Abstract
Fine-tuning DNA replication and transcription is crucial to prevent collisions between their machineries1. This is particularly important near promoters, where RNA polymerase II (RNAPII) initiates transcription and frequently arrests, forming R-loops2,3,4. Arrested RNAPII can obstruct DNA replication, which often initiates near promoters5,6. The mechanisms that rescue arrested RNAPII during elongation to avoid conflicts with co-directional replisomes remain unclear. Here, using genome-wide approaches and genetic screens, we identify CFAP20 as part of a protective pathway that salvages arrested RNAPII in promoter-proximal regions, diverting it from the path of co-directional replisomes. CFAP20-deficient cells accumulate R-loops near promoters, which leads to defects in replication timing and dynamics. These defects stem from accelerated replication-fork speeds that cause a secondary reduction in origin activity. Co-depletion of the Mediator complex or removal of R-loop-engaged RNAPII restores normal replication. Our findings suggest that transcription-dependent fork stalling in cis induces accelerated fork progression in trans, generating single-stranded DNA gaps. We propose that CFAP20 facilitates RNAPII elongation under high levels of Mediator-driven transcription, thereby preventing replisome collisions. This study provides a transcription-centred view of transcription–replication encounters, revealing how locally arrested transcription complexes propagate genome-wide replication phenotypes and defining CFAP20 as a key factor that safeguards genome stability.
Similar content being viewed by others
Main
The intricate dance between the replication and transcription processes, both of which operate on the same DNA template, must be tightly regulated to maintain genome integrity7. Collisions between these processes occur across nearly all species8,9 and can be either head-on (HO) or co-directional (CD), depending on which DNA strand is transcribed. HO collisions arise when the transcription machinery moves opposite to the replisome, with the transcribed strand serving as the lagging-strand template. In CD collisions, transcription and replication proceed in the same direction, with the transcribed strand acting as the leading-strand template1,10. Protein-coding genes are transcribed by RNAPII, which initiates at promoter sequences11. After promoter escape, RNAPII frequently undergoes transient promoter-proximal pausing12. Multi-protein complexes regulate its release into productive elongation: Integrator terminates and removes paused RNAPII at promoter-proximal sites6,13 whereas the Mediator complex, comprising a core body and a kinase module, coactivates RNAPII-dependent transcription14. After release, RNAPII at first elongates slowly near promoters, accelerating over the first approximately 10 kb of genes until reaching peak speed15. This early acceleration is stimulated by diverse elongation factors that act through distinct mechanisms16,17. Slow elongation or pausing promotes re-annealing of nascent transcripts to the template DNA, forming R-loops—three-stranded structures composed of an RNA–DNA hybrid and displaced single-stranded DNA2,3. RNAPII engaged with R-loops can obstruct replisomes, leading to genome instability10. Most studies of transcription–replication conflicts have focused on HO collisions18,19, which are generally considered more deleterious. However, most human genes are oriented co-directionally6, and active replication origins, particularly those that fire early in S-phase, frequently lie near promoters5. Given that RNAPII often pauses at promoter-proximal sites of highly transcribed genes, CD collisions in this region are likely. How replisomes navigate CD RNAPII during productive elongation remains unknown.
Genome-wide transcription–replication
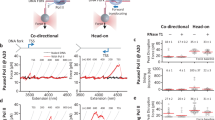
To investigate spatial connections between transcription, co-transcriptional R-loops and replication genome-wide, we mapped RNAPII occupancy by chromatin immunoprecipitation followed by sequencing (ChIP–seq); nascent transcription by bromouridine (BrU)–seq; and R-loops by DNA–RNA hybrid immunoprecipitation (DRIP)–seq in RPE1 cells (Fig. 1a). Replication origins were mapped using previously published Okazaki fragment (OK)–seq data from unperturbed RPE1 cells5 (Extended Data Fig. 1a), identifying 4,785 origins shared between two replicates. These aligned well with Origin-seq (Ori–seq) origins mapped in hydroxyurea (HU)-treated RPE1 cells20 and were enriched in transcriptionally active, early-to-mid-S-phase regions, as confirmed by single-cell 5-ethynyl-2′-deoxyuridine sequencing (scEdU–seq) in unperturbed cells21 (Extended Data Fig. 1b,c). For these origins, we calculated distances to the nearest transcription start site (TSS) and retained only those without another gene within 5 kb upstream, yielding 2,040 origins. RNAPII, BrU and DRIP profiles were overlaid with these coordinates and sorted by origin–TSS distance. Metaprofiles were generated by aligning all co-directionally (CO) oriented TSSs (n = 1,395) relative to origins and compared with HO TSSs (n = 408) (Fig. 1b and Extended Data Fig. 2a). As previously observed6, RNAPII binding and nascent transcription were higher at CD-oriented TSSs than at HO-oriented TSSs (Fig. 1c and Extended Data Fig. 2b,c). R-loop levels were modestly higher at CD-oriented TSSs, consistent with increased transcription (Fig. 1c). Extending this analysis, we plotted R-loop levels within 25-kb promoter windows adjacent to origins and up to 75 kb away in either orientation (Extended Data Fig. 2d). R-loop levels were markedly increased near origins, particularly in the CD orientation, suggesting that TSSs that are close to origins experience greater transcription stress (Fig. 1d). We propose that cells deploy mechanisms to mitigate transcription stress at these TSSs to minimize clashes with CD replisomes.
a, Heat maps of RNAPII ChIP–seq, BrU–seq and DRIP–seq in RPE1 cells, aligned around replication origins mapped by OK–seq5. b, Model showing RNAPII transcription on lagging-strand (HO) or leading-strand CD) templates relative to the replication fork. c, Metaprofiles of RNAPII ChIP–seq (green), BrU–seq (blue) and DRIP–seq (red) in RPE1 cells around TSSs oriented HO (n = 408) or CD (n = 1,395) relative to origins5. Data are averages after trimming the top and bottom 5% of data (a trim-mean of 0.1) to remove extreme values. d, Metaprofiles of DRIP–seq signals within a 25-kb window adjacent to origins extending up to 75 kb in HO and CD orientations (Trimmean 0.1). e, Schematic of CRISPR–Cas9 screens. NGS, next-generation sequencing. f, Correlation of normalized z-scores from CD437 and illudin S screens7; lowest and highest z-scores normalized to –1 and +1. g, Representative co-localization of GFP–CFAP20 with the primary cilium (arrowheads) marker acetylated α-tubulin. Scale bar, 20 μm. h, Representative image of immunofluorescent labelling of R-loops using S9.6 antibody. Scale bar, 10 μm. i, AlphaFold model of CFAP20, highlighting residue R100; positively charged residues are in blue. j, Quantification of nuclear R-loop signal from h for the indicated stable cell lines. Each coloured circle is one cell; black circles represent medians of independent experiments (more than 100 cells); black lines are means of all experiments; significance was calculated by one-way ANOVA with Šidák’s correction. P values from left to right: <0.0001, 0.9944, 0.0002, 0.9980, 0.0020 and 0.9980. NS, not significant. k, Schematic of sister fork symmetry principle. l, Representative sister fork symmetry observed by sequential CldU (red) and IdU (green) labelling. Scale bar, 5 μm. m, Quantification of sister fork symmetry from l. Data as in j (more than 100 fibres); significance by one-way ANOVA with Šidák’s correction. P values from left to right: <0.0001, 0.8851, <0.0001, >0.9999, <0.0001 and >0.9999.
CFAP20 in transcription–replication screens
To uncover mechanisms and factors that fine-tune the coexistence of transcription and replication, we performed two genome-wide CRISPR screens. Cells were transduced with 71,090 gRNAs targeting 18,053 protein-coding genes and left untreated, or exposed to illudin S to stall transcription17 or the DNA polymerase α inhibitor CD437 to stall replication22 (Fig. 1e). Genes at the intersection of these genetic screens encode proteins that respond to both transcription arrest and replication arrest. The genes with the highest scores in both screens encode three subunits of the 9-1-1 complex (RAD9–HUS1–RAD1), a known checkpoint complex that is strongly activated by transcription–replication encounters23. The fourth top hit at the intersection of these screens is the CFAP20 gene (Fig. 1f), which encodes a small (23 kDa) understudied protein that is currently known only as a ciliary protein24. In addition to its expected localization at the primary cilium of RPE1 cells, we observed that GFP–CFAP20 localized to the cell nucleus (Fig. 1g). This prompted us to investigate its nuclear function in more detail.
CFAP20 prevents R-loop accumulation
Although CFAP20 was previously suggested to be an essential gene25, we were able to generate a CFAP20 full knockout (KO) cell line in RPE1 TP53-KO cells (Extended Data Fig. 3a). In agreement with our CRISPR screens, clonogenic survival assays confirmed that CFAP20-KO cells are sensitive to illudin S and to CD437 (Extended Data Fig. 3b,c). Although our previous work revealed that many illudin S sensitizer genes are involved in transcription-coupled DNA repair (TCR)17, illudin S sensitization alone is not enough to unequivocally identify TCR genes26. In line with this, functional assays show that CFAP20 is fully dispensable for TCR (Extended Data Fig. 3d). Notably, illudin S treatment has been shown to cause R-loop accumulation independently of TCR27, which prompted us to investigate R-loop levels in CFAP20-deficient cells. Immunofluorescence experiments using the S9.6 antibody (Fig. 1h), recognizing the RNA–DNA hybrid of R-loops28,29, showed a twofold increase in R-loop levels in CFAP20-KO cells, similar to R-loop levels in BRCA1-KO cells30 (Extended Data Fig. 3e). While mining the COSMIC (Catalogue Of Somatic Mutations In Cancer) database, we observed a charge-loss substitution (R100C) in CFAP20, situated within a highly positively charged patch on the protein surface (Fig. 1i). This mutation is recurrent in a small number of tumour types, yet it has not been classified as a tumour driver (Extended Data Fig. 3f and Supplementary Table 1). Owing to its potential effect on CFAP20 function, we chose to characterize this mutant. Although the R-loop phenotype in CFAP20-KO cells was fully reversed by stable re-expression of GFP-tagged wild-type (WT) CFAP20, expression of the GFP–CFAP20(R100C) mutant did not rescue the R-loop phenotype (Fig. 1j). To demonstrate specificity, we lentivirally transduced CFAP20-KO cells with GFP–RNaseH1, which abolished the S9.6 signal (Fig. 1j). Moreover, imaging R-loops using catalytically inactive recombinant GFP-tagged RNaseH1(D210N) confirmed the accumulation of R-loops in CFAP20-KO cells31,32, which was fully reversed by re-expression of CFAP20 (Extended Data Fig. 3g). A consequence of R-loop accumulation is the asymmetry of sister forks progressing from single origins33 (Fig. 1k). Accordingly, we could detect a marked fork asymmetry in CFAP20-KO cells (Fig. 1l) which could be reversed by expression of WT CFAP20 and by lentiviral transduction of GFP–RNaseH1, but not by CFAP20(R100C) (Fig. 1m), indicating that this is an R-loop-driven phenotype.
CFAP20 limits R-loops beyond cilia
We next investigated whether the accumulation of R-loops is connected to the ciliary function of CFAP20. To test this, we exploited the observation that homozygous cfap20−/− zebrafish larvae develop anterior–posterior ventral axis curvature, which has been attributed to the loss of motile ciliary function24. Micro-injecting human CFAP20 mRNA into cfap20-deficient zebrafish embryos fully rescued the body-axis-curvature defect. A similar rescue was observed when micro-injecting the CFAP20R100C variant (Fig. 2a,b). Consistent with these findings, GFP–CFAP20(R100C) localized to the primary cilium of RPE1 cells (Extended Data Fig. 4a). These findings suggest that the R-loop phenotype in CFAP20-KO cells is unrelated to its ciliary function.
a, Representative micrographs of cfap20−/− zebrafish embryos two days after fertilization, with severe ventral anterior–posterior curvature rescued by microinjection of 25 pg human CFAP20 mRNA (WT or R100C). Scale bar, 0.2 mm. n = 2 biological replicates. b, Percentage of cfap20−/− homozygotes with curvature defects, either uninjected or rescued by 25 pg human CFAP20 mRNA (WT or R100C). Sample sizes are indicated next to the bars. Significance by two-tailed Fisher’s exact test. ****P < 0.0001. c, Quantification of competition assays between the indicated conditions. NLS, nuclear localization signal. Each coloured circle represents the mean of an independent experiment (more than 30,000 cells). The coloured line represents the mean of n = 3 biological independent experiments. Significance by one-way ANOVA with Šidák’s correction. ****P < 0.0001. d, Additional competition assay quantification as in c. P values from left to right: 0.0064, <0.0001 and <0.0001. e, Colony formation assay for the indicated cell lines. f, Cell-cycle profiles analysed by quantitative image-based cytometry in the indicated RPE1 lines. g, Quantitative image-based cytometry after cyclin A staining; red box highlights G2-phase cells with low levels of cyclin A. Green indicates the mean intensity of cyclin A per nucleus (0–250). h, Quantification of G2 cells with low levels of cyclin A from g. Data are mean (three technical replicates from three independent experiments). Significance by unpaired two-tailed t-test. P = 0.0131. i, Genome-wide CRISPR screen in CFAP20-KO cells. Genes are ranked by z-score, showing synthetic-viable (blue) interactions with CFAP20. j, Colony formation assay for the indicated cell lines. k, Quantification of sister fork symmetry for the indicated stable cell lines. Data are as in Fig. 1j; significance by one-way ANOVA with Dunnett’s correction. P values from left to right: 0.0006, 0.8623 and 0.7491. l, Quantification of nuclear R-loop signal from the indicated stable cell lines. Data are as in Fig. 1j; significance as in k. P values from left to right: 0.0185, 0.9987, 0.0486 and >0.9999. m, Averaged spike-in normalized metaplots around TSS of RNAPII ChIP–seq for the same 3,000 BrU–seq-positive genes >3 kb in the indicated RPE1 cells.
