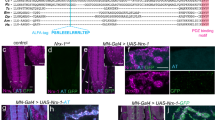
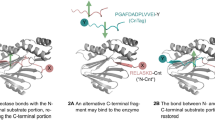
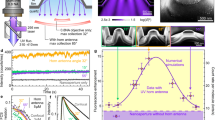
Abstract
Small peptide tags offer advantages as their compact size reduces target protein interference, making them valuable for labeling endogenous proteins. However, the lack of inherent fluorescence poses challenges for post-genome knockin monoclonal clone screening. Here we report an adaptable approach leveraging antigen-stabilizing fluorescent protein-fused nanobodies (Nbs) to selectively illuminate cells with successful ALFA tag knockins, streamlining high-throughput cell screening using fluorescence-activated cell sorting. Through targeted mutations and screening of ALFA Nbs (NbALFA), the fluorescently labeled Nb can be selectively degraded in the absence of the ALFA peptide. Conversely, successful insertion of the ALFA peptide into the genome results in a substantial increase in the fluorescence intensity of the Nb. This technique, termed ALFA Nb-guided endogenous labeling (ANGEL), enables a wide array of versatile applications within the native cellular environment. These applications include precise protein labeling and signal amplification through the tandem arrangement of ALFA tags, dynamic monitoring of protein behavior, initiation of protein degradation processes and analysis of protein interactome.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD052562. Further information on cloned constructs and additional data is available in the Supplementary Information. Source data are provided with this paper.
References
Muñoz, J. & Heck, A. J. R. From the human genome to the human proteome. Angew. Chem. Int. Ed. 53, 10864–10866 (2014).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, (2015).
Stangherlin, A., Seinkmane, E. & O’Neill, J. S. Understanding circadian regulation of mammalian cell function, protein homeostasis, and metabolism. Curr. Opin. Syst. Biol. 28, 100391 (2021).
Vermeulen, K., Van Bockstaele, D. R. & Berneman, Z. N. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 36, 131–149 (2003).
Shibata, Y., Shemesh, T., Prinz, W. A., Palazzo, A. F., Kozlov, M. M. & Rapoport, T. A. Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788 (2010).
Nixon-Abell, J. et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354, (2016).
Jozsef, L. et al. Reticulon 4 is necessary for endoplasmic reticulum tubulation, STIM1-Orai1 coupling, and store-operated calcium entry. J. Biol. Chem. 289, 9380–9395 (2014).
Li, K., Wang, G., Andersen, T., Zhou, P. Z. & Pu, W. T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS ONE 9, (2014).
Kamiyama, D. et al. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, (2016).
Schwinn, M. K., Steffen, L. S., Zimmerman, K., Wood, K. V. & Machleidt, T. A simple and scalable strategy for analysis of endogenous protein dynamics. Sci. Rep. 10, (2020).
Jin, B. K., Odongo, S., Radwanska, M. & Magez, S. Nanobodies: a review of generation, diagnostics and therapeutics. Int. J. Mol. Sci. 24, (2023).
Götzke, H. et al. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 10, (2019).
Li, X. H. et al. Transposase tools for genome engineering. Proc. Natl Acad. Sci. USA 110, 2279–2287 (2013).
Tang, J. C. Y. et al. Detection and manipulation of live antigen-expressing cells using conditionally stable nanobodies. eLife 5, e15312 (2016).
Sriram, S. M., Kim, B. Y. & Kwon, Y. T. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 12, 735–747 (2011).
Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011).
Martinez-Fonts, K. et al. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 11, 477 (2020).
Jaunky B. B. Shortening the Half-Life of the CRISPR/dCas9 System (Concordia University, 2023).
Xu, J. et al. N-end rule–mediated proteasomal degradation of ATGL promotes lipid storage. Diabetes 72, 210–222 (2023).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149 (2019).
Van Zwam, M. C. et al. IntAct: a non-disruptive internal tagging strategy to study actin isoform organization and function. PLoS Biol. 22, e3002551 (2024).
Shin, Y. J. et al. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 5, 14269 (2015).
Fei, J. Y. et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 130, 4180–4192 (2017).
Spector, D. L. & Lamond, A. I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646 (2011).
Koeppel, J. et al. Prediction of prime editing insertion efficiencies using sequence features and DNA repair determinants. Nat. Biotechnol. 41, 1446 (2023).
Ilik, A., Malszycki, M., Lübke, A. K., Schade, C., Meierhofer, D. & Aktas, T. SON and SRRM2 are essential for nuclear speckle formation. eLife 9, e60579 (2020).
Kim, J., Han, K. Y., Khanna, N., Ha, T. & Belmont, A. S. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 132, jcs226563 (2019).
