Abstract
Maternal exercise can improve the metabolic health of the offspring. However, the molecular mechanisms underlying the beneficial effects of maternal exercise on the offspring remain unclear. Here, we show that maternal exercise during pregnancy alleviates high-fat diet (HFD)-induced adipose inflammation and glucose intolerance in offspring mice, accompanied by upregulation of the adipokine serine protease inhibitor A3C (SERPINA3C) both in maternal adipose tissues and the fetal circulation. Adipose SERPINA3C knockdown impairs, but its overexpression in dams mimics, maternal exercise-mediated metabolic benefits in HFD-fed offspring. Maternal SERPINA3C is transported into the fetal circulation and promotes Krüppel-like factor 4 (Klf4) gene promoter demethylation in fetal preadipocytes to increase KLF4 expression, which inhibits adipose inflammation in HFD-fed offspring mice. The SERPINA3C–cathepsin G–integrin β1 axis activates phosphatidylinositol 3-kinase signalling in preadipocytes. This promotes nuclear translocation of the p110β subunit to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3) in the nucleus. O-linked β-N-acetylglucosamine (O-GlcNAc) transferase then binds to PIP3 to promote ten–eleven translocation methylcytosine dioxygenase 1 (TET1) O-GlcNAcylation, thereby enhancing TET1 activity to facilitate Klf4 gene promoter demethylation. These results provide mechanistic insights into maternal exercise-mediated improvement of offspring metabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data are available in the article and supplementary materials. The illustrations of mouse models and cell experiments (Figs. 3g, 4a and 8a) and the graphical abstract (Extended Data Fig. 10) were created with BioRender.com. The BioRender agreement number is MD27KTS36P for Figs. 3g, 4a and 8a and NZ27KTRYU8 for Extended Data Fig. 10. The MS proteomics data have been submitted to the ProteomeXchange consortium (https://proteomecentral.proteomexchange.org) through the iProX partner repository under the dataset identifier PXD055427. DIA data were processed using DIA-NN (version 1.8) against the Mus_musculus_10090_SP_20231220.fasta database. Source data are provided with this paper.
References
Tan, D. J. H. et al. Rising global burden of cancer attributable to high BMI from 2010 to 2019. Metabolism 152, 155744 (2024).
Xu, H. et al. Maternal antibiotic exposure enhances ILC2 activation in neonates via downregulation of IFN1 signaling. Nat. Commun. 14, 8332 (2023).
Taibl, K. R. et al. Newborn metabolomic signatures of maternal per- and polyfluoroalkyl substance exposure and reduced length of gestation. Nat. Commun. 14, 3120 (2023).
Feng, H. et al. Association of nutrients intake during pregnancy with the risk of allergic disease in offspring: a meta-analysis of prospective cohort studies. Food Sci. Hum. Wellness 12, 711–719 (2023).
Gao, J. et al. Maternal obesity exacerbates the responsiveness of offspring BALB/c mice to cow’s milk protein-induced food allergy. Food Sci. Hum. Wellness 12, 920–928 (2023).
Son, J. S. et al. Maternal exercise via exerkine apelin enhances brown adipogenesis and prevents metabolic dysfunction in offspring mice. Sci. Adv. 6, eaaz0359 (2020).
Kusuyama, J. et al. Placental superoxide dismutase 3 mediates benefits of maternal exercise on offspring health. Cell Metab. 33, 939–956 (2021).
Jeon, Y. G., Kim, Y. Y., Lee, G. & Kim, J. B. Physiological and pathological roles of lipogenesis. Nat. Metab. 5, 735–759 (2023).
Palani, N. P. et al. Adipogenic and SWAT cells separate from a common progenitor in human brown and white adipose depots. Nat. Metab. 5, 996–1013 (2023).
Mishra, G. & Townsend, K. L. The metabolic and functional roles of sensory nerves in adipose tissues. Nat. Metab. 5, 1461–1474 (2023).
Hornburg, D. et al. Dynamic lipidome alterations associated with human health, disease and ageing. Nat. Metab. 5, 1578–1594 (2023).
