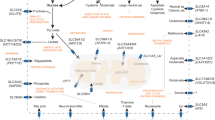
Abstract
Taurine plays a crucial role in mitochondrial translation. Mammalian cells obtain taurine via exogenous uptake mediated by the plasma membrane transporter SLC6A6 or via cytosolic biosynthesis. However, it remains unclear how taurine enters mitochondria and impacts cellular metabolism. Here we show that SLC6A6, but not exogenous taurine, is essential for mitochondrial metabolism and cancer cell growth. We discover that SLC6A6 also localizes to mitochondria and imports taurine for mitochondrial transfer RNA modifications. SLC6A6 deficiency specifically reduces mitochondrial taurine abundance and abrogates mitochondrial translation and cell proliferation. We identify protein kinase A as a regulator of SLC6A6 subcellular localization, as it promotes SLC6A6 presence at the plasma membrane while inhibiting its mitochondrial localization. Furthermore, we identify NFAT5 as a key regulator of mitochondrial function through SLC6A6 and demonstrate that targeting the NFAT5–SLC6A6 axis markedly impairs mitochondrial translation and tumour growth. Together, these findings suggest that SLC6A6 is a mitochondrial taurine transporter and an exploitable metabolic dependency in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Original data and images and western blots are available via Zenodo at https://doi.org/10.5281/zenodo.17853672 (ref. 68). The metabolomics data are available from MetaboLights under identifiers MTBLS13422 and MTBLS13424. The proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier: PXD071408. All other data supporting the findings of this study are also available from the corresponding authors upon request. Source data are provided with this paper.
Code availability
No custom code was used in this study.
References
Stipanuk, M. H., Dominy, J. E. Jr, Lee, J. I. & Coloso, R. M. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr. 136, 1652S–1659S (2006).
Ripps, H. & Shen, W. Review: taurine: a ‘very essential’ amino acid. Mol. Vis. 18, 2673–2686 (2012).
Espe, M. & Holen, E. Taurine attenuates apoptosis in primary liver cells isolated from Atlantic salmon (Salmo salar). Br. J. Nutr. 110, 20–28 (2013).
Baliou, S. et al. Protective role of taurine against oxidative stress (review). Mol. Med. Rep. 24, 605 (2021).
Falany, C. N., Johnson, M. R., Barnes, S. & Diasio, R. B. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J. Biol. Chem. 269, 19375–19379 (1994).
Schaffer, S. W., Azuma, J. & Madura, J. D. Mechanisms underlying taurine-mediated alterations in membrane function. Amino Acids 8, 231–246 (1995).
De Luca, A., Pierno, S. & Camerino, D. C. Taurine: the appeal of a safe amino acid for skeletal muscle disorders. J. Transl. Med. 13, 243 (2015).
Tappaz, M. L. Taurine biosynthetic enzymes and taurine transporter: molecular identification and regulations. Neurochem. Res. 29, 83–96 (2004).
Ueki, I. et al. Knockout of the murine cysteine dioxygenase gene results in severe impairment in ability to synthesize taurine and an increased catabolism of cysteine to hydrogen sulfide. Am. J. Physiol. Endocrinol. Metab. 301, E668–E684 (2011).
Cao, T. et al. Cancer SLC6A6-mediated taurine uptake transactivates immune checkpoint genes and induces exhaustion in CD8+ T cells. Cell 187, 2288–2304 e2227 (2024).
Sharma, S. et al. Taurine from tumour niche drives glycolysis to promote leukaemogenesis. Nature 644, 263–272 (2025).
Singh, P. et al. Taurine deficiency as a driver of aging. Science 380, eabn9257 (2023).
Suzuki, T. et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11, 4269 (2020).
Morscher, R. J. et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 554, 128–132 (2018).
Kopajtich, R. et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 95, 708–720 (2014).
Ghezzi, D. et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 90, 1079–1087 (2012).
Lu, J. L. et al. Taurine hypomodification underlies mitochondrial tRNATrp-related genetic diseases. Nucleic Acids Res. 52, 13351–13367 (2024).
Peng, G. X. et al. The human tRNA taurine modification enzyme GTPBP3 is an active GTPase linked to mitochondrial diseases. Nucleic Acids Res. 49, 2816–2834 (2021).