CFAP20 and Mediator are synthetic viable
We noticed that CFAP20-KO cells grow more slowly than parental cells. Flow-cytometry-based competitive cell-growth assays confirmed that CFAP20-KO cells are rapidly outcompeted by WT cells (Fig. 2c and Extended Data Fig. 4b) and GFP–CFAP20 rescue –cells (Fig. 2d). This led to markedly decreased colony formation in CFAP20-KO cells, which was reversed by re-expression of WT CFAP20 but not by CFAP20(R100C) (Fig. 2e). Quantitative image-based cytometry revealed no obvious differences in cell-cycle profiles between WT and CFAP20-KO cells (Fig. 2f), but showed an increase in the percentage of cyclin A-negative G2 cells in the CFAP20-KO cell population (Fig. 2g,h), suggestive of cell-cycle exit34. To gain genetic insight into the cause of the poor-growth phenotype, we performed a genome-wide CRISPR screen to identify genes whose knockout would improve the fitness of CFAP20-KO cells (Extended Data Fig. 4c). sgRNAs targeting multiple subunits of the Mediator coactivator complex (Fig. 2i) were strongly enriched in our screen, which suggests that Mediator is a driver of the poor fitness in CFAP20-KO cells. To validate these results, we knocked out CCNC (encoding cyclin C, a subunit of the Mediator kinase module) in CFAP20-KO cells (Extended Data Fig. 4d,e). We observed a marked increase in colony formation in CFAP20/CCNC-double-knockout (dKO) cells, compared with single CFAP20-KO cells (Fig. 2j). Knockout of CCNC in a CFAP20-KO background also reversed the increase in cyclin A-negative G2 cells (Extended Data Fig. 4f,g). Thus, inactivation of the Mediator kinase function greatly improves the fitness of human CFAP20-KO cells. Notably, transient knockdown of ccnc in zebrafish larvae did not rescue the anterior–posterior body-axis curvature of the cfap20−/− mutant, and resulted in the development of additional microphthalmia and pericardial oedema (Extended Data Fig. 4h,i). These findings indicate that loss of CCNC does not rescue the ciliary dysfunction caused by the loss of CFAP20 function, but rather that CCNC loss rescues a function of CFAP20 that is unrelated to cilia.
Mediator-dependent R-loops in CFAP20-KO cells
Because inactivation of the Mediator subunit CCNC in a CFAP20-KO background could rescue the poor cell growth, we wondered whether this could also rescue the R-loop phenotype. Immunofluorescence experiments using either the S9.6 antibody (Extended Data Fig. 5a) or purified GFP–RNaseH1(D210N) (Extended Data Fig. 5b) showed a full reversal of the R-loop phenotype in CFAP20/CCNC-dKO cells. Moreover, bidirectional fork asymmetry was also fully reversed in dKO cells, whereas single CCNC-KO cells showed no R-loop phenotype (Fig. 2k). Of note, knockout of CCNC did not rescue the R-loop phenotype of cells deficient in the Integrator subunit INTS9 (Extended Data Fig. 5c,d). To determine whether the Mediator-dependent function of cyclin C drives these phenotypes, we used a CCNC point mutant (CCNC(D182A)) that is defective in binding the Mediator complex35 (Extended Data Fig. 5e). Proteomic and co-immunoprecipitation analyses confirmed that WT GFP–CCNC associated with CDK8, CDK19 and fifteen Mediator subunits, whereas GFP–CCNC(D182A) still associated with CDK8 and CDK19 but did not associate with Mediator (Extended Data Fig. 5f–h). Immunofluorescence experiments showed that re-expression of WT GFP–CCNC in CFAP20/CCNC-dKO cells restored R-loop accumulation to the level of CFAP20-deficient cells. However, expression of the GFP–CCNC(D182A) Mediator-binding mutant in CFAP20/CCNC-dKO cells did not increase R-loop levels (Fig. 2l). Previous studies have shown that inactivation of the Mediator kinase module, by knockout of the CDK8 counterpart of cyclin C, leads to global repression of transcription by reducing RNAPII occupancy at promoters36. Consistently, RNAPII occupancy measured by ChIP–seq was similar between WT and CFAP20-KO cells but was reduced in CFAP20/CCNC-dKO cells (Fig. 2m and Extended Data Fig. 5i). Together, these results show that CFAP20 specifically suppresses R-loops induced by Mediator-dependent transcription.
R-loops accumulate at TSSs in CFAP20-KO cells
We next set out to map where R-loops accumulate in the absence of CFAP20 by using genome-wide DRIP–seq. R-loops mapped mainly to promoters (TSSs) and terminators (transcription termination sites; TTSs), and treatment with recombinant RNaseH1 consistently abolished DRIP signals (Fig. 3a). More R-loops accumulated at promoters in CFAP20-KO cells than in WT cells across 508 genes (Fig. 3b and Extended Data Fig. 6a), whereas this was not the case at terminators (Fig. 3c and Extended Data Fig. 6b). Mapped regions with increased R-loops were found in transcriptionally active, early-replicating areas of the genome (Extended Data Fig. 6c). When selecting promoters with the strongest R-loop increase in CFAP20-KO cells (a greater than 1.5-fold increase in signal from −5 kb to +5 kb around TSSs in CFAP20-KO over WT), the terminators of these same genes still did not show an increase (Fig. 3b,c). Metaprofiles of around 1,800 aligned TSSs, sorted on the basis of their directionality relative to origins of replication, revealed that CD-oriented TSSs exhibited a stronger increase in R-loop levels in CFAP20-KO cells than did HO-oriented TSSs (Fig. 4d,e). The magnitude of R-loop accumulation at CD-oriented promoters in CFAP20-KO cells did not correlate with the level of anti-sense transcription (Extended Data Fig. 6d,e), suggesting that transcription in the CD orientation relative to replication is responsible for this phenomenon. To further strengthen these findings, we used a defined episomal system in HEK293T cells, with a doxycycline (DOX)-inducible gene oriented either in the same direction (CD) or the opposite direction (HO) relative to a nearby unidirectional replication origin1 (Fig. 4f). After DOX induction, cells transfected with a control short interfering RNA (siRNA) exhibited R-loops on the HO plasmid but not on the CD plasmid, as previously reported1. Knockdown of CFAP20 in these cells triggered a strong accumulation of R-loops after transcription induction, which was selectively observed at the CD-oriented promoter (Fig. 4g and Extended Data Fig. 6f,g), and was accompanied by ATM-dependent phospho-CHK2 activation (Extended Data Fig. 6h), consistent with the characteristic DNA damage response that is associated with CD conflicts. Thus, CFAP20 prevents the accumulation of R-loops specifically at CD-oriented promoters, consistent with its genetic interaction with the promoter-associated Mediator complex.
a, Distribution of DRIP–seq reads across indicated regions of the ITGA11 gene in the specified cell lines and conditions. b, Metaprofiles of DRIP–seq aligned around 508 promoters (TSSs) of genes >50 kb (top) or 135 TSSs with ≥1.5-fold higher DRIP signal from –2 kb to +3 kb around TSSs (bottom) in CFAP20-KO relative to WT. c, Metaprofiles of DRIP–seq in the indicated cell lines and conditions aligned around 508 terminators (TTSs) of genes >50 kb (top) or 135 TTSs with ≥1.5-fold higher DRIP signal from −2 kb to +3 kb around TSSs (bottom) in CFAP20-KO relative to WT. d, Metaprofiles of DRIP–seq in WT and CFAP20-KO cells aligned around HO (n = 408) TSSs relative to origins mapped by OK–seq5. Data are averages after trimming the top and bottom 5% of data (a trim-mean of 0.1) to remove extreme values. e, Metaprofiles of DRIP–seq in WT and CFAP20-KO cells aligned around CD (n = 1,395) TSSs relative to origins mapped by OK–seq5. Data as in d. f, Schematic of episomal system for transcription–replication conflicts. The unidirectional EBV replication origin (oriP; red) is placed either behind (HO) or in front (CD) of the EBNA1 gene containing the R-loop-forming mAIRN segment (blue) under TetON control. g, DRIP–quantitative PCR (qPCR) in HEK293T cells with mAIRN HO or CD plasmids, transfected with control or CFAP20 siRNAs ± DOX for 48 h. DRIP signals around the mAIRN TSS are shown as % input; data represent n = 3 biological independent experiments. Significance by one-way ANOVA with Šidák’s correction. P values from left to right: 0.4616 and <0.0001. h, Heat maps of CFAP20 ChIP–seq in TY-CFAP20 cells aligned around origins mapped by OK–seq5. i, Metaprofiles of DRIP–seq (red), CFAP20 ChIP–seq (green) and BrU–seq (blue) in the indicated cell lines within a 25-kb window adjacent to origins extending up to 75 kb. Data are Trimmean 0.1 to remove extreme values.
a, Heat map of scEdU–seq from a single 15-min EdU pulse. Maximum-normalized log counts for WT and CFAP20-KO cells, ordered by S-phase progression (x axis) and binned per 400 kb (y axis) for a 60-Mb region of chromosome 2. b, Representative images (top) and quantification (bottom) of origins per Mb derived from inter-origin distances, using sequential CldU (red) and IdU (green) labelling of nascent DNA. Scale bar, 5 μm. Significance by one-way ANOVA with Dunnett’s correction. P values from left to right: 0.0134, 0.5742, 0.0079 and 0.6897. c, Number of replication forks per cell (y axis) versus S-phase progression (x axis) in WT and CFAP20-KO cells; LOESS fit (line) with standard error ribbon. d, Representative images (top) and quantification (bottom) of replication-fork speed in the indicated cells using sequential CldU (red) and IdU (green) labelling. Scale bar, 5 μm. Significance by one-way ANOVA with Dunnett’s correction. P values from left to right: <0.0001, 0.5175, <0.0001 and 0.5893. e, DNA replication speed over S-phase (y axis) in WT (n = 402) and CFAP20-KO (n = 331) cells, measured as median replication width. Each dot is one cell, the line indicates the fit using a LOESS fit and the ribbon indicates the 95% standard error for the fit. f, Outline of S1 nuclease experimental set-up. g, Representative DNA fibres in the indicated cell lines without or with S1 nuclease. Scale bar, 10 μm. h, Quantification of fibres ± S1 nuclease in the indicated cells using sequential CldU (red) and IdU (green) labelling. Data representation as in Fig. 1j (more than 100 fibres); black and blue lines represent means of all experiments. Significance between –S1 and +S1 conditions was determined by one-way ANOVA with Šidák’s correction. P values from left to right: 0.9982, <0.0001, >0.9999, <0.0001, 0.9992, 0.0003 and >0.9999.
CFAP20 limits Mediator-dependent stress
We next overlayed the difference in R-loops between CFAP20-KO cells and WT cells with origins of replication mapped using published OK–seq datasets5 (Extended Data Fig. 6i). To correlate this to the binding of CFAP20 in the genome, we also performed genome-wide ChIP–seq on TY1-tagged CFAP20 (Fig. 4h). This analysis revealed a particular increase in R-loops at TSSs close to origins in the CD orientation, which did not correlate with an increase in nascent transcription in these genomic regions in CFAP20-deficient cells (Fig. 4i). Moreover, we detected CFAP20 binding mainly to gene promoters, with a strong preference for CD-oriented promoters (Fig. 4h,i). These findings suggest that CFAP20 acts locally at promoters to prevent transcription stress and R-loop accumulation. The marked accumulation of R-loops at TSSs close to origins in CFAP20-KO cells raises the possibility that this affects DNA replication dynamics. We performed scEdU–seq, which showed that S-phase progression is mostly unaltered between WT and CFAP20-KO cells21 (Fig. 4a and Extended Data Fig. 7a–d), in line with our quantitative image-based cytometry analysis (Fig. 2f). To investigate an effect on replication-fork progression, we used DNA fibre assays to measure the distance between origins37 (Extended Data Fig. 7e), and used this to calculate the number of origins firing per megabase. Origin firing was suppressed in CFAP20-KO cells, and re-expressing WT GFP–CFAP20 rescued this phenotype, whereas GFP–CFAP20(R100C) did not (Fig. 4b). The additional knockout of CCNC also restored origin activity to WT levels. In line with this finding, scEdU–seq analysis showed that origin usage was less efficient in CFAP20-KO cells than in WT cells (Extended Data Fig. 7f). In addition, quantification of the number of replication forks from scEdU–seq showed that, compared with WT cells, CFAP20-KO cells exhibited a decreased number of forks throughout S-phase and across all human chromosomes21 (Fig. 4c and Extended Data Fig. 7g). This is consistent with a decrease in origin firing and a general increase in fork stalling in CFAP20-KO cells.
