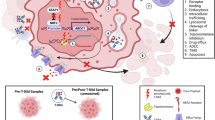
Abstract
Trastuzumab deruxtecan (T-DXd), an anti-HER2–drug conjugate, has transformed treatment for HER2-expressing solid tumors. However, heterogeneous intratumoral HER2 expression, particularly high densities of HER2-immunohistochemistry score 0 (HER2-IHC 0) cells, limits its clinical efficacy. Here, we discovered that targeted inhibition of F-box protein FBXL2 elevates HER2 expression on the plasma membrane of HER2-IHC 0 triple-negative breast cancer (TNBC) cells, thereby sensitizing them to T-DXd. Mechanistically, FBXL2 promotes HER2 polyubiquitination at K747 and proteasomal degradation. Notably, small molecules GGTi-2418 and ketoconazole effectively elevate HER2 expression via blocking FBXL2 membrane localization. We further developed lipid nanoparticles (LNPs) to encapsulate GGTi-2418 and ketoconazole, enabling their enrichment in TNBC tumors. Strikingly, GGTi-2418@LNP or ketoconazole@LNP combined with T-DXd induced robust and durable tumor regression in HER2-IHC 0 TNBC xenograft models in female mice. Together, this study highlights that targeted inhibition of FBXL2 combined with T-DXd may be a viable therapeutic strategy for treating HER2-IHC 0 solid tumors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All relevant data are available within the article and Supplementary Information. Source data are provided with this paper.
References
Ogitani, Y. et al. DS-8201a, a novel HER2-targeting ADC with a novel DNA topoisomerase I inhibitor, demonstrates a promising antitumor efficacy with differentiation from T-DM1. Clin. Cancer Res. 22, 5097–5108 (2016).
Meric-Bernstam, F. et al. Efficacy and safety of trastuzumab deruxtecan in patients with HER2-expressing solid tumors: primary results from the DESTINY-PanTumor02 phase II trial. J. Clin. Oncol. 42, 47–58 (2024).
Ogitani, Y., Hagihara, K., Oitate, M., Naito, H. & Agatsuma, T. Bystander killing effect of DS-8201a, a novel anti-human epidermal growth factor receptor 2 antibody–drug conjugate, in tumors with human epidermal growth factor receptor 2 heterogeneity. Cancer Sci. 107, 1039–1046 (2016).
Andre, F. et al. Trastuzumab deruxtecan versus treatment of physician’s choice in patients with HER2-positive metastatic breast cancer (DESTINY-Breast02): a randomised, open-label, multicentre, phase 3 trial. Lancet 401, 1773–1785 (2023).
Modi, S. et al. Trastuzumab deruxtecan in previously treated HER2-low advanced breast cancer. N. Engl. J. Med. 387, 9–20 (2022).
Shitara, K. et al. Trastuzumab deruxtecan in previously treated HER2-positive gastric cancer. N. Engl. J. Med. 382, 2419–2430 (2020).
Raghav, K. et al. Trastuzumab deruxtecan in patients with HER2-positive advanced colorectal cancer (DESTINY-CRC02): primary results from a multicentre, randomised, phase 2 trial. Lancet Oncol. 25, 1147–1162 (2024).
Mosele, F. et al. Trastuzumab deruxtecan in metastatic breast cancer with variable HER2 expression: the phase 2 DAISY trial. Nat. Med. 29, 2110–2120 (2023).
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Orrantia-Borunda, E., Anchondo-Nuñez, P., Acuña-Aguilar, L. E., Gómez-Valles, F. O. & Ramírez-Valdespino, C. A. in Breast Cancer (ed Mayrovitz, H.C.) Ch. 3 (Exon Publications, 2022).
Yang, R. et al. Therapeutic progress and challenges for triple negative breast cancer: targeted therapy and immunotherapy. Mol. Biomed. 3, 8 (2022).
Gampenrieder, S. P. et al. Influence of HER2 expression on prognosis in metastatic triple-negative breast cancer-results from an international, multicenter analysis coordinated by the AGMT Study Group. ESMO Open 8, 100747 (2023).
