DBI, whereas striatal MGE_CRABP1/TAC3 cells in pigs were enriched for SPATS2L and the cyclic AMP pathway gene ADCY8 (Extended Data Fig. 4f). Previous studies have described CRABP1 cells migrating to the cortex in the developing human and linked these to a cortical PVALB population, a derivative of the MGE_CRABP1/MAF class17. When examining differentially expressed genes in the MGE_CRABP1/MAF class, we observed increased expression of cortical migration markers such as CXCR4 and HCN1, with similar trends for these genes in the TAC3 class (Extended Data Fig. 4g,h). Given the presence of the MGE_CRABP1/TAC3 initial class in the cortex of both pigs and ferrets, we further examined the distribution of this initial class in humans and macaques. Using a combination of RNAscope and analysis of published datasets, we found no evidence of this population in the primate neocortex (Extended Data Fig. 7a–o). However, we cannot rule out the possibility of a rare migrating population, because discovering such a population might require additional cell coverage or closer spatial analysis. Together, these results indicate that MGE_CRABP1/TAC3 interneurons represent an ancestral population, migrate to the cortex in pigs and ferrets and exhibit lineage-specific marker-gene turnover.
Conserved TAC3 initial class is modified in rodents
Our previous studies examining both adult and developing mouse data did not detect TAC3 interneurons nor a MGE_CRABP1/TAC3 initial class, despite the fact that we analysed more than 140,000 inhibitory neurons in adult mice and 76,000 inhibitory neurons in developing mice6. However, the observation of a conserved MGE_CRABP1/TAC3 initial class in other mammals in this study raised the question of whether this class has been lost in mice. To examine striatal interneuron development more thoroughly in mice, we performed high-cellular-coverage analysis across multiple developmental time points by dissecting the MGE, LGE and developing striatum of Nkx2-1-Cre/Ai14 mouse embryos, and sorting tdTomato+ cells (Fig. 3a).
a, Schematic of experimental design. b, UMAP coloured by inhibitory neuron initial classes recovered from the MGE, LGE and striatum of developing mice. c, Heat map of selected initial class markers, scaled and normalized. d, UMAP coloured by markers of Lhx6/Crabp1-expressing initial classes, scaled and normalized. Top row contains markers of MGE_CRABP1/MAF; bottom row contains markers of MGE_CRABP1/TAC3. e, Pearson correlation based on PCA of mouse MGE_CRABP1 initial classes to integrated classes for all species except mice. f, Marker-gene expression in Glires from the integrated MGE_CRABP1/TAC3 cluster, scaled and normalized. g, High-magnification (100×) maximum intensity projection of tdTomato reporter expression and RNAscope of markers Crabp1 and Th in the E17 Nkx2-1-Cre;Ai14 mouse brain. Arrowheads indicate the MGE_CRABP1/TH initial class. Scale bar, 25 μm. h, High-magnification (100×) maximum intensity projection of RNAscope with markers Lhx6, Crabp1 and Th in the adult rat striatum. Arrowheads indicate the MGE_CRABP1/TH initial class. Scale bar, 25 μm.
We recovered 71,036 cells, which were predominantly MGE-derived inhibitory neurons, marked by Lhx6 and Nkx2-1. We detected two distinct Crabp1 populations of comparable abundance, each containing cells from all three developmental stages surveyed (Fig. 3b,c and Extended Data Fig. 3a). One Crabp1 population (2,353 cells) co-expressed the markers Maf, Mafb, Rbp4 and Col19a1, indicating that it represented the conserved MGE_CRABP1/MAF initial class. The second Crabp1-positive population (1,393 cells) expressed markers consistent with the MGE_CRABP1/TAC3 class, including Chrna3, Stxbp6 and Trhde. However, this class also expressed high levels of Th, with a small subset (seven cells) that co-expressed Tac2, the mouse orthologue of TAC3 (Fig. 3d). Pearson correlation analysis revealed that this Th-expressing population was most highly correlated with the MGE_CRABP1/TAC3 initial class across all developing species surveyed (Fig. 3e and Extended Data Fig. 1g). Integrated clustering further supported homology, because the mouse Th-expressing population co-clustered with the MGE_CRABP1/TAC3 class (Extended Data Fig. 1c,f), despite the fact that TH was not detected in this population in primates or in Laurasiatheria (pigs and ferrets) (Fig. 2c). This modification of the mouse MGE_CRABP1/TAC3 class to express Th in place of Tac2 is consistent with the turnover of other signalling genes observed among placental mammals. RNAscope in developing (E17) and adult mouse tissue confirmed the presence of Nkx2-1, Crabp1 and Chrna3 co-expressing cells specifically in the striatum, and the expression of Th in this population (Fig. 3g and Extended Data Fig. 3p–u).
To examine the phylogenetic origin of a TAC3 initial class with reduced Tac2 and increased Th expression, we next surveyed further Glires species (rats and rabbits) using scRNA-seq without enrichment. We did not recover distinct TAC3- or TH-expressing clusters when examining these species independently; however, co-embedding revealed a clear TAC3-expressing CRABP1/ETV1 population in rabbits, which was confirmed using RNAscope (Fig. 3f and Extended Data Fig. 3v). In rats, we observed Crabp1-expressing cells in the species-integrated TAC3 cluster marked by enriched expression of the class marker genes Trhde, Stxbp6 and Lhx8, but we could not detect Tac3 or Th transcripts (Fig. 3f). We thus next used RNAscope to assess the presence of Th or Tac3 in the MGE_CRABP1/TAC3 initial class in rats. We found that, similar to what was seen in mice, the MGE_CRABP1/TAC3 initial class expressed Th in place of Tac3, suggesting a modification within the rodent lineage (Fig. 3h and Extended Data Fig. 3w,x). Supporting the loss of Tac3 early in the rodent lineage, we could not detect Tac3 in Crabp1- and Lhx6-expressing cells in the striatum of naked mole rats (Extended Data Fig. 3y). These findings indicate that both initial classes of MGE_CRABP1 interneurons observed in mammals are conserved in the Glires clade, with rodents showing a derived reduction of Tac3 or Tac2 expression and a gain of Th expression early in neuronal development.
Comparative analysis of terminal classes
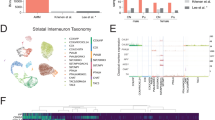
In adult mice, striatal Th interneurons represent a morphologically and physiologically diverse class that connects to principal SPNs and to a subset of other striatal interneuron classes25. Despite expressing Th, which encodes tyrosine hydroxylase, Th interneurons do not appear to synthesize dopamine25,26. Given the evidence for homology between initial classes (Fig. 3), we next examined potential homology between adult Th interneurons in mice and TAC3 cell types in primates. We used the recently published Allen Brain Cell (ABC) atlas of around four million sequenced cells, which provides an order of magnitude greater resolution than do previous studies of the adult mouse brain27. Using the ABC atlas taxonomy and metadata, we isolated 3,586 mouse striatal inhibitory interneurons, and reclustered and classified the populations on the basis of known markers (Extended Data Fig. 8a–c; Methods).
Mouse striatal Th interneurons (676 cells) were found mostly in ABC subclass ‘055 STR Lhx8 Gaba’ (clusters 0839–0844) (Extended Data Fig. 8d,e). These cells express markers of the MGE_CRABP1/CHRNA3 class (Crabp1, Chrna3, Trhde and Stxbp6), but Th was most highly expressed in clusters 0839–0842 (Extended Data Fig. 8f). Notably, we observed a small population of 51 Tac2-expressing cells within ABC clusters 0840–0842 (Extended Data Fig. 8b,f), a pattern resembling the small number of Tac2-expressing cells observed in developing mouse data (Fig. 3d). We confirmed the presence of rare Nkx2-1, Crabp1 and Tac2 co-expressing cells in the adult mouse striatum by RNAscope (Extended Data Fig. 8g). By contrast, in striatal interneurons of adult humans28 and marmosets24, TH was detected mainly in a small subset of the PTHLH/PVALB population (Extended Data Fig. 8i,l), a derivative of the CRABP1/MAF class, and had limited expression in TAC3 interneurons. Coupled with the expression of TH in the primate MGE_CRABP1/MAF initial class and the projection of TH gene modules between species using Hotspot29 (Extended Data Figs. 2c, 7i and 9), these observations suggest that TH marks different striatal interneuron subtypes in rodents and primates6,13.