Baliou, S. et al. Significance of taurine transporter (TauT) in homeostasis and its layers of regulation (review). Mol. Med. Rep. 22, 2163–2173 (2020).
Yoo, H. C. et al. A variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 31, 267–283 (2020).
Luongo, T. S. et al. SLC25A51 is a mammalian mitochondrial NAD+ transporter. Nature 588, 174–179 (2020).
Kory, N. et al. MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci. Adv. 6, eabe5310 (2020).
Kory, N. et al. SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science 362, eaat9528 (2018).
Papalazarou, V. et al. Phenotypic profiling of solute carriers characterizes serine transport in cancer. Nat. Metab. 5, 2148–2168 (2023).
Hewton, K. G., Johal, A. S. & Parker, S. J. Transporters at the interface between cytosolic and mitochondrial amino acid metabolism. Metabolites 11, 112 (2021).
Wang, Y. et al. SLC25A39 is necessary for mitochondrial glutathione import in mammalian cells. Nature 599, 136–140 (2021).
Yoneshiro, T. et al. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature 572, 614–619 (2019).
Verkerke, A. R. P. et al. SLC25A48 controls mitochondrial choline import and metabolism. Cell Metab. 36, 2156–2166 e2159 (2024).
Yousefi, R. et al. Monitoring mitochondrial translation in living cells. EMBO Rep. 22, e51635 (2021).
Aich, A. et al. Defective mitochondrial COX1 translation due to loss of COX14 function triggers ROS-induced inflammation in mouse liver. Nat. Commun. 15, 6914 (2024).
Fendt, S. M. et al. Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236 (2013).
Mullen, A. R. et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 (2011).
Metallo, C. M. et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2011).
Chen, W. W., Freinkman, E. & Sabatini, D. M. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat. Protoc. 12, 2215–2231 (2017).
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Du, B., Cheng, L., Xie, J., Chen, L. & Yan, K. Molecular basis of human taurine transporter uptake and inhibition. Nat. Commun. 16, 7394 (2025).
Li, F. et al. FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome. Nat. Cell Biol. 22, 728–739 (2020).
Voss, J. W., Pedersen, S. F., Christensen, S. T. & Lambert, I. H. Regulation of the expression and subcellular localization of the taurine transporter TauT in mouse NIH3T3 fibroblasts. Eur. J. Biochem. 271, 4646–4658 (2004).
Han, X., Budreau, A. M. & Chesney, R. W. Role of conserved peptide in taurine transporter inactivation modulated by protein kinase C. J. Am. Soc. Nephrol. 7, 2088–2096 (1996).
Lambert, I. H. & Hoffmann, E. K. Regulation of taurine transport in Ehrlich ascites tumor cells. J. Membr. Biol. 131, 67–79 (1993).
Go, C. D. et al. A proximity-dependent biotinylation map of a human cell. Nature 595, 120–124 (2021).
Ito, T. et al. Expression of taurine transporter is regulated through the TonE (tonicity-responsive element)/TonEBP (TonE-binding protein) pathway and contributes to cytoprotection in HepG2 cells. Biochem. J. 382, 177–182 (2004).
Woo, S. K., Lee, S. D., Na, K. Y., Park, W. K. & Kwon, H. M. TonEBP/NFAT5 stimulates transcription of HSP70 in response to hypertonicity. Mol. Cell. Biol. 22, 5753–5760 (2002).
Han, E. J. et al. Suppression of NFAT5-mediated inflammation and chronic arthritis by novel κB-binding inhibitors. EBioMedicine 18, 261–273 (2017).
Delaunay, S. et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature 607, 593–603 (2022).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Minton, D. R. et al. Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol. Cell 69, 610–621 (2018).
Suzuki, T., Suzuki, T., Wada, T., Saigo, K. & Watanabe, K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 21, 6581–6589 (2002).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Liu, Y. et al. A novel chemical-defined medium with bFGF and N2B27 supplements supports undifferentiated growth in human embryonic stem cells. Biochem. Biophys. Res. Commun. 346, 131–139 (2006).
Lee, J. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006).