Differences in origin activity are often compensated for by changes in replication-fork speed38. For instance, PARP inhibitors have been shown to trigger fork acceleration at first, followed by a secondary reduction in origin activity39,40. To test this possibility in the context of CFAP20 deficiency, we performed DNA fibre assays, which revealed that CFAP20-KO cells had an increased replication-fork speed (Fig. 4d). This phenotype was fully rescued by expressing WT GFP–CFAP20 but not by GFP–CFAP20(R100C). Meanwhile, the CFAP20/CCNC-dKO cells exhibited a normal fork speed, similar to that of WT cells (Fig. 4d). Treatment with a PARP inhibitor indeed caused fork speeding in WT cells41, but did not further accelerate forks in CFAP20-KO cells (Extended Data Fig. 8a). To extend these findings, we quantified replication-fork speed from scEdU–seq data, which confirmed the increased fork speed in CFAP20-KO cells and revealed that fork speeding occurs throughout S-phase (Fig. 4e). To identify the main cause of the replication defect in CFAP20-KO cells, we performed DNA fibre assays in combination with chemical inhibitors of replication-fork progression (aphidicolin) and origin activity38 (CDC7 kinase inhibitor, XL413) (Extended Data Fig. 8b). As expected, treating WT cells with the CDC7 kinase inhibitor reduced origin activity, which was accompanied by an acceleration of fork speed, whereas treatment with aphidicolin reduced fork speed and led to increased origin activity (Extended Data Fig. 8c,d). Whereas untreated CFAP20-KO cells already exhibited an increased fork speed and decreased origin activity (Fig. 4b–e), decreasing the fork speed with aphidicolin fully rescued origin activity in these cells (Extended Data Fig. 8c). If reduced origin activity were the main cause, this phenotype should be resilient to aphidicolin treatment38, which is not what we observed (Extended Data Fig. 8d). These experiments therefore reveal that the main cause of replication stress in CFAP20-KO cells is the accelerated fork rate, which consequently triggers a secondary decrease in origin activity.
CFAP20 limits Mediator-dependent gaps
Fork acceleration induced by PARP inhibition was shown to be associated with the formation of single-stranded DNA (ssDNA) gaps39. We therefore tested whether this is also the case in CFAP20-KO cells. To detect ssDNA gaps in the genome of CFAP20-KO cells, we used the DNA fibre assay in the presence of the ssDNA-specific S1 nuclease42 (Fig. 4f,g). Measurements of 5-iodo-2’-deoxyuridine (IdU) tracks showed a marked accumulation of ssDNA gaps in CFAP20-KO cells, which was reversed by expression of WT GFP–CFAP20 but not by the GFP–CFAP20(R100C) mutant (Fig. 4h). BRCA1-deficient cells also accumulate gaps, which were suggested43 to underlie their sensitivity to PARP inhibition. Notably, cell viability assays with increasing concentrations of PARP inhibitor showed that, in contrast to BRCA1-KO cells included in parallel, CFAP20-KO cells are not sensitive to PARP inhibition (Extended Data Fig. 8e). To investigate whether the accumulation of ssDNA gaps in CFAP20-KO cells is a consequence of Mediator-driven transcription, we performed S1 nuclease assays on different CCNC mutants. The ssDNA gap phenotype was fully reversed in CFAP20/CCNC-dKO cells. Re-expression of WT GFP–CCNC in these cells restored the ssDNA gap phenotype, whereas expression of the GFP–CCNC(D182A) Mediator-binding mutant did not (Fig. 4h).
During DNA replication, ssDNA gaps can arise from two main sources: incomplete lagging-strand processing and PRIMPOL-dependent repriming on the leading strand44,45,46. To assess the contribution of each mechanism, we first inhibited DNA polymerase α with the inhibitor CD437, which initiates replication at each Okazaki fragment on the lagging strand. Fork speeding in CFAP20-KO cells treated with CD437 was completely reversed (Extended Data Fig. 9a,b), and ssDNA gaps persisted (Extended Data Fig. 9c,d). Next, we used either siRNAs or CRISPR RNAs (crRNAs) to knock down or acutely knock out PRIMPOL. Under these conditions, we confirmed that loss of PRIMPOL fully reversed the fork speeding induced by PARP inhibitor treatment, as previously shown39. By contrast, the increased fork speed and fork asymmetry observed in CFAP20-KO cells were unaffected by the loss of PRIMPOL (Extended Data Fig. 9e–h). However, ssDNA gap accumulation was strongly suppressed by knockout of PRIMPOL (Extended Data Fig. 9i–k). The preferential accumulation of R-loops at CD-oriented promoters, where the transcribed strand for RNAPII serves as the leading-strand template during DNA replication (see Fig. 1b), is consistent with a repriming mechanism mediated by PRIMPOL. Supporting this model, fork asymmetry in CFAP20-KO cells was fully reversed by S1 nuclease treatment (Extended Data Fig. 9l). Together, these results suggest that CFAP20 suppresses Mediator-driven transcription stress at promoters to maintain replication fidelity.
CFAP20 salvages promoter-proximal RNAPII
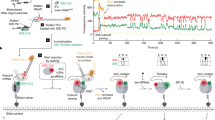
We next asked whether regions with increased R-loops in CFAP20-KO cells show differences in replication timing in our scEdU–seq dataset. By quantifying the replication timing of forks with the earliest, median and latest replication timing per 10-kb bin, we found that regions with increased R-loops in CFAP20-KO cells exhibited a delay in early DNA replication relative to WT cells, consistent with increased fork stalling at CD promoters in cis. By contrast, later-replicating regions completed DNA replication earlier in CFAP20-KO cells than in WT cells, consistent with the acceleration of replication forks observed in both DNA fibre and scEdU–seq experiments in trans (Extended Data Fig. 10a). Together, our results suggest that local Mediator-driven transcriptional stress at promoters, when not mitigated by CFAP20, culminates in global replication defects by increasing fork speed, which ultimately leads to reduced origin activity. To investigate R-loop dynamics in the promoter-proximal region, where R-loops specifically accumulate, we treated cells with the reversible transcription elongation inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB); this strongly suppressed R-loop accumulation in WT and CFAP20-KO cells (Fig. 5a,b). After DRB washout and release, R-loops returned rapidly to the original levels in both backgrounds (Fig. 5b), suggesting that R-loops are continuously formed in the promoter-proximal region. To extend these findings, we directly measured the RNAPII elongation rate by releasing cells after DRB elongation arrest and immediately pulse-labelling nascent transcripts using 4-thiouridine (4-SU) ribonucleoside (Fig. 5c). Isolation and sequencing of nascent transcripts revealed that CFAP20-KO cells showed a transcription elongation defect after DRB release (Fig. 5d and Extended Data Fig. 10b), which was also observed in CFAP20R100C cells (Extended Data Fig. 10c). Although the wave-front of RNAPII elongation was not different, suggesting that there was no effect on RNAPII processivity, we observed decreased elongation, consistent with an increased fraction of arrested RNAPII molecules (Fig. 5d). CFAP20 thus seems to salvage slowly elongating or arrested RNAPII molecules, thereby removing them from the path of CD replisomes. In support of such a role, we found that RNAPII co-immunoprecipitated with GFP–CFAP20 (Extended Data Fig. 10d,e). To further corroborate this model, we investigated whether replication phenotypes in CFAP20-deficient cells could be restored either by removing R-loops or by removing arrested RNAPII through α-amanitin degradation47 (Extended Data Fig. 10f). Whereas transient treatment with α-amanitin had no effect on fork speed (Fig. 5e) and only a marginal effect on fork symmetry (Extended Data Fig. 10g), overexpression of RNaseH1 led to a subtle but reproducible rescue of replication-fork speed in CFAP20-KO cells (Fig. 5e), along with full restoration of symmetric fork progression. However, degradation of RNAPII through transient treatment with α-amanitin combined with RNaseH1 overexpression fully restored both replication-fork speed and symmetry in CFAP20-deficient cells (Fig. 5e and Extended Data Fig. 10g). These data suggest that neither R-loops nor arrested RNAPII individually are sufficient to cause replication stress. Instead, CFAP20 acts on arrested RNAPII engaged with an R-loop to suppress Mediator-driven transcription stress, thereby minimizing interference with DNA replication (Fig. 5f).
a, Cartoon showing DRB mechanism of action. b, Quantification of nuclear R-loop signal in the indicated stable cell lines and conditions. Data as in Fig. 1j (more than 100 cells). Significance by one-way ANOVA with Šidák’s correction comparing WT and CFAP20-KO to respective conditions. P values from left to right: 0.8069, >0.9999, 0.9994, >0.9999, 0.0044, 0.0164, 0.7249 and >0.9999. c, Schematic of DRB treatment and release experiment. d, Averaged metaplots of TTchem–seq signal 60 min after DRB release for 508 genes ≥50 kb in WT or CFAP20-KO cells. e, Quantification of replication-fork speed in the indicated cells using sequential CldU (red) and IdU (green) labelling. Data as in Fig. 1j (more than 150 fibres). Significance by one-way ANOVA with Šidák’s correction comparing WT and CFAP20-KO to respective conditions. P values from left to right: 0.9292, 0.5810, 0.3237, 0.0040, >0.9999 and <0.0001. f, Model illustrating how CFAP20 acts on arrested RNAPII engaged with R-loops to suppress Mediator-driven transcription stress, thereby minimizing interference with DNA replication.
Discussion
Our findings support a model6 in which transcription–replication encounters are minimized by diverting RNAPII from the path of CD replisomes. During S-phase, promoter-proximally paused RNAPII is terminated by the Integrator complex, which clears the path for CD replisomes6. Once RNAPII transitions into productive elongation, Integrator no longer acts, and CFAP20 becomes essential to prevent Mediator-driven transcription–replication conflicts. Consistent with this, simultaneous loss of CFAP20 and INTS9 additively increases R-loops, with Integrator-dependent R-loops remaining Mediator-independent (Extended Data Fig. 5c,d). CFAP20 thus functions in a salvaging pathway that removes RNAPII stalled on R-loops, which otherwise obstruct replisomes10. Combined R-loop removal and RNAPII degradation reverses replication defects in CFAP20-deficient cells.
At CD genes, promoting elongation ensures that RNAPII remains ahead of replisomes that initiate near promoters. The Mediator complex and its kinase module enhance RNAPII promoter occupancy and transcriptional output36, increasing elongating RNAPII flux and transcriptional stress, which CFAP20 counterbalances. This explains the synthetic viability between Mediator loss and CFAP20 deficiency. Without CFAP20, impaired elongation after promoter release leads to fork stalling that is compensated for by the acceleration of neighbouring forks.
We propose a ‘block–trigger’ mechanism that involves R-loops and stalled RNAPII. R-loop accumulation induces asymmetric fork progression and modestly increases fork speed in CFAP20-KO cells. R-loops act as ‘blocks’ to fork movement in cis, whereas stalled RNAPII acts as a ‘trigger’ for fork acceleration in trans. RNAPII alone is a poor obstacle, consistent with evidence that DNA polymerases can bypass it through transient interaction with proliferating cell nuclear antigen (PCNA)7. However, RNAPII engaged with an R-loop combines both block and trigger functions, producing fork asymmetry and an increased trans fork speed. Thus, transcription-dependent fork stalling in cis drives trans acceleration and the accumulation of single-stranded DNA (ssDNA) gaps (Fig. 5f). Restoration of fork symmetry by S1 nuclease, which degrades leading-strand ssDNA, supports this model (Extended Data Fig. 9l). ssDNA gap accumulation in CFAP20-KO cells strongly depends on PRIMPOL, implicating leading-strand repriming45. PRIMPOL depletion did not affect fork speed, whereas inhibition of DNA polymerase α did (Extended Data Fig. 9a–k), explaining the hypersensitivity of CFAP20-deficient cells to the polymerase α inhibitor CD437 (Fig. 1f and Extended Data Fig. 3c). Fork acceleration induced by PARP inhibition also depends on polymerase α activity39, paralleling our observations, although PRIMPOL contributes under PARP inhibition. Fork acceleration was restored by PRIMPOL loss and PARP inhibition in WT but not in CFAP20-KO cells (Extended Data Fig. 9g), indicating that there are dual effects in CFAP20-KO cells: PRIMPOL-dependent repriming that generates ssDNA gaps, and polymerase α-driven acceleration of neighbouring forks.