Kuchay, S. et al. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI3K signalling cascade. Nat. Cell Biol. 15, 472–480 (2013).
Chen, B. B., Glasser, J. R., Coon, T. A. & Mallampalli, R. K. Skp–Cullin–F box E3 ligase component FBXL2 ubiquitinates Aurora B to inhibit tumorigenesis. Cell Death Dis. 4, e759 (2013).
Chen, B. B. et al. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood 119, 3132–3141 (2012).
Chen, B. B., Glasser, J. R., Coon, T. A. & Mallampalli, R. K. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene 31, 2566–2579 (2012).
Niu, M. et al. FBXL2 counteracts Grp94 to destabilize EGFR and inhibit EGFR-driven NSCLC growth. Nat. Commun. 12, 5919 (2021).
Kuchay, S. et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature 546, 554–558 (2017).
Ren, Y. et al. JAC1 suppresses proliferation of breast cancer through the JWA/p38/SMURF1/HER2 signaling. Cell Death Discov. 7, 85 (2021).
Gu, Y. et al. TRAF4 hyperactivates HER2 signaling and contributes to trastuzumab resistance in HER2-positive breast cancer. Oncogene 41, 4119–4129 (2022).
Luan, H. et al. CHIP/STUB1 ubiquitin ligase functions as a negative regulator of ERBB2 by promoting its early post-biosynthesis degradation. Cancers 13, 3936 (2021).
Levkowitz, G. et al. c-Cbl is a suppressor of the neu oncogene. J. Biol. Chem. 275, 35532–35539 (2000).
Denkert, C. et al. Clinical and molecular characteristics of HER2-low-positive breast cancer: pooled analysis of individual patient data from four prospective, neoadjuvant clinical trials. Lancet Oncol. 22, 1151–1161 (2021).
Ergun, Y., Ucar, G. & Akagunduz, B. Comparison of HER2-zero and HER2-low in terms of clinicopathological factors and survival in early-stage breast cancer: a systematic review and meta-analysis. Cancer Treat. Rev. 115, 102538 (2023).
Hong, B. et al. Consensus on clinical diagnosis and medical treatment of HER2-low breast cancer (2022 edition). J. Natl Cancer Cent. 3, 266–272 (2023).
Horisawa, N. et al. The frequency of low HER2 expression in breast cancer and a comparison of prognosis between patients with HER2-low and HER2-negative breast cancer by HR status. Breast Cancer 29, 234–241 (2022).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Akimov, V. et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640 (2018).
Mason, B. & Laman, H. The FBXL family of F-box proteins: variations on a theme. Open Biol. 10, 200319 (2020).
Mungamuri, S. K., Murk, W., Grumolato, L., Bernstein, E. & Aaronson, S. A. Chromatin modifications sequentially enhance ERBB2 expression in ERBB2-positive breast cancers. Cell Rep. 5, 302–313 (2013).
Karasic, T. B., Chiorean, E. G., Sebti, S. M. & O’Dwyer, P. J. A phase I study of GGTI-2418 (geranylgeranyl transferase I inhibitor) in patients with advanced solid tumors. Target Oncol. 14, 613–618 (2019).
Knox, C. et al. DrugBank 6.0: the DrugBank Knowledgebase for 2024. Nucleic Acids Res. 52, D1265–D1275 (2024).
Sinawe, H. & Casadesus, D. Uring culture. StatPearls https://www.ncbi.nlm.nih.gov/books/NBK557569/ (2025).
Patel, V., Liaw, B. & Oh, W. The role of ketoconazole in current prostate cancer care. Nat. Rev. Urol. 15, 643–651 (2018).
Agnihotri, S. et al. Ketoconazole and posaconazole selectively target HK2-expressing glioblastoma cells. Clin. Cancer Res. 25, 844–855 (2019).
Chen, Y. et al. Ketoconazole exacerbates mitophagy to induce apoptosis by downregulating cyclooxygenase-2 in hepatocellular carcinoma. J. Hepatol. 70, 66–77 (2019).
