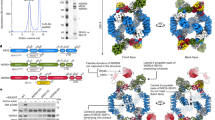
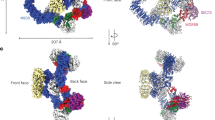
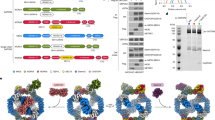
Abstract
The human KICSTOR complex, comprising KPTN, ITFG2, C12orf66 and the scaffolding protein SZT2, anchors the mTORC1 inhibitor GATOR1 to lysosomes. Mutations affecting KICSTOR subunits are associated with severe neurodevelopmental and epileptic disorders. Loss of KICSTOR mimics GATOR1 inactivation, resulting in constitutive mTORC1 activation, highlighting its critical role in nutrient sensing. Here, we used cryo-electron microscopy and computational modeling to determine the architectures of KICSTOR and the GATOR1–KICSTOR supercomplex. We show that SZT2 forms a crescent-shaped scaffold with repetitive tandem units, binding the ITFG2–KPTN heterodimer and C12orf66 at its C terminus. Structural and biochemical analyses revealed that GATOR1 binds the SZT2 N-terminal domain through NPRL3; disruption of this interaction hyperactivates mTORC1 and mislocalizes TFE3 independently of nutrient status. We further demonstrate the membrane-binding ability of KICSTOR, with SZT2 and C12orf66 preferentially interacting with negatively charged lipids—a requirement for lysosomal localization. These findings identify how KICSTOR positions GATOR1 on lysosomes to regulate nutrient-dependent mTORC1 signaling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The cryo-EM density maps were deposited to the Electron Microscopy Data Bank under accession codes EMD-64799 (KICSTOR in state 1), EMD-64902 (KICSTOR in state 2), EMD-64831 (KICSTOR CCC in state 3), EMD-64827 (KICSTOR CCC in state 4), EMD-64877 (KICSTOR CCC in state 5) and EMD-64665 (KICSTOR–GATOR1 complex), with corresponding atomic coordinates available in the PDB under accession numbers 9V6E (KICSTOR in state 1), 9VAN (KICSTOR in state 2), 9V86 (KICSTOR CCC in state 3), 9V80 (KICSTOR CCC in state 4), 9V9N (KICSTOR CCC in state 5) and 9V0J (KICSTOR–GATOR1), respectively. MS proteomics data are accessible from ProteomeXchange (PXD060391) through the PRIDE repository. Source data are provided with this paper.
References
Lim, C. Y. & Zoncu, R. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 214, 653–664 (2016).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017).
Schurmann, A., Brauers, A., Massmann, S., Becker, W. & Joost, H. G. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the RAS-related GTPases. J. Biol. Chem. 270, 28982–28988 (1995).
Sekiguchi, T., Hirose, E., Nakashima, N., Ii, M. & Nishimoto, T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 276, 7246–7257 (2001).
Inoki, K., Li, Y., Xu, T. & Guan, K. L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Gene Dev. 17, 1829–1834 (2003).
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008).
Sancak, Y. et al. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Anandapadamanaban, M., et al. Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210 (2019).
Rogala, K. B. et al. Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366, 468–475 (2019).
Bar-Peled, L. et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013).
Panchaud, N., Peli-Gulli, M. P. & De Virgilio, C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6, ra42 (2013).
Valenstein, M. L. et al. Structure of the nutrient-sensing hub GATOR2. Nature 607, 610–616 (2022).
Su, M. Y., et al. Cryo-EM structures of amino acid sensors bound to the human GATOR2 complex. Cell Rep. 44, 116088 (2025).
Peng, M., Yin, N. & Li, M. O. SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437 (2017).
Wolfson, R. L. et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017).
Baple, E. L. et al. Mutations in KPTN cause macrocephaly, neurodevelopmental delay, and seizures. Am. J. Hum. Genet 94, 87–94 (2014).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Holm, L., Laiho, A., Toronen, P. & Salgado, M. DALI shines a light on remote homologs: one hundred discoveries. Protein Sci. 32, e4519 (2023).
Wu, H. et al. Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism. J. Biol. Chem. 283, 16135–16146 (2008).
Sette, P. et al. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure 19, 1485–1495 (2011).