CFAP20 mutations have been linked to retinitis pigmentosa through ciliary dysfunction24. The R100C mutation, which distinguishes RNAPII-related from ciliary functions, is not associated with retinitis pigmentosa but is found across several tumour types in the COSMIC database (Extended Data Fig. 3f and Supplementary Table 1). The Cancer Genome Atlas (TCGA) analyses across 33 tumour types identified CFAP20 hotspot mutations48, and CRISPR screens in cyclin E1-amplified ovarian and uterine cancer cells revealed a specific vulnerability to CFAP20 loss49. Although not investigated further in those studies, our results highlight CFAP20’s role in maintaining transcription–replication homeostasis, suggesting that tumour cells depend on CFAP20 to mitigate transcriptional stress. Further investigation of the functions of CFAP20 will elucidate how human cells coordinate transcription with replication and might uncover therapeutic opportunities in cancers that rely on this safeguard.
Methods
Cell lines
All cell lines are listed in Supplementary Table 2 and were cultured at 37 °C with 5% CO2 in DMEM GlutaMAX (Gibco), supplemented with 10% fetal calf serum (Avantor, VWR; Supplementary Table 3) and penicillin–streptomycin (Sigma; Supplementary Table 3).
Compounds and materials
All compounds and instruments are listed in Supplementary Table 3.
Generation of knockout cells
Cells were transfected with Cas9-2A-GFP (Addgene, 48138) containing a guide RNA targeting the gene of interest (sgRNAs are listed in Supplementary Table 4 and plasmids in Supplementary Table 5) using Lipofectamine 2000 (Life Technologies, 11668027). Cells were sorted by fluorescence-activated cell sorting on BFP and GFP and were plated at a low density, after which individual clones were isolated. Isolated knockout clones were verified by Sanger sequencing and/or western blot analysis (primers and antibodies are listed in Supplementary Tables 6 and 7, respectively).
PGK plasmids with GFP-tagged protein
The CFAP20 gene was amplified from cDNA by PCR and inserted in PGK-EGFP-C1-IRES-PURO, thereby tagging CFAP20 at its N terminus with GFP (primers and plasmids are listed in Supplementary Tables 5 and 6). The CFAP20R100C mutant was generated using site-directed mutagenesis PCR. The CCNC gene was amplified from the CMV-EGFP-CCNC plasmid and inserted in pLenti-PGK-GFP-puro, thereby tagging CCNC at its N terminus with GFP. The CCNCD182A mutant was generated using site-directed mutagenesis PCR. A region spanning the CMV promoter was amplified by PCR and used to replace the PGK promoter in pLenti-PGK-GFP-puro. A fragment encoding RNaseH1 from plasmid pFRT-TO-EGFP-RNaseH1 was amplified by PCR and inserted in pLenti-CMV-GFP-puro. All sequences and plasmids were verified by Sanger sequencing.
Generation of stable cell lines
Cells were transfected with Lipofectamine 2000 (Life Technologies, 11668027) or polyethyleneimine reagent (Brunschwig Chemie, 23966-2) according to the manufacturer’s instructions. All plasmids are listed in Supplementary Table 5. Lentiviral particles were produced by co-transfecting pLenti plasmids with pMDLg-pRRE, pRSV-REV and pCMV-VSVG in a 2:1:1:4 ratio in HEK293T cells by using polyethylenimine reagent. After production, lentivirus was filtered through a 0.44-µm filter and added to RPE1 cells in a complete DMEM medium supplemented with 4 µg ml−1 polybrene and 10 mM HEPES. After overnight incubation, the medium was removed, and fresh medium was added. The expression of GFP was verified three days after lentiviral transduction.
CRISPR–Cas9 gene editing
For CRISPR–Cas9 gene editing, we used a previously described approach50. In brief, Cas9 expression was induced by 200 ng ml−1 DOX followed by transfection with 20 nM equimolar crRNA:tracrRNA duplexes with 1:1,000 RNAiMAX (Life Technologies).
RNA interference
For RNA interference (Supplementary Table 4), cells were transfected with 50 nM siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen). Cells were transfected twice with siRNAs at 0 h and 24 h and were typically analysed 60 h after the first transfection.
Zebrafish lines and husbandry
All adult zebrafish strains are listed in Supplementary Table 8 and were raised at 28.5 °C under a 14-h–10-h light–dark cycle. Larvae were raised to 48 hours post-fertilization (hpf) at 28.5 °C in an incubator in E3 medium. The mutant cfap20 line (ua5025)24 was a gift from W. Ted Allison. All fish were on an AB background and staged as previously described51. Anaesthesia for live imaging was achieved with 60 mg l−1 of eugenol. All rescue experiments were performed on at least two clutches. All animal experiments were performed with the approval of the University of Toronto Animal Care Committee in accordance with the guidelines from the Canadian Council for Animal Care (CCAC).
mRNA and morpholino microinjection in zebrafish
All oligonucleotides used in zebrafish strains are listed in Supplementary Table 8. Microinjection into the cell (mRNA) or the yolk syncytial layer (morpholino) before the four-cell stage was done using pulled (P-97; Sutter Instrument) glass capillary tubes (TW100F-4; World Precision Instruments). Unfertilized eggs or embryos stalled during gastrulation were removed at 12 hpf. WT and CFAP20R100C variant mRNAs were transcribed from linearized pCS2+ vectors. The WT-containing plasmid was a gift from W. T. Allison. The R100C variant sequence was ordered as a gBlock (Integrated DNA Technologies) and directionally cloned into pCS2+ using BamHI or XbaI restriction enzymes. In vitro transcription was performed using the SP6 mMessage mMachine kit (Thermo Fisher Scientific, AM1340), followed by phenol:chloroform purification. A dose response using the WT mRNA diluted with ddH2O (into cfap20+/− incross clutches) was performed (25, 100 pg) to ensure that a rescue efficiency higher than 90% was achieved (data not shown). Embryos from cfap20+/− incrosses were microinjected with WT or R100C CFAP20 mRNA and larvae were then raised to 48 hpf and groups were blinded. Larvae were scored on the basis of a straight extension of the anterior–posterior axis (normal) or ventral curling of the body (curvature). Embryos were then processed for DNA lysis and genotyped as below, and groups were then unblinded. Only scores from cfap20−/− homozygotes were analysed. Standard control and ccnc splice morpholino oligonucleotides (Gene Tools), as in a previous study51, were used to knock down ccnc. A dose response of 1, 2 and 4 ng MO was performed in AB incross (2 clutches; more than 20 animals) as in Extended Data Fig. 5f and larvae were scored at 48 hpf on the basis of the severity of the phenotype. An optimal dose of 1.5 ng was chosen for subsequent experiments. Cfap20+/− heterozygote incross embryos were injected, groups were blinded and larvae were raised to 48 hpf. Larvae were scored (as above) on the basis of anterior–posterior curvature (2 clutches; more than 50 animals), then processed for genotyping before unblinding. Only scores from injected or uninjected cfap20−/− homozygotes were analysed.
Zebrafish cfap20 genotyping
A genomic DNA template for PCR was generated by adding tissue to 50 µl 50 mM NaOH, heating at 95 °C for 20 min and then neutralizing with 5.5 µl 1 M Tris-HCl. The template was diluted 50-fold and PCR genotyping was performed using GoTaq 2 (Promega). Primer sequences are listed in Supplementary Table 8.
Microscopic analysis of zebrafish larvae
Larval zebrafish were anaesthetized as above and transferred to 1% agar-lined Petri dishes for imaging. Representative bright-field images were taken using ZEN 3.7 (Zeiss) at 32× magnification on a Lumar V12 (Zeiss) stereomicroscope with an Axiocam 712 mono (Zeiss) camera. All graphing of and statistical tests on zebrafish data were done in Prism 10 (GraphPad), as described in Supplementary Table 9. The absolute number of normal versus axis-curvature defects was compared statistically using Fisher’s exact test. Raw images were cropped, and brightness and contrast were adjusted in Photoshop 2024 (Adobe). Identical transformations were performed on control and experimental images.
Western blotting
Proteins were separated on 4–12% Criterion XT Bis-Tris gels (Bio-Rad, 3450124) in NuPAGE MOPS running buffer (Thermo Fisher Scientific, NP0001–02), or on 3–8% Criterion XT Tris-Acetate protein gel (Bio-Rad, 3450131) in Tris/Tricine/SDS Running Buffer (Bio-Rad, 1610744), followed by blotting onto PVDF membranes (EMD Millipore, IPFL00010). Membranes were blocked with 5% milk powder in phosphate-buffered saline (PBS) with 0.1% Tween for one hour at room temperature. Protein expression was analysed by immunoblotting with the designated primary antibodies (listed in Supplementary Table 7) and corresponding secondary antibodies at 1:10,000. For detection, the Odyssey infrared imaging scanning system (LICORbio) was used.
Immunoprecipitation
Cell pellets were solubilized in EBC-1 (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40 and 2 mM MgCl2 with protease inhibitor cocktails (Roche)) supplemented with 500 U benzonase for one hour at 4 °C under rotation. The lysates were cleared from insoluble chromatin by centrifugation and were subjected to immunoprecipitation with GFP Trap beads (Chromotek, GTA-200) for 1.5 h at 4 °C under rotation. The beads were then washed four to six times with EBC-2 buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40 and 1 mM EDTA) and boiled in Laemmli buffer. Bound proteins were resolved by SDS–PAGE and immunoblotted with the indicated antibodies (Supplementary Table 7). For endogenous immunoprecipitation, 2 µg of antibody was incubated with the samples in EBC-1 buffer and benzonase, and they were subjected to immunoprecipitation with protein A agarose beads (Millipore, 16-157).
Mass-spectrometry sample preparation
After pull-down, the GFP beads were washed three times with 50 mM ammonium bicarbonate, followed by overnight digestion using 2.5 μg trypsin at 37 °C under constant shaking. Digested peptides were separated from the beads by a 0.45-µm filter column (Meck, UFC30HV00) that was prewashed with 50 mM ammonium bicarbonate. Trypsin activity was quenched by acidifying the sample with trifluoroacetic acid to a final concentration of 1%. Peptides were desalted and concentrated using in-house assembled triple-disc C18 stage-tip columns (serial number 66883-U; Sigma-Aldrich) as previously described52.
Mass-spectrometry data acquisition
The GFP–CCNC and GFP–CCNC(D182A) samples with their corresponding GFP-NLS controls were analysed by on-line C18 nano-high performance liquid chromatography (HPLC) MS/MS with a system consisting of an UltiMate3000 nano gradient HPLC system (Thermo Fisher Scientific) and an Exploris480 mass spectrometer (Thermo Fisher Scientific). Digested peptides were injected onto a cartridge precolumn (300 μm × 5 mm, C18 PepMap, 5 μm) in 100% solvent A (0.1 % formic acid in milli-Q), with a flow of 10 μl per min for 3 min (Thermo Fisher Scientific), and eluted using a homemade analytical nano-HPLC column (30 cm × 75 μm; Reprosil-Pur C18-AQ 1.9 μm, 120 A (Dr. Maisch). The chromatography gradient length was 60 min from 2% to 40% solvent B, followed by a 5-min increase to 95% solvent B, another 5 min of 95% solvent B and back to 2% solvent B for chromatography column reconditioning. The mass spectrometer was operated in positive polarity data-dependent MS/MS mode with a cycle time between master scans of 3 s. Full-scan MS spectra were obtained with a resolution of 60,000, a normalized automatic gain control (AGC) target of 300% and a scan range of 350–1,600 m/z. Precursors were fragmented by higher-energy collisional dissociation (HCD) with a normalized collision energy of 28%. Tandem mass spectra (MS/MS) were recorded with a resolution of 30,000 and a normalized AGC target value of 75%. Precursor ions selected for MS/MS analysis were subsequently dynamically excluded from MS/MS analysis for 30 s and only precursors with a charge state of 2–6 triggered MS/MS events.
Mass-spectrometry data analysis
RAW data were analysed using MaxQuant (v.1.6.14.0) as previously described53,54.
Mass-spectrometry data availability
The mass-spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE55 partner repository with the dataset identifier PXD051449 (GFP–CFAP20(R100C) and GFP–CCNC sample sets).
CRISPR screens
For every screen, three populations of RPE1-iCas9 were transduced at a multiplicity of infection (MOI) of around 0.2 with a 1:1,000 dilution of TKOv3-in-pLCKO lentiviral library in medium containing 8 µg ml−1 hexadimethrine bromide (Sigma-Aldrich). The library was a gift from K. Chan, A. Tong and J. Moffat. Twenty-four hours after transduction, puromycin (Sigma-Aldrich) was added to 5 μg ml−1 to select for transduced cells. After all cells in non-transduced control populations had died and dishes with transduced populations had reached 90% confluence, a t = 0 sample was taken for each of the three populations. From the remaining cells of each population, 30 × 106 (corresponding to a library representation of more than 400) were grown as a control population. To screen for replication stress genes, RPE1-iCas9 parental cells were grown in the presence of the DNA pol α inhibitor CD437 at a concentration of 200 nM. The illudin S screen has been described previously17. To screen for synthetic-viable genes, RPE1-iCas9 CFAP20-KO cells were grown without drugs or inhibitors. DOX was added to the medium of all replicates from t = 0 onwards to induce expression of Cas9, at a concentration of 200 ng ml−1. After 3 doublings, 30 × 106 cells of each population were passed. After 12 doublings, all populations were collected.