To examine adult striatal interneuron homologies, we co-embedded adult human, marmoset and mouse striatal interneurons using scVI to account for species and dataset differences (Fig. 4a). We expanded the mouse data to all cells from ABC subclass 055, which includes cells from pallidum and hypothalamus dissections, and we supplemented the primate data with a pre-release of marmoset data from a forthcoming Human and Mammalian Brain Atlas (HMBA)30 (Extended Data Fig. 10a; Methods). Co-embedding revealed the expected conservation of the major striatal interneuron classes SST/NPY, PVALB/PTHLH and CHAT, and provided insight into previously uncharacterized homologies between mouse and primate cell types (Fig. 4b,c and Extended Data Fig. 10b). For example, in humans, we recovered CCK/VIP interneurons, a TAC3-expressing CGE-derived population that is distinct from MGE-derived TAC3 neurons (Extended Data Fig. 8m). In mice, we observed apparent homologues of both the CCK/VIP and the CCK primate populations, which the ABC atlas had annotated as cortical (Fig. 4c and Extended Data Fig. 10c).
a, UMAP coloured by adult striatal GABA interneuron datasets integrated using scVI. b, UMAP coloured by primate cell-type annotations (refs. 24,28,30). c, UMAP coloured by mouse cell-type annotations (ref. 27). d, UMAP coloured by clusters in mouse ABC subclass 055. e, Pearson correlations of gene expression across pre-integration clusters for refs. 24,27,28. Striatum-only mouse cells from the Th cluster (subclass 055) are broken into their ABC clusters. Marmoset TAC3/LHX8 cells were separated using pre-integration clusters. Inset highlights mouse–primate correlations for selected mouse clusters. f, SAMap homology scores for the same clusters and datasets as in e. g, Cell-type abundances of TAC3 populations in primate datasets and mouse striatal cells from parent clusters 0840–0842. h, Distribution of highlighted ABC clusters in the adult mouse brain and Th, Tac2 and Lhx8 expression (in counts per million reads (CPM)) found within those populations27. ABC atlas: mouse whole-brain transcriptomic cell type atlas—MERSCOPE v1 (available at https://doi.org/10.35077/g.610).
Among striatal interneurons, the primate TAC3 type formed an integrated cluster with mouse cells (Fig. 4c,d). Cells belonging to the mouse ABC subclass 055 that were obtained from extra-striatal dissections in the pallidum and hypothalamus clustered closely adjacent to primate TAC3 neurons, but mostly did not overlap with them (Fig. 4c). Within primates, the integrated TAC3 cluster contained an LHX8-enriched subpopulation. High Lhx8-expressing mouse striatal Th interneurons from ABC clusters 0841 and 0842 co-embedded mainly with this LHX8+ primate TAC3 subtype, whereas the lower Lhx8-expressing Th cluster 0840, with reduced Zic1 and Zic4 expression, was most associated with the primate LHX8− TAC3 type (Fig. 4c,d and Extended Data Figs. 10d and 8f).
Separately from integration, we used several other methods to assess cell-type homologies. Pearson correlations of gene expression show that human and marmoset homologous types consistently have the highest correlations to each other, whereas mouse cell-type correlations are higher within species than they are to their primate counterparts (Fig. 4e). Nonetheless, the strongest cross-species correlations to mouse types are the putative homologous types (Fig. 4e, inset). Similarly, Hotspot revealed conserved expression of gene modules in homologous cell types, but also highlighted differences in TH expression (Extended Data Fig. 9 and Supplementary Table 1). Finally, we used SAMap, which is an alternative method for co-embedding cross-species data by weighted sequence similarity and co-expression31. SAMap also predicted strong homology for the mouse clusters 0840–0842 and primate TAC3 (mainly the non-LHX8 primate subtype) (Fig. 4f and Extended Data Fig. 10e).
We next considered the abundance and spatial localization of mouse Th interneuron populations. In contrast to primates, in which TAC3 interneurons represent around 25–40% of striatal inhibitory interneurons, the homologous ABC clusters 0840–0842 contain less than 9% of mouse striatal inhibitory interneurons (of which 51 cells, or 1.4% of striatal interneurons, expressed Tac2) (Fig. 4g and Extended Data Fig. 10f). The mouse ABC 0840 cluster with lower expression of Lhx8, Zic1 and Zic4 was enriched in the dorsal striatum, whereas the 0842 cluster with higher expression of Tac2, Lhx8, Zic1 and Zic4 was enriched in the ventromedial striatum, mirroring the sparse ventromedial expression of Tac2 within Th striatal interneurons (Fig. 4h and Extended Data Fig. 8f). By contrast, the 0841 cluster, which did not integrate as well with primate striatal interneurons, was localized to the ventral lateral septum, basal forebrain and hypothalamus, and shared Tac2 and Lhx8 expression with 0842 but had lower Crabp1 expression. Together, these findings demonstrate the homology of primate TAC3 and mouse Th striatal interneurons, and reveal gene-expression divergence and spatial heterogeneity among mouse populations.
Discussion
By investigating developmental and adult cell-type diversity across a wide range of taxa, here we show that TAC3 striatal interneurons—which were previously considered to be primate-specific—are found broadly across placental mammals, albeit with clade- or species-specific modifications. In rodent evolution, TAC3 expression was reduced and replaced by Th, with a rare subpopulation in mice retaining remnant Tac2 in the ventromedial striatum. Recognition of this homology enables connecting insights from the functions of Th interneurons in tractable rodent models to our understanding of TAC3 interneurons in primates. Similarly, conserved markers across 160 million years of placental mammal evolution, such as acetylcholine receptors and thyrotropin-releasing hormone genes, could inform shared functions of this population. Future studies can investigate the functional consequences of the lineage-specific alterations of gene expression and redistribution of TAC3 populations reported here.
The conservation of initial classes followed by the modification of their gene expression and migratory destinations suggests that brain evolution among mammals acts on a conserved repertoire of initial classes to modify neural circuits, rather than by generating new cell types early in development. Supporting this model, recent studies examining the vertebrate retina at high cell coverage have identified rare mouse orthologues of populations that were previously considered to be primate-specific2,32. Although adult cell atlases might reveal novel populations33,34, developmental comparisons are required to address whether these emerge from conserved initial classes. Repeated instances of marker-gene turnover, including that of genes involved in developmental signalling pathways, highlight the importance of considering gene expression collectively, evaluating a wide range of species and regions and examining multiple developmental stages for homology assignments. The modification and redistribution of the TAC3 initial class also raises the possibility that some initial classes are more labile than others, representing preferred substrates for the evolutionary emergence of novel neuronal function.
Methods
Mouse experimental model and details of other animal experiments
All animal procedures in this study followed experimental protocols approved by the respective institutions.
Mouse procedures in this study followed experimental protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Francisco (UCSF). The Nkx2-1-Cre;Ai14 mouse line is the result of crossing the C57BL/6J-Tg(Nkx2-1-cre)2Sand/J (Nkx2-1-Cre; Jackson Laboratory stock no. 008661) and B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Ai14; Jackson Laboratory stock no. 007914) strains. Mouse housing and husbandry were performed in accordance with the standards of the Laboratory Animal Resource Center (LARC) at UCSF. Mice were group housed in a 12-h light–dark cycle, with access to food and water ad libitum. Nkx2-1Cre;Ai14 mice were crossed, and the date of a positive vaginal plug was considered as E0. Pregnant dams were euthanized, and the brains of embryos from their litters were extracted and assessed for tdTomato fluorescence at the following developmental stages: E15 (n = 16 embryos from 2 litters), E17 (n = 7 embryos from one litter) and E18 (n = 5 embryos from 2 litters).
