Abstract
Type 2 inflammation at barrier surfaces is an evolutionarily conserved response that promotes immunity to helminth parasites, allergic inflammation and tissue repair1,2,3,4. Direct sensing of environmental triggers by epithelial cells initiates type 2 inflammation, and signals derived from neurons can modulate immune responses5,6,7,8. However, how diverse sensory inputs from epithelial, neuronal and immune cells are coordinated and integrated remains unclear. Here we identify that TRPV1+ pain-sensing nociceptors co-opt chemosensory epithelial tuft cells to initiate a cascade of tissue responses that drive type 2 inflammation. Chemogenetic silencing or chemical ablation of TRPV1+ nociceptors results in a significant reduction in intestinal tuft cells and defective anti-helminth type 2 immunity. By contrast, chemogenetic activation of TRPV1+ nociceptors leads to remodelling of CGRP+ nerve fibres, significantly increased CGRP expression, enhanced tuft cell accumulation and protective anti-helminth type 2 immunity. Using spatial transcriptomic and single-cell RNA sequencing analyses, we reveal that nociceptor activation promotes rapid epithelial progenitor cell proliferation and differentiation. Mechanistically, intestinal epithelial cell-intrinsic and tuft cell-intrinsic expression of CGRP receptor subunits are required for tuft cell responses and type 2 immunity to helminth infection. Together, these results identify sensory convergence of a neuronal–epithelial tuft cell circuit as a critical upstream determinant of type 2 immunity and tissue adaptation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data necessary to understand and evaluate the conclusions of this paper are provided in this Article or the Supplementary Information. The bulk RNA-seq and the scRNA-seq data are available at the European Nucleotide Archive under accession number PRJEB101609. The spatial transcriptomic data are available at the BioImage Archive under accession number S-BIAD2351. Other data are available from the corresponding authors upon appropriate and reasonable request. Source data are provided with this paper.
Code availability
Code used for sequencing and spatial transcriptomic analyses will be made available upon request.
References
Gieseck, R. L. 3rd, Wilson, M. S. & Wynn, T. A. Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 18, 62–76 (2018).
Hammad, H., Debeuf, N., Aegerter, H., Brown, A. S. & Lambrecht, B. N. Emerging paradigms in type 2 immunity. Annu. Rev. Immunol. 40, 443–467 (2022).
Locksley, R. M. Asthma and allergic inflammation. Cell 140, 777–783 (2010).
Palm, N. W., Rosenstein, R. K. & Medzhitov, R. Allergic host defences. Nature 484, 465–472 (2012).
Hammad, H. & Lambrecht, B. N. Barrier epithelial cells and the control of type 2 immunity. Immunity 43, 29–40 (2015).
O’Leary, C. E., Schneider, C. & Locksley, R. M. Tuft cells—systemically dispersed sensory epithelia integrating immune and neural circuitry. Annu. Rev. Immunol. 37, 47–72 (2019).
Kotas, M. E., O’Leary, C. E. & Locksley, R. M. Tuft cells: context- and tissue-specific programming for a conserved cell lineage. Annu. Rev. Pathol. 18, 311–335 (2023).
Yano, H. & Artis, D. Neuronal regulation of innate lymphoid cell responses. Curr. Opin. Immunol. 76, 102205 (2022).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41.e7 (2018).
Luo, X. C. et al. Infection by the parasitic helminth Trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc. Natl Acad. Sci. USA 116, 5564–5569 (2019).
Howitt, M. R. et al. Tuft cells, taste–chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016).
McGinty, J. W. et al. Tuft-cell-derived leukotrienes drive rapid anti-helminth immunity in the small intestine but are dispensable for anti-protist immunity. Immunity 52, 528–541.e7 (2020).
Scholz, J. & Woolf, C. J. Can we conquer pain? Nat. Neurosci. 5, 1062–1067 (2002).
Basbaum, A. I., Bautista, D. M., Scherrer, G. & Julius, D. Cellular and molecular mechanisms of pain. Cell 139, 267–284 (2009).