Jacobsen, J. G. & Smith, L. H. Biochemistry and physiology of taurine and taurine derivatives. Physiol. Rev. 48, 424–511 (1968).
Stipanuk, M. H. Role of the liver in regulation of body cysteine and taurine levels: a brief review. Neurochem. Res. 29, 105–110 (2004).
Lai, Y., Tse, C. M. & Unadkat, J. D. Mitochondrial expression of the human equilibrative nucleoside transporter 1 (hENT1) results in enhanced mitochondrial toxicity of antiviral drugs. J. Biol. Chem. 279, 4490–4497 (2004).
Martinez-Reyes, I. & Chandel, N. S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11, 102 (2020).
Zong, W. X., Rabinowitz, J. D. & White, E. Mitochondria and cancer. Mol. Cell 61, 667–676 (2016).
Pavlova, N. N., Zhu, J. & Thompson, C. B. The hallmarks of cancer metabolism: still emerging. Cell Metab. 34, 355–377 (2022).
Vyas, S., Zaganjor, E. & Haigis, M. C. Mitochondria and cancer. Cell 166, 555–566 (2016).
Vasan, K., Werner, M. & Chandel, N. S. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 32, 341–352 (2020).
Yap, T. A. et al. Complex I inhibitor of oxidative phosphorylation in advanced solid tumors and acute myeloid leukemia: phase I trials. Nat. Med. 29, 115–126 (2023).
Bock, F. J. & Tait, S. W. G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 21, 85–100 (2020).
Li, F. et al. Blocking methionine catabolism induces senescence and confers vulnerability to GSK3 inhibition in liver cancer. Nat. Cancer 5, 131–146 (2024).
Mi, W. et al. α-Ketoglutarate dictates AMPK protein synthesis for energy sensing in human cancers. Nat. Chem. Biol. https://doi.org/10.1038/s41589-025-02013-z (2025).
Yang, L. et al. ACSS2 drives senescence-associated secretory phenotype by limiting purine biosynthesis through PAICS acetylation. Nat. Commun. 16, 2071 (2025).
Wen, M. et al. Li Fuming BET inhibition induces GDH1-dependent glutamine metabolic remodeling and vulnerability in liver cancer. Life Metab. 3, loae016 (2024).
Su, X., Lu, W. & Rabinowitz, J. D. Metabolite spectral accuracy on orbitraps. Anal. Chem. 89, 5940–5948 (2017).
Tang, Z. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017).
Li, L. et al. SLC6A6 imports taurine into mitochondria to sustain mitochondrial translation and tumour growth. Zenodo https://doi.org/10.5281/zenodo.17853672 (2025).
Acknowledgements
We are grateful to R. Cheng (Core Facility of Metabolomics, Institute of Metabolism and Integrative Biology, Fudan University) and N. Zhu (Core Facility of Molecular Biology, CAS Centre for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology) for technical assistance. This work was supported by the National Key R&D Program of China (grant nos. 2022YFA1103900 to Fuming Li, 2021YFA1300800 to X.-L.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB0570000 to X.-L.Z.); CAS Project for Young Scientists in Basic Research (grant no. YSBR-075) and National Natural Science Foundation of China (grant nos. 82273223 and 82573026 to Fuming Li, 32422048 to Li Chen and 32271300 to X.-L.Z.).
Author information
Authors and Affiliations
Contributions
L.L. and Fuming Li conceived the project and designed the experiments. Fuming Li, Li Chen, X.-L.Z. and P.L. supervised the project. L.L., J.Y., Z.-Q.C., L.W., F.Y., L.Z., W.M., X.X., H.Y. and J.W. performed the experiments. X.C. and Fei Li helped with the bioinformatic analysis. J.Y., X.L., Z.-Q.C., Li Chen and X.-L.Z. performed LC–MS and data analysis. L.L., J.Y., X.L., Z.-Q.C., Li Chen, X.-L.Z. and Fuming Li analysed the data. L.L. and Fuming Li wrote the paper. T.-J.Z., Ligong Chen and H.J. revised the manuscript provided. All authors revised and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Katsuhisa Inoue and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Alfredo Gimenez-Cassina and Jean Nakhle, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Exogenous taurine is dispensable for liver and lung cancer cell growth.