Rat procedures in this study followed experimental protocols approved by the IACUC at UCSF. Rat housing and husbandry were performed in accordance with the standards of the LARC at UCSF. Rats were group housed in a 12-h light–dark cycle, with access to food and water ad libitum. Long Evans outbred rats were purchased from Charles River (strain code 006). After rat crossing, the day of positive vaginal plug was considered E0. Pregnant dams were euthanized, the brains of embryos from their litters were extracted at E18 and brains from two litters were pooled for sequencing.
Pig samples (E62, E73 and one year and two months of age) were collected at the Swine Teaching and Research Center at the University of California, Davis. Pig procedures were performed in accordance with the standards of the Animal Welfare Act and under conditions approved by the Association of Assessment and Accreditation of Laboratory Animal Care (AAALAC), and followed experimental protocols approved by the IACUC at the University of California, Davis.
Opossum samples (n = 2, P20) come from a pedigreed, breeding colony of grey short-tailed opossums (Monodelphis domestica) that was established by founder individuals purchased from the Southwest Foundation for Biomedical Research, and is maintained by the laboratory of K.E.S. at the University of California, Los Angeles (UCLA). Opossums were euthanized by CO2 inhalation, followed by decapitation. These procedures are in accordance with the AVMA Guidelines for the Euthanasia of Animals 2013, and all animal procedures were approved by the IACUC at UCLA.
Sugar glider (n = 2, P40) (Petaurus breviceps) experiments were performed with the approval of the IACUC committee at Princeton University. Captive-born, adult sugar gliders were purchased through the US pet trade and subsequently housed in a breeding colony at Princeton University. Sugar gliders were fed a diet of dried food, fruits and protein daily, and housed in breeding pairs or trios. The colony was kept under a 12-h light–dark cycle (temperature, 20–27 °C; humidity, 30–70%). Adult female sugar gliders were checked for pouch young by manual palpation of the maternal pouch and visual inspection. Pouch young identified during inspection were collected by briefly anaesthetizing the mother with isoflurane and gently everting the pouch to expose the neonate. Joeys were gently detached from the nipple, euthanized and processed in the laboratory. More details about the sugar glider colony and husbandry can be found a previous study35.
Ferret samples (P1, P14, P22 and 33 months of age) were a gift from the University of Iowa National Ferret Resource and Research Center and were collected under experimental protocols approved by the IACUC at the University of Iowa. New Zealand white rabbit embryos (n = 3, PCD23) were obtained from BioIVT (USDA NY-TEC-0004) and procedures were performed in accordance with the standards of the Animal Welfare Act and under the AAALAC.
Harbour porpoise (Phocoena phocoena) brain samples were collected from stranded individuals by the Marine Mammal Center and obtained under the National Marine Fisheries Service (NMFS) permit no. 23922.
Dissections and dissociation
Dissections of embryonic and postnatal brain samples were performed in Hibernate-E (Gibco A1247601) under a stereo dissection microscope (Olympus SZ61 or Leica MZ10). The entire dissection process was done on ice, using Hibernate-E culture medium. The LGE, the striatum and the mantle zone of the MGE were manually dissected and collected as a striatal sample. The motor and somatosensory cortex was collected independently from ventral regions and collected as a cortical sample. The sex of the embryos was not determined, and thus the results reported are assumed to include animals of both sexes.
For the E73 pig sample, the brain was embedded in low-melting-point agarose and vibratome-sectioned at 300 μm in artificial cerebrospinal fluid (125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 1.25 mM NaH2PO4) before dissection under stereo dissection microscope.
For single-cell dissociation of samples, dissected regions were cut into small pieces and incubated with a prewarmed solution of papain (Worthington Biochemical) prepared according to the manufacturer’s instructions. After 30–60 min of incubation, samples were gently triturated with wide-orifice pipette tips. Once the samples were dissociated to a single-cell suspension, DMEM + 0.1% bovine serum albumin (BSA) was added to quench papain and cells were pelleted at 300g for 5 min at 4 °C. Finally, cells were resuspended in phosphate-buffered saline (PBS) supplemented with 0.04%–0.1% BSA. Mouse samples were then sorted for tdTomato expression on a BD FACSAria Fusion.
Single-cell sequencing of developing species
With the exception of sugar gliders, scRNA-seq of all developing species was completed using the 10x Genomics Chromium X controller and v.3.1 high-throughput RNA capture kits. Sugar glider sequencing was completed using the 10x Genomics Chromium Next GEM Single Cell 3’ Reagent Kit v.3.1 (10x Genomics, CG000315). Samples were loaded at approximately 30,000–100,000 cells per well (10,000 cells per well for sugar gliders) and library preparation was completed following the manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq and NovaSeq X platforms. Sequencing was performed at the UCSF Center for Advanced Technology, supported by UCSF PBBR, RRP IMIA and NIH 1S10OD028511-01 grants.
Alignments and gene models
Illumina BCL files were converted to Fastq files using bcl2fastq2. Genes were quantified with the CellRanger v.7.0.1 count function, using the mRatBN7.2 genome for rats, mOrycun1.1 for rabbits, mMonDom1.pri genome for opossums, ASM1176430v1.1 genome for ferrets, SusScrofa11.1 for pigs and mm10-2020A for mice. For sugar gliders, we used an early draft assembly of the ‘SugarGlider_lib1_testN_pseudohap’ genome36 (an updated but unannotated version of this is now available: https://www.dnazoo.org/assemblies/petaurus_breviceps). Annotations were lifted over from Trichosurus vulpecula (GCF_011100635.1) using Liftoff (https://github.com/agshumate/Liftoff; -a 05 -s 05 -d 5). To improve the recovery of missing transcripts, we used the ReferenceEnhancer37 package to update the SusScrofa11.1 and mMonDom1.pri gene annotations by manually examining read alignment to inhibitory interneuron marker-gene boundaries to improve 3′ end annotation and incorporating intergenic read pile-ups. For sugar gliders, we observed an excessive number of genes with read pile-ups just downstream of the 3′ UTR. Consequently, we applied a uniform 3-kb extension downstream of all genes, but extensions were truncated wherever they would overlap other genes.
Quality control
CellRanger output was input into CellBender v0.2.138 to remove ambient RNA and the CellBender filtered output matrix was used for downstream processing. Additional filtering was performed on individual sequencing lanes according to the distribution of read counts. Droplets with low gene counts and high gene counts (greater than 6,000 genes) were removed from the datasets. Droplets with high mitochondrial and ribosomal reads were removed on the basis of species-specific distributions. Doublets were removed using Scrublet (v.0.2.3)39.
Clustering and assignment of cell types
Analysis of developmental data was based on the Scanpy package and tutorial40 and to be consistent with a previous report6. Counts were normalized and log-transformed. The data were then scaled for each gene and PCA was performed. Batch correction was applied to species with more than one age collected using Harmony integration41. Leiden clustering was applied with resolution set to 1. Inhibitory neurons were then isolated as previously described6. The data were reset to raw, and normalization, log transformation, scaling, PCA and Leiden clustering at a resolution of 1 or 0.5 were repeated on the isolated inhibitory neurons. Remaining clusters containing excitatory neurons and/or doublets were removed when applicable and clustering was recalculated. Individual Leiden clusters were subdivided by increasing resolution for selected clusters or merged to initial classes. Initial classes and nomenclature were assigned on the basis of markers in previous work6.
