Abstract
Recalled memories become transiently labile and require stabilization1,2,3. The mechanism for stabilizing memories of survival-critical experiences, which are often emotionally salient and repeated, remains unclear4. Here we identify an astrocytic ensemble that is transcriptionally primed by emotional experience and functionally triggered by repeated experience to stabilize labile memory. Using a novel brain-wide Fos tagging and imaging method, we found that astrocytic Fos ensembles were preferentially recruited in regions with neuronal engrams5 and were more widespread during fear recall than during conditioning. We established the induction mechanism of the astrocytic ensemble, which involves two steps: (1) an initial fear experience that induces day-long, slow astrocytic state changes with noradrenaline receptor upregulation; and (2) enhanced noradrenaline responses during recall, a repeated experience, enabling astrocytes to integrate coincident signals from local engrams and long-range noradrenergic projections, which induce secondary astrocytic state changes, including the upregulation of Fos and the neuromodulatory molecule IGFBP2. Pharmacological and genetic perturbation of the astrocytic ensemble signalling modulate engrams, and memory stability and precision. The astrocytic ensemble thus acts as a multiday trace in a subset of astrocytes after experience-dependent neural activity, which are eligible to capture future repeated experiences for stabilizing memories.
Similar content being viewed by others
Main
Recall transiently destabilizes memories, which require re-stabilization to become long-lasting1,2,3. Despite its importance in human cognition and neuropsychiatric disorders6,7, the mechanisms that specifically stabilize memories of critical experiences—those that are essential for survival and frequently marked by emotional salience and repetition—remain incompletely understood. Memory traces are linked to specialized neuronal ensembles (neuronal engrams), which include neuronal populations that become Fos+ during both initial and repeated experiences8. However, neuronal Fos alone, a plasticity marker that indicates circuit remodelling9, is insufficient for memory stabilization, implying the need for additional cellular substrates.
By tiling the entire mammalian central nervous system (CNS), astrocytes structurally and functionally interact with neurons and other glia. Their functions contribute to information processing and animal behaviour, and their dysfunctions are widely implicated in CNS disorders10,11. Astrocytes are known to be developmentally diverse12,13,14 and can adaptively change their molecular programmes in response to physiology11,15,16,17,18 and pathology19,20,21—hereafter referred to as state—in specific ways that suggest flexibly altered functions in a context-dependent manner. This adaptability is akin to experience-dependent neuronal responses (such as Fos upregulation)22 and the subsequent formation of inter-regional neuronal engram network8. However, the manifold nature of astrocyte responses through an extensive array of cell-surface receptors for neuroactive biomolecules19,23,24 have made it challenging to determine whether, when and how multiple inputs are integrated by a subset of astrocytes to generate specific states that form astrocyte ensembles that are responsible for particular cellular outputs15,18,24,25. This is because the field has lacked adequate tools to explore behaviourally relevant astrocyte ensembles (BAEs) in an unbiased manner at the single-cell level as well as in a brain-wide manner, which would enable ensemble-specific molecular dissection and functional perturbation.
Brain-wide BAE tagging
To agnostically and holistically identify BAEs, we developed a brain-wide astrocyte Fos tagging tool that enables permanent genetic access to BAEs that express Fos due to a specific experience. Although behaviourally relevant stimuli-induced astrocyte Fos mRNA22 and c-Fos protein18,26 have been observed in selected brain regions, its brain-wide profiling remains largely unexplored. We systematically validated a novel genetic and imaging strategy for whole-brain BAE tagging, demonstrating astrocyte selectivity, Fos sensitivity and temporal regulation (Extended Data Figs. 1–3). First, using a CNS-wide-transducing adenovirus (AAV) vector (AAV-PHP.eB) with an astrocyte-specific GfaABC1D promoter27, we achieved widespread, astrocyte-selective (97.3–99.5%) expression (Extended Data Fig. 1). Second, we tested whether using AAV-PHP.eB to express Cre-dependent mNeonGreen (mNG) can serve as a marker of transient astrocytic Fos induction in Fos-iCreERT2 (TRAP2) mice28. This intersectional approach aimed to drive astrocytic mNG expression only when the Fos promoter was active during fast-acting 4-hydroxytamoxifen (4-OHT) administration, enabling behaviour epoch-specific astrocyte Fos tagging across the CNS. As a positive control, we virally expressed the astrocytic Gαq-coupled designer receptor exclusively activated by designer drugs (DREADD) hM3Dq, a strong inducer of astrocyte c-Fos26,29,30, combined with 4-OHT and the hM3Dq agonist clozapine N-oxide (CNO) intraperitoneal injection. hM3Dq stimulation yielded approximately 282 mNG+ astrocytes per mm2, and significantly lower tagging in controls (Extended Data Fig. 2). Third, to probe the temporal dynamics of Fos-mNG tagging, dark-adapted mice were exposed to light at various intervals relative to 4-OHT administration28. Tagging peaked within a 6-h window around 4-OHT administration, with an approximately 22-fold increase in astrocytes in visual cortex compared with those in somatosensory cortex, indicating regional specificity and dependence on local neuronal activity (Extended Data Fig. 3). Collectively, these experiments validated our behaviour epoch-specific, brain-wide BAE tagging strategy.
Recall induces astrocyte ensembles
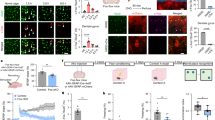
To broadly explore BAEs in a learning paradigm, we used a contextual fear conditioning and recall task5,28 (Fig. 1a). TRAP2::AAV-PHP.eB-GfaABC1D-DIO-mNG mice underwent three days of habituation to minimize contextual novelty and isolate associative components of fear learning and recall28,31. On day 4, one group received 3 foot shocks with immediate 4-OHT (FC), and another received 4-OHT immediately after recall 24 h post-FC (FR). Controls were non-shocked mice (NoFC) or fear-conditioned mice kept in the home cage for 24 h and given 4-OHT without recall (NoFR). Shocked groups exhibited robust freezing in the conditioned chamber, indicating learning and recall (Fig. 1b). One week later, whole-brain serial two-photon tomography (STPT)32 enabled the detection of individual mNG+ astrocytes (BAEs) (Supplementary Videos 1 and 2). After excluding ventricular niches for bona fide astrocyte counts (Methods), BAE counts across 677 regions covered approximately 80.7% of the brain (Supplementary Table 2).
a, Fos+ astrocytes were tagged with Cre-dependent mNG AAV in four cohorts (NoFC, FC, NoFR and FR), followed by STPT, 3D reconstruction, segmentation and single-cell counting. i.p., intraperitoneal injection. Scale bar, 1 mm. b, Freezing of shocked (FC, FR and NoFR: n = 12) and non-shocked (NoFC: n = 6) mice (left), and recall freezing shocked (FC and FR: n = 8) and non-shocked (NoFC: n = 6 mice) (right). c, 3D rendering of whole brain, the hippocampus (HPC), amygdala and primary visual cortex (VISp) with Fos-mNG astrocytes. A, anterior; D, dorsal; L, lateral; M medial; P. posterior; V, ventral. Scale bars, 500 µm. d,e, Heat map of fold changes in BAE count (versus NoFC (d) and FR versus FC (e); NoFC: n = 6; FC: n = 4; NoFR: n = 4; FR: n = 4 mice). Abbreviations for brain regions are listed in Extended Data Fig. 6. *False discovery rate (FDR) < 0.05. Levene’s test followed by one-way analysis of variance (ANOVA) or Kruskal–Wallis; post hoc Tukey or Dunn test with Bonferroni correction. FDR controlled by Benjamini–Hochberg procedure. Full data in Supplementary Table 2. f–h, FC- and FR-induced Fos counts in neurons versus astrocytes across engram areas for FC versus NoFC (f), FR versus NoFC (g) and FR versus NoFC with log scales, showing correlation Pearson’s correlation coefficient (r) (h). h, Amygdalar subregions are indicated with yellow dots. BMA, basomedial amygdala; MA, medial amygdala. i, Schematic of AAV tagging of Fos+ neurons and BAEs. j, Images of tagged Fos+ neurons and BAEs (representative of 10–22 sections from 3–5 mice per group). Scale bar, 200 μm. k, Quantification of Fos+ neurons (NoFC: n = 11 slices from 4 mice; FC: n = 10 slices from 3 mice; NoFR: n = 12 slices from 4 mice; FR: n = 13 slices from 4 mice) and BAE density (NoFC: n = 22 slices from 4 mice; FC: n = 18 slices from 3 mice; NoFR: n = 17 slices from 3 mice; FR: n = 15 slices from 5 mice). One-way ANOVA, Tukey’s test. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significantly different. Sample sizes and replicates in Supplementary Table 1.
Whereas the number of Fos+ neurons increased during both FC and FR, forming networks causally linked to fear memory traces—commonly referred to as engram neurons5,8,28, the density of Fos+ astrocytes during FC (FC-BAE; Fig. 1c) was seldom distinguishable from the NoFC control (NoFC-BAE). However, we found that the number of BAEs during recall (FR-BAE) was augmented across multiple regions (Fig. 1c). This was not simply reflecting delayed Fos upregulation after FC, because Fos induction was significantly higher in FR than in NoFR. Normalized comparisons across engram regions5 (Fig. 1d) showed a significantly greater number of FR-BAEs compared with FC-BAEs, particularly in the amygdala (Fig. 1e), the region long implicated in fear memory across species3,4,33,34,35. Consistently, in the lateral and basolateral amygdala (LA/B), astrocytic c-Fos protein was unchanged after FC or in NoFR versus NoFC, but FR induced a marked increase at 1.5 h with partial decay by 6 h (Extended Data Fig. 4). Consistently, the number of of FR-BAEs in the LA/B (FR-BAELA/B) was robust whether 4-OHT was given immediately or 3 h post-FR (Extended Data Fig. 5). These data indicate that astrocyte Fos promoter activity, and the associated surge in Fos mRNA and c-Fos protein expression, peaks within a biologically plausible window of 0–3 h following FR, but not following FC.
A recent study18 reported an increase in hippocampal astrocytic Fos after FC without habituation, contrasting with our findings (Fig. 1c–f). We hypothesized that this reflects contextual novelty: without prior habituation, novelty strongly drives astrocytic Fos. Supporting this, our ‘FC + novelty’ group (FC in novel context with no habituation) showed widespread astrocytic Fos, including in CA1 (Extended Data Fig. 6). Thus, the CA1 astrocytic Fos is likely to signal novelty, whereas our habituated FC protocol reveals astrocytic ensembles that are more selectively engaged in learning and recall.
To holistically compare the anatomical positioning of BAEs with previously reported brain-wide Fos+ neuron distribution during FC and FR5, we analysed subregions from previously defined regions in which Fos+ neuron counts of FC and FR were significantly higher than in non-shocked controls, referred to as neuronal ‘engram significant brain regions’5 (Fig. 1e–g and Methods). The number of Fos+ neurons increased during FC versus non-shocked controls, whereas there was no apparent change in astrocytic Fos counts (Fig. 1f). By contrast, FR elicited a strong correlation between neuronal and astrocytic Fos induction (r = 0.56; Fig. 1g,h), suggesting that local astrocyte–neuron interactions may underlie FR-BAE formation22. Notably, amygdala subregions that displayed selective astrocytic Fos increases during FR (Fig. 1e) also showed parallel neuronal Fos increases (Fig. 1h, yellow dots).
To test whether astrocytic and neuronal ensembles are co-engaged within the same regions, we performed dual-colour Fos tagging in the amygdala of TRAP2 mice (Fig. 1i–k). Neuron-specific DIO-mCherry and astrocyte-specific DIO-mNG AAVs were co-injected, and four groups were analysed: NoFC, FC, NoFR and FR. Neuronal Fos increased after FC or FR, consistent with past reports5,8,22. By contrast, astrocytic tagging was minimal following FC and NoFR but was robustly induced after FR (Fig. 1i–k), replicating our brain-wide observations (Fig. 1c–h). These results confirm that astrocyte Fos induction in the amygdala is recall-specific and distinct from neuronal ensemble dynamics.
Convergence induces astrocyte Fos
To identify mechanisms underlying astrocyte Fos induction, we combined pharmacological screening, circuit genetics, in vivo imaging and transcriptomics (Figs. 2–4). Growth factors that activate c-Fos in neurons36 did not trigger c-Fos signals in primary astrocytes (Extended Data Fig. 7), indicating that the induction mechanisms of c-Fos in neurons and astrocytes are distinct. Given the accumulating evidence that astrocyte G-protein-coupled receptor (GPCR) activation upregulates c-Fos expression26,29,30, we then stimulated primary astrocytes with a variety of neurotransmitters that bind to GPCRs expressed in astrocytes11,22,23, observing that noradrenaline (NA) increased c-Fos signals (Fig. 2b,c). The astrocyte c-Fos signals induced by NA at 0.1 µM and 1 µM were both abolished by propranolol (Fig. 2d), which antagonizes Gαs-coupled β-adrenergic receptors (β-receptors), but not by prazosin or atipamezole, which antagonize Gαq-coupled α1-adrenoreceptors (α1-receptors) or Gαi-coupled α2-adrenoreceptors, respectively (Fig. 2d). The astrocyte c-Fos induction was resistant to the Ca2+ chelator 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA-AM)37. The membrane-permeable Ca2+ ionophore ionomycin did not increase astrocyte c-Fos signals (Fig. 2d). However, the inhibition of cAMP production by the adenylate cyclase inhibitor SQ22536 attenuated astrocyte c-Fos signals induced by the adenylate cyclase–cAMP activator forskolin, the β-receptor agonist isoproterenol or NA (Fig. 2d). Together, these data show that NA induces c-Fos signals in primary astrocytes mainly through Gαs-coupled β-receptors and subsequent increases in cAMP.
a, Schematic of pharmacological screening. b, NA increased c-Fos intensity in GFAP+ astrocytes (n = 27–50 cells per condition). One-way ANOVA, Dunnett’s test versus vehicle (Veh). 5-HT, serotonin; a.u., arbitrary units; ACh, acetylcholine; DA, dopamine; GABA, γ-aminobutyric acid. c, Image showing NA (1 µM)-induced c-Fos increase (vehicle: n = 40; NA 1 μM: n = 41 cells). Scale bar, 10 μm. d, NA-induced c-Fos was blocked by inhibition of β-receptors or cAMP but not by Ca2+ mobilization (n = 28–55 cells per condition). One-way ANOVA, Dunnett’s or Tukey’s test. AC, adenylate cyclase; Atip, atipamezole; Frsk, forskolin; Isop, isoproterenol; Praz, prazosin; Prop, propranolol. e, Schematic of AAV-mediated Grm3 (encoding mGluR3) expression. f,g, Convergent NA and glutamate stimulation enhanced c-Fos in mGluR3+ astrocytes, but not in mGluR3− astrocytes (n = 98–131 cells per condition). One-way ANOVA, Tukey’s test. Scale bar, 20 μm. h, Schematic of signalling cascade. α1R, α1-receptor; βR, β-receptor. i, Ionomycin (Iono)-induced increase in Ca2+ increased c-Fos when combined with NA, isoproterenol (β-receptor agonist) or forskolin (n = 41–59 cells per condition). One-way ANOVA, Tukey’s test. j, Schematic of AAV injection into amygdala (LA/B) and LC in TRAP2 mice. Right, hM3Dq expression. Scale bars, 1 mm. k–m, Dual stimulation of LA/B and LC increased astrocyte density (LC–/LA/B–: n = 5; LC+/LA/B–: n = 4; LC–/LA/B+: n = 6; LC+/LA/B+: n = 5 mice). k, Images of hM3Dq-expressing cells. Scale bar, 200 μm. l, Fos co-expression in hM3Dq+ cells. Scale bar, 10 μm. m, Quantification of Fos-tagged astrocytes (mNG+) localized near hM3Dq+ LA/B neurons and NA fibres in l (representative of five biological replicates from the LC+/LA+ group). One-way ANOVA, Dunnett’s test. n–q, FR-BAE tagging with β-receptor blockade (propranolol, 10 mg kg−1) (n), LC silencing (o), amygdala (Amy) silencing (p) or RAM-based engram silencing (q) reduced BAE density. Scale bars (n,o–q, top and middle), 1 mm. o–q, Bottom image, magnified view showing local hM4Di-mCherry in neurons. Scale bars, 40 μm. Dox, doxycycline. r, FR-BAE density was reduced in all perturbation groups (Prop–/hM4Di–: n = 4; Prop+/hM4Di–: n = 4; LC: n = 4; amygdala (Amy): n = 5; engram: n = 4 mice). One-way ANOVA, Dunnett’s test. Data are mean ± s.e.m. Sample sizes and replicates in Supplementary Table 1.