Ramamurthy, V. et al. Structures of adnectin/protein complexes reveal an expanded binding footprint. Structure 20, 259–269 (2012).
Su, M. Y. et al. Hybrid structure of the RagA/C–Ragulator mTORC1 activation complex. Mol. Cell 68, 835–846 e833 (2017).
Egri, S. B. et al. Cryo-EM structures of the human GATOR1–Rag–Ragulator complex reveal a spatial-constraint regulated GAP mechanism. Mol. Cell 82, 1836–1849 (2022).
Shen, K. et al. Architecture of the human GATOR1 and GATOR1–Rag GTPases complexes. Nature 556, 64–69 (2018).
Qu, A. & Leahy, D. J. Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, αLβ2) integrin. Proc. Natl Acad. Sci. USA 92, 10277–10281 (1995).
Liu, M. et al. Structure of the DDB1–AMBRA1 E3 ligase receptor complex linked to cell cycle regulation. Nat. Commun. 14, 7631 (2023).
Merkley, E. D. et al. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine–lysine distances. Protein Sci. 23, 747–759 (2014).
Tafur, L. et al. Cryo-EM structure of the SEA complex. Nature 611, 399–404 (2022).
Wolfson, R. L. & Sabatini, D. M. The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 26, 301–309 (2017).
Kim, W., Underwood, R. S., Greenwald, I. & Shaye, D. D. OrthoList 2: a new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetics 210, 445–461 (2018).
Cattelani, C. et al. The SZT2 interactome unravels new functions of the KICSTOR complex. Cells 10, 2711 (2021).
Tan, S. M. The leucocyte β2 (CD18) integrins: the structure, functional regulation and signalling properties. Biosci. Rep. 32, 241–269 (2012).
Sbrissa, D., Ikonomov, O. C., Filios, C., Delvecchio, K. & Shisheva, A. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am. J. Physiol. Cell Physiol. 303, C436–C446 (2012).
Nakatsu, F. & Kawasaki, A. Functions of oxysterol-binding proteins at membrane contact sites and their control by phosphoinositide metabolism. Front. Cell Dev. Biol. 9, 664788 (2021).
Wallroth, A. & Haucke, V. Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem. 293, 1526–1535 (2018).
Hasegawa, J., Strunk, B. S. & Weisman, L. S. PI5P and PI(3,5)P: minor, but essential phosphoinositides. Cell Struct. Funct. 42, 49–60 (2017).
Fitzian, K., et al. TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation. Mol. Cell 81, 2705–2721 (2021).
Ebner, M., et al. Nutrient-regulated control of lysosome function by signaling lipid conversion. Cell 186, 5328–5346 (2023).
Hong, Z. et al. PtdIns3P controls mTORC1 signaling through lysosomal positioning. J. Cell Biol. 216, 4217–4233 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Cianfrocco, M. A. COSMIC: a global resource for structure prediction and structural analysis. Biophys. J. 123, 334a–335a (2024).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Su, M. Y., Fromm, S. A., Remis, J., Toso, D. B. & Hurley, J. H. Structural basis for the ARF GAP activity and specificity of the C9orf72 complex. Nat. Commun. 12, 3786 (2021).
Bolte, S. & Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 53, D543–D553 (2024).
Acknowledgements
We are grateful to H. Rao and F. Rao for their valuable comments on the manuscript. M.-Y.S. is an investigator of SUSTech Institute for Biological Electron Microscopy. We thank the following facilities for their technical support: the Cryo-EM Center at SUSTech, the Cryo-EM Facility and the Advanced MS Facility of the Kobilka Institute of Innovative Drug Discovery at the Chinese University of Hong Kong (Shenzhen), Shenzhen Medical Academy of Research and Translation (SMART) for the assistance in MST data collection. This work was supported by funding from the Shenzhen Medical Research Fund (B2402014 to G.S. and B2502011 to M.-Y.S.), National Natural Science Foundation of China (32571412 to M.-Y.S., 32170779 to C.-Y.L.), the Natural Science Foundation of Guangdong Province (2024A1515011683 to M.-Y.S.), Shenzhen Science and Technology Program (20231120103446003 to M.-Y.S.), National High-level Talents Program (HJJH22-003 to C.-Y.L.), CUHK-Shenzhen University Development Fund (to G.S.), Medical Research Innovation Project G030410001 and start-up funding from SUSTech (to M.-Y.S.).