Doheny, D. et al. An FDA-approved antifungal, ketoconazole, and its novel derivative suppress tGLI1-mediated breast cancer brain metastasis by inhibiting the DNA-binding activity of brain metastasis-promoting transcription factor tGLI1. Cancers 14, 4256 (2022).
Wang, H. et al. Activated pregnenolone X-receptor is a target for ketoconazole and its analogs. Clin. Cancer Res. 13, 2488–2495 (2007).
Liu, K. et al. A review of the clinical efficacy of FDA-approved antibody‒drug conjugates in human cancers. Mol. Cancer 23, 62 (2024).
Fu, Z., Li, S., Han, S., Shi, C. & Zhang, Y. Antibody drug conjugate: the ‘biological missile’ for targeted cancer therapy. Signal Transduct. Target. Ther. 7, 93 (2022).
Lewis Phillips, G. D. et al. Targeting HER2-positive breast cancer with trastuzumab–DM1, an antibody–cytotoxic drug conjugate. Cancer Res. 68, 9280–9290 (2008).
Lambert, J. M. & Chari, R. V. Ado-trastuzumab emtansine (T-DM1): an antibody–drug conjugate (ADC) for HER2-positive breast cancer. J. Med. Chem. 57, 6949–6964 (2014).
Bardia, A. et al. Trastuzumab deruxtecan after endocrine therapy in metastatic breast cancer. N. Engl. J. Med. 391, 2110–2122 (2024).
Tsao, L. C. et al. Effective extracellular payload release and immunomodulatory interactions govern the therapeutic effect of trastuzumab deruxtecan (T-DXd). Nat. Commun. 16, 3167 (2025).
Liu, Q. et al. A novel HER2 gene body enhancer contributes to HER2 expression. Oncogene 37, 687–694 (2018).
Marx, C., Held, J. M., Gibson, B. W. & Benz, C. C. ERBB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer Res. 70, 3709–3717 (2010).
Zhang, J. et al. A comprehensive review of small interfering RNAs (siRNAs): mechanism, therapeutic targets, and delivery strategies for cancer therapy. Int. J. Nanomedicine 18, 7605–7635 (2023).
Xianzhou, H. et al. Advances and applications of nanoparticles in cancer therapy. MEDCOMM–Oncology 3, e67 (2024).
Dasgupta, A., Sofias, A. M., Kiessling, F. & Lammers, T. Nanoparticle delivery to tumours: from EPR and ATR mechanisms to clinical impact. Nat. Rev. Bioeng. 2, 714–716 (2024).
Kon, E., Ad-El, N., Hazan-Halevy, I., Stotsky-Oterin, L. & Peer, D. Targeting cancer with mRNA–lipid nanoparticles: key considerations and future prospects. Nat. Rev. Clin. Oncol. 20, 739–754 (2023).
Bianchini, G., Balko, J. M., Mayer, I. A., Sanders, M. E. & Gianni, L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13, 674–690 (2016).
Kazi, A. et al. Blockade of protein geranylgeranylation inhibits CDK2-dependent p27Kip1 phosphorylation on Thr 187 and accumulates p27Kip1 in the nucleus: implications for breast cancer therapy. Mol. Cell. Biol. 29, 2254–2263 (2009).
Oo, C. & Chen, Y. C. The need for multiple doses of 400 mg ketoconazole as a precipitant inhibitor of a CYP3A substrate in an in vivo drug–drug interaction study. J. Clin. Pharmacol. 49, 368–369 (2009).
Small, E. J. et al. Antiandrogen withdrawal alone or in combination with ketoconazole in androgen-independent prostate cancer patients: a phase III trial (CALGB 9583). J. Clin. Oncol. 22, 1025–1033 (2004).
Nakabayashi, M. et al. Response to low-dose ketoconazole and subsequent dose escalation to high-dose ketoconazole in patients with androgen-independent prostate cancer. Cancer 107, 975–981 (2006).
Kelly, C. M. & Janjigian, Y. Y. The genomics and therapeutics of HER2-positive gastric cancer-from trastuzumab and beyond. J. Gastrointest. Oncol. 7, 750–762 (2016).
