Abstract
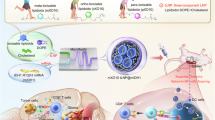
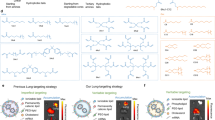
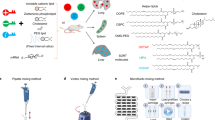
Organ-selective delivery of messenger RNA (mRNA) is critical for fulfilling the therapeutic potential of mRNA-based gene and protein replacement technologies. Despite clinical advances in the hepatic delivery of mRNA using lipid nanoparticles (LNPs), current strategies for extrahepatic-organ-selective mRNA delivery still have limitations. Here we report a peptide-encoded organ-selective targeting (POST) method for the delivery of mRNA to extrahepatic organs after systemic administration, which is based on the modular tuning of LNPs through surface engineering with specific amino acid sequences (POST codes). Molecular dynamics simulations and in vitro and in vivo testing show that the organ-selective targeting of POST results from the specific protein corona of the peptide-decorated LNPs, which is established from the mechanical optimization of the binding affinities between peptides with a particular sequence and plasma proteins. This approach can be used for the organ-selective delivery of different ribonucleic acids and multiple gene editing machinery. Overall, the POST platform creates a modular repertoire for LNP surface engineering for directing organ tropism, broadening the scope and versatility of organ-selective delivery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this work are available within the Article and its Supplementary Information. Nuclear magnetic resonance data shown in this study have been deposited in BMRbig (accession code bmrbig89). Proteomics data shown in this study have been deposited in iProX (accession code PXD043932). The scRNA-seq matrix data of this study have been deposited in the CNGB Sequence Archive (CNSA) of the China National GeneBank DataBase (CNGBdb) with accession number CNP0006703. Source data are provided with this paper.
Code availability
All custom code or mathematical algorithms used in this study are available from the corresponding authors on request. Molecular simulations were performed using the open-source software NAMD 2.14 package, and the Python v3.0 code used for the analysis and other post-processing tools used for this study are available from the corresponding authors upon request. The code for bioinformatics analysis is available via GitHub at https://github.com/shushu-Lee/ChangT-NM.
References
Barbier, A. J., Jiang, A. Y., Zhang, P., Wooster, R. & Anderson, D. G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 40, 840–854 (2022).
Miao, L. et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019).
Liu, C. et al. mRNA-based cancer therapeutics. Nat. Rev. Cancer 23, 526–543 (2023).
Rohner, E., Yang, R., Foo, K. S., Goedel, A. & Chien, K. R. Unlocking the promise of mRNA therapeutics. Nat. Biotechnol. 40, 1586–1600 (2022).
Chabanovska, O., Galow, A. M., David, R. & Lemcke, H. mRNA—a game changer in regenerative medicine, cell-based therapy and reprogramming strategies. Adv. Drug Deliv. Rev. 179, 114002 (2021).
Hou, X. C., Zaks, T., Langer, R. & Dong, Y. Z. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 7, 65–65 (2022).
Huang, X. G. et al. Nanotechnology-based strategies against SARS-CoV-2 variants. Nat. Nanotechnol. 17, 1027–1037 (2022).
Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 14, 1084–1087 (2019).
Dahlman, J. E. & Loughrey, D. Non-liver mRNA delivery. Acc. Chem. Res. 55, 13–23 (2022).
Dilliard, S. A. & Siegwart, D. J. Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs. Nat. Rev. Mater. 8, 282–300 (2023).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 15, 313–316 (2020).
Qiu, M. et al. Lung-selective mRNA delivery of synthetic lipid nanoparticles for the treatment of pulmonary lymphangioleiomyomatosis. Proc. Natl Acad. Sci. USA 119, e2116271119 (2022).
Riley, R. S. et al. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci. Adv. 7, eaba10 (2021).
Mitchell, M. J. et al. Ionizable lipid nanoparticles for in vivo mRNA Delivery to the placenta during pregnancy. J. Am. Chem. Soc. 145, 4691–4706 (2023).
Liu, S. et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 20, 701–710 (2021).
Dilliard, S. A., Cheng, Q. & Siegwart, D. J. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl Acad. Sci. USA 118, e2109256118 (2021).