Blandin, A. et al. Lipidomic analysis of adipose-derived extracellular vesicles reveals specific EV lipid sorting informative of the obesity metabolic state. Cell Rep. 42, 112169 (2023).
Rashid, M., Kondoh, K., Palfalvi, G., Nakajima, K.-I. & Minokoshi, Y. Inhibition of high-fat diet-induced inflammatory responses in adipose tissue by SF1-expressing neurons of the ventromedial hypothalamus. Cell Rep. 42, 112627 (2023).
Hildreth, A. D. et al. Adipose cDC1s contribute to obesity-associated inflammation through STING-dependent IL-12 production. Nat. Metab. 5, 2237–2252 (2023).
Boutari, C., Hill, M. A., Procaccini, C., Matarese, G. & Mantzoros, C. S. The key role of inflammation in the pathogenesis and management of obesity and CVD. Metabolism 145, 155627 (2023).
Almond, M. et al. Obesity dysregulates the pulmonary antiviral immune response. Nat. Commun. 14, 6607 (2023).
de Oliveira, M. C. et al. Eosinophils protect from metabolic alterations triggered by obesity. Metabolism 146, 155613 (2023).
Blandin, A. et al. Extracellular vesicles are carriers of adiponectin with insulin-sensitizing and anti-inflammatory properties. Cell Rep. 42, 112866 (2023).
Zhang, X. et al. Polysaccharide extract from Rosa laevigata fruit attenuates inflammatory obesity by targeting redox balance and gut interface in high-fat diet-fed rats. Food Sci. Hum. Wellness 12, 442–453 (2023).
Li, B.-Y., Guo, Y.-Y., Xiao, G., Guo, L. & Tang, Q.-Q. SERPINA3C ameliorates adipose tissue inflammation through the cathepsin G/integrin/AKT pathway. Mol. Metab. 61, 101500 (2022).
Lim, A. I. et al. Prenatal maternal infection promotes tissue-specific immunity and inflammation in offspring. Science 373, eabf3002 (2021).
Chae, S. A., Son, J. S., Zhu, M.-J., De Avila, J. M. & Du, A. M. Treadmill running of mouse as a model for studying influence of maternal exercise on offspring. Bio Protoc. 10, e3838 (2020).
Tokarz, V. L., MacDonald, P. E. & Klip, A. The cell biology of systemic insulin function. J. Cell Biol. 217, 2273–2289 (2018).
Beals, J. W. et al. Dietary weight loss-induced improvements in metabolic function are enhanced by exercise in people with obesity and prediabetes. Nat. Metab. 5, 1221–1235 (2023).
Nigro, P. et al. Exercise training remodels inguinal white adipose tissue through adaptations in innervation, vascularization, and the extracellular matrix. Cell Rep. 42, 112392 (2023).
Arumugasaamy, N., Rock, K. D., Kuo, C.-Y., Bale, T. L. & Fisher, J. P. Microphysiological systems of the placental barrier. Adv. Drug Deliv. Rev. 161–162, 161–175 (2020).
Fang, H. & Judd, R. L. Adiponectin regulation and function. Compr. Physiol. 8, 1031–1063 (2018).
Cui, B. et al. Exercise alleviates neovascular age-related macular degeneration by inhibiting AIM2 inflammasome in myeloid cells. Metabolism 144, 155584 (2023).
Ouchi, N. et al. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science 329, 454–457 (2010).
Shi, L. et al. Brown adipose tissue-derived Nrg4 alleviates endothelial inflammation and atherosclerosis in male mice. Nat. Metab. 4, 1573–1590 (2022).
Wang, C. et al. The effect and mechanism of TLR9/KLF4 in FFA-induced adipocyte inflammation. Mediators Inflamm. 2018, 6313484 (2018).
Mattei, A. L., Bailly, N. & Meissner, A. DNA methylation: a historical perspective. Trends Genet. 38, 676–707 (2022).
