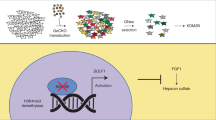
Abstract
Heparan sulfate proteoglycans (HSPGs) have been recognized as key plasma membrane-tethered co-receptors for a broad range of growth factors and cytokines containing cationic heparan-binding domains1,2. However, how HSPGs mechanistically mediate signalling at the cell surface—particularly in the context of cell surface RNA—remain poorly understood. During developmental and disease processes, vascular endothelial growth factor (VEGF-A), a heparan sulfate-binding factor, regulates endothelial cell growth and angiogenesis3. The regulatory paradigm for endothelial cell-mediated selectively of VEGF-A binding and activity has largely been focused on understanding the selective sulfation of the anionic heparan sulfate chains4,5,6,7,8. Here we examine the organizational rules of a new class of anionic cell surface conjugates, glycoRNAs9,10, and cell surface RNA-binding proteins (csRBPs11,12). Leveraging genome-scale knockout screens, we discovered that heparan sulfate biosynthesis and specifically the 6-O-sulfated forms of heparan sulfate chains are critical for the assembly of clusters of glycoRNAs and csRBPs (cell surface ribonucleoproteins (csRNPs)). Mechanistically, we show that these clusters antagonize heparan sulfate-mediated activation of ERK signalling downstream of VEGF-A. We demonstrate that the heparan sulfate-binding domain of VEGF-A165 is responsible for binding RNA, and that disrupting this interaction enhances ERK signalling and impairs vascular development both in vitro and in vivo and is conserved across species. Our study thus uncovers a previously unrecognized regulatory axis by which csRNPs negatively modulate heparan sulfate-mediated signalling in the context of angiogenesis driven by VEGF-A.
Similar content being viewed by others
Main
The extracellular matrix is a key interface that actively regulates cell–cell interactions, host–pathogen interactions, and autocrine and paracrine signalling through growth factor binding (reviewed in refs. 13,14). HSPGs are a central component of the extracellular matrix and consist of a core protein and one or more covalently linked heparan sulfate (HS) chains1. The protein interactome of HSPGs is well established and includes chemokines, cytokines, growth factors, morphogens, cell-adhesion proteins and viral components such as the SARS-CoV-2 spike protein5,15,16,17,18. HSPGs are conceptually considered as coreceptors that facilitate the formation of ligand–receptor complexes. Protein–HS interactions occur via sulfated domains of the chains, and high-level and/or site-specific sulfation of HS control the various activities ascribed to HSPGs16, which span development, physiology and disease.
In addition to HS, RNAs represent another class of negatively charged biopolymers on the cell surface (cell surface RNAs (csRNAs))9,19,20,21. Among them are glycoRNAs9, which contain sialylated and fucosylated N-glycans on small RNAs and can be detected by the sialic acid-binding immunoglobulin-type lectins (Siglec) family of receptors9. A covalent linkage between small non-coding RNA and N-glycans has been identified as the modified RNA base 3-(3-amino-3-carboxypropyl)uridine (acp3U)10. Functionally, N-glycosylation of RNA at acp3U has been shown to chemically shield this RNA modification from sending by endosomal TLRs22. GlycoRNAs have also been found to interact with lectins (for example, P-selectin) and control neutrophil recruitment in vivo23. In addition to glycoRNAs, the surface of cells also contains a selective suite of RBPs that are in physical proximity to glycoRNAs, form precise nanosized clusters and are sites of cell-penetrating peptide (for example, TAT) entry, regulated by RNA11. These csRBPs have been demonstrated on many different cell types and certain csRBPs can be selectively presented on cancer cells, in the cases of csNPM1 (ref. 12) and csU5 snRNP200 (ref. 24). Together, these reports highlight that csRNAs are not merely discarded passenger molecules but important biological players in cell surface biology. However, the specific roles of these csRNP clusters in cell signalling and development remain poorly understood.
Although insights into csRNPs are emerging, extracellular RNA (exRNA) and extracellular RNases have long been characterized, there is long-standing evidence that exRNA biology may contribute to angiogenesis. It is thought to modulate VEGF signalling; however, the precise mechanisms by which exRNA and its associated RNPs exert this regulatory effect remain unclear. For example, exRNA has been proposed to modulate VEGF activity by directly interacting with both the C terminus25 and the N terminus26 of the VEGF-A165 isoform. This RNA-mediated process appears to be sensitive to RNase treatment, as it can abrogate the observed VEGF stimulation25,27. Separately, angiogenin (ANG), an extracellular RNase, is known to promote angiogenesis (reviewed in ref. 28), yet whether it targets exRNAs is poorly defined. Of note, pharmacological inhibition of ANG orthologues in zebrafish broadly impairs vascular development, suggesting that exRNA degradation itself may have an essential regulatory role in angiogenesis during development29. Thus, despite decades of research generally on angiogenesis30,31 and RNase-mediated or exRNA-mediated angiogenic pathways27,28, it remains unresolved which specific RNA species are involved, through what receptors or molecular partners they act, and the exact mechanisms on the cell surface by which they modulate angiogenesis.
Here we uncovered a new layer of regulation of csRNPs as potential antagonist of HS-mediated signalling. We found that csRNP assembly on living cells depends on intact HS chains modified with N-sulfate and 6-O-sulfate. Functionally, VEGF-A165 can directly bind to RNA on the surface of endothelial cells, which negatively regulates its intracellular signalling cascade. The heparan-binding C terminus of VEGF-A165 can bind to glycoRNAs. Mutating key arginine residues to lysine in this domain decouples HS binding from glycoRNA binding and enabled on cell and in vivo experiments, revealing hyper-vascularization of the mouse retina and the developing vasculature in zebrafish embryos in this mutant. Together, our work nominates csRNPs as an antagonist of VEGF-A165-mediated signal transduction in endothelial cells and during angiogenesis.
Siglec-11 binds to csRNA
We previously found that Siglec-11, a member of the Siglec receptor family32, which are cell surface receptors, binds to the surface of HeLa cells in an RNA-dependent manner9, suggesting that Siglec-11 is potentially a csRNA receptor. To explore whether other members of the Siglec family are also RNA-dependent cell surface binders, we used confocal imaging and screened the 13 commercially available Siglec–Fc fusion proteins for binding to suspension (MOLM-13) and adherent (U2OS) cell lines. Only Siglec-4, Siglec-7, Siglec-9 and Siglec-11 specifically bound both cell types and only Siglec-11 was reduced after live-cell RNase treatment11 (RNase A and RNase III; Extended Data Fig. 1a–c). By contrast, although Siglec-7 and bulk cell surface sialic acids (periodate labelling33) were lost after live-cell sialidase treatment, Siglec-11 binding was not impacted (MOLM-13) or only mildly impacted (U2OS; Extended Data Fig. 1d). To test whether Siglec-11 was indeed binding near RNA, we used an anti-double-stranded RNA (dsRNA) antibody (9D5) that detects csRNA11. Co-staining of U2OS and MOLM-13 cells with 9D5 and Siglec-11 showed that 42% and 61% of 9D5 puncta overlapped with Siglec-11 on MOLM-13 and U2OS cells, respectively (Fig. 1a and Extended Data Fig. 2a,b), 34.5% and 24% of Siglec-11 puncta overlapped with 9D5 on MOLM-13 and U2OS cells, respectively. Co-staining 9D5 with Siglec-7 and separately with Siglec-9, both of which are not sensitive to RNases, showed that only 2.5% and 1.4% of 9D5 puncta overlapped with Siglec-7 or Siglec-9 in MOLM-13, and 0% and 2.8% of 9D5 puncta overlapped with Siglec-7 or Siglec-9 in U2OS, respectively. Finally, although the cell surface binding of Siglec-11 is sensitive to RNase, we next assessed whether Siglec-11 can directly interact with sialic acid-modified glycoRNAs. Using highly purified RNA (Methods), we evaluated enriched material from immunoprecipitations with Siglec-11 or an Fc control and chemically labelled for glycoRNAs (rPAL10). We found that Siglec-11 captured sialoglycoRNAs above the Fc and that this signal was sensitive to RNase treatment (Extended Data Fig. 2c). The well-correlated binding of 9D5 and Siglec-11 suggests that Siglec-11 ligands are near glycoRNA on the cell surface.
a, Representative images of U2OS cells co-stained with Siglec-11 (yellow) and 9D5 (magenta). An enlargement of the hatched box is shown, and a bright field (BF) is shown to represent the outline of the cell membrane. The nearest neighbour distance analysis of the Siglec pairs is also imaged (bottom). For each pair, the distance (µm) from that anchor (left side protein name in the figure key) to the other pair was calculated across the indicated number of cells. These values were plotted in a density histogram. b, Dot plot of genes identified in the genome-wide CRISPR-KO screen for the loss of Siglec-11 cell surface binding ranked by CRISPR score. The top 15 gene names are displayed, with cut-off shown. The inset illustrates how Siglec-11 could interact with a glycoRNA. c, Dot plot of genes identified in the genome-wide KO screen as in panel b, for the loss of 9D5 cell surface binding. The inset illustrates how 9D5 could interact with a glycoRNA. d, Upset plot analysis of genes with a score cut-off of −0.8 from the Siglec-11, 9D5 and MAA-I genome-wide KO screens. The common overlapping hits between 9D5 and Siglec-11 are highlighted in blue. The total number of hits for each intersection is noted. Statistical assessment was performed with a hypergeometric test and P values are shown without adjustment. e, Representative images of WT and KO U2OS cells stained live with 10E4, Siglec-11–Fc, 9D5 and Siglec-7 (all in red). Ab, antibody; dsRNA, double-stranded RNA; mE, mEmerald. f, Quantification of the indicated signal dot numbers and intensity per cell from panels e,g from three independent experiments. Mut, mutant. g, Representative images of the indicated cell lines stained live with anti-DDX21 and anti-hnRNP-U (both in red).
HSs organize csRNPs
To identify what mediates Siglec-11 and 9D5 binding, a genome-wide CRISPR–Cas9 gene-knockout (KO) approach34 was developed based on flow cytometry of MOLM-13 cells using Siglec-11, 9D5 and the plant lectin MAA-I, which is well characterized to selectively bind to sialic acid35 (Fig. 1b,c and Extended Data Fig. 3a). We identified 154, 187 and 246 hits in the Siglec-11, 9D5 and MAA-I screens, respectively (Fig. 1d and Supplementary Table 1). The top hits enriched in the MAA-I screen were related to sialic acid biosynthesis (for example, CMAS (#1) and NANS (#11)36; Supplementary Table 1), confirming the specificity of the screen; however, both of these genes scored poorly in the Siglec-11 and 9D5 screen. Intersectional and pairwise analysis of these enriched genes showed a more robust overlap between 9D5 and Siglec-11 than MAA-I with either of the other two probes (Fig. 1d), consistent with Siglec-11 association to the cell surface in an RNA-dependent manner. Direct inspection of the 41 genes commonly scoring between 9D5 and Siglec-11 showed consistent robust scores for each hit, whereas these genes often scored poorly in the MAA-I screen (Extended Data Fig. 3b), highlighting the overlap and common effect of these genes on both 9D5 and Siglec-11 cell surface binding. Gene ontology analysis revealed terms enriched in membrane compartments and processes including heparin biosynthetic process (Extended Data Fig. 3c). Inspection of the top hits of Siglec-11 revealed EXT1, EXT2 and UXS1 as the top three genes, which are key enzymes of HS biogenesis (Fig. 1b). The 9D5 screen also revealed EXT1 as a highly scoring hit, whereas UXS1 and EXT2 were present but below the cut-off.
To verify that HS chains are required for the tethering of Siglec-11 and RNA to the cell surface, we generated KO clones in U2OS cells. Within the Golgi, HS chains are initiated by xylosylation of proteoglycan core proteins, which depends on UDP-xylose formation catalysed by UXS1 (refs. 1,37). This is subsequently elongated by a hetero-dimeric complex formed by EXT1 and EXT2 by adding alternating residues of D-glucuronic acid (GlcA) and GlcNAc to the nascent polymer38,39,40, providing the eventual substrate for sulfation and HS-interacting proteins to bind. We stained KO cells (Extended Data Fig. 4a,b) with 10E4, a specific antibody recognizing mature HS41,42, and verified that HS was not produced (Fig. 1e,f). Moreover, EXT1 KO, EXT2 KO and UXS1 KO cells all show complete loss of both Siglec-11 and 9D5 binding. The binding of Siglec-7 and Siglec-9 was not affected on EXT2 KO cells (Fig. 1e,f and Extended Data Fig. 4c,d). The loss of 9D5 signal was selective to the cell surface as intracellular staining revealed no change inside of cells upon EXT2 KO (Extended Data Fig. 4e). To understand whether this effect was due to the enzymatic activity of EXT2, EXT2 KO cells were transfected with wild-type (WT) EXT2 or a catalytically inactive (D517N/D573N40) mutant cDNA (Extended Data Fig. 4f). Reintroduction of WT EXT2 rescued the loss of 9D5 and Siglec-11, whereas the catalytically inactive mutant did not (Fig. 1e–g). These effects were not restricted to EXT2 KO, as EXT1 KO and UXS1 KO also resulted in the loss of HS, 9D5 and Siglec-11 binding (Extended Data Fig. 4g–i). These results show the dependency of clustered csRNA on HS biosynthesis.