Poon, W., Kingston, B. R., Ouyang, B., Ngo, W. & Chan, W. C. W. A framework for designing delivery systems. Nat. Nanotechnol. 15, 819–829 (2020).
Kedmi, R. et al. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 13, 214–219 (2018).
Levin, A. et al. Biomimetic peptide self-assembly for functional materials. Nat. Rev. Chem. 4, 615–634 (2020).
Huang, J. J. et al. Identification of potent antimicrobial peptides via a machine-learning pipeline that mines the entire space of peptide sequences. Nat. Biomed. Eng. 7, 797–810 (2023).
Xie, J. et al. Cell-penetrating peptides in diagnosis and treatment of human diseases: from preclinical research to clinical application. Front. Pharmacol. 11, 697 (2020).
Fosgerau, K. & Hoffmann, T. Peptide therapeutics: current status and future directions. Drug Discov. Today 20, 122–128 (2015).
Foss, D. V. et al. Peptide-mediated delivery of CRISPR enzymes for the efficient editing of primary human lymphocytes. Nat. Biomed. Eng. 7, 647–660 (2023).
Zhang, Z. et al. Efficient engineering of human and mouse primary cells using peptide-assisted genome editing. Nat. Biotechnol. 42, 305–315 (2023).
Rhym, L. H., Manan, R. S., Koller, A., Stephanie, G. & Anderson, D. G. Peptide-encoding mRNA barcodes for the high-throughput in vivo screening of libraries of lipid nanoparticles for mRNA delivery. Nat. Biomed. Eng. 7, 901–910 (2023).
Rossler, S. L., Grob, N. M., Buchwald, S. L. & Pentelute, B. L. Abiotic peptides as carriers of information for the encoding of small-molecule library synthesis. Science 379, eadf1354 (2023).
Huang, W. M. et al. Maleimide-thiol adducts stabilized through stretching. Nat. Chem. 11, 310–319 (2019).
Stroock, A. et al. Chaotic mixer for microchannels. Science 295, 647–651 (2002).
Li, S. D. & Huang, L. Stealth nanoparticles: high density but sheddable PEG is a key for tumor targeting. J. Control. Release 145, 178–181 (2010).
Wang, S. et al. The role of protein corona on nanodrugs for organ-targeting and its prospects of application. J. Control. Release 360, 15–43 (2023).
Oh, J. Y. et al. Cloaking nanoparticles with protein corona shield for targeted drug delivery. Nat. Commun. 9, 4548 (2018).
Bashiri, G. et al. Nanoparticle protein corona: from structure and function to therapeutic targeting. Lab Chip 23, 1432–1466 (2023).
Ngo, W. et al. Identifying cell receptors for the nanoparticle protein corona using genome screens. Nat. Chem. Biol. 18, 1023–1031 (2022).
Singh, B., Fu, C. & Bhattacharya, J. Vascular expression of the αvβ3-integrin in lung and other organs. Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L217–L226 (2000).
Teoh, C., Tan, S. & Tran, T. Integrins as therapeutic targets for respiratory diseases. Curr. Mol. Med. 15, 714–734 (2015).
Shanker, V. R., Bruun, T. U. J., Hie, B. L. & Kim, P. S. Unsupervised evolution of protein and antibody complexes with a structure-informed language model. Science 385, 46–53 (2024).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Schoenmaker, L. et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 601, 120586 (2021).
Huang, X. G. et al. The landscape of mRNA nanomedicine. Nat. Med. 28, 2273–2287 (2022).
Zhang, Y. B., Sun, C. Z., Wang, C., Jankovic, K. E. & Dong, Y. Z. Lipids and lipid derivatives for RNA delivery. Chem. Rev. 121, 12181–12277 (2021).
Hajj, K. A. et al. A potent branched-tail lipid nanoparticle enables multiplexed mRNA delivery and gene editing in vivo. Nano Lett. 20, 5167–5175 (2020).
Roberts, T. C., Langer, R. & Wood, M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694 (2020).
Paunovska, K., Loughrey, D. & Dahlman, J. E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 23, 265–280 (2022).
Yang, L. et al. Efficient delivery of antisense oligonucleotides using bioreducible lipid nanoparticles in vitro and in vivo. Mol. Ther. Nucleic Acids 19, 1357–1367 (2020).