Author information
Authors and Affiliations
Contributions
M.-Y.S. conceptualized and supervised the project, designed the experimental strategy and performed the biochemical and structural studies. F.T. and H.Z. conducted the biochemical assays and cellular experiments. X.M. performed the cryo-EM sample screening, data collection and liposome-binding assays. S.C., L.W. and Z.F. executed the cell biological experiments. S.T. and S.W. provided technical assistance. G.S., C.-Y.L. and M.-Y.S. designed the study, interpreted the results and wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Volker Haucke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM structure determination of the full-length KICSTOR complex.
a, Representative motion-corrected cryo-EM micrograph (of 30,943 micrographs from 4 datasets) of the full-length human KICSTOR complex. b, Representative 2D class averages of the full-length KICSTOR complex. c, Workflow for cryo-EM data processing of full-length KICSTOR datasets. NU-refinement: non-uniform refinement. d, FSC plots for the consensus reconstruction of the full-length KICSTOR in state 1 are between two independently refined half-maps with no mask (blue), spherical mask (orange), loose mask (green), and tight mask (red). A cutoff value of 0.143 was used to estimate the resolution. e, Angular particle distribution for projection views of full-length KICSTOR in state 1, calculated using CryoSPARC. The heat map shows the number of particles for each viewing angle. f, Full-length KICSTOR in state 1 density map, color-coded according to local resolution estimation. g-i, Results of NU-refinement for the consensus reconstruction of full-length KICSTOR in state 2. (g) FSC curves, (h) angular distribution heatmap, (i) local resolution estimation, with the map color-coded to reflect the resolution. j, Masking and local refinement applied to the SZT2SZ1-SZ2-SZ3 and SZT2SZ7-C12orf66 regions of full-length KICSTOR in state 1. k, Masking and local refinement applied to the SZT2SZ1-SZ2-SZ3 or SZT2SZ7-C12orf66 regions of full-length KICSTOR in state 2.
Extended Data Fig. 2 Overview of cryo-EM processing of the KICSTOR C-terminal core complex (CCC).
a, Superose 6 size-exclusion chromatogram profile of the human KICSTOR CCC. The peak corresponding to KICSTOR CCC is marked with a black asterisk. Coomassie blue-stained SDS-PAGE analysis of the purified KICSTOR. MW, molecular weight. mAU, milliabsorbance. Data in a is representative of at least three independent experiments. b, Representative motion-corrected cryo-EM micrograph (of 8,829 micrographs from 1 dataset) of the KICSTOR C-terminal core complex. c, Representative 2D class averages of the KICSTOR CCC. d, Workflow for cryo-EM data processing of the KICSTOR CCC. NU-refinement: non-uniform refinement. e-g. Half-map FSC plots, angular particle distributions and local resolution estimation from consensus reconstructions of the KICSTOR CCC for state 3 (class 1), state 4 (class 2), and state 5 (class 3). h-i. Masking and local refinement applied to the SZT2SZ7-C12orf66 regions of the KICSTOR CCC for state 3 and state 5. j, Individual composite maps generated for different states of the KICSTOR CCC, prepared for subsequent model building.
Extended Data Fig. 3 Structural comparison of full-length KICSTOR and KICSTOR CCC complex.
a-b, Fitting of the fulllength KICSTOR models into cryo-EM reconstructions for state 1 and state 2. Zoomed-in views of the SZT2SZ7-C12orf66 region are shown on the right. c-e, Fitting of the KICSTOR CCC models into cryo-EM reconstructions for state 3, state 4 and state 5. Corresponding zoomed-in views of the SZT2SZ7-C12orf66 portions are displayed on the right. f, Superimposition of the structural models from all states to illustrate conformational differences on SZT2SZ7-C12orf66. Dotted boxes indicate regions that are magnified in panels g-i. RMSD values for the superimposed structures are shown in Supplementary Table 2. g, Structural comparison of the SZT2SZ7-C12orf66 region between state 1 and state 2. The arrow indicates the rotation observed in the subunits. h, Structural comparison of the SZT2SZ7-C12orf66 region between state 1 and state 3. i, Structural comparison of the SZT2SZ7-C12orf66 region between state 1 and state 5.