Hao, M., Tang, J., Ge, S., Li, T. & Xia, N. Bacterial-artificial-chromosome-based genome editing methods and the applications in herpesvirus research. Microorganisms 11, 589 (2023).
Dion, W. et al. Four-dimensional nuclear speckle phase separation dynamics regulate proteostasis. Sci. Adv. 8, eabl4150 (2022).
Rivera, C. et al. Revealing RCOR2 as a regulatory component of nuclear speckles. Epigenetics Chromatin 14, 51 (2021).
Zhang, P. et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18, 35 (2017).
Doman, J. L., Sousa, A. A., Randolph, P. B., Chen, P. J. & Liu, D. R. Designing and executing prime editing experiments in mammalian cells. Nat. Protoc. 17, 2431–2468 (2022).
Acknowledgements
This project was supported by the National Key R&D Program of China (2022YFC3400600 to L.Y.; 2024YFC3406600 and 2022ZD0211900, both to P.X.), the National Natural Science Foundation of China (92254306, 21927813 and T2394513, all to P.X.; 32227802 and 31970704, both to L.Y.) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB37040301 to P.X.). We thank Y. Tian (Institute of Biophysics, Chinese Academy of Sciences) for plasmids pCMV-PE2 and pU6-Sp-pegRNA-HEK3_CTT_ins. We thank J. Jia (Core Facility, Institute of Biophysics, Chinese Academy of Sciences) for the technical support in flow cytometry analyses. We thank H. Deng and M. Han in the Technology Center for Protein Sciences, Tsinghua University, for protein MS analysis.
Author information
Authors and Affiliations
Contributions
P.X. and L.Y. conceived the study and wrote the paper with contributions from all authors. Z.W. and F.H. performed experiments. F.X. and W.H. analyzed data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Screening of unstable NbALFA.
a, Sequence alignment of the amino acids of NbALFA with other Nbs susceptible to degradation, including αCA dNb6mut, αDHFR dNb6mut, and dGBP1. Amino acid positions are numbered according to the ImMunoGeneTics information system (IMGT), FR: framework, CDR: complementarity determining region. b, Normalized fluorescence intensities of NbALFA-EGFP and 6mutNbALFA-EGFP, with or without the ALFA tag, were measured in U2OS cells transiently transfected with either NbALFA-EGFP or 6mutNbALFA-EGFP, along with mCherry-FLAG or mCherry-FLAG-ALFA plasmids. Quantitative analysis from three biologically independent experiments using Fiji software normalized fluorescence intensities to that of NbALFA-EGFP with mCherry-FLAG transfection. Data are expressed as the mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test; *p < 0.05 (p = 0.0120),***p < 0.0001 (p = 0.0009). c, Validation of the ubiquitin fusion technique in 293T cells transiently transfected with either Ub(G76V)-(M)NbALFA-mEmerald or Ub-(M)NbALFA-mEmerald. Whole-cell lysates were prepared, and protein samples were separated by SDS-PAGE, transferred to PVDF membranes, and probed with anti-EGFP and anti-β-actin antibodies. Protein expression was visualized with enhanced luminescence reagents.
Extended Data Fig. 2 (R)-NbALFA does not affect protein degradation.
Parental cells (U2OS with (R)NbALFA-mEmerald stably integrated into the genome), U2OS-ALFA-CKAP4, and U2OS-ALFA-LMNA cell lines were treated with 50 µg/ml CHX for 0, 12 and 24 h. a,c, Total protein from these cells was extracted using RIPA lysis buffer, separated by SDS-PAGE, and immunoblotted with anti-CKAP4, anti-LMNA and anti-GAPDH antibodies. P53 as positive controls. b,d, Quantitative analysis of CKAP4 (b) and LMNA (d) levels, normalized to GAPDH. The average values from three biologically independent experiments were calculated using Fiji software, with CKAP4 and LMNA amounts normalized to the group without CHX treatment. Data are expressed as the mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test CHX: cycloheximide.
Extended Data Fig. 3 ANGEL-mediated degradation of native proteins.