Chen, Y. et al. Epigenetic modification of nucleic acids: from basic studies to medical applications. Chem. Soc. Rev. 46, 2844–2872 (2017).
Li, H.-J. et al. Roles of ten–eleven translocation family proteins and their O-linked β-N-acetylglucosaminylated forms in cancer development. Oncol. Lett. 21, 1 (2021).
Vella, P. et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 (2013).
Hrit, J. et al. OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development. eLife 7, e34870 (2018).
Ong, Q., Han, W. & Yang, X. O-GlcNAc as an integrator of signaling pathways. Front. Endocrinol. (Lausanne) 9, 599 (2018).
Wang, R. et al. Switch of phosphorylation to O-GlcNAcylation of AhR contributes to vascular oxidative stress induced by benzo[a]pyrene. Food Sci. Hum. Wellness 12, 2263–2275 (2023).
Kebede, M. et al. Glucose activates free fatty acid receptor 1 gene transcription via phosphatidylinositol-3-kinase-dependent O-GlcNAcylation of pancreas–duodenum homeobox-1. Proc. Natl Acad. Sci. USA 109, 2376–2381 (2012).
Martelli, A. M. et al. Phosphatidylinositol 3-kinase translocates to the nucleus of osteoblast-like MC3T3-E1 cells in response to insulin-like growth factor I and platelet-derived growth factor but not to the proapoptotic cytokine tumor necrosis factor α. J. Bone Miner. Res. 15, 1716–1730 (2000).
Kumar, A. et al. Nuclear but not cytosolic phosphoinositide 3-kinase β has an essential function in cell survival. Mol. Cell. Biol. 31, 2122–2133 (2011).
Birsoy, K., Chen, Z. & Friedman, J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 7, 339–347 (2008).
Luo, H.-Y. et al. Hepatic Klf10–Fh1 axis promotes exercise-mediated amelioration of NASH in mice. Metabolism 155, 155916 (2024).
Moore, T. M. et al. Conserved multi-tissue transcriptomic adaptations to exercise training in humans and mice. Cell Rep. 42, 112499 (2023).
Jin, L. et al. The muscle-enriched myokine Musclin impairs beige fat thermogenesis and systemic energy homeostasis via Tfr1/PKA signaling in male mice. Nat. Commun. 14, 4257 (2023).
Gu, X., Wang, L., Liu, S. & Shan, T. Adipose tissue adipokines and lipokines: functions and regulatory mechanism in skeletal muscle development and homeostasis. Metabolism 139, 155379 (2023).
Chen, M. et al. Cdo1–Camkk2–AMPK axis confers the protective effects of exercise against NAFLD in mice. Nat. Commun. 14, 8391 (2023).
Zhu, J.-Y., Chen, M., Mu, W.-J., Luo, H.-Y. & Guo, L. Higd1a facilitates exercise-mediated alleviation of fatty liver in diet-induced obese mice. Metabolism 134, 155241 (2022).
Gasbarrino, K. et al. Relationship between circulating adipokines and cholesterol efflux in subjects with severe carotid atherosclerosis. Metabolism 140, 155381 (2023).
Son, J. S. et al. Maternal exercise intergenerationally drives muscle-based thermogenesis via activation of apelin–AMPK signaling. eBioMedicine 76, 103842 (2022).
Kong, W. et al. Soluble ST2, a preeclampsia-related cytokine receptor, is transported bi-directionally across the placenta. Placenta 63, 21–25 (2018).
Letterio, J. J. et al. Maternal rescue of transforming growth factor-β1 null mice. Science 264, 1936–1938 (1994).
Borovecki, F. et al. Bone morphogenetic protein-7 from serum of pregnant mice is available to the fetus through placental transfer during early stages of development. Nephron Exp. Nephrol. 97, e26–e32 (2004).
Popliker, M. et al. Onset of endogenous synthesis of epidermal growth factor in neonatal mice. Dev. Biol. 119, 38–44 (1987).
Qian, L. L. et al. Serpina3c deficiency induced necroptosis promotes non-alcoholic fatty liver disease through β-catenin/Foxo1/TLR4 signaling. FASEB J. 36, e22316 (2022).