Sequencing and analysis of CRISPR screens
Genomic DNA was isolated from each population using the Blood and Cell Culture DNA Maxi Kit (QIAGEN). Then, 3 µg of gDNA from each population was amplified using the KAPA HiFi ReadyMix PCR Kit (Roche) with the TKO outer Fw and Rv primers (primers are listed in Supplementary Table 5), followed by a second PCR reaction using reverse primers with different Illumina i7 index sequences for each sample to identify the sample after pooled sequencing as described56. The second PCR products of each pool were purified using the QIAquick PCR Purification Kit (QIAGEN). Samples were sequenced on a NovaSeq 6000 and reads were mapped to the TKOv3 library sequences, not allowing any mismatches. To compare the illudin S to the CD437 screen (Fig. 1f), the lowest z-score for each screen was normalized to −1 (sensitizer genes:UVSSA for illudin S; HUS1 for CD437), and the highest score was normalized to +1 (resistance genes: PTGR1 for illudin S; CDAN1 for CD437). The synthetic-lethal and synthetic-viable interactions were analysed by comparing the CFAP20-KO line with the parental RPE1-iCas9 WT by first normalizing end-point reads based on t = 0 reads, as described previously50. We used an adapted version of DrugZ, termed IsogenicZ, which can be found at https://github.com/kdelint/IsogenicZ.
Immunostaining
Cells were grown on coverslips and fixed with 4% formaldehyde. By incubating with 0.5% Triton X-100 in PBS for 5 min, cells were permeabilized, followed by blocking with 100 mM glycine for 10 min. After washing with WB buffer (0.5% bovine serum albumin (BSA) and 0.05% Tween 20 in PBS), coverslips were incubated with the primary antibody (Supplementary Table 7) in WB buffer for two hours at room temperature. Cells were then washed extensively and labelled with their corresponding secondary antibody (Supplementary Table 7) in WB buffer containing 0.1 μg ml−1 DAPI for one hour at room temperature. Finally, the coverslips were washed extensively with PBS and mounted in Polymount (Brunschwig, 18606).
Immunostaining for detection of RNA–DNA hybrids
Indirect immunofluorescence with S9.6 antibody against RNA–DNA hybrids was performed as previously described57. Imaging of RNA–DNA hybrids using GFP–RNaseH1(D210N) was performed as described previously31.
Recovery of RNA synthesis
Cells were irradiated with UV-C light (12 J m−2), allowed to recover for the indicated periods and pulse-labelled with 400 μM 5-ethynyl-uridine (EU; Jena Bioscience) for one hour, followed by a 15 min medium-chase with DMEM without supplements. Cells were fixed with 3.7% formaldehyde in PBS for 15 min, permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature and blocked in 1.5% BSA (Thermo Fisher Scientific) in PBS. Nascent RNA was visualized by click-iT chemistry, labelling the cells for one hour with a mix of 60 μM Atto azide–Alexa 594 (ATTO-TEC), 4 mM copper sulfate (Sigma), 10 mM ascorbic acid (Sigma) and 0.1 μg ml−1 DAPI in a 50 mM Tris-buffer (pH 8). Cells were washed extensively with PBS and mounted in Polymount (Brunschwig).
Microscopic analysis of fixed human cells
Images of fixed samples were acquired on a Zeiss AxioImager M2 wide-field fluorescence microscope equipped with 63× Plan-Apo (1.4 NA) oil-immersion objectives (Zeiss) and an HXP 120 metal-halide lamp was used for excitation. Fluorescent probes were detected using the following filters for DAPI (excitation filter, 350/50 nm; dichroic mirror, 400 nm; emission filter, 460/50 nm), Alexa 488 (excitation filter, 470/40 nm; dichroic mirror, 495 nm; emission filter, 525/50 nm) or Alexa 647 (excitation filter, 640/30 nm; dichroic mirror, 660 nm; emission filter, 690/50 nm). Images were recorded using ZEN 2012 (blue edition, v.1.1.0.0) and analysed in Image J (v.1.47–1.48). Graphs were plotted and analysed using GraphPad Prism 10 (10.2.3), Microsoft Excel 365 and Adobe Illustrator 2022, as described in Supplementary Table 9.
Quantitative image-based cytometry
Quantitative image-based cytometry was performed as described previously58. Colour-coded scatter plots and bar charts of asynchronous cell populations were generated with Spotfire data visualization software (v.10.10.1; TIBCO). Representative scatter plots and bar charts are shown.
Pairwise fluorescent competitive growth assay
Cell lines stably expressing either GFP or mCherry were seeded in a 1:1 ratio (30,000 cells per 6 wells). Cells were grown as usual and split every three days. During trypsinization, samples were taken at each time point. Cell pellets were washed with PBS followed by incubation in 2% formaldehyde in PBS for 15 min. Samples were quenched with glycine, washed with PBS, fixed in ice-cold methanol and stored at −20 °C. On the day of analysis, pellets were washed once with PBS and resuspended in 350 µl PBS. An AECE NovoCyte flow cytometer and NovoExpress software (Agilent) were used for analysis. For immunostaining, cells were grown simultaneously on coverslips and fixed in 4% formaldehyde at the corresponding time points. After permeabilization with 0.5% Triton X in PBS, cells were mounted with ProLong Gold Antifade Mountant with DNA Stain DAPI (Invitrogen, P36935).
Clonogenic growth assays
Cells were plated at low density in 6-cm culture dishes and allowed to attach, and were grown for ten days in growth medium supplemented with the indicated concentrations of the drugs. To visualize clones, cells were fixed with NaCl and stained with methylene blue. Formed clones were manually counted.
CellTiter-Glo assays
In a Costar black, clear-bottom 96-well plate, cells were seeded (WT and CFAP20-KO, 200 per well; BRCA1-KO, 400 per well) in medium containing increasing doses of olaparib or dimethyl sulfoxide (DMSO; 0.1% final DMSO concentration). Wells with no cells were included as a background luminescence control. After six days, the viability measurement was performed according to the manufacturer’s protocol. In brief, CellTiter-Glo substrate was dissolved in CellTiter-Glo buffer (Promega), and 100 µl of this was added to 100 µl fresh medium per well. The plate was briefly shaken and after equilibration, luminescence was recorded on a SpectraMax iD3 microplate reader (Molecular Devices). Luminescence values were corrected for background and for each cell line, normalized to wells treated with DMSO. Data were exported to GraphPad Prism 9.3.1 for further analysis.
DNA fibre spreading assay
Treatments with different compounds are shown in each experiment. Cells were labelled with 25 µM 5-chloro-2’-deoxyuridine (CldU) (Merck; Supplementary Table 2) for 20 min and washed three times with PBS, followed by labelling with 250 µM IdU (Merck; Supplementary Table 3) for 20 min. Labelled cells were collected and resuspended in 1× cold PBS. Two microlitres of the cell suspension was spotted on a positively charged slide (VWR) and then mixed with 7 µl of lysis buffer (200 mM Tris-HCl pH 7.4, 50 mM EDTA and 0.5% (w/v) SDS). The cells were incubated in lysis buffer horizontally for 5 min and then tilted at about 45°, allowing the drop to run by gravity. The DNA spreads were air-dried at room temperature and were then fixed in methanol/acetic acid (3:1) at room temperature for 10 min and stored at 4 °C overnight. Slides were processed as previously described59. Fibres were visualized and imaged using a Zeiss Axio Imager-M2 wide-field fluorescence microscope equipped with 40× Plan-Apo (1.4 NA) oil-immersion objectives (Zeiss) and an HXP 120 metal-halide lamp was used for excitation. Images were recorded and analysed with ZEN 2012 (blue edition, v.1.1.0.0) and analysed in Image J (v.1.53). Replication-fork speed (kb min−1) was calculated on the basis of the assumption that 1 µm of DNA fibre corresponds to 2.59 kb, as previously shown60.
DNA fibre assay with S1 nuclease
For the DNA fibre assay with the ssDNA nuclease (S1 nuclease), cells were labelled with 25 µM CldU for 15 min, washed three times with PBS and labelled again with 250 µM IdU for one hour. Cells were treated and processed as previously shown42,59.
scEdU–seq
The scEdU–seq procedure was performed according to a method described previously21. RPE1 WT and CFAP20-KO were labelled with 15-min pulses of EdU (10 μM). The cells were trypsinized, fixed in 70% ethanol and kept at −20 °C for 24 h. Then, the samples were resuspended and washed in 1 ml wash buffer (47.5 ml RNAse-free H2O, 1 ml 1 M HEPES pH 7.5, 1.5 ml 5 M NaCl, 3.6 µl pure spermidine solution, with an additional 0.05% Tween, and 4 µl ml−1 0.5 M EDTA). Next, biotin-PEG3-azide was conjugated to the EdU molecules through a CuAAC click reaction, followed by staining with DAPI. Single S-phase RPE1 cells were then sorted into 384-well plates for scEdU–seq processing. After sorting, libraries were prepared as follows: proteinase K digestion, NlaIII genome digestion, DNA blunt ending, A-tailing and adapter ligation incorporating cell barcodes and unique molecular identifiers (UMIs). Single-cell libraries were pooled and bound to MyOneC1 streptavidin beads to capture DNA replication fragments. These fragments were released by heat denaturation and filled in using the Klenow enzyme. The libraries underwent amplification through in vitro transcription, reverse transcription and PCR, followed by Illumina sequencing (NextSeq1000 P3 2×100 bp). The code for analysis and plotting can be accessed on GitHub21.
DRIP–qPCR
Approximately 1 ×107 cells per condition were lysed in 1.6 ml TE buffer supplemented with 82 μl of 10% SDS and 10 μl of 10 mg ml−1 proteinase K and incubated at 37 °C overnight. DNA was isolated by phenol:chloroform:isoamyl alcohol (25.24:1, v/v) extraction and isopropanol precipitation. DNA was reconstituted in 130 μl TE buffer, transferred to AFA microTUBEs with snap caps and sonicated for 4 min using a Covaris E220 sonicator (140 peak incident power, 10% duty factor and 200 bursts per cycle). Sonicated DNA was quantified on a NanoDrop 2000c spectrophotometer. For immunoprecipitation, 4 μg of DNA was resuspended in 150 μl 1× binding buffer (10 mM Na3PO4 pH 7, 140 mM NaCl and 0.05% Triton X-100), 10% removed as input DNA and the remaining sample bound to 6 μg of S9.6 antibody in 1× binding buffer overnight at 4 °C. Protein A/G agarose beads were added for two hours. Bound beads were washed three times in 1× binding buffer for 10 min at 4 °C. Elution was performed in elution buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 0.5% SDS and proteinase K) for 45 min at 55 °C with agitation. Eluted DNA was purified by phenol:chloroform:isoamyl alcohol (25.24:1, v/v) extraction and ethanol precipitation. Enrichment analysis of RNA–DNA hybrids in input and immunoprecipitation samples was performed by qPCR using the primers listed in Supplementary Table 6.
DRIP–seq
DRIP–seq was performed as previously described61 with minor modifications. Samples were sequenced using an Illumina NextSeq500 or HiSeq X, using paired-end sequencing with 42 bp or 151 bp from each end.
BrU–seq
Cells were grown to 80–90% confluency in three 15-cm plates per condition and incubated for 30 min with 2 mM BrU (Sigma, 850187). After incubation, cells were lysed in Trizol (Thermo Fisher Scientific, 15596018) and BrU-containing RNA was isolated as previously described62. cDNA libraries were made from the BrU-labelled RNA using the Illumina TruSeq library kit and paired-end 151-bp sequenced using the Illumina NovaSeq platform at the University of Michigan Advanced Genomics Core. Single-end or paired-end sequencing data were used for downstream analyses.