Moktefi, A. et al. Reappraisal of HER2 status in the spectrum of advanced urothelial carcinoma: a need of guidelines for treatment eligibility. Mod. Pathol. 31, 1270–1281 (2018).
Kim, Y. N., Chung, Y. S., Park, E., Lee, S. T. & Lee, J. Y. Human epidermal growth factor receptor-2 expression and subsequent dynamic changes in patients with ovarian cancer. Sci. Rep. 14, 7992 (2024).
Xu, J. et al. FBXO3 stabilizes USP4 and Twist1 to promote PI3K-mediated breast cancer metastasis. PLoS Biol. 21, e3002446 (2023).
Tilley, W. D. et al. Detection of discrete androgen receptor epitopes in prostate cancer by immunostaining: measurement by color video image analysis. Cancer Res. 54, 4096–4102 (1994).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Hast, M. A. et al. Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase. Chem. Biol. 16, 181–192 (2009).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016).
Lindahl, E., Hess, B. & Van Der Spoel, D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. Mol. Mod. Ann. 7, 306–317 (2001).
Gan, J. -h et al. DrugRep: an automatic virtual screening server for drug repurposing. Acta Pharmacol. Sin. 44, 888–896 (2023).
Eberhardt, J., Santos-Martins, D., Tillack, A. F. & Forli, S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 61, 3891–3898 (2021).
Wolff, A. C. et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 36, 2105–2122 (2018).
Boehm, O. et al. Clinical chemistry reference database for Wistar rats and C57/BL6 mice. Biol. Chem. 388, 547–554 (2007).
Mazzaccara, C. et al. Age-related reference intervals of the main biochemical and hematological parameters in C57BL/6J, 129SV/EV and C3H/HeJ mouse strains. PLoS ONE 3, e3772 (2008).
Jing, W. et al. An in situ engineered chimeric IL-2 receptor potentiates the tumoricidal activity of proinflammatory CAR macrophages in renal cell carcinoma. Nat. Cancer 6, 838–853 (2025).
Bishara, A. J. & Hittner, J. B. Testing the significance of a correlation with nonnormal data: comparison of Pearson, Spearman, transformation, and resampling approaches. Psychol. Methods 17, 399–417 (2012).
Acknowledgements
We thank Z. Li (Sichuan University) and members of the Z.-X.X. laboratory for stimulating discussions during the study. Human HER2-0 TNBC PDO 1 was provided by the Department of Breast Surgery of Sichuan Provincial People’s Hospital. We would like to thank H. Kang in Life Science Core Facilities (College of Life Science) for technical support. This work was supported by The National Key R&D Program of China (2022YFA1103701 to Z.-X.J.X.), the National Natural Science Foundation of China (32471314 to M.N.; 32371446 to P.M.), the Prevention and Control of Emerging and Major Infectious Disease-National Science and Technology Major Project (2025ZD01904100 to Y.C.), Sichuan Science and Technology Program-Central Government Guiding Local Fund (2023ZYD0127 to M.N.), the National Natural Science Foundation of China (82573859 to Y.Y., 32401162 to J.L. and 32271348 to Q.L.), the Science and Technology Department of Sichuan Province (2024NSFSC0731 to Q.L.), the 1.3.5 Project for disciplines of excellence (ZYYC23019 to P.M.) and the Postdoctor Research Fund (2024HXBH085 to J.L.) by West China Hospital, Sichuan University. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
M.N. and Z.-X.J.X. conceived the project and performed the project planning. J.X., J.L., T.H., Y.Y., R.B., H.Z., X.C., J.G., X.L., R.G., C.X. and Y.Z. performed the research. P.M. and J.L. performed LNP design and preparation. Y.C. and Y.L. performed protein–protein interaction prediction and virtual screening for inhibitors of GGTase I. M.N., Z.-X.J.X., J.X., Q.L. and P.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks Heikki Joensuu, Gail Lewis, Michele Pagano and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Silencing of FBXL2 protein expression upregulates HER2 expression on the plasma membrane and sensitizes HER2-0 TNBC cells to T-DXd treatment.