a, AAV injection of GRABNE2h with optical fibre above LA/B to monitor bulk NA response (representative image from 6 mice; scale bar, 200 µm). b, ΔF/F traces showing NA signals in home cage (HC) and CtxA on fear conditioning (FC) day. NA signal increased with CtxA exposure and foot shocks, and showed larger, longer responses during recall (FR). Right, summary plots (n = 5–6 mice). AUC, area under the curve. c, Top, AAV injection of astrocyte-selective cAMPinG1 and RCaMP3 with optical fibre above LA/B to monitor astrocyte cAMP and Ca2+. Bottom, co-expression of cAMPinG1 and RCaMP3 in LA/B astrocytes (representative image from 8 mice; scale bars: 200 µm (main image), 20 µm (magnified views, bottom)). d, Astrocyte cAMP signals increased with context exposure and shocks, and during FR they increased gradually from HC baseline and were further enhanced in CtxA. Propranolol (10 mg kg−1, β-receptor antagonist) abolished these effects. Right, summary plot (n = 8 mice). e, Astrocyte Ca2+ signals also increased with context exposure and shocks. During FR, Ca2+ amplitude was reduced but event frequency was significantly higher. Right, summary plots (n = 7 mice). Wilcoxon signed-rank test. Data are mean ± s.e.m; in some cases, error bars are smaller than symbols. Sample sizes and number of replicates in Supplementary Table 1.
a, t-Distributed stochastic neighbour embedding (tSNE) clustering of scRNA-seq data from amygdalar astrocytes across NoFC, FC, NoFR and FR conditions (n = 4 mice per group). b, Dot plots showing Fos expression. Dot colour indicates z-score (grey, depletion; green, enrichment). c, Left, dot plots showing adrenoreceptor gene expression. Undetected receptors (Adra1d, Adra2c and Adrb3) are not shown. Right, Adra1a and Adrb1 upregulation in NoFR versus FC. Horizontal lines represent median. Mann–Whitney test. d, RNAscope timeline. e, RNAscope images and violin plots of Adra1a and Adrb1 in Slc1a3+ astrocytes (18–28 cells from 5 mice per group). One-way ANOVA, Dunnett’s test versus NoFC. f, Co-expression index for Adra1a and Adrb1 (Methods) peaks at 1 day post-FC. One-way ANOVA, Dunnett’s test versus NoFC. n = 18–28 cells, 5 mice per group. g, BAE tagging timeline. h, Representative images and quantification of tagged FR-BAE, peaking at 1 day post-FC. n = 4–5 mice per group. One-way ANOVA, Dunnett’s test versus NoFC. i, RNAscope of Adrb1 and Fos in Slc1a3+ astrocytes (FR). Arrows indicate a Fos and Adrb1-high cell (representative images from 18 sections, 6 mice). Scale bar, 10 μm. j, Pearson’s correlation coefficient (r) for Fos and Adrb1 expression (z-scores). Fos-high: z > 2 (3.1% cells); Adrb1-high: z > 0.5 (26.3%). Rationale in Methods. n = 814 cells from 6 mice. k, Adrb1 expression is increased in Fos-high astrocytes (25 Fos-high cells, 789 Fos-low cells) and Fos is enriched in Adrb1-high astrocytes (215 Adrb1-high cells, 600 Adrb1-low cells). Mann–Whitney test. l, Schematic of Adra1a or Adrb1 conditional knockout (cKO). m, RNAscope of Adra1a and Adrb1 in Slc1a3+ astrocytes. Ctrl, control. n, Violin plots showing reduced targeted receptor after conditional knockout (77–117 cells, 3 mice per group). Horizontal lines represent the median. o,p, Images (o) and quantification (p) of FR-BAEs, showing decreased density following Adra1a or Adrb1 conditional knockout. Scale bar, 200 μm. n = 5–9 mice, One-way ANOVA, Dunnett’s test versus FR controls. Data are mean ± s.e.m. Sample sizes and replicates in Supplementary Table 1.
Astrocytes in vivo respond to both local circuits and long-range neuromodulators15. Our data (Fig. 1g and Extended Data Fig. 3d) suggest that local activity also contributes to Fos induction in vivo. Given the transcriptional differences between in vitro and in vivo astrocytes, notably the reduced expression of metabotropic glutamate receptor 3 (mGluR3, encoded by Grm3) in cultured astrocytes17,38, which is abundantly expressed in adult in vivo astrocytes39, we heterologously expressed mGluR3 in primary astrocytes via AAVs to mimic the in vivo context (Fig. 2e). In mGluR3-expressing astrocytes, glutamate alone was ineffective, but in combination with NA it strongly enhanced c-Fos intensity (Fig. 2f,g). No such synergy was seen in mGluR3-negative astrocytes. Because Gαi-coupled GPCRs such as mGluR3 can enhance Gβγ–Ca2+ signalling when Gαq–phospholipase Cβ is co-activated29,40, NA may recruit α1-receptors (Gαq) and mGluR3 (Gαi) to cooperate with β-receptor–cAMP pathways (Fig. 2h). Supporting this, ionomycin-induced Ca2+ signals enhanced c-Fos when combined with NA, isoproterenol or forskolin (Fig. 2i). Thus, astrocytic c-Fos is driven primarily by β-receptor–cAMP and is further potentiated by convergent glutamate and NA signalling.
To test this convergence in vivo, we expressed neuron-specific hM3Dq in LA/B and locus coeruleus (LC), which mainly consists of noradrenergic neurons to stimulate them with CNO (Fig. 2j–m). The number of Fos-mNG+ astrocytes remained low when only LA/B or LC neurons were activated, but increased markedly when both were stimulated (Fig. 2j–m). This provides in vivo evidence that coincident local and noradrenergic inputs induce astrocytic Fos.
We next tested the necessity of local and far-reaching NA neural signals during FR-BAE tagging. TRAP2::AAV-PHP.eB-GfaABC1D-DIO-mNG mice received one of four perturbations at the time of tagging: β-receptor antagonism with propranolol (Fig. 2n) or hM4Di-mediated silencing of LC neurons (Fig. 2o), amygdalar neurons (Fig. 2p) or amygdalar engram neurons tagged with the robust activity marking (RAM) system41,42 whose hM4Di-based silencing (Extended Data Fig. 8) is known to reduce fear memory recall41 (Fig. 2q). In all conditions, FR-BAE density decreased significantly (Fig. 2r), demonstrating dependence on both LC-NA and local engram activity.
To further test whether FR-BAEs reside in anatomical contexts where they could receive convergent inputs from both noradrenergic projections and local engram neurons, we performed dual-colour Fos tagging with neuron-specific DIO-mCherry and astrocyte-specific DIO-mNG AAVs in TRAP2 mice, combined with noradrenaline transporter (NET) immunostaining to visualize noradrenergic projections (Extended Data Fig. 9). Territories of FR-BAEs showed greater overlap with noradrenergic projections and FR engram neurons than those of non-FR-BAEs, with no change in astrocyte territory size. These findings indicate that FR-BAEs are positioned to integrate convergent inputs from noradrenergic projections and local engram neurons, supporting the model that their coincidence drives FR-BAE induction in the amygdala.
Recall-evoked NA–astrocyte signalling
Our data (Fig. 2) support a convergence model in which NA and engram activity induce astrocyte Fos. However, since both activities occur during FC and FR8,43, the stronger astrocyte Fos induction during FR than FC (Fig. 1) raises the question of how astrocytes generate this selective response. To address this, we examined NA release and astrocyte cAMP–Ca2+ signalling dynamics across fear learning and recall using in vivo fibre photometry (Fig. 3 and Extended Data Fig. 8). We deployed AAV2/9-hSyn1-GRABNE2h44 to monitor bulk NA signals in the LA/B of freely behaving mice (Fig. 3a,b). During habituation 24 h prior to FC, NA signals were observed when the mice were placed into context A (CtxA; foot shock chamber) (peak ΔF/F = 0.142 ± 0.017), probably reflecting mild stressors and/or locomotion44 (NoFC; Fig. 3b). On top of the contextual NA signals, single foot shocks evoked enhanced and relatively fast (peak ΔF/F = 0.206 ± 0.04; half-decay = 4.3 min) NA responses in LA/B (FC; Fig. 3b). In addition, further enhanced (peak ΔF/F = 0.211 ± 0.022) and prolonged (half-decay = 13.5 min) NA signals were observed during the contextual FR (FR; Fig. 3b). The prolonged NA signals were absent in context B (CtxB) (Extended Data Fig. 10c,d), a neutral, non-conditioned context where mice show no freezing (Extended Data Fig. 10b), indicating that recall of contextual fear drives sustained NA signals.
We next used dual-colour fibre photometry (AAV2/5-GfaABC1D-cAMPinG1 and AAV2/5-GfaABC1D-RCaMP345) to measure astrocyte cAMP and Ca2+ signals (Fig. 3c). Consistent with the NA signals (Fig. 3a,b), cAMPinG1 signals increased during habituation and foot shocks, which fully decayed within about 2 min (NoFC and FC; Fig. 3d). On the FR day, however, astrocytes displayed a progressive increase in cAMP (FR; Fig. 3d), which was absent in CtxB (Extended Data Fig. 10e,f). β-receptor antagonism (propranolol) in vivo abolished the astrocytic cAMP signalling during FR (Fig. 3d), which suppressed FR-BAE induction (Fig. 2n,r) and freezing (Extended Data Fig. 10b′). Foot shock elicited astrocyte Ca2+ signals as reported previously46, and Ca2+ event frequency increased during FR (Fig. 3e). These data collectively indicate that fear experiences induce the enhancement of NA signals within the amygdala, which strongly trigger astrocyte cAMP signals through β-receptors during FR.
Multiday astrocyte state changes
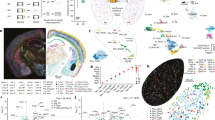
The increased astrocyte cAMP–Ca2+ signalling during FR raised the question of how astrocytes mount stronger responses during recall than during conditioning, despite NA release and engram neural activity occurring in both fear experiences. To address this, we examined experience-dependent molecular changes in astrocytes under NoFC, FC, NoFR and FR conditions. We dissected amygdala tissue 90 min after behavioural testing and isolated astrocytes by fluorescence-activated cell sorting (FACS) using the ATPase Na+/K+ transporting subunit β2 (ATP1B2)47 antibody ACSA2 (Extended Data Fig. 11a) and performed single-cell RNA sequencing (scRNA-seq). After quality control, we obtained profiles of 1,903 astrocytes (Fig. 4a and Extended Data Fig. 11b,c) and identified 4 transcriptionally distinct astrocyte clusters (AST1–4; Fig. 4a, Extended Data Fig. 11d,e and Supplementary Table 3). We did not observe any noticeable shifts in the subcluster composition across conditions, suggesting that FC and FR do not alter cluster identity. Fos-positive astrocytes comprised 2.9–4.2% of total astrocytes (Fig. 4b), consistent with earlier data (Extended Data Fig. 4). Of note, FR astrocytes showed the highest Fos levels (Fig. 4b), in line with BAE tagging (Fig. 1). Whereas scRNA-seq provides a snapshot at a single time point (approximately twofold increase in FR versus FC; Fig. 4b), the Fos-TRAP system integrates promoter activity over hours, which is likely to account for the greater fold differences observed with tagging (more than 30-fold in FR versus FC; Fig. 1d,e).
Subsequent analyses revealed that Adra1a (α1-receptor) and Adrb1 (β1-adrenoreceptor) as the two dominant adrenoreceptor genes expressed in amygdalar astrocytes. Notably, both receptors were upregulated in the FC group (1.5 h post-FC) and further elevated in the NoFR group (24 h post-FC without recall), suggesting continued priming of astrocytes over time (Fig. 4c). This data suggests a model in which FC induces a molecular state change in astrocytes, with a latency, increasing adrenoreceptor expression to potentiate NA signalling and enable future repeated fear-evoked Fos induction during FR. The enhancement in astrocytic adrenoreceptor expression is likely to underlie the NA–β-receptor-driven cAMP signals during FR (Fig. 3c,d) and Fos induction in response to recall of strong fear memory a day after conditioning (Fig. 1). To test this model, we explored temporal changes in adrenoreceptor expression, their link to FR-BAE induction, and causal roles in Fos induction.
We examined the time course of Adra1a and Adrb1 expression following FC, using single-cell RNAscope on amygdalar sections collected at 1.5 h, 1 day, 2 days, 3 days and 14 days post-conditioning (Fig. 4d–f). Adrb1 expression was significantly increased from 1.5 h to 3 days post-conditioning (approximately twofold versus NoFC; Fig. 4e), but returned to baseline by 14 days. By contrast, Adra1a expression was increased at 1 day post-conditioning (Fig. 4e), showed partial decay by 2 to 3 days, and peaked again at 14 days. We calculated a co-expression index for each cell (relative Adrb1 expression × relative Adra1a expression; Methods) and found a peak at 1 day post-FC (Fig. 4f), followed by gradual decline. To test whether this molecular profile predicts the degree of astrocyte Fos induction during recall, we performed FR-BAE tagging at multiple time points post-FC (1 day, 2 days, 3 days and 14 days; Fig. 4g,h). Consistent with the adrenoreceptor co-expression time course (Fig. 4f), FR-BAE density peaked at 1 day and declined thereafter (Fig. 4h), suggesting a defined window for FR-BAE induction. Moreover, RNAscope further revealed strong Fos–Adrb1 correlation in FR astrocytes (Fig. 4i–k).
To determine whether these adrenoreceptors are causally required for FR-BAE induction, we performed astrocyte-specific knockdown of Adra1a or Adrb1 in the amygdala (Fig. 4l) using floxed mouse lines48,49,50, AAV2/5-GfaABC1D-Cre-4x6T51 and Flp-based Fos-tagging AAVs (AAV2/5-Fos-FlpoERT2 and AAV2/5-GfaABC1D-fDIO-mNG). Knockdown of either receptor (Fig. 4m,n) significantly reduced FR-BAE density (Fig. 4o,p), indicating that both Gαq-coupled α1-receptors and Gαs-coupled β1-receptors are necessary for FR-BAE induction in vivo. Our earlier finding that the α1-receptor antagonist prazosin did not block NA-induced c-Fos induction in cultured astrocytes (Fig. 2d) probably reflects the downregulation of Adra1a in vitro17, highlighting the importance of in vivo validation. Collectively, our data support a model in which initial fear experience primes astrocytes to upregulate adrenoreceptor expression over time, creating a multiday molecular trace that allows future NA release upon repeated experiences (that is, recall) to selectively recruit the astrocyte ensemble.
The astrocyte ensemble stabilizes memory
Transcriptomics enabled us to probe molecular changes of FR-BAEs linked to functional outputs. We mined differentially expressed genes between Adrb1-positive and Adrb1-negative astrocytes (Extended Data Fig. 12a) and identified Igfbp2 as highly upregulated in Adrb1-positive astrocytes (Extended Data Fig. 12a,b). Our single-cell RNAscope revealed highly correlated expression of Adrb1 and Igfbp2 (Extended Data Fig. 12c–e), suggesting that astrocytic Adrb1 upregulation could promote Igfbp2 induction. Igfbp2 encodes the secreted protein IGFBP2, which was recently reported to be enriched in ‘peri-neuronal engram’ Fos+ astrocytes of the amygdala after fear recall22.