Chu, C., Artis, D. & Chiu, I. M. Neuro-immune interactions in the tissues. Immunity 52, 464–474 (2020).
Ansaldo, E., Farley, T. K. & Belkaid, Y. Control of immunity by the microbiota. Annu. Rev. Immunol. 39, 449–479 (2021).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Blander, J. M., Longman, R. S., Iliev, I. D., Sonnenberg, G. F. & Artis, D. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 18, 851–860 (2017).
Veiga-Fernandes, H. & Mucida, D. Neuro-immune interactions at barrier surfaces. Cell 165, 801–811 (2016).
Li, W., Germain, R. N. & Gerner, M. Y. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl Acad. Sci. USA 114, E7321–E7330 (2017).
Klementowicz, J. E., Travis, M. A. & Grencis, R. K. Trichuris muris: a model of gastrointestinal parasite infection. Semin. Immunopathol. 34, 815–828 (2012).
Else, K. J., Finkelman, F. D., Maliszewski, C. R. & Grencis, R. K. Cytokine-mediated regulation of chronic intestinal helminth infection. J. Exp. Med. 179, 347–351 (1994).
Owyang, A. M. et al. Interleukin 25 regulates type 2 cytokine-dependent immunity and limits chronic inflammation in the gastrointestinal tract. J. Exp. Med. 203, 843–849 (2006).
Bancroft, A. J., McKenzie, A. N. & Grencis, R. K. A critical role for IL-13 in resistance to intestinal nematode infection. J. Immunol. 160, 3453–3461 (1998).
Cliffe, L. J. & Grencis, R. K. The Trichuris muris system: a paradigm of resistance and susceptibility to intestinal nematode infection. Adv. Parasitol. 57, 255–307 (2004).
Hockley, J. R. F. et al. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 68, 633–644 (2019).
Guo, T. et al. Optical recording reveals topological distribution of functionally classified colorectal afferent neurons in intact lumbosacral DRG. Physiol. Rep. 7, e14097 (2019).
Ferguson, S. M. et al. Transient neuronal inhibition reveals opposing roles of indirect and direct pathways in sensitization. Nat. Neurosci. 14, 22–24 (2011).
Urban, D. J. & Roth, B. L. DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu. Rev. Pharmacol. Toxicol. 55, 399–417 (2015).
Zhang, W. et al. Gut-innervating nociceptors regulate the intestinal microbiota to promote tissue protection. Cell 185, 4170–4189 (2022).
Zhu, H. et al. Cre-dependent DREADD (designer receptors exclusively activated by designer drugs) mice. Genesis 54, 439–446 (2016).
Elekes, K. et al. Role of capsaicin-sensitive afferents and sensory neuropeptides in endotoxin-induced airway inflammation and consequent bronchial hyperreactivity in the mouse. Regul. Pept. 141, 44–54 (2007).
Mishra, S. K. & Hoon, M. A. Ablation of TrpV1 neurons reveals their selective role in thermal pain sensation. Mol. Cell. Neurosci. 43, 157–163 (2010).
Gerbe, F. et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016).
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016).
Matsumoto, I., Ohmoto, M., Narukawa, M., Yoshihara, Y. & Abe, K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat. Neurosci. 14, 685–687 (2011).
Guo, C. J. et al. Kallikrein 7 promotes atopic dermatitis-associated itch independently of skin inflammation. J. Invest. Dermatol. 140, 1244–1252 (2020).
Bian, Z. et al. High-throughput functional characterization of visceral afferents by optical recordings from thoracolumbar and lumbosacral dorsal root ganglia. Front. Neurosci. 15, 657361 (2021).
Ualiyeva, S. et al. Tuft cell-produced cysteinyl leukotrienes and IL-25 synergistically initiate lung type 2 inflammation. Sci. Immunol. 6, eabj0474 (2021).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Oyesola, O. O. & Tait Wojno, E. D. Prostaglandin regulation of type 2 inflammation: from basic biology to therapeutic interventions. Eur. J. Immunol. 51, 2399–2416 (2021).