(a) MDA-MB-231 cells stably expressing shRNA targeting FBXL2 (#1 or #2), Smurf1 (#1 or #2), Smurf2 (#1 or #2), CHIP (#1 or #2), c-Cbl (#1 or #2) or control (shCtrl) were subjected to Western blot analyses. The blots are representative of two independent experiments with similar results. (b-d) The replicated experiments of Fig. 1f. MDA-MB-231 and MDA-MB-468 cells stably expressing shRNA targeting FBXL2 (#1 or #2) or control (-) were subjected to Western blot analyses. MDA-MB-453 (HER2-2 + ) or MDA-MB-175 (HER2-1 + ) cells were used as positive control. (e-f) MDA-MB-231, MDA-MB-468, or MDA-MB-453 cells were subjected to flow cytometry analyses. Cells were gated based on forward and side scatter properties (FSC-A and SSC-A). A plot showing 1×104 events was presented. (g) The MDA-MB-468 stable cells were subjected to immunofluorescence staining (IF) assays for HER2 (red) and counterstained with DAPI (blue). Scale bar = 50 μm. (h-k) MDA-MB-231 or MDA-MB-468 stable cells were subjected to colony formation assays. Cells were treated with 10 μg/mL T-DXd or IgG-DXd (-) for 14-21 days (h-i). Data from three independent experiments were presented as mean ± SD (j-k). Scale bar = 1 cm. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test (j, k). The blots (b-d) and images (e-g) are representative of 3 independent experiments with similar results.
Extended Data Fig. 2 Silencing of FBXL2 enhances the sensitivity of HR-positive HER2-low breast cancer cells to T-DXd.
(a) HER2-0 TNBC cells (Hs578T, SUM159 or HCC1806) stably expressing shRNA specific for FBXL2 (#1 or #2) or control (shCtrl) were subjected to Western blot analyses. (b-d) Hs578T, SUM159 or HCC1806 stable cells were treated with IgG-DXd or T-DXd for 7 days and were then subjected to MTS assays for determination of IC50. Data from three independent experiments were presented as mean ± SD. (e) MCF-7 (ER/PR-positive, HER2-1 + /0) or T47D (ER/PR-positive, HER2-1 + ) breast cancer cells stably expressing shRNA specific for FBXL2 (#1 or #2) or control (-) were subjected to Western blot analyses. MDA-MB-453 (HER2-2 + ) or MDA-MB-175 (HER2-1 + ) cells were used as control. (f-g) MCF-7 or T47D stable cells were treated with IgG-DXd or T-DXd for 7 days and were then subjected to MTS assays for determination of IC50. Data from three independent experiments were presented as mean ± SD. Statistical analysis was conducted using two-way ANOVA with Tukey’s multiple-comparison test (b-d, f, g). The blots (a, e) are representative of 3 independent experiments with similar results.
Extended Data Fig. 3 Characterization of lipid nanoparticles (LNPs) delivering siFBXL2, and siFBXL2@LNP can silence FBXL2 expression and elevate HER2 protein expression.
(a) A schematic illustration for the preparation of siFBXL2@LNP. (b-e) Characterization of LNPs by measuring the size distributions (b-c), morphology with transmission electron microscope (TEM, d), and storage stability at 4 °C (e). PDI, polydispersity index. Scale bar = 50 nm. (f-h) MDA-MB-231 or MDA-MB-468 cells were transfected by Cy7-siFBXL2@LNP (siFBXL2#2) or LNP (Ctrl, nanoparticle alone) for 24 h, followed by flow cytometry analyses (f). Cells were gated using FSC-A and SSC-A with a representative plot showing 1×104 events (g). Data from 3 independent experiments were presented as mean ± SD (h). (i-j) Representative confocal laser scanning microscopy (CLSM) images of MDA-MB-231 cells upon incubation with Cy5-siFBXL2@LNP (red) for an indicated time and stained with LysoTracker (green) and DAPI (blue) (i). Scale bar = 10 μm or 2 μm. The co-localization between Cy5-siFBXL2@LNP and LysoTracker (as analyzed by two-tailed Pearson’s correlation coefficient) was quantified, and data from 30 cells were presented as mean ± SD (j). (k-l) The replicated experiments of Fig. 2a. MDA-MB-231 or MDA-MB-468 cells transfected by LNP (-), siCtrl@LNP, or siFBXL2@LNP were subjected to Western blot analyses. Statistical analysis was conducted using an unpaired two-tailed t-test (h). Three independent experiments were performed (a-d) with similar results. The blots (k-l) are representative of 3 independent experiments.