Cross-species integration of developmental data
Individual species were randomly downsampled to include a maximum of 1,000 cells per initial class from original clustering to account for rare populations and unequal sampling across species. Mouse and rat gene names were converted to human orthologues. Subsetted data were reset to raw and datasets were combined. Genes expressed in all species and CRABP1 (a total of 10,709 genes) were retained for downstream analyses. Counts were normalized, log-transformed and scaled. PCA was performed and Harmony41 was used to integrate the data across species on the basis of batch and species. Leiden clustering was applied with resolution set to 1. Individual Leiden clusters were subdivided by increasing resolution for individual clusters or merged to initial classes. Initial classes and nomenclature were assigned on the basis of markers in previous work6. The Pearson correlation coefficient of integrated clusters and original clusters across species was calculated based on the basis of the average PCA of the initial class in each species. The Jaccard index was calculated to determine the similarity between the original and the integrated class labels for each cell in the integrated dataset.
Differential expression of cortical and striatal TAC3 populations
Differential expression analysis was performed according to the decoupleR tutorial42. Pseudobulking was first performed using decoupleR based on batch and class. Differential expression of MGE_CRABP1 initial classes was performed using the pyDEseq2 package43. Contrast was set between cortical and striatal regions.
Pseudotime calculation
For pseudotime and trajectory inference, we converted anndata objects to R object with Zellkonverter and used the R package Slingshot44. Scanpy-based UMAP dimensional reduction and interneuron classes were used as input for all datasets. Lineage reconstruction was performed with the slingshot() function, with the ‘Progenitor’ class being designated as the beginning of pseudotime with start.clus = “Progenitor”. All initial classes were set as end points with the end.clus parameter. Averaged pseudotime values per cell were calculated with slingAvgPseudotime(). Differential expression analysis across pseudotime between species was calculated using TradeSeq45.
Analysis of published dataset of developing macaques
Macaque data were taken from a previous study46. We first isolated the LHX6_CRABP1 population, normalized, log-transformed, scaled, batch-corrected and performed Leiden clustering. Clusters were labelled on the basis of gene expression of known markers. Owing to the presence of MGE_CRABP1 populations in the A1C and insula dissections, we next isolated cells from A1C and insula dissections. We again normalized, log-transformed, scaled, batch-corrected and performed Leiden clustering. We examined marker-gene expression and renamed dissections A1C and insula to include striatum owing to the presence of medium spiny neurons in these dissections.
Analysis of published human dataset
Human data were taken from a previous study47. We normalized, log-transformed and scaled raw counts to examine gene expression in pre-labelled clusters.
Individual analyses of published datasets of adult marmosets, humans and mice
Adult mouse striatal interneurons were curated from the previously published ABC atlas27 class ‘08 CNU-MGE GABA’ (n = 18,849 cells), which we subsetted further to include only striatal cells (anatomical_division_label = “STR”), comprising 4,785 cells derived from a single ABC dataset (WMB-10Xv3-STR). This dataset initially contained cells from ABC subclasses ‘054 STR Prox1 Lhx6 Gaba’, ‘055 STR Lhx8 Gaba’, ‘056 Sst Chodl Gaba’, ‘057 NDB-SI-MA-STRv Lhx8 Gaba’ and ‘058 PAL-STR Gaba-Chol’, but after further examination, we found that most of the cells from ABC subclass 057 originated from outside the striatum. Excluding subclass 057 resulted in a final dataset of 3,586 cells.
Adult human striatal interneurons were curated from the neuron dataset of a previously published28 atlas by first selecting for the ROI terms ‘Basal Nuclei (BN) – Body of the Caudate – CaB’, ‘Basal Nuclei (BN) – Putamen – Pu’ and ‘Basal Nuclei (BN) – Nucleus Accumbens – NAC’, totalling 88,044 cells. These cells were further subsetted to include only cells from the supercluster terms ‘CGE interneuron’ (n = 711), ‘MGE interneuron’ (n = 238) or ‘Splatter’ (n = 6,716). The remaining 7,665 cells were re-embedded (UMAP from 30 PCs) using 2,000 variable features (Seurat ‘vst’) and reclustered using Louvain (at arbitrary resolution 0.8) into 22 clusters. These clusters were used to further exclude putative medium spiny neurons on the basis of MEIS2 expression, producing a final dataset of 6,617 striatal interneurons.
Adult marmoset striatal interneurons were curated from the previously published24 Marmoset Census dataset of 6,249 striatal GABAergic neurons (‘striatum.GAD’). Again, we excluded MEIS2-expressing cells (in this case using marmoset census cluster 02). Using the same method as for the adult human data, the remaining 3,930 cells were re-embedded and clustered into 22 clusters, from which we identified 6 small clusters (totalling 551 cells) that appeared to be contamination from excitatory (SLC17A6- and SLC17A-expressing) neurons (possibly from doublets or rare co-transmitting neurons); we excluded these, leaving 3,379 cells in the final dataset.
After cell selection in each species, the data were reprocessed using a typical Seurat (v.5.0.3) pipeline: NormalizeData (natural log of [1 + reads per 10,000 cells]), FindVariableFeatures (2,000 features using Seurat ‘vst’), ScaleData/PCA, FindNeighbors (using 30 PCs), FindClusters (Louvain resolution 0.8) and RunUMAP (using 30 PCs). In all cases, we found our re-embeddings were consistent or coherent with the previously published cell groupings and labels.
Integration analysis of published adult datasets
For cross-species integrations, we used scVI (ref. 48; scvi-tools v1.2.0). We used the same set of striatal inhibitory interneurons from ref. 24 (marmoset) and ref. 28 (human) as selected above. However, after a preliminary version of the analyses described below, we dropped a mixed cluster of 98 marmoset cells and a cluster of 575 putatively hypothalamic or mixed human cells, because these never showed cross-species homologies and are unlikely to be striatal, leaving a final set of 3,281 marmoset and 6,042 human cells. We used a similar set of cells from ref. 27 (mouse) to that detailed above, but for the sake of comparative analysis, we also expanded the set of mouse cells to include any cell found in the ABC class ‘08 CNU-MGE GABA’ subclass ‘055 STR Lhx8 Gaba’, which included cells from hypothalamus, pallidum and striatum-like amygdala nuclei (sAMY) dissections. We also included in the starting set any GABAergic or cholinergic cells that were derived from striatal dissections, which included cells that the ABC atlas had annotated as being non-striatal, although we excluded cells from class ‘09 CNU-LGE GABA’ and ‘05 OB-IMN GABA’. These additional inclusions produced a set of 3,770 mouse cells (or 184 more cells than the original mouse set). However, to allow proper modelling of donor effects in our scVI model, we also removed any cells from donors that contributed fewer than 30 cells, which left 3,553 cells (and removed nearly all cells from sAMY dissections).
During revision of this manuscript, a new large and high-quality dataset of marmoset cells became publicly available as an unannotated pre-release of the HMBA. This early pre-release contained 282,806 high-quality curated cells, which we included in our initial set as well. To find homologous cells in this dataset specifically, we performed an initial scVI integration with all of the starting sets. For all scVI integrations, we used scanpy (v.1.10.3) to find highly variable genes (flavor = “seurat_v3”, n_top_genes = 4,000, batch_key = “experiment”, subset = True). We used an scVI model to integrate across ‘experiment’ (so ignoring species commonality across the two marmoset datasets) with a covariate for the individual donor marmosets to model donor effects, and we trained with n_hidden = 256, n_latent = 64, n_layers = 3 and dispersion = “gene_batch” for 250 epochs. To annotate HMBA cells that did not integrate with the homologous types, we found k = 10 nearest neighbours on the scVI latent space, and clustered using scanpy leiden (flavor = “igraph”, resolution = 1, n_iterations = 2), and we calculated the ‘experiment’ entropy for each cluster (scipy.stats.entropy; scipy v.1.11.2). After manually examining the consequences of using different cut-offs, we removed any HMBA cells that were present in clusters with entropy < 0.1 (which were clusters that generally failed to incorporate non-HMBA cells). This pre-filtering removed 267,945 cells (94.7% of the total), leaving 27,737 cells.