Qian, L.-L. et al. Protective role of serpina3c as a novel thrombin inhibitor against atherosclerosis in mice. Clin. Sci. (Lond.) 135, 447–463 (2021).
Marco, A., Kisliouk, T., Tabachnik, T., Weller, A. & Meiri, N. DNA CpG methylation (5-methylcytosine) and its derivative (5-hydroxymethylcytosine) alter histone posttranslational modifications at the Pomc promoter, affecting the impact of perinatal diet on leanness and obesity of the offspring. Diabetes 65, 2258–2267 (2016).
Kusuyama, J., Alves-Wagner, A. B., Makarewicz, N. S. & Goodyear, L. J. Effects of maternal and paternal exercise on offspring metabolism. Nat. Metab. 2, 858–872 (2020).
Laker, R. C. et al. Exercise prevents maternal high-fat diet-induced hypermethylation of the Pgc-1α gene and age-dependent metabolic dysfunction in the offspring. Diabetes 63, 1605–1611 (2014).
Wang, Q. et al. Integrin β1 in adipose-derived stem cells accelerates wound healing via activating PI3K/AKT pathway. Tissue Eng. Regen. Med. 17, 183–192 (2020).
Ye, Q. et al. Deficiency of gluconeogenic enzyme PCK1 promotes metabolic-associated fatty liver disease through PI3K/AKT/PDGF axis activation in male mice. Nat. Commun. 14, 1402 (2023).
Liu, Z. et al. Lignans from Patrinia scabiosaefolia improve insulin resistance by activating PI-3K/AKT pathway and promoting GLUT4 expression. Food Sci. Hum. Wellness 12, 2014–2021 (2023).
Palmieri, M. et al. PI3Kα translocation mediates nuclear PtdIns(3,4,5)P3 effector signaling in colorectal cancer. Mol. Cell. Proteomics 22, 100529 (2023).
Yang, X. et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 (2008).
Palmeira, P., Quinello, C., Silveira-Lessa, A. L., Zago, C. A. & Carneiro-Sampaio, M. IgG placental transfer in healthy and pathological pregnancies. Clin. Dev. Immunol. 2012, 985646 (2012).
Guo, Y.-Y. et al. Cdo1 promotes PPARγ-mediated adipose tissue lipolysis in male mice. Nat. Metab. 4, 1352–1368 (2022).
Li, B.-Y., Peng, W.-Q., Liu, Y., Guo, L. & Tang, Q.-Q. HIGD1A links SIRT1 activity to adipose browning by inhibiting the ROS/DNA damage pathway. Cell Rep. 42, 112731 (2023).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 50, D1522–D1527 (2022).
Pennington, K. A., Schlitt, J. M. & Schulz, L. C. Isolation of primary mouse trophoblast cells and trophoblast invasion assay. J. Vis. Exp. 7, e3202 (2012).
Lin, A. et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 19, 238–251 (2017).
Peng, W.-Q. et al. l-Theanine activates the browning of white adipose tissue through the AMPK/α-ketoglutarate/Prdm16 axis and ameliorates diet-induced obesity in mice. Diabetes 70, 1458–1472 (2021).
Acknowledgements
This work was supported by the Program for Overseas High-level Talents at Shanghai Institutions of Higher Learning (TP2022100 to L.G.), the National Natural Science Foundation of China (nos. 32070751 and 31871435 to L.G.) and the Research and Innovation Grant for Graduate Students, Shanghai University of Sport (project no. YJSCX-2024-020 to Y.L.).
Author information
Authors and Affiliations
Contributions
Y.L. was involved in the study design, conducted the experiments, analysed the data and drafted the paper. R.-Y.L., J.-Y.Z., M.C., W.-J.M., H.-Y.L., L.-J.Y., Y.L., S.L., M.-T.Y., X.L. and H.-M.C. performed the experiments. L.G. conceived the idea, designed and supervised the study, obtained the funding and cowrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Joji Kusuyama, Emma Borgeson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Jean Nakhle and Ashley Castellanos-Jankiewicz, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 2 Maternal exercise increases circulating Serpina3c level, which potentially crosses the placental barrier.