Extended Data Fig. 4 siFBXL2@LNP can significantly silence FBXL2 protein expression and increase HER2 expression in the HER2-0 TNBC xenograft mouse model, and the combined siFBXL2@LNP and T-DXd treatment does not yield observable alteration of body weights or tissue injury.
(a) MDA-MB-231 tumor-bearing mice were i.v. administrated with Cy7-siFBXL2@LNP (0.5 mg/kg). Mice were euthanized at different times (6 h, 12 h or 24 h) post-injection. The fluorescence (NIRF) images and quantified fluorescent intensity of tumors and major organs were shown. Data were presented as mean ± SEM (n = 3/group). (b-d) Mice bearing MDA-MB-231 tumors (CDX) were intravenously (i.v.) administrated with siFBXL2@LNP or siCtrl@LNP (0.5 mg/kg) once two days for three times. The experimental procedure was shown (b). Mice were euthanized by day 5, and tumors were subject to Q-PCR assays (c) and Western analyses (d). Data were presented as mean ± SEM (n = 4/group). (e-f) Mice bearing MDA-MB-231 tumors were i.v. administered with indicated drugs, as shown in Fig. 3a. Mice were euthanized 18 days after treatment. Tissues were dissected and subjected to H&E staining (e). Body weight of mice was measured (f). Data were presented as mean ± SEM (n = 5/group). Scale bar = 100 μm. (g-j) Mice bearing HER2-0 TNBC patients-derived tumors (PDX) were intravenously (i.v.) administrated with siFBXL2@LNP or siCtrl@LNP (0.5 mg/kg) three times every other day. Mice were euthanized by day 5. Tumors were subjected to Q-PCR assays (g), Western analyses (h), or IHC analyses (i-j). Data were quantified by AOD and presented as mean ± SEM (n = 4/group). Scale bar = 50 μm. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test (a) or an unpaired two-tailed t-test (c, g, j).
Extended Data Fig. 5 FBXL2 facilitates HER2 protein polyubiquitination on K747 site, and E3 ubiquitin ligase FBXL20 is a negative regulator of HER2.
(a) The replicated experiments of Fig. 4c. SKBR3 or MDA-MB-453 cells stably expressing HA-FBXL2 or vector (Vec) were subjected to protein half-life assays. Data from two independent experiments were presented (Fig. 4d). (b) Four potential interaction models between FBXL2 (colored in magenta) and the HER2 kinase domain (colored in cyan) were selected from the protein-protein interaction prediction by ZDOCK 3.0.2. (c) HEK293T cells were co-transfected with indicated expressing plasmids for 24 h. Cells were then treated with MG132 (10 μM) for 10 h prior to IP-Western analyses. (d) The replicated experiments of Fig. 5j. HEK293T cells were co-transfected with indicated expressing plasmids for 24 h, and cells were then subjected to protein half-life assays. Data from two independent experiments were presented (Fig. 5k). (e) MDA-MB-231 cells stably expressing Flag-HER2 or Vector (-) were treated with 10 μM MG132 for 10 h prior to IP-Western analyses. (f) MDA-MB-231 cells stably expressing shRNA targeting FBXL20 (#1 or #2), or control (shCtrl) were subjected to Western blot analyses. (g) MDA-MB-231 cells stably silencing of FBXL20 expression were infected with a recombinant lentivirus expressing specific shRNA targeting FBXL2 or control (shCtrl). Cells were then subjected to Western blot analyses. The blots (c, e-g) are representative of 3 independent experiments.