After pre-filtering HMBA cells (‘first pruning’), we found new variable genes, redid scVI integration and redid clustering with the same parameters. We used these clusters to identify, select and remove MEIS2-expressing cells from the expanded mouse data and HMBA data, removing a further 3,296 (mostly HMBA) cells from the data. After this ‘second pruning’, we repeated variable gene selection, integration and Leiden clustering with the same parameters. These Leiden clusters for this final integration were used to annotate the HMBA data on the basis of the co-clustering cells from the other datasets, and the UMAP from this integration is used in the integration figures.
During editing of this manuscript, an expanded and newly annotated version of the marmoset HMBA data became publicly available, with annotations from a draft cross-species Basal Ganglia taxonomy (https://alleninstitute.github.io/HMBA_BasalGanglia_Consensus_Taxonomy). We verified that the striatal cells were included in the initial release that we used in our analyses, and we found a reassuring correspondence between our clustering/annotations and the taxonomy at the ‘Group’ level (the finest annotation present). We added those labels in Extended Data Fig. 10b. We applied the taxonomy Group labels to our initial dataset to check the extent to which our integrated cluster entropy-based pre-filtering had selected cells that the taxonomy annotated as striatal inhibitory interneurons, finding that non-targeted groups were efficiently removed and that the fraction kept was between 98.5% and 100% for all desired groups except for STR SST-ADARB2 GABA and STR SST-RSPO2 GABA. We also attempted an integration that included those excluded groups, but those groups still failed to integrate with cells from the other datasets.
Pearson correlation of gene expression in published adult datasets
For correlation analysis, we used the same curated cell sets that were used for the individual dataset analysis and annotation, but we dropped several clusters for clarity. As in the integration analysis, marmoset ‘Mixed’ and human ‘Hypothalamus’ and ‘Mixed’ clusters were dropped, and mouse Th-type cells (almost entirely ABC subclass 055) were split into their ABC cluster identities. The small number of cells from subclass ‘054 STR Prox1 Lhx6 Gaba’ that we annotated as Th were dropped for clarity. Pearson correlations were calculated on the metacells (cell-averaged normalized expression by species and cluster) using all 1:1 homologues, and so correlations are among log-transformed data.
SAMap analysis of published adult datasets
For SAMap, we followed the authors’ tutorial with only minor variations. We used protein-coding transcript (CDS) sequences matching the original reference assemblies (GENCODE 35 for human, GENCODE M21 for mouse), although we used only the more complete marmoset reference that was used for the HMBA data (RefSeq GCF_011100555.1, mCalJa1.2.pat.X) for either marmoset dataset. For all six pairwise directional comparisons between the three species, we ran tblastx (NCBI BLAST v.2.9.0). For each gene pair in the tblastx results, we chose the transcript pair with the highest mapping score, and we used that for the gene pair mapping score (the ‘maps’ for SAMap). We ran SAMap (v.1.0.15; sam-algorithm v.1.0.2), specifying our own clusters and using pairwise = True, and we calculated scores using samap.analysis.get_mapping_scores with n_top = 0 (to use all cells in each cluster). We used the same datasets and clusterings as were used for the Pearson correlation (with the same additional clusters being dropped). For the version with marmoset HMBA data, we used the data subset and annotations we generated after scVI integration. For aesthetic reasons, we set the self-self SAMap scores to 1 (which are 0 by default).
Gene-module discovery and projection using Hotspot
Hotspot (hotspotsc v1.1.1; https://github.com/YosefLab/Hotspot) was used for unsupervised partitioning of genes into modules following the authors’ recommendations29. For each dataset, only genes present with non-zero counts were used, and Hotspot was run using the model ‘danb’ on the PCA embedding. Genes were selected from Hotspot autocorrelations with a false discovery rate less than 0.05 and partitioned using agglomerative clustering, as previously shown by the authors using the default ‘min_gene_threshold = 30’.
To address computational constraints on hotspot module discovery in the larger developing macaque (109,111 cells) and developing mouse (71,023 cells) datasets, we subsetted 50% of the cells. To make the subsets representative, we sampled within high-resolution clusters: for macaque, we used the 124 ‘hires_leiden’ clusters already present in the data; for mouse, we reclustered (Leiden at resolution = 10) to obtain 157 distinct clusters used only for this purpose. All other analyses (including module scores/projections) use the full datasets.
Gene-module scores were calculated using PC1 following PCA using only the genes within the module, min–max scaled from 0 to 1. For cross-species module projections, we found the unsupervised gene module most correlated (Spearman) with a given marker gene of interest. We then found the gene orthologues in other species (subsetting genes if necessary) and calculated module scores.
Immunohistochemistry tissue processing
Samples were fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4 °C with constant agitation. The PFA was then replaced with fresh PBS (pH 7.4) and samples were cryopreserved by incubation for 24 h in 10% sucrose diluted in PBS (pH 7.4), followed by a 24-h incubation in 20% sucrose, and finally a 24-h incubation in 30% sucrose before being embedded in OCT (Tissue-Tek, VWR). Tissue was then frozen at –80 °C, cryosectioned at 20 μm and stored at −80 °C until use. Slides from P22 ferret were a gift from A. Kriegstein. Sections from P2 naked mole rat were a gift from S. Villeda.
Adult Nkx2-1-Cre;Ai14 mice were transcardially perfused with PBS followed by 4% PFA in PBS; their brains were dissected out and post-fixed overnight at 4 °C in 4% PFA in PBS with constant agitation. Fifty-micrometre sections were obtained on a vibratome, and preserved in freezing buffer (30% ethylene glycol, 28% sucrose in 0.1 M sodium phosphate buffer) at −20 °C until use.
Rhesus macaque sections from previous studies were provided by the Primate Center at the University of California, Davis and prepared for histology as stated above. All animal procedures conformed to the requirements of the Animal Welfare Act, and protocols were approved before implementation by the IACUC at the University of California, Davis.
RNAscope was performed following the manufacturer’s instructions for the Advanced Cell Diagnostics RNAscope Multiplex Fluorescent Reagent Kit V2 Assay (ACD, 323120). For immunostaining of the Nkx2-1-Cre;Ai14 line, rabbit anti-RFP (Rockland 600-401-379) was diluted 1:1,000 in blocking buffer (PBS + 5% BSA + 0.3% Triton X-100) and incubated for one hour at 37 °C. Alexa-dye-conjugated goat secondary 594 antibody was diluted 1:500 in blocking buffer and incubated for 30 min at 37 °C.
After RNA in situ hybridization, TrueBlack lipofuscin autofluorescence quencher (Biotium, 23007) was applied to adult ferret, adult pig and seven-month-old macaque samples according to the manufacturer’s directions. Slides were mounted using Prolong Gold Antifade plus DAPI (Invitrogen, P36931).
Imaging
Entire tissue sections were imaged single plane in widefield mode on a Leica DMI8 inverted microscope (Leica Microsystems) connected to a Flash4 V3 camera (Hamamatsu), with a 5× (0.12 NA), 10× (0.4 NA) or 20× (0.8) Plan Apochromat objective. Stitching was performed automatically in LAS X. Regions of interest were imaged at higher resolution using a laser scanning confocal microscope (Stellaris, Leica) using a 100× oil (1.4 NA) Plan Apochromat objective with a pixel size of 116.25 µm × 116.25 µm. Z-stacks of the entire volume of cells were acquired using optimal sections. Images from all samples were acquired under the same imaging settings with minimal adjustment of illumination intensity.
Image processing
The 100× images are maximum intensity projections produced from volumes acquired on the confocal microscope. Brightness and contrast were manually adjusted in Fiji for publication. Image processing of scans was performed using Imaris v.10.1.0 (Bitplane). Files were converted using the Imaris v.10 Converter and imported into Surpass mode for interaction. Images were saved as 3,500-dpi PNG files for publication.