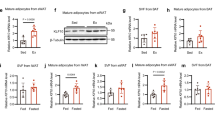
a, Heatmap of differentially expressed serum protein between Ex and Sed pregnant dams, as assessed by LC-MS/MS (n = 3 mice/group) (P < 0.05). b, Differentially expressed proteins identified by the intersection of secreted proteins and inflammation-related proteins based on serum proteomics results (P < 0.05). c, RT-qPCR analysis of Serpina3c mRNA expression in the placentas of dams. d, Mice were treated as described in Fig. 3f. RT-qPCR analysis of Serpina3c mRNA expression in iWAT, liver and Gas of dams was shown. e, Pregnant dams were injected with GFP or Flag-Serpina3c adenoviruses (Ad) in iWAT at embryonic day 2.5 (E2.5), which is equal to gestation day 18.5 (d18.5) for mother. At E18.5 (equal to gestation d18.5 for mother), maternal iWAT and fetal liver and inguinal region were collected. Flag antibody-mediated IP followed by western blotting with Serpina3c antibody was performed, n = 3 mice for each group. f, Immunofluorescence (IF) staining of CK7 protein in primary mouse trophoblast cells, with MAEC cells as a negative control. CK7 serves as the marker for trophoblast cells. scale bar: 50 μm, n = 3 mice for each group. g, Flag antibody-mediated IP in primary trophoblast cells extracted from dams injected locally in iWAT with Ad-GFP or Ad-Flag-Serpina3c, followed by western blotting with a Serpina3c antibody, n = 3 mice for each group. h, CM from Ad-GFP or Ad-Flag-Sfrp5-infected adipocytes was added to the apical chamber (AC) of transwell inserts cultured with BeWo cells. IP was performed using the Flag antibody, followed by western blotting with Sfrp5 antibody to detect the presence of Flag-Sfrp5 in the supernatant of apical and basolateral chambers, n = 3 independent biological replicates. For statistical analyses, unpaired two-tailed Student’s t tests between Sed and Ex groups were performed in c, d. All values are represented as means with error bars representing S.D. n = 6 for each group unless otherwise mentioned. iWAT, inguinal white adipose tissue; Gas, gastrocnemius; IP, immunoprecipitation; CM, culture medium.
Extended Data Fig. 3 The impact of WATs-specific knockdown of Serpina3c on the pregnant dams and fetuses.
Dams were treated as in Fig. 4. a, Body weight change curve and body weight gain during pregnancy of the dams. b, The serum Serpina3c level at gestation day 18.5 (d18.5) in dams. c, Serpina3c mRNA levels in iWAT, POAT, liver and Gas of dams at gestation d18.5. d, mRNA levels of the indicated genes were determined by RT-qPCR in placenta from dams at gestation d18.5. e, The serum Serpina3c level in fetuses at E18.5. f, g, Western blotting (f) was conducted to detect Serpina3c protein levels in placenta from dams, as well as in fetal liver and inguinal region. Quantitative analysis (g) by Serpina3c/HSP90 ratio was performed. n = 3 mice for each group. h, The number of pups per litter from dams. i,j, Western blotting (i) was conducted to detect Serpina3c protein levels in iWAT and POAT from dams after the weaning period, and quantitative analysis (j) by Serpina3c/HSP90 ratio was performed. n = 3 mice for each group. For statistical analyses, two-way ANOVA with Bonferroni’s post hoc tests were performed in a,b,h,j, and unpaired two-tailed Student’s t tests were performed in c-e,g. All values are represented as means with error bars representing S.D. n = 6 for each group unless otherwise mentioned. WATs, white adipose tissues; E18.5, embryonic day 18.5; RT-qPCR, reverse transcription-quantitative PCR; POAT, peri-ovarian adipose tissue.