Given the colocalization of glycoRNA and csRBPs11, we next tested whether csRBPs are also organized by HS. On WT U2OS cells, both cs-DDX21 and cs-hnRNP-U colocalize with 9D5 (Extended Data Fig. 4j). Consistently, no observable DDX21 and hnRNP-U antibody binding occurred on the cell surface in EXT2 KO cells (Fig. 1f,g). This loss was not due to bulk changes in DDX21 or hnRNP-U as whole-cell lysates had no changes between the WT and KO (Extended Data Fig. 4k). The formation of csRBP clusters is also dependent on EXT2 enzymatic activities as reintroduction of the WT, but not the catalytically inactive (D517N/D573N)40, rescued the formation of csRBP clusters in EXT2 KO cells (Fig. 1f,g). These findings indicate that HS is a critical scaffold in modulating the recruitment and retention of RNA and their respective binding proteins (for example, csRBP), csRNPs, to the cell surface.
csRNPs depend on intact HS polymers
To determine whether intact HS chains on the cell surface facilitate csRNP clustering, we performed cell staining on U2OS cells after heparin lyase treatment (30 min, heparin lyases I/II/III). Treated cells lost the binding of 10E4, Siglec-11, 9D5, anti-DDX21 and anti-hnRNP-U, whereas Siglec-7 and Siglec-9 binding was not impacted (Extended Data Fig. 5a,b). Given the colocalization of 9D5 and Siglec-11 and sensitivity to heparin lyases, we sought to understand the spatial and temporal relationship between 9D5, Siglec-11 and HS. We therefore performed a three-colour co-staining experiment on U2OS cells and found that Siglec-11 puncta are highly correlated in localization with 10E4 puncta (Fig. 2a and Extended Data Fig. 5c,d, green line). This relationship was also true when examining 10E4, Siglec-11 and anti-DDX21 (Extended Data Fig. 5e), supporting the clustering model of csRNPs near to sites of HSPGs. We next assessed the temporal regulation on csRNP clustering by conducting a recovery experiment in cells after heparin lyase treatment, removal of the enzymes and incubation of the cells with fresh media for 0, 45, 90 or 180 min to access de novo HS production and cell surface presentation (Fig. 2a and Extended Data Fig. 5c,d). HS puncta reappeared 45 min after a recovery from heparinase treatment (9.4%). At this early time point, Siglec-11 and 9D5 clustering was not detected. After 90 min, however, 9D5 and Siglec-11 started to recover together at sites of large HS clusters, suggesting that a critical concentration of HS is needed for proper csRNP clustering. After 180 min, the levels of HS, Siglec-11 and 9D5 binding recovered to native levels, confirming that intact HS chains are vital for csRNP cluster formation on the live cell surface.
a, Representative images of U2OS cells treated with a heparinase pool for 30 min, recovering for the indicated times and co-stained with 10E4 (cyan), Siglec-11 (yellow) and 9D5 (magenta). An enlargement of the hatched box is shown, and a bright field is shown to represent the outline of the cell membrane. Three independent experiments were performed. b, Schematic of a HSPG with the regions of activity for the various enzymes and HS oligos perturbed in the following experiments. 2S, 2-O-sulfation; 6S, 6-O-sulfation; NS, N-sulfation. c, Representative images of WT, NDST1-KO-treated, HS6ST1-KO-treated, HS2ST1-KO-treated, Sulf1 stably overexpressing (OE)-treated, Sulf2 stably OE-treated, Tega HS #09-treated and Tega HS #37-treated U2OS cells stained with 10E4, Siglec-11, 9D5, anti-DDX21 and anti-hnRNP-U (all in red) separately. d, Quantification of data in panel c with the number of cells counted from three independent experiments. Data are mean ± s.e.m.
6-O-sulfated HS facilitates csRNPs
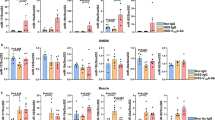
The complex but precise sulfation of HS chains16 is initiated by one or more members of the NDST family of N-deacetylases-N-sulfotransferases (NDSTs) acting on a subset of GlcNAc residues (Fig. 2b). NDST1 KO in U2OS cells (Extended Data Fig. 6a–c) resulted in a 74% loss of N-sulfoglucosamine residues as measured by reduction of 10E4 staining (Fig. 2c,d). NDST1 deficiency caused a 77%, 72%, 62% and 63% loss of binding intensity per cell in Siglec-11, 9D5, cs-DDX21 and cs-hnRNP-U, respectively (Fig. 2c,d), suggesting that sulfation of HS chains is required for tethering RNA and associated proteins to the cell surface. To broadly and non-genetically inhibit sulfation, we treated cells with sodium chlorate, a metabolic inhibitor of sulfation43, and found near complete loss of 10E4, Siglec-11, 9D5, cs-DDX21 and cs-hnRNP-U, whereas sodium chloride (control) had no effect (Extended Data Fig. 6d). Correspondingly, 3G10 antibody specific for cut HS41, only bound cells after heparin lyase treatment (Extended Data Fig. 6e).
Following N-sulfation, partial epimerization of adjacent GlcA residues to L-iduronic acid (IdoA) can lead to 2-O-sulfation (catalysed by Hs2st), 6-O-sulfation of GlcNAc and GlcNS residues (by HS6ST1–3)37,44. We therefore generated HS2ST1-KO and HS6ST1-KO U2OS cells (Extended Data Fig. 6a–c). Loss of 2-O-sulfation via HS2ST1 KO did not significantly alter the binding of the antibody panel, whereas loss of 6-O-sulfation through HS6ST1 KO reduced Siglec-11, 9D5, cs-DDX21 and cs-hnRNP-U binding by 76%, 67%, 56% and 53%, respectively (Fig. 2c,d). To complement this result, we individually overexpressed Sulf1 or Sulf2, two extracellular sulfatases that can remove 6-O-sulfation from intact HS45,46 (Fig. 2b and Extended Data Fig. 6f,g). Indeed, overexpression of these sulfatases removed total Siglec-11, 9D5 binding, cs-DDX21 and cs-hnRNP-U on U2OS cells (Fig. 2c,d). Finally, we performed a competition experiment by adding exogenous HS chains with high N-sulfation, 6-O-sulfation and 2-O-sulfation (rHS09) to the media. rHS09 caused a loss of clustering of Siglec-11, 9D5, cs-DDX21 and cs-hnRNP-U, whereas the addition of HS chains with only high N-sulfation and 6-O-sulfation (rHS37) increased the average level of binding twofold to threefold (Fig. 2b–d). Although both rHS37 and rHS09 are highly sulfated, rHS37 merely lacks the 2-O-sulfate groups. Combined with the genetic evidence, this indicates that N-sulfation and 6-O-sulfation of HS promotes csRNP formation.
csRNAs regulate VEGF-A165 signalling
Functionally, HSPGs bind to extracellular growth factors, facilitating signal transduction by forming a growth factor–growth factor receptor complex on the cell surface16. VEGF-A is a prototypical pro-angiogenesis factor causing endothelial cell migration and proliferation, with certain forms dependent on 6-O-sulfated HS for cell binding7,47. Upon activation by VEGF-A, the VEGF receptor (VEGFR), a receptor tyrosine kinases expressed on endothelial cells48,49, signals through activation of RAS–RAF1–MEK, which in turn phosphorylates ERK1/215. Given this, we tested the presence and enzyme sensitivity of csRNPs on human umbilical vein endothelial cells (HUVECs). HUVECs generally stain positively for the same ligands tested on cancer cells and heparin lyase treatment resulted in a complete loss of binding across the antibody panel (Fig. 3a and Extended Data Fig. 7a). Treatment of HUVECs with RNases reduced binding of Siglec-11, 9D5, cs-DDX21 and cs-hnRNP-U puncta by 73%, 83%, 96% and 93%, respectively, without affecting 10E4 staining. These features of csRNP assembly, including heparin lyase and RNase sensitive, were also consistent on primary cultures of keratinocytes (Extended Data Fig. 7b,c), confirming that control of csRNP clustering by HSPGs is conserved on primary cells such as endothelial cells and keratinocytes.
a, Quantification of 10E4, Siglec-11, 9D5, cs-DDX21 and cs-hnRNP-U intensity per cell from three independent staining experiments on HUVECs. Data are mean ± s.e.m. a.u., arbitrary units. b, Western blot analysis of whole-cell lysates isolated from HUVECs after starvation and treatment with an RNase pool followed by 3 ng ml−1 VEGF-A165, VEGF-A121 or EGF stimulation (top). Quantification of the ratio of pERK to total ERK was also calculated across the biological triplicates (bottom). Statistical assessment was performed with a two-sided Student’s t-test and P values are shown. Data are mean ± s.e.m. c, Representative images of starved HUVECs treated with an RNase pool, and subsequently with VEGF-A165 or VEGF-A121, finally stained with anti-VEGF-A165 (red) or anti-VEGF-A (red). Statistical assessment was performed with a Student’s t-test and P values are shown. Data are mean ± s.e.m. NS, not significant. d, Representative images of HUVECs treated with an RNase pool and stained with anti-VEGFR2 (red). Statistical assessment was performed with a Student’s t-test and P values are shown. Data are mean ± s.e.m. e, Representative images of starved HUVECs treated with VEGF-A165 and then co-stained with anti-VEGF-A165 (purple) and Siglec-11 (yellow). Three independent experiments were performed. f, Schematic of the microfluidic chip (top left) used to grow HUVECs without (top middle) and with (top right) RNase A for 6 days. Representative images of BFP expressed in the HUVECs. Statistical assessment of the total migration area was performed using an unpaired two-sided Student’s t-test (bottom). Data are mean ± s.e.m. Four independent experiments were performed. g, Representative image (maximum z-projection view (left) and z-projection slice view (right)) of sprouts from a +RNase A device for the cells (blue), F-actin (red) and PECAM1 (green). Four independent experiments were performed.
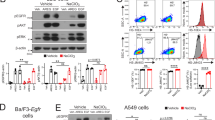
Various proteoforms of VEGF-A, including VEGF-A121 and VEGF-A165, bind to the VEGFR via their N-terminal domains; however, VEGF-A165 contains an extended C terminus that enables interactions with HS and neuropilin-1 (a VEGFR coreceptor)50,51. We hypothesized that this domain could also interact with glycoRNAs. To test this, we serum starved HUVECs and used western blotting to detect total and phosphorylated ERK (pERK) after VEGF-A stimulation. Upon stimulation with 3 ng ml−1 of VEGF-A165 or VEGF-A121 for 5 min, we observed an increase in pERK levels in serum-starved HUVECs (Fig. 3b, brown bars). Pre-treatment of HUVECs with RNases before the addition of VEGF-A165 resulted in a 3-fold and 2.1-fold increase in pERK levels compared with cells without RNase treatment (Fig. 3b, green bars), and VEGF-A121 activity was RNase insensitive. The RNase-dependent effect of VEGF-A165 (and the insensitivity of VEGF-A121) is also seen at 25 ng ml−1 (Extended Data Fig. 8a–c). The RNase-dependent enhancement occurred through VEGFR2 signalling (at tyrosine 1175), as we observed detectable pVEGFR2 that was enhanced by 2.69-fold upon RNase treatment of the HUVECs (Extended Data Fig. 8d). Consistently, in vitro binding analysis with microscale thermophoresis showed that only VEGF-A165, but not VEGF-A121, was capable of interacting with small RNA (Extended Data Fig. 8e). These findings suggest that csRNA antagonizes VEGF-A165 activation of ERK signalling at least in part through VEGFR2. This effect was specific to VEGF-A165 as epidermal growth factor (EGF), which also signals through the ERK pathway but does not have a HS-binding domain52, was able to fully induce pERK in the presence of RNase (Fig. 3b). These data suggest that growth factors bearing a HS-binding domain are probably subject to RNase modulation.
We next assessed whether VEGF-A165 is recruited to csRNP clusters and whether they impact cell surface association of VEGF-A165. Serum starvation and pre-treatment with the RNases led to approximately two times more binding of VEGF-A165 to the cell surface of HUVECs, whereas VEGF-A121 was unchanged (Fig. 3c). The observed increase in VEGF-A165 cell binding was not due to changes in its receptor VEGFR2 (ref. 3), as we found no changes in the receptor abundance on the cell surface after RNase treatment (Fig. 3d). Co-staining of VEGF-A165 and Siglec-11 on HUVECs revealed 78% of Siglec-11 puncta overlapped with VEGF-A165 and 40% of VEGF-A165 puncta overlapped with Siglec-11 (Fig. 3e and Extended Data Fig. 8f,g). These data support the binding of VEGF-A165 at assembled csRNPs and that RNase treatment selectively increases VEGF-A165 cell binding.
RNase-dependent changes in pERK occur quickly (within 5 min) after VEGF-A165 addition, suggesting that downstream gene expression and cell activation could be impacted in the HUVECs. To test this, we leveraged a multi-channel microfluidic chip to assess the effect of long-term RNase A treatment on HUVECs. Blue fluorescent protein (BFP)-labelled HUVECs were pre-treated with or without RNase A and loaded into the left channel with polymerized collagen in the central channel (Fig. 3f, left). Growth into the cell-free collagen region was evaluated after 6 days (fresh RNase A added daily). RNase A treatment significantly enhanced the total migration area (BFP signal) of the HUVECs into the 3D matrix (Fig. 3f and Extended Data Fig. 8h). Staining the extended HUVEC protrusions for F-actin (red) and PECAM1 (green) demonstrated cytoskeleton and cell–cell junctions, respectively, highlighting productive expansion and growth into the collagen (Fig. 3g and Extended Data Fig. 8i). 3D reconstructions of the finger-like protrusions revealed hollow, lumen-like, tubular formations, providing evidence that the RNase A-treated HUVECs can generate structures reminiscent of vessels. The enhanced growth with RNase A treatment observed in 3D further suggests that csRNA is antagonistic to VEGF-A activity.
VEGF-A165 selects RNA using arginines
Although previous reports have suggested that exRNA could interact with either the C terminus25 or N terzminus26 of VEGF-A165, our data (Fig. 3) point to the C-terminal HS-binding domain (Fig. 4a). To test whether VEGF-A165 directly interacts with RNA on the surface of cells, we starved HUVECs for 4 h, added VEGF-A165 and UV-C (254 nm) crosslinked the culture to produce covalent bonds between any RNA and VEGF-A165. We then immunoprecipitated the VEGF-A165 with a validated anti-VEGF-A antibody and found that without UV-C, no RNA was crosslinked to VEGF-A165; however, upon crosslinking, the native migrating band disappears (Extended Data Fig. 9a,b). We predicted that this was due to covalent UV crosslinking of VEGF-A165 to RNA; consistent with this, RNase treatment of the lysate after UV exposure restored the native migrating VEGF-A165 band (Extended Data Fig. 9b). Next, we performed RNA immunoprecipitation sequencing using VEGF-A165 to determine the specificity of RNA binding. Using small RNAs from HUVECs and capturing with the anti-VEGF-A antibody, we found that VEGF-A165 enriched 235 RNAs over the input and a control omitting the VEGF-A165 (Extended Data Fig. 9c,d and Supplementary Table 2). Small nuclear RNA, rRNA, small nucleolar RNA and tRNAs were most abundant in the VEGF-A165 immunoprecipitate (Extended Data Fig. 9e). Comparing these hits to the initial set of glycoRNAs (defined in HeLa cells), we found significant overlap of 34 RNAs (Extended Data Fig. 9f and Supplementary Table 2). These data demonstrate specific preference for VEGF-A165 binding to certain RNAs, which overlap with known glycoRNA transcripts.
a, Protein structure of VEGF-A165 (left), and the amino acid sequence of VEGF-A165 WT (top right) and the arginine-to-lysine mutant (bottom right). b, Western blot analysis of whole-cell lysates isolated from HUVECs after starvation and stimulated by 25 ng ml−1 VEGF-A165HS WT or VEGF-A165HS(R/K) (left). Quantification of the ratio of pERK to total ERK was also calculated across the biological triplicates (right). Statistical assessment was performed with a two-sided Student’s t-test and P values are shown. Data are mean ± s.e.m. c, Western blot analysis of whole-cell lysates isolated from HUVECs after starvation and stimulated by 25 ng ml−1 VEGF-A165HS WT or VEGF-A165HS(R/K) with or without RNase treatment (top). Quantification of the ratio of pERK to total ERK was also calculated across the biological triplicates (bottom). Statistical assessment was performed with a two-sided Student’s t-test and P values are shown. Data are mean ± s.e.m. d, Representative images of postnatal day 6 retinas of mice 4 h post-intraocular administration of human VEGF-A165HS WT (100 ng per retina), VEGF-A165HS(R/K) (100 ng per retina) or vehicle (PBS) were immunostained with anti-ERG (red) and IB4 (white). e, Quantification of ERG numbers (left) and IB4 intensity (right) in the vascular front regions (blue dashed line area in panel d). Statistical analysis was performed using an unpaired two-sided Student’s t-test and P values are shown without adjustment. Data are mean ± s.e.m. f, Representative images of fli1a–eGFP zebrafish larvae at 36 h post-fertilization with the indicated injection of mRNA. g, Quantification of the normalized vessel length in panel f was calculated across three independent experiments. The boxplots show the median, the 25th and 75th percentiles (bounds of the box), and minimum and maximum values (whiskers). Statistical assessment was performed with a two-sided Student’s t-test and P values are shown. h, Schematic of HS–RNP complex regulation of VEGF-A signalling and angiogenesis. The schematic was created in BioRender. Flynn, R. (2025) https://BioRender.com/m98s373.