Given that IGFBP2 and its binding partners, insulin-like growth factors (IGFs), are crucial for synaptic plasticity52,53 and that chronic deletion of IGFBP2 disrupts synaptic transmission, circuit maturation and memory22,52,54, we hypothesized that IGFBP2 expression in FR-BAELA/B contributes to circuit de-stabilization and re-stabilization upon recall2,3. We tested this hypothesis with a spatiotemporally specific in vivo pharmacological perturbation of IGFBP2 (Extended Data Fig. 12f–h). We bilaterally infused an IGFBP2-neutralizing antibody52 (IGFBP2-Ab; 1 mg ml−1) immediately after FR in CtxA—that is, within the reconsolidation window during which memories become labile to be weakened or strengthened2. Mice received repeated daily infusions and FR sessions for five days (Extended Data Fig. 12f). IGFBP2-Ab treatment reduced freezing compared with controls (Extended Data Fig. 12g,h), indicating impaired memory stabilization; controls infused with IgG showed no change. Combined, our transcriptomics, RNAscope and pharmacological perturbation data indicate that a subset of astrocytes increases adrenoreceptor expression upon fear experiences, enhancing NA responsiveness and recall-driven induction of Fos and Igfbp2 to stabilize fear memory.
To directly examine the causal role of FR-BAE signalling in memory stabilization, we performed a loss-of-function experiment selectively targeting FR-BAEs in LA/B. We used iβARK, a genetically encoded 122-amino acid peptide that binds Gαq–GTP to suppress GPCR signalling26 (Extended Data Fig. 13a). In ex vivo Ca2+ imaging in amygdalar astrocytes, iβARK reduced α1-receptor-evoked Ca2+ responses relative to its mutant control, although some residual oscillatory activity persisted (Extended Data Fig. 13c,f,g), consistent with our prior findings in the striatum26. To improve efficacy, we developed iβARK2 by fusing the original peptide to a membrane-anchoring Lck domain, enabling more effective proximity-based inhibition (Extended Data Fig. 13a,b). iβARK2 showed robust membrane localization across astrocytic compartments, including fine processes (Extended Data Fig. 13d). Astrocytes expressing iβARK2 displayed significantly attenuated Ca2+ responses relative to both iβARK and the control iβARK2mut (Extended Data Fig. 13e–g). We next generated Cre-dependent AAV versions of iβARK2 and iβARK2mut for use in Fos-iCreERT2 TRAP mice to selectively target FR-BAEs (Fig. 5a,b). Remarkably, silencing FR-BAEs with iβARK2 impaired memory stabilization upon repeated recall (Fig. 5c), consistent with the behavioural effects observed upon neutralization of astrocyte-derived IGFBP2 (Extended Data Fig. 12f–h). Moreover, astrocytic IGFBP2 levels were significantly reduced in iβARK2-expressing mice compared with iβARK2mut controls (Fig. 5d), suggesting that suppression of GPCR signalling in FR-BAEs disrupts the recall-dependent transcriptional induction of IGFBP2 and compromises memory stabilization.
a, Schematic of behavioural and RNAscope analysis. b, Fos-iβARK2+ astrocytes in LA/B. Inset, enlarged view of an iβARK2+ astrocyte (representative of 12 mice). Scale bar, 200 μm. c, Reduced freezing at second recall in iβARK2 versus sustained freezing in iβARK2mut control (iβARK2mut: n = 13; iβARK2: n = 11 mice). Wilcoxon matched-pairs signed rank test. d, RNAscope images of Igfbp2 with mCherry–iβARK2 or mCherry–iβARK2mut and violin plots showing lower Igfbp2 expression in the presence of iβARK2 (iβARK2mut: n = 23; iβARK2: n = 24 cells; 4 mice per group). Mann–Whitney test. Horizontal lines represent the median. Scale bar, 10 μm. e, FR-BAE tagging with astrocytic Adrb1 overexpression. f, RNAscope images of Adrb1 and Igfbp2 in Slc1a3+ astrocytes with or without Adrb1 overexpression. Graphs showing increased expression with Adrb1-OE (ctrl: 501; Adrb1-OE: 1,810 cells; 18 sections from 6 mice per group). Mann–Whitney test. Scale bar, 10 μm. g,h, Images (g) and quantification (h) showing higher FR-BAE density with Adrb1 overexpression. Mann–Whitney test. Scale bar, 200 μm. i, Memory tests under weak (one-shock) versus strong (three-shock) conditioning. j, Adrb1 overexpression has no effect on freezing behaviour under strong conditioning (ctrl: n = 5; Adrb1-OE: n = 7 mice). Wilcoxon matched-pairs signed rank test. k, Under weak conditioning, Adrb1 overexpression enhanced freezing at second recall (n = 13 mice per group). Wilcoxon matched-pairs signed rank test. l, After strong conditioning, Adrb1 overexpression caused greater freezing in CtxB versus control (ctrl: n = 13; Adrb1-OE: n = 14 mice). Mann–Whitney test. m, FR engram labelling with mCherry and c-Fos immunohistochemistry 90 min after freezing test with or without Adrb1 overexpression. n,o, FR engram neurons exhibit similar density but higher reactivation with Adrb1 overexpression (ctrl: n = 15; Adrb1-OE: n = 12 slices; 3 mice per group). Scale bar, 50 μm. Mann–Whitney test. Data are mean ± s.e.m. Sample sizes and replicates in Supplementary Table 1.
Astrocyte–neuron engrams modulate memory
To further explore the causal impact of BAE signalling on neuronal engram and memory formation, we performed a gain-of-function experiment targeting astrocytic β1-receptors in LA/B. Astrocytes transduced with AAV2/5-GfaABC1D-Adrb1 for Adrb1 overexpression (Adrb1-OE) showed significantly increased Igfbp2 expression compared with controls (Fig. 5f). Adrb1 overexpression also increased the density of FR-BAEs (Fos-mNG+ astrocytes) by around threefold relative to controls (Fig. 5g,h), indicating that astrocyte β1-receptor overexpression amplifies the recall-induced astrocyte ensemble.
We next functionally assessed the behavioural effect of the augmented astrocyte ensembles (Fig. 5i–l and Extended Data Fig. 14). We detected no differences in ambulation in an open arena or time spent in the centre zone during habituation between control and Adrb1-OE mice (Extended Data Fig. 14a,b). We then tested whether Adrb1 overexpression affected memory stabilization. With three-shock conditioning, both groups showed equivalent freezing during initial recall and second recall after reconsolidation (Fig. 5j), potentially owing to a ceiling effect. To create parametric space to detect enhancements in memory, we trained mice to a weak (one-shock) conditioning paradigm (Fig. 5i,k). Adrb1-OE mice did not show altered freezing during FR one day after FC; however, Adrb1-OE mice exhibited significantly increased freezing during the second recall (Fig. 5k), indicating enhanced memory stabilization following the reconsolidation after initial recall. In another cohort with three-shock conditioning, initial recall showed no significant difference in freezing between the groups (Extended Data Fig. 14i,j). However, we observed significantly enhanced freezing in an altered context (CtxB) in mice with Adrb1 overexpression compared with controls (Fig. 5l). These data suggest that overstabilization of the initial fear memory leads to generalization or a loss of memory precision41,55,56,57.
Finally, we explored whether fear generalization is accompanied by activity of engram neurons. We bilaterally injected AAV2/8-hSyn1-DIO-mCherry with and without AAV2/5-GfaABC1D-Adrb1, and tagged engram neurons during FR. Mice were euthanized 1.5 h after the CtxB freezing test and the reactivation of FR engram neurons was assessed by c-Fos immunoreactivity (Fig. 5m–o). The overall number of mCherry+ neurons (FR engram neurons) was not significantly different between groups, but the c-Fos levels in FR engram neurons in the Adrb1-OE mice, which showed fear generalization, were significantly higher than those in control mice (Fig. 5o). These findings suggest that the larger astrocyte ensembles contribute to enhance subsequent reactivation of engram neurons, consistent with the view that memory retrievability is not determined during memory formation, but may be regulated after memory retrieval8, and demonstrating that astrocytes are involved in that crucial process.
Discussion
These data demonstrate that astrocytes can link discrete fear experiences across at least a day-long timescale. As postulated more than a century ago58 and recently implicated in brain computation11,25,59,60, astrocytes may integrate information on slower timescales than neurons. Whereas neurons and astrocytes have been traditionally compared over milliseconds versus seconds-to-minutes, our findings reveal a functionally significant mismatch on the scale of days. Unlike neurons that express the immediate early genes during both FC and FR5,8,28, astrocytes respond to fear conditioning (FC) with rapid Ca2+ and cAMP increases61, but undergo slower transcriptional state changes62. Our single-cell transcriptomics and RNAscope revealed that multiple days post-FC, without recall, astrocytes upregulated Adra1a and Adrb1, marking a learning-dependent state that facilitates heightened responses during repeated experiences (FR). This in turn drives expression of Fos and Igfbp2, supporting memory stabilization. The astrocyte ensembles identified in this study may partially meet the criteria for engram cells8 that undergo enduring biochemical changes upon experiences. However, our findings suggest that the astrocytic ensemble may not serve as a direct memory trace encoding experience-specific information like neurons because its perturbation did not alter initial fear recall. Instead, it may function as an eligibility trace for memory stabilization over days, creating a permissive microenvironment within circuits that supports the stabilization of memories related to repeated experiences. From a computational and bioenergetic perspective, astrocytes may underlie retrospective linking63,64, which benefits from spaced intervals over days between learning sessions65 by providing a slow modulatory trace that complements fast neuronal activity64, thereby offloading the energetically costly task of maintaining firing patterns in neurons across days and enabling more stable circuit transformations60.
Mounting evidence from animal3,4,57 and human imaging6 studies has highlighted the amygdala and NA in modulating long-term memory. Early pharmacological studies targeting protein synthesis66, β-receptors67, PKA35 or cAMP–CREB34 in the amygdala disrupted memory re-stabilization, which our findings indicate could partially be mediated by astrocytes. We show that astrocytes undergo learning-dependent increases in adrenoreceptor expressions, and manipulating these receptors alters memory stability, precision and neuronal engram representations. Notably, both NA input and reactivated neuronal engram activity are required to recruit astrocytic ensembles that stabilize labile memories. These findings imply that astrocyte ensembles may not simply be passive bulk substrates of NA signals, but instead exert behaviourally relevant, circuit-specific regulations through astrocyte-defined microcircuitry15,68. Disrupting the precise NA–astrocyte signalling through β1R overexpression led to fear generalization, suggesting physiological astrocytic roles in selective memory stability that constrain maladaptive updating. In this regard, targeting these astrocyte–neuron interaction mechanisms could be beneficial in treating cognitive and neuropsychiatric dysfunctions, including post-traumatic stress disorder, in which hyperactivity of the NA system is a prominent feature4,69.
Deciphering how diverse astrocyte responses link to multiple circuit outputs has long been a challenge15. Considerable effort has been devoted to probe24 or perturb11 astrocyte Ca2+ signals since they were first reported70, yet causal relationships are usually inferred from separate subjects, leaving input–output links unclear. Astrocyte perturbation in a bulk manner is also incongruent with the heterogeneity of astrocyte identity49,71, state62,72 and response59,73. Our approaches bridge this gap: by exploiting context-specific astrocyte state changes18,20,22, the brain-wide BAE tagging method enables broad exploration of ensembles causal for neural processing and behaviour. This framework facilitates research into astrocytic ensembles across diverse behaviours and disease conditions. Expanding such efforts will accelerate discoveries in computation and pathology that require understanding of multicellular interactions as a core attribute of brain biology.
Methods
Mouse models
Young adult (8- to 12-week-old) wild-type C57BL/6NCrSlc mice obtained from the Japan SLC, Fos2A-iCreERT2 (TRAP228, JAX 030323) mice obtained from the Jackson Laboratory, Adra1a flox mice49,74 obtained from RIKEN (RBRC11837) and Adrb1 flox mice50 (accession number CDB1068K, generated by RIKEN Center for Biosystems Dynamics Research) were used for in vivo experiments. TRAP2 mice were bred and maintained in our animal facility, backcrossed with C57BL/6NCrSlc at least eight times before use for experiments. Wild-type C57BL/6NCrSlc female mice were obtained from the Japan SLC to obtain postnatal day 0–1 (P0–P1) pups. Experiments were done using both male and female mice. Number of mice used in each experiment is listed in the main text and/or figure legends accordingly. Mice were housed in groups of 2–5 in a temperature- and humidity-controlled room (23 ± 3 °C, 45 ± 5% humidity) under a 12-h light:dark cycle (lights on from 07:00 to 19:00) and given ad libitum access to water and laboratory mouse diet at all times. After fibre implantation, the mice were single housed under a 12-h light:dark cycle (lights on from 07:00 to 19:00) and given ad libitum access to water and laboratory mouse diet at all times. After fibre implantation, the mice were singly housed. All experiments were approved by the RIKEN Animal Care and Use Committee.
Cell lines
AAVpro 293T cells (Clontech, 632273) were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, 11995073) with fetal bovine serum (FBS; Cosmo Bio, CCP-FBS-BR-500) and 0.1% antibiotic-antimycotic (Gibco, 15240062). Cells were grown in a humidified cell culture incubation with 95% air, 5% CO2 at 37 °C.
Viral vector construction
All plasmid constructs were generated using standard molecular biology techniques and the In-Fusion HD Cloning Kit (Clontech). The pZac2.1 GfaABC1D-Rpl22-HA plasmids (Addgene plasmid #111811) were digested with XhoI and NotI, which removed the Rpl22-HA sequence. Multiple cloning sequences (MCS) comprised of restriction enzyme sites that were absent from the backbone pZac2.1 plasmid were generated via gene synthesis (Eurofins Genomics) and incorporated to produce pZac2.1-GfaABC1D-MCS plasmids. The PCR-amplified DIO-GFP sequence from Addgene plasmid #28304 was cloned into the mammalian expression vector pCAG-tdTomato (Addgene #83029) between EcoRI sites to generate pCAG-DIO-EGFP-tdTomato. The PCR-amplified DIO-EGFP-tdTomato was inserted into the XbaI and NotI sites of the pZac2.1-GfaABC1D-MCS plasmid to generate pZac2.1-GfaBC1D-DIO-EGFP-tdTomato plasmids. The PCR-amplified WPRE sequence from pFBAAV-CAG-XCaMP-R-WPRE (a gift from H. Bito12) was inserted into the NotI site of the resulting plasmid to generate pZac2.1-GfaABC1D-DIO-EGFP-tdTomato-WPRE plasmids. The plasmids were digested with NotI and AgeI, which removed the tdTomato sequence, and a PCR-amplified mScarlet-I sequence (Addgene #98816) was incorporated, and the resulting plasmids were digested with XhoI and KpnI, which removed the EGFP sequence, and the mNG sequence from Addgene #99135 was ligated using DNA ligation kit (Takara) to generate pZac2.1-GfaABC1D-DIO-mNG-mScarlet-I-WPRE plasmids. The mScarlet-I sequence was removed with restriction enzymatic digestion with PspOMI and NotI to generate pZac2.1-GfaABC1D-DIO-mNG-WPRE plasmids. The PCR-amplified hM4Di-mCherry sequence from Addgene #92286 was cloned into the PmeI site of pAAV-RAM-d2TTA::TRE-MCS-WPRE (Addgene #63931) to generate pAAV-RAM-d2TTA::TRE-hM4Di-mCherry-WPRE plasmids. pAAV-GfaABC1D-Flag-mAdrb1-WPRE, pAAV-GfaABC1D-mGrm3-mCherry-WPRE, pAAV-GfaABC1D-Lck-mCherry-iβARK(D110A), pAAV-GfaABC1D-Lck-mCherry-iβARK, pAAV-GfaABC1D-DIO-Lck-mCherry-iβARK(D110A), pAAV-GfaABC1D-DIO-Lck-mCherry-iβARK pAAV-Fos-FlpoERT2 and pAAV-GfaABC1D-FRT-mNG plasmids were designed by the authors and constructed by VectorBuilder. The PCR-amplified DIO-Lck-mCherry-iβARK(D110A) and DIO-Lck-mcherry-iβARK sequence from pAAV-GfaABC1D-DIO-Lck-mCherry-iβARK(D110A) and pAAV-GfaABC1D-DIO-Lck-mCherry-iβARK was clone into the EcoRI site of pAAV-GfaABC1D-MCS-4x6T-WPRE (Addgene #196417) to generate pAAV-GfaABC1D-DIO-Lck-mCherry-iβARK(D110A)-4x6T and pAAV-GfaABC1D-DIO-Lck-mCherry-iβARK-4x6T. pAAV-GfaABC1D-mGrm3-mCherry-WPRE was digested with EcoNI and AccI to remove mGrm3-mCherry, and PCR-amplified mGrm3 was cloned back to the backbone to generate AAV-GfaABC1D-mGrm3-WPRE plasmids. The PCR-amplified XCaMP-Gf-WPRE from the pFBAAV-CAG-XCaMP-Gf-WPRE plasmid (a gift from H. Bito75) was cloned into the NcoI and HindIII site of pAAV-hSyn1-cyto-mRuby3-iATPSnFR1.0 (Addgene #102557) to generate pAAV-hSyn1-XCaMP-Gf-WPRE plasmids. The mCherry-CAAX sequence digested with EcoRI of the pZac2.1-GfaABC1D-mCherry-CAAX plasmid (a gift from T. Takano76) was ligated into the EcoRI and EcoRV the resultant plasmids to generate pAAV-hSyn1-mCherry-CAAX-WPRE plasmids. All constructs were sequenced before use. All our constructs have been deposited to Addgene in the Nagai Lab repository for unrestricted distribution (http://www.addgene.org/Jun_Nagai, Supplementary Table 4).