Stanbery, A. G., Shuchi, S., Jakob von, M., Tait Wojno, E. D. & Ziegler, S. F. TSLP, IL-33, and IL-25: not just for allergy and helminth infection. J. Allergy Clin. Immunol. 150, 1302–1313 (2022).
Salic, A. & Mitchison, T. J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA 105, 2415–2420 (2008).
Gore, R., Riedl, M. S., Kitto, K. F., Fairbanks, C. A. & Vulchanova, L. AAV-mediated gene delivery to the enteric nervous system by intracolonic injection. Methods Mol. Biol. 1950, 407–415 (2019).
Skorput, A. G. J. et al. Targeting the somatosensory system with AAV9 and AAV2retro viral vectors. PLoS ONE 17, e0264938 (2022).
Gerbe, F., Legraverend, C. & Jay, P. The intestinal epithelium tuft cells: specification and function. Cell. Mol. Life Sci. 69, 2907–2917 (2012).
Gerbe, F. & Jay, P. Intestinal tuft cells: epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol. 9, 1353–1359 (2016).
de Klerk, N., Saroj, S. D., Wassing, G. M., Maudsdotter, L. & Jonsson, A. B. The host cell transcription factor EGR1 Is induced by bacteria through the EGFR–ERK1/2 pathway. Front. Cell Infect. Microbiol. 7, 16 (2017).
Malagola, E. et al. Isthmus progenitor cells contribute to homeostatic cellular turnover and support regeneration following intestinal injury. Cell 187, 3056–3071 (2024).
Capdevila, C. et al. Time-resolved fate mapping identifies the intestinal upper crypt zone as an origin of Lgr5+ crypt base columnar cells. Cell 187, 3039–3055 (2024).
Doods, H. et al. Pharmacological profile of BIBN4096BS, the first selective small molecule CGRP antagonist. Br. J. Pharmacol. 129, 420–423 (2000).
Yang, D. et al. Nociceptor neurons direct goblet cells via a CGRP–RAMP1 axis to drive mucus production and gut barrier protection. Cell 185, 4190–4205 (2022).
Nagashima, H. et al. Neuropeptide CGRP limits group 2 innate lymphoid cell responses and constrains type 2 inflammation. Immunity 51, 682–695 (2019).
Wallrapp, A. et al. Calcitonin gene-related peptide negatively regulates alarmin-driven type 2 innate lymphoid cell responses. Immunity 51, 709–723 (2019).
Xu, H. et al. Transcriptional atlas of intestinal immune cells reveals that neuropeptide α-CGRP modulates group 2 innate lymphoid cell responses. Immunity 51, 696–708 (2019).
Tsou, A. M. et al. Neuropeptide regulation of non-redundant ILC2 responses at barrier surfaces. Nature 611, 787–793 (2022).
Saunders, C. J., Christensen, M., Finger, T. E. & Tizzano, M. Cholinergic neurotransmission links solitary chemosensory cells to nasal inflammation. Proc. Natl Acad. Sci. USA 111, 6075–6080 (2014).
Ndjim, M. et al. Tuft cell acetylcholine is released into the gut lumen to promote anti-helminth immunity. Immunity 57, 1260–1273 (2024).
Billipp, T. E. et al. Tuft cell-derived acetylcholine promotes epithelial chloride secretion and intestinal helminth clearance. Immunity 57, 1243–1259 (2024).
Krasteva, G. et al. Cholinergic chemosensory cells in the trachea regulate breathing. Proc. Natl Acad. Sci. USA 108, 9478–9483 (2011).
Bankova, L. G. et al. The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Sci. Immunol. 3, eaat9453 (2018).
Westphalen, C. B. et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest. 124, 1283–1295 (2014).
Workman, M. J. et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23, 49–59 (2017).
Artis, D. et al. The IL-27 receptor (WSX-1) is an inhibitor of innate and adaptive elements of type 2 immunity. J. Immunol. 173, 5626–5634 (2004).