We next examined more closely the HS-binding domain of VEGF-A165, which is an arginine-rich protein domain (HS WT; Fig. 4a). Arginine-rich domains are well established to facilitate interactions with HS16 and RNA53. By introducing mutations converting arginine residues to lysine residues (R-to-K mutations) within the HS-binding domain of VEGF-A165, we hypothesized that we could selectively perturb RNA binding while preserving the overall positive charge of the domain. To this end, we generated an R/K mutant VEGF-A165 HS domain by changing all eight arginine residues in the VEGF-A165 HS domain to lysine (HS(R/K); Fig. 4a and Extended Data Fig. 9g). We expressed a WT and R/K version of VEGF-A165 and evaluated their ability to stimulate pERK after serum starvation of HUVECs. The addition of the same amount of VEGF-A165 HS(R/K) resulted in approximately twofold more pERK activation than the HS WT (Fig. 4b) as well as 2.67-fold more spots per cell (Extended Data Fig. 9h). A similar effect was seen when examining the levels of pVEGFR2 after HS WT or HS(R/K) addition to starved HUVECs (Extended Data Fig. 9i). This increase in pERK and pVEGFR2 levels is reminiscent of the twofold increase in VEGF-A165 activation after RNase treatment (Fig. 3b), suggesting that HS(R/K) has reduced detection of RNA.
We next evaluated the sensitivity of the VEGF-A165 HS(R/K) mutant to RNase treatment. The VEGF-A165 HS(R/K) was less sensitive to RNase treatment and showed high pERK levels regardless of the addition of RNase (Fig. 4c; R/K was 51% less sensitive to RNase than WT). To test whether VEGF-A interacts with sialoglycoRNA and whether this is defective in the HS(R/K) form, we performed an RNA immunoprecipitation followed by rPAL analysis. HS WT was able to enrich for the sialoglycoRNA signal that was RNase sensitive, whereas the HS(R/K) was not (Extended Data Fig. 9j). Microscale thermophoresis analysis of small RNA treated with and without sialidase also revealed a reduction (approximately 35%) in interaction with HS WT after the removal of sialic acids (Extended Data Fig. 9k). To establish whether the HS(R/K) still binds to HS, we selected rHS29, which has N-sulfation, 2-O-sulfation and 6-O-sulfation; on the basis of expression data in HUVECs54, NDST1, HS2ST1 and HS6ST1 are expressed (Extended Data Fig. 9l) and VEGF-A165 has been reported to interact with N-sulfated, 2-O-sulfated and 6-O-sulfated HS4,6. In vitro, the VEGF-A165 HS(R/K) has a similar interaction with rHS29 as the HS WT VEGF-A165 (Extended Data Fig. 9m), in line with the model that the R/K mutation has an RNA-selective phenotype.
VEGF-A165 (R/K) mutant enhances angiogenesis
In the neonatal mouse retina, intravitreal injection of exogenous VEGF-A165 leads to excessive endothelial cell accumulation, loss of vascular branching and aberrant tip cell morphology55. Using this model, we performed intravitreal injection of recombinant VEGF-A165HS WT and VEGF-A165 HS(R/K) into the eyes of postnatal day 6 mice. Retinal vasculature was assessed 4 h post-injection using isolectin B4 (IB4) staining to visualize vessels and the endothelial nuclear marker ETS-related gene (ERG) immunostaining to quantify endothelial cells. Consistent with previous findings55, both forms of VEGF-A165 significantly reduced the number of filopodia in tip cells compared with PBS controls, indicating altered tip cell morphology (Extended Data Fig. 10a). As expected, VEGF-A165HS WT induced an increase in IB4 coverage and ERG + endothelial cell abundance at the vascular front (Fig. 4d,e). Injection of HS(R/K) elicited a further enhancement of VEGF activity, marked by significantly elevated IB4 intensity and increased endothelial cell density in the vascular front (Fig. 4d,e). Staining with anti-VEGF-A in the mouse retina across the three examined conditions showed similar patterns of VEGF-A expression confirming that the intravitreal protein injection did not generally disrupt the normal expression pattens of VEGF-A (Extended Data Fig. 10b). These in vivo results are consistent with our HUVEC data where HS(R/K) potentiates VEGF-driven angiogenesis.
We next investigated the conservation of this effect in vivo using zebrafish (Danio rerio) embryos. The role of Vegfa during zebrafish development has been extensively studied (reviewed in ref. 56). Vegfa is expressed and secreted by the somites57,58, and binds to it receptors Kdrl in endothelial cells, inducing its phosphorylation and activating the Mapk cascade, which leads to Erk phosphorylation in specific endothelial cells59. We therefore leveraged Tg(fli1a–eGFP), a well-validated zebrafish transgenic model of vascular development60,61, to assess vascular effects during development. We injected in vitro-transcribed mRNA encoding either haemagglutinin (HA)–vegfaHS WT or HA–vegfaHS(R/K) at the one cell stage of Tg(fli1a–eGFP) embryos and monitored vasculature formation by eGFP fluorescence after 24 h post-fertilization. We verified that both mRNAs were expressed at similar levels by western blotting against the HA tag from protein extracts derived from the injected embryos (Extended Data Fig. 11a). Although HA–vegfaHS WT-injected embryos showed no vascularization defects in the trunk, the vegfaHS(R/K) had a significant defect in the vasculature in the zebrafish trunk displaying underdeveloped trunk vessels (Fig. 4f,g and Extended Data Fig. 11b). We also found that the vegfaHS(R/K) significantly increased the number of endothelial cells in the dorsal aorta and posterior cardinal vein region, whereas vegfaHS WT slightly reduced the number in the same area (Extended Data Fig. 11c).
Finally, to assess whether the activity of the VEGF-A165 HS domain is sufficient to alter growth factor signalling, we designed a fusion system between the HS domain and Wnt3a. Wnt3a is a signalling protein and mitogen important for embryonic development among many other processes62,63. We hypothesized that fusing Wnt3a to the VEGF-A165HS WT (Wnt3a–HS WT) would target it to csRNPs and therefore quench its bioactivity, whereas a native Wnt3a or Wnt3a–HS(R/K) fusion would result in reduced anterior development and overall axial shortening of the embryo, as previously reported64,65. As compared with uninjected embryos, Wnt3a-injected embryos had significantly shorter axial length due to hyperactivation of WNT signalling in the embryo64,65 (Extended Data Fig. 11d,e). This phenotype was ameliorated if the embryos were injected with an mRNA encoding Wnt3a–HS WT, but the axial length was shortened again after injecting Wnt3a–HS(R/K) (Extended Data Fig. 11d,e); each fusion protein was expressed (Extended Data Fig. 11f). These developmental phenotypes were accompanied by upregulation of WNT signalling downstream targets66, with the levels of 6 out of 8 targets elevated in Wnt3a and Wnt3a–HS(R/K), but not in Wnt3a–HS WT (Extended Data Fig. 11g). These data provide evidence that the enhanced activity of HS(R/K) (RNA-binding deficient) is sufficient to drive cell surface ligand binding to enable cellular and organismal phenotypes.
Discussion
HSPGs are widely appreciated to govern critical processes at the cell surface in health and disease. Here we have began to refine this view with the characterization of how glycoRNAs and csRBPs assemble into clustered RNPs nucleated around HSPGs. Through genetic and enzymatic perturbation, we have demonstrated that HS chains are important for csRNP clustering and if removed, can repopulate the cell surface within hours. Molecularly, the sulfation of the HS chains is critical, with N-sulfation and 6-O-sulfation responsible for promoting the csRNP clustering. Finally, we established a new mechanistic pathway for information to be transferred from outside to inside of a cell: through the direct binding and regulation of growth factors such as VEGF-A165 to RNAs including glycoRNAs at csRNPs on endothelial cells as demonstrated with 2D, 3D and in vivo models.
Mechanistically, we propose a new role for RNA binding to VEGF-A on the cell surface. VEGFA is composed of 8 exons and has 16 distinct mRNA isoforms reported67,68,69. VEGF165a was the first characterized and is the prototypical pro-angiogenic isoform70. Amino acids encoded in exons 6a and 7 enable HS binding and are important for bioavailability of the VEGF-A isoforms71,72,73. Lacking the HS domain (for example, 111 and 121) results in free diffusible forms, whereas incorporation of the HS-binding domain results in extracellular matrix tethering73,74. Although previous reports have suggested that exRNA operates to positively influence VEGF-A activity25,26,27, our data have demonstrated that cell-bound csRNPs are negative regulators of VEGF-A through the HS-binding C-terminal domain, and that this domain can interact with sialoglycoRNAs. This nominates a hypothesis: that features of endothelial cells themselves (which present specific types of csRNPs) can differentially regulate and precisely control the activity of VEGF-A to enable physiological angiogenesis. A central molecular paradigm that cells use to communicate with the extracellular environment is the extracellular matrix and, in particular, the assembly of complexes on HS-modified proteins. It will be of great interest to examine the mechanistic and functional role of csRNPs in regulating other well-studied factors with HS-binding domains. Consistent with our data, another recent study has also found a genetic dependency of csRBPs and csRNA (not assayed for glycans) on HSs for cell surface organization75. Furthermore, the examination of these mechanisms in more complex and diverse tissue and microenvironment settings could provide new views on intercellular communication.
To contextualize the physiological role of csRNP–VEGF-A association and move away from bulk destruction of csRNA with live-cell RNase treatment, we developed a mutant form of the HS domain of VEGF-A165 (R/K) that maintains its cationic nature and binding to HS but reduces its responsiveness to RNase-mediated destruction of csRNPs as well as losing specific interactions with sialoglycoRNA. This enabled the examination of csRNP-dependent and csRNP-independent signalling of VEGF-A on cells, in mice and in zebrafish. In all cases, the R/K mutant mirrored the RNase effect, suggesting a concerted negative action of presented csRNPs on VEGF-A165 activity. These data are also consistent with other work described in zebrafish, which have demonstrated that inhibiting secreted RNase activity resulted in defects in trunk vasculature (and spinal neuron aberrations)29. This highlights csRNPs as a ‘counterbalance’ domain that can modulate the magnitude of activity by VEGF-A.
Finally, the role of RNA in regulating biological processes is expansive and spans its catalytic, scaffolding and information-carrying capabilities76,77,78; however, these functions have traditionally been restricted to intracellular pathways. Our data have demonstrated that canonical signal transduction by VEGF-A165 across the plasma membrane is modulated by the presence of csRNPs. Together, our data establish a new type of RNA regulation where the abundance or organization of topologically csRNPs can directly modulate intracellular signalling cascades through selective interaction with growth factors, impacting cellular decision making. Understanding the evolution of HS-binding proteins, glycoRNAs and csRBPs could enable better modelling of developmental and homeostatic processes in complex cellular environments.
Methods
Cell culture
MOLM-13 cells were cultured in Roswell Park Memorial Institute 1640 Medium (Thermo Fisher Scientific). U2OS (American Type Culture Collection (ATCC)) were cultured in McCoy’s 5A Medium (Thermo Fisher Scientific). HEK293T cells were cultured in DMEM media (Thermo Fisher Scientific). MOLM-13, U2OS and HEK293T cells were supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and 1× penicillin–streptomycin (Thermo Fisher Scientific). Primary umbilical vein endothelial cells (HUVECs; ATCC) were cultured in vascular cell basal medium (ATCC) supplemented with endothelial cell growth kit-VEGF (ATCC). Keratinocyte cells (ATCC) were cultured in dermal cell basal medium (ATCC) supplemented with keratinocyte growth kit (ATCC). All the experiments on HUVECs are performed before passage 10. All cells were cultured at 37 °C with 5% CO2 and maintained as mycoplasma negative. U2OS cells were transfected with Avalanche-Omni Transfection Reagent (EZ Biosystems). Immortalized HUVECs expressing BFP (Im-HUVEC-BFP; a gift from R. Kamm at MIT)79 were cultured in VascuLife VEGF-Mv endothelial medium (Lifeline Cell Technology) and passage p15 was used for experiments.
Live-cell enzyme and chemical treatments
For RNase treatment, RNase A (Sigma) and ShortCut RNase III (New England Biolabs (NEB)) were added directly to the cell culture at a final concentration of 18 µM and 100 U ml−1, respectively. MOLM-13, U2OS, HUVECs and keratinocytes were all treated for 45 min.
For heparinase treatment, heparinase I (NEB), heparinase II (NEB) and heparinase III (NEB) were added directly to the cell culture at a final concentration of 4 units per millilitre (each) for 30 min.
For sodium chloride (NaCl) and sodium chlorate (NaClO3) treatment, NaCl and NaClO3 were added to the cell culture at a final concentration of 50 mM for 24 h.
For exogenous Tega HS chain treatment, rHS09 (TEGA Therapeutics) and rHS37 (TEGA Therapeutics) were added directly to the cell culture at a final concentration of 4 µM for 60 min.