a, Relative growth of Hepa1-6 and HepaMP9-1 cells in D10 and D-N2B27 medium for 72 h. n = 4 biological replicates. b, Relative 293 T cell growth in D10 and D-N2B27 medium. c, d, Relative growth of of 293 T (c) and IMR90 (d) cells in DMEM containing indicated supplements for 72 h. n = 4 biological replicates. e, Relative growth of different cancer cells cultured in D10 or D-dFBS medium supplemented with indicated concentrations of taurine. n = 3 biological replicates. f, g, Representative crystal violet staining (f) and quantification (g) of clones from cancer cells cultured in D10 or D-dFBS medium supplemented with indicated concentrations of taurine. n = 3 biological replicates (g). h, Immunoblot analysis of taurine biosynthetic enzymes in different cell lines. HSP90 serves as loading control. All statistical graphs show the mean ± s.e.m. P values were calculated using a two-tailed Student’s t-test (a-c) or one-way ANOVA (d, e, g). Experiments were repeated at least three times independently, with similar results (a-h).
Extended Data Fig. 2 SLC6A6 is required for liver and lung cancer cell growth.
a, qPCR analysis of SLC6A6 mRNA levels from control (shCtrl) and SLC6A6 (sh6A6)/Slc6a6 (sh6a6) knockdown cancer cells. Representative results from three independent experiments are shown. b, c, Representative crystal violet staining (b) and quantification (c) of clonal formation from shCtrl and sh6A6 cancer cells. n = 4 biological replicates (c). d, Representative TMRE staining of shCtrl and sh6A6 cancer cells. Scale bar: 10 µm. e, Immunoblots of indicated mitochondrial proteins from shCtrl and sh6A6 cancer cells cultured in D-N2B27 medium. HSP90 serves as loading control. f, Relative growth of indicated groups of cancer cells cultured in D10 or D-dFBS medium. n = 3 biological replicates. g, Immunoblots of mitochondrial proteins from indicated groups cells cultured in D10 medium. HSP90 serves as loading control. All statistical graphs show the mean ± s.e.m. P values were calculated using one-way ANOVA (a, c, f). Experiments were repeated at least three times independently, with similar results (a-g).
Extended Data Fig. 3 SLC6A6 is required for mitochondrial translation and tRNA modifications.
a, Immunoblot analysis of puromycin incorporation into mitochondrial encoded proteins from In Organello Translation assay. TOM20 serves as loading control. b, Secondary structures of hmtRNATrp with indicated tm5U modification. Arrows indicate specific RNase T1 cleavage sites generating U34‑containing fragments. c, Intensity fractions (%) of U34-containing fragments with different modification states in the indicated A549 cells. Sequences are annotated with their mass-to-charge ratio (m/z), charge state, and relative modification frequencies. Experiments were repeated three times independently, with similar results (a).
Extended Data Fig. 4 SLC6A6 deficiency alters glucose and glutamine metabolism in cancer cells.
a, A scheme of [U-13C] glucose isotope labeling and flux analysis. b, [U-13C] glucose-derived citrate M + 2 labeling percentage (% of total pool) of A549 and Huh7 cells in control and SLC6A6 knockdown groups. n = 3 biological replicates. c, A scheme of [U-13C] glutamine isotope labeling and flux analysis. d, e, [U-13C] glutamine-derived citrate M + 4 (d) and M + 5 (e) labeling percentage (% of total pool) of A549 and Huh7 cells in control and SLC6A6 knockdown groups. n = 3 biological replicates. f, [U-13C] glutamine-derived Malate M + 3 and M + 4 labeling percentage (% of total pool) of A549 and Huh7 cells in control and SLC6A6 knockdown groups. n = 3 biological replicates. All statistical graphs show the mean ± s.e.m. P values were calculated using two-tailed Student’s t-test (b) and one-way ANOVA (d-f). Experiments were repeated three times independently, with similar results (b, d-f).