Camberis, M., Le Gros, G. & Urban, J. Jr. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Immunol. 55, 19.12.1–19.12.27 (2003).
Tamari, M. et al. Sensory neurons promote immune homeostasis in the lung. Cell 187, 44–61 (2024).
Krashes, M. J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011).
Lin, X. et al. IL-17RA-signaling in Lgr5+ intestinal stem cells induces expression of transcription factor ATOH1 to promote secretory cell lineage commitment. Immunity 55, 237–253 (2022).
Yaniv, Z. & Lowekamp, B. SimpleITK Imaris extensions. Zenodo https://doi.org/10.5281/zenodo.7854019 (2023).
Reina-Campos, M. et al. Tissue-resident memory CD8 T cell diversity is spatiotemporally imprinted. Nature 639, 483–492 (2025).
Covert, I. et al. Predictive and robust gene selection for spatial transcriptomics. Nat. Commun. 14, 2091 (2023).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
Varrone, M., Tavernari, D., Santamaria-Martinez, A., Walsh, L. A. & Ciriello, G. CellCharter reveals spatial cell niches associated with tissue remodeling and cell plasticity. Nat. Genet. 56, 74–84 (2024).
Acknowledgements
We thank members of the Artis laboratory for their discussions and critical reading of the manuscript; A. Alonso and other members of the Epigenomics Core of Weill Cornell Medicine for performing scRNA-seq; B. He and L. Dizon at the Translational Research Core of Weill Cornell Medicine for their assistance and staff at the High Resolution Translational Immunology team at the Allen Institute for Immunology for running Xenium samples. This research was supported by the Crohn’s and Colitis Foundation (Research Fellowship Awards 901000 to W.Z., 937437 to H.Y., 935259 to M.L. and 1455492 to X.H.); the Allen Discovery Center program, a Paul G. Allen Frontiers Group advised program of the Allen Family Philanthropies (to D.A. and B.S.K.); the Kenneth Rainin Foundation (to D.A.); Cure for IBD (to D.A.); Weill Cornell Medicine Jill Roberts Institute (to D.A.); the Sanders Family (to D.A.); Linda and Glenn Greenberg (to D.A.); the Rosanne H. Silbermann Foundation (to D.A.); the Parker Institute for Cancer Immunotherapy at Weill Cornell Medicine (to D.A.); the Doris Duke Charitable Foundation (to B.S.K.); the National Institutes of Health NIAID Division of Intramural Research (to R.N.G.); and extramural grants: K99DK138295 and U01AI095608-15S1 (to W.Z.); K99AI180354 (to H.Y.); K99CA290052 (to M.L.); AR070116, AR077007, AR080392, AI167933 and AI167047 (all to B.S.K.); R01DK103901, R01AR077183 and R01DK134773 (all to H.H.); and DK126871, AI151599, AI095466, AI095608, AR070116, AI172027 and DK132244 (all to D.A).
Author information
Authors and Affiliations
Contributions
W.Z. and E.R.E. carried out most of the experiments and analysed the data. H.Y. helped with bone marrow transplantation and various experiments. S.G. carried out intestinal organoid experiments. J.U. and A.G. performed bioinformatic analysis for bulk and scRNA-seq experiments. Z.X. and H.H. carried out intra-intestinal AAV microinjection experiments. H.I. and R.N.G. carried out the 3D imaging experiments and analyses. Z.W. and B.S.K. helped with intrathecal injection experiments. N.E., J.R.I., S.A.-S., P.J.S. and M.H. carried out tissue quality control and Xenium experiments. M.N.C., M.H., A-M.G. and A.W.G. performed Xenium analyses. T.M. and R.J.X. helped with scRNA-seq experiments. M.L., X.H., P.Z., E.H. and V.R.d.G. helped with various experiments. D.A., W.Z. and E.R.E. conceived the project, designed experiments, analysed data and wrote the manuscript with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
D.A. has contributed to scientific advisory boards at Boehringer-Ingelheim, Pfizer, Takeda and the Kenneth Rainin Foundation. B.S.K. is a founder of Alys Pharmaceuticals; he has served as a consultant for 23andMe, ABRAX Japan, AbbVie, Attovia Therapeutics, Almirall, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Cara Therapeutics, Clexio Biosciences, Eli Lilly and Company, Escient Pharmaceuticals, Evommune, Galderma, Genentech, GlaxoSmithKline, LEO Pharma, Novartis, Pfizer, Recens Medical, Regeneron Pharmaceuticals, Sanofi, Septerna, Triveni Bio, and Vial; he has stock in ABRAX Japan, Alys Pharmaceuticals, Attovia Therapeutics, Locus Biosciences, Recens Medical, and Triveni Bio; and he holds a patent for the use of JAK1 inhibitors for chronic pruritus. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
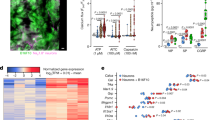
Extended Data Fig. 1 TRPV1+ nociceptors regulate anti-helminth type 2 immunity.