For serum starvation, HUVECs were cultured in vascular cell basal medium (PCS100030, Bioresource Center) without endothelial cell growth kit-VEGF (Bioresource Center) after being washed briefly three times with phosphate buffer saline (PBS). VEGF-A165 (Thermo Fisher Scientific), VEGF-A121 (Thermo Fisher Scientific), EGF (Thermo Fisher Scientific), VEGF-A165HS WT (Supplementary Table 3, cell-free Escherichia coli protein production by Liberum Biotech) and VEGF-A165 HS(R/K) (Supplementary Table 3, cell-free E. coli protein production by Liberum Biotech) were then added to starved HUVECs at a final concentration of 3 ng ml−1 and 25 ng ml−1 for 5 min, separately. The WT and R/K proteoforms differ by approximately 400 Da (or approximately 1% total mass) so equal mass was added in each experiment, which is approximately equal moles.
Live-cell labelling and microscopy analysis
Adherent cells were cultured on glass coverslips #1.5 (Bioscience Tools) 24 h before labelling. MOLM-13 cells were counted and then blocked as per the manufacturer’s protocol with Human TruStain FcX (Fc block, BioLegend) for 15 min on ice before labelling. For Siglecs staining in live cells, 1 µg ml−1 of recombinant human IgG1 Fc (R&D Systems), Siglec-1–Fc chimera protein (R&D Systems), Siglec-2–Fc chimera protein (R&D Systems), Siglec-3–Fc chimera protein (R&D Systems), Siglec-4–Fc chimera protein (R&D Systems), Siglec-5–Fc chimera protein (R&D Systems), Siglec-6–Fc chimera protein (R&D Systems), Siglec-7–Fc chimera protein (R&D Systems), Siglec-8–Fc chimera protein (R&D Systems), Siglec-9–Fc chimera protein (R&D Systems), Siglec-10–Fc chimera protein (R&D Systems), Siglec-11–Fc chimera protein (R&D Systems), Siglec-14–Fc chimera protein (R&D Systems) and Siglec-15–Fc chimera protein (R&D Systems) were precomplexed with 0.5 µg ml−1 of donkey anti-human IgG AF647 (ImmunoResearch) secondary antibody in FACS buffer (0.5% BSA (Sigma) in 1× PBS) for 45 min on ice. For 9D5, 10E4 and 3G10 staining, 2.5 µg ml−1 9D5 (Absolute Antibody), 1 µg ml−1 anti-HS 10E4 (Amsbio), 1 µg ml−1 anti-HS 3G10 (Amsbio) and anti-VEGF (R&D Systems) were precomplexed with 1.25 µg ml−1 of goat anti-rabbit AF647 secondary antibody (Thermo Fisher Scientific), 0.5 µg ml−1 of goat anti-mouse AF647 secondary antibody (Thermo Fisher Scientific) or donkey anti-goat IgG AF647 (Thermo Fisher Scientific) secondary antibodies in FACS buffer for 45 min on ice, separately. For VEGF-A165, VEGF-A121 and VEGFR2 staining, 1 µg ml−1 of anti-VEGF-A165 (R&D Systems), 1 µg ml−1 of anti-VEGF-A (Proteintech), and 1 µg ml−1 anti-VEGFR2 (R&D Systems) were precomplexed with 0.5 µg ml−1 of goat anti-rabbit AF647 secondary antibody and donkey anti-goat AF647 secondary antibody (Thermo Fisher Scientific). Precomplexed antibodies were then incubated with cells for 45 min on ice. For DDX21 and hnRNP-U staining, 2.5 µg ml−1 of anti-DDX21 (Novus Biological) and anti-hnRNP-U (Proteintech) were incubated with cells for 45 min on ice. Cells were gently washed twice by FACS buffer and then stained with 2.5 µg ml−1 of goat anti-rabbit AF647 for 30 min on ice.
For 9D5 and Siglecs co-staining, recombinant human Siglec–Fc chimera proteins were precomplexed with donkey anti-human IgG AF488 (ImmunoResearch), 9D5 was precomplexed with goat anti-rabbit AF647 secondary antibody. For 9D5, Siglec-11 and 10E4 co-staining, 10E4 was precomplexed with goat anti-mouse AF568 (Thermo Fisher Scientific). For 10E4, Siglec-11 and DDX21 co-staining, 10E4 was precomplexed with goat anti-mouse AF568 (Thermo Fisher Scientific), Siglec-11 was precomplexed with donkey anti-human IgG AF488 (ImmunoResearch) and anti-DDX21 was incubated with precomplexed 10E4 and Siglec-11 on cells for 45 min on ice. Cells were gently washed twice by FACS buffer and then stained with goat anti-rabbit AF647 for 30 min on ice. For VEGF-A165 and Siglec-11 co-staining, anti-VEGF-A165 was precomplexed with donkey anti-goat AF647 secondary antibody, recombinant human Siglec-11–Fc chimera protein was precomplexed with donkey anti-human IgG AF488. For 9D5 and DDX21 or hnRNP-U co-staining, mouse host 9D5 (Absolute Antibody) was precomplexed with goat anti-mouse AF488 secondary antibody (Thermo Fisher Scientific). Sequential labelling was used for cs-DDX21 and hnRNP-U labelling as described above. Antibodies or regents were precomplexed in separate tubes with the same concentration as used in single-channel staining and then mixed before adding to cells. After staining, cells were washed three times with ice-cold PBS and a fixation was performed with 3.7% formaldehyde for 15 min at room temperature in the dark. Nuclei were stained with 0.1 µg ml−1 DAPI in PBS. MOLM-13 cells were applied to glass slides using a CytoSpin centrifuge (Thermo Fisher Scientific) at 500g for 5 min.
For mEmerald-positive U2OS cell imaging, cells were fixed with 3.7% formaldehyde for 15 min at room temperature and then stained with DAPI. Finally, all samples above were mounted in ProLong Diamond Antifade Mountant (Thermo Fisher Scientific) and a coverglass was sealed over the samples with nail polish. All samples were then imaged on a Leica SP8 STED ONE microscope with ×63 oil lens. Images were acquired using Leica LAS X software. The DAPI channel was acquired with a PMT detector while all other channels were imaged using hybrid detectors.
For quantification and statistical analysis, at least three random regions of interest (ROIs) from three independent samples were acquired across one or more z-slices. To analyse the colocalization, images were processed using Imaris Microscopy Image Analysis software (Oxford Instruments). A single z-slice from each ROI was taken, selected to be near the middle of the cells (with respect to their z-thickness), and the spot-finder function was used to identify spots of roughly 0.5 µm. In-software background subtraction was used as the default settings, and spots were selected by thresholding spot quality at the elbow of the distribution. This resulted in a series of x and y positions for each spot from each channel, which were then exported for quantitative analysis. Colocalization of spots from paired channels were analysed by implementing a custom Python script (https://github.com/FlynnLab/jonperr) to identify the nearest neighbours of each spot (in nm) with a k-d tree algorithm (scipy.spatial.KDTree). Then, the distances between nearest neighbours were calculated for each pair of targets across all ROIs and plotted in a histogram. To assess the relative fraction of each spot type (channel #1) within the spots of the other pair (channel #2), we calculated a Manders’ colocalization coefficient using the aforementioned Python script. We performed this calculation in both directions: spots of channel #1 in total channel #2 spots, and the reverse.
To quantify and compare the intensities of spots on the cell surface, Leica LAS X software was used to identify ROIs throughout the entire 4× slice z-stack. To quantify and compare the spot numbers of spots on the cell surface, Imaris was used to identify spots throughout the entire 4× slice z-stack. The mean intensities and numbers of ROIs were then divided by the cell numbers and compared across groups. Statistical analysis and data plotting were performed using GraphPad Prism 10.
Live-cell flow cytometry
MOLM-13 cells were directly counted. U2OS cells were gently lifted with Accutase (Sigma-Aldrich) for 3 min at 37 °C, quenched with growth media and then counted. For each condition, 50,000 cells were used. For antibody staining, MOLM-13 cells were blocked as per the manufacturer’s protocol with Human TruStain FcX in FACS buffer for 15 min on ice. Of recombinant human IgG1–Fc, Siglec-7–Fc chimera protein and Siglec-11–Fc chimera protein, 1 µg ml−1 was precomplexed with 0.5 µg ml−1 of donkey anti-human IgG AF647 secondary antibody in FACS buffer for 45 min on ice. Precomplexed antibodies were then added to bind cells on ice for 45 min. For live-cell periodate labelling of cell surface glycans33, cells were washed twice with cold PBS + Ca + Mg and then incubated at 4 °C in cold PBS + 1 mM sodium periodate for 5 min at 1 million cells per millilitre. Cells were then quenched with 1 mM glycerol added to the PBS, and then cells were washed twice with cold PBS. Cells were then incubated at 4 °C in cold FACS buffer + 25 µM aminooxy-biotin (Cayman Chemical) + 10 mM aniline for 30 min at 1 million cells per millilitre. Cells were mixed via pipetting halfway through the incubation. Cells were then washed once with cold 1× PBS and blocked as per the manufacturer’s protocol with Human TruStain FcX in FACS buffer for 15 min on ice. Cells were then stained for 30 min on ice with streptavidin–AF647 at 1 µg ml−1. After staining, all cells were spun at 4 °C for 3 min at 400g and supernatant was discarded. Cells were washed once with 150 µl of FACS buffer, spun under the same conditions, and finally resuspended in FACS buffer containing 0.1 µg ml−1 DAPI. Data collection occurred on a BD Biosciences LSRFortessa 3, and a gating strategy was used to isolate live, single cells, to examine antibody binding using FlowJo Software (FlowJo LLC).
Western blot
Cells were quickly rinsed with ice-cold PBS and directly lysed with sample buffer (150 mM NaCl, 50 mM Tris and 0.5% Triton X-100, pH 7.4) containing phosphatase inhibitor cocktail (Cell Signaling Technology) on ice for 15 min. Zebrafish samples were sonicated in samples buffer. After centrifugation at 12,000g for 15 min at 4 °C, lysates were heated at 95 °C for 10 min in 1× NuPAGE LDS loading buffer (Thermo Fisher Scientific) containing 5 mM dithiothreitol. Samples were then resolved by SDS–PAGE using AnyKD Criterion TGX Precast Midi Protein Gels (Bio-Rad Laboratories) and transferred to nitrocellulose membranes. Membranes were blocked in blocking buffer and incubated with primary antibodies (diluted in blocking buffer) at 4 °C overnight. After washing three times for 3 min each in 1× PBS with 0.1% Tween-20 (PBST), membranes were incubated with secondary antibodies at room temperature for 45 min, followed by the same three times PBST washing. Membranes were finally rinsed in 1× PBS and scanned on a LI-COR Odyssey CLx scanner. Images and intensity of bands were acquired using LI-COR Image Studio software.
The primary antibodies used were: mouse monoclonal anti-EXT2 (sc-514092, Santa Cruz; immunoblot 1:1,000), anti-EXT1 (sc-515144, Santa Cruz; immunoblot 1:1,000), mouse monoclonal anti-GAPDH (sc-47724, Santa Cruz; immunoblot 1:1,000), rabbit polyclonal anti-GFP (A11122, Invitrogen; immunoblot 1:1,000), mouse monoclonal anti-NDST1 (sc100790, Santa Cruz; immunoblot 1:1,000), mouse monoclonal anti-HS6ST1 (sc-398231, Santa Cruz; immunoblot 1:1,000), mouse monoclonal anti-HS2ST1 (sc-376530, Santa Cruz; immunoblot 1:1,000), rabbit monoclonal anti-phospho-p44/42 MAPK (Erk1/2(Thr202/Tyr204); 4370S, Cell Signaling Technology, immunoblot 1:500), mouse monoclonal anti-p44/42 MAPK (Erk1/2; 4696S, Cell Signaling Technology; immunoblot 1:500), rabbit monoclonal anti-phospho-VEGFR2(Tyr1175) (2478S, Cell Signaling Technology; immunoblot 1:500), rabbit polyclonal anti-VEGFR2 (26415-AP, Proteintech; immunoblot 1:500), mouse monoclonal anti-HA (sc-7392, Santa Cruz; immunoblot 1:1,000), rabbit polyclonal anti-DDX21 (NB100-1718, Novus Biologicals; immunoblot 1:500), rabbit polyclonal anti-hnRNP-U (14599-1-AP, Proteintech; immunoblot 1:500) and rabbit polyclonal anti-WNT3A (26744-1-AP, Proteintech; immunoblot 1:500). The secondary antibodies used were: IRDye 800CW goat anti-rabbit IgG secondary antibody (926032211, LI-COR Biosciences; immunoblot 1:1,000) and IRDye 800CW goat anti-mouse IgG secondary antibody (926-32210, LI-COR Biosciences; immunoblot 1:1,000).
Genome-wide CRISPR–Cas9 screening
MOLM-13 cells expressing Cas9 under blasticidin selection23 were grown as above and selected with 10 µg ml−1 blasticidin for 3 days to ensure a homogenous starting population. Starting after selection on day 0, 30 million cells were infected with 1:150 the genome-wide single-guide RNA (sgRNA) lentivirus23 in 60 ml of fresh media with 8 µg ml−1 polybrene. On day 3, cells were spun down and resuspended in 70 ml of fresh media with 1 µg ml−1 puromycin to select for sgRNA-infected cells. On day 5, the media were exchanged for fresh media with 1 µg ml−1 puromycin. On day 6, the cells were switched to normal media without puromycin for expansion. From days 7 to 18, the cells were counted and passaged as needed in fresh media to maintain a cell density between 750,000 and 2,000,000 cells per millilitre. On day 18, cells were counted: 40 million cells were saved before sorting for an input population reference and 200 million cells for each Siglec-11, 9D5 and MAA-I were saved for staining and sorting. To stain, 1,000 µg Siglec-11 was precomplexed with 270 µg secondary antibody, 450 µg 9D5 was precomplexed with 225 µg secondary antibody, and 266 µg of MAA-I was precomplexed with 266 µg streptavidin–AF647. Live-cell staining was performed as noted above with Fc blocking; however, before FACS sorting, cells were filtered over a 40-µm strainer (Corning) and stained with DAPI. Cells were selected for DAPI negative (live) and then the bottom 5% intensity of cells stained with each Siglec-11, 9D5 or MAA-I were sorted into tubes. Cells were collected in FACS buffer after sorting and spun down into pellets at 500g for 4 min at room temperature; input cells were processed in a similar manner to obtain a cell pellet. After removing the supernatant, cell pellets were frozen at −80 °C for later processing. Cells were then processed by resuspending in 200 µl 1× PBS + 5 µl RNaseA + 20 µl proteinase K and incubated at 25 °C for 5 min. Then, 200 µl of Buffer AL (Qiagen) was added and samples were carefully vortexed to mix without shearing genomic DNA (gDNA). The samples were then headed to 56 °C for 60 min. After heating, 200 µl of 100% ethanol was added, gentle vortexing was again used, and then the material was purified over Zymo columns. All spins were performed at 6,000g for 20 s; after spinning the sample through, the columns were washed twice with 80% ethanol and then the DNA was eluted with 2 × 15 µl water. For input samples the initial volumes were scaled up from 200 µl to 1,000 µl of digestion and Qiagen buffers.