a, Quantitative analysis of fluorescence intensity of SON. The average SON intensity was measured using Fiji software, with relative fluorescence SON intensity normalized to that in the HA-SPOP-transfected group. The number of cells analyzed was: HA-SPOP, n = 16; HA-NbALFA-SPOPmut, n = 17; HA-NbALFA-SPOP, n = 20. Three biologically independent replicates were performed and data are expressed as mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test. ****p < 0.001 (p = 1.36e-8 (HA-SPOP vs. HA-NbALFA-SPOPmut), 2.01e-8 (HA-NbALFA-SPOPmut vs. HA-NbALFA-SPOP)). b, Representative images of U2OS-2XALFA-SON cells with or without (R)NbALFA-HaloTag. Cells were stained with DAPI (cyan) and HaloTag TMR ligand (magenta), following the commercial protocol. The cells were then fixed and subjected to immunofluorescence staining using SON (green) antibody. c, U2OS-2XALFA-SON cells stably expressing (R)NbALFA-HaloTag were transiently transfected with pcDNA3.1-hyPBase-excision plasmid. After 1 week, cells were stained with HaloTag TMR ligand, and monoclonal cells with reduced fluorescence were sorted for further analysis. d, Representative images of SON degradation in the excised (R)NbALFA-HaloTag U2OS-2XALFA-SON cell line. (R)NbALFA-HaloTag in U2OS-2XALFA-SON cells were excised using the PB transposon system, followed by transient transfected with HA-SPOP, HA-NbALFA-SPOPmut, or HA-NbALFA-SPOP. After a 6-h incubation period, doxycycline (1 μg/ml) was added to induce SPOP expression for 24 h. The cells were subsequently stained with DAPI (cyan), fixed with 4% paraformaldehyde, and subjected to immunofluorescence staining using HA (green) and SON (magenta) antibodies. e, Quantitative analysis of fluorescence intensity of SON. The average SON intensity from three biologically independent experiments was calculated using Fiji software, with relative fluorescence intensity normalized to that in the HA-SPOP-transfected group. Number of cells: HA-SPOP, n = 22; HA-NbALFA-SPOPmut, n = 23; HA-NbALFA-SPOP, n = 16. Data are expressed as mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test. ****p < 0.001 (p = 1.14e-17 (HA-SPOP vs. HA-NbALFA-SPOP), 2.22e-18 (HA-SPOPmut vs. HA-NbALFA-SPOP)). Representative cells were indicated with dotted circles, and the zoomed-in regions of these circles were displayed in the lower left corner of the images. Scale bars: 50 µm.
Extended Data Fig. 4 Mass spectrometry data-based analysis of the protein interactome.
a,b, Gene Ontology (GO) terms (a) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (b) of CKAP4 immunoprecipitated proteins reveal a 10-fold enrichment. The U2OS-ALFA-CKAP4 cell line, which lacks (R)NbALFA-HaloTag and wild-type (WT) U2OS cells, used as controls, were lysed. The supernatants underwent immunoprecipitation according to the manufacturer’s instructions. c,d, GO terms (c) and KEGG (d) analysis of SON immunoprecipitated proteins also show a 10-fold enrichment. The U2OS-2XALFA-SON cell line, which lacks the (R)NbALFA-HaloTag, and WT U2OS cells, used as controls, were lysed. The supernatants were then subjected to immunoprecipitation following the manufacturer’s instructions.
Extended Data Fig. 5 Movement and dynamics of nuclear speckles in the cell cycle.
a, Dual-color imaging of SON (magenta) and Lamin (green) during mitosis was performed using the U2OS-2XALFA-SON cell line, stably expressing mEmerald-LMNA. Cells were synchronized using a thymidine-nocodazole block, and images were captured with a FV3000 confocal laser scanning microscope. Gaussian filter (σ = 1) was applied via ImageJ. Scale bars: 5 µm. b, Dual-color imaging of SON (magenta) and H2B (green) during mitosis was performed using the U2OS-2XALFA-SON cell line, transiently transfected with H2B-sfGFP. Cells were synchronized using a double thymidine block, and images were captured with a SpinSR10 microscope. Scale bars: 5 µm. c, Nuclear speckles fusion and fission in U2OS-2XALFA-SON cells stained with HaloTag TMR ligand, showing the fusion (0–7 min, 19–22 min) and fission (8–18 min) evens. Scale bars: 5 µm.
Extended Data Fig. 6 Effects of DRB on nuclear speckles.
a, Quantification of nuclear speckles with or without DRB treatment. U2OS-2XALFA-SON cells stained with HaloTag TMR ligand. Nuclear speckles in five regions across three biological independent experiment were calculated using Fiji software. Data are expressed as the mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test. ***p < 0.001 (p = 0.0002). b, Imaging of nuclear speckles in U2OS-2XALFA-SON cells stained with Hoechst (cyan) and HaloTag TMR ligand (magenta), with or without DRB treatment. Scale bars: 10 µm. c–e, Analysis of (c) roundness, (d) Feret’s diameter, and (e) brightness of nuclear speckles in U2OS-2XALFA-SON cells with or without DRB treatment (control: n = 1054, DRB: n = 699), using Fiji software.