a-b, Calca and Calcb expression were assessed in sorted ILC2s (n = 5), Th2 cells (n = 3), purified DRGs (n = 6), and intestinal muscularis layers (n = 6) from wild-type B6 mice on day 17 post Trichuris infection. c, Schematic of nociceptor silencing during Trichuris infection. d, Schematic of RTX-mediated nociceptor ablation and Trichuris infection. e-f, Representative images (e) and tuft cell quantification (f) from mice treated with DMSO (n = 7) and RTX (n = 7). DAPI (blue), EpCAM (green), DCLK1 (red). Scale bar=50 µm. g-i, Representative flow cytometry plots (g), frequency (h) and total number (i) of Lineage– GATA3+ ILC2s in mLNs from mice treated with DMSO (n = 4) and RTX (n = 5). j, Frequency of GATA3+ CD4+ T cells in mLNs from mice in g-i. k-l, Representative histology and goblet cell quantification of the proximal colon by AB-PAS staining from mice in g-i. Scale bar=50 µm. m, Trichuris worm counts from mice treated with DMSO (n = 7) and RTX (n = 7). Data are representative of 2 independent experiments (h-j, l) or pooled from 2 (a, b, f, m) independent experiments. Data are mean ± s.e.m. Statistics were calculated by two-way ANOVA test (a, b) or unpaired two-tailed t-test (f, h-j, l, m). The illustrations in c–d were created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
Extended Data Fig. 2 Gating strategies for immune cells and epithelial cells in the mouse mesenteric lymph nodes and intestines.
Gating strategy for different immune cells with surface markers. Lineage markers included: CD3ε, CD5, TCRγδ, CD11b, CD11c, B220, CD19, F4/80, FcεRIα and TER-119. a, Eosinophils were gated as Live CD45+ SiglecF+. b, ILC2s were gated as Live CD45+ Lineage− CD127+ CD90.2+ KLRG1+ GATA3+. c, CD4+ T cells were gated as Live CD45+ CD3ε+ CD4+; and CD4+ Th2 cells were further gated as GATA3+ FoxP3−; CD4+ Th1 cells were further gated as T-bet+ FoxP3−. d, Epithelial cells were gated as Live CD45−/lo EpCAM+ cells. Among those cells, tuft cells were gated as CD24hi SiglecF+. For experiments using Lgr5eGFP mice, non-tuft epithelial cells (ECs) were gated as CD24–/lo SiglecF−. Among the non-tuft ECs, intestinal stem cells (ISCs) were gated as GFPhi, progenitor cells were gated as GFPlo, and other ECs were gated as GFP−.