To amplify the sgRNAs out of the gDNA, we performed real-time PCR and for each sample performed 12 parallel reactions each with 1 µg of input gDNA. The PCRs were 50 µl final with 200 nM forward and reverse primers with 1× Q5 PCR Master Mix (NEB); 23 cycles of 98 °C for 20 s, 65 °C for 20 s and 72 °C for 90 s were completed. After PCR, the 12 reactions were pooled and purified over a Zymo column following the manufacturer’s recommended protocol for PCR DNA. To add a final index primer for sequencing, a final round of PCR was performed by taking 100 ng of amplified sgRNA library, and five cycles of the above PCR program was run followed by Zymo column clean up. The finally indexed libraries were assessed for size and concentration on a BioAnalyzer High Sensitivity DNA Chip (Agilent). Libraries were pooled equimolar and then sequencing on the NextSeq platform (Illumina) with a 19-bp read 1 and two 8-bp index reads. For the read 1, a custom sequencing primer 5′-TCTTCCGATCTCTTGTGGAAAGGACGAAACACCG-3′ was used. Enrichment of guides and genes were analysed using the MAGeCK statistical package80 by comparing read counts from each cell line with counts from matching plasmid as the initial population.
Generation of KO and stably expressing cell lines
All CRISPR–Cas9 KO assays used PX459 (ref. 81). The target oligonucleotides used are listed in Supplementary Table 3. U2OS cells were transfected with gDNA vectors. Two days later, puromycin (ant-pr-1, Invivogen) was added to the cell culture at a final concentration of 2 µg ml−1 and the live cells were selected by flow cytometry (FACS Calibur 2, BD Science) for isolation of single clones. The expanded individual clones were screened by gDNA sequencing and western blot analysis.
cDNA for human EXT2 was amplified from the cDNA library (Takara Bio); cDNAs for Sulf1 and Sulf2 were gifts from S. Rosen (Addgene plasmid)46. All three cDNA were inserted into mEmerald-C1 (Addgene). All plasmids were verified by DNA sequencing. To generate U2OS cells stably expressing mEmerald–EXT2, mEmerald–EXT2(D517N/D573N), mEmerald–Sulf1 or mEmerald–Sulf2, cells were transfected with the indicated plasmids and selected using 200–1,000 µg µl−1 (gradually increasing) G418 (Invivogen) for 2 weeks; green-positive cells were sorted into mono-clones by flow cytometry and cultured in the presence of 200 µg µl−1 G418 for 2 weeks. Proliferated clones were verified by immunoblotting and fluorescence imaging.
UV crosslinking
25 ng ml−1 of VEGF-A165 was added to starved HUVECs for 5 min. Cells were treated by UV (60,000 µJ, approximately 2 min) on ice and then directly lysed with samples buffer. For RNase treatment, RNase A and RNase III were added to the samples at a final concentration of 1,000 ng ml−1 and 20 U ml−1, resepectively. Samples were all incubated at 37 °C for 10 min and then lysed on ice for another 10 min. After centrifugation at 12,000g for 15 min at 4 °C, lysates were incubated with 5 µl Protein-G bead (Thermo Fisher Scientific) pre-conjugated with 1 µg of anti-VEGF-A (Proteintech) at 4 °C overnight. The beads were washed three times with PBS and heated at 95 °C for 10 min in 1× NuPAGE LDS loading buffer containing 5 mM dithiothreitol. Samples were then analysed by western blot described above. Anti-VEGF-A165 (AF293-NA, R&D Systems; immunoblot 1:1,000) and donkey anti-goat IgG secondary antibody (92632214, LI-COR Biosciences; immunoblot 1:1,000) were used as primary and secondary antibodies, separately.
Microfluidic device setup
IdenTx3 microfluidic chips were purchased from AIM Biotech (#DAX-1). Each chip consists of a central 3D gel channel flanked by two media channels. Type I collagen gel was prepared using a collagen kit (Nitta Gelatin) consisting of Cellmatrix type I-A (3 mg ml−1), 10× Ham’s F12 medium and reconstitution buffer in an 8:1:1 ratio, resulting in a final collagen concentration of 2.4 mg ml−1. The chips were cultured using vascular cell basal medium supplemented with endothelial cell growth kit-VEGF, with or without 10 µM RNase A (0.121 mg ml−1 final concentration).
The central channel of the devices was first filled with 18 µl of collagen. After collagen polymerization for 30 min at 37 °C, one of the medium channels was injected with 15 µl of Im-HUVEC-BFP cell suspension (±RNase A supplementation) at a concentration of 8 million cells per millilitre. The other medium channel was filled with 15 µl of medium ± RNase A. The microfluidic chip was then tilted by 90° for 1–2 min to allow cells to adhere to the collagen gel surface. Subsequently, the four reservoirs in each chip were filled with 50 µl of medium ± RNase A. After 2 h, the medium ± RNase A in the reservoirs was replaced. The devices were cultured for 6 days, and all four reservoirs were replenished daily with 50 µl of medium ± RNase A.
Use and analysis of microfluidic devices
Cells in the idenTx3 chips were fixed with 4% paraformaldehyde in 1× PBS for 20 min and permeabilized with 0.2% Triton X-100–PBS for 5 min. Then, the cells were incubated with 1% BSA in 0.2% Triton X-100–PBS for 30 min to block nonspecific binding. The cells were rinsed with 0.2% Triton X-100–PBS two times for 5 min each and incubated overnight at 4 °C with Alexa Fluor 488-conjugated mouse anti-human platelet/endothelial cell adhesion molecule-1 (PECAM-1; Cell Signaling Technology) at a 1:300 dilution in 0.2% Triton X-100–PBS. Afterwards, the cells were rinsed with 0.2% Triton X-100–PBS three times for 5 min each and incubated with Alexa Fluor Plus 555 Phalloidin (Thermo Fisher Scientific) at a 1:400 dilution in PBS for 20 min at room temperature. Finally, the cells were rinsed three times with PBS and stored in PBS for immunofluorescence imaging.
Images were acquired using a Nikon AXR point scanning confocal microscope with ×10 and ×20 objectives (Nikon), and z-stack images were processed using the Nikon Denoise.ai software and ImageJ. The total migration area was determined by manually measuring the total area of endothelial migration starting from the original gel interface in ImageJ. Statistical analysis (unpaired t-test) was performed in GraphPad Prism 10.
In vitro immunoprecipitation and rPAL
For the small RNA fraction used for immunoprecipitation, TRIzol extractions were performed as previously described in detail9. For specifics: TRIzol was added to cultured HEK293T cells. After the initial lysis, the TRIzol’ed samples were incubated at 37 °C to further denature non-covalent interactions. Phase separation was initiated by adding 0.2× volumes of 100% chloroform, vortexing to mix, and finally spinning at 12,000g for 15 min at 4 °C. The aqueous phase was carefully removed, transferred to a fresh tube and mixed with 2× volumes of 100% ethanol. The RNA was then purified over a Zymo column. For all column cleanups, we followed the following protocol. First, 350 µl of pure water was added to each column and spun at 10,000g for 30 s, and the flowthrough was discarded. This step was repeated until all the precipitated RNA was passed over the column once. The column was then washed three times total: once using 400 µl of RNA Prep Buffer (3 M GuHCl in 80% ethanol) and twice with 400 µl of 80% ethanol. The RNA was then treated with proteinase K (Ambion) on the column. Proteinase K was diluted 1:19 in water and added directly to the column matrix and then allowed to incubate on the column at 37 °C for 45 min. Next, eluted RNA was spun out into fresh tubes and a second elution with water was performed. To the eluate, 1.5 µg of the mucinase StcE (Sigma-Aldrich) was added for every 50 µl of RNA and placed at 37 °C for 30 min to digest. The RNA was then cleaned up again using the Zymo column. Of RNA-binding buffer, 2× volumes were added and vortexed, and then 2× (samples + buffer) of 100% ethanol were added and vortexed. After total RNA extraction, the RNA can be further processed to fractionate small RNA (approximately 17–200 nt) and large RNA (more than 200 nt) using Zymo columns, as recommended by the manufacturer. First, an adjusted RNA-binding buffer was made by mixing equal volumes of RNA-binding buffer and 100% ethanol. Two volumes of the adjusted buffer were added to the total RNA and were vortexed thoroughly to mix. The sample was then bound to the column, but the flow through (small RNA fraction) was saved. One volume of 100% ethanol was added to the small RNA and vortexed to mix. The small RNA was then bound to new columns, cleaned up using 2 × 400 µl 80% ethanol, and finally eluted using water. Lyophilized RNA was resuspended in the size-exclusion chromatography buffer (6 M guanidine hydrochloride (Sigma) in 1× PBS) and loaded onto a superose 6 (5/150, Cytiva) column. The sample was run at a flow rate of 0.3 ml min−1 and finally collected into 150-µl fractions. The RNA fractions were cleaned on Zymo column before immunoprecipitation.
For Siglec-11 immunoprecipitation, 5 µl Protein-A bead was pre-conjugated with 10 pmol of IgG–Fc or Siglec-11 in samples buffer for 1 h at room temperature. After washing three times with samples buffer, beads were incubated with 1 µg of small RNA for 3 h at 4 °C. After washing three times with samples buffer, the beads were suspended in 100 µl RNA-binding buffer and heated for 5 min at 55 °C. The beads were removed and the RNA extract solution was transferred to a new tube. Here, 50 µl of pure water was added and vortexed for 10 s, and then 300 µl of 100% ethanol was added and vortexed for 10 s. The RNAs were purified over a Zymo column.
For VEGF immunoprecipitation, 5 µl Protein-G bead was pre-conjugated with 1 µg of anti-VEGF-A (Proteintech) antibodies in samples buffer for 1 h at 4 °C. After washing three times with samples buffer, beads were incubated with 5 µg of VEGF-A165HS WT and VEGF-A165HS(R/K) immunoprecipitation for 2 h at 4 °C. The beads were washed three times with samples buffer and then incubated with small RNA for 2 h at 4 °C. After washing three times with samples buffer, the beads were suspended in 50 µl RNA-binding buffer and heated for 5 min at 55 °C. The beads were removed and the RNA extract solution was transferred to a new tube. Here, 100 µl of pure water was added and vortexed for 10 s, and then 300 µl of 100% ethanol was added and vortexed for 10 s. The RNAs were purified over a Zymo column.
For rPAL labelling, experiments were performed as previously described10. Specifically, lyophilized RNAs were suspend with 28 µl blocking buffer (1 µl 16 mM mPEG3-Ald (BroadPharm), 15 µl 1 M MgSO4 and 12 µl 1 M NH4OAc pH5 (with HCl)) and then incubated for 45 min at 37 °C. One microlitre of 30 mM aldehyde reactive probe (ARP/aminooxy biotin, Cayman Chemicals) was added first, then 2 µl of 2 mM NaIO4 (periodate) was added. The periodate was allowed to perform oxidation for exactly 10 min at room temperature in the dark. The periodate was then quenched by adding 3 µl of 22 mM sodium sulfite. The reaction was allowed to proceed for 5 min at 25 °C and then moved to 35 °C for 90 min. The reaction was then cleaned up by the Zymo column. The RNAs were eluted from the column using 2 × 6.2 µl water. For in vitro RNase treatment, the RNase cocktail enzyme mix (Thermo Scientific) was added directly to RNA samples with RNase buffer (200 mM Tris pH 7.5, 1 M KCl and 1 mM MgCl2) at a final concentration of 100 U ml−1 and 4,000 U ml−1, separately. The digestion was incubated at 37 °C for 1 h. After RNase digestion, the samples (digested or mock treated) were directly combined with Gel Loading Buffer II (95% formamide, 18 mM EDTA and 0.025% SDS) with a final concentration of 1× SybrGold (Thermo Fisher Scientific) and denatured at 55 °C for 10 min. Immediately after this incubation, the RNA was placed on ice for at least 2 min. The samples were then loaded into a 1% agarose, 0.75% formaldehyde and 1.5× MOPS buffer (Lonza) denaturing gel. RNA was electrophoresed in 1× MOPS at 115 V for 34–45 min, depending on the length of the gel. Subsequently, the RNA was visualized on a UV gel imager. The RNA was transferred with 3 M NaCl pH 1 (with HCl) to a nitrocellulose membrane for 90 min at 25 °C. Post-transfer, the membrane was rinsed in 1× PBS and dried on Whatman Paper (GE Healthcare). Dried membranes were rehydrated in Intercept Protein-Free Blocking buffer (LI-COR Biosciences) for 30 min at room temperature. After the blocking, the membranes were stained using IRDye IR800 streptavidin for 30 min at 25 °C. Excess streptavidin–IR800 was washed from the membranes using three washes with 0.1% Tween-20 in 1× PBS for 3 min each at 25 °C. The membranes were scanned on a LI-COR Odyssey CLx scanner (LI-COR Biosciences). Images were acquired using LI-COR Image Studio software.
VEGF immunoprecipitation sequencing
Of Protein-G beads, 5 µl was pre-conjugated with 1 µg of anti-VEGF-A antibody (Proteintech) in samples buffer for 1 h at 4 °C. After washing three times with samples buffer, beads were incubated with 1 µg of VEGF-A165 (Thermo Fisher Scientific) for 2 h at 4 °C. The beads were then incubated with 1 µg of biological triplicates small RNA extracted from HUVECs for 2 h at 4 °C. After washing three times with samples buffer, the beads were suspended in 50 µl RNA-binding buffer and heated for 5 min at 55 °C. The beads were removed and the RNA extract solution was transferred to a new tube. Of pure water, 100 µl was added and vortexed, and then 300 µl of 100% ethanol was added and vortexed for 10 s. The RNAs were purified over a Zymo column and then lyophilized overnight. Of HUVEC small RNA, 100 ng served as input. Sequencing libraries were generated largely as reported in ref. 82, with the following new 3′ ligation linker, RT primer and cDNA ligation linker ordered from IDT (Supplementary Table 3). Libraries were sequenced on the Illumina NovaSeq X Plus in single-end mode (read 1, 100 bp). Raw reads were demultiplexed, unique molecular identifiers (UMI) extracted and adapter trimmed using Cutadapt (v4.9)83. Reads were aligned to a custom reference of human small non-coding RNAs84 (https://github.com/y9c/m6A-SACseq/tree/main/db) using Bowtie2 (v2.5.4)85 with -k 10 to retain up to 10 alignments per read. Aligned reads were deduplicated by UMI and position using a published Python script86. In brief, reads with the same UMI and alignment position were grouped; within each group, if one alignment had the highest score, it was retained, otherwise one of the tied top-scoring alignments was selected at random.