Liang, X. H. et al. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 34, 875–880 (2016).
Katti, A., Diaz, B. J., Caragine, C. M., Sanjana, N. E. & Dow, L. E. CRISPR in cancer biology and therapy. Nat. Rev. Cancer 22, 259–279 (2022).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Kim, G. B. et al. Rapid generation of somatic mouse mosaics with locus-specific, stably integrated transgenic elements. Cell 179, 251–267 (2019).
Finn, J. D. et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Wang, S. et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221 (2003).
Rothgangl, T. et al. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat. Biotechnol. 39, 949–957 (2021).
Donati, Y., Blaskovic, S., Ruchonnet-Métrailler, I., Lascano Maillard, J. & Barazzone-Argiroffo, C. Simultaneous isolation of endothelial and alveolar epithelial type I and type II cells during mouse lung development in the absence of a transgenic reporter. Am. J. Physiol. Lung Cell. Mol. Physiol. 318, L619–L630 (2020).
Toulmin, S. A. et al. Type II alveolar cell MHCII improves respiratory viral disease outcomes while exhibiting limited antigen presentation. Nat. Commun. 12, 3993 (2021).
Olesch, C. et al. Picturing of the lung tumor cellular composition by multispectral flow cytometry. Front. Immunol. 13, 827719 (2022).
Sun, Y. et al. In vivo editing of lung stem cells for durable gene correction in mice. Science 384, 1196–1202 (2024).
Modulation of CD19 surface expression in B cell acute lymphoblastic leukemia. Nat. Immunol. 23, 1410–1411 (2022).
Swingle, K. L. et al. Placenta-tropic VEGF mRNA lipid nanoparticles ameliorate murine pre-eclampsia. Nature https://doi.org/10.1038/s41586-024-08291-2 (2024).
Lian, X. et al. Bone-marrow-homing lipid nanoparticles for genome editing in diseased and malignant haematopoietic stem cells. Nat. Nanotechnol. 19, 1409–1417 (2024).
Adolfsson, J. et al. Upregulation of Flt3 expression within the bone marrow Lin−Sca1+c-kit+ stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 15, 659–669 (2001).
Engelhard, S. et al. Endomucin marks quiescent long-term multi-lineage repopulating hematopoietic stem cells and is essential for their transendothelial migration. Cell Rep. 43, 114475 (2024).
Mayle, A., Luo, M., Jeong, M. & Goodell, M. A. Flow cytometry analysis of murine hematopoietic stem cells. Cytom. Part A 83A, 27–37 (2013).
Rodeheffer, M. S., Birsoy, K. & Friedman, J. M. Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249 (2008).
Lee, Y.-H., Petkova, A. P., Mottillo, E. P. & Granneman, J. G. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480–491 (2012).
Garritson, J. D. et al. BMPER is a marker of adipose progenitors and adipocytes and a positive modulator of adipogenesis. Commun. Biol. 6, 638 (2023).
Zomer, H. D. & Reddi, P. P. Characterization of rodent Sertoli cell primary cultures. Mol. Reprod. Dev. 87, 857–870 (2020).
Song, Y., Zhang, X., Desmarais, J. A. & Nagano, M. Postnatal development of mouse spermatogonial stem cells as determined by immunophenotype, regenerative capacity, and long-term culture-initiating ability: a model for practical applications. Sci. Rep. 14, 2299 (2024).
Omo-Lamai, S. et al. Physicochemical targeting of lipid nanoparticles to the lungs induces clotting: mechanisms and solutions. Adv. Mater. 36, 2312026 (2023).
Witten, J. et al. Artificial intelligence-guided design of lipid nanoparticles for pulmonary gene therapy. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02490-y (2024).
Li, B. et al. Accelerating ionizable lipid discovery for mRNA delivery using machine learning and combinatorial chemistry. Nat. Mater. 23, 1002–1008 (2024).
Dobrowolski, C. et al. Nanoparticle single-cell multiomic readouts reveal that cell heterogeneity influences lipid nanoparticle-mediated messenger RNA delivery. Nat. Nanotechnol. 17, 871–879 (2022).
