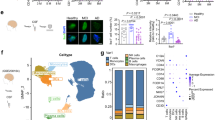
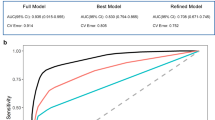
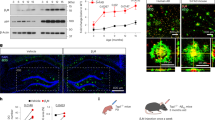
Abstract
Astrocytes play essential roles in the brain, and their dysfunction is associated with nearly every form of neurological disease. Despite their ubiquity, knowledge of how astrocytes contribute to disease pathogenesis is incomplete; accordingly, harnessing their biology toward therapeutics remains a major challenge. Here we show that the transcription factor Sox9 plays a context-specific role in maintaining astrocyte function and circuit activity in the aging hippocampus and Alzheimer’s disease (AD) models. We found that Sox9 overexpression in astrocytes in AD models clears existing amyloid beta (Aβ) plaques and preserves cognitive function. Mechanistically, Sox9 promotes the phagocytosis of Aβ plaques by astrocytes through the regulation of the phagocytic receptor MEGF10, which is sufficient to preserve cognitive function in AD models. Collectively, these studies highlight a role for astrocytic Sox9 during aging and AD while identifying Sox9−MEGF10 signaling as a prospective astrocyte-based therapeutic approach to ameliorate cognitive decline in neurodegenerative disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All primary antibodies used in this study are listed in the Methods. Sequencing data were deposited in the Gene Expression Omnibus database under accession number GSE294900 for RNA-seq datasets and GSE294933 for the ChIP−seq datasets. Source data are provided with this paper.
Code availability
No custom software codes were used to analyze the data.
References
Khakh, B. S. & Deneen, B. The emerging nature of astrocyte diversity. Annu. Rev. Neurosci. 42, 187–207 (2019).
Allen, N. J. Astrocyte regulation of synaptic behavior. Annu. Rev. Cell Dev. Biol. 30, 439–463 (2014).
Cheng, Y. T., Woo, J. & Deneen, B. Sculpting astrocyte diversity through circuits and transcription. Neuroscientist 29, 445–460 (2023).
Cheng, Y. T. et al. Inhibitory input directs astrocyte morphogenesis through glial GABABR. Nature 617, 369–376 (2023).
Huang, A. Y. et al. Region-specific transcriptional control of astrocyte function oversees local circuit activities. Neuron 106, 992–1008 (2020).
Ung, K. et al. Olfactory bulb astrocytes mediate sensory circuit processing through Sox9 in the mouse brain. Nat. Commun. 12, 5230 (2021).
Sardar, D. et al. Induction of astrocytic Slc22a3 regulates sensory processing through histone serotonylation. Science 380, eade0027 (2023).
Cheng, Y. T. et al. Social deprivation induces astrocytic TRPA1-GABA suppression of hippocampal circuits. Neuron 111, 1301–1315 (2023).
Escartin, C. et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325 (2021).
Patani, R., Hardingham, G. E. & Liddelow, S. A. Functional roles of reactive astrocytes in neuroinflammation and neurodegeneration. Nat. Rev. Neurol. 19, 395–409 (2023).
Brandebura, A. N., Paumier, A., Onur, T. S. & Allen, N. J. Astrocyte contribution to dysfunction, risk and progression in neurodegenerative disorders. Nat. Rev. Neurosci. 24, 23–39 (2023).
Verkhratsky, A. Astroglial calcium signaling in aging and Alzheimer’s disease. Cold Spring Harb. Perspect. Biol. 11, a035188 (2019).
Habib, N. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Allen, W. E., Blosser, T. R., Sullivan, Z. A., Dulac, C. & Zhuang, X. Molecular and spatial signatures of mouse brain aging at single-cell resolution. Cell 186, 194–208 (2023).
Clarke, L. E. et al. Normal aging induces A1-like astrocyte reactivity. Proc. Natl Acad. Sci. USA 115, E1896–E1905 (2018).
Boisvert, M. M., Erikson, G. A., Shokhirev, M. N. & Allen, N. J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269–285 (2018).
Deneen, B. et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968 (2006).
Lozzi, B., Huang, T. W., Sardar, D., Huang, A. Y. & Deneen, B. Regionally distinct astrocytes display unique transcription factor profiles in the adult brain. Front. Neurosci. 14, 61 (2020).