Statistics and reproducibility
All RNAscope and immunofluorescence experiments were independently reproduced at least twice on non-consecutive sections. The images are representative of the results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE269902, and the data are browsable at https://dev-mammal-inhibneurons.cells.ucsc.edu. The developing macaque data used in this study are available through a previous study6, and are browsable at https://dev-inhibitory-neurons.cells.ucsc.edu/. The adult marmoset data used in this study were from the HMBA30 and are available at https://assets.nemoarchive.org/col-vdk0cmw. The adult mouse and human data used in this study are accessible from the ABC atlas (RRID:SCR_024440): https://portal.brain-map.org/atlases-and-data/bkp/abc-atlas.
Code availability
Scripts and annotation files for the study have been deposited at GitHub: https://github.com/mdeber/Corrigan2024.
Change history
11 December 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41586-025-09993-x
References
Katz, P. S. & Harris-Warrick, R. M. The evolution of neuronal circuits underlying species-specific behavior. Curr. Opin. Neurobiol. 9, 628–633 (1999).
Hahn, J. et al. Evolution of neuronal cell classes and types in the vertebrate retina. Nature 624, 415–424 (2023).
Striedter, G. F. Principles of Brain Evolution (Sinauer Associates, 2005).
Ma, S. et al. Molecular and cellular evolution of the primate dorsolateral prefrontal cortex. Science 377, eabo7257 (2022).
Krienen, F. M. et al. Innovations present in the primate interneuron repertoire. Nature 586, 262–269 (2020).
Schmitz, M. T. et al. The development and evolution of inhibitory neurons in primate cerebrum. Nature 603, 871–877 (2022).
DeFelipe, J. et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat. Rev. Neurosci. 14, 202–216 (2013).
Ramón y Cajal, S. Recuerdos de mi Vida: Historia de mi Labor Científica (Alianza Editorial, 1981).
Flames, N. et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 27, 9682–9695 (2007).
Lim, L., Mi, D., Llorca, A. & Marín, O. Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018).
Fishell, G. & Kepecs, A. Interneuron types as attractors and controllers. Annu. Rev. Neurosci. 43, 1–30 (2020).
Gokce, O. et al. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep. 16, 1126–1137 (2016).
Garma, L. D. et al. Interneuron diversity in the human dorsal striatum. Nat. Commun. 15, 6164 (2024).
Marin, O., Anderson, S. A. & Rubenstein, J. L. Origin and molecular specification of striatal interneurons. J. Neurosci. 20, 6063–6076 (2000).
Muñoz-Manchado, A. B. et al. Diversity of Interneurons in the dorsal striatum revealed by single-cell RNA sequencing and PatchSeq. Cell Rep. 24, 2179–2190.e7 (2018).
Kocaturk, S., Guven, E. B., Shah, F., Tepper, J. M. & Assous, M. Cholinergic control of striatal GABAergic microcircuits. Cell Rep. 41, 111531 (2022).
Shi, Y. et al. Mouse and human share conserved transcriptional programs for interneuron development. Science 374, eabj6641 (2021).
Charvet, C. J., de Sousa, A. A. & Vassilopoulos, T. Translating time: challenges, progress, and future directions. Brain Res. Bull. 221, 111212 (2025).
Suárez, R. et al. Development of body, head and brain features in the Australian fat-tailed dunnart (Sminthopsis crassicaudata; Marsupialia: Dasyuridae); a postnatal model of forebrain formation. PLoS ONE 12, e0184450 (2017).
Kim, J. et al. An expanded subventricular zone supports postnatal cortical interneuron migration in gyrencephalic brains. Nat. Neurosci. 28, 1598–1609 (2025).
Hansen, D. V. et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 16, 1576–1587 (2013).
Su-Feher, L. et al. Single cell enhancer activity distinguishes GABAergic and cholinergic lineages in embryonic mouse basal ganglia. Proc. Natl Acad. Sci. USA 119, e2108760119 (2022).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Krienen, F. M. et al. A marmoset brain cell census reveals regional specialization of cellular identities. Sci. Adv. 9, eadk3986 (2023).
Tepper, J. M. et al. Heterogeneity and diversity of striatal GABAergic interneurons: update 2018. Front. Neuroanat. 12, 91 (2018).
Xenias, H. S., Ibáñez-Sandoval, O., Koós, T. & Tepper, J. M. Are striatal tyrosine hydroxylase interneurons dopaminergic? J. Neurosci. 35, 6584–6599 (2015).
Yao, Z. et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332 (2023).
Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
DeTomaso, D. & Yosef, N. Hotspot identifies informative gene modules across modalities of single-cell genomics. Cell Syst. 12, 446–456 (2021).
Caceres, C. et al. Human and mammalian brain atlas: multiome analysis of single nuclei from marmoset brain. NeMO https://assets.nemoarchive.org/col-vdk0cmw (2024).
Tarashansky, A. J. et al. Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 10, e66747 (2021).
Peng, Y.-R. et al. Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237 (2019).
Niepoth, N. et al. Evolution of a novel adrenal cell type that promotes parental care. Nature 629, 1082–1090 (2024).
Kamath, T. et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat. Neurosci. 25, 588–595 (2022).
Feigin, C. Y. et al. Convergent deployment of ancestral functions during the evolution of mammalian flight membranes. Sci. Adv. 9, eade7511 (2023).
Moreno, J. A. et al. Emx2 underlies the development and evolution of marsupial gliding membranes. Nature 629, 127–135 (2024).
Pool, A.-H., Poldsam, H., Chen, S., Thomson, M. & Oka, Y. Recovery of missing single-cell RNA-sequencing data with optimized transcriptomic references. Nat. Methods 20, 1506–1515 (2023).
Fleming, S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods 20, 1323–1335 (2023).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Badia-I-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2, vbac016 (2022).
Muzellec, B., Teleńczuk, M., Cabeli, V. & Andreux, M. PyDESeq2: a python package for bulk RNA-seq differential expression analysis. Bioinformatics 39, btad547 (2023).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Van den Berge, K. et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11, 1201 (2020).
Micali, N. et al. Molecular programs of regional specification and neural stem cell fate progression in macaque telencephalon. Science 382, eadf3786 (2023).
Velmeshev, D. et al. Single-cell analysis of prenatal and postnatal human cortical development. Science 382, eadf0834 (2023).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Acknowledgements
We thank A. Kriegstein, D. Glawe, M. Starkenburg, E. Maga, P. Ji, S. Sopocy, J. Shea, S. Villeda, C. Sehnert, A. Taranatal, J. Isbell, A. Fox and the Ferret National Resource and Research Center (NHLBI 75N92025C00007) for samples for genomic and histological analysis; and M. Mui, C. Mrejen and E. Gaylord for assistance with sample preparation, imaging and image analysis, respectively. Imaging was performed at the Innovation Core at the Weill Institute for Neurosciences. This material is based on work supported by the National Science Foundation under grant number 2034836 (E.K.C.). This study was supported by the following funding sources: National Institutes of Health R21OD022988-03 (K.E.S.); National Institute of Neurological Disorders and Stroke R01NS123516 (J.D.B.); National Institutes of Health UM1MH130981, DP2MH140136 and the Klingenstein–Simons Fellowship (F.M.K.); and National Institutes of Health DP2MH122400, U01MH114825, UM1MH130991, the W.M. Keck Foundation and the Schmidt Futures Foundation (A.A.P.). A.A.P. is a New York Stem Cell Foundation Robertson Investigator.