Phillips, J. C. et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Sali, A. & Blundell, T. L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Abdin, O., Nim, S., Wen, H. & Kim, P. M. PepNN: a deep attention model for the identification of peptide binding sites. Commun. Biol. 5, 503 (2022).
Kozlovskii, I. & Popov, P. Protein-peptide binding site detection using 3D convolutional neural networks. J. Chem. Inf. Model. 61, 3814–3823 (2021).
Valdés-Tresanco, M. S., Valdés-Tresanco, M. E., Valiente, P. A. & Moreno, E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 17, 6281–6291 (2021).
Miller, B. R. III et al. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321 (2012).
Acknowledgements
Y.S.’s work is supported by the National Key Research and Development Program of China (2021YFA0719301), the National Natural Science Foundation of China (grant numbers U21A20203, 12102229 and 11921002), Tsinghua University Dushi Program, the Overseas High-level Scholar Introduction Program, and the Tsinghua University Startup Funding. S.J. thanks the National Natural Science Foundation of China (grant number 32400944) and the Postdoctoral Science Foundation of China (grant number 2023M740152). Z.Q. thanks the National Science Foundation grant (CMMI-2145392) for supporting this work. X.L. acknowledges support from the National Natural Science Foundation of China (grant number 22175188) and start-up funding from the Institute of Chemistry, Chinese Academy of Sciences. L.M. thanks the Beijing Natural Science Foundation (Z220022) for financial support. We thank X. Shao, J. Ji, P. Jiao and the Core Facility of the Center of Biomedical Analysis, Tsinghua University, for assistance with the flow cytometry experiments. For this work, we used the resources of the Center of High-Performance Computing of Tsinghua University.
Author information
Authors and Affiliations
Contributions
T.C. and Y.S. conceived the project and designed the experiments. T.C., Y.Z., S.J., J.B., J.G., Yue Wang, Yiting Wang, H.L., J.L., L.N., X.C., Shuai Liu, H.Z., W.P., F.L., Shiyi Liu, W.W., G.W., L.W., L.M., X.L. and Z.D. performed the experiments. M.J. and Z.Q. developed the full-atom molecular model and performed the SMD simulation. Z.Z. and H.G. developed the AI-driven framework for POST code design. S.L. and B.B. performed the bioinformatics analysis. T.C. and Y.S. analysed the data and wrote the manuscript. Y.S. supervised the project. All authors contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Y.S. and T.C. have filed a patent in the China National Intellectual Property Administration (patent no. 202310977908.7) based on the POST platform developed in this study. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Materials thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Evaluation of the performance of POST-LNPs.
(a) Ex vivo organ luminescence images showing the expression level of luciferase in mouse liver, lung, spleen, heart, and kidney, respectively, at 6, 8, and 12 h. n = 3 biologically independent animals for each condition. (b) Consistent performance of organ-selectivity by POST-LNPs after storage at 4°C for one month. Ex vivo organ luminescence images indicate expected organ selectivity from each kind of POST-LNPs. n = 3 biologically independent animals for each condition. (c) Representative fluorescence images showing the localization of Cy5-labeled mRNA and Cy7-labeled peptides. Scale bar: 10 μm. Similar results were seen in n = 3 animals for each experimental group.
Extended Data Fig. 2 SMD simulation results for polyR-Vtn assembly.
(a, f, k, p) The diagrams of force-extension curves of SMD simulations for polyR-Vtn assembly with 4 R (a), 5 R (f), 7 R (k), and 8 R (p), respectively. The black curve in each of the plots gives the curve of an exemplary simulation, while the gray shade gives the range of the average force ± standard deviation given by n = 5 SMD simulations under the same conditions. (b-e, g-j, l-o, q-t) The snapshots of trajectories in exemplary SMD simulations for polyR-Vtn assembly with 4 R (b-e), 5 R (g-j), 7 R (l-o), and 8 R (q-t), respectively. The points at which the snapshots are taken are marked in corresponding force-extension diagrams of SMD simulations.
Extended Data Fig. 3 Vtn adsorption selectively facilitates cellular uptake of 6R-LNP in vitro.