Extended Data Fig. 7 Nuclear speckle morphology under compound treatments.
a, U2OS-2XALFA-SON cells were stained with HaloTag TMR ligand and treated with 100 nM BafA1, 20 μM CCCP, 4 mM DTT, 100 μM H2O2, 1 μM TG or 2.5 μg/ml TM for 2 h. Images were acquired using an FV3000 confocal laser scanning microscope. b,c, Quantitative analysis of the roundness (b) and the Feret’s diameters (c) of nuclear speckles in control and treatment groups. Speckle counts: Control, n = 657; BafA1, n = 479; CCCP, n = 686; DDT, n = 475; H2O2, n = 948; Oligomycin A, n = 559; Rapamycin, n = 682; TG, n = 436; TM, n = 524. Data are expressed as the mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test. CCCP: Carbonyl cyanide 3-chlorophenylhydrazone, DTT: dithiothreitol, H2O2: hydrogen peroxide, TG: thapsigargin, TM: tunicamycin.
Extended Data Fig. 8 Movement and dynamics of nuclear speckles with DRB treatment.
a, Movements of nuclear speckles in U2OS-2XALFA-SON cells with or without DRB treatment (control: n = 1054, DRB: n = 699), using Fiji software. Data are expressed as the mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test. b, Kinetics of SON fluorescence recovery in U2OS-2XALFA-SON cells with (n = 10) or without DRB treatment (n = 20). c, Images show SON fluorescence recovery in U2OS-2XALFA-SON cells, with or without DRB treatment, taken before (0 min) and after bleaching. Square: region of interest is labeled by photobleaching, Scale bars: 10 µm. d, Quantitative analysis of SON recovery half-time of control and DRB-treaded group in (c) control, n = 20; DRB, n = 10. Data are expressed as the mean ± s.e.m. Significance was calculated by two-tailed Student’s t-test.**p < 0.001 (p = 0.0080).
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Tables 1–4
Supplementary Table 1: Mass spectrometric data of endogenous CKAP4 interactome. Endogenous CKAP4 immunoprecipitated proteins in U2OS-ALFA-CKAP4 cells with (R)NbALFA-HaloTag excision, using WT U2OS cells as control. Supplementary Table 2: Mass spectrometric data of endogenous SON interactome. Endogenous SON immunoprecipitated proteins in U2OS–2×ALFA–SON cells with (R)NbALFA-HaloTag excision, using WT U2OS cells as control. Supplementary Table 3: Relationship among MIG, nuclear speckles and nuclear membrane. During long-term imaging of SON and LMNA, the sequences of mitotic interchromatin granule (MIG) disappearance, nuclear speckle formation, and nuclear membrane reconstruction were statistically analyzed. Supplementary Table 4: Sequences of plasmids, gRNAs and epegRNA. Sequences of gRNA and epegRNA for ALFA tag knockins across various genes using ANGEL, along with the key plasmid sequences.
Supplementary Video 1
Long-term dual-color imaging of SON and Lamin U2OS–2×ALFA–SON cells (magenta) stably expressing mEmerald-lamin A/C (LMNA; green) were synchronized by thymidine–nocodazole block. Images were captured with FV3000 confocal laser scanning microscope at 2-min intervals.
Supplementary Video 2
Long-term dual-color imaging of SON with H2B U2OS–2×ALFA–SON cells (magenta), transiently transfected with H2BC21-sfGFP (green) were synchronized by double thymidine block. Images were captured with the SpinSR10 microscope at 10-min intervals.
Supplementary Video 3
SON tracking imaging U2OS–2×ALFA–SON cell line stained with HaloTag TMR ligand. Images were captured with LSM980 microscope at 1-min intervals.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1b,d,f.
Source Data Fig. 1
Unprocessed western blots of Fig. 1c,e.
Source Data Fig. 2
Statistical source data for Fig. 2b–f.
Source Data Fig. 6
Unprocessed western blots of Fig. 6a–c.
Source Data Fig. 6
Statistical source data for Fig. 6e.
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1b.
Source Data Extended Data Fig. 1
Unprocessed western blots for Extended Data Fig. 1c.
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2b,d.
Source Data Extended Data Fig. 2
Unprocessed western blots for Extended Data Fig. 2a,c.
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3a,e.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4a–d.
Source Data Extended Data Fig. 6
Statistical source data for Extended Data Fig. 6a–e.
Source Data Extended Data Fig. 7
Statistical source data for Extended Data Fig. 7b,c.
Source Data Extended Data Fig. 8
Statistical source data for Extended Data Fig. 8a,b,d.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Z., Hu, F., Xue, F. et al. ALFA nanobody-guided endogenous labeling. Nat Chem Biol 21, 1992–2001 (2025). https://doi.org/10.1038/s41589-025-02019-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41589-025-02019-7