Extended Data Fig. 3 Tuft cell-deficient mice exhibit defective type 2 immunity to Trichuris infection.
a, Schematic of Trichuris infection in Pou2f3−/− mice (n = 4-5) compared to Pou2f3+/− littermate controls (n = 4). b-d, Representative flow cytometry plots (b), frequency (c) and total number (d) of Lineage– GATA3+ ILC2s in the mLNs. e, Frequency of GATA3+ CD4+ T cells in the mLNs. f-g, Representative histology and goblet cell quantification of the proximal colon by AB-PAS staining. Scale bar=50 µm. h, Trichuris worm counts from the ceca of Pou2f3−/− (n = 9) versus Pou2f3+/− littermates (n = 9). Data are representative of 4 independent experiments (c-e, g) or pooled from 2 experiments (h). Data are mean ± s.e.m. Statistics were calculated by unpaired two-tailed t-test. The illustration in a was created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
Extended Data Fig. 4 Acute activation of nociceptors promotes anti-helminth type 2 inflammation.
a, Representative images of phospho-ERK+ T12/T13/L1 DRGs isolated from TRPV1hM3Dq (n = 4) or their littermate control mice (n = 4) treated with CNO. pERK (red), DAPI (blue). Scale bar=100 µm. b, Intestinal CGRP in TRPV1hM3Dq (n = 4) and littermate control (n = 4) mice in the steady state. c, Schematic of nociceptor activation during Nippostrongylus infection. d-e, Representative images (d) and tuft cell quantification (e) of duodenum from littermate controls (n = 10) and TRPV1hM3Dq mice (n = 15). DAPI (blue), E-Cadherin (green), DCLK1 (red). Scale bar=100 µm. f-g, Representative flow cytometry plots (f) and frequency (g) of Lineage– GATA3+ ILC2s in mLNs from littermate controls (n = 6) and TRPV1hM3Dq mice (n = 8). h, Frequency of CD45+ SiglecF+ eosinophils in mLNs from mice in f-g. i, Nippostrongylus worm counts from mice in f-g. Data are representative of 2 independent experiments (b) or pooled from 2 (g-i) or 3 (e) independent experiments. Data are mean ± s.e.m. Statistics were calculated by unpaired two-tailed t-test (b, e, g-i). The illustration in c was created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
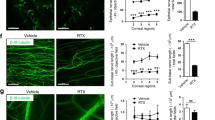
Extended Data Fig. 5 TRPV1+ nociceptors regulate intestinal tuft cell homeostasis.
a, Schematic of steady-state chemogenetic TRPV1+ nociceptor silencing. b-d, Representative images (b) and tuft cell quantification from the ileum (c) and proximal colon (d) from littermate controls (n = 9) and TRPV1hM4Di mice (n = 12). DAPI (blue), EpCAM (green), DCLK1 (red). Scale bar=100 µm. e, Schematic of RTX-mediated TRPV1+ nociceptor ablation. f-h, Representative images and tuft cell quantification from the ileum and proximal colon from DMSO-treated mice (n = 7) and RTX-treated mice (n = 7). DAPI (blue), EpCAM (green), DCLK1 (red). Scale bar=100 µm. i, Schematic of steady-state intrathecal injection of RTX to ablate DRG TRPV1+ nociceptors. j-l, Representative images and tuft cell quantification from the ileum and proximal colon from DMSO-treated mice (n = 16) and RTX-treated mice (n = 13). DAPI (blue), EpCAM (green), DCLK1 (red). Scale bar=100 µm. Data are pooled from 2 experiments. Data are mean ± s.e.m. Statistics were calculated by unpaired two-tailed t-test. The illustrations in a, e and i were created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
Extended Data Fig. 6 Nociceptor activation promotes intestinal tuft cell accumulation independent of ILC2s.