Extended Data Fig. 4 Maternal exercise-mediated improvement of offspring metabolic health is blunted by WATs-specific knockdown of Serpina3c in dams.
Mice were treated as indicated in Fig. 4. a, Glucose concentration during the i.p. GTT in male and female offspring after 7 weeks of HFD feeding. b, HE staining of iWAT and VAT from male and female offspring, respectively. VAT includes male eWAT and female POAT. Scale bar: 50 μm. c,d, Quantitative analyses of adipocyte size distribution of iWAT and VAT from male (c) and female (d) offspring. e,f, Liver TG levels from female (e) and male (f) offspring. g,h, Quantitative analysis of p-AKT/AKT ratio in Fig. 4i. n = 3 mice for each group. i,j, Quantitative analyses of the F4/80 IF staining in Fig. 4l. k, Flow cytometry for detecting CD11b+/F4/80+ macrophages from CD45+ cells in SVF isolated from offspring iWAT. For statistical analyses, two-way ANOVA with Bonferroni’s post hoc tests were performed in e-j. All values are represented as means with error bars representing S.D. n = 6 for each group unless otherwise mentioned. i.p., intraperitoneal injection; GTT, glucose tolerance test; TG, triglyceride; IF, immunofluorescence.
Extended Data Fig. 5 The impact of WATs-specific overexpression of Serpina3c on the dams and fetuses.
Mice were treated as in Fig. 5. a, Body weight change curve and body weight gain during pregnancy of the dams. b, The serum Serpina3c level at gestation day 18.5 (d18.5) in dams. c, Serpina3c mRNA levels in iWAT, POAT, liver and Gas of dams at gestation d18.5. d, The serum Serpina3c level in fetuses at E18.5. e, f, Western blotting (e) was conducted to detect Serpina3c protein levels in placenta from dams, as well as in fetal liver and inguinal region. Quantitative analysis (f) by Serpina3c/HSP90 ratio was performed. n = 3 mice for each group. g,h, Western blotting(g) was conducted to detect Serpina3c protein levels in iWAT and POAT from dams after the weaning period, and quantitative analysis (h) by Serpina3c/HSP90 ratio was performed. n = 3 mice for each group. For statistical analyses, two-way ANOVA with Bonferroni’s post hoc tests were performed in a,b,h, and unpaired two-tailed Student’s t tests were performed in c,d,f,. All values are represented as means with error bars representing S.D. n = 6 for each group unless otherwise mentioned. E18.5, embryonic day 18.5; Gas, gastrocnemius.
Extended Data Fig. 6 Maternal exercise-mediated improvement of offspring metabolic health can be mimicked by WATs-specific overexpression of Serpina3c in dams.
Mice were treated as indicated in Fig. 5. a,b, Glucose concentration during an i.p. GTT in male (a) and female (b) offspring after 7 weeks of HFD feeding. c, HE staining of iWAT and VAT from male and female offspring, respectively, Scale bar: 50 μm. d,e, Quantitative analyses of adipocyte size distribution of iWAT and VAT from male (d) and female (e) offspring. f,g, Quantitative analysis of p-AKT/AKT ratio in Fig. 5h, with (f) for male offspring and (g) for female offspring. n = 3 mice for each group. h,i, Quantitative analyses of the IF staining of F4/80 in Fig. 5l. j, Flow cytometry for detecting CD11b+/F4/80+ macrophages from CD45+ cells in SVF isolated from offspring iWAT. For statistical analyses, two-way ANOVA with Bonferroni’s post hoc tests were performed in f-i. All values are represented as means with error bars representing S.D. n = 6 for each group unless otherwise mentioned. i.p., intraperitoneal injection; GTT, glucose tolerance test; SVF, stromal vascular fraction; IF, immunofluorescence.