Extended Data Fig. 4 Model overlays of SZT2 and C12orf66.
a, Overlay of SZT2SZ1 (light gray) with spermine synthase (light sky blue, PDB 3C6K), SZT2NTD, Depdc5NTD-SABA domains (forest green, PDB 7T3B), and CD11a I domain (rosy brown, PDB 1LFA). RMSD values for the superimposed structures are shown in Supplementary Table 2. b, Overlay of C12orf66NTD (orange) with BroxBro1 (cornflower blue, PDB 3R9M), SRP68 (pink, PDB 7QWQ), C12orf66roadblock (orange), and Lamtor2 (purple, PDB 6B9X). RMSD values for the superimposed structures are shown in Supplementary Table 2. c, Close-up view of the SZT2-C12orf66 interface. W3428 of SZT2 is required to interact with C12orf66 L216 and F269, as shown by the pulldown assay in d-e. d, Pulldown experiment of wild-type or mutated SZT2 with KPTN, ITFG2, and C12orf66. TSF, twin-strep-flag. e, Pulldown experiment of wild-type or mutated C12orf66 with KPTN, ITFG2, and SZT2. TSF, twin-strep-flag. Data in d and e are representative of three independent experiments.
Extended Data Fig. 5 Cryo-EM structure determination of the human GATOR1-KICSTOR supercomplex.
a, EM image of the negatively stained GATOR1-KICSTOR supercomplex. The shape of the supercomplex can be recognized and circled in the raw image. Data in a is representative of at least three independent experiments. b, Representative motion-corrected cryo-EM micrograph (of 9,936 micrographs from 1 dataset) of the human GATOR1-KICSTOR complex. c, Representative 2D class averages for the GATOR1-KICSTOR supercomplex. d, Workflow for the cryo-EM data processing of human GATOR1-KICSTOR complex. NU-refinement: non-uniform refinement. e-g, FSC plots (e), angular distribution heatmap (f) and local resolution estimation (g) for the consensus reconstruction of GATOR1-KICSTOR supercomplex. h-j, Masking and local refinement results of Nprl3-SZT2. Half-map FSC plots (h), angular particle distributions (i) and local resolution estimation (j) from local refinement on Nprl3-SZT2 regions. k, Composite map of GATOR1-KICSTOR supercomplex was generated for subsequent model building.
Extended Data Fig. 6 Model versus map FSC curves.
a-b, Map versus model FSC curves with masked and unmasked were calculated for full-length KICSTOR in state 1 and state 2 against the consensus full map in Phenix. c-e, Map versus model FSC curves with masked and unmasked were calculated for KICSTOR CCC in state 3, state 4 and state 5 against the consensus full map in Phenix. f, FSC between the model and map for the KICSTOR-GATOR1 complex against the cryo-EM map. g, Representative cryo-EM densities fitted to the model.
Extended Data Fig. 7 Structural comparison of GATOR1 in GAP mode and KICSTOR-bound conformations.
a, Structural alignment of GATOR1 in this study with the reported GAP-mode conformation (PDB 7T3B). Depdc5, Nprl2, and Nprl3 are colored in forest green/light-sky blue, dark salmon/yellow, and thistle/yellow-green, respectively, and presented in two different orientations. b, Close-up view of the Nprl3 longin-TINI domains (PDB 7T3B). c, Close-up view of the Nprl3 longin-TINI domains bound to SZT2NTD. d, Cryo-EM densities fitted to the Nprl3TINI model. e, The map covering the interface between Nprl3-SZT2. f, Structural alignment of Nprl2 with its GAP-mode conformation (PDB 7T3B). RMSD values for the superimposed structures are shown in Supplementary Table 2.
Extended Data Fig. 8 Hypothetical full-length KICSTOR model.
The model for residues 352-1713 of SZT2 was predicted from AlphaFold2 and then combined with the two refined models in this study.