Viral production
Recombinant AAVs were generated by triple transfection of AAVpro 293T cells (Clontech, 632273) using polyethylenimine (PEI; Polysciences inc., 24765-100). Cells were cultured up to 50–70% confluency in DMEM (Gibco, 11995073) with FBS (Cosmo Bio, CCP-FBS-BR-500) and 0.1% antibiotic-antimycotic (Gibco, 15240062). The plasmids and PEI were mixed with Opti-MEM (31985-088) and kept for 5 min at room temperature. The mixture was combined with a DNA mix which contained pAAV Capsids, pHelper (Agilent Technologies, 240071-54) and pAAV transgenes. All these plasmids had a concentration of 4.2 μg per dish. The DNA mix was then applied with PEI/Opti-MEM, and incubated for 20 min at room temperature. Then, this solution was added to the cells and incubated for 24 h. After incubation, the medium was changed to DMEM with 2% FBS and 0.1% antibiotic-antimycotic. Viral particles were collected from the medium at 120 h post-transfection. Cell pellets were centrifuged (3,500 rpm, 30 min, 4 °C). The supernatant was filtered by Setricup (Merc, S2HVU02RE) and pellets were resuspended in FBS-free DMEM and freeze-thawed three times. In the case of PHP.eB capsid virus production, the supernatant was resuspended in DMEM and fractured by harshly pipetting. Thawed pellets were centrifuged (3,500 rpm, 10 min, 4 °C) again and the supernatant was filtered by Setricup (Merc, S2HVU02RE). Media containing viral particles were centrifuged (22,000 rpm, 2 h, 4 °C) by Optima XE-100 (Beckman Coulter, A94516). The pellets were collected with 10–50 μl PBS and clarified by centrifugation (4,000 rpm, 10 min, 4 °C). The supernatant-containing viral particles were stored at 4 °C. AAV titre was analysed by quantitative PCR (QuantStudio 12 K; Applied Biosystems) using primers designed to selectively bind AAV2 inverted terminal repeats (forward: 5′-AACATGCTACGCAGAGAGGGAGTGG-3′, reverse: 5′-CATGAGACAAGGAACCCCTAGTGATGGAG-3′).
Viral injections
All surgical procedures were conducted under general anaesthesia using continuous isoflurane (induction at 5%, maintenance at 1.5% to 2% vol/vol). Depth of anaesthesia was monitored continuously and adjusted when necessary. Intravenous administration of AAV-PHP.eB (50 µl to deliver 2.5 × 1012 genome copies (GCs) per mouse) was performed by injection into the retro-orbital sinus of TRAP2 mice. Two weeks after AAV-PHP.eB GfaABC1D-DIO-mNG-WPRE or AAV-PHP.eBGfaABC1D-DIO-mNG-mScaret-I-WPRE injections, mice were administered 4-OHT for inducing Cre recombination under the Fos promoter (Fos tagging) and then returned to their home cage until they were euthanized 7 days after tagging. In some cases, Fos tagging was performed using the FLP–FRT system (fDIO), and mice were euthanized one day after tagging. For stereotaxic microinjection of AAVs, mice were fitted into a stereotaxic frame (David Kopf Instruments) with their heads secured by blunt ear bars and their noses placed into a veterinary grade anaesthesia and ventilation system (VetEquip 911103). The surgical incision site was then cleaned with 70% ethanol (vol/vol). Craniotomies (1–2 mm in diameter) were made powered by a high-speed drill at specific coordinates based on the bregma position. Bilateral viral injections were conducted using a stereotaxic apparatus (David Kopf Instruments) to guide the placement of beveled glass pipettes (World Precision Instruments, 1B100-4) into the regions of interest: the basolateral amygdala (anterior–posterior (AP): −1.48 mm, medial–lateral (ML): 3.3 mm, dorsal–ventral (DV): 5.00 mm to bregma) and LC (AP: −5.4 mm, ML: 0.85 mm, DV: 3.7 mm to bregma). Total of 0.3 μl of AAV was delivered at 0.1 μl min−1 using a syringe pump (SmartTouch Pump, World Precision Instruments). The glass pipette was withdrawn 5–10 min after the infusion to prevent the backflow. Mice were then sutured, returned to their home cage, and allowed to rest for at least two to three weeks before any assessments were conducted. AAV titres (GCs per ml) were adjusted with sterile saline to deliver the indicated genome copies into the regions of interest: AAV2/5-GfaABC1D-DIO-mNG-WPRE (3.0 × 1012), AAV2/5-GfaABC1D-hM3Dq-mCherry (3.0 × 1012), AAV2/DJ8-RAM-d2TTA::TRE-hM4Di-WPRE (3.0 × 1013), AAV2/5-GfaABC1D-Flag-mAdrb1-WPRE (3.0 × 1012), AAV2/9-rTH-Cre (3.0 × 1012), AAV2/8-hSyn1-DIO-hM3Dq (1.0 × 1013), AAV2/8-hSyn1-DIO-hM4Di (1.0 × 1013), AAV2/8-hSyn1-DIO-mCherry (3.0 × 1012), AAV2/DJ8-Ef1a-hM3Dq (4.8 × 1011), AAV2/DJ8-Ef1a-hM4Di (4.0 × 1012), AAV2/5-hSyn1-mCherry-CAXX (3.0 × 1012) AAV2/9-hSyn1-GRABNE2h-WPRE (2.7 × 1012), AAV2/1 GfaABC1D-cAMPinG1-NE (1.3 × 1013), and AAV2/1 GfaABC1D-RCaMP3 (1.5 × 1013), GfaABC1D-GCaMP6f (1.1 × 1013), AAV2/5-GfaABC1D-iβARK-mCherry (1.3 × 1012), AAV2/5-GfaABC1D-iβARK(D110A)-mCherry (1.3 × 1012), AAV2/5-GfaABC1D-Lck-mCherry-iβARK (1.3 × 1012), AAV2/5-GfaABC1D-Lck-mCherry-iβARK(D110A) (1.1 × 1013), AAV2/5-GfaABC1D-DIO-Lck-mCherry-iβARK-4x6T (3.0 × 1013), AAV2/5-GfaABC1D-Lck-mCherry-iβARK(D110A) (3.0 × 1013), AAV2/5-Fos-FlpoERT2 (3.0 × 1012), AAV2/5-GfaABC1D-fDIO-mNG (1.0 × 1012), AAV2/5-GfaABC1D-Cre-4x6T (1.2 × 1013).
Drug administration in vivo
4-OHT (Sigma, H6278) was dissolved at 25 mg ml−1 in 100% ethanol with agitation at 37 °C for 15 min and was then aliquoted and stored at −20 °C for up to several weeks until use. Before use, 4-OHT was redissolved in ethanol by shaking at 37 °C for 15 min and corn oil (Sigma, 259853 and S5007) was added to give a final concentration of 3 mg ml−1 4-OHT. The ethanol was evaporated by incubation at 50 °C. The final 3 mg ml−1 4-OHT solutions were always used on the day they were prepared. All 4-OHT injections were delivered intraperitoneally at 50 mg kg−1 2–4 weeks after AAV injection into TRAP2 mice for inducing Cre recombination under the Fos promoter.
CNO (Tocris, 4936) was administered intraperitoneally 0.5 h before the initiation of behavioural assessments. CNO was administered intraperitoneally 20 min before 4-OHT intraperitoneal injection. Propranolol (MedChemExpress, HY-B0573) was dissolved in dH2O at 100 mM and stored at −20 °C. The final solutions were always used on the day they were prepared.
Mice were placed on doxycycline (40 mg kg−1, Bio-Serv, F4159) chow 24 h prior to and for at least 1 week after viral injection, switched to doxycycline-free chow 48 h before recall, and euthanized 24 h later. IGFBP2 antibody was infused bilaterally through implanted guide cannulas targeting basolateral amygdala. Infusions were performed using an internal cannula connected to a 10-µl Hamiltion syringe. One microgram of IGFBP2 antibody (1 mg ml−1, 1 µl per site) was infused into mice immediately after a fear recall paradigm at 400 nl min−1 using a syringe pump (SmartTouch Pump, World Precision Instruments).
Immunohistochemical analysis
Mice were deeply anaesthetized with isoflurane and transcardially perfused with PBS, followed by 4% paraformaldehyde (PFA) in PBS for fixation. The dissected brains were post-fixed overnight at 4 °C in 4% PFA in PBS, and cryoprotected in 30% sucrose in PBS. The brains were embedded with OCT compound (Sakura Finetek Japan, 4583) and sliced into coronal sections with a thickness of 50 μm at −20 °C using cryostat (CryoStar NX70, Thermo Fisher Scientific, 956960). The sections were washed with PBS 3 times for 5 min each and stored in 50% glycerol (Fujifilm, 075-00616) in PBS at −20 °C. For staining with c-Fos antibody, the sections were permeabilized with PBS containing 1% Triton X-100 (Nacalai tesque, 35501), incubated at room temperature for 3 h, and then incubated with 0.3% Triton X-100 in PBS containing 1% normal donkey serum (NDS, Sigma-Aldrich, D9663) at room temperature for 1 h for blocking, and then incubated at 4 °C overnight in blocking buffer. With the following primary antibodies, Triton X-100 was used at 0.1% for permeabilization and PBS containing 1% NDS was used for blocking and antibody reactions: rat anti-GFAP (1:500, Thermo Fisher Scientific, 13-0300), mouse anti-S100β (1:500, Sigma-Aldrich, S2532), goat-Sox9 (1:100, R&D systems, AF3075) mouse anti-RFP (1:500, MBL, M155-3), rabbit anti-RFP (1:500, Rockland, 600-401-379), mouse anti-NET (1:2,000, MAb technologies, NET05-2) and rabbit anti-c-Fos (1:5,000, Synaptic Systems, 226003). After washing with PBS 3 times for 10 min each, the sections were incubated at room temperature for 2 h in blocking buffer with the following secondary antibodies: Alexa Fluor 488 donkey anti-rat, Alexa Fluor 488 donkey anti-chicken, Alexa Fluor 568 donkey anti-mouse, Alexa Fluor 568 donkey anti-rabbit, Alexa Fluor 568 donkey anti-goat, Alexa Fluor 647 donkey anti-rabbit, Alexa Fluor 647 donkey anti-mouse, Alexa Fluor 647 donkey anti-chicken (1:1,000, Abcam, ThermoFisher and Jackson ImmunoResearch). The sections were incubated at 4 °C overnight in blocking buffer with secondary antibodies when staining with anti-c-Fos antibody. After washing with PBS 3 times for 10 min each, the sections were mounted on slides (Fluoromount-G, Southern bio, 0100-35) and visualized under a confocal laser microscope (Olympus, FV3000) and a fluorescent microscope (Olympus, VS200). Immunoreactivities were analysed using ImageJ software (NIH). The numbers of c-Fos+ neurons (Fig. 5o and Extended Data Fig. 8b) and astrocytes (Extended Data Fig. 4b) were quantified based on a consistent c-Fos intensity threshold for each experiment. This threshold was defined such that the number of c-Fos+ neurons or astrocytes corresponded to the values previously reported18,42,77. Number of cells, field of views, sections and mice used in each analysis is listed in the main text and/or figure legends accordingly.
Visual stimulation
Mice were singly housed in a light-proof chamber for 48 h. On the day of tagging, mice were exposed to 1 h of light inside the chamber and injected intraperitoneally with 50 mg kg−1 4-OHT either 24 h, 12 h, 6 h or 3 h before or 0 h, 3 h, 6 h, 12 h or 24 h after the termination of light exposure. Mice were returned to the dark chamber for an additional two days and then returned to a regular light-dark cycle until the time of euthanasia seven days after tagging.
Contextual fear conditioning
Behavioural tests were performed during the light cycle between 07:00 and 19:00. Temperature and humidity of the experimental rooms were maintained at 23 ± 2 °C and 55 ± 5%, respectively. Background noise (55 dB) was generated by a white noise generator (Ohara). All the experimental mice were transferred to the behaviour testing room at least 1 h before the tests to acclimatize to the environment and to reduce stress. Mice were habituated by freely exploring the conditioning chamber (CtxA) that consisted of a square partition (30 × 30 × 15 cm; 100 lux and no odour) with a grid floor wired to a shock generator (Ohara) for 15 min per day for 3 days. On the fourth day, they were either fear conditioned (1 time or 3 times shock; 2 s, 0.3 mA) (FC group) or presented with no shocks (NoFC group) in the conditioning chamber. One day after the conditioning, mice were either returned to CtxA for 3 min for fear memory recall (FR group) or kept in their home cages (NoFR group). Freezing durations exceeding 70% in the conditioned chamber 1 day post-conditioning with 3 foot shocks or exceeding 50% 1 day post-conditioning with a single foot shock, are considered indicative of sufficient conditioning. All the experimental mice received intraperitoneal injections of 4-OHT (50 mg kg−1) immediately after the task for Fos tagging. Freezing was automatically measured throughout the testing trial by the TimeFZ4 software (version 2021_7_29, Ohara) and subsequently processed by a blinded investigator. The chambers were cleaned with 70% ethanol between tests. In some cases, 10 mg kg−1 Propranolol (MedChemExpress, HY-B0573) was administered by intraperitoneal injection to mice 0.5 h before, and 2.5 h and 5.5 h after 4-OHT-based Fos tagging in order to block β-adrenergic signalling throughout the tagging timeframe. In some experiments using DREADDs, mice were injected with CNO (1 mg kg−1) 10 min before the task in CtxA. For experiments in Fig. 2n in which CNO was used for silencing neuronal activity or propranolol was used for antagonizing β-adrenoreceptor signalling to test their roles in astrocyte Fos induction during fear memory recall, both were administered with consideration of their pharmacokinetics. Propranolol was given both before and after the behavioural session to ensure effective β-receptor blockade throughout the several-hour Fos-tagging window (Extended Data Fig. 3), given its short half-life78 (~1.5–2 h), whereas CNO was given once before the session owing to its prolonged effect79 (>6 h). Freezing was measured for 3 min in the CtxB chamber, which consisted of a rhombic partition (30 cm × 60 cm × 15 cm, 10 lux and 1% acetic acid odour) with a flat floor (Ohara).