Sun, W. et al. SOX9 is an astrocyte-specific nuclear marker in the adult brain outside the neurogenic regions. J. Neurosci. 37, 4493–4507 (2017).
Alamoudi, A. A. SOX9 expression is increased in Alzheimer’s disease (AD) and is associated with disease progression and APOE4 genotype: a computational approach. Cureus 15, e36129 (2023).
Choi, D. J. et al. A Parkinson’s disease gene, DJ-1, repairs brain injury through Sox9 stabilization and astrogliosis. Glia 66, 445–458 (2018).
Choi, D. J., An, J., Jou, I., Park, S. M. & Joe, E. H. A Parkinson’s disease gene, DJ-1, regulates anti-inflammatory roles of astrocytes through prostaglandin D2 synthase expression. Neurobiol. Dis. 127, 482–491 (2019).
Lachmann, A. et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010).
Keenan, A. B. et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224 (2019).
Lachmann, A. et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 9, 1366 (2018).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Ximerakis, M. et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 22, 1696–1708 (2019).
Popov, A. et al. Mitochondrial malfunction and atrophy of astrocytes in the aged human cerebral cortex. Nat. Commun. 14, 8380 (2023).
Rodriguez, J. J. et al. Complex and region-specific changes in astroglial markers in the aging brain. Neurobiol. Aging 35, 15–23 (2014).
Lee, E. et al. A distinct astrocyte subtype in the aging mouse brain characterized by impaired protein homeostasis. Nat. Aging 2, 726–741 (2022).
Shigetomi, E., Tong, X., Kwan, K. Y., Corey, D. P. & Khakh, B. S. TRPA1 channels regulate astrocyte resting calcium and inhibitory synapse efficacy through GAT-3. Nat. Neurosci. 15, 70–80 (2011).
Shigetomi, E., Jackson-Weaver, O., Huckstepp, R. T., O’Dell, T. J. & Khakh, B. S. TRPA1 channels are regulators of astrocyte basal calcium levels and long-term potentiation via constitutive D-serine release. J. Neurosci. 33, 10143–10153 (2013).
Bazargani, N. & Attwell, D. Astrocyte calcium signaling: the third wave. Nat. Neurosci. 19, 182–189 (2016).
Adamsky, A. et al. Astrocytic activation generates de novo neuronal potentiation and memory enhancement. Cell 174, 59–71 (2018).
Soreq, L., Bird, H., Mohamed, W. & Hardy, J. Single-cell RNA sequencing analysis of human Alzheimer’s disease brain samples reveals neuronal and glial specific cells differential expression. PLoS ONE 18, e0277630 (2023).
Park, H. et al. Single-cell RNA-sequencing identifies disease-associated oligodendrocytes in male APP NL-G-F and 5XFAD mice. Nat. Commun. 14, 802 (2023).
Hassiotis, S. et al. Lysosomal LAMP1 immunoreactivity exists in both diffuse and neuritic amyloid plaques in the human hippocampus. Eur. J. Neurosci. 47, 1043–1053 (2018).
Gowrishankar, S. et al. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl Acad. Sci. USA 112, E3699–E3708 (2015).
Jung, H. et al. Anti-inflammatory clearance of amyloid-β by a chimeric Gas6 fusion protein. Nat. Med. 28, 1802–1812 (2022).
Lee, S. Y. & Chung, W. S. The roles of astrocytic phagocytosis in maintaining homeostasis of brains. J. Pharm. Sci. 145, 223–227 (2021).
Kono, R., Ikegaya, Y. & Koyama, R. Phagocytic glial cells in brain homeostasis. Cells 10, 1348 (2021).
Chung, W. S. et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 (2013).
Mockenhaupt, K. et al. Yin Yang 1 controls cerebellar astrocyte maturation. Glia 71, 2437–2455 (2023).
Welle, A. et al. Epigenetic control of region-specific transcriptional programs in mouse cerebellar and cortical astrocytes. Glia 69, 2160–2177 (2021).
Lardenoije, R. et al. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 131, 21–64 (2015).
Neal, M. & Richardson, J. R. Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 432–443 (2018).