ChIP–seq
Cells were grown to 80–90% confluency and cross-linked with 0.5 mg ml−1 disuccinimidyl glutarate (Thermo Fisher Scientific) in PBS for 45 min at room temperature. Cells were washed once with PBS, followed by incubation with 1% formaldehyde for 20 min at room temperature. Fixation was stopped by adding glycine in PBS to a final concentration of 0.1 M for 3 min at room temperature. This was followed by washing with cold PBS and collection of the cells in 0.25% Triton X-100, 10 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0) and 20 mM HEPES (pH 7.6) in milli-Q. Chromatin was pelleted by centrifugation for 5 min at 400g and incubated in 150 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0) and 50 mM HEPES (pH 7.6) in milli-Q for 10 min at 4 °C. Chromatin was again pelleted by centrifugation and resuspended in ChIP buffer (0.15 % SDS, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0) and 20 mM HEPES (pH 7.6) in milli-Q) to a final concentration of 15 × 106 cells per ml. Chromatin was sonicated to approximately one nucleosome using the Bioruptor Pico (Diagenode), with 8–15 cycles of 30 s on and 30 s off in a 4 °C water bath. RNAPII ChIP was performed using 28 µg of chromatin (22 µg for CCNC-KO and CFAP20/CCNC-dKO) + 40 ng of Drosophila spike-in chromatin (32.6 ng for CCNC-KO and CFAP20/CCNC-dKO; Active Motif, 53083) with 3 µl of RNAPII antibody and 1 µg spike-in antibody (Supplementary Table 6) by overnight incubation at 4 °C. TY ChIP was performed using 84 µg of WT chromatin and 60 µg of CFAP20-KO + TY-CFAP20 chromatin + 74 ng and 53 ng of Drosophila spike-in chromatin, respectively (Active Motif, 53083) with 5.7 µg of TY antibody (Diagenode, C15200054) and 1 μg spike-in antibody (Active Motif, 61686) (Supplementary Table 6) by overnight incubation at 4 °C. Protein–chromatin pull-down followed, with a 1:1 mix of protein A and protein G Dynabeads for RNAPII ChIPs, and protein A Dynabeads for TY ChIPs (Thermo Fisher Scientific, 10001D and 10003D). ChIP samples were washed extensively and purified using the QIAGEN MinElute kit. Sample libraries were prepared using the HiFi KAPA sample preparation kit and A–T-mediated ligation of NEXTflex adapters or xGen UDI-UMI adapters. Samples were sequenced using an Illumina NextSeq500 or HiSeq X, using paired-end sequencing with 42 bp or 151 bp from each end.
TTchem–seq
TTchem–seq was performed as described previously63. For TTchem experiments in WT or CFAP20-KO cells, this included depletion of rRNAs using the QIAseq FastSelect rRNA depletion kit (QIAGEN), followed by library preparation using the TruSeq Stranded Total RNA kit (Illumina, 20020596). For CFAP20-KO cells expressing WT GFP–CFAP20 or GFP–CFAP20(R100C), no ribosomal RNA was performed. The libraries were amplified according to the manufacturer’s instructions, pooled and paired-end sequenced on a DNBSEQ-G400 (BGI) system.
Definition of replication origins
OK–seq data in untreated RPE1 cells were downloaded from a previous report5 (datasets GSM3130725 and GSM3130726). Sequences were trimmed using TrimGalore (v.0.6.5) and aligned to hg38 using STAR (v.2.7.7a) with the genome file GCA_000001405.15_GRCh38. Duplicate reads were removed using SAMtools (v.1.11) with fixmate -m and markdup -r settings. Replication initiation zones were subsequently defined using the replication fork directionality analysis R toolkit (OKseqHMM v.2.0.0; available at https://github.com/CL-CHEN-Lab/OK-Seq; ref. 64), with read coverage threshold 6 for GSM3130725 and 1 for GSM3130726, and smoothing window size 15 kb. Initiation zones present in both datasets were identified using mergePeaks of HOMER tools (v.4.8.2)65, with -d given. Origins were defined as the centre of initiation zones. For all origins, their nearest TSS was defined using annotatePeaks of HOMER tools, together with the distance between the TSS and the origin. Here, a negative distance represents an origin upstream of the TSS (CD transcription relative to replication), whereas a positive distance represents an origin downstream of a TSS (HO transcription relative to replication). To allow for clean transcription versus replication analyses, we further selected only origins for which the nearest TSS was not preceded by another gene within 5 kb upstream of the TSS (Extended Data Fig. 2a). This resulted in a list of 2,040 origins.
ChIP–seq, DRIP–seq, BrdU–seq and TTchem–seq data analysis
For all sequencing data, a sequencing quality profile was generated using FastQC (v.0.11.9). Sequences were trimmed using TrimGalore (v.0.6.5). For ChIP–seq, reads were aligned to the human genome 38 GCA_000001405.15_GRCh38 and Drosophila genome BDGP6 using bwa-mem tools (BWA, v.0.7.17)66. For DRIP–seq, reads were aligned to the human genome 38 GCA_000001405.15_GRCh38 using bwa-mem tools (BWA, v.0.7.17)66. Only uniquely or primary mapping and high-quality reads (>q30) were included in the analyses. For BrU–seq and TTchem–seq, reads were aligned to hg38 using STAR (v.2.7.7a)67 with the genome file GCA_000001405.15_GRCh38. For ChIP–seq, BrU–seq and TTchem–seq data, duplicate reads were removed using SAMtools (v.1.11) with fixmate -m and markdup -r settings. Bam files were converted into stranded TagDirectories (with fixed fragment length 150–200 when automated fragment length definitions varied extensively) and UCSC genome tracks using HOMER tools (v.4.8.2)65. Example genome tracks were generated in IGV (v.2.4.3). A list of 2,040 origin coordinates was defined using data derived from a previous study5, as described in ‘Definition of replication origins’. A list of 49,948 gene coordinates was obtained from the UCSC genome database selecting the ‘knownCanonical’ table containing the canonical TSSs per gene68. To prevent contamination of binding profiles, genes were selected to be non-overlapping with at least 2 kb between genes and a minimal size of 3 kb (n = 9,944). From this, a set of 3,000 actively transcribed genes was selected by calculating gene-size-corrected read densities of BrU–seq data in WT cells, using the AnnotatePeaks.pl tool of HOMER with default settings. These 3,000 actively transcribed genes were used in downstream analyses, unless stated otherwise. For all DRIP–seq, ChIP–seq, BrU–seq and TTchem–seq experiments, read-density profiles around origin or TSS/TTS coordinates were defined using the AnnotatePeaks.pl tool of HOMER, using the default normalization to 10 million reads. For ChIP–seq experiments around transcribed genes, reads were normalized to the number of identified spike-in reads. Individual datasets were subsequently processed into heat maps or binding profiles using R (v.4.0.5) and Rstudio (v.1.1.423)69. Where indicated, average read-density profiles were generated after trimming 10% of the data (trim-mean 0.1; removing the top 5% and bottom 5% of datapoints) to remove extreme values.
Metaprofiles of TSSs in CD and HO orientations
We aligned TSSs with either a negative distance (CD) or a positive distance (HO) relative to the nearest origin. We subsequently generated average read-density profiles of RNAPII ChIP–seq, BrU–seq and DRIP–seq for all 1,395 CD and 408 HO genes at a maximum distance of 75 kb from the origin. We also sub-selected HO TSSs into those at 75–50 kb (n = 37), 50–25 kb (n = 80) or 25–0 kb (n = 291) upstream of the origin, and CD TSSs into those at 0–25 kb (n = 1,199), 25–50 kb (n = 143) or 50-75 kb (n = 53) downstream of the origin. These analyses provide a transcription-centred view of replication.
Statistics and reproducibility
Experimental data were plotted for statistical analysis in GraphPad Prism 10.2.3 (GraphPad). In figures showing all data points, each coloured circle represents a single cell, and the black circles represent the median of each independent biological repeat—of which there were at least two—as indicated for each experiment. More information on the n of each experiment is provided in the source data. Statistical analyses were performed on the median of each independent biological repeat per experiment using one-way ANOVA after Dunnett’s or Šidák’s correction where appropriate, unpaired two-tailed t-test or Fisher’s exact test, as indicated in the figure legends. All experiments were independently repeated at least twice, with similar results obtained. All micrographs are representative images of experiments that were performed at least twice, with similar results. In the figures, the notation NS = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 is used, and precise P values are provided in the figure legends and the source data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
BrU–seq, ChIP–seq, DRIP–seq, and TTchem–seq data have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE266575. scEdU–seq data have been deposited under the accession number GSE276603. The mass-spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD051449. CFAP20 mutations in human cancer were analysed by COSMIC (https://cancer.sanger.ac.uk/cosmic). The CFAP20 mutation R100C/H and its occurrence in cancer were analysed using cBioPortal (http://cBioportal.org). Source data are provided with this paper.
References
Hamperl, S., Bocek, M. J., Saldivar, J. C., Swigut, T. & Cimprich, K. A. Transcription–replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170, 774–786 (2017).
Shivji, M. K. K., Renaudin, X., Williams, C. H. & Venkitaraman, A. R. BRCA2 regulates transcription elongation by RNA polymerase II to prevent R-loop accumulation. Cell Rep. 22, 1031–1039 (2018).
Zatreanu, D. et al. Elongation factor TFIIS prevents transcription stress and R-loop accumulation to maintain genome stability. Mol. Cell 76, 57–69 (2019).
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I. & Chedin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012).
Chen, Y. H. et al. Transcription shapes DNA replication initiation and termination in human cells. Nat. Struct. Mol. Biol. 26, 67–77 (2019).
Bhowmick, R., Mehta, K. P. M., Lerdrup, M. & Cortez, D. Integrator facilitates RNAPII removal to prevent transcription–replication collisions and genome instability. Mol. Cell 83, 2357–2366 (2023).
Fenstermaker, T. K., Petruk, S., Kovermann, S. K., Brock, H. W. & Mazo, A. RNA polymerase II associates with active genes during DNA replication. Nature 620, 426–433 (2023).
Lang, K. S. et al. Replication–transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 170, 787–799 (2017).
Bruno, F., Coronel-Guisado, C. & Gonzalez-Aguilera, C. Collisions of RNA polymerases behind the replication fork promote alternative RNA splicing in newly replicated chromatin. Mol. Cell 84, 221–233 (2024).
Bruning, J. G. & Marians, K. J. Replisome bypass of transcription complexes and R-loops. Nucleic Acids Res. 48, 10353–10367 (2020).
Osman, S. & Cramer, P. Structural biology of RNA polymerase II transcription: 20 years on. Annu. Rev. Cell Dev. Biol. 36, 1–34 (2020).
Richter, W. F., Nayak, S., Iwasa, J. & Taatjes, D. J. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 23, 732–749 (2022).
Stein, C. B. et al. Integrator endonuclease drives promoter-proximal termination at all RNA polymerase II-transcribed loci. Mol. Cell 82, 4232–4245 (2022).
Malik, S. & Roeder, R. G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 (2010).
Danko, C. G. et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50, 212–222 (2013).
Fitz, J., Neumann, T. & Pavri, R. Regulation of RNA polymerase II processivity by Spt5 is restricted to a narrow window during elongation. EMBO J. 37, e97965 (2018).
van der Weegen, Y. et al. ELOF1 is a transcription-coupled DNA repair factor that directs RNA polymerase II ubiquitylation. Nat. Cell Biol. 23, 595–607 (2021).
Alzu, A. et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151, 835–846 (2012).
Chappidi, N. et al. Fork cleavage-religation cycle and active transcription mediate replication restart after fork stalling at co-transcriptional R-loops. Mol. Cell 77, 528–541 (2020).
Macheret, M. & Halazonetis, T. D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 555, 112–116 (2018).
van den Berg, J. et al. Quantifying DNA replication speeds in single cells by scEdU-seq. Nat. Methods 21, 1175–1184 (2024).
Han, T. et al. The antitumor toxin CD437 is a direct inhibitor of DNA polymerase alpha. Nat. Chem. Biol. 12, 511–515 (2016).
Barroso, S. et al. The DNA damage response acts as a safeguard against harmful DNA–RNA hybrids of different origins. EMBO Rep. 20, e47250 (2019).
Chrystal, P. W. et al. The inner junction protein CFAP20 functions in motile and non-motile cilia and is critical for vision. Nat. Commun. 13, 6595 (2022).
Wang, T. et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015).
van den Heuvel, D. et al. STK19 facilitates the clearance of lesion-stalled RNAPII during transcription-coupled DNA repair. Cell 187, 7107–7125 (2024).
Goulielmaki, E. et al. The splicing factor XAB2 interacts with ERCC1–XPF and XPG for R-loop processing. Nat. Commun. 12, 3153 (2021).
Bou-Nader, C., Bothra, A., Garboczi, D. N., Leppla, S. H. & Zhang, J. Structural basis of R-loop recognition by the S9.6 monoclonal antibody. Nat. Commun. 13, 1641 (2022).
Stoy, H. et al. Direct visualization of transcription-replication conflicts reveals post-replicative DNA:RNA hybrids. Nat. Struct. Mol. Biol. 30, 348–359 (2023).
Crossley, M. P. et al. R-loop-derived cytoplasmic RNA–DNA hybrids activate an immune response. Nature 613, 187–194 (2023).
Crossley, M. P. et al. Catalytically inactive, purified RNase H1: a specific and sensitive probe for RNA–DNA hybrid imaging. J. Cell Biol. 220, e202101092 (2021).
Teloni, F. et al. Efficient pre-mRNA cleavage prevents replication-stress-associated genome instability. Mol. Cell 73, 670–683 (2019).
Tuduri, S. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009).
Cornwell, J. A. et al. Loss of CDK4/6 activity in S/G2 phase leads to cell cycle reversal. Nature 619, 363–370 (2023).
Park, M. J. et al. Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J. Biol. Chem. 293, 4870–4882 (2018).