(a) The schematic of the experiment procedures. (b) Representative confocal images showing cellular uptake of LNPs that are labeled with Cy5-DSPE. Scale bar: 50 μm. (c) Quantification of cells containing Cy5-LNPs under indicated conditions. n = 5 confocal images analyzed for each condition. n = 3 independent experiments for each condition. (d) Quantification of luciferase expression following delivery of luciferase mRNA using LNPs with or without adsorption of Vtn protein. n = 3 independent experiments for each condition. One-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test were used for statistical analysis. All data were plotted as the mean ± s.d. Schematic in a created using BioRender.com.
Extended Data Fig. 4 Depletion of Vtn from serum reduces cellular uptake of 6R-LNP.
(a) The schematic of the experiment procedures. (b) Quantitative analysis of Vtn protein concentration in untreated serum and Vtn-depleted serum, respectively. n = 3 biologically independent samples for each condition. (c) Quantitative analysis of Vtn adsorbed onto 6R-LNP after incubation in untreated serum or Vtn-depleted serum, respectively. n = 3 biologically independent samples for each condition. (d) Representative confocal images showing cellular uptake of 6R-LNP labeled by Cy5-DSPE, after pre-incubation of 6R-LNP in untreated or Vtn-depleted serum. U-87 MG and A498 cell lines with high αvβ3 integrin-expressing were used. n = 5 independent experiments. Scale bar: 50 μm. (e) Quantification of cells that uptake Cy5-LNPs under indicated conditions. n = 5 confocal images analyzed for each condition. n = 5 independent experiments. (f) Quantification of Luc expression in cells following delivery of Luc mRNA using 6R-LNP pre-incubated in untreated serum or Vtn-depleted serum, respectively. n = 3 independent experiments. Unpaired, two-sided student’s t-test was used for statistical analysis between two groups. ns: p > 0.05. All data were plotted as the mean ± s.d. Schematic in a created using BioRender.com.
Extended Data Fig. 5 Blocking Vtn receptors reduces cellular uptake of 6R-LNP.
(a) The schematic of the experiment procedures that pre-incubates cells with freely dissolved Vtn proteins. (b) Representative confocal images showing cellular uptake of Vtn-coated 6R-LNP (labeled by Cy5-DSPE). n = 5 independent experiments. Scale bar: 50 μm. (c) Quantification of cells that uptake Cy5-LNPs under indicated conditions. n = 5 confocal images analyzed for each condition. n = 5 independent experiments. (d) Quantification of Luc expression in cells following delivery of Luc mRNA using Vtn-coated 6R-LNP. n = 3 independent experiments. (e) The schematic of the experiment procedures that pre-incubate cells with Cilengitide. (f) Representative confocal images showing cellular uptake of Vtn-coated 6R-LNP (labeled by Cy5-DSPE) pre-incubated with small molecule inhibitor Cilengitide. n = 5 independent experiments. Scale bar: 50 μm. (g) Quantification of cells that uptake Cy5-LNPs under indicated conditions. n = 5 confocal images analyzed for each condition. n = 5 independent experiments. (h) Quantification of Luc expression in cells following delivery of Luc mRNA using Vtn-coated 6R-LNP pre-incubated with Cilengitide. n = 3 independent experiments. Unpaired, two-sided student’s t-test was used for statistical analysis between two groups. ns: p > 0.05. All data were plotted as the mean ± s.d. Schematics in a and e created using BioRender.com.
Extended Data Fig. 6 Modulation of Itgav/Itgb3 expression changes cellular uptake of 6R-LNP.
(a & b) qPCR analysis of the knockdown of Itgav and/or Itgb3 expression in U87-MG cell line (a) and A498 cell line (b), respectively. n = 3 biologically independent samples for each condition. (c) Representative confocal images showing the cellular uptake of Vtn-coated 6R-LNP (labeled by Cy5-DSPE) in U87-MG (upper) and A498 (lower) cell lines under indicated conditions. n = 5 independent experiments. Scale bar: 50 μm. (d) Quantification of cells that uptake Cy5-LNPs in U87-MG (left) and A498 (right) cell lines under indicated conditions. n = 5 confocal images analyzed for each condition. n = 5 independent experiments. (e) Quantification of Luc expression in U87-MG (left) and A498 (right) cells, respectively, following delivery of Luc mRNA using Vtn-coated 6R-LNP under indicated conditions. n = 3 independent experiments. (f) qPCR analysis of the overexpression of Itgav and/or Itgb3 in 3T3 cell line. n = 3 biologically independent samples for each condition. (g) Representative confocal images showing the cellular uptake of Vtn-coated 6R-LNP (labeled with Cy5-DSPE) under indicated conditions. n = 5 independent experiments. Scale bar: 50 μm. (h) Quantification of cells that uptake Cy5-LNPs under indicated conditions. n = 5 confocal images analyzed for each condition. n = 5 independent experiments. (i) Quantification of Luc expression in cells following delivery of Luc mRNA using Vtn-coated 6R-LNP under indicated conditions. n = 3 independent experiments. One-way analysis of variance (ANOVA) and Dunnett’s multiple comparisonstest were used for statistical analysis. ns: p > 0.05. All data were plotted as the mean ± s.d.