Extended Data Fig. 7 The effect of maternal exercise on offspring mice at weaning stage and the role of Serpina3c and SERPINA3 in the epigenetic regulation of Klf4.
a-e, Mice were treated as indicated in Fig. 1. Offspring mice were sacrificed for analysis after weaning. a, Body weight of male and female offspring mice at weaning. b, Body composition of male and female offspring mice at weaning. c,d, mRNA levels of the indicated genes were determined by RT-qPCR in iWAT and VAT from male (c) and female (d) offspring mice at weaning. e, SVF isolated from embryonic day18.5 (E18.5) fetuses were induced to adipogenic differentiation into mature adipocytes. Light microscope images of the cells at Day 0 (D0), Day 2 (D2), and Day 4 (D4) of differentiation and Oil Red O staining images at Day 6 (D6) of differentiation were shown. Scale bar: 50 μm, n = 3 mice for each group. f, Diagram showing three regions in the mouse (mus) Klf4 gene promoter and two regions in human (homo) KLF4 gene promoter. g, Klf4 mRNA levels were determined by RT-qPCR in 3T3-L1 preadipocytes, C2C12 myotubes, and primary hepatocytes treated with 20 ng/ml rSerpina3c for 24 h. h-j, 5mc and 5hmc enrichment on mouse Klf4 gene promoter in 3T3-L1 preadipocytes (h), C2C12 myotubes (i), and mice primary hepatocytes (j) treated with 20 ng/ml rSerpina3c for 24 h. NC, nonspecific control. k, KLF4 mRNA levels were determined by RT-qPCR in human primary adipocytes treated with 20 ng/ml rSERPINA3 for 24 h. l, 5mc and 5hmc enrichment on human Klf4 gene promoter in human primary adipocytes treated with 20 ng/ml rSERPINA3 for 24 h. For statistical analyses in a-d, g-l, unpaired two-tailed Student’s t tests were performed. All values are represented as means with error bars representing S.D. n = 6 for each group. rSerpina3c, recombinant Serpina3c protein; rSERPINA3, recombinant SERPINA3.
Extended Data Fig. 8 The positive feedback loop between Serpina3c and Klf4.
a,b, 3T3-L1 preadipocytes were treated by 20 ng/ml rSerpina3c for 24 h. The mRNA levels of the indicated genes. (a) and α-KG levels (b) were determined. c, 3T3-L1 adipocytes were infected with Ad-Klf4 or treated with 10 μM JNKi (SP600125) for 24 h. Western blotting and quantitative analysis were done. n = 3 independent biological replicates. d,e, 3T3-L1 adipocytes were infected with Ad-Klf4 or treated with 10 μM JNKi for 24 h, followed by treatment with 10 μM CHX. Western blotting was then performed (d) and quantified (e). n = 3 independent biological replicates. f, Serpina3c mRNA levels in iWAT and VAT of offspring mice at weaning. g, Serum Serpina3c level of offspring mice at weaning. h, Activity of TETs in 3T3-L1 mature adipocytes after 20 ng/ml rSerpina3c treatment for 6 h. i, O-GlcNAcylation (O-GlcNAc) of Tet1 in 3T3-L1 adipocytes treated with 20 ng/ml rSerpina3c for 12 h. n = 3 independent biological replicates. j, 5mc and 5hmc enrichment on mouse Klf4 gene promoter in 3T3-L1 adipocytes after 20 ng/ml rSerpina3c treatment for 24 h. k. The indicated mRNA levels in 3T3-L1 adipocytes treated with siKlf4 or siAdiponectin. l, 3T3-L1 adipocytes were transfected with siNC or siKlf4. 24 h later, cells were treated with 20 ng/ml rSerpina3c for 24 h. Adiponectin mRNA levels were determined. m, 3T3-L1 adipocytes were transfected with siNC or siAdiponectin. 24 h later, cells were treated with 20 ng/ml rSerpina3c for 24 h. Klf4 mRNA levels were determined. n, 3T3-L1 cells were treated as in (l). Tet1 enrichment on the Adiponectin gene promoter in 3T3-L1 adipocytes was determined. o, Diagram showing two regions on the mouse (mus) Adiponectin gene promoter. p, 3T3-L1 cells were treated as in (l). 5mc and 5hmc enrichment on mus Adiponectin gene promoter was shown. q, 3T3-L1 adipocytes with/out Klf4 knockdown were pretreated with TNFα (10 ng/ml) for 12 h and then treated with rSerpina3c for 30 h. Then the mRNA levels of the indicated genes were determined. Unpaired two-tailed Student’s t tests were performed in a, b, f-h, j, k. One-way ANOVA tests plus Bonferroni’s post hoc tests were performed in c, l-n, p, q. All values are represented as means with error bars representing S.D. n = 6 for each group unless otherwise mentioned. α-KG, α-ketoglutarate; JNKi, JNK inhibitor; CHX, cycloheximide.