Singh, T. D. et al. MEGF10 functions as a receptor for the uptake of amyloid-β. FEBS Lett. 584, 3936–3942 (2010).
Saito, T. et al. Single App knock-in mouse models of Alzheimer’s disease. Nat. Neurosci. 17, 661–663 (2014).
Gedam, M. et al. Complement C3aR depletion reverses HIF-1α-induced metabolic impairment and enhances microglial response to Aβ pathology. J. Clin. Invest. 133, e167501 (2023).
Jankowsky, J. L. & Zheng, H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol. Neurodegener. 12, 89 (2017).
John Lin, C. C. et al. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405 (2017).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Acknowledgements
This work was supported by National Institutes of Health grants R35-NS132230, R01-AG071687 and R01-CA284455 to B.D.; K01-AG083128 to D.-J.C; and R56-MH133822 to J.W. We are thankful for support from the David and Eula Wintermann Foundation. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under award number P50HD103555 for the use of the Microscopy Core facilities and the Animal Phenotyping & Preclinical Endpoints Core facilities. This study was jointly supported by resources from Houston Methodist and Baylor College of Medicine.
Author information
Authors and Affiliations
Contributions
D.-J.C. and B.D. conceptualized the project, designed experiments and wrote the paper. D.-J.C. performed and analyzed main experiments and bioinformatics datasets. S.M., W.K., E.-A.C.S., Y.K., D.S., B.L., Y.-T.C., M.R.W., K.S. and T.-W.H. performed the experiments. J.W. performed the electrophysiology studies. J.J. discussed the animal model and results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Peer review
Peer review information
Nature Neuroscience thanks Won-Suk Chung and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sox9 expression and gene regulation in aged astrocytes.
a. UMAP plot of Sox9 expression in the total cell clusters at 2-3 months and 21-23 months. b. % of cell types in the Sox9-expressed total cells at 2-3 months and 21-23 months. c. Representative confocal Z-stack images of DAPI (blue), Sox9 (red), and GFP (green) in hippocampal astrocytes of 4 month-old mice and 18 month-old mice. Scale bars, 5 µm. d. Histogram analysis of Sox9 (red) and DAPI (blue) intensities in hippocampal astrocytes of each group. e. Violin plot of Sox10 in Sox9-expressed cell types (Astrocytes and non-astrocytes) of 2-3 months and 21-23 months. f. Representative confocal Z-stack images of Sox9 (white), Sox10 (red), and GFP (green) in hippocampal astrocytes. Scale bars, 10 µm. The graph reveals Sox9 expression level between Sox9-positive and Sox10-negative cells (Sox10 negative), and Sox9-positive and Sox10-positive cells (Sox10 positive). n = 5 mice of each group. g, h. Representative confocal Z-stack images of Sox9 (red) and GFP (green) in hippocampal astrocytes on CA1, CA2, and CA3 regions of 4 month-old mice and 18 month-old mice. Scale bars, 50 µm. i. Quantification of Sox9 intensities in hippocampal astrocytes of 4 month-old mice and 18 month-old mice. n = 6 mice of each group. j. Representative confocal Z-stack images of Sox9 (red) and GFP (green) in hippocampal astrocytes of 4 week-old, 6 month-old, 12 month-old, and 24 month-old mice. Scale bars, 20 µm. k. Quantification of Sox9 intensities in hippocampal astrocytes of each group. n = 6 mice of each group. l. Schematic illustration for RNAseq from sorted astrocytes in the hippocampus of Con and Sox9cKO at 4 months and 18 months. n = 3 mice for each group. m, n. GO analysis (m) and GO circle plot (n) of DEGs between Con and Sox9cKO at 4 months and 18 months: p-value < 0.05. o. A Venn diagram reveals the number of specific DEGs between Con and Sox9cKO at 4 months and 8 months, with DEGs between 4 month-old and 18 month-old WT mice. p. Top 5 molecular functions of DEGs between Con and Sox9cKO at 4 months and 18 months: p-value < 0.005. Two-sided unpaired Student’s t-test (e, f), One-way ANOVA followed by Tukey’s comparisons test (k), and Two-way ANOVA with Sidak’s correction (i) were used for analysis. The mean ± s.e.m. All illustrations were created using BioRender.com.