Deduplicated transcript-level UMI counts were used for differential analysis in DESeq2 (v1.42.1). Two DESeq2 models were built: one comparing immunoprecipitation versus input (pI), and one comparing control versus input (cI). Size factors were estimated from all transcripts excluding long rRNAs (18S, 28S and 45S). To detect transcripts specifically enriched in the immunoprecipitation condition, we defined a centred Z-score:
Where
is the average size-factor correction across replicate-matched samples. An empirical null was estimated from Z-scores with ∣Z∣ < 0.5, assuming a Gaussian distribution. Wald tests were performed, and P values were adjusted using the Benjamini–Hochberg procedure.
RIP hits were defined by the following criteria:
-
DESeq2 filter: adjused PpI < 0.05 and log2FCpI > 0.5
-
True positives: passing the DESeq2 filter and adjused Pcentred < 0.05 and Zcentred > 0
Plots were generated using R (v4.3.3).
Electrophoretic mobility shift assay
Of rHS29 (TEGA Therapeutics), 2 µM was incubated with 1 µg of VEGF-A165 HS WT or VEGF-A165 HS(R/K) in PBS for 30 min at 25 °C. Of 50% glycerol, 4 µl was added to 10 µl of sample and the mix was then loaded into anyKD PAGE gel without SDS in the running buffer. Electrophoresis was conducted at constant voltage (150 V) at 4 °C. Gel was stained with Bulldog blue dye for 30 min at room temperature, washed three times with pure water for 5 min each, and finally scanned on a LI-COR Odyssey CLx scanner.
Microscale thermophoresis assay
Fluorescent HUVEC small RNA served as the target and VEGF-A served as the ligand in the microscale thermophoresis assay. For small RNA end repair, 10 µg of small RNA was incubated with 2.5 µl of FastAP (Thermo Fisher Scientific), 2.5 µl of T4 PNK (NEB) and 10 µl of reaction buffer (NEB) for 45 min at 37 °C. After 3′ end repair, the RNAs were purified over a Zymo column and eluted in 2 × 10 µl of pure water. For ligation, 2.5 µg of small RNA was incubated with 1.5 µl of 1 mM ATP, 3 µl of DMSO, 4.86 µl of 10 µM AF647 pCp (Jena Bioscience), 2 µl of 30 U T4 RNA ligase (NEB), 9 µl of 15% PEG8000 and 3 µl of 10× reaction buffer (NEB) overnight at 16 °C. The RNAs were then purified over a Zymo column. The binding of labelled small RNA to VEGF-A was measured on a Monolith NT.115 machine (NanoTemper) with the software MO.Control. Small RNA concentration was set as 5 nM (estimated from the average molecular weight of the small RNA pool). The VEGF-A165 gradient was set from 5.5 µM to 0.000336 µM; the VEGF-A121 gradient was set from 15 µM to 0.000458 µM. Excitation power was 4% and microscale thermophoresis power was 40% at 25 °C. Data from two independent biological replicates were analysed by GraphPad Prism.
Animal models
Zebrafish (D. rerio) lines were housed in AAALAC-accredited facilities and maintained in accordance with protocols approved by the Massachusetts Institute of Technology’s Committee on Animal Care. All experiments were conducted using zebrafish of the AB/Tübingen (TAB5/14) genetic background. The experiments involved zebrafish embryos at stages ranging from 1 to 5 days post-fertilization. As sex determination in zebrafish occurs after these stages, sex was not considered a variable in this study. Embryos were raised in clutches under standard growth conditions, without allocation into specific experimental groups or isolation.
Mouse work was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the US National Institutes of the Health. All of the animals were handled according to the approved institutional animal care and use committee protocols (#00001771) of the Boston Children’s Hospital. Sex was not considered a variable in this study. Every effort was made to minimize suffering.
Blinding strategy was applied in all animal experiments. Animals were all picked randomly.
Intraocular injection and retinal analysis
Intraocular injection of human recombinant WT VEGF-A165 HS WT and VEGF-A165 HS(R/K) was performed in postnatal day 6 C57BL/6 pups based on previous publications with minor modifications55,87. Pups were first anaesthetized by hypothermia. The eyelids were carefully separated by a 30-G needle and eyeballs were lifted by a forceps for injection. Of VEGF-A165 HS WT and VEGF-A165 HS(R/K) both at a concentration of 100 ng μl−1 or saline (vehicle control), 1 µl was injected into the vitreous humor using a Nanofil syringe attached with a 35-G needle (World Precision Instrument). Pups were placed on a heating pad and returned to the mother’s care. After 4 h, eyeballs were collected and processed for retina immunohistochemistry.
Retina immunohistochemistry is based on a publication with minor modifications87. In brief, isolated eyeballs from postnatal day 6 pups were fixed in 4% paraformaldehyde in PBS at room temperature for 30 min. Retinas were isolated under dissection microscopy, permeabilized in 0.1% Triton X-100 and 0.1% PBST at room temperature for 30 min, and blocked with 1% BSA in PBST at room temperature for 30 min. Primary antibodies were added to the sections with indicated dilution in PBST (anti-ERG from Abcam, dilution 1:500; anti-mVEGF165 from R&D system, dilution 1:500) and Alexa Fluor 647 isolectin GS-IB4 conjugate (Invitrogen, dilution 1:500) at 4 °C overnight. Retinas were further washed three times with PBST for 10 min at room temperature and incubated with fluorescent-conjugated secondary antibodies (Alexa 546 donkey anti-rabbit IgG, Alexa 488 donkey anti-goat IgG) with 1:500 dilution in PBST for 1 h at room temperature. Stained retinas were washed three times with PBST for 10 min at room temperature and mounted using Fluoromount-G slide mounting medium (Southern Biotech) on microscope slides (Thermo Fisher Scientific). Flat mount retina slides were proceeded to image with confocal microscopy.
Quantification of retinal images
Quantifications were based on previous publications87,88. Images were acquired using a LSM810 or LSM910 confocal microscope (Zeiss) equipped with an EC Plan-Neofluar ×10/0.3, a Plan-Apochromat ×20/0.8 or a Plan-Apochromat ×63/1.4 Oil DIC objective. Images were taken using Zen2.1 software (Zeiss) and processed and quantified with Fiji (NIH).
For vascular front and endothelial cell number quantification, tile-scanned images of flat-mounted retinas stained with IB4 and ERG were used. Vascular front areas were manually lassoed based on the first three rows of ERG+ cell number and IB4 signal. Numbers of ERG+ cells were manually counted. A value of average signals from a single retina is represented as n = 1.
For filopodia number quantification, tile-scanned images of flat-mounted retinas stained with IB4 were used. Filopodia numbers were manually counted from images taken with ×10, ×20 or ×63 objective. A value of average signals from a single retina is represented as n = 1.
Wnt3a–VEGF-A–HS fusions
gBlocks (double-strand DNA) of Wnt3a, Wnt3a–HS WT and Wnt3a–HS(R/K) (Supplementary Table 3) were purchased from IDT, and then used as templates to perform PCR with primers (GGATCCTAATACGACTCACTATAGGGAGA served as the forward primer for all three DNAs; GAATTCTTACTTGCAGGTGTGCACGT served as the reverse primer for Wnt3a; GAATTCTTACCGCCTCGGCTTGTCACATC served as the reverse primer for Wnt3a–HS WT; and GGATTCTTATTTTTTCGGCTTGTCACATT served as the reverse primer for Wnt3a–HS(R/K)). All PCR products were digested by EcoRI (R3101M, NEB) and BamHI (R3136M, NEB), and then ligated into pcDNA3.1 (Invitrogen). All plasmids were verified by DNA sequencing. Double-strand DNAs of Wnt3a, Wnt3a–HS WT and Wnt3a–HS(R/K) were generated through EcoRI/BamHI digestion, agarose gel separation and purification. For the in vitro transcription reaction, 500 ng and 300 ng of linearized, digested DNA were used in an overnight (2-h) setup with the mMESSAGE mMACHINE T7 ULTRA Transcription Kit (AM1345, Invitrogen). The resulting in vitro transcription product was treated with TURBO DNase, included in the same kit, for 15 min. Following DNase treatment, the samples underwent poly-A tailing using the tailing reaction provided by the mMESSAGE mMACHINE Kit. Before adding the ePAP (NEB) enzyme, 2 μl of the sample was set aside for later analysis via RNA agarose gel to confirm band size and serve as a control for the post-poly-A tailing reaction. The samples were then purified using the Zymo RNA Clean and Concentrator kit (Zymo Research).
Zebrafish embryo analysis
Tg(fli1a–eGFP) zebrafish line and WT zebrafish were applied for Vegfa and Wnt3a injection, separately. Zebrafish were crossed by placing a male and female in a shared container, keeping them physically separated until the following morning when they were combined for fertilization. Embryos were collected for injection 30 min after combining the males and females. The injection mix, consisting of 35 ng μl−1 in vitro-transcribed RNA encoding Wnt3a or 17.5 ng µl−1 in vitro-transcribed RNA encoding VEGF-A165 and 0.07% Phenol Red solution (Sigma), was prepared and loaded into needles made from glass capillary tubing (64-0766). Each embryo was injected with 1 nl of the injection mix using a picolitre injector (Warner Instruments). Injected embryos, along with uninjected controls, were incubated in fish water at 28 °C until collection at 24–36 h post-fertilization.
Zebrafish embryos at 24 h post-fertilization were collected, dechorionated using ethanol-sterilized forceps, and transferred to fresh fish water. Individual embryos were anaesthetized by transferring them into 1X Tricaine solution, prepared by diluting a 25X Tricaine stock solution (pH 7, containing 4 mg ml−1 Tricaine-S (Syndel) and 20 mM Tris) to 1X in fish water. Following anaesthesia, embryos were fixed in 4% paraformaldehyde in fish water. Tg(fli1a–eGFP) zebrafish samples were permeabilized in Triton X-100 and then immunolabelled with anti-GFP antibody. For imaging, embryos were embedded in 1.5% low-melt agarose (50100, Lonza) and transferred to a clear dish suspended in low-melt agarose. Images were captured using a Leica M205 FCA fluorescence stereo microscope at ×5.5 magnification, with the same settings applied to all embryos. For vessel quantification in fli1a–eGFP line, the longitudinal length of all vessels (termed ‘L’) along axial with the same length (termed ‘l’ = 200 μm, anterior to posterior) was measured in ImageJ. The ratio L:l was quantified.
qRT–PCR
Quantitative reverse transcriptase PCR (qRT–PCR) was performed using Brilliant II SYBR Green qRT–PCR one-step Master Mix kit (600825, Agilent) on a CFX Connect Real-Time System (Bio-Rad). Of RNA, 60 ng was used in the reaction with 1.25 µM primers. The data were analysed using the ΔΔCt method. 18S rRNA was used as a control to normalize the expression of target genes89. The primer sequences used for RT–PCR in this study are listed in Supplementary Table 3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw sequencing data have been deposited in the Gene Expression Omnibus database (GSE279647). Any additional information is available from the corresponding author on request. Source data are provided with this paper.
Code availability
This paper does not report original code.
References
Sarrazin, S., Lamanna, W. C. & Esko, J. D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952 (2011).
Xie, M. & Li, J.-P. Heparan sulfate proteoglycan — a common receptor for diverse cytokines. Cell Signal. 54, 115–121 (2019).
Apte, R. S., Chen, D. S. & Ferrara, N. VEGF in signaling and disease: beyond discovery and development. Cell 176, 1248–1264 (2019).
Robinson, C. J., Mulloy, B., Gallagher, J. T. & Stringer, S. E. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J. Biol. Chem. 281, 1731–1740 (2006).
Xu, D., Fuster, M. M., Lawrence, R. & Esko, J. D. Heparan sulfate regulates VEGF165- and VEGF121-mediated vascular hyperpermeability. J. Biol. Chem. 286, 737–745 (2011).
Corti, F. et al. N-terminal syndecan-2 domain selectively enhances 6-O heparan sulfate chains sulfation and promotes VEGFA165-dependent neovascularization. Nat. Commun. 10, 1562 (2019).
Ferreras, C. et al. Endothelial heparan sulfate 6-O-sulfation levels regulate angiogenic responses of endothelial cells to fibroblast growth factor 2 and vascular endothelial growth factor. J. Biol. Chem. 287, 36132–36146 (2012).
Jakobsson, L. et al. Heparan sulfate in trans potentiates VEGFR-mediated angiogenesis. Dev. Cell 10, 625–34 (2006).
Flynn, R. A. et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184, 3109–3124.e22 (2021).
Xie, Y. et al. The modified RNA base acp3U is an attachment site for N-glycans in glycoRNA. Cell https://doi.org/10.1016/j.cell.2024.07.044 (2024).
Perr, J. et al. RNA-binding proteins and glycoRNAs form domains on the cell surface for cell-penetrating peptide entry. Cell 188, 1878–1895.e25 (2025).
George, B. et al. Targeting cell surface NPM1 as a therapeutic strategy against acute myeloid leukemia. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02648-2 (2025).
Huang, J. et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct. Target. Ther. 6, 153 (2021).
Clause, K. C. & Barker, T. H. Extracellular matrix signaling in morphogenesis and repair. Curr. Opin. Biotechnol. 24, 830–833 (2013).
Roskoski, R. VEGF receptor protein-tyrosine kinases: structure and regulation. Biochem. Biophys. Res. Commun. 375, 287–291 (2008).
Xu, D. & Esko, J. D. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83, 129–157 (2014).
Nelson, R. M. et al. Heparin oligosaccharides bind L- and P-selectin and inhibit acute inflammation. Blood 82, 3253–3258 (1993).
Clausen, T. M. et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183, 1043–1057.e15 (2020).