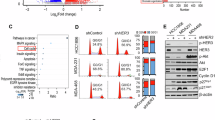
Extended Data Fig. 6 Wild-type FBXL2, but not FBXL2C420S, reduces the steady-state levels of HER2 mRNA.
(a) HEK293T cells were co-transfected with indicated expressing plasmids for 24 h. Cells were then treated with 10 μM MG132 for 10 h prior to IP-Western analyses. (b-d) MDA-MB-231 or MDA-MB-468 cells stably expressing shFBXL2 (#1 or #2) or control (-) were subjected to Q-PCR assays or Western blot analyses. Data from three independent experiments were presented as mean ± SD. (e-f) MDA-MB-231 or MDA-MB-468 cells were treated with an indicated dose of GGTi-2418 for 16 h prior to Q-PCR assays. Data from three independent experiments were presented as mean ± SD. (g-h) The replicated experiments of Fig. 6f, g. MDA-MB-231 or MDA-MB-468 cells were treated with 15 μM GGTi-2418 or DMSO (-) for 16 h, followed by Western blot analyses. HER2-2+ breast cancer cell line MDA-MB-453 and HER2-1+ breast cancer cell line MDA-MB-175 was used as positive controls. (i-j) MDA-MB-231, MDA-MB-468, or MDA-MB-453 cells were subjected to flow cytometry assays. Cells were gated based on FSC-A and SSC-A. A plot showing 1×104 events was presented. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test (b-f). Three independent experiments were performed with similar results (i-j). The blots (a, g-h) are representative of 3 independent experiments.
Extended Data Fig. 7 The characterization of GGTi-2418@LNP delivery system.
(a) High-performance liquid chromatography (HPLC) spectra of GGTi-2418 and GGTi-2418@LNP at 214 nm. (b) HPLC stand curves of GGTi-2418. (c-f) Characterization of GGTi-2418@LNP by measuring the size distributions (c-d), morphology with transmission electron microscope (TEM, e), and storage stability at 4 °C (f). PDI, polydispersity index. Scale bar = 50 nm. (g-i) Plasma concentration-time curves of GGTi-2418 after intravenous (i.v.) injection of free GGTi-2418 or GGTi-2418@LNP (20 mg/kg) in BALB/c nude mice bearing MDA-MB-231 tumors by HPLC analysis (g). Pharmacokinetic parameters of GGTi-2418 were calculated using a non-compartmental model by DAS2.0 (h). t1/2z: half-life; AUC0-∞: the area under the plasma concentration versus time curve; CLz: clearance; Cmax: the highest plasma concentration of the drug in the body. In addition, the concentration of GGTi-2418 in major organs and tumours was shown (i). Data were presented as mean ± SEM (n = 4/group). (j-m) Mice bearing MDA-MB-231 tumors were i.v. administration of GGTi-2418@LNP (20 mg/kg) or LNP (equal amount) according to the procedure (j). Tumors were then subjected to IHC analyses and representative images were shown (k). The expression levels of HER2 and IP3R3 were quantified by AOD (l) and the numbers of p27-positive cells was quantified (m). Data are presented as mean ± SEM (n = 3 /group). Scale bar = 50 μm. Statistical analysis was conducted using an unpaired two-tailed t-test (i, l, m). Three independent experiments were performed with similar results (a-f).
Extended Data Fig. 8 The combined GGTi-2418@LNP and T-DXd treatment does not yield observable alterations of body weight or tissue injury in the HER2-0 TNBC xenograft mouse model.