Author information
Authors and Affiliations
Contributions
E.K.C., M.F.P., F.M.K. and A.A.P. designed the methodology. E.K.C., A.P., M.T.G., M.S., H.S., S.H., M. Duhne and S.L. performed the experiments. E.K.C. performed microscopic analysis. E.K.C., A.P., M.T.G. and S.H. performed the scRNA-seq processing. E.K.C performed scRNA-seq analysis of developmental datasets. S. de la O performed pseudotime analysis. M. DeBerardine performed scRNA-seq analysis of published adult datasets. E.K.C. and A.A.P. wrote the manuscript with input from all authors. E.K.C., C.C.H., M.F.P., J.D.B., K.E.S., F.M.K. and A.A.P. acquired the funding. E.K.C., M.F.P., F.M.K. and A.A.P. conceptualized the study. M.T.S., R.M., J.D.B., K.E.S., C.C.H., M.F.P., F.M.K. and A.A.P. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Integrated clustering of inhibitory neurons across placental mammal telencephalon development.
a. Taxonomy of surveyed species. b. Heat map of selected marker expression of integrated inhibitory initial class classes, scaled and normalized. c. UMAP coloured by the original classification of cells for each species from Extended Data Figs. 2−7 and key for f,g, highlighting general correspondence of individual species class labels. d. UMAP coloured by species (macaque: n = 14,971 inhibitory neurons, rabbit: n = 8,396 inhibitory neurons, rat: n = 10,052 inhibitory neurons, mouse: n = 8,492 inhibitory neurons, pig: n = 7,912 inhibitory neurons, ferret: n = 9,530 inhibitory neurons, opossum: n = 7,757 inhibitory neurons, sugar glider: n = 7,890 inhibitory neurons) and key for e-g. e. Bar plot of proportion of downsampled cells contributing to integrated initial class cluster. f. Jaccard index of original class label and integrated class label by species. Most distinctions between subclasses are clear from original single species clustering, while the integrated clustering is required to partition some subclasses in select species. g. Pearson correlation of initial classes calculated using original class labels. h. Hierarchical clustering of initial classes and species. i. Slingshot pseudotime calculation. j. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot.
Extended Data Fig. 2 Survey of inhibitory neuron initial classes in the developing macaque, rat and rabbit brain.
a. UMAP coloured by inhibitory initial classes in developing macaque scRNA-seq data from Schmitz et al.6. (n = 109,111 inhibitory neurons). b. Heat map of selected markers of inhibitory initial class markers, scaled and normalized. c. UMAP coloured by markers of MGE_CRABP1 initial classes, scaled and normalized. d. Slingshot pseudotime calculation. e. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot. f. UMAP coloured by inhibitory neuron initial classes in the E18 developing rat brain (n = 35,923 inhibitory neurons). g. UMAP coloured by region of dissection. h. Heat map of selected markers of inhibitory initial class markers, scaled and normalized. Bar plot indicates proportion of cells from cortical and striatal dissection in each initial class. i. UMAP coloured by markers of MGE_CRABP1 initial classes, scaled and normalized. j. Slingshot pseudotime calculation. k. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot. l. UMAP coloured by inhibitory neuron initial classes in the E23 developing rabbit brain (n = 36,580 inhibitory neurons. m. UMAP coloured by region of dissection. n. Heat map of selected markers of inhibitory initial class markers, scaled and normalized. Bar plot indicates proportion of cells from cortical and striatal dissection in each initial class. o. UMAP coloured by markers of MGE_CRABP1 initial classes, scaled and normalized. p. Slingshot pseudotime calculation. q. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot.
Extended Data Fig. 3 Survey of inhibitory neuron initial classes in the developing mouse and investigation of the TAC3 initial class in Glire brains.
a. UMAP coloured by developmental timepoint. b. Slingshot pseudotime calculation of developing mouse inhibitory neurons. c. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot. Owing to the enrichment of MGE precursors through use of a Nkx2-1-Cre;Ai14 reporter line, our mouse inhibitory neuron dataset contains more immature cell types compared to other samples and required more granular clustering for pseudotime and lineage analysis. d–o. Line graphs of gene expression within integrated MGE_CRABP1/TAC3 class across pseudotime for all developmental species. p. Image acquired at 20X of E17 Nkx2-1-Cre;Ai14 mouse brain, showing tdTomato reporter expression and Crabp1, Chrna3 RNAscope with inset indicating the highlighted region in the striatum (q). q. High-magnification (100X) image with arrowhead indicating the MGE_CRABP1/TH initial class. r. Image acquired at 20X of adult Nkx2-1-Cre;Ai14 mouse brain, showing tdTomato reporter expression and Crabp1, Chrna3 RNAscope with inset indicating highlighted region in the striatum (s). s. High-magnification (100X) image with arrowhead indicating MGE_CRABP1/TH initial class. t. Image acquired at 20X of adult Nkx2-1-Cre;Ai14 mouse brain, showing tdTomato reporter expression and Chrna3, Th RNAscope with inset indicating highlighted region in the striatum (u). u. High-magnification (100X) image with arrowhead indicating Th interneuron. v. High-magnification (100X) image of rabbit RNAscope with arrowhead indicating MGE_CRABP1/TAC3 class. w. Image acquired at 10X of adult rat RNAscope with markers Lhx6, Crabp1, and Tac3 and inset indicating highlighted region in the striatum (x). x. High-magnification (100X) image with arrowhead indicating no Tac3 expression in Lhx6/Crabp1-expressing cells. y. Image of naked mole rat striatum acquired at 40x with white arrowheads indicating no Tac3 expression in Lhx6/Crabp1-expressing cells and blue arrowhead indicating Tac3-expressing cell.
Extended Data Fig. 4 Survey of inhibitory neuron initial classes in the developing pig and ferret brain.
a. UMAP coloured by inhibitory neuron initial classes in the E73 and E83 developing pig brain (n = 80,278 inhibitory neurons). b. UMAP coloured by batch. c. UMAP coloured by region of dissection. d. Heat map of selected markers of inhibitory initial class markers, normalized. Bar plot indicates proportion of cells from cortical and striatal dissection in each initial class. e. UMAP coloured by markers of MGE_CRABP1 initial classes, scaled and normalized. f,g. Differential expression analysis using pyDEseq2 two-sided Wald test, with adjustments for multiple comparisons using Benjamini-Hochberg method for (f) MGE_CRABP1/TAC3 initial class from cortical and striatal dissections (striatum: n = 1,430 cells, cortex: n = 246 cells), (g) MGE_CRABP1/MAF initial class from cortical and striatal dissections (striatum: n = 1,965 cells, cortex: n = 518 cells). h. Heat map of MGE_CRABP1/TAC3 and MGE_CRABP1/MAF initial classes by region of dissection, gene expression is normalized and scaled. i. Slingshot pseudotime calculation. j. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot. k. UMAP coloured by inhibitory neuron initial classes in the P1, P5, and P14 developing ferret brain (n = 80,278 inhibitory neurons). l. UMAP coloured by batch. m. UMAP coloured by region of dissection. n. Heat map of selected markers of inhibitory initial class markers, scaled and normalized. Bar plot indicates proportion of cells from cortical and striatal dissection in each initial class. o. UMAP coloured by markers of MGE_CRABP1 initial classes, scaled and normalized. p. Matrix plot of selected MGE_CRABP1 markers and initial class annotation of ferret within integrated dataset. q. Dot plot of MGE_CRABP1/TAC3 initial class gene expression, by region of dissection (striatum: n = 724, cortex: n = 58). r. Slingshot pseudotime calculation. s. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot.
Extended Data Fig. 5 Survey of inhibitory neuron initial classes in the developing sugar glider and opossum brain.
a. UMAP coloured by inhibitory neuron initial classes in the P40 developing sugar glider brain (n = 52,228 inhibitory neurons). b. UMAP coloured by region of dissection. c. UMAP coloured by markers of MGE_CRABP1 initial classes, scaled and normalized. d. Heat map of selected markers of inhibitory initial class markers, scaled and normalized. Bar plot indicates proportion of cells from cortical and striatum dissection in each initial class. e. Slingshot pseudotime calculation. f. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot. g. UMAP coloured by inhibitory neuron initial classes in the P20 developing opossum brain (n = 74,024 inhibitory neurons). h. UMAP coloured by region of dissection. i. UMAP coloured by markers of MGE_CRABP1 initial classes, scaled and normalized. j. Heat map of selected markers of inhibitory initial class markers, scaled and normalized. Bar plot indicates proportion of cells from cortical and striatal dissection in each initial class. k. Slingshot pseudotime calculation. l. Reconstruction of lineage trajectories with circles visualizing cell-type connections calculated using Slingshot.