Extended Data Fig. 2 Altered astrocyte morphology in the aging brain.
a. UMAP plot of astrocyte clusters from the whole brain of 2-3 month-old mice and 21-23 month-old mice. b. Violin plot of Aldh1l1 expression in each group. c. Count value of Aldh1l1 RNA expression from RNAseq of hippocampal astrocytes of 4 month-old mice and 18 month-old mice. n = 3 mice for each group d. Representative confocal Z-stack images of GFP in the hippocampus of 4 month-old mice and 18 month-old mice. Scale bars, 20 µm. e. Quantification of the number of interactions by Sholl analysis of hippocampal astrocytes of 4 month-old mice and 18 month-old mice. n = 6 mice for each group. f. Schematic illustration for WT; Aldh1l1-creER; LSL; tdTomato mouse. g. Representative confocal Z-stack images of tdTomato-astrocytes in the hippocampus of 4 month-old mice and 18 month-old mice. Scale bars, 20 µm. h. Quantification of the number of interactions by Sholl analysis of hippocampal tdTomato-astrocytes of 4 month-old mice (n = 13 mice) and 18 month-old mice (n = 14 mice). i. UMAP plot of total cell clusters from the whole brain of 2-3 month-old mice and 21-23 month-old mice. j, k. Atp1b2 (ACSA-2 antigen) expression (j) and Aldh1l1 (k) from the total cell clusters at 2-3 months and 21-23 months. l. Representative confocal Z-stack images of Sox9 (red) and GFP (green) in the hippocampus of 4 month-old mice and 18 month-old mice. Scale bars, 100 µm. m. Percentage of Aldh1l1-expressed astrocytes in the total Sox9-expressed cells. n = 6 mice for each group. Two-sided unpaired Student’s t-test (c) and Two-way ANOVA with Sidak’s correction (e, h) were used for analysis. The mean ± s.e.m. ns: not significant. All illustrations were created using BioRender.com.
Extended Data Fig. 3 Overexpression of Sox9 in aged astrocytes.
a. Schematic illustration for AAV-GFAP-Sox9-Flag injection (AAV-Sox9ov) or AAV-GFAP-lck-smV5-4x6T (AAV-Con) on the hippocampus in 16 month-old WT mice. b. Representative confocal Z-stack images of Sox9 (red) and GFP (green) in hippocampal astrocytes in the AAV-Con with AAV-Sox9ov. Scale bars, 20 µm. c. Quantification of the number of interactions by Sholl analysis of hippocampal astrocytes in each group. n = 5 mice for each group. d. Schematic illustration for AAV-Con or AAV-Sox9ov with AAV-GFAP-Gcamp6 (Gcamp6) in the hippocampus of 18 month-old WT mice. e. Representative traces between AAV-Con and AAV-Sox9ov at 18 months showing spontaneous GCaMP6 activity in astrocytes from the hippocampus. f. Quantification of somatic ∆F/F GCaMP6 signal amplitude and frequency derived from 5 mice of each group. g. Schematic illustration for AAV-Con or AAV-Sox9ov in the hippocampus in 16 month-old WT mice. h. LTP traces from AAV-Con (n = 9 mice) and AAV-Sox9ov (n = 8 mice) hippocampal slices at 18 months. i. Quantification of LTP. n = 7 of each group. The normalized fEPSPs were calculated by averaging responses for the last 5 min. Two-sided unpaired Student’s t-test (f, i) and Two-way ANOVA with Sidak’s correction (c) were used for analysis. The mean ± s.e.m. ns: not significant. All illustrations were created using BioRender.com.
Extended Data Fig. 4 Sox9 expression and gene regulation in APPNLGF astrocytes.