Extended Data Fig. 7 Knockdown of Itgav or Itgb3 reduces the lung delivery of 6R-LNP in vivo.
(a) Percentage of VtnR+ cells in mouse heart, liver, spleen, lung, and kidney, respectively. n = 3 animals for each condition. (b) Experiment design of EGFP mRNA delivery in C57/BL6 wild-type mice using 6R-LNP. (c) Quantification of VtnR+ and VtnR- cell populations, respectively, within EGFP+ cells in the lung after EGFP mRNA delivery by 6R-LNP. n = 3 animals for each condition. (d) Experiment design for the knockdown of Itgav and/or Itgb3 in the lung of C57/BL6 wild-type mice using CRISPR/Cas9 system with 6R-LNP. (e) qPCR analysis of the knockdown of Itgav and/or Itgb3 expression in the lung under indicated conditions. n = 3 animals for each condition. (f) Flow cytometry analysis of the knockdown of Itgav and/or Itgb3 receptor expression in the lung under indicated conditions. n = 3 animals for each condition. (g) Quantification of Itgav+Itgb3+ (that is, VtnR + ) cell population in the lung after gene editing under indicated conditions. n = 3 animals for each condition. (h) The workflow of Luc mRNA delivery using 6R-LNP. (i) Ex vivo organ luminescence images showing the expression level of luciferase in the liver, lung, spleen, heart, and kidney, respectively, of mice with lung-edited knockdown of Itgav and/or Itgb3 as indicated. n = 3 animals for each condition. (j) Quantification of luminescence of the lung of gene-edited mice as indicated, after Luc mRNA delivery by 6R-LNP. n = 3 animals for each condition. One-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test were used for statistical analysis. ns: p > 0.05. All data were plotted as the mean ± s.d. Schematics in b, d and h created using BioRender.com.
Extended Data Fig. 8 Ectopic expression of Itgav and Itgb3 in the liver could re-direct the delivery of 6R-LNP in vivo.
(a) Experiment design of the hepatic overexpression of Itgav and/or Itgb3 in mouse liver using 3R-LNP. (b) qPCR analysis of the overexpression of Itgav and/or Itgb3 in the liver under indicated conditions. n = 3 animals for each condition. (c) Flow cytometry analysis of the overexpression of Itgav and/or Itgb3 in the liver under indicated conditions. n = 3 animals for each condition. (d) Quantification of Itgav+Itgb3+ (that is, VtnR + ) cell population in liver after overexpression of Itgav and/or Itgb3 as indicated. n = 3 animals for each condition. (e) The workflow of Luc mRNA delivery using 6R-LNP. (f) Ex vivo organ luminescence images showing the expression level of luciferase in the liver, lung, spleen, heart, and kidney, respectively, of mice with ectopic hepatic overexpression of Itgav and/or Itgb3 as indicated. n = 3 animals for each condition. (g) Quantification of luminescence of the liver (left) and lung (right), respectively, of mice with ectopic hepatic overexpression of Itgav and/or Itgb3 as indicated, after Luc mRNA delivery by 6R-LNP. n = 3 animals for each condition. (h) Stacked bar chart showing the relative expression of luciferase in the liver, lung, and spleen, respectively, of C57/BL6 wild-type mice with ectopic hepatic overexpression of Itgav and/or Itgb3 as indicated, after Luc mRNA delivery by 6R-LNP. n = 3 animals for each condition. One-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test were used for statistical analysis. ns: p > 0.05. All data were plotted as the mean ± s.d. Schematics in a and e created using BioRender.com.