Extended Data Fig. 6 Spatial distribution of the MGE_CRABP1/TAC3 class in Laurasiatheria using RNAscope.
a. High-magnification (100X) image of adult porpoise striatum with arrows indicating MGE_CRABP1/TAC3 class. b. Image acquired at 20X of E35 pig with inset highlighting striatum (c). c. High-magnification (100X) image with arrows indicating MGE_CRABP1/TAC3 class. d. Image acquired at 10X of E50 pig with insets highlighting striatum (e) and cortex (f). e,f. High-magnification (100X) image with arrows indicating MGE_CRABP1/TAC3 class. g. Image acquired at 10X of E56 pig sagittal section and insets highlighting striatum (h) and migrating cells (i). h,i. High-magnification (100X) image with arrows indicating MGE_CRABP1/TAC3 class. j. Image of pig E62 coronal migration stream acquired at 20X with inset highlighting migrating cells (k). k. High-magnification (100X) image with arrows indicating MGE_CRABP1/TAC3 class. l. Image acquired at 10X of E73 pig coronal section with inset highlighting striatum (m). m. High-magnification (100X) image with arrows indicating MGE_CRABP1/TAC3 class. n. Image acquired at 10X of E73 pig sagittal section with inset highlighting cortex (o). o. High-magnification (100X) image, with arrows indicating MGE_CRABP1/TAC3 class. p. Image acquired at 10X of P14 ferret with insets highlighting striatum(q) and cortex (r). q,r. High-magnification (100X) image with arrows indicating MGE_CRABP1/TAC3 class. s. Image acquired at 5X of the adult pig section, with insets highlighting regions in the cortex (t), and striatum (u). t,u. High-magnification (100X) image with arrows indicating MGE_CRABP1/TAC3 class. v. Image acquired at 20x of adult ferret with insets indicating highlighted regions in the cortex (w) and striatum (x). w,x. High-magnification (100X) image with arrowheads indicating the MGE_CRABP1/TAC3 class.
Extended Data Fig. 7 Spatial distribution of the MGE_CRABP1/TAC3 class in primates.
a. Whole-section image acquired at 20X of macaque PCD65 RNAscope with markers LHX6, CRABP1, and TAC3 and inset indicating the highlighted region in the striatum (b). b. High-magnification (100X) maximum intensity projection with arrowheads indicating the MGE_CRABP1/TAC3 class. c. Whole-section image of macaque PCD80 RNAscope with markers LHX6, CRABP1, and TAC3, and inset indicating the highlighted region in the striatum (d). d. High-magnification (100X) maximum intensity projection with arrowheads indicating the MGE_CRABP1/TAC3 class. e. Whole-section image acquired at 10X of 7 months old macaque RNAscope with markers LHX6, CRABP1, and TAC3, with insets indicating the highlighted region in the striatum (f). f. High-magnification (100X) maximum intensity projection with arrowheads indicating the MGE_CRABP1/TAC3 class. g. UMAP coloured by MGE_CRABP1 initial class. h. UMAP coloured by age of sample. i. UMAP coloured by selected markers of MGE_CRABP1 classes. j. Bar plot representing the percentage of MGE_CRABP1/TAC3 population isolated from specified dissection regions, with A1C and Insula dissections renamed to include striatum owing to the presence of medium spiny neurons in these dissections. k. A1C and Insula samples isolated and clustered, coloured by region of dissection as originally stated. l. Gene expression of medium spiny neurons and MGE_CRABP1/TAC3 initial class in A1C and insula dissection regions, indicating contaminating striatum. m. UMAP coloured by inhibitory interneuron class. n. UMAP coloured by age of sample. o. UMAP coloured by markers of MGE_CRABP1/TAC3 class. Among presumptive MGE origin cells, TAC3 was observed in SST_RELN cells, however these cells exhibit low expression of CRABP1 and ETV1 and are presumed to be a derivative of MGE_LHX6/MAF initial class6.
Extended Data Fig. 8 Adult inhibitory neuron terminal classes.
a. UMAP coloured by striatal inhibitory neuron terminal classes in adult mouse. b. UMAP coloured by Pthlh/Pvalb and Th interneuron class markers in adult mouse, normalized. c. Heat map of selected markers of inhibitory neuron terminal classes in adult mouse, scaled and normalized. d. UMAP coloured by ABC atlas subclass annotation. e. UMAP coloured by ABC atlas clusters which comprise 055 subclass. f. Dot plot of selected Th/TAC3 class marker expression in ABC atlas clusters. g. High-magnification (100X) maximum intensity projection of adult Nkx2-1-Cre;Ai14 mouse brain tdTomato reporter expression with Crabp1 and Tac2 RNAscope and arrowheads indicating a rare Tac2 interneuron in the mouse. h. UMAP coloured by striatal inhibitory neuron terminal classes in adult marmoset. i. UMAP coloured by markers of inhibitory neuron terminal classes in adult marmoset, normalized. j. Heat map of selected markers of striatal inhibitory neuron terminal classes in adult marmoset, scaled and normalized. k. UMAP coloured by striatal inhibitory neuron terminal classes in adult human. l. UMAP coloured by markers of inhibitory neuron terminal classes in adult human, normalized. m. Heat map of selected markers of inhibitory neuron terminal classes in adult human, scaled and normalized.
Extended Data Fig. 9 Hotspot gene modules across adult datasets.
To further examine the relationship between the terminal classes across species, we projected gene co-expression modules that mark interneuron populations in one species onto other species using Hotspot29. Non-overlapping gene modules were discovered in each species, and the modules with the highest correlations to individual marker genes were selected for cross-species projection (Supplementary Table 1; see Methods). Marker genes are also plotted for reference. The species in which the module was discovered is always plotted on the left. a,b, Mouse Chat and Sst modules showed strong enrichment in the corresponding primate cell types. c–f, Human and marmoset CHAT and SST modules showed strong enrichment in the corresponding cell types across species. g,h, Mouse Tac2 and Th modules showed enrichment in primate Tac3 clusters, but with variable selectivity. i–l. Marmoset and human TAC3 modules (i,k) showed enrichment in primate TAC3 clusters and expression in mouse Th and Sst/Npy clusters, while Marmoset and human TH clusters (j,l) showed enrichment in mouse PTHLH/PVALB, supporting a distinction between primate and rodent TH populations.
Extended Data Fig. 10 Comparison of adult inhibitory neuron terminal classes.
a. Schematic demonstrating the cross-species scVI integration workflow, starting with how striatal inhibitory interneurons were identified and annotated from previously unlabelled marmoset HMBA data (see Methods). b. Integrated UMAP embeddings coloured by clusters for each primate species. New labels from HMBA Basal Ganglia taxonomy (from taxonomy’s “Group” level) are grey in far right panel. c. Dot plot showing the adult mouse cells from the ABC, now with the inclusion of CCK-expressing types which were detected in striatal dissections but annotated as cortical. This Cck type was predominantly annotated as class 06 CTX-CGE GABA subclass 049 Lamp5 Gaba, and Cck/Vip was predominantly class 06 subclass 046 Vip Gaba. d. Integrated UMAP embedding coloured by LHX8/Lhx8 expression in each species. Small numbers of LHX8-expressing TAC3 types from Krienen et al. and Siletti et al. datasets co-embed with the HMBA TAC3/LHX8 cluster. e. SAMap homology as in Fig. 4, but using marmoset HMBA data. f. Abundance of the TAC3 type in each species, shown as a cell count ratio to each of the other major inhibitory interneuron types.
Supplementary information
Supplementary Table 1
Hotspot gene modules in adult datasets across species. Adult mouse, human, and marmoset Hotspot gene modules for genes of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Corrigan, E.K., DeBerardine, M., Poddar, A. et al. Conservation and alteration of mammalian striatal interneurons. Nature 647, 187–193 (2025). https://doi.org/10.1038/s41586-025-09592-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09592-w