a. Top 5 related phenotype-genotype integrator association (PhenGenI) analyses of the DEGs between 18 month-old WT astrocytes and Sox9cKO astrocytes. b. UMAP plot of total cell clusters from human anterior hippocampal cortex of age-matched controls (Human Con) and human AD patients (Human AD). c, d. ALDH1L1 expression (c) and SOX9 (d) in the total cell clusters of Human Con and Human AD. e. Table for information on human AD samples with age-matched controls. f. Representative confocal Z-stack images of GFAP (green) and Aβ (red) in the hippocampus of human AD and Human Con. Scale bars, 20 µm. g. Representative confocal Z-stack images of Aβ (green) and Lamp1 (red) in the hippocampus of the WT mouse and APPNLGF mouse from 2 month-old to 8 month-old. Scale bars, 500 µm. h, i. Representative confocal Z-stack images of GFP (green), Aβ (purple), and Sox9 (red) in the hippocampus of 4 month-old WT (h) and 4 month-old APPNLGF mouse (i). Scale bars, 50 µm (full image) and 20 µm (each region). j. Quantification graphs of Sox9 intensities (from each area; left panel, by distance from plaque; right panel) in astrocytes of each group. n = 5 mice of each group. k. Heatmaps comparing Sox9-ChIP at 4 kb from the peak center in hippocampal astrocytes from 6-month-old WT; Aldh1l1-GFP (Young_Sox9) and 18-month-old WT; Aldh1l1-GFP (Old_Sox9), and 6-month-old APPNLGF; Aldh1l1-GFP (APPNLGF_Sox9). l. A Venn diagram reveals the number of shared genes from each ChIP dataset. m. GO analysis for specific genes of each group. n, o. UMAP plot of total cell cluster (n) and astrocyte clusters (o) from scRNAseq between WT and APPNLGF at 6 months. One-way ANOVA followed by Tukey’s comparisons test (j) was used for analysis. The mean ± s.e.m. ns: not significant.
Extended Data Fig. 5 Analysis of pathogenesis after Sox9cKO in APPNLGF mice.
a. Representative confocal Z-stack images of GFP (green) and Sox9 (red) in the APPNLGF; Sox9Con and APPNLGF; Sox9cKO. Quantification graphs of Sox9 intensity in each group (lower panel). n = 5 mice for each group. b. Brains from APPNLGF; Sox9Con (n = 4 mice) and APPNLGF; Sox9cKO (n = 4 mice) were homogenized, fractionated into 1% Triton X-100 soluble fraction, and processed for Western blot using antibodies against APP (Y188 clone). The levels of full length of APP, CTF99 and CTF83 were examined through Western blot. Actin was used as a loading control. Quantification graphs of APP intensities. Full length APP (upper panel), CTF99 (middle panel) and CTF 83 (lower panel) of each group. c, d. Brains from WT (n = 3 mice) and APPNLGF (n = 3 mice) (c) and APPNLGF; Sox9Con (n = 4 mice) and APPNLGF; Sox9cKO (n = 4 mice) (d) were homogenized, fractionated into 1% Triton X-100 soluble and insoluble fractions. The levels of Aβ were examined through Western blot. Actin was used as a loading control. Quantification graphs for the levels of soluble and insoluble Aβ (right panel). e. Representative confocal Z-stack images of Aβ (green) and NeuN (red), in the cortex (CTX, left panel) and hippocampus (HC, right panel) of WT, APPNLGF; Sox9Con, and APPNLGF; Sox9cKO. Scale bars, 100 µm. f. Quantification graphs for the number of NeuN-positive cells in each group. n = 5 mice of each group. g. Representative confocal Z-stack images of Aβ (blue), vGlut1 (red), and PSD95 (green) in the cortex (CTX, left panel) and hippocampus (HC, right panel) of WT, APPNLGF; Sox9Con, and APPNLGF; Sox9cKO. Scale bars, 10 µm. h. Quantification graphs of the number of vGlu1, PSD95, and synapse formation in each group. n = 5 mice of each group. i. Representative confocal Z-stack images of DAPI (blue), Aβ (violet), Sox9 (red), and GFP (green) on the hippocampus of WT, APPNLGF; Sox9Con, and APPNLGF; Sox9cKO. Scale bars, 10 µm. j. Quantification of the number of interactions by Sholl analysis of hippocampal astrocytes in each group with WT (e) or without WT (f). n = 5 mice for each group. k. Representative confocal Z-stack images of DAPI (blue), Sox9 (red), and GFAP (green) on the hippocampus of WT, APPNLGF; Sox9Con, and APPNLGF; Sox9cKO. Scale bars, 20 µm. l. Quantification graphs for GFAP intensities. n = 5 mice for each group. m. Representative traces of WT, APPNLGF; Sox9Con, and APPNLGF; Sox9cKO at 4 months showing spontaneous GCaMP6 activity in astrocytes from the hippocampus. n. Quantification of somatic ∆F/F GCaMP6 signal amplitude and frequency derived from 5 mice of each group. Two-sided unpaired Student’s t-test (a, b, d), One-way ANOVA followed by Tukey’s comparisons test (f, h, l), and Two-way ANOVA with Sidak’s correction (j) were used for analysis. The mean ± s.e.m.