Extended Data Fig. 5 Mitochondrial localization of SLC6A6.
a, Partial colocalization of SLC6A6-EGFP with PM marker CellMaskTM stain (red) in different cancer cell lines. Scale bar: 10 µm. b, Quantification of colocalization between SLC6A6-EGFP and CellMaskTM stain in (a). c, Colocalization of mouse SLC6A6-EGFP with Mitotracker in HepaMP9-1 cells. Scale bar: 10 µm. d, Quantification of colocalization between mouse SLC6A6-EGFP and Mitotracker. e, Representative IF staining of V5 and COX4 in A549 and Huh7 cells expressing SLC6A6-V5. Scale bar: 10 µm. f, Quantification of colocalization between SLC6A6-5 and COX4 in (e). g, Immunoblots of SLC6A6 from input, PM and mitochondrial lysates isolated from A549 and Huh7 cells and loaded at same percentages. h, Conserved MTS sequence across different species. Positively charged residues and mutated residues in MTS are shown in red. i, j, Representative EGFP and Mitotracker staining (i), and quantification of colocalization (j) in Huh7 cells expressing EGFP (CMV-EGFP), or EGFP fused to wild type (WT) or mutant (MT) SLC6A6 MTS. k, Immunoblots of SLC6A6-HA from input, PM and Mito lysates from Huh7 cells expressing SLC6A6-HA-WT or SLC6A6-HA-MT MTS. l, Relative growth of control (shCtrl) and SLC6A6 knockdown (sh6A6) A549 and Huh7 cells expressing shRNA-resistant SLC6A6-WT or SLC6A6-MT. n = 4 biological replicates. All statistical graphs show the mean ± s.e.m. P values were calculated using one-way ANOVA (l). Experiments were repeated at least three times independently, with similar results (a-g, i-k).
Extended Data Fig. 6 SLC6A6 mediates mitochondrial taurine transport.
a, Relative taurine abundance in control (shCtrl) and SLC6A6 knockdown (sh6A6) A549 and Huh7 cells. n = 3 biological replicates. b, Representative immunofluorescent imaging of CoroNaTM Green and Mitotracker staining in different cancer cells. Scale bar: 10 µm. c, Relative mitochondrial D4 taurine abundance after incubating purified mitochondria with indicated concentrations of D4 taurine, NaCl and P4S for 1 h. n = 3 LC-MS detections. n = 3 biological replicates. d, Relative mitochondrial D4 taurine abundance after incubating purified mitochondria with 10 mM taurine and 10 mM NaCl and indicated concentrations of P4S for 1 h. n = 3 LC-MS detections. All statistical graphs show the mean ± s.e.m. P values were calculated using one-way ANOVA (a, c, d). Experiments were repeated three times independently, with similar results (a-d).
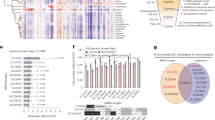
Extended Data Fig. 7 PKA promotes SLC6A6 localization to plasma membrane.
a, Fold change in cell number after culturing A549 and Huh7 cells with indicated concentrations of Fsk for 72 h. n = 4 biological replicates. b, Immunoblots of A549 and Huh7 cells treated with indicated concentrations of Fsk for 72 h. HSP90 serves as loading control. c, A volcano plot showing adjusted p values and fold change in intensities of SLC6A6-binding proteins from proteomic analysis. n = 3 biological replicates. d, KEGG analysis of 589 SLC6A6-binding proteins. e, Immunoblots of input and PM lysates from control (shCtrl) and RAB2A knockdown (shRAB2A) A549 and Huh7 cells. All statistical graphs show the mean ± s.e.m. P values were calculated using one-way ANOVA (a) and two-tailed Student’s t-test (c). Experiments were repeated three times independently, with similar results (a, b, e).
Extended Data Fig. 8 NFAT5 is required for cancer cell growth and mitochondrial function.
a, Kaplan–Meier overall survival plots stratified by SLC6A6 high (red) or low (blue) mRNA levels from TCGA database. b, Kaplan–Meier overall survival plots stratified by NFAT5 high (red) or low (blue) mRNA levels from TCGA database. A log-rank Mantel–Cox test was performed between the groups of each plot (a, b). c, Positive correlation between SLC6A6 and NFAT5 mRNA levels from pan-cancer analysis. d, e, Representative crystal violet staining (d) and quantification (e) of clonal formation from shCtrl and shNFAT5 cancr cells. n = 4 biological replicates (e). f, Quantification of shCtrl and shNFAT5 Huh7 and H1299 cell death (% trypan blue+) in galactose-containing medium (10 mM) for 48 h. n = 4 biological replicates. g, Relative taurine abundance in shCtrl and shNFAT5 A549 and Huh7 cells. n = 3 biological replicates. Data presented as mean ± s.e.m. of three independent experiments; statistical significance was determined by one-way ANOVA (e-g). Experiments were repeated three times independently, with similar results (d-g).