Extended Data Fig. 9 The effects of Klk1 and Klk1b26 on the DNA methylation level of the Klf4 gene promoter in adipocytes, the effects of Serpina3c on the DNA methylation level of the Cebpb gene promoter in adipocytes and the effects of Serpina3c knockdown or overexpression on glucose uptake and inflammation-related genes expression in maternal WATs.
a, Activity of TETs in 3T3-L1 adipocytes after being treated with 50 ng/ml rKlk1 or rKlk1b26 for 6 h. b, 5mc and 5hmc enrichment on Klf4 gene promoter in 3T3-L1 adipocytes after being treated by 50 ng/ml rKlk1 or rKlk1b26 for 24 h. c, 5mc and 5hmc enrichment on mouse Cebpb gene promoter in 3T3-L1 adipocytes treated with 20 ng/ml rSerpina3c for 24 h. d,e, 2-NBDG uptake was determined in iWAT and POAT from the chow diet-fed sedentary dams (at gestation d18.5) with/out Serpina3c knockdown in the WATs (d) and with/out Serpina3c overexpression in the WATs (e). 2-NBDG uptake was measured after in vitro stimulation of the iWAT and POAT with/out insulin. f,g, The mRNA levels of the indicated genes were determined by RT-qPCR in iWAT and POAT from the chow diet-fed sedentary dams (at gestation d18.5) with/out Serpina3c knockdown in the WATs (f) and with/out Serpina3c overexpression in the WATs (g). For statistical analyses in a,b, one-way ANOVA tests plus Bonferroni’s post hoc tests were performed, and unpaired two-tailed Student’s t tests were performed in c-g. All values are represented as means with error bars representing S.D. n = 6 for each group. rKlk1, recombinant Klk1 protein; rKlk1b26, recombinant Klk1b26 protein; WATs, white adipose tissues.
Extended Data Fig. 10 A graph model depicting the role of adipokine Serpina3c in maternal exercise-mediated amelioration of HFD-induced metabolic disorders in offspring mice.
During maternal exercise in pregnancy, increased expression and secretion of Serpina3c in maternal WAT were observed. Maternal Serpina3c can be transported to enter the fetal circulation and further act on fetal preadipocytes, facilitating the demethylation of the Klf4 gene promoter through the Cathepsin G/Integrin β1/PI3K/OGT/Tet1 signaling axis to increase Klf4 gene expression. Moreover, Klf4 can also maintain the protein stability of Serpina3c in adipocytes. This establishes a positive feedback loop between Serpina3c and Klf4 in the white adipose tissues of offspring mice, effectively suppressing WATs inflammation induced by HFD feeding and enhancing glucose tolerance and insulin sensitivity. Consequently, the above process mitigates metabolic disturbances caused by HFD feeding in offspring mice. WATs, white adipose tissues. Figure created with BioRender.com.
Supplementary information
Supplementary Information
Supplementary Table 2. Primers used in qPCR.
Supplementary Table 1
Raw data for serum proteomics in Extended Data Fig. 2a,b.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Li, RY., Zhu, JY. et al. Maternal exercise prevents metabolic disorders in offspring mice through SERPINA3C. Nat Metab 7, 401–420 (2025). https://doi.org/10.1038/s42255-024-01213-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42255-024-01213-6
This article is cited by
-
SERPINA3C as a mediator of metabolic health in offspring
Nature Metabolism (2025)