Extended Data Fig. 6 Alteration of microglia after astrocyte-specific Sox9 manipulation in APPNLGF mouse.
a-d. Representative confocal Z-stack images of DAPI (blue) co-stained with Iba1 (green) and Aβ (red) in the WT, APPNLGF; Sox9Con, and APPNLGF; Sox9cKO at 4 months (a), WT, AAV-con, and AAV-Sox9ov in APPNLGF at 8 months (c). Scale bars, 20 µm. Quantification graphs of microglia coverage. =5 mice for each group (b, d). e. Representative confocal Z-stack images of Iba1 (blue) co-stained with CD68 (green) and Aβ (red) in the AAV-con and AAV-Sox9ov in APPNLGF at 8 months. Scale bars, 10 µm. f. Quantification of phagocytic index for engulfed Aβ normalized by microglia cell body area. n = 5 mice for each group. Two-sided unpaired Student’s t-test (f) and One-way ANOVA followed by Tukey’s comparisons test (b, d) were used for analysis. The mean ± s.e.m.
Extended Data Fig. 7 Astrocyte-specific overexpression of Sox9 in APPNLGF mouse.
a. Schematic illustration for AAV-GFAP-Sox9-Flag (AAV-Sox9ov) with AAV-GFAP-lck-smV5-4x6T (AAV-Con) in the cortex and hippocampus of the APPNLGF mouse line. b. Representative confocal Z-stack images of Sox9 (green) and DAPI (blue) in the cortex and hippocampus of the APPNLGF mouse. Scale bars, 500 µm and 100 µm. c. Quantification graphs of Sox9 intensity in each group. n = 5 mice of each group. d. Representative confocal Z-stack images of Sox9 (white) and GFP (green) with NeuN (red, upper panel) and Iba1 (red, lower panel) in the hippocampus of the APPNLGF mouse. Scale bars, 20 µm. e. Representative confocal Z-stack images of Aβ (green) and NeuN (red), in the cortex (CTX, upper panel) and hippocampus (HC, lower panel) of WT, AAV-Con, and AAV-Sox9. Scale bars, 50 µm. f. Quantification graphs for the number of NeuN-positive cells in each group. n = 5 mice of each group. g. Representative confocal Z-stack images of Aβ (blue), vGlut1 (red), and PSD95 (green) in the cortex (CTX, left panel) and hippocampus (HC, right panel) between AAV-Con and AAV-Sox9. Scale bars, 5 µm. h. Quantification graphs of the number of vGlu1, PSD95, and synapse formation in each group. n = 5 mice of each group. i. Schematic illustration of AAV-GFAP-mCherry-P2A vector in the hippocampus of APPNLGF; Aldh1l1-GFP mice. j. Representative confocal Z-stack images of GFP (green) and mCherry (red) with NeuN (blue, middle panel) and Iba1 (blue, right panel) in the hippocampus of the APPNLGF; Aldh1l1-GFP mouse. Scale bars, 200 µm, 50 µm (GFP, mCherry only), 20 µm (GFP, mCherry with NeuN or Iba1). k. Schematic illustration of AAV-GFAP-mCherry-P2A (AAV-mCh-Con) and AAV-GFAP-mCherry-P2A-Sox9 (AAV-mCh-Sox9ov) in the hippocampus of APPNLGF; Aldh1l1-GFP mice. l. Fluorescence-activated cell sorting (FACS) gating strategy for mCherry and GFP double-positive astrocytes. Two-sided unpaired Student’s t-test (c) and One-way ANOVA followed by Tukey’s comparisons test (f, h) were used for analysis. The mean ± s.e.m. All illustrations were created using BioRender.com.