(a-b) Mice bearing HER2-0 TNBC PDX tumors (PDX#1 or PDX#2) were i.v. administrated with LNP (-) or GGTi-2418@LNP (20 mg/kg, once every day for 2 times) and T-DXd or IgG-DXd (4 mg/kg, one time) alone or in combination. The experimental procedure was shown. (c-d) MDA-MB-231 tumors shown in Fig. 7e were sectioned and subjected to IHC analyses and TUNEL staining assays (c). The respective staining positive cells were quantified and presented as mean ± SEM (d) (n = 6 for the group of control, siFBXL2@LNP or T-DXd, or n = 5 for the group of siFBXL2@LNP + T-DXd). Scale bar = 50 μm. (e-h) Mice bearing MDA-MB-231 tumors (e, n = 6/group), MDA-MB-468 tumors (f, n = 6/group), or TNBC PDX#1 (g, n = 5/group) or PDX#2 (h, n = 6/group) tumors (g & h) were treated with indicated drugs as shown in Fig. 7d or Extended Data Fig. 8a, b. The body weights of mice were shown. Data were presented as mean ± SEM. (i) Mice bearing MDA-MB-231 tumors treated with indicated drugs (as shown in Fig. 7d) were euthanized by day 18 after treatment. Tissues were dissected and subjected to H&E staining (n = 6). Scale bar = 100 μm. Statistical analysis was conducted using one-way ANOVA with Tukey’s multiple-comparison test (d).
Extended Data Fig. 9 Virtual screening for GGTase I inhibitors.
(a) A workflow of virtual screening for GGTase I inhibitors. (b) The chemical structures and the Docking models of nilotinib, fexinidazole, pilocarpine or zoledronic acid and GGTase I. Imidazolyl was marked with a red circle. (c) MDA-MB-231 cells were treated with 15 μM GGTi-2418 for 16 h, or were treated with 12.5 μM of ketoconazole (KCZ), nilotinib (NOB), fexinidazole (FDZ), pilocarpine (PCP), or zoledronic acid (ZDA) for 48 h prior to Western blot analysis. (d) The replicated experiments of Fig. 8d. MDA-MB-231 or MDA-MB-468 cells were treated with 12.5 μM KCZ or DMSO (-) for 48 h, followed by Western blot analyses. HER2-2+ breast cancer cell line MDA-MB-453 and HER2-1+ breast cancer cell line MDA-MB-175 were used as the positive control. (e) MDA-MB-231 cells stably silencing FBXL2 or MDA-MB-231 cells treated with12.5 μM KCZ for 48 h were subjected to Western blot analyses. (f) MDA-MB-231 or MDA-MB-468 cells were pretreated with or without 12.5 μM KCZ for 48 h, followed by incubation with either 10 μg/mL T-DXd or IgG-DXd for 14-21 days prior to colony formation assays. Scale bar = 1 cm. (g-i) Mice bearing MDA-MB-231 tumors (h, n = 6/group) or MDA-MB-468 tumors (i, n = 5/group) were i.v. administrated with LNP (-) or KCZ@LNP (10 mg/kg, once every day for 2 times) and T-DXd or IgG-DXd (4 mg/kg, one time). The experimental procedure (g) and the body weights of mice (h & i) were shown. Data were presented as mean ± SEM. All blots (c-e) and the images (f) are representative of 3 independent experiments.
Extended Data Fig. 10 No sign of acute liver or kidney injury are observed after i.v. injection KCZ@LNP or GGTi-2418@LNP.
(a-c) C57BL/6 mice were i.v. administrated with GGTi-2418@LNP (20 mg/kg, once every day for 2 times) or KCZ@LNP (10 mg/kg, once every day for 2 times). At defined time points post-injection, the blood was collected for blood biochemistry examination (a-b), including liver function markers (ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; TP, total protein) and kidney function markers (CREA, creatinine; UREA, urea). The grey area is the normal range. The mice body weight was recorded every two days (c). Data are presented as mean ± SEM (n = 4/group). Statistical analysis was conducted using two-way ANOVA with Tukey’s multiple-comparison test (c).
Supplementary information
Supplementary Tables
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 8
Statistical source data.
Source Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, J., Liu, J., Liu, Y. et al. Targeted inhibition of FBXL2 confers susceptibility of HER2-negative breast cancer to trastuzumab deruxtecan. Nat Cancer (2026). https://doi.org/10.1038/s43018-025-01112-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43018-025-01112-z
This article is cited by
-
Preventing HER2 degradation improves efficacy of trastuzumab deruxtecan
Nature Cancer (2026)