Huang, N. et al. Natural display of nuclear-encoded RNA on the cell surface and its impact on cell interaction. Genome Biol. 21, 225 (2020).
Rieber, M. & Bacalao, J. An “external” RNA removable from mammalian cells by mild proteolysis. Proc. Natl Acad. Sci. USA 71, 4960–4964 (1974).
Juliano, R., Ciszkowski, J., Waite, D. & Mayhew, E. RNA associated with plasma membranes of ehrlich ascites carcinoma cells. FEBS Lett. 22, 27–30 (1972).
Graziano, V. R. et al. RNA N-glycosylation enables immune evasion and homeostatic efferocytosis. Nature 645, 784–792 (2025).
Zhang, N. et al. Cell surface RNAs control neutrophil recruitment. Cell 187, 846–860.e17 (2024).
Knorr, K. et al. Systematic evaluation of AML-associated antigens identifies anti-U5 SNRNP200 therapeutic antibodies for the treatment of acute myeloid leukemia. Nat. Cancer 4, 1675–1692 (2023).
Fischer, S. et al. Extracellular RNA mediates endothelial-cell permeability via vascular endothelial growth factor. Blood 110, 2457–2465 (2007).
Qin, W.-P. et al. Effect of extracellular ribonucleic acids on neurovascularization in osteoarthritis. Adv. Sci. 10, e2301763 (2023).
Fischer, S. et al. Signaling mechanism of extracellular RNA in endothelial cells. FASEB J. 23, 2100–2109 (2009).
Sheng, J. & Xu, Z. Three decades of research on angiogenin: a review and perspective. Acta Biochim. Biophys. Sin. 48, 399–410 (2016).
Ferguson, R., Holloway, D. E., Chandrasekhar, A., Acharya, K. R. & Subramanian, V. The catalytic activity and secretion of zebrafish RNases are essential for their in vivo function in motor neurons and vasculature. Sci. Rep. 9, 1107 (2019).
Folkman, J. Angiogenesis. Annu. Rev. Med. 57, 1–18 (2006).
Senger, D. R. & Davis, G. E. Angiogenesis. Cold Spring Harb. Perspect. Biol. 3, a005090 (2011).
Macauley, M. S., Crocker, P. R. & Paulson, J. C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 (2014).
Zeng, Y., Ramya, T. N. C., Dirksen, A., Dawson, P. E. & Paulson, J. C. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 6, 207–209 (2009).
Tzelepis, K. et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 (2016).
Bojar, D. et al. A useful guide to lectin binding: machine-learning directed annotation of 57 unique lectin specificities. ACS Chem. Biol. 17, 2993–3012 (2022).
Lewis, A. L., Chen, X., Schnaar, R. L. & Varki, A. in Essentials of Glycobiology (eds Varki, A. et al.) Ch. 15 (Cold Spring Harbor Laboratory Press, 2015).
Merry, C. L. R., Lindahl, U., Couchman, J. & Esko, J. D. in Essentials of Glycobiology (eds Varki, A. et al.) Ch. 17 (Cold Spring Harbor Laboratory Press, 2022).
McCormick, C. et al. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat. Genet. 19, 158–161 (1998).
Lind, T., Tufaro, F., McCormick, C., Lindahl, U. & Lidholt, K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J. Biol. Chem. 273, 26265–26268 (1998).
Leisico, F. et al. Structure of the human heparan sulfate polymerase complex EXT1-EXT2. Nat. Commun. 13, 7110 (2022).
David, G., Bai, X. M., Van der Schueren, B., Cassiman, J. J. & Van den Berghe, H. Developmental changes in heparan sulfate expression: in situ detection with mAbs. J. Cell Biol. 119, 961–975 (1992).
van den Born, J. et al. Novel heparan sulfate structures revealed by monoclonal antibodies. J. Biol. Chem. 280, 20516–20523 (2005).
Farley, J. R., Nakayama, G., Cryns, D. & Segel, I. H. Adenosine triphosphate sulfurylase from Penicillium chrysogenum equilibrium binding, substrate hydrolysis, and isotope exchange studies. Arch. Biochem. Biophys. 185, 376–390 (1978).
Esko, J. D. & Lindahl, U. Molecular diversity of heparan sulfate. J. Clin. Invest. 108, 169–173 (2001).
Lai, J. et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J. Biol. Chem. 278, 23107–23117 (2003).
Morimoto-Tomita, M., Uchimura, K., Werb, Z., Hemmerich, S. & Rosen, S. D. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 277, 49175–49185 (2002).
Ono, K., Hattori, H., Takeshita, S., Kurita, A. & Ishihara, M. Structural features in heparin that interact with VEGF165 and modulate its biological activity. Glycobiology 9, 705–711 (1999).
Chen, Y., Short, C., Halász, A. M. & Edwards, J. S. The impact of high density receptor clusters on VEGF signaling. Electron. Proc. Theor. Comput. Sci. 2013, 37–52 (2013).
Güven, E., Wester, M. J., Edwards, J. S. & Halász, ÁM. Modeling the cluster size distribution of vascular endothelial growth factor (VEGF) receptors. Bioinform. Biol. Insights 16, 11779322221085078 (2022).
Migdal, M. et al. Neuropilin-1 is a placenta growth factor-2 receptor. J. Biol. Chem. 273, 22272–22278 (1998).
Soker, S., Takashima, S., Miao, H. Q., Neufeld, G. & Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 92, 735–745 (1998).
Higashiyama, S., Abraham, J. A. & Klagsbrun, M. Heparin-binding EGF-like growth factor stimulation of smooth muscle cell migration: dependence on interactions with cell surface heparan sulfate. J. Cell Biol. 122, 933–940 (1993).
Corley, M., Burns, M. C. & Yeo, G. W. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell 78, 9–29 (2020).
Madugundu, A. K. et al. Integrated transcriptomic and proteomic analysis of primary human umbilical vein endothelial cells. Proteomics 19, e1800315 (2019).
Eilken, H. M. et al. Pericytes regulate VEGF-induced endothelial sprouting through VEGFR1. Nat. Commun. 8, 1574 (2017).
Lagendijk, A. K., Yap, A. S. & Hogan, B. M. Endothelial cell–cell adhesion during zebrafish vascular development. Cell Adh. Migr. 8, 136–145 (2014).
Liang, D. et al. Cloning and characterization of vascular endothelial growth factor (VEGF) from zebrafish, Danio rerio. Biochim. Biophys. Acta 1397, 14–20 (1998).
Liang, D. et al. The role of vascular endothelial growth factor (VEGF) in vasculogenesis, angiogenesis, and hematopoiesis in zebrafish development. Mech. Dev. 108, 29–43 (2001).
Shin, M. et al. Vegfc acts through ERK to induce sprouting and differentiation of trunk lymphatic progenitors. Development 143, 3785–3795 (2016).
Zeidan, B. S., Waltzer, W. C., Monheit, A. G. & Rapaport, F. T. Anemia associated with pregnancy in a cyclosporine-treated renal allograft recipient. Transplant. Proc. 23, 2301–2303 (1991).
Lawson, N. D. & Weinstein, B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 (2002).
Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012).
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Robertson, J. K. et al. Targeting the Wnt pathway in zebrafish as a screening method to identify novel therapeutic compounds. Exp. Biol. Med. 239, 169–176 (2014).
Kumar, S. et al. Molecular dissection of Wnt3a-Frizzled8 interaction reveals essential and modulatory determinants of Wnt signaling activity. BMC Biol. 12, 44 (2014).
Ramakrishnan, A.-B. & Cadigan, K. M. Wnt target genes and where to find them. F1000Research https://doi.org/10.12688/f1000research.11034.1 (2017).
Gu, F. et al. VEGF111b, a new member of VEGFxxxb isoforms and induced by mitomycin C, inhibits angiogenesis. Biochem. Biophys. Res. Commun. 441, 18–24 (2013).
Eswarappa, S. M. et al. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 157, 1605–1618 (2014).
Woolard, J., Bevan, H. S., Harper, S. J. & Bates, D. O. Molecular diversity of VEGF-A as a regulator of its biological activity. Microcirculation 16, 572–592 (2009).
Olsson, A.-K., Dimberg, A., Kreuger, J. & Claesson-Welsh, L. VEGF receptor signalling — in control of vascular function. Nat. Rev. Mol. Cell Biol. 7, 359–371 (2006).
Fairbrother, W. J., Champe, M. A., Christinger, H. W., Keyt, B. A. & Starovasnik, M. A. Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure 6, 637–648 (1998).
Lee, T.-Y., Folkman, J. & Javaherian, K. HSPG-binding peptide corresponding to the exon 6a-encoded domain of VEGF inhibits tumor growth by blocking angiogenesis in murine model. PLoS ONE 5, e9945 (2010).
Krilleke, D. et al. Molecular mapping and functional characterization of the VEGF164 heparin-binding domain. J. Biol. Chem. 282, 28045–28056 (2007).
Houck, K. A., Leung, D. W., Rowland, A. M., Winer, J. & Ferrara, N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 267, 26031–26037 (1992).
Li, Z. et al. Cell-surface RNA forms ternary complex with RNA-binding proteins and heparan sulfate to recruit immune receptors. Mol. Cell 85, 4633-4650.e11 (2025).
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011).
Cech, T. R. & Steitz, J. A. The noncoding RNA revolution — trashing old rules to forge new ones. Cell 157, 77–94 (2014).
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018).
Wan, Z. et al. A robust vasculogenic microfluidic model using human immortalized endothelial cells and Thy1 positive fibroblasts. Biomaterials 276, 121032 (2021).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Watkins, C. P., Zhang, W., Wylder, A. C., Katanski, C. D. & Pan, T. A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat. Commun. 13, 2491 (2022).
Kechin, A., Boyarskikh, U., Kel, A. & Filipenko, M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24, 1138–1143 (2017).
Ge, R. et al. m6A-SAC-seq for quantitative whole transcriptome m6A profiling. Nat. Protoc. 18, 626–657 (2023).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Smith, T., Monti, M., Willis, A. E. & Kalmár, L. Benchmarking tRNA-seq quantification approaches by realistic tRNA-seq data simulation identifies two novel approaches with higher accuracy. eLife 13, RP96955 (2024).
Kuo, A. et al. Murine endothelial serine palmitoyltransferase 1 (SPTLC1) is required for vascular development and systemic sphingolipid homeostasis. eLife 11, e78861 (2022).
Yanagida, K. et al. Sphingosine 1-phosphate receptor signaling establishes AP-1 gradients to allow for retinal endothelial cell specialization. Dev. Cell 52, 779–793.e7 (2020).
Wang, J., Yang, Z. & Xiao, A. Double-check the zebrafish 18S rRNA qPCR primers: they may be wrong. Preprint at bioRxiv https://doi.org/10.1101/2021.10.23.464780 (2021).
Acknowledgements
We thank P. A. Sharp, K. Pedram, C. R. Bertozzi, R. Caldwell, R. Savani, K. Godula, D. Xu and other members of the Flynn laboratory for helpful comments and discussions. This work was supported by grants from the Wellcome Trust (RG83195 and G106133 to K.T.), the UKRI Medical Research Council (RG83195 to K.T.) and Leukaemia UK (G117699 to K.T.), the National Science Foundation Graduate Research Fellowship Program (2141064 to J.S.), the Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (PDF-578200–2023 to S.K.), the Burroughs Wellcome Fund Career Award for Medical Scientists (to R.A.F.), the Sontag Foundation Distinguished Scientist Award (to R.A.F.), the Rita Allen Foundation (to R.A.F.), Yosemite (to R.A.F.), the Scleroderma Research Foundation (to R.A.F.), the G. Harold and Leila Y. Mathers Charitable Foundation (to R.A.F.), the National Institutes of Health (NIH) National Institute of General Medical Sciences under award number R35GM151157 (to R.A.F.), the National Institute Environmental Health Sciences under award number R01ES037423 (to R.A.F. and E.C.), the National Heart, Lung, and Blood Institute under award number P01HL131474, Project 3 (to J.D.E.) and the National Eye Institute under award number R01EY031715 (to T.H.).
Author information
Authors and Affiliations
Contributions
R.A.F. conceived and supervised the project. P.C. designed and performed the bulk of the experiments. L.K. and C.G.L. performed the flow cytometry experiments and analysis. J.Perr designed the csRBP experiments. S.K.R. and J.Porat performed the size-exclusion chromatography. J.Porat and P.C. performed the biochemical and glycoRNA assays. K.T., E.Y., J.M.L.D. and R.A.F. designed, performed and analysed the knockout screens. P.C., R.G., C.P.W. and P.H. performed the sequencing experiment and data analysis. J.D.E. designed the inhibitor and KO experiments surrounding HS pathways. S.K., J.S. and R.R. designed, implemented and analysed the 3D HUVEC assays. E.C. designed, implemented and analysed the zebrafish experiments. A.K. and T.H. designed, implemented and analysed the mouse experiments. R.A.F., P.C. and E.C. wrote the manuscript. All authors discussed the results and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.A.F. is a stockholder of ORNA Therapeutics; and a board of directors member and stockholder of Blue Planet Systems. K.T. is a shareholder of and has received research funding from Storm Therapeutics. R.A.F., E.C. and P.C. have filed a patent related to HS-targeting domains. J.D.E. is a co-founder of TEGA Therapeutics. J.D.E. and The Regents of the University of California have licensed a University invention to and have an equity interest in TEGA Therapeutics. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. R.A.F. and P.C. have filed a patent related to the use of RNase for the modulation of growth factor activity. R.A.F., P.C. and E.C. have filed a patent related to the development and use of RNA-binding mutant growth factors. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Larisa Haupt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Binding profile and RNase dependency of 13 human Siglec receptors.
a. Representative images of U2OS and MOLM-13 cells stained live with Fc-IgG1 or the indicated Siglec-Fc reagents (red). Scale bar, 10 µm. b. Representative images of U2OS and MOLM-13 cells treated with RNase A and RNase III (RNase pool) for 30 min and stained live with the indicated Siglec-Fc reagents (red). Scale bar, 10 µm. c. Quantification of the indicated Siglec dot numbers in U2OS and MOLM-13 cells treated with RNase A and RNase III for 30 min from 3 independent experiments with the number (n) of cells analyzed noted. Statistical assessment was performed with a Student's t-test and P values are shown. Data are mean ± s.e.m. d. Representative histograms (left) from flow cytometry experiments of U2OS or MOLM-13 cells treated live with or without a sialidase pool and then analyzed for surface binding of Fc-IgG (control), Siglec-7, Siglec-11, or total surface glycan (periodate) signal. Triplicate experiments are quantified with the mean fluorescence intensity (MFI) on the right for each cell line, and two-sided Student's t-tests were used to assess statistical differences, with the fold changes noted. Data are mean ± s.e.m.