a, Schematic of bulk RNA sequencing of sorted intestinal tuft cells upon nociceptor activation. b, Schematic of IL-4Rα blockade upon steady-state chemogenetic nociceptor activation. c, Schematic of bone marrow (BM) chimera set up for NMUR1+ ILC2 depletion and chemogenetic nociceptor activation. d-e, Frequency of Lineage– GATA3+ ILC2s in small intestinal lamina propria (SILP, d) and large intestinal lamina propria (LILP, e) in ROSA26LSL.DTR (littermate control) BM to hM3Dq mice (n = 8 and 10), ROSA26LSL.DTR (littermate control) BM to TRPV1hM3Dq mice (n = 9 and 10), Nmur1iCre-eGFP.ROSA26LSL.DTR (ILC2DTR) BM to hM3Dq mice (n = 4), and Nmur1iCre-eGFP.ROSA26LSL.DTR (ILC2DTR) BM to TRPV1hM3Dq mice (n = 10). f, Representative images from the ileum and proximal colons from the indicated bone marrow chimera in ROSA26LSL.DTR (littermate control) BM to hM3Dq mice (n = 11), ROSA26LSL.DTR (littermate control) BM to TRPV1hM3Dq mice (n = 10), Nmur1iCre-eGFP.ROSA26LSL.DTR (ILC2DTR) BM to hM3Dq mice (n = 4), and Nmur1iCre-eGFP.ROSA26LSL.DTR (ILC2DTR) BM to TRPV1hM3Dq mice (n = 10). DAPI (blue), EpCAM (green), DCLK1 (red). Scale bar=100 µm. g-h, Tuft cell quantification of the ileum (g) or the proximal colon (h) from the indicated bone marrow chimera from mice in f. i, Trpv1-Cre mice were infected with AAV9 viruses carrying the empty vector (mCherry) (n = 8) versus excitatory DREADD (hM3D(Gq)-mCherry) (n = 8) via intra-intestinal injection, given CNO in drinking water for 10 days and analyzed for intestinal tuft cells by immunofluorescence microscopy. Data are pooled from 3 independent experiments. Data are mean ± s.e.m. Statistics were calculated by two-way ANOVA test. The illustrations in a–c and i were created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
Extended Data Fig. 7 Spatial transcriptomic analysis of IECs in response to acute nociceptor activation.
a, Schematic of chemogenetic nociceptor activation for Xenium analysis. b, Overview of the Xenium-based spatial transcriptomics data structure. c, Overview of the processed Xenium data of mouse ileum with UMAP and all cell clusters colored by cell class. d, Gene expression for Epcam and top 5 marker genes per IEC population, determined by Wilcoxon rank-sum test. e, Visualization of CellCharter zones. f, IEC composition of CellCharter zones. g, Spatial embedding of IECs along the crypt-villus spatial axis. h, Crypt-villus axis distribution in each spatial zone, with IQR ranges as vertical dashed lines. i-j, IEC gene expression (i) and ISC-related gene expression (j) along the crypt-villus axis with epithelial global statistical analysis. Statistical significance of gene expression changes was determined via using a %ΔAUC threshold of ± 10. k, Visualization of Ramp1 gene expression in Xenium dataset. Scale bar=100 µm. l, CGRP receptor gene expression (Ramp1/Calcrl) along the crypt-villus axis with epithelial global statistical analysis. Shown images are representative of 3 biological replicates in each group from 1 independent experiment in k. The illustration in a was created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
Extended Data Fig. 8 scRNA-sequencing analysis of IEC changes in response to acute nociceptor activation.
a, Schematic of scRNA-sequencing analysis of the ileal epithelium upon chemogenetic nociceptor activation. b, UMAP of scRNA-seq data from IECs from mice in a. c, Dot plots showing the mean ileal expression of the indicated genes in each cluster. The dot size represents the proportion of cells in a cluster with the gene detected. d, Visualization of Slc34a2, Ccl25, Stmn1 and Olfm4 gene expression in Xenium dataset. Scale bar=100 µm. e-f, UMAPs of Slc34a2, Ccl25, Stmn1, Olfm4 and Fgfbp1 gene expression from the scRNA-seq analysis. g-h, Volcano plot showing differentially expressed genes (P < 0.05) in indicated IEC clusters. i, GO pathway analysis of the indicated cluster in TRPV1hM3Dq group compared to their littermate controls. Shown images are representative of 3 biological replicates in each group from 1 independent experiment in d. Statistics were calculated by two-tailed likelihood-ratio test used on pseudobulked expression values (g-h) or by one-tailed Fisher’s exact test (i). Multiple test correction was performed using the Benjamini-Hochberg procedure (g-i). The illustration in a was created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
Extended Data Fig. 9 TRPV1+ nociceptors regulate intestinal tuft cells in a CGRP-dependent manner.