Whole-brain imaging
Mice were deeply anaesthetized with isoflurane and transcardially perfused with PBS, followed by 4% PFA in PBS for fixation of brain tissues. The dissected brains were post-fixed overnight at 4 °C in 4% PFA in PBS and stored in PBS at 4 °C until embedding. To oxidize agarose, 2.25 g agarose (A6013, Sigma-Aldrich) was added to 100 ml of 10 mM sodium periodate (S1878, Sigma-Aldrich)/50 mM phosphate buffer and stirred gently for 2.5 h at room temperature in the dark. The oxidized agarose was filtered on a bottle-top vacuum filter (0.2 μm, 569-0020, Thermo Scientific) washed 3 times with phosphate buffer and resuspended in 50 ml of phosphate buffer to make 4.5% oxidized agarose solution. The oxidized agarose solution was boiled with a microwave oven to melt the agarose and kept at 60–65 °C. The brains were pat-dried and embedded in oxidized agarose solution with a plastic mould (18646A, Polysciences). For covalent crosslinking between oxidized agarose and the surface of the brain, borate/borohydride solution was prepared the day before use by adding 0.2 g sodium borohydride (452882, Sigma-Aldrich) in 100 ml of prewarmed (40 °C) borate buffer (50 mM borax, 221732, Sigma-Aldrich; 50 mM boric acid, B6768, Sigma-Aldrich; pH 9.0–9.5) in the chemical fume hood. The agarose block was removed from the mould, soaked in borate/borohydride solution to initiate crosslinking and incubated at 4 °C overnight. The block was washed and kept in phosphate buffer at 4 °C until imaging.
The agarose block was mounted on a glass slide using Loctite 401 glue immediately before imaging. STPT was performed with the TissueCyte 1400FC system (TissueVision) equipped with Coherent Chameleon Ultra II laser (Coherent) or Spectra-Physics Insight X3+ laser (MKS Spectra-Physics) and a Nikon 16× water-immersion objective (MRP07220, NA: 0.8). The laser wavelength was tuned at 1,000 nm. Brains were coronally sectioned with a built-in vibrating microtome at 50-μm thickness with a frequency of 60 Hz and a speed of 0.5 mm s−1. Four optical z-planes were taken at 50 μm, 62.5 μm, 75 μm and 87.5 μm below the cut surface of the tissue block. To scan an entire coronal plane, individual tile images were acquired (832 × 832 pixels per frame, resolution at 1.382 μm per pixel) by moving the stage between each tile. Following the full scanning of a coronal plane, a 50-μm section was cut away by vibrating microtome and imaging of a new cut surface repeated to acquire optical sections covering the entire brain from the caudal to the rostral side (~320 coronal sections). The raw image tiles were first trimmed and subjected to flat field correction, then stitched into 2D mosaic coronal section images using the Autostitcher software (TissueVision). The signals from the green channel (500–550 nm, mNG signal) and from the far-red channel (>600 nm, background) were simultaneously acquired, and the latter was used to subtract background from the green channel to enhance the signal to noise ratio. Each region of the whole brain was aligned and segmented according to the Mouse Brain Atlas (NeuroInfo, MBL Bioscience, BN-200). Owing to the high quality preservation of tissue architecture during imaging, we semi-automatically performed alignment to a standard mouse brain atlas, segmentation of 839 brain regions, and detection of single Fos-tagged mNG astrocytes (BAEs), defined by their territories that typically displayed diameters of approximately 40 to 60 μm with ~10-µm-wide cell bodies68 (Fig. 1a and Supplementary Videos 1 and 2). Consistent with our intersectional genetic approach, we rarely observed neuron-like mNG-positive cells, with larger cell bodies (>~20-μm diameter) and elongated processes (>~50-μm). We only found these cells in regions nearby ventricles and the midline where well-characterized GFAP-positive neural progenitor cells reside48,80, and they were excluded in subsequent analyses. As a result, using this brain-wide analysis approach we successfully quantified the density of BAEs in 677 brain regions that accounted for 80.7% of the whole brain.
Correlational analysis of Fos + cell counts
To observe correlation between Fos+ astrocytes and neurons, we used the same TRAP2 (Fos-iCreERT2) system28 used by Roy et al.5, but instead with a Cre-dependent mNG reporter expressed in Fos+ astrocytes, allowing comparison of Fos+ cell counts from anatomically matched regions between studies. The regions of interest were first narrowed to the 117 engram-signifiant regions defined by Roy et al. Then, to ensure analysis of bona fide astrocyte-tagged regions, areas with possible neuronal leakage (mNG+ neuron-like cells), particularly near ventricles were excluded (see above). In addition, high-level hierarchical structure areas (such as midbrain or thalamus) with extremely high total counts that skew correlation analysis were also excluded. In the end, this selection yielded 75 regions for subsequent correlational analyses. Changes in Fos+ astrocyte numbers (FC–NoFC and FR–NoFC) were calculated using the non-foot shocked control group as the baseline, consistent with Roy et al. Supplementary table 1 from Roy et al. provided region-by-region counts of Fos+ neurons in non-foot shocked controls, FC and FR groups, enabling us to extract the corresponding changes in Fos+ neuron numbers for each of the 75 regions. Pearson correlation analyses between these neuronal and astrocytic Fos+ cell count changes were performed.
Whole-brain visualization
For image visualization in 3D, low-resolution image stacks were created across all four channels. These stacks were downsampled to 25 μm per pixel resolution in the x and y axes, while the inter-slice distance was maintained at 50 μm, resulting in a voxel resolution of 25 × 25 × 50 μm3. To minimize high-frequency artifacts, median filtering and Gaussian smoothing were applied to the coronal images prior to downscaling. The processed 3D image stacks were stored in the NIfTI format, and the world coordinates were adjusted to align roughly with the Allen Mouse Brain Common Coordinate Framework v.3 (CCFv3)81. For registration to the CCFv3, the Allen STP-1675 template (filename: P56_Atlas.nii.gz; available at https://scalablebrainatlas.incf.org/mouse/ABA_v3#downloads) was used as the reference image. This template, generated from STPT averaged over 1,675 specimens, was combined with corresponding atlas annotations (filename: P56_Annotation.nii.gz). Image alignment was performed using the Advanced Normalization Tools (ANTs)82. An affine registration was first applied at 3 scales (downscaling factors of 4, 2 and 1) with Gaussian smoothing of 2, 1 and 1 voxels and 250, 200 and 50 iterations, respectively. This was followed by deformable (SyN) registration at four scales (downscaling factors of 8, 4, 2 and 1) with Gaussian smoothing of 4, 2, 1 and 1 voxels and 1,000, 500, 100 and 50 iterations, respectively, using mutual information as the metric. After computing the transformation, individual image stacks and atlas labels were mapped to and from the CCFv3 image space. After aligning with the NIfTI world coordinates, cell positions were mapped to the Allen template using the antsApplyTransformsToPoints function from ANTs. Brain regions from the Allen atlas annotations were then assigned to individual cells based on their locations in the CCFv3 image space.
Primary astrocyte culture
Astrocyte primary cultures were obtained as previously described with some modifications83. In brief, cerebral cortical tissues were isolated from P0–P1 neonatal mice and dissociated with a scalpel, gently agitated in 0.05% Trypsin-EDTA (Gibco, 25300-054), and rapidly strained with a Falcon 100 µm cell strainer (352360). Dissociated cells were seeded at a rate of four brains per bottle on poly-d-lysine-coated bottles. The cells were cultured in DMEM (Gibco, 11995073) supplemented with FBS (Cosmo Bio, CCP-FBS-BR-500) and penicillin-streptomycin. When the cells reached confluence after several weeks, the culture bottles were gently shaken at 120 rpm overnight at 37 °C to remove non-astrocytic cells. Astrocytes were then detached using Trypsin-EDTA, replated onto poly-d-lysine-coated bottles, and after 1–2 weeks, cultures were shaken again at 120 rpm for 4 h. Astrocytes were then re-seeded on coverslips in 12-well plates at a density of 1.0 × 105 cells per well, which were maintained in 5% CO2 at 37 °C, with the medium being changed every 2 to 3 days.
Primary astrocyte c-Fos analysis
Five days after re-seeding, primary astrocytes were incubated with tetrodotoxin (TTX at 1 µM, Fujifilm, 207-15901) to eliminate the potential effect of neuronal firing, for at least 5 min to allow adequate equilibration, together with the following inhibitors, blockers or chelators indicated for each experiment unless otherwise described: prazosin hydrochloride (MedChemExpress, HY-B0193A), atipamezole hydrochloride (Tokyo Chemical Industry, A2956), propranolol hydrochloride (MedChemExpress, HY-B0573), BAPTA-AM (SIGMA, A1076), and SQ 22536 (Tokyo chemical industry, Q0105). The cultures were then incubated with the following growth factors, neurotransmitters, neuromodulators or neuropeptides in 5% CO2 at 37 °C for 1.5 h: l-glutamic acid monosodium salt hydrate (Sigma-Aldrich, G5889), GABA (Sigma-Aldrich, A2129), ATP disodium salt hydrate from yeast (Nacalai tesque, 01072-24), l-noradrenaline hydrochloride (Sigma-Aldrich, 74480), dopamine hydrochloride (Sigma-Aldrich, H8502), acetylcholine chloride (Sigma-Aldrich, A6625), serotonin hydrochloride (Alfa Aesar, B21263.03), brain derived neurotrophic factor (BDNF, Fujifilm, 028-16451), neurotrophin-3 (Fujifilm, 146-09231, 50 ng ml−1), neurotrophin-4 (Fujifilm, 142-06634, 100 ng ml−1), forskolin (Fujifilm, 067-02191, 10 μM), nerve growth factor-β (NGFβ, Fujifilm, 141-07601, 100 ng ml−1), epidermal growth factor (EGF, Fujifilm, 059-07873, 100 ng ml−1), PDGF-AA (Fujifilm, 163–19731, 100 ng ml−1), PDGF-BB (Fujifilm, 164–24031, 100 ng ml−1), Ciliary Neurotrophic Factor (CNTF, Fujifilm, 032-23501, 100 ng ml−1), IGF1 (Fujifilm, 099-04511, 100 ng ml−1). The following agonist and ionophore were used: isoproterenol (MedChemExpress, HY-B0468) and ionomycin (LKT laboratories, I5752). Ninety minutes after incubation, primary astrocytes were fixed in 4% PFA (Nacalai Tesque) for 10 min, washed with PBS 3 times, permeabilized with 0.1% Triton X-100 (Nacalai Tesque) for 10 min and blocked with 1% NDS (Sigma-Aldrich, D9663) for 30 min. Primary astrocytes were then incubated overnight at 4 °C with the following primary antibodies: rabbit anti-c-Fos (1:1,000, Synaptic Systems, 226003), rat anti-GFAP (1:1,000, Thermo Fisher Scientific, 13-0300), rabbit anti-IBA1 (1:1,000, Fujifilm, 019-19741), chicken anti-TUJ1 (1:1,000 Novus Biologicals, NB100-1612), mouse anti-OLIG2 (1:100, Millipore, MABN50). The cultures were washed 3 times in PBS for 10 min each, then incubated with DAPI (1:10,000, Sigma-Aldrich, D9542) and the following secondary antibodies (1:1,000) in PBS containing 1% NDS for 1 h at room temperature: Alexa Fluor 488 donkey anti-rat (abcam, ab150153), Alexa Fluor 568 donkey anti-mouse (abcam, ab175700), Alexa Fluor 568 donkey anti-rabbit (Abcam, ab175692), Alexa Fluor 647 donkey anti-rabbit (abcam, ab150067), Alexa Fluor 647 donkey anti-chicken (Jackson ImmunoResearch, 703-605-155). Fluorescent images were taken using a 20× objective lens (UPlanXApo 20×, Olympus) with a confocal laser-scanning microscope (Olympus, FV3000 with Fluoview). Immunoreactivity quantification of c-Fos was performed using ImageJ. The c-Fos brightness of GFAP-positive astrocytes were measured by the intensity of ROIs that encompassed the cell nuclei. In some experiments involving a heterologous expression of mGluR3, we applied AAV2/5-GfaABC1D-Grm3 (800,000 multiplicity of infection) 5 h after re-seeding for expressing mGluR3 in primary astrocytes and incubated for 5 days before pharmacological experiments. Ninety minutes after stimulation with 0.1 µM NA and/or 100 µM glutamate in the presence of 1 µM TTX, primary astrocytes were fixed and stained as described above using primary antibodies: rabbit anti-mGluR3 (1:500, abcam, ab166608) and mouse anti-c-Fos (1:50, Santa Cruz Biotechnology, sc166940). It is noteworthy that the mouse anti-c-Fos antibody, different from the rabbit anti-c-Fos antibody used in other experiments, exhibited overall low immunoreactivity. However, this did not affect the quantification of c-Fos mobilization. Fluorescent imaging and quantification of c-Fos intensity were performed as described above.
Fibre photometry
AAVs for expressing the noradrenaline biosensor GRABNE2h on the neuronal cell surface, or calcium indicator RCaMP3 in astrocytes with cAMP sensor cAMPinG1 in astrocytes were unilaterally injected into the basolateral amygdala. Optical fibres were implanted above the injection site two weeks post-injection to capture emission of fluorescence signals, with recordings conducted one week after fibre implantation. For fluorescence signal collection, mice were tethered to a patch cable, and a 465 nm LED light (Doric, 40–50 µW at the tip of patch cable) or a 560 nm LED light (Doric, 40–50 µW at the tip of patch cable) was used to excite the signal, with an isosbestic 405 nm LED light used to correct for movement artifacts. Mice tethered to the patch cable were habituated in a testing chamber for 5 min on the day of recording. A time-division multiplexing scheme was used, where the 465 nm, 560 nm and 405 nm LED lights were emitted alternately at 20 Hz (turned on for 24 ms and off for 26 ms), with the timing precisely controlled by a programmable pulse generator (Master-9, AMPI). Each excitation light was reflected by a dichroic mirror and coupled into an optical fibre cable (Doric, 200 μm in diameter, 2 m in length, NA 0.57) through a pinhole (200 μm in diameter). The fluorescence signals were detected by a photomultiplier tube with a GaAsP photocathode (H16722-40; Hamamatsu Photonics) and digitized by a data acquisition module (USB-6341, National Instruments). A custom-written LabVIEW program extracted signals at a sampling frequency of 1 kHz in real time. Fluorescence signals collected during each light pulse were averaged, after dropping the first and last 2 samples (1 ms each) to eliminate potential noise caused during the alternation of LED lights. The data were smoothed using moving average and corrected for bleaching correction by fitting to least-squares linear. The fluorescence response was calculated (fitted 465 + 1)/(fitted 405 + 1) − baseline. The baselines of GRABNE2h and astrocyte cAMPinG1 signals were identified during periods of no spontaneous signals in home cages for 1–2 min before the context exposure. Signal analyses were performed in Python (v.3.0.0) using JupyterLab (v.3.6.7, Project Jupyter). Numerical computations and data handling were conducted using NumPy84 (v.1.26.4) and Pandas (v.2.1.4, NumFOCUS). The area under the curve of fluorescence traces was calculated with scikit-learn (sklearn, v.1.2.2, RRID:SCR_002577), and half-decay times of signal responses were extracted. Astrocytic RCaMP3 signals were bandpass filtered between 0.0003 and 1 Hz with SciPy85 (v.1.11.4). Peaks of Ca2+ signals were defined as data points exceeding 4× s.d. above the baseline mean, with baseline noise estimated from ≥20 s intervals lacking spontaneous activity in home cage recordings before context exposure. Signal traces were visualized with Matplotlib86 (v.3.8.0).