Extended Data Fig. 2 Siglec-11 is a major RNA binding Siglec on the cell surface.
a. Representative images of U2OS and MOLM-13 cells co-stained with the indicated Siglec-Fc reagents (yellow) and 9D5 (magenta). An enlargement of the hatched box is shown, and a bright field (BF) is shown to represent the outline of the cell membrane. Scale bar, 10 µm. b. Nearest neighbor distance analysis of the Siglec pairs imaged was shown below. For each pair, the distance (nanometers, nm) from that anchor (left side protein name in the figure key) to the other pair was calculated across the indicated number of cells. These values were plotted in a density histogram. c. 10 pmol of Fc-IgG (control) or Siglec-11 was immunoprecipitated to Protein A beads. 1 µg of small RNA was then incubated to beads. RNA was extracted from the beads, purified, rPAL labeled, treated with or without RNase, and finally analyzed by Northern blot. Quantification of the ratio of RNase-sensitive sialoglycoRNA that Siglec-11 binds to IgG control is calculated across the biological triplicates. Statistical assessment was performed with a two-sided Student's t-test and P values are shown. Data are mean ± s.e.m.
Extended Data Fig. 3 Analysis of genome-wide CRISPR knockout screen.
a. Dot plot of genes identified in the genome-wide CRISPR knockout (KO) screen for loss of MAA-I cell surface binding ranked by CRISPR score. The top 15 gene names are displayed with a line drawn at the −0.8 score cut-off. The inset cartoon illustrates how MAA-I could interact with a cell surface glycoRNA. b. Heatmap plot of 41 genes overlapping between 9D5 and Siglec-11 KO screen. c. Gene ontology (GO) cellular compartment (top) and biological process (bottom) analysis of KO screen hits from MAA-I (blue), 9D5 (red), and Siglec-11 (green). The top 4 terms across the three screens were intersected and the union is displayed with the significance of each term represented by circle size and plotted on the x-axis by their fold enrichment.
Extended Data Fig. 4 EXT2 knockouts and rescues modulate csRNP clusters.
a. Sequence alignment of partial EXT2 coding sequences from WT and EXT2 knockout (KO) U2OS cells. Protospacer adjacent motif (PAM), green; sgRNA target, blue; mutated sequence, red. b. Western blot analysis of whole cell lysate. (left) from wild-type (WT) and (right) EXT2 KO U2OS cell lines. 2 independent experiments were performed. c. Representative images of WT and EXT2 KO U2OS cells stained with Siglec-9 (red). Scale bar, 10 µm. 3 independent experiments were performed. d. Number of cells quantified in Fig. 1e,g, and EDF4C across 3 independent experiments. e. Representative images of WT and EXT2 KO U2OS cells fixed, permeabilized, and stained with 9D5. Scale bar, 10 µm. Quantification of 9D5 intensity counted from 3 independent experiments. Data are mean ± s.e.m. f. Western blot analysis of whole cell lysate from the indicated cell lines. 2 independent experiments were performed. g. Western blot analysis of whole cell lysate (right) and sequence alignment of partial EXT1 coding sequences (left) from WT and EXT1 KO U2OS cells. 2 independent experiments were performed. h. Sequence alignment of partial UXS1 coding sequences from WT and UXS1 KO U2OS cells. i. Representative images of WT, EXT1 KO, and UXS1 KO U2OS cells stained with 10E4, Siglec-11, 9D5, DDX21, and hnRNP-U (all in red), separately. Scale bar, 10 µm. 3 independent experiments were performed. j. Representative images of U2OS cells co-stained live with 9D5 (green) and anti-DDX21 (red) or anti-hnRNP-U (red). An enlargement of the hatched box is shown, and a bright field (BF) is shown to represent the outline of the cell membrane. Scale bar, 10 µm. 2 independent experiments were performed. k. Western blot analysis of whole cell lysate from WT and EXT2 KO U2OS cell lines. Quantification of the ratio of total DDX21 and hnRNP-U to GAPDH is calculated across the biological triplicates. Statistical assessment was performed with a two-sided Student's t-test and P values are shown. Data are mean ± s.e.m.
Extended Data Fig. 5 Quantification of cell surface ligands after heparinase treatment.
a. Representative images of U2OS cells treated with heparinase pool for 30 min, and stained live with 10E4, Siglec-11, 9D5, anti-DDX21, anti-hnRNP-U, Siglec-7-Fc, and Siglec-9-Fc (all in red), separately. Scale bar, 10 µm. b. Quantification of data in A with the number of cells per condition from 3 independent experiments is shown. Statistical assessment was performed with a two-sided Student's t-test and P values are shown. Data are mean ± s.e.m. c. Nearest neighbor distance analysis of the 10E4 and Siglec-11 in Fig. 2a. For each pair, the nm distance from 10E4 to Siglec-11 was calculated across each time point. These values were plotted in a density histogram. d. Quantification fraction of 10E4-occupied, Siglec-11 dots in Fig. 2a from 3 independent experiments. e. Representative images of U2OS cells co-stained with the indicated 10E4 (Cyan), Siglec-11 (yellow), and DDX21 (magenta). An enlargement of the hatched box is shown, and a bright field (BF) is shown to represent the outline of the cell membrane. Scale bar, 10 µm. 2 independent experiments were performed.
Extended Data Fig. 6 Effects of cellular sulfation and sulfatases on csRNP clustering.
a. Representative zoom-out images of Fig. 2c. Scale bar, 10 µm. 3 independent experiments were performed. b. Sequence alignment of partial NDST1, HS6ST1, and HS2ST1 coding sequences from wild-type (WT) and the indicated knock-out (KO) U2OS cell lines. Protospacer adjacent motif (PAM), green; sgRNA target, blue; mutated sequence, red. c. Western blot analysis of whole cell lysate from WT, NDST1, HS6ST1, and HS2ST1 KO U2OS cells. 2 independent experiments were performed. d. Representative images of WT and NaCl or NaClO3 treated U2OS cells stained with 10E4, Siglec-11, 9D5, anti-DDX21, and anti-hnRNP-U (all in red). Scale bar, 10 µm. 3 independent experiments were performed. e. Representative images of WT and NaCl or NaClO3 treated U2OS cells stained with 3G10 (red). Scale bar, 10 µm. 1 independent experiment was performed. f. Representative images of mEmerald-Sufl1 and mEmerald-Sulf2 stably overexpressing U2OS cells. Scale bar, 10 µm. 2 independent experiments were performed. g. Western blot analysis of whole cell lysate isolated from mEmerald-Sufl1 and mEmerald-Sulf2 stably overexpressing U2OS cells. 2 independent experiments were performed.
Extended Data Fig. 7 Live cell labeling of HS-csRNP clusters in HUVECs and keratinocytes with RNase and heparinase treatment.
a. Representative images of HUVECs treated with the RNase pool or heparinase pool, and stained live with 10E4, Siglec-11, 9D5, anti-DDX21, and anti-hnRNP-U (all in red). Scale bar, 10 µm. b. Representative images of keratinocytes with the RNase pool or heparinase pool, and stained live with 10E4, Siglec-11, 9D5, anti-DDX21, and anti-hnRNP-U (all in red). Scale bar, 10 µm. c. Quantification of 10E4, Siglec-11, 9D5, cs-DDX21, and cs-hnRNP-U intensity per cell from 3 independent staining experiments on keratinocytes. Data are mean ± s.e.m.
Extended Data Fig. 8 HUVEC response to live cell enzymes, VEGF-A, and EGF addition.
a. Western blot analysis of whole cell lysate isolated from HUVECs after starvation and treatment with RNase pool followed by 25 ng/mL VEGF-A165 stimulation. Quantification of the ratio of phosphorylated ERK (pERK) to total ERK is calculated across the biological triplicates. Statistical assessment was performed with a two-sided Student's t-test and P values are shown. Data are mean ± s.e.m. b. Western blot analysis as in (A) with HUVECs stimulated with 25 ng/mL of VEGF-A121. Statistical assessment and data are as in (a). c. Western blot analysis as in (A) with HUVECs stimulated with 25 ng/mL of EGF. Statistical assessment and data are as in (a). d. Western blot analysis as in (A) with HUVECs stimulated with 3 ng/mL of VEGF-A165. Quantification of the ratio of phosphorylated VEGFR2 (pVEGFR2) to total VEGFR2 is calculated across the biological triplicates. Statistical assessment and data are as in (a). e. MST assay of VEGF-A and small RNA binding. n = 3 biologically independent repeats. Data are mean ± s.e.m. f. Representative images of HUVECs after serum starvation and treated with or without VEGF-A165 and then co-stained with anti-VEGF-A165 (purple) and Siglec-11 (yellow). Zoomed region shown as an inset. Scale bar, 10 µm. 3 independent experiments were performed. g. Nearest neighbor distance analysis of the VEGF-A165 and Siglec-11 in Figure S5E. For each pair, the nm distance from VEGF-A165 to Siglec-11 was calculated across. These values were plotted in a density histogram. h. The four biological replicate microfluidic chip without (left) and with (right) 10 µM RNase A for 6 days. Scale bar, 200 µm. i. Immunofluorescence confocal image (maximum z-projection view) of HUVEC structures after 6 days of +RNase treatment on microfluidic chip. Cells (blue), F-actin (red), and PECAM1 (green). Scale bar, 100 µm. 4 independent experiments were performed.
Extended Data Fig. 9 Heparan sulfate binding domain control of RNA-VEGFA165 binding.
a. Western blot analysis of the indicated amount of VEGF-A165. 2 independent experiments were performed. b. Lysates from HUVECs after serum starvation, treatment with or without 25 ng/mL VEGF-A165, and UV-crosslinking, were treated with or without RNase pool and immunoprecipitated (IP) with an anti-VEGF-A antibody (Proteintech). Immunoprecipitated samples were analyzed by Western blot using an anti-VEGF-A165 antibody (R&D system). 3 independent experiments were performed. c. Principal component analysis (PCA) of VEGF-A165 RIP-seq results. d. Enriched transcripts in VEGF-A165 RIP-seq. Each dot represents a unique small non-coding RNA (ncRNA) transcript, colored by biotype as indicated in the legend. The red dotted line marks the filtering citeria: log2FoldChange (IP/Input) > 0.5 and adjusted p-value < 0.05. Statistical assessment was performed with a two-sided Wald test and P values are adjusted for multiple comparisons using the Benjamini-Hochbery procedure. Solid circles indicate true positive hits that were also significantly enriched relative to the IgG control (see Methods for details). e. Abundance of enriched transcripts grouped by biotype. Color scheme matches (d). f. Overlap between enriched transcripts and glycoRNA defined by ManNAz-seq, grouped by ncRNA family. The p-value was calculated using a two-sided hypergeometric test with all human small ncRNAs as the background. g. Western blot analysis of the beads pre-conjugated with 5 µg of VEGF-A165. h. Representative images of the indicated HUVECs stained with anti-VEGF-A165 (red). Scale bar, 10 µm. Quantification of the images with number of cells noted per biological triplicate. Statistical assessment was performed with a two-sided Student's t-test and P values are shown. Data are mean ± s.e.m. i. Western blot analysis of whole cell lysate isolated from the indicated HUVECs. Quantification of the ratio of phosphorylated VEGFR2 (pVEGFR2) to total VEGFR2 is calculated across the biological triplicates. Statistical assessment was performed with a two-sided Student's t-test and P values are shown. Data are mean ± s.e.m. j. In vitro IP of VEGF-A165 HS WT or HS(R/K) with small RNA and rPAL. 3 independent experiments were performed. k. MST assay of VEGF-A and small RNA treated with sialidase binding. n = 3 biologically independent repeats. Data are mean ± s.e.m. l. RNA-seq analysis of the sulfotransferases expressed from HUVECs and their FPKM values from ref. 54. m. EMSA analysis of the indicated VEGF-A proteins with or without the addition of rHS29.
Extended Data Fig. 10 Activity of VEGF-A165 variants on and visualization of filopodia in murine retinas.
a. Representative images of P6 retinas of mice 4 h post intraocular administration of human VEGF-A165 HS WT (100 ng/retina), HS(R/K) (100 ng/retina) or vehicle (PBS) were immunostained with anti-ERG (red) and IB4 (white). Scale bar, 10 µm. Filopodia are manually annotated in yellow. Quantification of filopodia numbers in the vascular front regions (as shown in yellow line). n = 20 (PBS), 9 (HS-WT), and 10 (HS-RK) from 4 biological independent experiments. Statistical analysis was performed using an unpaired two-sided Student's t-test. P values are shown without adjustment. Data are mean ± s.e.m. b. Representative images of P6 retinas of mice 4 h post intraocular administration of human VEGF-A165 HS WT (100 ng/retina), HS(R/K) (100 ng/retina) or vehicle (PBS) were immunostained with anti-mVEGF164 (green), anti-ERG (red) and IB4 (white). Scale bar, 100 µm.
Extended Data Fig. 11 Activity of VEGF-A165 HS WT and HS(R/K) in zebrafish.
a. Western blot analysis of whole cell lysate isolated from zebrafish. b. Representative images of fli1a:EGFP zebrafish larvae at 36 h post fertilization (hpf) with the indicated injection of mRNA. DNA was stained with DAPI (blue). Scale bar, 0.3 mm. c. Quantification of GFP-positive endothelial cells numbers in the hatched box is calculated across three independent experiments. Boxes plots show the median, the 25th and 75th percentiles (bounds of the box), and minimum and maximum values (whiskers). Statistical assessment was performed with a two-sided Student's t-test and P values are shown. d. Representative images of zebrafish larvae at 36 hpf with the indicated injection of mRNA. Scale bar, 0.3 mm. e. Quantification of larvae length in (C) is calculated across three independent experiments. Boxes plots show the median, the 25th and 75th percentiles (bounds of the box), and minimum and maximum values (whiskers). Statistical assessment was performed with a two-sided Student's t-test and P values are shown. f. Western blot analysis of whole cell lysate isolated from zebrafish. g. RT-PCR analyses of Wnt3a-target gene expression in zebrafish cells post fertilization with the indicated injection of mRNA. Statistical assessment was performed with a two-sided Student’s t-test and P values are shown. Data are mean ± s.e.m.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chai, P., Kheiri, S., Kuo, A. et al. GlycoRNA complexed with heparan sulfate regulates VEGF-A signalling. Nature (2026). https://doi.org/10.1038/s41586-025-10052-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41586-025-10052-8