Extended Data Fig. 8 Astrocyte-specific Aβ phagocytosis after manipulation of Sox9.
a-c. Representative confocal Z-stack images of GFP-positive astrocytes (blue) co-stained with lysosome membrane marker (Lamp1, red) and Aβ (green) in the hippocampus of WT and APPNLGF at 4 months (a), in the cortex of APPNLGF; Sox9Con, and APPNLGF; Sox9cKO at 4 months (b), and in the hippocampus of APPNLGF; Sox9Con, and APPNLGF; Sox9cKO at 4 months (c). Scale bars, 10 µm. Quantification of phagocytic index for engulfed Aβ that was normalized by astrocyte cell body area. n = 5 mice of each group (lower panel). d, e. Representative confocal Z-stack images of GFP-positive astrocytes (blue) co-stained with lysosome membrane marker (CathD, red) and Aβ (green) in the hippocampus of APPNLGF; Sox9Con, and APPNLGF; Sox9cKO at 4 months (d), and AAV-Con and AAV-Sox9ov at 8 months (e). Scale bars, 5 µm. Quantification of phagocytic index for engulfed Aβ t normalized by astrocyte cell body area. n = 5 mice of each group (lower panel). f. Schematic illustration for injection of AAV-Synapsin-APP (APP NL-F) in the hippocampus of WT; Aldh1l1-GFP mice. g. Representative confocal Z-stack images of GFP-positive astrocytes with Aβ (red) in the hippocampus of the Con and APPNL-F mice. Scale bars, 100 µm. Quantification graphs of the Aβ intensity in each group (right panel). n = 5 mice of each group. h. Representative confocal Z-stack images of GFP-positive astrocytes (blue) co-stained with lysosome membrane marker (Lamp1, purple) and Aβ (green) in the hippocampus of Con and APPNL-F mice. Scale bars, 10 µm. i. Representative confocal images of Thioflavin S staining in the hippocampus of the Con and APPNL-F mice. Scale bars, 100 µm and 20 µm. Quantification graphs of the Thioflavin S intensity in each group (right panel). n = 5 mice of each group. j. Representative confocal images of Thioflavin S staining in the hippocampus of the APPNL-F/Con and APPNL-F/Sox9ov mice. Scale bars, 100 µm and 20 µm. Quantification graphs of Thioflavin S intensity in each group (right panel). n = 5 mice of each group. k. Representative confocal Z-stack images of GFP (blue), Aβ (red), and vGlut1 (green, upper panel) or PSD95 (green, lower panel) in the hippocampus of APPNL-F mice. n = 5 mice. Scale bars, 2 µm. A two-sided unpaired Student’s t-test (a-e, g-j) was used for analysis. The mean ± s.e.m. All illustrations were created using BioRender.com.
Extended Data Fig. 9 Regulation of MEGF10 expression by Sox9.
a-d. UMAP plot of astrocyte clusters from human anterior hippocampal cortex of human AD patients (Human AD) (a) and APPNLGF mice (c). Violin plot of each phagocytosis-related receptor from Sox9 high-expressing astrocytes and Sox9 low-expressing astrocytes in Human AD (b) and APPNLGF mice (d). Increased genes were marked blue, decreased genes were marked red, and unchanged genes were marked black (b, d). e-g. Representative confocal Z-stack images of GFP-positive astrocytes (green) co-stained with LRP1 (red, e), CD36 (red, f), and RAGE (red, g) in the APPNLGF; Sox9Con, and APPNLGF; Sox9cKO at 4 months. Scale bars, 5 µm. Quantification graphs of the LRP1 (a), CD36 (b), and RAGE (c) intensity in each group (right panel). n = 5 mice of each group. h. Representative confocal Z-stack images of GFP-positive astrocytes (green) co-stained with MEGF10 in the WT, 4 months-old APPNLGF, and 8 months-old APPNLGF. Scale bars, 10 µm. Quantification graphs of the MEGF10 intensity in each group. n = 5 mice of each group (right panel). i, j. ChIP-PCR screen identifying Sox9 association with the Megf10 promoter. Quantification graphs (right panel) were normalized in triplicate with individual qPCR (Each sample was created by combining tissues obtained from 6 mice of each group, 3 technical repeats). k. Representative confocal Z-stack images of Aβ (blue), vGlut1 (red), and PSD95 (green) in the hippocampus of WT, AAV-Con, and AAV-MEGF10. Scale bars, 5 µm. l. Quantification graphs of the number of vGlu1, PSD95, and synapse formation in each group. n = 5 mice of each group. m-p. Representative confocal Z-stack images of GFP (green) and MEGF10 (red) in the hippocampus of 4 month-old WT, 18 month-old WT, and 18 month-old Sox9cKO (m), AAV-Con, and AAV-MEGF10 at 18 months (o). Scale bars, 10 µm. Quantification graphs of MEGF10 intensity in each group. n = 5 mice of each group (n, p). Two-sided unpaired Student’s t-test (e-g, i, p) and One-way ANOVA followed by Tukey’s comparisons test (h, l, n) were used for analysis. The mean ± s.e.m.