a, Schematic of BIBN4096 administration upon acute nociceptor activation. b, Schematic of CGRP administration in wild-type mice in the steady state. c, Intestinal CGRP levels in DMSO-treated mice (n = 8) versus RTX-treated mice (n = 6). d, Schematic of CGRP administration post RTX-mediated nociceptor ablation. e-f, Representative images and tuft cell quantification from the ileum in mice treated with DMSO and PBS (n = 9), RTX and PBS (n = 8), DMSO and CGRP (n = 9), or RTX and CGRP (n = 9). DAPI (blue), EpCAM (green), DCLK1 (red). Scale bar=50 µm. g, Schematic of CGRP administration in either wild-type mice, tuft cell-sufficient Pou2f3+/− mice, or tuft cell-deficient Pou2f3−/− mice during Trichuris infection. h, Schematic of in vitro small intestinal organoid culture system with IL-13. i-j, Gene expression analysis of Dclk1 and Pou2f3 in the intestinal organoid cultures treated with vehicle control (n = 10) versus CGRP (n = 10) in the presence of IL-13. k, Schematic of in vitro small intestinal organoid culture system with CGRP alone. l, Calcrl expression was assessed by qRT-PCR in sorted intestinal tuft cells (n = 4-5), Lgr5hi intestinal stem cells (ISCs) (n = 7), Lgr5low intestinal progenitor cells (Progenitors) (n = 7), and all the other epithelial cells (Other ECs) (n = 7) from Lgr5eGFP reporter mice. m, Ramp1 expression was assessed by qRT-PCR in sorted intestinal epithelial cells as shown in l. Data are pooled from 2 independent experiments (c, f, i, j) or 3 independent experiments (l, m). Data are mean ± s.e.m. Statistics were calculated by unpaired two-tailed t-test in c, two-way ANOVA test in f, l, or paired two-tailed t-test in i, j. The illustrations in a, b, d, g, h and k were created in BioRender. Emanuel, E. (2025) https://biorender.com/b1xymo3.
Extended Data Fig. 10 Convergence of neuronal-epithelial-immune sensing promotes type 2 inflammation.
Model: Upon activation, TRPV1+ nociceptors release sensory neuropeptide CGRP, leading to (1) epithelial progenitor differentiation into intestinal tuft cells as well as (2) tuft cell activation with upregulated Tuft-2 gene signatures and effector molecules such as cysteinyl leukotrienes, prostaglandins and alarmin cytokines, via the epithelial CGRP receptor complex CALCRL and RAMP1. These tuft cell effector molecules expand and activate (3) ILC2s and CD4+ Th2 cells, that release cytokines including IL-4, IL-5 and IL-13 that further drive type 2 inflammation and immunity, goblet cell mucus secretion, epithelial turnover and smooth muscle contractions (4). In the context of helminth infection, these processes help drive expulsion of gastrointestinal nematodes (5). This ilustration was created in BioRender.Emanuel, E. (2025) https://biorender.com/b1xymo3.
Supplementary information
Supplementary Information
Supplementary Data 1–5.
Supplementary Table 1
Ce3D imaging antibody list.
Supplementary Table 2
Xenium gene panel list.
Supplementary Video 1
Nociceptors promote intestinal tuft cell accumulation independent of helminth infection. Representative videos of multiplex 3D images of DCLK1+ tuft cells in the ileum from littermate controls (n = 4) and TRPV1hM3Dq mice (n = 4). βIII-tubulin (cyan), DCLK1 (yellow), nucleus (purple).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Emanuel, E.R., Yano, H. et al. Neuro-epithelial circuits promote sensory convergence and intestinal immunity. Nature (2026). https://doi.org/10.1038/s41586-025-09921-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41586-025-09921-z