Acute brain slice preparation for astrocyte intracellular Ca2+ imaging
Coronal amygdalar slices were prepared from adult mice with AAV injection26,87. In brief, animals were deeply anaesthetized with isoflurane and decapitated with sharp shears. The brains were placed and sliced in ice-cold modified artificial cerebrospinal fluid (ACSF) containing the following (in mM): 194 sucrose, 30 NaCl, 4.5 KCl, 1 MgCl2, 26 NaHCO3, 1.2 NaH2PO4 and 10 d-glucose, saturated with 95% O2 and 5% CO2. A vibratome (DSK NLS-MT) was used to cut 300-µm brain sections. The slices were allowed to equilibrate for 30 min at 32–34 °C in normal ACSF containing (in mM); 124 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 26 NaHCO3, 1.2 NaH2PO4 and 10 d-glucose continuously bubbled with 95% O2 and 5% CO2. Slices were then stored at 21–23 °C in the same buffer until use. All slices were used within 2–6 h of slicing. Cells for all the experiments were imaged with a 16× water-immersion objective lens (Nikon, N16XLWD-PF, MRP07220, NA: 0.8) with Nikon two-photon laser-scanning microscope (A1R MP) equipped with a 920 nm wavelength laser (Alcor, Spark Lasers). To image GCaMP6f signals, astrocytes located in the LA/B and typically ∼20 to ∼30 µm below the slice surface were selected for imaging. Images were acquired at 1 frame per second. Slices were maintained in ACSF (124 mM NaCl, 4.5 mM KCl, 1 mM MgCl2, 1.2 mM NaH2PO4, 26 mM NaHCO3, 10 mM d-glucose and 2.0 mM CaCl2) through a perfusion system. Phenylephrine (PE, Tocris Bioscience 2838) was applied in the bath in the presence of 300 nM tetrodotoxin (Cayman Chemical 14964) were applied in the bath at least 5 min prior to recording to allow adequate equilibration. A constant flow of fresh buffer perfused the imaging chamber at all times. Spontaneous Ca2+ activity was recorded for 3 min (0 µM PE), followed by a 2-minute bath application of 1 µM PE, a 5-min washout, a 2-min application of 3 µM PE, and a 5-min washout. Ca2+ signals were processed in ImageJ (NIH) and presented as the relative change in fluorescence (ΔF/F).
scRNA-seq and analysis
Mice were anaesthetized with isoflurane 90 min after the initiation of behavioural paradigms and transcardially perfused with PBS (30 ml for 2 min). The amygdala was freshly dissected out using a Brain Matrix (Ted Pella, 15067) and was placed into cold Hanks’ balanced salt solution (HBSS) (14025092, Gibco) containing 10 mM HEPES (17514-15, Naclai Tesque), 0.54% glucose (16806-25, Naclai Tesque), 5.0 µg ml−1 actinomycin D (A1410, Sigma-Aldrich), 10 µM triptolide (T3652, Sigma-Aldrich) and 27.1 µg ml−1 anisomycin (A9789, Sigma-Aldrich), as previously described88,89. Then, the tissue block was incubated for 15 min at 37 °C in 3 ml enzyme solution (HBSS, 10 mM HEPES, 0.54% glucose, 0.5 mM EDTA (15575020, Invitrogen), 1.0 mM l-cysteine (10309-41, Naclai Tesque), 5.0 µg ml−1 actinomycin D, 10 µM triptolide and 27.1 µg ml−1 anisomycin) containing 20 U papain per mouse (LS003127, Worthington Biochemical). The tissue was then homogenized by passing through a 18 G needle and was further incubated for 15 min at 37 °C. Following homogenization using a 20 G needle, the resulting cell suspension was centrifuged at 500g for 5 min at 4 °C. The homogenate was separated by gradient centrifugation with 30% Percoll (P1644, Sigma-Aldrich) in PBS at 500g for 25 min at 4 °C (no brake). The pellet containing astrocytes at the bottom of the tube was then collected and washed once with PBS containing 2% FBS and 10 mM EDTA before staining. Fc receptors were blocked with Fc block (2.4G2, BD Bioscience) for 10 min at 4 °C before incubation with primary antibodies. Cells were stained with antibodies directed against CD11b-BV786 (1:200, M1/70, BD Bioscience), CD45-APC-Cy7 (1:200, 30-F11, BioLegend), O1-eFluor660 (1:200, 50-6506-82, eBioscience) and ASCA2-PE-Cy7 (1:200, 130-116-246, Miltenyi Biotec) for 40 min at 4 °C. Additionally, cells were treated with hashtag antibodies to label source samples (TotalSeqB0301-B0310, B0312 and B0314, BioLegend). After washing, cells were sorted using a CytoFLEX SRT (Beckman Coulter). Data were acquired with CytExpert software (Beckman Coulter). Post-acquisition analysis was performed using FlowJo software, version 10.9.0. Single-cell libraries were generated and sequenced on the Illumina NovaSeq 6000 sequencer. Sequence reads were processed and aligned to the mouse genome (mm10, GENCODE vM23/Ensembl98; https://cf.10xgenomics.com/supp/cell-exp/refdata-gex-mm10-2020-A.tar.gz) using CellRanger 3.0. Initially, genes expressed in <3 cells and cells with <100 genes and mitochondrial reads <6% were excluded from the analysis. Pericytes were removed from amygdalar cells based on Pdgfrb. Hashtag oligos (HTOs) were demultiplexed using HTODemux and singlet cells were used for subsequent analysis. Principal component analysis was performed on the expression data matrix using the Seurat. Thirty principal components were used for generating tSNE plots. Transcriptomic clusters were identified from a total 1,903 amygdalar astrocytes with resolution set to 0.4. Differentially expressed gene candidates from a comparison of Adrb1-positive and Adrb1-negative astrocytes were estimated using Mann–Whitney–Wilcoxon test.
RNAscope
Cryosections were prepared as described above and stored at −80 °C. RNAscope in situ hybridization was performed using RNAscope Multiplex Fluorescent Reagent Kit v.2 with TSA Vivid Dyes (ACDBio, 323270), according to the manufacturer’s instructions. The sections were washed for 5 min with PBS, and then heated at 60 °C for 30 min before post-fix. After dehydration in an ethanol series, hydrogen peroxide was applied for 10 min at room temperature. The sections were washed with ddH2O twice, and then incubated in Target Retrieval Reagents for 5 min at 90–100 °C. A wash with ddH2O for 15 s was followed by a dehydration with 100% ethanol for 3 min and dried at room temperature. Then, the sections were incubated with Protease III for 30 min at 40 °C. The sections were washed with ddH2O twice and then incubated with probes Mm-Fos (ACDBio, 316921), Mm-Adra1a (ACDBio, 408611), Mm-Adrb1-C2 (ACDBio, 449761-C2), Mm-Igfbp2 (ACDBio, 405951) and Mm-Slc1a3-C3 (ACDBio, 430781-C3) for 2 h at 40 °C. Thereafter, the sections were washed twice with wash buffer for 2 min between incubations at 40 °C. The sections were incubated in AMP1 for 30 min, AMP2 for 30 min and AMP3 for 15 min at 40 °C, then incubated in horseradish peroxidase (HRP) for 15 min, fluorophore for 30 min and HRP-blocker for 15 min at 40 °C for each channel. In the staining of mCherry, slices were washed in 0.1 M PBS 3 times for 10 min each, followed by immunohistochemistry that was performed as described above. Following primary and secondary antibodies used: rabbit anti-RFP (1:250, Rockland, 600-401-379), Alexa Fluor 568 donkey anti-rabbit (1:1,000, Abcam, ab175692). Finally, the sections were incubated with DAPI at room temperature, removed DAPI, and mounted in Prolong Gold Antifade Reagent (Invitrogen, P36934). Images were obtained in the same way as immunohistochemistry described above except with a step size of 0.5 µm. Images were processed with ImageJ. Astrocyte somata were delineated using Slc1a3 signals. The intensity of Fos, Adra1a, Adrb1 and Igfbp2 signals within the somata were measured. To account for differences in somata sizes, the signals were normalized using Slc1a3. Subsequently, the normalized signals were z-scored within each sample to evaluate gene expression across a heterogeneous cell population. This approach facilitated the robust identification of cells exhibiting high gene expression relative to the population mean. In Fig. 4e,f, adrenoreceptor-positive cells were identified based on scRNA-seq data, selecting the top 50% of cells ranked by Adra1a expression and the top 20% of cells ranked by Adrb1 expression. Co-expression index of Adra1a and Adrb1, calculated by relative expression level of Adra1a to NoFC median multiplied by relative expression level of Adrb1 for each cell. In Fig. 4j, the correlation analysis of Fos and Adrb1, expression levels were evaluated using z-scores of RNAscope signal intensity to assess gene expression across a heterogeneous population of cells, enabling robust identification of cells with high gene expression relative to the population mean. Specifically, we identified astrocytes with z-scores greater than 2 in RNAscope Fos signal intensity as Fos-high astrocytes, capturing 3% of the astrocyte population with significantly elevated Fos expression, consistent with scRNA-seq data in Fig. 4c where Fos-positive astrocytes occupied 4% of the total astrocytes analysed. Similarly, Adrb1-high astrocytes were identified using z-scores greater than 0.5, capturing 26% of astrocytes, also in line with scRNA-seq data in Fig. 4c where Adrb1-positive astrocytes occupied 20% of the total astrocytes analysed. In addition, Igfbp2-high astrocytes were identified using z-scores greater than 0.5, capturing 24% of all astrocytes in Extended Data Fig. 12d,e, which is consistent with scRNA-seq data indicating that Igfbp2-positive astrocytes occupy 24% of the total astrocytes analysed.
Statistical analysis
Most statistical tests were run in Prism 10.1.2 (GraphPad). The specific tests used are indicated in the figure legends and Supplementary Table 1. All tests were two-sided unless otherwise noted. Astrocyte Fos count analyses across groups for each brain region were run with R v.4.3.2 on RStudio v.2023.12.1.402 and relied on the tidyverse, car, dunn.test and dplyr packages and base R functions aov(), TukeyHSD() and p.adjust(). The data were first analysed with Levene’s test for homogeneity of variance. Brain regions that passed Levene’s test (P > 0.05) were followed by a one-way ANOVA test with Tukey’s honest significant difference test and regions that failed Levene’s test (P < 0.05; no homogeneity of variances) were followed by the Kruskal–Wallis test with Dunn’s post hoc test and Bonferroni adjustment. False discovery rates were calculated across groups and brain regions from post hoc P values using the Benjamini–Hochberg method.
Mice were randomly assigned to experimental groups, and mice assigned to different experimental conditions were run in parallel. Mouse experiments and analyses were done blinded to group allocation. Tissue samples that yielded insufficient cell counts (less than 30 cells) during scRNA-seq quality control were excluded from the study. The results of statistical comparisons, n numbers and P values are shown in the figure panels or figure legends along with averages. n is defined as the numbers of cells, sections or mice throughout on a case-by-case basis; the unit of analysis is stated in the text or in each figure legend. In the figures, summarized data are shown as mean ± s.e.m. with a scatter plot and P values are indicated by asterisks: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the single-cell transcriptomics data are available at Gene Expression Omnibus with accession identifier GSE272414. All raw replicate data used to generate the figures and the associated statistical tests are provided in Supplementary Table 1. Fos-mNG astrocyte counts in 677 brain regions in NoFC, FC, NoFR and FR groups are provided in Supplementary Table 2. All raw data supporting the findings of this study are available on the RIKEN CBS data sharing platform (https://neurodata.riken.jp/id/20250831-001; https://doi.org/10.60178/cbs.20250831-001).
Code availability
Custom code for 3D rendering of the brain-wide Fos-mNG astrocyte map is available on GitHub (https://github.com/BrainImageAnalysis/Astrocyte3DMapping/). Custom code for statistical analysis brain-wide Fos-mNG astrocyte counts, fibre photometry data analysis and single-cell transcriptomic analysis is available on GitHub (https://github.com/Jun-Nagai-Lab/Dewa-et-al).
References
Misanin, J. R., Miller, R. R. & Lewis, D. J. Retrograde amnesia produced by electroconvulsive shock after reactivation of a consolidated memory trace. Science 160, 554–555 (1968).
Przybyslawski, J. & Sara, S. J. Reconsolidation of memory after its reactivation. Behav. Brain Res. 84, 241–246 (1997).
Dudai, Y. The restless engram: consolidations never end. Annu. Rev. Neurosci. 35, 227–247 (2012).
Lee, J. L. C., Nader, K. & Schiller, D. An update on memory reconsolidation updating. Trends Cogn. Sci. 21, 531–545 (2017).
Roy, D. S. et al. Brain-wide mapping reveals that engrams for a single memory are distributed across multiple brain regions. Nat. Commun. 13, 1799 (2022).
Agren, T. et al. Disruption of reconsolidation erases a fear memory trace in the human amygdala. Science 337, 1550–1552 (2012).
Finnie, P. S. B. & Nader, K. Amyloid beta secreted during consolidation prevents memory malleability. Curr. Biol. 30, 1934–1940.e4 (2020).
Josselyn, S. A. & Tonegawa, S. Memory engrams: recalling the past and imagining the future. Science 367, eaaw4325 (2020).
Flavell, S. W. & Greenberg, M. E. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 31, 563–590 (2008).
Allen, N. J. & Lyons, D. A. Glia as architects of central nervous system formation and function. Science 362, 181–185 (2018).
Nagai, J. et al. Behaviorally consequential astrocytic regulation of neural circuits. Neuron 109, 576–596 (2021).
Endo, F. et al. Molecular basis of astrocyte diversity and morphology across the CNS in health and disease. Science 378, eadc9020 (2022).
Molofsky, A. V. et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194 (2014).
Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
Murphy-Royal, C., Ching, S. & Papouin, T. A conceptual framework for astrocyte function. Nat. Neurosci. 26, 1848–1856 (2023).
Sardar, D. et al. Induction of astrocytic Slc22a3 regulates sensory processing through histone serotonylation. Science 380, eade0027 (2023).
Sardar, D. et al. Mapping astrocyte transcriptional signatures in response to neuroactive compounds. Int. J. Mol. Sci. 22, 3975 (2021).
Williamson, M. R. et al. Learning-associated astrocyte ensembles regulate memory recall. Nature 637, 478–486 (2025).
Yu, X. et al. Context-specific striatal astrocyte molecular responses are phenotypically exploitable. Neuron 108, 1146–1162.e10 (2020).
Lee, H.-G. et al. Disease-associated astrocyte epigenetic memory promotes CNS pathology. Nature 627, 865–872 (2024).
Escartin, C. et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325 (2021).
Sun, W. et al. Spatial transcriptomics reveal neuron-astrocyte synergy in long-term memory. Nature 627, 374–381 (2024).
Porter, J. T. & McCarthy, K. D. Astrocytic neurotransmitter receptors in situ and in vivo. Prog. Neurobiol. 51, 439–455 (1997).
Semyanov, A., Henneberger, C. & Agarwal, A. Making sense of astrocytic calcium signals - from acquisition to interpretation. Nat. Rev. Neurosci. 21, 551–564 (2020).
Mu, Y. et al. Glia accumulate evidence that actions are futile and suppress unsuccessful behavior. Cell 178, 27–43.e19 (2019).
Nagai, J. et al. Specific and behaviorally consequential astrocyte Gq GPCR signaling attenuation in vivo with iβARK. Neuron 109, 2256–2274.e9 (2021).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
DeNardo, L. A. et al. Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat. Neurosci. 22, 460–469 (2019).
Nagai, J. et al. Hyperactivity with disrupted attention by activation of an astrocyte synaptogenic cue. Cell 177, 1280–1292.e20 (2019).
Adamsky, A. et al. Astrocytic activation generates de novo neuronal potentiation and memory enhancement. Cell 174, 59–71.e14 (2018).
Fanselow, M. S. Factors governing one-trial contextual conditioning. Anim. Learn. Behav. 18, 264–270 (1990).
Ragan, T. et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258 (2012).
Piantadosi, S. C. et al. Holographic stimulation of opposing amygdala ensembles bidirectionally modulates valence-specific behavior via mutual inhibition. Neuron 112, 593–610.e5 (2024).
Josselyn, S. A. et al. Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. J. Neurosci. 21, 2404–2412 (2001).
Tronson, N. C., Wiseman, S. L., Olausson, P. & Taylor, J. R. Bidirectional behavioral plasticity of memory reconsolidation depends on amygdalar protein kinase A. Nat. Neurosci. 9, 167–169 (2006).