Extended Data Fig. 9 POST-LNPs enable organ-selective delivery to other extrahepatic organs.
(a) Ex vivo organ imaging results showing the fluorescence level of EGFP in placenta, liver, lung, spleen, heart, and kidney of pregnant mice (E16.5). (b) Quantification of absolute radiance of EGFP in the placenta, liver, lung, and spleen under indicated conditions. (c) Stacked bar chart showing the percentage of EGFP radiance change in the placenta, liver, lung, and spleen. (d) Ex vivo organ imaging results showing the fluorescence level of EGFP in bone marrow, liver, lung, spleen, heart, and kidney of mice. (e) Quantification of absolute radiance of EGFP in the bone marrow, liver, lung, and spleen. (f) Stacked bar chart showing the percentage of EGFP radiance change in the bone marrow, liver, lung, and spleen. (g) Ex vivo organ imaging results showing the fluorescence level of EGFP in adipose tissue, liver, lung, spleen, heart, and kidney of mice. (h) Quantification of absolute radiance of EGFP in the adipose tissue, liver, lung, and spleen. (i) Stacked bar chart showing the percentage of EGFP radiance change in the adipose tissue, liver, lung, and spleen. (j) Ex vivo organ imaging results showing the fluorescence level of EGFP in testis, liver, lung, spleen, heart, and kidney of mice. (k) Quantification of absolute radiance of EGFP in the testis, liver, lung, and spleen. (l) Stacked bar chart showing the percentage of EGFP radiance change in the testis, liver, lung, and spleen. One-way analysis of variance (ANOVA) and Dunnett’s multiple comparisonstest were used for statistical analysis. ns: p > 0.05. All data are from n = 3 animals for each condition. All data were plotted as the mean ± s.d. Schematics in a, d, g and j created using BioRender.com.
Extended Data Fig. 10 POST-LNP could deliver mRNA into parenchymal cells in target organs.
(a) UMAP plotting showing cell identity annotations in the lung. Cell types are colored according to the dot representative of each cluster. (b) Featured UMAP plotting showing single-cell distribution of Luc mRNA in the lung after delivery by lung-specific 6R-LNP. (c) UMAP plotting showing cell identity annotations in the spleen. (d) Featured UMAP plotting showing single-cell distribution of Luc mRNA in the spleen after delivery by spleen-specific 6D-LNP. (e) Percentage of cells with exogenously delivered Luc mRNA by 6R-LNP in the lung. (f) Percentage of cells with exogenously delivered Luc mRNA by 6D-LNP in the spleen. n = 4 animals for each condition. (g) Quantification of the percentage of EGFP+ cells in the fibroblasts, alveolar type I cells, alveolar type II cells, and basal progenitor cells, respectively. (h) Quantification of the percentage of EGFP+ cells in the B cells, macrophage, and T cells respectively. (i) Quantification of the percentage of EGFP+ cells in the trophoblast cells. (j) Quantification of the percentage of EGFP+ cells in the long-term hematopoietic stem cells (LT-HSC) and multi-lineage reconstituting CD150 + LT-HSC. (k) Quantification of the percentage of EGFP+ cells in the adipocyte progenitor cells. (l) Quantification of the percentage of EGFP+ cells in the spermatogonia cells. n = 3 animals for each condition. P-values were calculated using unpaired Student’s t-test. All data were plotted as the mean ± s.d.
Supplementary information
Supplementary Information
Supplementary Figs. 1–42 and Tables 1–19.
Supplementary Video 1
SMD simulation trajectory showing the failure of 6R-Vtn assembly under externally applied tensile force. The direction of the force is illustrated by the red cone.
Supplementary Data 1
Source data for Supplementary Figures 13, 19, 21, 23, 25, 26, 27, 29, 31 and 32.
Supplementary Data 2
Coding sequences of the mRNA used in this manuscript.
Source data
Source Data Fig. 1–6 and Source Data Extended Data Fig. 2–10
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chang, T., Zheng, Y., Jiang, M. et al. Peptide codes for organ-selective mRNA delivery. Nat. Mater. 25, 146–159 (2026). https://doi.org/10.1038/s41563-025-02331-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41563-025-02331-6