Extended Data Fig. 9 Liposome-binding assays of different recombinant KICSTOR proteins with LUVs.
a, b,d, Liposome-binding assays of KICSTOR complex with LUVs of different compositions (a: 80% DOPC: 20% DOPE (neutral mix); b: 50% DOPC: 20% DOPE: 30% DOPS; d: 70% DOPC: 20% DOPE: 10% PtdIns3P). c, Quantification of different recombinant proteins to their interactions with LUVs containing 50% DOPC: 20% DOPE: 30% DOPS. f, g, i, Liposome-binding assays of KPTN/ITFG2/SZT2 complex with LUVs of different compositions (f: 80% DOPC: 20% DOPE (neutral mix); g: 50% DOPC: 20% DOPE: 30% DOPS; i: 70% DOPC: 20% DOPE: 10% PtdIns3P). k, l, n, Liposome-binding assays of GST-C12orf66 with LUVs of different compositions (k: 80% DOPC: 20% DOPE (neutral mix); l: 50% DOPC: 20% DOPE: 30% DOPS; n: 70% DOPC: 20% DOPE: 10% PtdIns3P). p, q, s, Liposome-binding assays of KPTN/ITFG2 complex with LUVs of different compositions (p: 80% DOPC: 20% DOPE (neutral mix); q: 50% DOPC: 20% DOPE: 30% DOPS; s: 70% DOPC: 20% DOPE: 10% PtdIns3P). e, Quantification of different recombinant proteins to their interactions with LUVs containing 70% DOPC: 20% DOPE: 10% PtdIns3P. h, m, r, Quantification of liposome-bound KPTN/ITFG2/SZT2 complex, GST-C12orf66, or KPTN/ITFG2 complex to LUVs containing DOPC and DOPE and additionally, the amount of DOPS, indicated in the figures. j, o, t, Quantification of liposome-bound KPTN/ITFG2/SZT2 complex, GST-C12orf66, or KPTN/ITFG2 complex to LUVs with different composition. Results were presented as mean ± s.e.m. Quantified data of liposome-binding assays in (c, e, h, j, m, o, r, t) represent mean ± s.e.m. of three independent experiments. % P was quantified as described in the Methods section. Statistical analysis was carried out using one-way analysis of variance (ANOVA). S, supernatant; P, pellet. The input lanes indicated 20% input of the proteins used in the experiment.
Extended Data Fig. 10 Hypothetical model of KICSTOR-GATOR1-GATOR2-Rag-Ragulator complex tethered on the lysosomal membrane.
a, Model was assembled using the reported structures of GATOR2 (PDB 7UHY), an AlphaFold2 prediction for WDR59 protein, and dual mode of GATOR1 which is bound to two copies of Rag-Ragulator, one in the “GAP conformation” and another one in the “inhibitory conformation” (PDB 7T3C) and KICSTOR from this study. b, Color code for the KICSTOR1, GATOR1, Rag-Ragulator and GATOR2 subunits. c, The reported structures of GATOR2 (PDB 7UHY), dual mode of GATOR1, which is bound to two copies of Rag-Ragulator, one in the “GAP conformation” and another one in the “inhibitory conformation” (PDB 7T3C), and an AlphaFold2 prediction for WDR59 protein, were assembled and superimposed on SEA complex (PDB 8ADL). The dotted line box indicates the interface between GATOR1 and GATOR2. d, KICSTOR from this study was superimposed on the GATOR1-dual mode. The dotted line box indicates the interface between GATOR1 and KICSTOR. e, Zoomed-in view on the clashing of SZT2SZN1-SZN2 and Sec13.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Tables 1–4 and source data for supplementary figures.
Supplementary Data
Statistical source data for Supplementary Information.
Source data
Source Data Figs. 3–7 and Extended Data Fig. 9
Statistical source data.
Source Data Fig. 1
Unprocessed gels.
Source Data Fig. 2
Unprocessed western blots and gels.
Source Data Fig. 3
Unprocessed western blots and gels.
Source Data Fig. 4
Unprocessed western blots and gels.
Source Data Fig. 6
Unprocessed gels.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 9
Unprocessed gels.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Teng, F., Zeng, H., Mai, X. et al. Architecture of the human KICSTOR and GATOR1–KICSTOR complexes. Nat Struct Mol Biol 32, 2587–2600 (2025). https://doi.org/10.1038/s41594-025-01693-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01693-4