Lin, Y. et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455, 1198–1204 (2008).
Smith, N. A. et al. Fluorescent Ca2+ indicators directly inhibit the Na,K-ATPase and disrupt cellular functions. Sci. Signal. 11, eaal2039 (2018).
Takano, T. et al. Rapid manifestation of reactive astrogliosis in acute hippocampal brain slices. Glia 62, 78–95 (2014).
Sun, W. et al. Glutamate-dependent neuroglial calcium signaling differs between young and adult brain. Science 339, 197–200 (2013).
Pfeil, E. M. et al. Heterotrimeric G protein subunit Gαq is a master switch for Gβγ-mediated calcium mobilization by Gi-coupled GPCRs. Mol. Cell 80, 940–954.e6 (2020).
Sun, X. et al. Functionally distinct neuronal ensembles within the memory engram. Cell 181, 410–423.e17 (2020).
Sørensen, A. T. et al. A robust activity marking system for exploring active neuronal ensembles. eLife 5, e13918 (2016).
Uematsu, A. et al. Modular organization of the brainstem noradrenaline system coordinates opposing learning states. Nat. Neurosci. 20, 1602–1611 (2017).
Feng, J. et al. Monitoring norepinephrine release in vivo using next-generation GRABNE sensors. Neuron 112, 1930–1942 (2024).
Yokoyama, T. et al. A multicolor suite for deciphering population coding of calcium and cAMP in vivo. Nat. Methods 21, 897–907 (2024).
Suthard, R. L. et al. Basolateral amygdala astrocytes are engaged by the acquisition and expression of a contextual fear memory. J. Neurosci. 43, 4997–5013 (2023).
Batiuk, M. Y. et al. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J. Biol. Chem. 292, 8874–8891 (2017).
Furube, E. et al. Neural stem cell phenotype of tanycyte-like ependymal cells in the circumventricular organs and central canal of adult mouse brain. Sci. Rep. 10, 2826 (2020).
Kohro, Y. et al. Spinal astrocytes in superficial laminae gate brainstem descending control of mechanosensory hypersensitivity. Nat. Neurosci. 23, 1376–1387 (2020).
Numata, G. et al. In vivo effects of cardiomyocyte-specific β-1 blockade on afterload- and frequency-dependent cardiac performance. Am. J. Physiol. Heart. Circ. Physiol. 328, H543–H549 (2025).
Gleichman, A. J., Kawaguchi, R., Sofroniew, M. V. & Carmichael, S. T. A toolbox of astrocyte-specific, serotype-independent adeno-associated viral vectors using microRNA targeting sequences. Nat. Commun. 14, 7426 (2023).
Caldwell, A. L. M. et al. Aberrant astrocyte protein secretion contributes to altered neuronal development in multiple models of neurodevelopmental disorders. Nat. Neurosci. 25, 1163–1178 (2022).
Tu, X. et al. Local autocrine plasticity signaling in single dendritic spines by insulin-like growth factors. Sci. Adv. 9, eadg0666 (2023).
Khan, S. et al. IGFBP2 plays an essential role in cognitive development during early life. Adv. Sci. 6, 1901152 (2019).
Han, J.-H. et al. Increasing CREB in the auditory thalamus enhances memory and generalization of auditory conditioned fear. Learn. Mem. 15, 443–453 (2008).
Ramsaran, A. I. et al. A shift in the mechanisms controlling hippocampal engram formation during brain maturation. Science 380, 543–551 (2023).
McCall, J. G. et al. Locus coeruleus to basolateral amygdala noradrenergic projections promote anxiety-like behavior. eLife 6, e18247 (2017).
Ramón y Cajal, S. Algunas conjeturas sobre el mecanismo anatómico de la ideación, asociación y atención. Rev. Med. Cir. Pract. 36, 497–508 (1895).
Cahill, M. K. et al. Network-level encoding of local neurotransmitters in cortical astrocytes. Nature 629, 146–153 (2024).
Gong, L., Pasqualetti, F., Papouin, T. & Ching, S. Astrocytes as a mechanism for contextually-guided network dynamics and function. PLoS Comput. Biol. 20, e1012186 (2024).
Oe, Y. et al. Distinct temporal integration of noradrenaline signaling by astrocytic second messengers during vigilance. Nat. Commun. 11, 471 (2020).
Sapkota, D. et al. Activity-dependent translation dynamically alters the proteome of the perisynaptic astrocyte process. Cell Rep. 41, 111474 (2022).
Choucry, A., Nomoto, M. & Inokuchi, K. Engram mechanisms of memory linking and identity. Nat. Rev. Neurosci. 25, 375–392 (2024).
Zaki, Y. et al. Offline ensemble co-reactivation links memories across days. Nature 637, 145–155 (2025).
Smolen, P., Zhang, Y. & Byrne, J. H. The right time to learn: mechanisms and optimization of spaced learning. Nat. Rev. Neurosci. 17, 77–88 (2016).
Nader, K., Schafe, G. E. & Le Doux, J. E. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature 406, 722–726 (2000).
Debiec, J. & Ledoux, J. E. Disruption of reconsolidation but not consolidation of auditory fear conditioning by noradrenergic blockade in the amygdala. Neuroscience 129, 267–272 (2004).
Baldwin, K. T., Murai, K. K. & Khakh, B. S. Astrocyte morphology. Trends Cell Biol. 34, 547–565 (2024).
Naegeli, C. et al. Locus coeruleus activity mediates hyperresponsiveness in posttraumatic stress disorder. Biol. Psychiatry 83, 254–262 (2018).
Cornell-Bell, A. H., Finkbeiner, S. M., Cooper, M. S. & Smith, S. J. Glutamate induces calcium waves in cultured astrocytes: long-range glial signaling. Science 247, 470–473 (1990).
Ollivier, M. et al. Crym-positive striatal astrocytes gate perseverative behaviour. Nature 627, 358–366 (2024).
Rusakov, D. A., Bard, L., Stewart, M. G. & Henneberger, C. Diversity of astroglial functions alludes to subcellular specialisation. Trends Neurosci. 37, 228–242 (2014).
King, C. M. et al. Local resting Ca2+ controls the scale of astroglial Ca2+ signals. Cell Rep. 30, 3466–3477.e4 (2020).
Horii, T. et al. Efficient generation of conditional knockout mice via sequential introduction of lox sites. Sci. Rep. 7, 7891 (2017).
Inoue, M. et al. Rational engineering of XCaMPs, a multicolor GECI suite for in vivo imaging of complex brain circuit dynamics. Cell 177, 1346–1360.e24 (2019).
Takano, T. et al. Chemico-genetic discovery of astrocytic control of inhibition in vivo. Nature 588, 296–302 (2020).
Lesuis, S. L. et al. Stress disrupts engram ensembles in lateral amygdala to generalize threat memory in mice. Cell 188, 121–140.e20 (2025).
Levy, A., Ngai, S. H., Finck, A. D., Kawashima, K. & Spector, S. Disposition of propranolol isomers in mice. Eur. J. Pharmacol. 40, 93–100 (1976).
Roth, B. L. DREADDs for neuroscientists. Neuron 89, 683–694 (2016).
Yao, Z. et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332 (2023).
Wang, Q. et al. The Allen Mouse Brain Common Coordinate Framework: a 3D reference atlas. Cell 181, 936–953.e20 (2020).
Avants, B. B., Epstein, C. L., Grossman, M. & Gee, J. C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008).
Schildge, S., Bohrer, C., Beck, K. & Schachtrup, C. Isolation and culture of mouse cortical astrocytes. J. Vis. Exp. https://doi.org/10.3791/50079 (2013).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Yu, X. et al. Reducing astrocyte calcium signaling in vivo alters striatal microcircuits and causes repetitive behavior. Neuron 99, 1170–1187.e9 (2018).
Yamasaki, A. et al. IRF8 and MAFB drive distinct transcriptional machineries in different resident macrophages of the central nervous system. Commun. Biol. 7, 896 (2024).
Marsh, S. E. et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci. 25, 306–316 (2022).
Acknowledgements
We thank N. Fujimori, R. Ando, N. Miyasaka and N. Mataga for technical assistance; M. Motoyoshi for assisting with imaging; Y. Li for sharing GRAB sensors; and T. J. McHugh, J. P. Johansen, T. Kitamura, K. Tanaka, T. Kitazawa, T. Kanaya, B. Wulaer, E. Shigetomi, S. Okabe, B. Deneen and B. S. Khakh for valuable comments and discussions. STPT, cryosectioning and image acquisition were performed with common use equipment at the Support Unit for Bio-Material Analysis, RRD of RIKEN CBS. We thank the Division of Gene Expression Dynamics, the MEXT Cooperative Research Project Program, the Medical Research Center Initiative for High Depth Omics, and acknowledge support from CURE:JPMXP1323015486 for the Medical Institute of Bioregulation, Kyushu University for sequencing. Collaborations between the J.N. and T.M. groups are supported by Japan Agency for Medical Research and Development (AMED) grant JP24gm1910004. This work was supported in part by JSPS KAKENHI grant numbers 24K02128 (to J.N.), 24H02180 (to J.N.), 24KJ2074 (to K.K.), 23H04179 (to J.N.), JP22K15625 (to K.D.), JP21H00220 (to J.N.), JP21H02588 (to J.N.), JP21H05640 (to J.N.), JP21K15207 (to T.Y.), JP24H00861 (to M.S.), JP23H02782 (to M.S.), JP21H02752 (to T.M.), JP22H05062 (to T.M.), JP21H05044 (to M.I.), JP24K02264 (to M.I.), JSPS KAKENHI 24K02441 (E.T.), 25K19410 (G.N.), 24K19053 (S.N.) by Japan Science and Technology Agency (JST) under grant numbers JPMJFR2249 (to J.N.), JPMJAX211K (to T.Y.), JPMJPR1906 (to M.S.), by AMED under grant numbers JP21km0908001 (to J.N.), JP22dk0207063 (to J.N.), JP23jf0126004 (to J.N. and T.M.), JP21wm0425001 (to T.M.), JP23gm6510022 (to M.S.), JP21zf0127003h (to M.I.), JP22wm0425011 (to M.I.), Multidisciplinary Frontier Brain and Neuroscience Discoveries (Brain/MINDS 2.0) by AMED JP23wm0625001 (to H.S.), by RIKEN-Osaka University Science and Technology Hub Collaborative Research Program (to J.N.), and by the Takeda Science Foundation (to J.N.), the Mitsubishi Foundation (to T.M.), Daiichi Sankyo Foundation of Life Science (to T.M.), Mochida Memorial Foundation for Medical and Pharmaceutical Research (to T.M.), Astellas Foundation for Research on Metabolic Disorders (to T.M.), Ono Pharmaceutical Foundation for Oncology (to T.M.), Immunology and Neurology (to T.M.), the Nakajima Foundation (to J.N.), the Chemo-Sero-Therapeutic Research Institute (to M.I.) and the Uehara Memorial Foundation (to T.M.). K.K. and H.K. were partly supported by JSPS Research Fellowships (24KJ2074 and 22KJ3141, respectively).
Author information
Authors and Affiliations
Contributions
K.D. carried out vector constructions with help from A. Komori, performed mouse surgeries and behaviour tests, and with M.A.H. and J.N., analysed brain-wide imaging data. A. Kasai provided guidance on the analysis of brain-wide data. H.K. performed ex vivo astrocyte calcium imaging and analysis. H.S. performed 3D rendering and video creation. A. Kuwahara performed primary astrocyte experiments with help from K.D., who established the protocol. K.K. conducted in vivo fibre photometry and animal behaviour experiments, including associated surgeries with guidance from N.T. on coding for data analysis. T.Y. and M.S. provided guidance on astrocyte sensors. M.T., G.N., S.N. and E.T. provided key mouse lines and contributed intellectually to related analyses. Tissue preparation for single-cell transcriptomics was performed by K.D., K.K., H.K. and M.A.H., followed by cell sorting by A.Y. and analysis of sequencing data by K.K. with guidance from M.I. and T.M., who provided equipment. K.D., A. Komori and N.A. performed RNAscope analyses. K.D. and K.K. performed in vivo infusion of antibodies, for which K.K. performed mouse surgeries and histological analyses. J.N. conceptualized and directed the project, guided analyses and wrote the paper with help from K.D., K.K., A.K. and H.S. All authors commented on and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Christian Henneberger, Gertrudis Perea and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Validation of astrocyte Fos tagging approach: brain-wide astrocyte selectivity.
a, Schematic illustrating an AAV PHP.eB vector intravenous (i.v.) injection into retro orbital sinus of adult mice for expressing red fluorescence reporter mScarlet-I in astrocytes in whole brain. b, Representative images showing that whole-brain expression of mScarlet-I was observed in AAV injected mice, but not in saline injected mice. AAV PHP.eB-mediated mScarlet-I delivery resulted in expression within S100β/GFAP positive astrocytes. Subpanel b’ shows mScarlet-I expression in the orbitofrontal cortex (ORB), the prelimbic cortex (PL), the anterior insular cortex (AIC), the dorsomedial striatum (DMS), the nucleus accumbens (ACB), the mediodorsal thalamus (MD), and the lateral and basal amygdala (LA/B). Representative image from 3 mice. c, Bar plots showing that 97.3%–99.5% of astrocytes were positive for mScarlet-I, whose signals were enhanced with anti-RFP antibody, in the indicated 7 brain regions (left) and that those regions had few mScarlet-I-positive neurons (right). n = 3 mice. Data are mean ± SEM (error bars smaller than symbols in some cases). Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 2 Validation of astrocyte Fos tagging approach: Fos upregulation sensitivity.
a, Schematic illustrating AAV-PHP.eB vector i.v. injection into Fos-iCreERT2 (TRAP2) mice for expressing green fluorescence reporter mNeonGreen (mNG) in Fos+ astrocytes. AAV2/5-GfaABC1D-hM3Dq-mCherry was stereotaxically delivered into the orbitofrontal cortex (ORB) to induce Fos in astrocytes by 1 mg/kg CNO i.p. injection and tag those Fos+ astrocytes with mNG by 50 mg/kg 4-hydroxytamoxifen (4-OHT) i.p. injection. b, hM3Dq activation with CNO administration increased Fos expression in ORB astrocytes. hM3Dq-, CNO+: n = 12 slices, hM3Dq+, CNO-: n = 12 slices, hM3Dq+, CNO+: n = 12 slices, hM3Dq-, 3 mice/group. ****p < 0.0001. One-way ANOVA, Tukey’s test. Data are mean ± SEM. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 3 Validation of astrocyte Fos tagging approach: Temporal window of astrocyte Fos tagging with visual stimulation.
a, Schematic illustrating AAV-PHP.eB vector i.v. injection into Fos-iCreERT2 (TRAP2) mice for expressing green fluorescence reporter mNeonGreen (mNG) in Fos+ astrocytes. b, Timeline of visual stimulation experiment to determine effective time window of astrocyte Fos tagging. c, Example images of Fos-tagged (mNG-positive) astrocytes in the primary visual cortex (VISp) of TRAP2 mice that underwent the visual stimulation experiment (representative of 9–12 slices from 3-4 mice/group). d, Quantification of mNG astrocyte density in TRAP2 mice VISp and primary somatosensory cortex (SSp): NS: n = 12 slices from 4 mice, −24 h: n = 12 slices from 4 mice, −12 h: n = 12 slices from 3 mice, −6 h: n = 12 slices from 4 mice, −3 h: n = 12 slides from 4 mice, 0 h: n = 12 slices from 4 mice, 3 h: n = 9 slices from 3 mice, 6 h: n = 9 slices from 3 mice, 12 h: n = 9 slices from 3 mice, 24 h: n = 9 slices from 3 mice. Two-way ANOVA, Šídák’s multiple comparisons test. Data are mean ± SEM. *p < 0.05, ***p < 0.001, ****p < 0.0001. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 4 Astrocyte c-Fos protein levels following fear conditioning and recall.