Sooraj, D. et al. MED12 and BRD4 cooperate to sustain cancer growth upon loss of mediator kinase. Mol. Cell 82, 123–139 (2022).
Bellelli, R. et al. Synthetic lethality between DNA polymerase epsilon and RTEL1 in metazoan DNA replication. Cell Rep. 31, 107675 (2020).
Rodriguez-Acebes, S., Mouron, S. & Mendez, J. Uncoupling fork speed and origin activity to identify the primary cause of replicative stress phenotypes. J. Biol. Chem. 293, 12855–12861 (2018).
Machacova, Z., Chroma, K., Lukac, D., Protivankova, I. & Moudry, P. DNA polymerase α-primase facilitates PARP inhibitor-induced fork acceleration and protects BRCA1-deficient cells against ssDNA gaps. Nat. Commun. 15, 7375 (2024).
Sedlackova, H. et al. Equilibrium between nascent and parental MCM proteins protects replicating genomes. Nature 587, 297–302 (2020).
Maya-Mendoza, A. et al. High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284 (2018).
Quinet, A., Carvajal-Maldonado, D., Lemacon, D. & Vindigni, A. DNA fiber analysis: mind the gap!. Methods Enzymol. 591, 55–82 (2017).
Cong, K. et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell 81, 3128–3144 e3127 (2021).
Jahjah, T., Singh, J. K., Gottifredi, V. & Quinet, A. Tolerating DNA damage by repriming: gap filling in the spotlight. DNA Repair 142, 103758 (2024).
Quinet, A. et al. PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-deficient cells. Mol. Cell 77, 461–474 (2020).
Lange, S. S., Takata, K. & Wood, R. D. DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 (2011).
Nguyen, V. T. et al. In vivo degradation of RNA polymerase II largest subunit triggered by α-amanitin. Nucleic Acids Res. 24, 2924–2929 (1996).
Seiler, M. et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296 (2018).
Gallo, D. et al. CCNE1 amplification is synthetic lethal with PKMYT1 kinase inhibition. Nature 604, 749–756 (2022).
van Schie, J. J. M. et al. MMS22L-TONSL functions in sister chromatid cohesion in a pathway parallel to DSCC1-RFC. Life Sci. Alliance 6, e202201596 (2023).
He, B. et al. Lmx1b and FoxC combinatorially regulate podocin expression in podocytes. J. Am. Soc. Nephrol. 25, 2764–2777 (2014).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Hart, T. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015).
Davo-Martinez, C. et al. Different SWI/SNF complexes coordinately promote R-loop- and RAD52-dependent transcription-coupled homologous recombination. Nucleic Acids Res. 51, 9055–9074 (2023).
Lezaja, A. et al. RPA shields inherited DNA lesions for post-mitotic DNA synthesis. Nat. Commun. 12, 3827 (2021).
Gaggioli, V. et al. Dynamic de novo heterochromatin assembly and disassembly at replication forks ensures fork stability. Nat. Cell Biol. 25, 1017–1032 (2023).
Jackson, D. A. & Pombo, A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998).
Sanz, L. A. & Chedin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 14, 1734–1755 (2019).
Andrade-Lima, L. C., Veloso, A., Paulsen, M. T., Menck, C. F. & Ljungman, M. DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res. 43, 2744–2756 (2015).
Gregersen, L. H., Mitter, R. & Svejstrup, J. Q. Using TTchem-seq for profiling nascent transcription and measuring transcript elongation. Nat. Protoc. 15, 604–627 (2020).
Liu, Y., Wu, X., d’Aubenton-Carafa, Y., Thermes, C. & Chen, C. L. OKseqHMM: a genome-wide replication fork directionality analysis toolkit. Nucleic Acids Res. 51, e22 (2023).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arXiv.1303.3997 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2019).
Acknowledgements
We thank current and former members of the M.S.L. laboratory. We acknowledge A. J. L. de Groot for the purification of catalytically inactive GFP-tagged RNaseH1. The M.S.L. laboratory was supported by a Netherlands Scientific Organization Vici grant (VI.C.212.005), an ENW grant (OCENW.KLEIN.090) and an ERC consolidator grant (101043815; STOP-FIX-GO); M.S.L. and R.M.F.W. were jointly supported by the Netherlands Scientific Organization (ENW grant OCENW.M20.056); R.M.F.W. and K.d.L. were supported by the ADORE Foundation (project ADORE2024-9-01; ‘Cut-to-the Chase’); the S.M.N. laboratory was supported by the Netherlands Scientific Organization (NWO Vidi grant 192.039), the Dutch Cancer Foundation (KWF 13472) and the Oncode Institute; the H.v.A. laboratory was supported by a Netherlands Scientific Organization Vici grant (VI.C.182.052); the A.C.O.V. laboratory was supported by an ERC starting grant (310913; ‘Decoding SUMO’); the V.T. laboratory was supported by the Canadian Institutes of Health Research (grant PJT-173358) and the Ush1F Collaborative; the M. Ljungman laboratory was supported by the National Human Genome Research Institute (grant 5UM1HG009382) and the National Cancer Institute (grant 5R01CA213214); the M.A. laboratory was supported by the Swiss National Science Foundation (grants 197003 and 10000233); the A.v.O. laboratory was supported by an ERC advanced grant (101053581; scTranslatomics); and the S.H. laboratory was supported by an ERC starting grant (852798; ConflictResolution). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.U.: conceptualization, investigation (performed S9.6 and GFP–RNaseH1(D210N) R-loop staining; performed DNA fibre measurements of fork speed, symmetry, origin firing and S1 nuclease; generated TTchem–seq and ChIP–seq samples; and performed immunoprecipitation for western blot analysis), validation, formal analysis, writing (original draft, and review and editing) and visualization. D.E.C.B.: conceptualization, iInvestigation (generated and validated cell lines; generated plasmids and viruses; performed immunoprecipitation for mass spectrometry and western blot analysis; performed cilia co-localization by microscopy; performed competition assay by flow cytometry; and generated ChIP–seq, BrU–seq, DRIP–seq and TTchem–seq samples), methodology, validation, writing (original draft) and visualization. P.W.C.: investigation (performed mRNA and morpholino microinjection in zebrafish, cfap20 genotyping and microscopic analysis) and validation. M. Lalonde: investigation (performed DRIP–qPCR on HO and CD constructs and performed western blot analysis). A.P.: investigation (performed quantitative image-based cytometry experiments). G.Y.: investigation (performed CD437 CRISPR screen and colony survival assay). I.K.: investigation (performed CD437 CRISPR screen). K.d.L.: formal analysis (CRISPR screen). M.v.d.W.: methodology (creation of the TY1-CFAP20 cell line). T.J.W.: investigation (performed CellTiter-Glo cell viability assay). S.J.B.: validation. A.P.W.: investigation (performed recovery of RNA synthesis assay). N.K.v.O.: formal analysis (mass spectrometry). N.S.: investigation (fork speed DNA fibres upon CD437 treatment). J.L.: validation. M. Ljungman: supervision (BrU–seq samples) and funding acquisition. A.v.O.: supervision (scEdU–seq) and funding acquisition. H.v.A.: supervision and funding acquisition. A.C.O.V.: supervision (mass-spectrometry analysis) and funding acquisition. S.M.N.: supervision (CellTiter-Glo assay) and funding acquisition. R.M.F.W.: supervision (CRISPR screen) and funding acquisition. M.A.: supervision (quantitative image-based cytometry experiments) and funding acquisition. S.H.: supervision (DRIP–qPCR) and funding acquisition. V.T.: supervision (zebrafish experiments) and funding acquisition. J.v.d.B.: methodology, investigation (generated and analysed scEdU–seq data), software, formal analysis and visualization. D.v.d.H.: conceptualization, software, formal analysis (analysed ChIP–seq, DRIP–seq, BrU–seq and TTchem–seq data and performed alignment with OK–seq), visualization and writing (original draft). M.S.L.: conceptualization, software (AlphaFold models), formal analysis (CRISPR screen and mass spectrometry), writing (original draft, and review and editing), visualization, supervision, funding acquisition and project administration.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Marco Saponaro and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Comparison of origin mapping by OK–seq, Ori–seq-HU and scEdU–seq.
a, Comparison of analysis of published OK–seq data5 with Ori–seq in HU-treated RPE1 cells20 in a 5 Mbp region of chromosome 2. b, Mean replication timing based on scEdU–seq21 of origins mapped by OK–seq5 in unperturbed RPE1 cells. n = 402 cells. The box plot is defined by the median ± interquartile range (IQR) and whiskers are 1.5 x IQR. c, Comparison of Ori–seq from20, OK–seq from5, and scEdU–seq in unperturbed RPE1 cells from21. Histone marks (H3K36me3 and H3K9me3) and nascent transcription (scEU–seq) are shown for comparison.
Extended Data Fig. 2 Generation of S-curves and metaprofiles and alignment to TSSs.
a, Step-by-step generation of S-curves aligned to published OK–seq5 and distinction between HO and CD orientation. b, Heat map obtained from S-curves of RNAPII ChIP–seq (green), BrU–seq (blue) and DRIP–seq (red) from −5 kb before TSS to +25 kb downstream the TSS. c, Metaprofile analysis of the second repeat for each sequencing showed, as in Fig. 1c. Data are Trimmean 0.1 to remove extreme values. d, Workflow for the generation of metaprofiles within a 25-kb window starting immediately adjacent to origins and extending up to 75 kb away in HO and CD orientations starting from the S-curve aligned to OK–seq5.
Extended Data Fig. 3 Validation of CRISPR screens and bona fide R-loop signal.
a, Sanger sequencing around the CFAP20 sgRNA-targeting region of the indicated cell lines. b, Quantification of clonogenic survival assay after illudin S treatment between the indicated conditions. The coloured line represents the mean of all independent experiments. Error bars represent standard deviation of n = 2 independent experiments. Statistical significance between WT and CFAP20-KO was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.7449, 0.4192, 0.0173, 0.0056. c, Quantification of clonogenic survival assay after CD437 treatment between the indicated conditions. The coloured line represents the mean of all independent experiments. Error bars represent standard deviation of n = 4 independent experiments. Statistical significance between WT and CFAP20-KO was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.0831, <0.0001, <0.0001. d, Quantification of the RNA recovery synthesis of the indicated conditions. Cells were either untreated, or analysed at 3 and 24 h after irradiation with 12 J/m2 UV-C. Each coloured circle represents 1 cell. Each black circle represents the median of an independent experiment (>100 cells). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.6417, <0.0001. e, Left: immunofluorescent labelling of R-loops using the S9.6 antibody. Scale bar, 10 μm. Right: Quantification of the nuclear R-loop signal for the indicated stable cell lines and conditions. Each coloured circle represents 1 cell. Each black circle represents the median of an independent experiment (>100 cells). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Dunnett’s correction for multiple testing. P-values shown are respectively 0.0233, 0.0345. f, Visualization of a CFAP20 point mutational analysis across 231 non-redundant cancer genome sequencing studies. This revealed the presence of a recurrent p.R100C/H amino acid substitution in tumours derived from thirteen different patients. See Supplementary Table 1 for details on tumour types. g, Left: labelling of R-loops using purified GFP-tagged RNaseH1(D210N). The results are identical to results obtained using the S9.6 antibody. Scale bar, 10 μm. Right: quantification of the nuclear R-loop signal for the indicated stable cell lines and conditions. Each coloured circle represents 1 cell. Each black circle represents the median of an independent experiment (>100 cells). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Dunnett’s correction for multiple testing. P-values shown are respectively 0.0363, 0.9493.
Extended Data Fig. 4 Depletion of Mediator in CFAP20-KO cells rescues cell-cycle exit, but not body-axis curvature.
a, Co-localization of GFP–CFAP20(R100C) with primary cilium marker acetylated α-tubulin. Scale bar, 10 μm. b, Representative microscopy images of a flow-cytometry-based competition assay between WT cells (GFP-NLS) and CFAP20-KO cells (mCherry-NLS), or between CFAP20-KO rescued with GFP-CFAP20 or CFAP20-KO cells (mCherry-NLS). Scale bar, 20 μm. Example of gating strategy used in Supplementary Fig. 1. c, Outline of the CRISPR screen in CFAP20-KO cells. d, Sanger sequencing around the CCNC sgRNA-targeting region of the indicated cell lines. e, Western blot analysis for CCNC and HSPA4 as a loading control in the indicated cell lines. Raw blot available in Supplementary Fig. 2. f, Quantitative image-based cytometry in the indicated RPE1 cell lines after staining for cyclin A as shown in Fig. 2g,h. Red box indicates the population of cells in G2-phase with low cyclin A levels. g, Quantification of % of G2 cells with low cyclin A levels. n = 3 independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Dunnett’s correction for multiple testing. P-values shown are respectively 0.0037, 0.7235. h, Morpholino-mediated knockdown of ccnc causes microphthalmia, oedema and reduced trunk diameter in a dose-dependent manner. At 4 ng ccnc MO doses, many severe developmental deformities were observed and there was increased mortality. (Uninjected larvae n = 90, 1 ng ccnc n = 71, 2 ng ccnc n = 37, 4 ng ccnc n = 23) i, The knockdown of ccnc does not rescue the motile cilia mediated body curvature phenotypes of cfap20−/− homozygotes. Morphants display ventral body curvature in addition to microphthalmia and oedema. (Control n = 12, ccnc n = 11.)