Extended Data Fig. 10 Synaptic phagocytosis.
a, c, e. Representative confocal Z-stack images of GFP-positive astrocytes (blue) co-stained with CathD (green) and vGlut1 (red) in the APPNLGF; Sox9Con and APPNLGF; Sox9cKO at 4 months (a), AAV-con and AAV-Sox9ov in APPNLGF at 8 months (c), and AAV-con and AAV-Megf10ov in APPNLGF at 8 months (e). Scale bars, 5 µm. Quantification of phagocytic index for engulfed Aβ normalized by astrocyte cell body area. n = 5 mice for each group (right panel). b, d, f. Representative confocal Z-stack images of GFP-positive astrocytes (blue) co-stained with CathD (green) and PSD95 (red) in the APPNLGF; Sox9Con and APPNLGF; Sox9cKO at 4 months (b), AAV-con and AAV-Sox9ov in APPNLGF at 8 months (d), and AAV-con and AAV-MEGF10ov in APPNLGF at 8 months (f). Scale bars, 5 µm. Quantification of phagocytic index for engulfed Aβ normalized by astrocyte cell body area. n = 5 mice for each group (right panel). g. Schematic illustration for the AAV-hSyn-PSD-mCherry-pHluorin system. h-l. Representative confocal Z-stack images of GFP-positive astrocytes (blue) co-stained with mCherry (red) and pHluorin (green) in the WT (h), AAV-con and AAV-Sox9ov in APPNLGF at 8 months (i), and AAV-con and AAV-MEGF10ov in APPNLGF at 8 months (k). The yellow arrow shows mCherry only in astrocytes. Scale bars, 5 µm. Quantification of phagocytic index for engulfed Aβ normalized by astrocyte cell body area (j, l). n = 5 mice for each group. A two-sided unpaired Student’s t-test (a-l) was used for analysis. The mean ± s.e.m. All illustrations were created using BioRender.com.
Supplementary information
Supplementary Table 1
RNA-seq of FACS-sorted astrocytes from 4 months and 18 months.
Supplementary Table 2
RNA-seq of FACS-sorted astrocytes from WT and Sox9-cKO at 4 months and 18 months.
Supplementary Table 3
ChIP−seq results and scRNA-seq.
Supplementary Table 4
RNA-seq of FACS-sorted astrocytes between AAV–Con and AAV–Sox9ov.
Supplementary Table 5
Detailed breeding strategy.
Supplementary Table 6
Detailed statistical information.
Source data
Source Data Fig. 1
Statistical source.
Source Data Fig. 2
Statistical source.
Source Data Fig. 3
Statistical source.
Source Data Fig. 4
Statistical source.
Source Data Fig. 5
Statistical source.
Source Data Extended Data Fig.1
Statistical source.
Source Data Extended Data Fig. 2
Statistical source.
Source Data Extended Data Fig. 3
Statistical source.
Source Data Extended Data Fig. 4
Statistical source.
Source Data Extended Data Fig. 5
Statistical source.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source.
Source Data Extended Data Fig. 7
Statistical source.
Source Data Extended Data Fig. 8
Statistical source.
Source Data Extended Data Fig. 9
Statistical source.
Source Data Extended Data Fig. 10
Statistical source.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choi, DJ., Murali, S., Kwon, W. et al. Astrocytic Sox9 overexpression in Alzheimer’s disease mouse models promotes Aβ plaque phagocytosis and preserves cognitive function. Nat Neurosci 29, 88–99 (2026). https://doi.org/10.1038/s41593-025-02115-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-025-02115-w