a, Representative immunostaining of c-Fos expression in a subset of amygdalar astrocytes after fear conditioning (FC) and fear recall (FR). Arrowheads denote c-Fos+ Sox9+ cells (representative of 15 slices from 3 mice/group). b, Quantification of c-Fos+ astrocytes in the lateral and basolateral amygdala (LA/B) after FC and FR. n = 15 slices from 3 mice/group. One-way ANOVA, Tukey’s test. Data are mean ± SEM. **p < 0.01. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 5 Validation of astrocyte Fos tagging approach: Temporal window of astrocyte Fos tagging with fear recall.
a, Schematic illustrating AAV-PHP.eB vector i.v. injection into Fos-iCreERT2 (TRAP2) mice for expressing green fluorescence reporter mNeonGreen (mNG) in Fos+ astrocytes. b, Timeline of fear recall experiment to determine effective time window of astrocyte Fos tagging. c, Example images of Fos-tagged (mNG-positive) astrocytes in the lateral and basolateral amygdala (LA/B) of TRAP2 mice that underwent the fear recall experiment (representative of 17–48 slices from 3–8 mice/group). d, NS: n = 24 slices from 4 mice, −12 h: n = 30 slices from 5 mice, −6 h: n = 42 slices from 7 mice, −3 h: n = 23 slides from 4 mice, 0 h: n = 48 slices from 8 mice, 3 h: n = 30 slices from 5 mice, 6 h: n = 18 slices from 3 mice, 12 h: n = 17 slices from 3 mice. Two-way ANOVA, Šídák’s multiple comparisons test. Data are mean ± SEM. ****p < 0.0001. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 6 Fear conditioning with contextual novelty induces astrocyte Fos ensembles.
a, Left: Whole-brain mapping pipeline. Alongside NoFC and FC groups (Fig. 1a), an additional cohort received 4-OHT immediately after CtxA exposure with shocks but no habituation (FC+Novelty). Brains were fixed and processed for STPT, 3D reconstruction, regional segmentation, and single-cell counting as in Fig. 1a. b, 3D rendering of the whole brain and three brain regions - the hippocampus (HPC; CA1, CA2, CA3 and dentate gyrus), the amygdala (lateral, basolateral and basomedial amygdalar subregions), primary visual cortex (VISp) - showing that Fos-mNG astrocytes (white dots). D dorsal, V ventral, A anterior, P posterior, M medial, L lateral. Scale bars, 500 µm. c-d, Heatmap of BAE count fold changes (vs NoFC in c, vs FC in d, n = 4 mice/group). *FDR < 0.05. *FDR < 0.05; full stats in Supplementary Table 2. Levene’s test followed by one-way ANOVA or Kruskal-Wallis; post-hoc Tukey or Dunn with Bonferroni. FDR controlled by Benjamini-Hochberg. Full data in Supplementary Table 2. Primary visual area (VISp), primary somatosensory area barrel field (SSp-bfd), orbital cortex (ORB), prelimbic cortex (PL), infralimbic cortex (ILA), entorhinal cortex (ENT), anterior cingulate cortex (ACA), olfactory cortex (OLF), retrosplenial cortex (RSP), lateral amygdala (LA), basal amygdala (BA), central amygdala (CEA), mediodorsal thalamus (MD), dorsal geniculate thalamus (GENd), anterodorsal thalamus (AD), anteroventral thalamus (AV), ventroposterior medial nucleus of thalamus (VPM), caudoputamen (CP), nucleus accumbens (ACB), dentate gyrus (DG), hippocampal CA1 (CA1), hippocampal CA3 (CA3), para-subiculum (PAR), and pre-subiculum (PRE). e, Hippocampus/CA1 BAE counts higher in FC+Novelty. FC-, Novelty-: n = 6 mice, FC+, Novelty-: n = 4 mice, FC+, Novelty+: n = 4 mice. Data are mean ± SEM. *p < 0.05. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 7 Growth factors known to induce neuronal c-Fos did not induce astrocyte c-Fos.
a, Schematics illustrating the design of pharmacological screening. b, Representative images from 9 biological replicates showing high purity of primary astrocyte culture. The immunoreactivities of Iba1, a microglia marker, Tuj1, a neuronal marker and Olig2, an oligodendrocyte marker, were not evident, and most of the DAPI signal overlapped with GFAP-positive primary astrocytes. c, Graph showing that c-Fos intensity in GFAP-positive astrocytes was increased with noradrenaline (NA), but not with growth factors that are known to induce c-Fos expression in primary neurons. TTX (1 µM) was used throughout the culture experiments to eliminate the potential effect of neuronal firing. Veh: n = 41 cells, NA: n = 45 cells, BDNF: n = 29 cells, NT-3: n = 33 cells, NT-4: n = 41 cells, NGFβ: n = 35 cells, EGF: n = 36 cells, PDGF-AA: n = 39 cells, PDGF-BB: n = 35 cells, CNTF: n = 40 cells, IGF: n = 46 cells. One-way ANOVA, Dunnett’s test (vs Veh). Data are mean ± SEM. ****p < 0.0001; ns, not significantly different. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 8 Validation of Robust Activity Marking (RAM) technique for silencing engram neurons.
a, Schematic showing the injection of RAM-hM4Di-mCherry or RAM-NLS-mKate2 into the amygdala. Mice underwent fear conditioning two weeks later followed by i.p. CNO (1 mg/kg) injections the day after. Brain samples were intracardially fixed for immunohistochemistry analysis. b, Graphs showing the percentage of RAM-expressed neurons in LA/B (left), co-expression of c-Fos and RAM (center), and overall c-Fos+ neurons (right). mKate2: n = 15 slices, hM4Di: n = 14 slices, 5 mice per group. Mann-Whitney test. ****p < 0.0001. c, Representative images (from mKate2: n = 15 slices, hM4Di: n = 14 slices, 5 mice/group) showing expression of neuronal marker NeuN and RAM-tagged engram neurons (indicated by mCherry signals) in b. Overlapping c-Fos-expressing engram neurons were fewer in the RAM-hM4Di group. Data are mean ± SEM. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 9 FR-BAE proximity to NA projections and engram neurons.
a, Images (representative of n = 16 slices from 4 mice) showing FR-BAE (green) in the amygdala overlapping with astrocyte markers GFAP and S100β (cyan) and proximal to NA projections (white) and engram neurons (red). b, Graphs showing that engram neurons (left) and NA projections (center) are statistically more colocalized with FR-BAE than non-FR-BAE astrocytes. Astrocyte territory size is not different between BAE and non-FR-BAE astrocytes (right). n = 16 slices from 4 mice/group. Mann-Whitney test. *p < 0.05; ns, not significantly different. Data are mean ± SEM. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 10 NA, astrocyte cAMP and Ca2+ signals in a context relevant or irrelevant to fear.
a, Histological analysis of optical fiber placement in LA/B for fiber photometry. Termination sites were mapped onto a standard atlas (adapted from Paxinos, G. & Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates, 2nd ed. Academic Press, 2001). Circles show fiber tips from individual mice, color-coded by experimental group. n = 6 mice in GRABNE2h and 8 mice in cAMPinG1/RCaMP3 experiments. b, Freezing before, during, after FC, during FR, and in CtxB (distinct from CtxA). Freezing was lower in CtxB and in CtxA with Prop vs FR (FR: n = 14; CtxB: n = 13; Prop: n = 8 mice). c, dF/F traces: NA signals evoked by CtxA or CtxB (early) persisted longer in CtxA (late). n = 6 mice. d, Scatter plots: integrated areas of early and late NA signals in CtxA vs CtxB. n = 6 mice. e, Astrocyte cAMP traces: gradual buildup in HC, strong early responses to context exposure. FR further enhanced late cAMP in CtxA, not CtxB. n = 8 mice. f, Scatter plots: late cAMP signal areas higher in CtxA vs CtxB, consistent with GRABNE2h. n = 8 mice. g, Astrocyte Ca2+ traces: evoked by context exposure (early). At 7–15 min, signal frequency remained elevated in CtxA, not CtxB (late). n = 7 mice. h, Scatter plots: late Ca2+ frequency higher in CtxA vs CtxB. n = 7 mice. Wilcoxon signed-rank test or paired t test was used based on normality of the data distribution. *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significantly different. Data are mean ± SEM (error bars smaller than symbols in some cases). Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 11 scRNA-seq of amygdalar astrocytes.
a, Representative gating strategy of FACS-sorting of ACSA2+ astrocytes from the amygdala for scRNA-seq. b, Violin plot showing relative expression levels of cell type marker genes in scRNA-seq. Aldoc, an astrocyte marker, was the only gene showing high expression. c, tSNE map showing astrocyte genes Aldoc, Atp1a2 (coding astrocyte cell surface antigen-2, ACSA2 with which we aimed to FACS-sort astrocytes from the amygdala tissue), and Slc1a3 with which we identified high expression in astrocytes across amygdalar astrocytes in our RNAscope experiments. d, Left: heatmap showing the top 20 genes that were upregulated in each astrocyte cluster relative to the others. Right: Example cluster marker genes identified were plotted onto the tSNE map. See also Supplementary Table 3. e, Left: tSNE plot of astrocyte clusters to illustrate that the 4 clusters are found in every single mouse examined. Right: Bar graph showing the proportion of each astrocyte cluster in the four experimental groups. n = 16 mice in b-d, 4 mice/group in e.
Extended Data Fig. 12 Astrocytic Igfbp2 stabilizes memory during reconsolidation.
a, Top 10 differentially expressed gene candidates, ranked by p value comparing Adrb1-positive vs. -negative astrocytes. Heatmap shows decreased (blue) or increased (red) expression. b, Violin plot showing Igfbp2 enrichment in Adrb1-positive astrocytes. n = 293 Adrb1–, 87 Adrb1+ astrocytes. Mann–Whitney test. c, RNAscope images (representative of 588 cells from 6 mice) of Adrb1 and Igfbp2 in Slc1a3-expressing astrocytes in LA/B from FR. Arrow: astrocyte with high Adrb1 and Igfbp2. d, Correlation of Igfbp2 and Adrb1 levels. Igfbp2-high astrocytes (z > 0.5) = 24% of astrocytes. Rationale in Methods. e, Adrb1 expression enriched in Igfbp2-high astrocytes (n = 451 low, 137 high cells). Igfbp2 expression enriched in Adrb1-high astrocytes (n = 426 low, 162 high cells). Mann–Whitney test. Lines, median. f, Schematic of in vivo Igfbp2 perturbation. Mice implanted with bilateral cannulae into LA/B for Igfbp2-neutralizing antibody or control IgG infusion immediately after FR. g, Scatter graph: IgG infusion (n = 9 mice) showed no change in freezing between 1st and 2nd FR, whereas Igfbp2 antibody infusion (n = 12 mice) reduced freezing in 2nd FR, indicating destabilized memory. Wilcoxon signed-rank one-tail test. h, Freezing behaviour during 5-day Igfbp2 antibody infusions into LA/B within reconsolidation windows. n = 9 mice IgG, 12 mice Igfbp2 antibody. Two-way ANOVA with Šídák’s test. i, Cannula placement in LA/B. Sites mapped on a standard atlas (adapted from Paxinos, G. & Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates, 2nd ed. Academic Press, 2001). Each circle indicates cannula termination, coloured by group. n = 9 mice IgG, 12 mice Igfbp2 antibody. j, Representative LA/B image showing Alexa-labelled antibody infusion (representative of 36 sections from 21 mice). *p < 0.05, **p < 0.01, ****p < 0.0001; ns, not significant. Data are mean and SEM (error bars smaller than symbols in some cases). Full details in Supplementary Table 1.
Extended Data Fig. 13 Astrocytic iβARK2 validation in the amygdala.
a, Schematics showing the Gαq GPCR signalling cascade and the blocking of which with iβARK. Left: iβARK, a 122-residue inhibitory peptide from β-adrenergic receptor kinase26, directly binds to Gαq subunits and thus sequesters them to block subsequent Gαq-GPCR-induced signalling cascades. Right: iβARK2 with Lck allows it to localize to the plasma membrane, thus being more proximal to and binding Gαq subunits more efficiently. b, Top: AAV2/5 constructs for astrocytic iβARK or iβARKmut. Bottom: AAV2/5 constructs for iβARK2 and iβARK2mut. c, Schematic for Ca2+ imaging of iβARK-positive LA/B astrocytes. d, Representative images showing expression of iβARK and iβARK2 in LA/B. iβARK2 has improved expression near plasma membrane (iβARK: n = 5, iβARK2: n = 5 biological replicates). e, Representative pseudo-coloured images showing the high fluorescence of GCaMP6f in LA/B astrocytes (representative of n = 28 iβARK2mut and 13 iβARK2 astrocytes). f, dF/F traces of Ca2+ responses in a iβARK(D110A)- or iβARK-expressing astrocyte evoked by PE (phenylephrine), an agonist of Gαq-coupled α1A-adrenoceptor (coded by Adra1a) analyzed for soma and processes separately. g, Scatter graph showing the average Ca2+ responses to 0, 1, and 3 µM PE, where iβARK significantly reduced the PE-induced Ca2+ responses. iβARKmut: n = 29 cells, iβARK: n = 33 cells, iβARK2mut: n = 28 cells, iβARK2: n = 13 cells, 3-4 mice per group. Two-way ANOVA, Tukey’s multiple comparisons test. Data are mean ± SEM. **p < 0.01; ns, not significantly different. Sample sizes and replicates in Supplementary Table 1.
Extended Data Fig. 14 Additional assessments of the behavioural consequences of Adrb1-OE mice.
a, Graphs showing no alteration in locomotion of Adrb1-OE mice during the open field test. Ctrl: n = 13 mice, Adrb1-OE: n = 14 mice. Two-way ANOVA and Šídák’s multiple comparisons test were used. b, Graphs showing no alteration in time spent in center zone of Adrb1-OE mice during the open field test. Ctrl: n = 13 mice, Adrb1-OE: n = 14 mice. Mann-Whitney test. c-k, Minute-by-minute freezing behaviour of Ctrl and Adrb1-OE mice across conditioning and recall sessions. After three-shock FC (c), freezing was measured during the 1st (d) and 2nd FR (e). After one-shock FC (f), freezing was measured during the 1st (g) and 2nd FR (h), with Adrb1-OE mice showing significantly more freezing. In a separate three-shock FC cohort (i), freezing was measured during the 1st FR (j) and CtxB exposure (k), where Adrb1-OE mice also showed increased freezing. n = 5–7 mice per group (c–e), 13 per group (f–h), and 13–14 per group (i–k). Two-way ANOVA with Šídák’s test. Data are mean ± SEM. *p < 0.05; ns, not significantly different. Sample sizes and replicates in Supplementary Table 1.
Supplementary information
Supplementary Table 1
Details of raw replicates, statistical analyses, n numbers and relevant parameters, related to all figures.
Supplementary Table 2
Fos-mNG astrocyte counts in 677 brain regions in NoFC, FC, FC + novelty, NoFR and FR groups.
Supplementary Table 3
Single-cell transcriptomic data including the list of top 20 genes that differentiate astrocyte clusters and the differentially expressed genes estimated between Adrb1-positive and negative astrocytes.
Supplementary Table 4
AAV constructs and Addgene IDs.
Supplementary Video 1
STPT images, astrocyte marking, and brain region segmentation in the NoFC group.
Supplementary Video 2
STPT images, astrocyte marking, and brain region segmentation in the FR group.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dewa, Ki., Kaseda, K., Kuwahara, A. et al. The astrocytic ensemble acts as a multiday trace to stabilize memory. Nature 648, 146–156 (2025). https://doi.org/10.1038/s41586-025-09619-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09619-2
This article is cited by
-
Astroengrams: rethinking the cellular substrate for memory
Nature Reviews Neuroscience (2026)
-
Beyond neurons: astrocytic ensembles stabilize memories after recall
Signal Transduction and Targeted Therapy (2026)
-
How emotional memories are engraved on the brain, with surprising helper cells
Nature (2025)
-
The ‘silent’ brain cells that shape our behaviour, memory and health
Nature (2025)