Extended Data Fig. 9 Targeting NFAT5-SLC6A6 axis limits mitochondrial translation and tumour growth.
a, Relative number of cancer cells treated with indicated doses of KRN2 for 72 h. Cell numbers were normalized to vehicle control groups. n = 4 biological replicates. b, Relative cell number of vector control (Vector) and SLC6A6-overexpressing (6A6-OE) A549 and Huh7 cells treated with indicated doses of KRN2 for 72 h. Cell numbers were normalized to corresponding vehicle control groups. n = 3 biological replicates. c, Representative A549 and H1299 xenograft tumour images from vehicle control and KRN2 treatment groups. d, Quantification of A549 and H1299 xenograft tumour weight from vehicle control and KRN2 treatment groups. Ctrl: n = 8 for A549, n = 10 for H1299. KRN2: n = 8 for A549, n = 6 for H1299. e, Representative HepaMP9-1 allograft tumour images from vehicle control and KRN2 treatment groups. f, Quantification of HepaMP9-1 allograft tumour weight from vehicle control (n = 10) and KRN2 (n = 10) treatment groups. g, Q-PCR analysis of HepaMP9-1 allograft tumours from vehicle and KRN2 cohorts. n = 3 tumours. h, Immunoblot analysis of HepaMP9-1 allograft tumours from vehicle and KRN2 cohorts. n = 3 tumours. i, Quantification of relative mouse body weight change from vehicle and KRN2 treatment cohorts. n = 5 mice for each cohort. j, Representative HE staining of liver, lung and kidney sections from vehicle control and KRN2 treated mice. Scale bar: 10 µm. Data presented as mean ± s.e.m. of three independent experiments; statistical significance was determined by one-way ANOVA (a, b) and a two-tailed Student’s t-test (d, f, g, i). Experiments were repeated three times independently, with similar results (a, b, g, h, j).
Supplementary information
Supplementary Table 1
Antibody details.
Supplementary Table 2
Quantitative reverse transcription PCR primer sequences.
Supplementary Table 3
shRNA/sgRNA oligo sequences.
Supplementary Table 4
Proteomics data analysis.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1.
Source Data Fig. 1
Raw images from western blotting in Fig. 1.
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 2
Raw images from western blotting in Fig. 2.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 3
Raw images from western blotting in Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Fig. 4
Raw images from western blotting in Fig. 4.
Source Data Fig. 5
Statistical source data for Fig. 5.
Source Data Fig. 5
Raw images from western blotting in Fig. 5.
Source Data Fig. 6
Statistical source data for Fig. 6.
Source Data Fig. 6
Raw images from western blotting in Fig. 6.
Source Data Fig. 7
Statistical source data for Fig. 7.
Source Data Fig. 7
Raw images from western blotting in Fig. 7.
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1.
Source Data Extended Data Fig. 1
Raw images from western blotting in Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2.
Source Data Extended Data Fig. 2
Raw images from western blotting in Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Raw images from western blotting in Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5.
Source Data Extended Data Fig. 5
Raw images from western blotting in Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Statistical source data for Extended Data Fig. 6.
Source Data Extended Data Fig. 7
Statistical source data for Extended Data Fig. 7.
Source Data Extended Data Fig. 7
Raw images from western blotting in Extended Data Fig. 7.
Source Data Extended Data Fig. 8
Statistical source data for Extended Data Fig. 8.
Source Data Extended Data Fig. 9
Statistical source data for Extended Data Fig. 9.
Source Data Extended Data Fig. 9
Raw images from western blotting in Extended Data Fig. 9.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, L., You, J., Chai, ZQ. et al. SLC6A6 imports taurine into mitochondria to sustain mitochondrial translation and tumour growth. Nat Metab (2026). https://doi.org/10.1038/s42255-026-01455-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42255-026-01455-6