Extended Data Fig. 5 Depletion of Mediator, but not Integrator, in CFAP20-KO cells rescues R-loop accumulation.
a, Quantification of nuclear R-loop signal from indicated stable cell lines. Data as in Fig. 1j; significance by one-way ANOVA with Dunnett’s correction. P-values shown are respectively 0.0001, 0.4714, 0.9932. b, Quantification of the nuclear R-loop signal for the indicated stable cell lines and conditions using purified GFP-RNaseH1(D210N). Each coloured circle represents 1 cell. Each black circle represents the mean of an independent experiment (>100 cells). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Dunnett’s correction for multiple testing. P-values shown are respectively 0.0218, >0.9999, 0.9983. c, Top: western blot analysis 6 days post-transfection with crRNA for INTS9, using antibodies against INTS9 and Tubulin as a loading control in the indicated cell lines. Raw blot available in Supplementary Fig. 2. Bottom: representative images of immunofluorescent labelling of R-loops using the S9.6 antibody in the indicated conditions. Scale bar, 10 μm. d, Quantification of relative nuclear RNA:DNA hybrid intensity in the indicated cell lines from c. Each coloured circle represents 1 cell. Each black circle represents the median of an independent experiment (>100 cells). The black lines represent the mean of all independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.0037, 0.0054, 0.8483. e, Cartoon of interaction of CCNC WT (yellow) and mutant (blue) with core Mediator. f,g, Volcano plot depicting enriched proteins after pull-down of WT GFP–CCNC (f) or GFP–CCNC(D182A) (g) relative to GFP-NLS pull-down analysed by label-free MS in quadruplicate. Highlighted are subunits of the mediator complex (orange). Statistical analysis was performed using t-tests (FDR = 0.05, S0 = 0.1). h, Co-immunoprecipitation of WT GFP–CCNC or GFP–CCNC(D182A) stably expressed in CCNC-KO or CFAP20/CCNC-dKO cells. The input is 0.5% of the total protein lysate. Raw blot available in Supplementary Fig. 2. i, Heat maps around TSS of RNAPII ChIP–seq for 3,000 BrU–seq positive genes >3 kb in indicated RPE1 cells.
Extended Data Fig. 6 CFAP20-KO cells accumulate R-loops at CD collisions.
a,b, Heat maps of DRIP–seq in the indicated cell lines and conditions aligned around 508 TTSs of genes > 50 kb. c, Violin plot of the median replication time (based on scEdU–seq) of regions containing R-loops versus all other regions (based on DRIP–seq). n = 402 as in Extended Data Fig. 1b. The box plot is defined by the median ± interquartile range (IQR) and whiskers are 1.5 x IQR. Note that regions with R-loops replicate early. d, Analysis of DRIP–seq signal in sense and anti-sense transcription based on BrU–seq. Differences in amount of anti-sense transcripts does not affect the relative increase in DRIP–seq signal around TSSs in CFAP20-KO vs WT cells. Metaprofile data are Trimmean 0.1 to remove extreme values. e, Representation of sense and anti-sense transcripts around TSSs in HO and CD orientation within 25-kb from the origins. f,g, DRIP–qPCR analysis of HEK293T cells on mAIRN HO or CD plasmids (f) or the endogenous RPL13A locus (g), transfected with siCTRL or siCFAP20 siRNAs ± DOX for 48 h. DRIP signals around each locus are shown as % input. n = 3 independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown in f are respectively 0.4616, <0.0001 and in g are respectively 0.0015, 0.0132. h, Western blot analysis of HEK293T cells containing the mAIRN HO/CD episomal plasmids after DOX induction for 48 h with the indicated antibodies. Raw blot available in Supplementary Fig. 2. i, Heat maps of DRIP–seq in CFAP20-KO cells aligned around origins mapped by OK–seq5.
Extended Data Fig. 7 Quality control and validation of scEdU–seq analysis.
a, Coefficient of Variation (y axis) versus average reads per bin (x axis) for all single-pulse scEdU–seq cells. Each dot is a single cell and the shaded area between two straight lines contain selected cells for subsequent analysis. b, Pearson correlation matrix of replication timing for each pseudo-bulk and WT and CFAP20-KO over three S-phase fractions (early, mid and late), number and colours indicate Pearson correlation c, Dimensionally reduced distance between single cells by UMAP for WT and CFAP20-KO cells. Each dot is a single cell, dots are coloured by S-phase progression or DNA content (based on DAPI). d, DNA content (DAPI, y-axis) versus S-phase progression (x-axis) based on scEdU–seq signal (single EdU pulse for 15 min) from WT and CFAP20-KO cells. The line indicates the fit for a linear model and the ribbon indicates the 95% standard error for the fit. e, Quantification of inter-origin-distance for the indicated cell lines and conditions used to calculate origin firing in Fig. 5b. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Dunnett’s correction for multiple testing. P-values shown are respectively 0.0160, 0.8523, 0.0073, 0.8815. f, Heat map comparison of binned IZ timing (left, log2) and IZ efficiency (right, log2) for WT (y-axis) and CFAP20-KO (x-axis) RPE1 cells. Blue dashed line indicates the IZ efficiency in WT while continuous blue line indicates IZ efficiency in CFAP20-KO cells. g, Replication forks per cell for each individual chromosome quantified from the scEdU–seq experiment WT (y-axis) and CFAP20-KO RPE1 cells.
Extended Data Fig. 8 CFAP20-KO cells first exhibit an increased fork speed, leading to decreased origin activation, independently of PARP inhibition.
a, Schematics of the experimental set-up with the indicated treatments. PARP inhibitor (olaparib) was used at a 10 µM for 17 h before labelling with CldU and IdU for 20 min each. Bottom: quantification of replication-fork speed observed in the indicated cells and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.0020, 0.2884. b, Schematics of the experimental set-up with the indicated treatments. CDC7 kinase inhibitor (XL413) was used at 60 µM for 4 h, Aphidicolin was used at 5 µM for 2 h and each treatment was kept during the labelling of DNA with CldU and IdU. c, Quantification of origins (based on inter-origin distance) observed in the indicated cells and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.0042, 0.0385, 0.2876. d, Quantification of replication-fork speed observed in the indicated cells and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.0197, 0.3621, 0.0143. e, Relative survival of the indicated cell lines exposed to increasing concentration of PARP inhibitor (olaparib) measured by the CellTiter-Glo luminescent cell viability assay. Error bars represent standard deviation of n = 3 independent experiments. Statistical significance between WT and CFAP20-KO or BRCA1-KO was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown between WT and CFAP20-KO are respectively 0.9998, 0.7674, 0.9955, >0.9999, 0.9921, >0.9999, >0.9999, 0.9321, 0.9597, >0.9999 while P-values shown between WT and BRCA1-KO are respectively >0.9999, >0.9999, 0.8527, <0.0001, <0.0001, <0.0001, < 0.0001, <0.0001, <0.0001, <0.0001.
Extended Data Fig. 9 CFAP20-KO cells induce increased fork speed through DNA pol α and ssDNA gaps through a PRIMPOL.
a, Schematics of the experimental set-up with the indicated treatments. DNA polymerase α inhibitor (CD437) was used during the last 20 min of the IdU labelling at a concentration of 1 µM. b, Quantification of replication-fork speed observed in the indicated cells and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-value shown is 0.0009. c, Schematics of the experimental set-up with the indicated treatments. DNA polymerase α inhibitor (CD437) was used at a 1 µM for 1 h during the labelling with IdU, followed by ±S1 nuclease treatment to detect ssDNA gaps. d, Quantification of IdU track length observed in the indicated cells and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black (−S1) and blue (+S1) lines represent the mean of all independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively <0.0001, 0.0424. e, Schematics of the experimental set-up with the indicated treatments. PRIMPOL knockdown was obtained after 2 days of transfection with siRNA, followed by sequential labelling with CldU and IdU. f, Western blot analysis 2 days post-transfection with siRNA against PRIMPOL, using antibodies against PRIMPOL and HSPA4 as a loading control in the indicated cell lines. Raw blot available in Supplementary Fig. 2. g, PARP inhibitor (olaparib) was used at a 10 µM for 17 h before labelling with CldU and IdU for 20 min each. Quantification of replication-fork speed observed in the indicated cells and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively <0.0001, 0.9911, 0.1748. Of note, the samples treated only with PARP inhibitor have been plotted from Extended Data Fig. 8a. h, Quantification of the sister fork symmetry for the indicated stable cell lines and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-value shown is 0.1079. i, Schematics of the experimental set-up with the indicated treatments. PRIMPOL acute knockout was obtained after 6 days of transfection with crRNA. ±S1 nuclease treatment for 30 min was used to detect ssDNA gaps. j, Western blot analysis 6 days post-transfection with 2 combined crRNA against PRIMPOL, using antibodies against PRIMPOL and Tubulin as a loading control in the indicated cell lines. Raw blot available in Supplementary Fig. 2. k, Quantification of IdU track length observed in the indicated cells and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black (−S1) and blue (+S1) lines represent the mean of all independent experiments. Statistical significance between the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.9989, <0.0001. l, Quantification of the sister fork symmetry from Fig. 5g for the indicated stable cell lines and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-value shown is <0.0001.
Extended Data Fig. 10 CFAP20(R100C) binds to RNAPII and leads to impaired elongation.
a, Violin plots showing replication timing (y-axis) for the earliest, middle, and latest occurrence of DNA replication forks (left to right plots) within a 10 kb region (each point represents a bin) in WT and CFAP20-KO cells (x-axis), stratified by regions either positive (top) or negative (bottom) for DRIP–seq signal. We observe overall faster replication-fork progression in cis regions (positive for DRIP–seq signal) compared to trans regions (negative for DRIP–seq signal), based on both the median (adjusted p-values: positive DRIP = 1.51e−7; negative DRIP = 2.0e−18, pairwise t-test) and maximum (adjusted p-values: positive DRIP = 2.14e−31; negative DRIP = 1.11e−10, pairwise t-test) replication timing in CFAP20-KO compared to WT. Notably, replication forks in bins with the earliest timing show no significant difference between WT and CFAP20-KO (adjusted p-values: positive DRIP = 0.175; negative DRIP = 0.223, pairwise t-test), suggesting that the observed effects are not due to changes in early initiation zones, but rather reflect altered progression of DNA replication forks in cis. The box plot is defined by the median ± interquartile range (IQR) and whiskers are 1.5 x IQR. b, UCSC genome browser tracks showing the read density of TTchem–seq signal at 60 min after DRB release across the OPHN1 gene in WT or CFAP20-KO cells. c, Averaged metaplots of TTchem–seq signal at 60 min after DRB release of 508 genes of at least 50-kb in CFAP20-KO cells rescued by CFAP20WTor CFAP20R100C cells. d,e, Co-immunoprecipitation of GFP from RPE1 cells expressing GFP-NLS or GFP–CFAP20 (d) and GFP-NLS, GFP–CFAP20 or GFP–CFAP20(R100C) (e). Raw blot available in Supplementary Fig. 2. The input is 0.5% of the total protein lysate. f, Western blot analysis of RNAPII degradation after α-amanitin treatment for 4 h. Raw blot available in Supplementary Fig. 2. g, Quantification of the sister fork symmetry from Fig. 5e for the indicated stable cell lines and conditions. Each coloured circle represents 1 fibre. Each black circle represents the median of an independent experiment (>100 fibres). The black lines represent the mean of all independent experiments. Statistical significance between WT and the indicated conditions and CFAP20-KO cells and the indicated conditions was determined by one-way ANOVA analysis after Šidák’s correction for multiple testing. P-values shown are respectively 0.9992, 0.7270, >0.9999, <0.0001, 0.0020, <0.0001.
Supplementary information
Supplementary Information
Supplementary Fig. 1 shows the gating strategy used in flow cytometry assays. Supplementary Fig. 2 shows all raw blots of western blot analysis and immunoprecipitation. Supplementary Table 1 contains all tumour cases for CFAP20R100C/H; Supplementary Table 2 contains all cell lines used; Supplementary Table 3 contains all compounds and kits used; Supplementary Table 4 contains all sgRNA, siRNA and crRNAs used in the article; Supplementary Table 5 contains all plasmids used; Supplementary Table 6 contains all primers used; Supplementary Table 7 contains all antibodies used; Supplementary Table 8 contains zebrafish strains and oligos used in zebrafish experiments; Supplementary Table 9 contains all programs used; and Supplementary References lists the additional references cited.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Uruci, S., Boer, D.E.C., Chrystal, P.W. et al. CFAP20 salvages arrested RNAPII from the path of co-directional replisomes. Nature (2026). https://doi.org/10.1038/s41586-025-09943-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41586-025-09943-7